Abstract
Background
We performed an in-depth analysis of the ABC gene family in Aedes aegypti (Diptera: Culicidae), which is an important vector species of arthropod-borne viral infections such as chikungunya, dengue, and Zika. Despite its importance, previous studies of the Arthropod ABC family have not focused on this species. Reports of insecticide resistance among pests and vectors indicate that some of these ATP-dependent efflux pumps are involved in compound traffic and multidrug resistance phenotypes.
Results
We identified 53 classic complete ABC proteins annotated in the A. aegypti genome. A phylogenetic analysis of Aedes aegypti ABC proteins was carried out to assign the novel proteins to the ABC subfamilies. We also determined 9 full-length sequences of DNA repair (MutS, RAD50) and structural maintenance of chromosome (SMC) proteins that contain the ABC signature.
Conclusions
After inclusion of the putative ABC proteins into the evolutionary tree of the gene family, we classified A. aegypti ABC proteins into the established subfamilies (A to H), but the phylogenetic positioning of MutS, RAD50 and SMC proteins among ABC subfamilies—as well as the highly supported grouping of RAD50 and SMC—prompted us to name a new J subfamily of A. aegypti ABC proteins.
Keywords: Aedes aegypti; MutS, RAD50 and SMC proteins; MDR phenotype; ABC protein classification; ABC protein subfamily J
Background
The ATP-binding cassette (ABC) transporters constitute a diverse gene family consisting of proteins found in all cellular organisms and participating in several different biological pathways [1]. Among these processes, the ABC transporters are mostly involved in extra and intracellular trans membrane ATP energy driven traffic of molecules such as lipids, amino acids, hormones and xenobiotics [2, 3].
Members of this family are characterized by two trans membrane domains (TMD) and two nucleotide-binding domains (NBD) characterized by conserved motifs: Walker A, Walker B, ABC signature (LSGGQ-motif), Q loop, and H loop [1, 4]. The TMD domains of the ABC-transporters are composed of five to ten membrane-spanning regions that are involved in substrate translocation. The four domains (two TMD and two NBD) of a functional ABC transporter might be present in a single protein (full transporter) or in dimers of separate proteins that have at least one TMD and one NBD each (half transporter) [3, 5].
The traditional classification is based on sequence similarity and arranged the ABC protein diversity into eight subfamilies (A- H) [6]. The ABCE and ABCF subfamilies are unique among the ABC proteins because they exhibit a pair of linked nucleotide-binding domains while lacking trans membrane domains [3, 6]. The ABCH subfamily was described for protozoa [7] and insects [8, 9], but it has not yet been found in mammals, bacteria, and yeast genomes. Plants, besides presenting eukaryotic ABC subfamilies A to G, exhibit a heterogeneous and extensive group of ABC proteins that bear similarities to the components of prokaryotic multi-subunit ABC transporters. This group was named subfamily I and includes NBD and TMD domains and homologues of soluble cytosolic proteins that interact with NBDs as well as putative substrate-binding proteins similar to the periplasmic binding proteins [10].
Three other groups of proteins not assigned to the subfamilies mentioned above exhibit ABC transporter domains: (1) the MutS proteins that are responsible for DNA mismatch repair and maintenance of genomic stability [11, 12]; (2) the structural maintenance of chromosome proteins (SMC), which are mostly responsible for chromosome condensation and sister chromatid cohesion [13], and (3) the Rad 50 proteins that also function on DNA repair [8, 9, 14].
Although MutS, SMC, and Rad50 proteins show ABC protein characteristics, they have not yet been included in the standard ABC classification for humans, arthropods, and the Caenorhabditis elegans nematode [8, 9, 15, 16]. Nonetheless, in the complete inventory of ABC proteins of the Arabidopsis thaliana plant, SMC proteins were proposed as a new ABC protein subfamily [17].
Some ABC proteins have been associated with multidrug resistance (MDR) phenotype in a variety of organisms. This phenotype is associated with the overexpression of P-glycoproteins (P-gp/MDR/ABCB1), the multidrug resistance protein (MDR/ABCC), and the breast cancer resistance protein (BCRP/ABCG2) [5, 18, 19]. These act as efflux pumps that result in resistance to chemotherapeutics, antibiotics, and antiretroviral drugs [20, 21].
One important control mechanism of vector-borne diseases is vector control, which relies mainly on insecticide treatments of vector populations. In these populations, the insecticide-resistant phenotype arises due to the selection of genetically resistant individuals that exhibit higher fitness under special conditions [22, 23]. Multiple insecticide resistance can be separated into two main categories: cross-resistance—when a single mechanism confers resistance to a range of different insecticides; and multiple resistance—when several coexisting defense mechanisms act in the same organism [24, 25]. The involvement of ABC transporters in insecticide resistance and transport is poorly documented, but an increasing number of studies have shown that ABC transporters have been linked to insecticide and nicotine transport [26–28] and insect resistance to Bacillus thuringiensis toxins and pyrethroids [29, 30]. The high expression of P-gp in insecticide resistant pests such as Heliothis virescens and Helicoverpa armigera has been suggested to be a mechanism of resistance [31, 32].
Recent surveys of the ABC gene family in arthropods included the fruit fly Drosophila melanogaster, the mosquito Anopheles gambiae, the beetle Tribolium castaneum, the honey bee Apis mellifera, the silkmoth Bombyx mori, the water flea Daphia pulex, and the spider mite Tetranychus urticae [16]. Analyses focusing on crustaceans such as the sea lice Caligus rogercresseyi [33] and Lepeophtheirus salmonis [34] were also carried out. These studies left out the A. aegypti mosquito, which is an important vector species of arthropod-borne viral infections such as chikungunya, dengue, and Zika diseases [35]. In 2016, Lu et al. [36] conducted a comparative analysis of the ABC transporter family in three mosquito species (Anopheles gambiae, Aedes aegypti, and Culex pipiens quinquefasciatus) and found 55, 69, and 70 ABC genes, respectively. The search for Aedes aegypti ABC proteins, however, was carried out within a limited evolutionary range because only mosquito sequences were analyzed.
In this study, we surveyed the Aedes aegypti genome in a broader evolutionary spectrum, employing human and Drosophila ABC transporters as queries. By including all the putative proteins that exhibit the ABC domain into a phylogenetic analysis, we showed that SMC, Rad 50, and MutS proteins were part of the main ABC gene family diversification, which justifies the proposition of a new subfamily of the ABC proteins.
Results
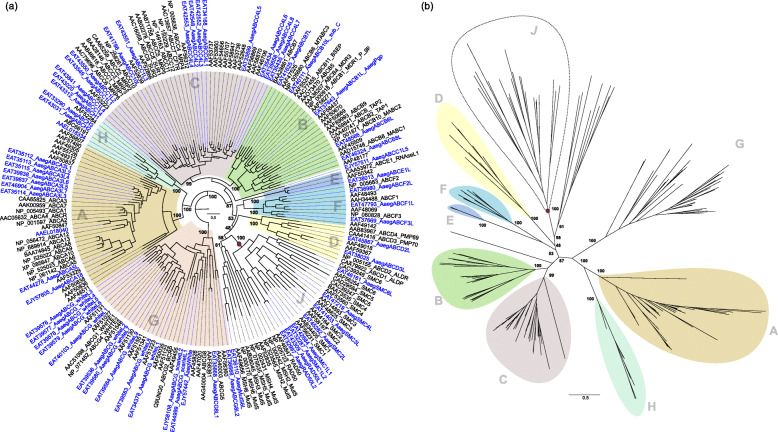
The BLASTp search on the A. aegypti genome retrieved 62 complete proteins that were identified as ABC transporters when submitted to the NCBI Conserved Domain Database. The ABC gene family phylogeny recovered subfamilies A-H with significant statistical support (Fig. 1). The sizes of gene subfamilies varied significantly with subfamilies A-C and G consisting of the larger groups. Sister group associations between ABC subfamilies were less resolved. The single exception was the clade with subfamilies ABCA and ABCH that were grouped with maximum statistical support. In all ABC subfamilies, A. aegypti proteins had a tendency to be positioned among human and Drosophila sequences suggesting that the duplication events that gave rise to current ABC diversity took place before the evolution of those lineages. Clusters containing ABC genes exclusively from A. aegypti were found in subfamilies ABCA, ABCC, and ABCG. These clusters indicate mosquito-specific duplication events.
Fig. 1.
a Maximum likelihood phylogeny of the ABC gene family including SMC, Rad50 and MutS genes. ABC subfamilies are shown with the new mosquito sequences highlighted in blue. b Numbers at branches indicate statistical support (ultra-fast bootstrap) for each subfamily A-J
The variation of the rate of evolution within each ABC subfamily as measured by the heterogeneity of the distance between the common ancestor of all members of the subfamily and the tips was higher in subfamily ABCA. In this subfamily, an interesting pattern of rate increase along lineages was observed (Fig. 1). As expected, deeper nodes exhibited lower statistical support demonstrating that the evolutionary relationships between these subfamilies were not fully resolved. Surprisingly, root placement using the minimal ancestor deviation (MAD) method suggested that subfamily ABCG is a sister to the remaining ABC transporters including the clades consisting of SMC and Rad50 proteins as well as the MutS proteins that were positioned as a sister to subfamily ABCD (Fig. 1).
Discussion
To investigate ABC transporters in the A. aegypti genome within a broader evolutionary context, we identified A. aegypti ABC homologs employing human and D. melanogaster as queries (Table 1). We also identified the conserved domains of all the putative A. aegypti ABC transporters to investigate the assignment of the putative proteins to the described subfamilies of these transporters. We identified ten members of the A. aegypti ABCA subfamily (Fig. 1 and Table 2). This subfamily contains longer proteins that ranged from 1419 to 1673 amino acid residues. Nine of these members have the topology of full transporters with two NBDs and two TMDS (Table 2). The A. aegypti ABCA subfamily was encoded by genes organized in tandem indicating specific gene duplication events (Table 2). Four members of this cluster have genes organized in tandem in the supercontig 1.726, two members belong to the supercontig 1.321, and four belong to other supercontigs (Table 2). The roles of arthropod ABCA members are unclear [16], but this subfamily has been described as involved with lipid transport in mammals [37].
Table 1.
Classification of ABC proteins subfamilies in Homo sapiens and Drosophila melanogaster
| Homo sapiens | Drosophila melanogaster | ||||
|---|---|---|---|---|---|
| Sub-family | Seq ID | GenBank accession | Sub-family | Seq ID | GenBank accession |
| A | ABCA1 | NP_005493 | A | AAF50836 | |
| A | ABCA2 | NP_001597 | A | AAF50837 | |
| A | ABCA3 | CAA65825 | A | AAF50838 | |
| A | ABCA4_ABCR | AAC05632 | A | AAF50847 | |
| A | ABCA5 | NP_061142 | A | AAF53329 | |
| A | ABCA6 | NP_525023 | A | AAF55726 | |
| A | ABCA7 | AAK00959 | A | AAF57490 | |
| A | ABCA8 | BAA74845 | B | AAF45509 | |
| A | ABCA9 | NP_525022 | B | AAF47525 | |
| A | ABCA10 | XP_085647 | B | AAF48177 | |
| A | ABCA12 | NP_056472 | B | AAF50669 | |
| A | ABCA13 | NP_689914 | B | AAF50670 | |
| B | ABCB1_MDR1_P_gp | NP_000918 | B | AAF53736 | |
| B | ABCB2_TAP1 | CAA40741 | B | AAF53737 | |
| B | ABCB_TAP2 | AAA59841 | B | AAF55241 | |
| B | ABCB4_MDR3 | AAA36207 | B | AAF58271 | |
| B | ABCB5 | AAO73470 | B | AAF58437 | |
| B | ABCB6_MTABC3 | NP_005680 | C | AAF46706 | |
| B | ABCB7 | BAA28861 | C | AAF52639 | |
| B | ABCB8_MABC1 | AAD15748 | C | AAF52648 | |
| B | ABCB9 | AAF89993 | C | AAF52866 | |
| B | ABCB10_MABC2 | XP_001871 | C | AAF53223 | |
| B | ABCB11_BSEP | AAC77455 | C | AAF53950 | |
| C | ABCC1_MRP1 | AAB46616 | C | AAF54656 | |
| C | ABCC2_MRP2 | CAA65259 | C | AAF55707 | |
| C | ABCC3_MRP3 | BAA28146 | C | AAF56312 | |
| C | ABCC4_MRP4 | NP_005836 | C | AAF56869 | |
| C | ABCC5_MRP5 | AAB71758 | C | AAF56870 | |
| C | ABCC6_MRP6 | AAC79696 | C | AAF58947 | |
| C | ABCC7_CFTR | AAC13657 | D | AAF49018 | |
| C | ABCC8_SUR1 | AAB02278 | D | AAF59367 | |
| C | ABCC9_SUR2 | AAC16058 | E | AAF50342 | |
| C | ABCC10 | NP_258261 | F | AAF48069 | |
| C | ABCC11 | NP_149163 | F | AAF48493 | |
| C | ABCC12 | NP_150229 | F | AAF49142 | |
| D | ABCD1_ALDP | CAA79922 | G | AAF45826 | |
| D | ABCD2_ALDR | NP_005155 | G | AAF47020 | |
| D | ABCD3_PMP70 | CAA41416 | G | AAF49455 | |
| D | ABCD4_PMP69 | AAB83967 | G | AAF50035 | |
| E | ABCE1_RNAseL1 | CAA53972 | G | AAF51027 | |
| F | ABCF1 | AAH34488 | G | AAF51122 | |
| F | ABCF2 | NP_005683 | G | AAF51130 | |
| F | ABCF3 | NP_060828 | G | AAF51131 | |
| G | ABCG1_WHITE1 | AAC51098 | G | AAF51223 | |
| G | ABCG2_BCRP | Q9UNQ0 | G | AAF51341 | |
| G | ABCG4_WHITE2 | NP_071452 | G | AAF51548 | |
| G | ABCG5 | AAG40003 | G | AAF51551 | |
| G | ABCG8 | AAG40004 | G | AAF52835 | |
| J | SMC1 | AAB34405 | G | AAF56360 | |
| J | SMC4 | BAA73535 | G | AAF56361 | |
| J | SMC3 | AAC14893 | H | AAF52284 | |
| J | SMC2 | AAI44164 | H | AAF56807 | |
| J | RAD50 | NP_005723 | H | ABC66191 | |
| J | SMC5 | CAC39247 | J | SMC1 | AAF56231 |
| J | SMC6 | CAC39248 | J | SMC4 | AAF53560 |
| J | MSH4_MutS | NP_002431 | J | SMC2 | AAF58197 |
| J | MSH3_MutS | AAB06045 | J | RAD50 | AAF46847 |
| J | MSH5_MutS | NP_079535 | J | SMC5 | CAD29584 |
| J | MSH2_MutS | NP_000242 | J | SMC6 | AAF56254 |
| J | MSH6_MutS | NP_000170 | J | MSH2_MutS | NP_523565 |
| J | MSH6_MutS | AAF49656 | |||
| J | SMC3 | AAF48625 | |||
Table 2.
Characterization of the 62 A. aegypti ABC proteins
| Sub-family | Name | VectorBase (accession number) | Size (amino acids) | Predicted topology | Location (gene) |
Orientation (gene) |
|---|---|---|---|---|---|---|
| A | ||||||
| AaegABCA3L1 | AAEL012702-PA | 1669 | TMD1-NBD1-TMD2-NBD2 | 1.726: 372101–377,726 | + | |
| AaegABCA3L2 | AAEL012700-PA | 1648 | TMD1-NBD1-TMD2-NBD2 | 1.726: 388899–394,375 | + | |
| AaegABCA3L3 | AAEL012701-PA | 1622 | TMD1-NBD1-TMD2-NBD2 | 1.726: 409854–439,050 | + | |
| AaegABCA3L4 | AAEL012698-PA | 1652 | TMD1-NBD1-TMD2-NBD2 | 1.726: 450626–459,977 | + | |
| AaegABCA3L5 | AAEL008388-PA | 1666 | TMD1-NBD1-TMD2-NBD2 | 1.321: 644618–664,804 | – | |
| AaegABCA3L6 | AAEL008384-PA | 1660 | TMD1-NBD1-TMD2-NBD2 | 1.321: 675803–697,600 | – | |
| AaegABCA3L7 | AAEL001938-PA | 1673 | TMD1-NBD1-TMD2-NBD2 | 1.46: 792516–818,527 | – | |
| AaegABCA5L | AAEL004331-PA | 1419 | TMD1-NBD1-TMD2-NBD2 | 1.115: 240545–271,476 | + | |
| AaegABCA5L | AAEL018040-PA | 1987 | TMD1-NBD1-TMD2-NBD2 | 3.322: 613800–714,818 | – | |
| AaegABCA18 | AAEL017572-PA | 347 | NBD | 1.176: 1628836–1,629,879 | – | |
| B | ||||||
| AaegABCB1L/AaegP-gp | AAEL010379-PA | 1307 | TMD1-NBD1-TMD2-NBD2 | 1.474: 313030–327,570 | + | |
| AaegABCB6L | AAEL000434-PA | 693 | TMD-NBD | 1.8: 3711414–3,730,662 | + | |
| AaegABCB7L | AAEL006717-PA | 734 | TMD-NBD | 1.219: 178589–203,717 | – | |
| AaegABCB8L | AAEL002468-PA | 703 | TMD-NBD | 1.58: 1203051–1,224,141 | – | |
| AaegABCB10L | AAEL008134-PA | 848 | TMD-NBD | 1.302: 73729–107,503 | + | |
| C | ||||||
| AaegABCC1L1 | AAEL005026-PA | 1384 | TMD0-TMD1-NBD1-TMD2-NBD2 | 1.139: 1168407–1,184,363 | + | |
| AaegABCC1L2 | AAEL005045-PA | 1514 | TMD0-TMD1-NBD1-TMD2-NBD2 | 1.139: 1184563–1,195,380 | – | |
| AaegABCC1L3 | AAEL005030-PA | 1396 | TMD0-TMD1-NBD1-TMD2-NBD2 | 1.139: 1233513–1,252,972 | – | |
| AaegABCC1L4 | AAEL004743-PA | 1089 | TMD0-TMD1-NBD1 | 1.129: 994901–1,030,978 | + | |
| AaegABCC1L5 | AAEL017209-PA | 903 | TMD0 -TMD1-NBD1 | 1.107: 820177–825,969 | – | |
| AaegABCC4L1 | AAEL013567-PA | 1311 | TMD1-NBD1-TMD2-NBD2 | 1.871: 281423–317,150 | + | |
| AaegABCC4L2 | AAEL005918-PA | 1312 | TMD1-NBD1-TMD2-NBD2 | 1.180: 664096–681,744 | – | |
| AaegABCC4L3 | AAEL005937-PA | 1300 | TMD1-NBD1-TMD2-NBD2 | 1.180: 724473–765,746 | + | |
| AaegABCC4L4 | AAEL005929-PA | 1413 | TMD1-NBD1-TMD2-NBD2 | 1.180: 786121–801,780 | + | |
| AaegABCC4L5 | AAEL013834-PA | 1235 | TMD1-NBD1-TMD2-NBD2 | 1.936: 291553–353,031 | – | |
| AaegABCC4L6 | AAEL012395-PA | 1357 | TMD1-NBD1-TMD2-NBD2 | 1.688: 67831–72,390 | – | |
| AaegABCC4L7 | AAEL012386-PA | 1351 | TMD1-NBD1-TMD2-NBD2 | 1.688: 87463–91,714 | + | |
| AaegABCC4L8 | AAEL012192-PA | 1345 | TMD1-NBD1-TMD2-NBD2 | 1.664: 660781–670,973 | – | |
| AaegABCC10L | AAEL006622-PA | 1540 | TMD0-TMD1-NBD1-TMD2-NBD2 | 1.213: 838086–915,438 | + | |
| AaegABCC14 | AAEL005499-PA | 1382 | TMD1-NBD1-TMD2-NBD2 | 1.160: 1362499–1,398,139 | – | |
| D | ||||||
| AaegABCD2L | AAEL002913-PA | 659 | TMD-NBD | 1.71: 1617561–1,676,168 | + | |
| AaegABCD3L | AAEL010047-PA | 753 | TMD-NBD | 1.449: 843528–895,566 | + | |
| E | ||||||
| AaegABCE1L | AAEL010059-PA | 609 | NBD1-NBD2 | 1.450: 713084–727,146 | + | |
| F | ||||||
| AaegABCF1L | AAEL001101-PA | 894 | NBD1-NBD2 | 1.23: 2941514–2,961,984 | – | |
| AaegABCF2L | AAEL010977-PA | 602 | NBD1-NBD2 | 1.529: 122943–143,748 | – | |
| AaegABCF3L | AAEL010359-PA | 609 | NBD1-NBD2 | 1.450: 713084–727,146 | + | |
| G | ||||||
| AaegWhite* | AAEL016999-PA | 692 | NBD-TMD | 1.71: 816675–879,820 | – | |
| AaegABCG/whiteL1 | AAEL008138-PA | 773 | NBD-TMD | 1.303: 412767–432,830 | + | |
| AaegABCG/whiteL2 | AAEL008672-PA | 689 | NBD-TMD | 1.340: 378892–469,513 | + | |
| AaegABCG/whiteL3 | AAEL008624-PA | 593 | NBD-TMD | 1.337: 23491–61,022 | – | |
| AaegABCG/whiteL4 | AAEL008632-PA | 607 | NBD-TMD | 1.337: 68512–71,099 | – | |
| AaegABCG/whiteL5 | AAEL008628-PA | 571 | NBD-TMD | 1.337: 85665–99,799 | – | |
| AaegABCG/whiteL6 | AAEL008625-PA | 606 | NBD-TMD | 1.337: 119628–131,014 | + | |
| AaegABCG/whiteL7 | AAEL008629-PA | 723 | NBD-TMD | 1.337: 131034–224,797 | – | |
| AaegABCG/whiteL8 | AAEL008631-PA | 759 | NBD-TMD | 1.337: 276979–394,542 | – | |
| AaegABCG/whiteL9 | AAEL008635-PA | 676 | NBD-TMD | 1.337: 470559–525,334 | – | |
| AaegABCG/whiteL10 | AAEL013372-PA | 599 | NBD-TMD | 1.830: 256110–301,680 | + | |
| AaegABCG/scarletL1 | AAEL003703-PA | 616 | NBD-TMD | 1.94: 984346–994,398 | – | |
| AaegABCG/scarletL2 | AAEL017106-PA | 689 | NBD-TMD | 1.1174: 142758–145,023 | – | |
| AaegABCG8L1 | AAEL012170-PA | 275 | NBD | 1.662: 13565–20,660 | + | |
| AaegABCG8L2 | AAEL011265-PA | 787 | NBD-TMD | 1.561: 434947–481,728 | + | |
| H | ||||||
| AaegABCH1 | AAEL005249-PA | 872 | NBD-TMD | 1.147: 1132338–1,176,150 | + | |
| AaegABCH2 | AAEL005491-PA | 783 | NBD-TMD | 1.159: 901905–920,193 | + | |
| AaegABCH3 | AAEL014428-PA | 727 | NBD-TMD | 1.1111: 126773–184,623 | + | |
| AaegABCH4L5 | AAEL018334-PA | 814 | NBD-TMD | 1.181:178199–420,930 | – | |
| J | ||||||
| AaegSMC1L1 | AAEL005802-PA | 1227 | NBD | 1.175: 1418947–1,437,720 | – | |
| AaegSMC1L2 | AAEL015592-PA | 594 | NBD | 1.3565: 4371–6283 | – | |
| AaegSMC2L | AAEL003449-PA | 1182 | NBD | 1.86: 847376–874,751 | – | |
| AaegSMC3L | AAEL006937-PA | 1201 | NBD | 1.229: 536438–586,285 | – | |
| AaegSMC4L | AAEL001655-PA | 1347 | NBD | 1.38: 1385452–1,432,324 | + | |
| AaegSMC6L | AAEL002581-PA | 1107 | NBD | 1.61: 165187–198,927 | + | |
| AaegRAD50L1 | AAEL005245-PA | 1034 | NBD | 1.147: 747831–755,196 | – | |
| AaegRAD50L2 | AAEL014748-PA | 1051 | NBD | 1.1248: 101280–115,288 | – | |
| AaegMutS6L | AAEL011780-PA | 1130 | NBD | 1.612: 217022–240,932 | + |
Five sequences retrieved from the A. aegypti genome were assigned to the ABCB subfamily (Fig. 1, Table 2). This subfamily is composed of putative homologs of the human P-glycoprotein, which plays key physiological roles such as the excretion of toxic compounds and the multidrug resistance phenotype [3, 26, 27, 37, 38]. The identified A. aegypti ABCB proteins are intimately related to the human mitochondrial transporters HsABCB6, HsABCB7, HsABCB8, and HsABCB10 leading us to suppose that these proteins have a similar role associated with the iron metabolism and the transport of Fe/S protein precursors from the mitochondria to the cytoplasm [37, 39]. We also note that one D. melanogaster protein classified as belonging to the ABCB (CG31792_B) subfamily was recovered in the ABCC clade. This may be due to misclassification or to recent duplication and functional change. In either case, this protein should be further investigated.
One of the most diverse subfamilies identified in the mosquito genome was the ABCC with 15 members—all full transporters (Table 2). This subfamily presents a high diversity of sequences as well as functional roles when compared with the human ABCC proteins. These functions are related to ion transport, cell surface receptors, toxin secretion, and multidrug resistance [38]. A sub-clade containing all the MRP from humans and D. melanogaster was recovered including four A. aegypti proteins (AaegABCC1L1, AaegABCC1L2, AaegABCC1L4, and AaegABCC1L5) suggesting that these proteins might also be responsible for protection against xenobiotics [40] and for the MDR phenotype [38, 41].
The ABCD and ABCE subfamilies were the least diverse of the groups identified in humans—the former is known to appear as half transporters forming homo or heterodimers in peroxisomes acting in lipid transport [3, 39, 42]. The ABCD subfamily has two members and the ABCE subfamily has only one protein described for most eukaryotes (Table 2) with the exception of A. thaliana [17]. This was consistent with the findings of a single ABCE gene in the A. aegypti genome. These proteins lack the TMD and were first described as the RNAseL protein participating in ribosome biogenesis and protein translation [37–39, 43–46]. Like ABCE proteins, the ABCF subfamily also lacks the TMD and is involved in the ribosome complex formation and activation [46–48]; only three of these proteins were found in the mosquito genome in our analysis.
Although only five members of the ABCG proteins were described in humans [3, 37], 15 proteins belonging to this group were identified for A. aegypti (Table 2). This number is greater than the 11 genes previously identified in An. gambiae [9]. This excessive number of ABCG proteins in A. aegypti mosquito is likely due to a series of duplication events that is supported by the tandem organization observed in the supercontig 1.337 of the A. aegypti genome (Table 2). In D. melanogaster, the white gene is the most studied gene from the ABCG subfamily, and the product of this gene can form dimers with the scarlet and brown proteins (scarlet and brown genes, respectively). These dimers are transporters of eye pigment precursors in D. melanogaster [49, 50]. Only one ortholog of the white and scarlet proteins was found in the A. aegypti genome but no ortholog of the brown protein was found. In humans, ABCG5 and ABCG8 are glycoproteins that also form obligate heterodimers. These are useful to limit the absorption of plant sterols and cholesterol from the diet and promote secretion of plant sterols and cholesterol from liver cells into the bile. Based on their head-to-head orientation and clear orthologous relationships with human ABCG5 and ABCG8, these arthropod ABCGs probably have a similar role as their human orthologues [37].
The ABCH subfamily was exclusively found in insects with no reports in mammals, plants, or yeast [9, 37]. Here, four members of the ABCH subfamily were identified in the A. aegypti genome (Fig. 1 and Table 2). This included the sequence AAEL018334, which has been previously assigned to ABCG subfamily. Although these are proteins with unknown function, topological similarities with the ABCG proteins have suggested that the ABCH might be involved in sterol transport and multidrug resistance [51, 52].
Insect P-glycoproteins and multidrug-resistance associated proteins are frequently associated with pesticide resistance as reported in Heliothis virescens and Helicoverpa armigera [30, 31] and insecticide transport. The expression of A. aegypti P-gp (AAEL010379) increases eightfold in the temephos-treated larvae, and silencing of this gene expression significantly increases temephos toxicity [27]. These findings suggested that ABC transport, which consists of ATP-dependent efflux pumps, might be involved with compound traffic and multidrug resistance phenotypes. New insights into insecticide efflux, ATP-dependent efflux pump inhibitors, and/or RNAi associated with pesticides will potentially assist in the development of control strategies for important vectors of infectious diseases like A. aegypti.
Rad50 shares topological and sequence features with SMC proteins [52]. Notably, Rad50 has a relatively well-conserved LSGG motif compared to the classic ABC proteins. Moreover, it has an extensive coiled region that facilities dimerization of large molecules restoring the close proximity of the Walker A and B motifs for nucleotide binding [53]. SMCs have more degenerated versions of this signature motif and contain minimal Walker A and B motifs (Supplemental material 1) [54]. Finally, perhaps a distant lineage but still within the ABC diversification [55], are the DNA repair enzymes such as MutS [56].
SMC proteins formed a highly supported clade with the Rad50 proteins. These proteins form dimers and have a conserved mechanism of conformational change observed in the classic ABC proteins. The ATP binding and NBD dimerization promote changes in the substrate-binding domains that are important for the function of the ABC-type ATPases. The substrate-binding domains of the SMC and Rad50 proteins are located in similar positions as the classic ABC proteins [52]. The ABC proteins subfamilies are grouped together based on sequence similarity and proteins belonging to the same subfamily usually have similar functions. Our results showed that ABC subfamilies were always strongly recovered in the gene family phylogeny and that the sequences of SMC and Rad50 proteins formed a well-supported clade (100 bootstrap support), sister to MutS proteins, and ABC transporters excluding ABCG. Functional similarities are also observed within the groups.
We know the following: (i) SMC and Rad50 proteins exhibit similar functions on DNA repair and chromosomal maintenance [8, 9, 11, 12, 14], (ii) they form a strongly supported clade with ABC transporters phylogeny, and (iii) they exhibit the structural and sequence characteristics of ABC proteins. Thus, we propose these proteins be included in the ABC gene family with the creation of a new subfamily called J (Fig. 1; Table 2) that includes ABC proteins involved in DNA repair and structural maintenance of the chromosomes.
Conclusions
In summary, we found 53 classic complete ABC proteins annotated in the A. aegypti genome that were classified in traditional ABC subfamilies (A-H) as reported in other species. We also found 9 sequences of the Rad, MutS, and SMC in the Aedes genome database that clustered with human and D. melanogaster orthologs in the same clade. Considering other similarities observed between these enzymes and the classic ABC proteins, we propose these proteins be included in the ABC gene family followed by creation of a new subfamily called J that includes ABC enzymes involved in DNA repair and the structural maintenance of the chromosome.
Methods
Sequence sampling and alignment
We selected protein sequences of ABC protein subfamilies from humans (46 sequences) and Drosophila melanogaster (50 sequences) including sequences from SMC, Rad50, and MutS genes ensuring a broad evolutionary diversity. These sequences were used as query for BLAST searches of A. aegypti ABC proteins. To identify A. aegypti ABC proteins, human and Drosophila sequences were used as queries to search sequences on the mosquito genome (VectorBase) and on the NCBI protein database using BLASTp [57]. Sequences resulting from these searches that had more than 30% identity were considered as homologous (Supplemental material 2).
All putative homologous sequences were submitted to the NCBI Conserved Domain Database [58, 59] to confirm the presence of the ATP-binding cassette domain. Datasets were aligned using the MUSCLE software [60] and then pruned for removal of regions with high frequency of indels using TrimAL using the “gappyout” command (Supplemental material 3) [61].
Inference of ABC gene genealogy
The maximum likelihood (ML) tree topology was inferred with the IqTree 1.6 [62] program employing the LG + R10 model of amino acid substitution that was chosen by the Bayesian information criterion. This model uses the LG amino acid replacement matrix [63] coupled with ten relative rate classes to accommodate among-site rate heterogeneity [64]. Branch support was assessed by the ultrafast bootstrap implementation of IqTree using 1000 replicates [65]. IqTree was executed via the command “iqtree -s infile -bb 1000”. Because no outgroup was included in our analysis, rooting of the ABC gene genealogy was performed using the minimal ancestor deviation method of Tria at al. [66]. Rooting is necessary for establishing the chronological direction of the ABC gene family evolution (Supplemental material 4).
Supplementary information
Additional file 1. Sequences.txt: Rad50, SMC and MutS sequences highlighted Walker A and Walker B motifs and ABC signature.
Additional file 2. ABC.txt: Unaligned ABC sequences used in this study.
Additional file 3. ABCalin.txt: Aligned and trimmed ABC sequences employed in evolutionary analyses.
Additional file 4. ABC.unrooted.txt: Unrooted maximum likelihood phylogenetic tree of the ABC gene family.
Acknowledgments
The authors are grateful to Dr. Silvia Andrade Justi (Walter Reed Biosystematics Unit, Smithsonian Institute and Walter Reed Army, Institute of Research Entomology branch Maryland, USA) for providing assistance in accomplishing this work. We dedicate this paper to the memory of our friend Dr. Mario Alberto Cardoso da Silva Neto for his interest in mosquitoes.
Abbreviations
- MDR
Multidrug resistance protein
- P-gp
P-glycoproteins
- BCRP
Breast cancer resistance protein
- SMC
Structural maintenance of chromosome proteins
- TMD
Trans membrane domains
- NBD
Nucleotide-binding domains
- NCBI
National Center for Biotechnology Information
- MAD
Minimal ancestor deviation
Authors’ contributions
The authors JFM, CGS and MFM conceptualization, writing-original draft preparation, JFM, TSS, BMV search genomics data. JFM, MFM, TSS, BMV and CGS did bioinformatics analyses. ESLA and ACAM critically revised the manuscript. All authors reviewed the manuscript and approved the final manuscript.
Funding
This work was supported by the Fundação Carlos Chagas Filho de Amparo.
à Pesquisa do Estado do Rio de Janeiro (FAPERJ-E-26/201.331/2016-REDE ZIKA), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Instituto Nacional de Ciência e Tecnologia em Entomologia Molecular (INCT-EM/CNPq),
and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Availability of data and materials
The datasets analysed during the current study are available in the GenBank and VectorBase repositories. All accession numbers were listed in Tables 1 and 2.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
One of the authors (CGS) is a member of the editorial board for the BMC Genomics journal.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Janaina Figueira-Mansur and Carlos G. Schrago contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-020-06873-8.
References
- 1.Higgins CF, Linton KJ. ABC Transporters: an Introduction and Overview. In: Holland IB, SPC C, Kuchler K, CFBT-ABCP H, editors. ABC Proteins. London: Academic Press; 2003. [Google Scholar]
- 2.Ling V. Multidrug resistance: molecular mechanisms and clinical relevance. Cancer Chemother Pharmacol. 1997;40:S3–S8. doi: 10.1007/s002800051053. [DOI] [PubMed] [Google Scholar]
- 3.Dean M, Hamon Y, Chimini G. The human ATP-binding cassette transporter superfamily. J Lipid Res. 2001;42(7):1007–1017. [PubMed] [Google Scholar]
- 4.Hyde SC, Emsley P, Hartshorn MJ, Mimmack MM, Gileadi U, Pearce SR, Gallagher MP, Gill DR, Hubbard RE, Higgins CF. Structural model of ATP-binding proteinsassociated with cystic fibrosis, multidrug resistance and bacterial transport. Nature. 1990;346:362–365. doi: 10.1038/346362a0. [DOI] [PubMed] [Google Scholar]
- 5.Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 6.Klein I, Sarkadi B, Váradi A. An inventory of the human ABC proteins. Biochim Biophys Acta Biomembr. 1999;1461:237–262. doi: 10.1016/s0005-2736(99)00161-3. [DOI] [PubMed] [Google Scholar]
- 7.Anjard C, Loomis WF. Evolutionary analyses of ABC transporters of Dictyostelium discoideum. Eukaryot Cell. 2002;1:643–652. doi: 10.1128/EC.1.4.643-652.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sturm A, Cunningham P, Dean M. The ABC transporter gene family of Daphnia pulex. BMC Genomics. 2009;10:170. doi: 10.1186/1471-2164-10-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth C, Holm I, Graille M, Dehoux P, Rzhetsky A, Wincker P, Weissenbach J, Brey P. Identification of the Anopheles gambiae ATP-binding cassette transporter superfamily genes. Mol Cell. 2003;15(2):150–158. [PubMed] [Google Scholar]
- 10.Verrier PJ, Bird D, Burla B, Dassa E, Forestier C, Geisler M, Klein M, Kolukisaoglu U, Lee Y, Martinoia E, Murphy A, Rea PA, Samuels L, Schulz B, Spalding EP, Yazaki K, Frederica L. Theodoulou: plant ABC proteins – a unified nomenclature and updated inventory. Trends Plant Sci. 2008;13(4):151–159. doi: 10.1016/j.tplants.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Eisen J. A.: a phylogenomic study of the MutS family of proteins. Nucleic Acids Res. 1998;26:4291–4300. doi: 10.1093/nar/26.18.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Culligan KM, Hays JB. Arabidopsis MutS homologs-AtMSH2, AtMSH3, AtMSH6, and a novel AtMSH7-form three distinct protein heterodimers with different specificities for mismatched DNA. Plant Cell. 2000;12(6):991–1002. doi: 10.1105/tpc.12.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strunnikov AV, Jessberger R. Structural maintenance of chromosomes (SMC) proteins: conserved molecular properties for multiple biological functions. Eur J Biochem. 1999;263:6–13. doi: 10.1046/j.1432-1327.1999.00509.x. [DOI] [PubMed] [Google Scholar]
- 14.Alani E, Subbiah S, Kleckner N. The yeast RAD50 gene encodes a predicted 153-kD protein containing a purine nucleotide-binding domain and two large heptad-repeat regions. Genetics. 1989;122(1):47–57. doi: 10.1093/genetics/122.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.J A S, Ralph S, Zhao ZY, Baillie DL, Ling V. The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome Biol. 2004;5:R15. doi: 10.1186/gb-2004-5-3-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dermauw W, Van Leeuwen T. The ABC gene family in arthropods: comparative genomics and role insecticide transport and resistance. Insect Biochem Mol Biol. 2014;45:89–110. doi: 10.1016/j.ibmb.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Sánchez-Fernández R, Emyr Davies TG, Coleman JOD, Rea PA. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J Biol Chem. 2001;276:30231–30244. doi: 10.1074/jbc.M103104200. [DOI] [PubMed] [Google Scholar]
- 18.Decottignies A, Goffeau A. Complete inventory of the yeast ABC proteins. Nat Genet. 1997;15:137–145. doi: 10.1038/ng0297-137. [DOI] [PubMed] [Google Scholar]
- 19.Allikmets R, Gerrard B. Hutchinson a, Dean M: characterization of the human ABC superfamily: isolation and mapping of 21 new genes using the expressed sequence tags database. Hum Mol Genet. 1996;5:1649–1655. doi: 10.1093/hmg/5.10.1649. [DOI] [PubMed] [Google Scholar]
- 20.Lage H. ABC-transporters: implications on drug resistance from microorganisms to human cancers. Int J Antimicrob Agents. 2003;22:188–199. doi: 10.1016/s0924-8579(03)00203-6. [DOI] [PubMed] [Google Scholar]
- 21.Biedler JL, Riehm H. Cellular resistance to actinomycin D in Chinese hamster cells in vitro: cross-resistance, radioautographic and cytogenetic studies. Cancer Res. 1970;30(4):1174–1184. [PubMed] [Google Scholar]
- 22.Montella IR, Martins AJ, Viana-Medeiros PF, Lima JBP, Braga IA, Valle D. Insecticide resistance mechanisms of Brazilian Aedes aegypti populations from 2001 to 2004. Am J Trop Med Hyg. 2007;77:467–477. [PubMed] [Google Scholar]
- 23.Albrieu Llinás G, Seccacini E, Gardenal CN, Licastro S. Current resistance status to temephos in Aedes aegypti from different regions of Argentina. Mem Inst Oswaldo Cruz. 2010;105:113–116. doi: 10.1590/s0074-02762010000100019. [DOI] [PubMed] [Google Scholar]
- 24.Milani R. Genetical aspects of insecticide resistance. Bull World Health Organ. 1963;29(Suppl):77–87. [PMC free article] [PubMed] [Google Scholar]
- 25.Busvine JR. Cross and multiple resistance in mosquitoes. Cah ORSTOM sér Ent méd. 1968;VI:215–219. [Google Scholar]
- 26.Porretta D, Gargani M, Bellini R, Medici A, Punelli F, Urbanelli S. Defence mechanisms against insecticides temephos and diflubenzuron in the mosquito Aedes caspius: the P-glycoprotein efflux pumps. Med Vet Entomol. 2008;22(1):48–54. doi: 10.1111/j.1365-2915.2008.00712.x. [DOI] [PubMed] [Google Scholar]
- 27.Figueira-Mansur J, Ferreira-Pereira A, Mansur JF, Franco TA, Alvarenga ES, Sorgine MH, Neves BC, Melo AC, Leal WS, Masuda H, Moreira MF. Silencing of P-glycoprotein increases mortality in temephos-treated Aedes aegypti larvae. Insect Mol Biol. 2013;22(6):648–658. doi: 10.1111/imb.12052. [DOI] [PubMed] [Google Scholar]
- 28.Murray CL, Quaglia M, Arnason JT, Morris CE. A putative nicotine pump at the metabolic blood–brain barrier of the tobacco hornworm. J Neurobiol. 1994;25:23–24. doi: 10.1002/neu.480250103. [DOI] [PubMed] [Google Scholar]
- 29.Gahan LJ, Pauchet Y, Vogel H, Heckel DG. An ABC transporter mutation is correlated with insect resistance to bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 2010;6:1–11. doi: 10.1371/journal.pgen.1001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pignatelli P, Ingham VA, Balabanidou V, Vontas J, Lycett G, Ranson H. The Anopheles gambiae ATP-binding cassette transporter family: phylogenetic analysis and tissue localization provide clues on function and role in insecticide resistance. Insect Mol Biol. 2018;27(1):110–122. doi: 10.1111/imb.12351. [DOI] [PubMed] [Google Scholar]
- 31.Lanning CL, Fine RL, Corcoran JJ, Ayad HM, Rose RL, Abou-Donia MB. Tobacco budworm P-glycoprotein: biochemical characterization and its involvement in pesticide resistance. Biochim Biophys Acta. 1996;1291(2):155–162. doi: 10.1016/0304-4165(96)00060-8. [DOI] [PubMed] [Google Scholar]
- 32.Srinivas R, Jayalakshmi SK. Sreeramulu K: hydrolysis of organophosphorus compounds by an esterase isozyme from insecticide resistant pest Helicoverpa armigera. Indian J Exp Biol. 2004;42(2):214–216. [PubMed] [Google Scholar]
- 33.Valenzuela-Muñoz V, Sturm A, Gallardo-Escárate C. Transcriptomic insights on the ABC transporter gene family in the salmon louse Caligus rogercresseyi. Parasit Vectors. 2015;8:209. doi: 10.1186/s13071-015-0801-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carmona-Antoñanzas G, Carmichael SN, Heumann J, Taggart JB, Gharbi K, Bron JE, Bekaert M, Sturm A. A Survey of the ATP-Binding Cassette (ABC) Gene Superfamily in the Salmon Louse (Lepeophtheirus salmonis) PLoS One. 2015;10(9):e0137394. doi: 10.1371/journal.pone.0137394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berry IM, Rutvisuttinunt W, Sippy R, Beltran-Ayala E, Figueroa K, Ryan S, Srikanth A, Stewart-Ibarra AM, Endy T, Jarman RG. The origins of dengue and chikungunya viruses in Ecuador following increased migration from Venezuela and Colombia. BMC Evol Biol. 2020;20(1):31. doi: 10.1186/s12862-020-1596-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu H, Xua Y, Cui F. Phylogenetic analysis of the ATP-binding cassette transporter family in three mosquito species. Pestic Biochem Physiol. 2016;132:118–124. doi: 10.1016/j.pestbp.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Dean M, Annilo T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet. 2005;6:123–142. doi: 10.1146/annurev.genom.6.080604.162122. [DOI] [PubMed] [Google Scholar]
- 38.Tapadia MG, Lakhotia SC. Expression of mdr49 and mdr65 multidrug resistance genes in larval tissues of Drosophila melanogaster under normal and stress conditions. Cell Stress Chaperones. 2005;10:7–11. doi: 10.1379/CSC-67R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitsuhashi N, Miki T, Senbongi H, Yokoi N, Yano H, Miyazaki M, Nakajima N, Iwanaga T, Yokoyama Y, Shibata T, Seino S. MTABC3, a novel mitochondrial ATP-binding cassette protein involved in iron homeostasis. J Biol Chem. 2000;275:17536–17540. doi: 10.1074/jbc.275.23.17536. [DOI] [PubMed] [Google Scholar]
- 40.Tarnay JN, Szeri F, Iliás A, Annilo T, Sung C, Le Saux O, Váradi A, Dean M, Boyd CD, Robinow S. The dMRP/CG6214 gene of Drosophila is evolutionarily and functionally related to the human multidrug resistance-associated protein family. Insect Mol Biol. 2004;13:539–548. doi: 10.1111/j.0962-1075.2004.00512.x. [DOI] [PubMed] [Google Scholar]
- 41.Szeri F, Iliás A, Pomozi V, Robinow S, Bakos É, Váradi A. The high turnover Drosophila multidrug resistance-associated protein shares the biochemical features of its human orthologous. Biochim Biophys Acta Biomembr. 1788;2009:402–409. doi: 10.1016/j.bbamem.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Shani N. D V: Peroxisomal ABC transporters. Methods Enzimol. 1998;292:753–793. doi: 10.1016/s0076-6879(98)92058-4. [DOI] [PubMed] [Google Scholar]
- 43.Chen ZQ, Dong J, Ishimura A, Daar I, Hinnebusch AG, Dean M. The essential vertebrate ABCE1 protein interacts with eukaryotic initiation factors. J Biol Chem. 2006;281:7452–7457. doi: 10.1074/jbc.M510603200. [DOI] [PubMed] [Google Scholar]
- 44.Dong J, Lai R, Nielsen K, Fekete CA, Qiu H, Hinnebusch AG. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J Biol Chem. 2004;279:42157–42168. doi: 10.1074/jbc.M404502200. [DOI] [PubMed] [Google Scholar]
- 45.Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T. Cloning and characterization of a RNase L inhibitor. A new component of the interferon-regulated 2-5A pathway. J Biol Chem. 1995;270:13308–13317. doi: 10.1074/jbc.270.22.13308. [DOI] [PubMed] [Google Scholar]
- 46.Marton MJ, Vazquez de Aldana CR, Qiu H, Chakraburtty K, Hinnebusch a G. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2alpha kinase GCN2. Mol Cell Biol. 1997;17:4474–4489. doi: 10.1128/mcb.17.8.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paytubi S, Morrice NA, Boudeau J, Proud CG. The N-terminal region of ABC50 interacts with eukaryotic initiation factor eIF2 and is a target for regulatory phosphorylation by CK2. Biochem J. 2008;409:223–231. doi: 10.1042/BJ20070811. [DOI] [PubMed] [Google Scholar]
- 48.Paytubi S, Wang X, Lam YW, Izquierdo L, Hunter MJ, Jan E, Hundal HS, Proud CG. ABC50 promotes translation initiation in mammalian cells. J Biol Chem. 2009;284:24061–24073. doi: 10.1074/jbc.M109.031625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sullivan D, Grillo S, Kitos R. Subcellular localization of the first three enzymes of the ommochrome synthetic pathway in Drosophila melanogaster. J Exp Zool. 1974;188:225–233. doi: 10.1002/jez.1401880210. [DOI] [PubMed] [Google Scholar]
- 50.Mackenzie SM, Brooker MR, Gill TR, Cox GB, Howells AJ, Ewart GD. Mutations in the white gene of Drosophila melanogaster affecting ABC transporters that determine eye colouration. Biochim Biophys Acta Biomembr. 1999;1419:173–185. doi: 10.1016/s0005-2736(99)00064-4. [DOI] [PubMed] [Google Scholar]
- 51.Popovic M, Zaja R, Loncar J, Smital T. A novel ABC transporter: the first insight into zebrafish (Danio rerio) ABCH1. Mar Environ Res. 2010;69(SUPPL.1):S11–S13. doi: 10.1016/j.marenvres.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 52.Hopfner KP. Tainer J a.: Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr Opin Struct Biol. 2003;13:249–255. doi: 10.1016/s0959-440x(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 53.Melby TE, Ciampaglio CN, Briscoe G, Erickson HP. The Symmetrical Structure of Structural Maintenance of Chromosomes (SMC) and MukB Proteins: Long, Antiparallel Coiled Coils, Folded at a Flexible Hinge. J Cell Biol. 1998;142(6):1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soppa J. Prokaryotic Structural Maintenance of Chromosomes (SMC) Proteins: Distribution, Phylogeny, and Comparison with MukBs and Additional Prokaryotic and Eukaryotic Coiled-Coil Proteins. Gene. 2001;278(1–2):253–264. doi: 10.1016/s0378-1119(01)00733-8. [DOI] [PubMed] [Google Scholar]
- 55.Aravind L, Walker DR, Kroonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, Wind N, Sixma TK. The Crystal Structure of DNA Mismatch Repair Protein MutS Binding to a G X T Mismatch. Nature. 2000;407(6805):711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 57.Altschul SF, Lipman DJ. Protein database searches for multiple alignments. Proc Natl Acad Sci U S A. 1990;87:5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2014;43(Database issue):D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marchler-Bauer A, Bryant SH. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004;32(WEB SERVER ISS):327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar RC. MUSCLE:multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: A fast and. effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 64.Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994;39(3):306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 65.Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tria FDK, Landan G, Dagan T. Phylogenetic rooting using minimal ancestor deviation. Nat Ecol Evol. 2017;1:193. doi: 10.1038/s41559-017-0193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Sequences.txt: Rad50, SMC and MutS sequences highlighted Walker A and Walker B motifs and ABC signature.
Additional file 2. ABC.txt: Unaligned ABC sequences used in this study.
Additional file 3. ABCalin.txt: Aligned and trimmed ABC sequences employed in evolutionary analyses.
Additional file 4. ABC.unrooted.txt: Unrooted maximum likelihood phylogenetic tree of the ABC gene family.
Data Availability Statement
The datasets analysed during the current study are available in the GenBank and VectorBase repositories. All accession numbers were listed in Tables 1 and 2.