Abstract
Background
Although tremendous improvement has been seen in cancer diagnosis and treatment, its morbidity and mortality is still high due to lack of ideal biomarkers. An increasing number of studies have demonstrated that the expression of lncRNA small nucleolar RNA host gene 6 (SNHG6) has significantly negative correlation with various cancer prognosis. The present meta-analysis was aimed to clarify the potential of clinical application of SNHG6 in cancers.
Methods
A detailed literature review was conducted by searching through PubMed and Web of Science databases. The expression level of SNHG6, clinicopathological features and survival outcomes were extracted from eligible studies. Pooled analysis was performed with a DerSimonian-Laird random-effect model. The results were further validated through the Cancer Genome Atlas (TCGA) dataset.
Results
Five studies with a total of 487 cases were finally included in this meta-analysis. The results demonstrated that a high expression of SNHG6 was significantly associated with an increased risk of poor overall survival (OS) in cancer patients (HR = 2.06, 95% CI 1.56–2.73). Similar results from the TCGA dataset further confirmed our findings.
Conclusions
Overexpressed SNHG6 was significantly associated with poor prognosis in various cancers. Therefore, SNHG6 may become a novel molecular target for treatment and prognostic evaluation.
Keywords: Cancer, Prognosis, Long no-coding RNA, SNHG6
Background
Cancer has become the leading cause of death globally. Over the past decades, although tremendous improvement has been achieved in its diagnosis and treatment, the prognosis is still poor, especially for advanced cancers [1]. In the United States, it was estimated that approximately 1,762,450 cancer cases would be diagnosed and an estimated 606,880 people would die from cancer in 2019 [2]. It can impose huge financial burden on patients’ families and society. Therefore, novel biomarkers are urgent to be discovered for early diagnosis, treatment and prognostic assessment.
Among ~ 90% of human genome DNA that are transcribed, only 2% of them encode protein. The others which encode non-protein are named as non-coding RNAs (ncRNAs) [3, 4]. Long non-coding RNAs (lncRNAs) is a category of endogenous ncRNAs with a length of more than 200 nucleotides, which accounting for > 70% of ncRNAs [5–7]. Increasing studies have demonstrated that lncRNAs is involved in various normal cellular processes including development, differentiation and metabolism by epigenetic regulation, transcription and post-transcriptional regulation [8–11]. Recently, dysregulation of lncRNAs has been reported to be associated with oncogenesis and cancer progression [12–15], which suggested the potential of lncRNAs to be a new biomarker for early diagnosis, prognostic value and therapeutic target.
Small nucleolar RNA host gene 6 (SNHG6), also known as U87HG, is a novel lncRNA located in chromosome 8q13.1. In 2016, Chang et al. firstly illustrated that SNHG6 was overexpressed in hepatocellular carcinoma (HCC) and promoted tumor growth and metastasis by inducing epithelial to mesenchymal transition (EMT) [16]. In recent years, accumulating evidence revealed that SNHG6 was aberrantly expression in various types of cancers and was significantly correlated with clinical stage and prognosis [16–18]. Whereas, due to small sample size in these researches, the prognostic value of SNHG6 is limited and controversial. Therefore, this meta-analysis was conducted to investigate the potential prognostic value of SNHG6 in human cancers.
Methods
Literature search
This study was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement [19]. Studies on the association between SNHG6 expression and prognosis of human cancers were identified from PubMed and Web of Science (WOS) database (up to October 30, 2019) with the following search strategy: (“small nuclear RNA host gene 6” or “SNHG6”) and (“cancer” or “carcinoma” or “neoplasm” or “tumor”). Reference lists of the identified articles and relevant reviews were manually examined for additional eligible studies. Potential eligible studies were selected by two independent authors (XX and HXS), and controversial articles were resolved by discussion and consensus.
Inclusion criteria
Studies included in this meta-analysis met all the following criteria: (i) study population was cancer patients; (ii) OS were analyzed according to SNHG6 expression pattern; (iii) multi-adjusted hazard ratios (HRs) and their 95% confidence intervals (CIs) were provided; (iv) articles were published in English or Chinese. Studies only reported risk estimates from univariate analysis were excluded from this meta-analysis.
Data extraction
The following information was extracted from each included study by two independent authors (XX and HXS): the surname of first author, publication year, country, number of patients, age of patients, methods to determine SNHG6 expression, cut-off value, prognostic data, and adjusted variables. Any discrepancies were solved by consensus.
Quality assessment
The methodological quality of each included study was evaluated by two independent reviewers (XX and HXS) using the Newcastle-Ottawa Scale (NOS) with reasonable modifications. NOS is an eight-item instrument that focused on the characteristics of study population, study comparability, follow-up and outcome of interest. The total score of NOS is 9. A study with a score of ≥ 7 was considered to be of high-quality.
Validation by reviewing public data
This study meets the publication guidelines provided by The Cancer Genome Atlas (TCGA). Gene Expression Profiling Interactive Analysis (GEPIA) [20] was used to verify the correlation between SNHG6 and OS and to assess the expression pattern of SNHG6 in human cancers.
RNA extraction and qRT-PCR
Total RNA was extracted from the human RCC cell lines 786-O and Caki-1, as well as one normal kidney cell line HK-2 using the RNAiso Plus (TaKaRa, Japan). Then the RNA was transcribed into cDNA using the PrimeScript RT Reagent Kit (TaKaRa, Japan). qRT-PCR assay was performed using ABI 7500 FAST Real-Time PCR System (Applied Biosystems, USA) and SYBR Green PCR Kit (Takara, China). The mRNA expression level was calculated using the 2−∆∆Ct method after normalization with β-actin. The primers involved were SNHG6 Forward 5′-ATACTTCTGCTTCGTTACCT-3′; Reverse 5′-CTCATTTTCATCATTTGCT-3′; β-actin Forward 5′-ATCATGAAGTGTGACGTGGAC-3′; Reverse 5′-GACTCGTCATACTCCTGCTTG-3′.
Statistical methods
The impact of SNHG6 expression on the OS of cancer patients was quantified by the summary HRs and their 95% CIs. A DerSimonian-Laird random-effect model [21] was used to calculate the summary risk estimates. Existence of heterogeneity among included studies was determined using the Q statistic (significant level set at 0.1) [22]. I2 statistic was further used to assess the degree of heterogeneity (low heterogeneity: I2 < 25%; moderate heterogeneity: I2 = 25–50%; high heterogeneity: I2 > 50%). Sensitivity analysis was performed by sequential omission of each included study. Publication bias was assessed using a visual funnel plot. All of the statistical analyses were performed with STATA 11.0 (StataCorp, College Station, Texas USA), using two-sided P values.
Results
Literature search and study characteristics
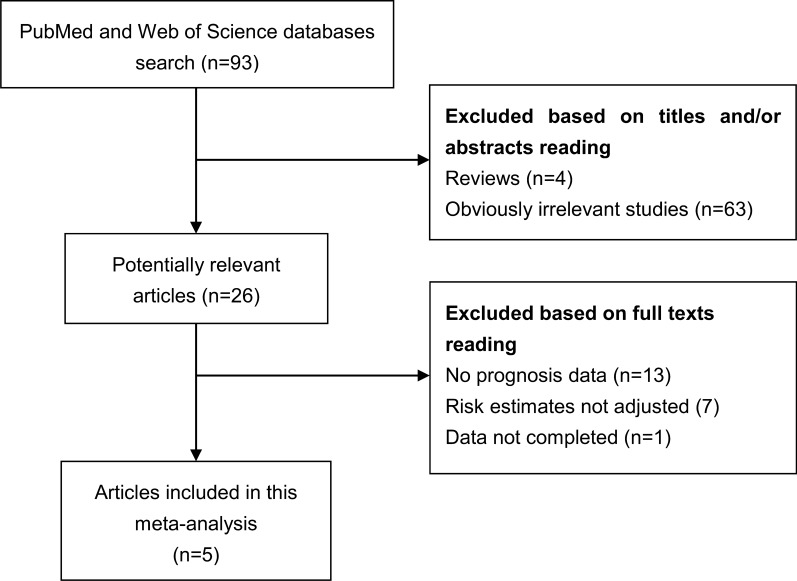
Literature search and selection has been shown in Fig. 1. Five studies [23–27] were finally included in this meta-analysis aimed to evaluate the association between SNHG6 expression and OS of cancer patients. Three studies were performed in colorectal cancer (CRC), one in renal cell carcinoma (RCC) and one in glioma. All of these studies were published between 2018 and 2019. These studies were performed in China and involved a total of 487 cases. SNHG6 expression data was obtained by real time PCR (RT-PCR). The methodological quality, as assessed by the NOS, ranged from 8 to 9 (with a mean of 8.4). The main characteristics of each study have been summarized in Table 1.
Fig. 1.
The diagram shows the procedure of literature search and study selection
Table 1.
Main characteristics of the studies included in the meta-analysis
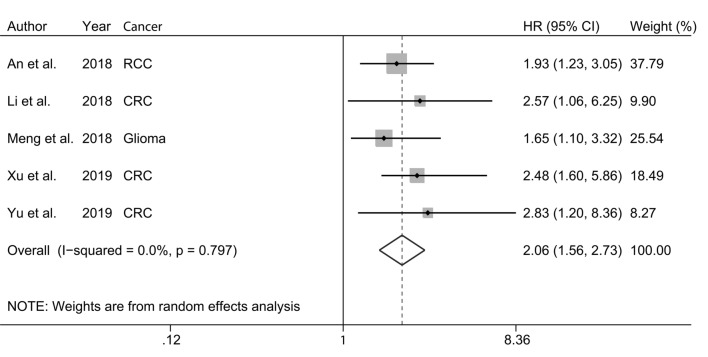
| Authors | Year | Cancer | HR (95% CI) | Method | No. | Follow | NOS |
|---|---|---|---|---|---|---|---|
| An et al. | 2018 | RCC | 1.93 (1.23–3.05) | RT-PCR | 81 | 80 m | 9 |
| Li et al. | 2018 | CRC | 2.57 (1.06–6.25) | RT-PCR | 74 | NR | 8 |
| Meng et al. | 2018 | Glioma | 1.65 (1.10–3.32) | RT-PCR | 71 | 60 m | 9 |
| Xu et al. | 2019 | CRC | 2.48 (1.60–5.86) | RT-PCR | 120 | NR | 8 |
| Yu et al. | 2019 | CRC | 2.83 (1.20–8.36) | RT-PCR | 141 | NR | 8 |
RCC renal cell carcinoma, CRC colorectal cancer, HR hazard ratio, 95% CI 95% confidence interval, NR no report
Systematic review
Three studies performed in CRC included 74, 120, and 141 subjects, respectively. The study by Li et al. [24] indicated that SNHG6 was generally up-regulated in CRC tissues and high level of SNHG6 expression was strongly associated with advanced tumor stage (P = 0.026) and poor prognosis (P = 0.0215). The study by Xu et al. [26] reported that SNHG6 expression was an independent prognostic biomarker (HR = 2.48, 95% CI = 1.60–5.86, P = 0.002) for CRC in the multivariate analysis. Yu et al. [27] found high expression of SNHG6 was positively related with tumor size, advanced TNM stage, and tumor metastasis. In addition, SNHG6 was an independent prognostic factor of poor OS (HR = 2.83, 95% CI 1.20–8.36, P = 0.018) and RFS (HR = 2.07, 95% CI 1.17–6.20, P = 0.020). The study by An et al. [23] reported that elevated SNHG6 was significantly associated with tumor progression and lymph node metastasis in a total of 81 cases of RCC. In addition, SNHG6 expression was associated with overall prognosis in RCC. Meng et al. [25] investigated the role of SNHG6 in glioma and found that the expression of SNHG6 was negatively associated with the OS (HR = 1.65; 95% CI 1.10–3.32; P = 0.0076).
Overall analysis
The HRs for each included study and for the combination of all studies are shown in Fig. 2. A high expression of SNHG6 was significantly associated with an increased risk of poor OS (HR = 2.06, 95% CI 1.56–2.73). There was no obvious heterogeneity among the studies (I2 = 0.0%; P = 0.797).
Fig. 2.
Forest plot for the correlation between expression level of SNHG6 and overall survival (OS)
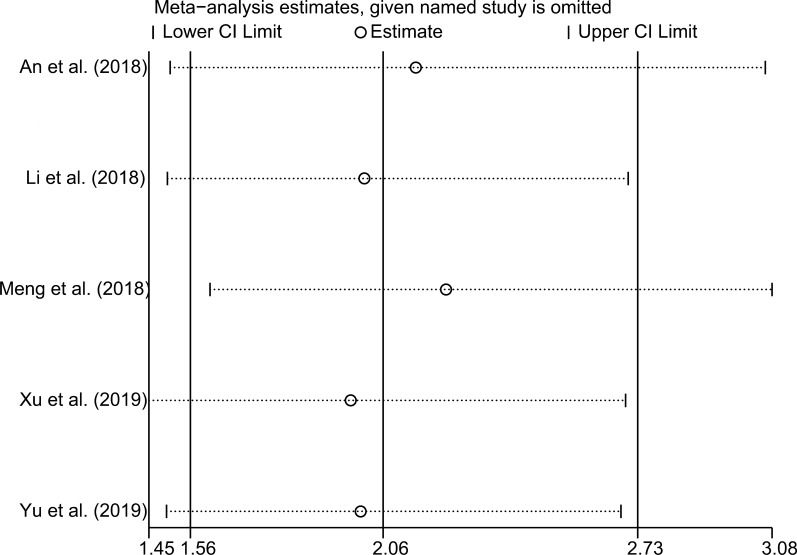
Sensitivity analysis and publication bias
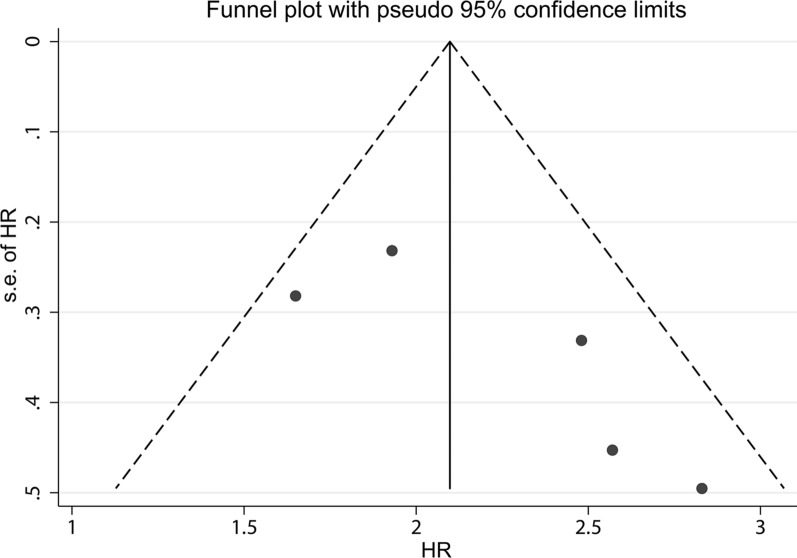
In order to assess the stability of the pooled risk estimate of the association between SNHG6 expression and OS, sensitivity analysis was performed by omitting each included study in turn. As shown in Fig. 3, the pooled result was not dominated by any single study. There was no evidence of publication bias with a visual funnel plot (Fig. 4).
Fig. 3.
Sensitivity analysis of the included studies concerning SNHG6 and overall survival (OS)
Fig. 4.
Funnel plot of SNHG6 for overall survival
SNHG6 and OS of CRC
Three studies reported data on the relationship between SNHG6 expression and OS of CRC. The summary data based on these studies indicated that SNHG6 status was significantly associated with the OS with a combined HR of 2.58 (95% CI 1.63–4.09). No obvious heterogeneity was observed across studies (I2 = 0.0%, P = 0.976).
Validation of the SNHG6 expression in RCC cells
The mRNA expression of SNHG6 in two RCC cell lines (786-O and Caki-1) and one normal kidney cell line (HK-2) was detected using qRT-PCR. As a result, the expression of SNHG6 in RCCs was significantly upregulated compared with that in HK-2 cells (Additional file 1: Figure S1).
Validation of the results in TCGA dataset
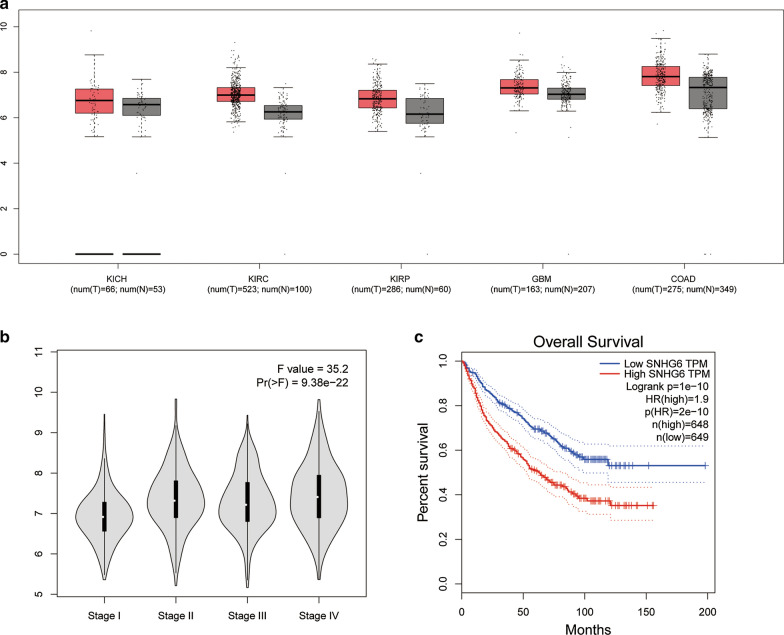
We further used TCGA dataset to investigate SNHG6 expression level in human cancers. As shown in Fig. 5a, SNHG6 was upregulated in kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), glioblastoma multiforme (GBM) and colon adenocarcinoma (COAD) when compared with non-cancer tissues. The violin plot indicated that SNHG6 expression was significantly associated with clinical stage in these human cancers (Fig. 5b). Finally, a survival plot merging SNHG6 expression data and OS data of RCC, GBM and CRC from the TCGA dataset were performed. As shown in Fig. 5c, the overexpression of SNHG6 was significantly associated with an unfavorable OS, which was consistent with our results in this meta-analysis.
Fig. 5.
Validation of SNHG6 expression of various cancers in the TCGA dataset. a The expression levels of SNHG6 in KICH, KIRC, KIRP, GBM and COAD. b Violin plot showing that SNHG6 expression was significantly associated with clinical stage in these human cancers. c Overall survival plot of SNHG16 in TCGA cohort (n = 1297, log-rank p < 0.001)
Discussion
In the past decades, a large amount of lncRNA transcripts were discovered by high throughput genome sequencing technologies. Along with the emerging evidence, the pivotal role of lncRNAs in oncogenesis was unveiled gradually [28, 29]. lncRNAs, including SNHG6, were also indicated as prognostic biomarkers in literature [23, 24]. However, the prognostic value of SNHG6 was limited and contentious due to the small sample size. Therefore, our study aimed to clarify the potential prognostic value in multiple cancers through a pooled analysis.
As a novel lncRNA, SNHG6 had been discovered overexpressed in various types of cancers, including CRC [30], HCC [16], breast cancer [31], gastric cancer [32], lung cancer [33], glioma [25] and osteosarcoma [18]. In addition, a negative correlation between SNHG6 expression and prognosis has been demonstrated in the literature [33, 34]. As for the carcinogenesis role of SNHG6, the underlying molecular mechanisms had been partly elucidated. Through the competing endogenous RNA (ceRNA) mechanism, SNHG6 can competitively sponge miRNAs and regulate their target genes. In 2016, Chang et al. firstly reported that SNHG6 facilitated tumor growth and metastasis in hepatocellular carcinoma by competitively binding miR-101-3p to regulate ZEB1 [16]. Wang et al. found that the up-regulation of SNHG6 significantly repressed the expression of miR-125b and increased the NUAK1 expression [35]. miR-26a-5p/ULK1 and miR-26a-5p/MAPK6 axis were also regulated by SNHG6 and participated in the development and progression of breast cancer and osteosarcoma, respectively [18, 31]. Another study by Jafari-Oliayi et al. [34] also found that the expression of SNHG6 was significantly upregulated in primary breast cancers. SNHG6 silencing led to G1 cell cycle arrest and suppressed cell proliferation. Several signaling pathways can be activated by SNHG6, including the MAPK and JNK pathway in gastric cancer [32], PI3K/AKT/mTOR pathway and TGF-β/Smad pathway in colorectal cancer [36, 37]. SNHG6 could also regulate cell cycle through interacting with crucial factors like p21 [32, 38]. In summary, elevated SNHG6 plays a vital role in cancer cell proliferation, migration and invasion.
In this meta-analysis, five original studies with a total of 487 cases were finally included and the results were pooled with adjustment for multiple confounding factors. The overall analysis showed that a high expression of SNHG6 was significantly correlated with an unfavorable OS (HR = 2.06, 95% CI 1.56–2.73). Moreover, the results of subgroup analysis were also demonstrated the negative correlation between expression level of SNHG6 and prognosis, which was consistent with overall analysis.
Several limitations of our study should be acknowledged. First, because we only included studies providing multi-adjusted results, the number of papers included in final analysis was relatively small. Secondly, all of the five included studies were performed in China with Chinese population. Therefore, the generalization of our findings is relatively limited. Thirdly, various therapies for different patients may have been used in included studies, which can lead to some bias. Finally, the definition for high SNHG6 expression was obscure and might be different in the included studies, which may also affect the final pooled results.
Conclusion
In summary, the results of this meta-analysis support that SNHG6 overexpression is significantly associated with a poor prognosis in human cancers. SNHG6 may become a novel molecular target for treatment and prognostic evaluation. However, due to the limitations of our study, more well-designed studies are warranted to validate the role of SNHG6 in human cancers.
Supplementary information
Additional file 1: Figure S1. The mRNA expression of SNHG6 in two RCC cell lines (786-O and Caki-1) and one normal kidney cell line (HK-2) was detected by qRT-PCR.
Acknowledgements
We thank all researchers and participants of included studies for their contributions. This study was supported by the Zhejiang Medical and Health Science and Technology Project (2020KY542) and the National Natural Science Foundation of China (81702500).
Abbreviations
- lncRNA
Long non-coding RNA
- WHO
World Health Organization
- HCC
Hepatocellular carcinoma
- EMT
Epithelial to mesenchymal transition
- HR
Hazard ratio
- CI
Confidence interval
- OS
Overall survival
- TCGA
The Cancer Genome Atlas
- NOS
Newcastle–Ottawa Scale
- CRC
Colorectal cancer
- RCC
Renal cell carcinoma
- KICH
Kidney chromophobe
- KIRC
Kidney renal clear cell carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- GBM
Glioblastoma multiforme
- COAD
Colon adenocarcinoma
- ceRNA
Competing endogenous RNA
Authors’ contributions
Conceived and designed the study: XX and BL. Performed the study: HXS, QWM, XX and BL. Analyzed the data: XX and QWM. Contributed analysis tools/materials: HXS, QWM, XX and BL. Wrote the paper: HXS, QWM and XX. All authors read and approved the final manuscript.
Availability of data and materials
All data and materials analyzed in this study are included in this published article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests in this study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xin Xu, Email: drxuxin@zju.edu.cn.
Ben Liu, Email: drliuben@zju.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12935-020-01383-9.
References
- 1.Binabaj MM, Bahrami A, Bahreyni A, Shafiee M, Rahmani F, Khazaei M, et al. The prognostic value of long noncoding RNA MEG3 expression in the survival of patients with cancer: a meta-analysis. J Cell Biochem. 2018;119(11):9583–9590. doi: 10.1002/jcb.27276. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA A Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65(6):1140–1151. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunham I, Kundaje A, Aldred S, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein LD. Human genome: end of the beginning. Nature. 2004;431(7011):915–916. doi: 10.1038/431915a. [DOI] [PubMed] [Google Scholar]
- 8.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 9.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16(2):71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao X, Su Z, Mookhtiar AK. Long non-coding RNA: a versatile regulator of the nuclear factor-kappaB signalling circuit. Immunology. 2017;150(4):379–388. doi: 10.1111/imm.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20(3):300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 12.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang L, Yuan Y, Li C, Guo T, Qi H, Xiao Y, et al. Upregulation of SNHG6 regulates ZEB1 expression by competitively binding miR-101-3p and interacting with UPF1 in hepatocellular carcinoma. Cancer Lett. 2016;383(2):183–194. doi: 10.1016/j.canlet.2016.09.034. [DOI] [PubMed] [Google Scholar]
- 17.Cao C, Zhang T, Zhang D, Xie L, Zou X, Lei L, et al. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene. 2017;36(8):1112–1122. doi: 10.1038/onc.2016.278. [DOI] [PubMed] [Google Scholar]
- 18.Zhu X, Yang G, Xu J, Zhang C. Silencing of SNHG6 induced cell autophagy by targeting miR-26a-5p/ULK1 signaling pathway in human osteosarcoma. Cancer Cell Int. 2019;19:82. doi: 10.1186/s12935-019-0794-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 22.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 23.An HX, Xu B, Wang Q, Li YS, Shen LF, Li SG. Up-regulation of long non-coding RNA SNHG6 predicts poor prognosis in renal cell carcinoma. Eur Rev Med Pharmacol Sci. 2018;22(24):8624–8629. doi: 10.26355/eurrev_201812_16626. [DOI] [PubMed] [Google Scholar]
- 24.Li M, Bian Z, Yao S, Zhang J, Jin G, Wang X, et al. Up-regulated expression of SNHG6 predicts poor prognosis in colorectal cancer. Pathol Res Pract. 2018;214(5):784–789. doi: 10.1016/j.prp.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 25.Meng Q, Yang BY, Liu B, Yang JX, Sun Y. Long non-coding RNA SNHG6 promotes glioma tumorigenesis by sponging miR-101-3p. Int J Biol Mark. 2018;33(2):148–155. doi: 10.1177/1724600817747524. [DOI] [PubMed] [Google Scholar]
- 26.Xu M, Chen X, Lin K, Zeng K, Liu X, Xu X, et al. lncRNA SNHG6 regulates EZH2 expression by sponging miR-26a/b and miR-214 in colorectal cancer. J Hematol Oncol. 2019;12(1):3. doi: 10.1186/s13045-018-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu C, Sun J, Leng X, Yang J. Long noncoding RNA SNHG6 functions as a competing endogenous RNA by sponging miR-181a-5p to regulate E2F5 expression in colorectal cancer. Cancer Manage Res. 2019;11:611–624. doi: 10.2147/CMAR.S182719. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Pecero ML, Salvador-Bofill J, Molina-Pinelo S. Long non-coding RNAs as monitoring tools and therapeutic targets in breast cancer. Cell Oncol. 2019;42(1):1–12. doi: 10.1007/s13402-018-0412-6. [DOI] [PubMed] [Google Scholar]
- 29.Calore F, Lovat F, Garofalo M. Non-coding RNAs and cancer. Int J Mol Sci. 2013;14(8):17085–17110. doi: 10.3390/ijms140817085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Lan Z, He J, Lai Q, Yao X, Li Q, et al. LncRNA SNHG6 promotes chemoresistance through ULK1-induced autophagy by sponging miR-26a-5p in colorectal cancer cells. Cancer Cell Int. 2019;19:234. doi: 10.1186/s12935-019-0951-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lv P, Qiu X, Gu Y, Yang X, Xu X, Yang Y. Long non-coding RNA SNHG6 enhances cell proliferation, migration and invasion by regulating miR-26a-5p/MAPK6 in breast cancer. Biomed Pharmacother. 2019;110:294–301. doi: 10.1016/j.biopha.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Li D, Zhao M, Huang S, Zhang Q, Lin H, et al. Long noncoding RNA SNHG6 regulates p21 expression via activation of the JNK pathway and regulation of EZH2 in gastric cancer cells. Life Sci. 2018;208:295–304. doi: 10.1016/j.lfs.2018.07.032. [DOI] [PubMed] [Google Scholar]
- 33.Liang R, Xiao G, Wang M, Li X, Li Y, Hui Z, et al. SNHG6 functions as a competing endogenous RNA to regulate E2F7 expression by sponging miR-26a-5p in lung adenocarcinoma. Biomed Pharmacother. 2018;107:1434–1446. doi: 10.1016/j.biopha.2018.08.099. [DOI] [PubMed] [Google Scholar]
- 34.Jafari-Oliayi A, Asadi MH. SNHG6 is upregulated in primary breast cancers and promotes cell cycle progression in breast cancer-derived cell lines. Cell Oncol. 2019;42(2):211–221. doi: 10.1007/s13402-019-00422-6. [DOI] [PubMed] [Google Scholar]
- 35.Wang C, Tao W, Ni S, Chen Q. Upregulation of lncRNA snoRNA host gene 6 regulates NUAK family SnF1-like kinase-1 expression by competitively binding microRNA-125b and interacting with Snail1/2 in bladder cancer. J Cell Biochem. 2019;120(1):357–367. doi: 10.1002/jcb.27387. [DOI] [PubMed] [Google Scholar]
- 36.Meng S, Jian Z, Yan X, Li J, Zhang R. LncRNA SNHG6 inhibits cell proliferation and metastasis by targeting ETS1 via the PI3K/AKT/mTOR pathway in colorectal cancer. Mol Med Rep. 2019;20(3):2541–2548. doi: 10.3892/mmr.2019.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Lai Q, He J, Li Q, Ding J, Lan Z, et al. LncRNA SNHG6 promotes proliferation, invasion and migration in colorectal cancer cells by activating TGF-beta/Smad signaling pathway via targeting UPF1 and inducing EMT via regulation of ZEB1. Int J Med Sci. 2019;16(1):51–59. doi: 10.7150/ijms.27359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruan J, Zheng L, Hu N, Guan G, Chen J, Zhou X, et al. Long noncoding RNA SNHG6 promotes osteosarcoma cell proliferation through regulating p21 and KLF2. Arch Biochem Biophys. 2018;646:128–136. doi: 10.1016/j.abb.2018.03.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. The mRNA expression of SNHG6 in two RCC cell lines (786-O and Caki-1) and one normal kidney cell line (HK-2) was detected by qRT-PCR.
Data Availability Statement
All data and materials analyzed in this study are included in this published article.