Abstract
Glycoproteins play a central role in many biological processes including disease mechanisms. Nevertheless, because glycoproteins are heterogeneous entities, it remains unclear how glycosylation modulates the protein structure and function. Here, we assess the ability of tandem-trapped ion mobility spectrometry – mass spectrometry (tandem-TIMS/MS) to characterize the structure and sequence of the homo-tetrameric glycoprotein avidin. We show that (1) tandem-TIMS/MS retains native-like avidin tetramers with deeply buried solvent particles; (2) applying high activation voltages in the interface of tandem-TIMS results in collision-induced dissociation (CID) of avidin tetramers into compact monomers, dimers, and trimers with cross sections consistent with x-ray structures and reports from surface-induced dissociation (SID); (3) avidin oligomers are best described as heterogeneous ensembles with (essentially) random combinations of monomer glycoforms; (4) native top-down sequence analysis of the avidin tetramer is possible by CID in tandem-TIMS. Overall, our results demonstrate that tandem-TIMS/MS has the potential to correlate individual proteoforms to variations in protein structure.
Graphical Abstract
The function of proteins is directly linked to the conformations they adopt and to the motions by which they interconvert.1 This causal relationship between the conformational heterogeneity2 of a protein and its biological activity3 has been documented even for species as small as ubiquitin.4 The link between the biological function of a protein and its structural heterogeneity takes on increased complexity when differentially modified variants (proteoforms) co-exist5 and, furthermore, when the protein forms a complex.6–9 Here, the conformational heterogeneity of the protein complex is convolved with structural effects caused by post-translational modifications and variations in the amino acid sequence.
One of the most common protein modifications is the post-/co-translational glycosylation of asparagine or arginine residues in the endoplasmic reticulum by glycosyltransferases (N-glycosylation).10,11 Glycoproteins are involved in most major diseases including cancer,12 cardiovascular diseases,13 and HIV/AIDS.8,9 Because glycosylation profoundly influences the physical and chemical properties of the protein,14,15 it is imperative to characterize the structures of glycoproteins.
The structural analysis of glycoproteins and their complexes remains challenging, however, because protein glycosylation is not a well-regulated process.11,16 Glycoproteins typically exist as a heterogeneous mixture of glycoforms that differ in both the isomer of the attached glycans (“micro-heterogeneity”) as well as in the location of the glycosylated amino acid residues (“macro-heterogeneity”). Furthermore, because the glycan moiety is structurally flexible,17 it is difficult to obtain accurate structural information by traditional biophysical methods that measure convoluted signals in bulk.18 For these reasons, the current structural understanding of glycoproteins often stems from studies where the glycan has been enzymatically released from the protein.19,20
Hence, to relate individual proteoforms to variations in protein structure, a method is required that (1) characterizes distinct conformations of (2) individual proteoforms from a heterogeneous mixture. The conformational heterogeneity of proteins and protein complexes can be characterized by native ion mobility spectrometry – mass spectrometry methods.21–25 These measurements preserve protein conformations close to their native structures and characterize them by their momentum transfer cross section. Top-down sequence analysis using (native) high-resolution mass spectrometry identifies amino acid sequence, and also the type and the location of post-translational modifications.26–35 However, the ability of currently available methods to carry out both tasks in one instrumental setup is limited.
We have recently developed a tandem-trapped ion mobility spectrometer (tandem-TIMS), incorporated into a Qq-TOF mass spectrometer.36 Here, we investigate the ability of tandem-TIMS/MS to characterize the structural heterogeneity and the sequence of glycoprotein complexes. We use avidin, a homo-tetramer of a 128-amino acid residue protein with a glycosylation site at Asn17.37 Several glycoforms and sequence variants have been reported for avidin.38–40 We first demonstrate the ability of tandem-TIMS/MS to retain intact avidin tetramers. Collision-induced unfolding (CIU) and dissociation (CID) for mobility-selected avidin tetramers reveal presence of buried solvent particles within the intact tetramer as well as the glycosylation patterns of various avidin assemblies. Our data suggest the presence of compact, folded avidin monomers, dimers, and trimers from CID of the tetramer. Finally, we demonstrate the ability of tandem-TIMS/MS to perform top-down sequencing on native, intact avidin tetramer by TIMS2 and TIMS2-MS2 workflows.
EXPERIMENTAL DETAILS
Tandem-TIMS/MS instrumentation and measurements
Tandem-TIMS36 is incorporated in a ESI-QqTOF mass spectrometer (Bruker Daltonics, Billerica, MA, USA, see Figures S1 and S2 in the Supporting Information). Tandem-TIMS comprises two TIMS analyzers,41–43 each composed of an entrance funnel, an analyzer tunnel, and an exit funnel. Samples were infused into the electrospray ionization (ESI) source in positive mode through a gastight syringe. Nitrogen was used as buffer gas.
A full description of the experimental details are found in the Supporting Information (Section S1). Briefly, ions are mobility-selected and collisionally-activated in the interface between TIMS-1 and TIMS-2. As described elsewhere,36 mobility-selection is achieved by applying a transmitting dc voltage at aperture-2 only when the ions of interest are eluting. Mobility-selected ions can be collisionally-activated by dc-only electric fields between aperture-2 and deflector-2. To increase the electric field strength in the interface of tandem-TIMS, two nickel micro grids were installed in aperture-2 and deflector-2 (200 lines per inch, mn32, Precision eForming, Cortland, NY). The opening and the wire line widths are 112 μm and 15 μm, respectively. The grids are attached to aperture-2 and deflector-2 of our original tandem-TIMS design by means of conductive silver epoxy (McMaster, Atlanta, GA). More details are found in Figure S3 (Supporting Information).
Materials and sample preparations
Avidin, ammonium acetate, and high concentration ESI tuning mix were obtained from Thermo Fisher Scientific (Waltham, MA), Sigma-Aldrich (St. Louis, MO), and Agilent (Santa Clara, CA). 30 μM avidin samples were prepared in 100 μM ammonium acetate solution. ESI tuning mix was used as obtained for ion mobility calibration.
Calibration of ion mobilities and momentum transfer cross sections
Ion mobilities in tandem-TIMS are determined via a calibration tion procedure. For calibration, we used reduced ion mobilities of per-fluorophosphazenes contained in ESI tuning mix as described elsewhere.41,42,44 The ion mobilities were previously converted44 from cross sections reported by Stow el at.45 All ion mobilities and cross sections are reported for nitrogen gas. More details on the calibration procedure are found in Section S7 (Supporting Information).
RESULTS AND DISCUSSION
Tandem-TIMS retains intact, native-like, folded avidin tetramers
Avidin is a non-covalent protein assembly37 and therefore potentially sensitive to collisional-activation. However, dissociation of peptide/protein assemblies can be avoided by applying low DC voltages (“soft settings”) throughout the instrument. Using this strategy, we previously demonstrated that TIMS and tandem-TIMS preserve weakly bound peptide assemblies46 and native-like structures of the protein ubiquitin.36,47
Figure 1A shows the mass spectrum recorded on our tandem-TIMS instrument under optimized, “soft” settings for avidin (see Section S1.2, Supporting Information, for details). The mass spectrum shows three dominant peaks between ~3200 to ~4000 m/z which correspond to avidin tetramers with charge states 17+ to 19+ (minor abundances of charge states 16+ and 20+ are also observed). Minor abundances between ~5000 – 6000 m/z suggest the presence of aggregated avidin tetramers in line with prior work.48,49
Figure 1. Tandem-TIMS/MS retains intact, native-like avidin tetramers.

(A) Tetramers with charge states 17+ to 19+ predominate in the native mass spectrum recorded for avidin by tandem-TIMS/MS. (B) Cross sections obtained for avidin tetramers by tandem-TIMS (black circles) agree with cross sections obtained on a drift-tube (red squares) and the x-ray structures (shaded). (C) Mass spectrum obtained for avidin tetramers 18+ and 19+ with calculated m/z for non-glycosylated and glycosylated tetramers for charge state 18+ (red arrows). (D) Broad tetramer peaks are observed in the ion mobility spectra, here shown for charge state 18+.
Figure 1B compares cross sections obtained by tandem-TIMS for charge states 16+ to 19+ (CCSN2 = 4089 – 4178 Å2) to cross sections recorded on a drift-tube (CCSN2 = 4150 – 4160 Å2, charge states 15+ to 17+)50 and those calculated by the PSA method51–53 for avidin x-ray structures (see Figure S4, Supporting Information). Figure 1B shows that the three data sets agree with one another. Furthermore, the cross sections observed by tandem-TIMS increase only marginally (<3%) with increasing charge state (16+ to 19+). These observations imply that avidin is kinetically trapped in a folded, native-like conformation.
In line with other reports on avidin48,54 and comparable protein complexes,49,55 the tetramer peaks observed in the mass- and ion mobility spectra are broad (Figures 1C and 1D). Additionally, the nested ion mobility / mass spectra reveal broad, asymmetric peaks (see Figure S5, Supporting Information), which implies that an increase in mass correlates with an increase in cross section. Thus, each charge state is probably composed of a number of unresolved avidin tetramer species that differ in their masses and ion mobilities. Further, the observed mass peak for tetramer 18+ is shifted to higher m/z when compared to the m/z calculated from the sequence mass for non-glycosylated avidin and nine possible homotetrameric glycoforms (Fig. 1C). As discussed previously,56 this m/z shift indicates adduction of salts or solvent in the native-like avidin tetramers.
Avidin tetramers are a heterogeneous ensemble of stable, non-interconverting species
Clemmer and co-workers previously used a tandem-drift tube ion mobility spectrometer to discuss the nature of broad ion mobility spectra of the protein ubiquitin from native electrospray.22 By selecting narrow ion mobility windows within a charge state, they demonstrated that the broad peak of native-like ubiquitin arises from structurally distinct, unresolved conformations that do not interconvert on the time-scale of the ion mobility measurement.
Figure 2 shows equivalent measurements carried out on our tandem-TIMS instrument for avidin. Figure 2A plots the arrival time distribution for charge state 18+ upon mobility separation in TIMS-1 but transmission-only in the interface region and TIMS-2. We then mobility-select several regions within this peak to allow only the selected ions to pass into TIMS-2 (see Figure 2B and Figures S6, S7, Supporting Information). As shown in Figure 2B, these mobility-selected “slices” reconstruct the shape of the peak, demonstrating the ability of tandem-TIMS to probe ions with well-defined ion mobilities.
Figure 2. Tandem-ion mobility experiments for avidin tetramer 18+.

(A) A broad peak is observed for avidin tetramers 18+ upon mobility analysis in TIMS-1. (B) Four mobility windows (“slices”) within tetramer 18+ are selected in the interface of tandem-TIMS and transmitted through TIMS-2. The mobility-selected regions reconstruct the shape of the full tetramer 18+ peak shown in (A). (C) Nine mobility “slices” are selected in the interface and mobility-analyzed in TIMS-2. The overlay of mobility-selected peaks reconstructs the full tetramer 18+ peak. For clarity, the abundances of the mobility-selected traces are scaled by a constant factor of 1.56.
Next, we probe structural changes of the mobility-selected ions by conducting a second mobility analysis in TIMS-2. Figure 2C compares the complete avidin tetramer peak to nine selected “slices” after mobility-analysis in TIMS-2. The plot shows that the full tetramer peak can be represented as a sum of individual mobility-selected regions. The data show that the selected ions retain their mobilities and relative abundances (see Figure S7, Supporting Information). Furthermore, the corresponding mass spectra of mobility-selected ions show that an increase in cross section correlates with an increase in mass (Figure S7, Supporting Information). Thus, the mobility-selected regions reproduce the asymmetry noticed in the nested ion mobility / mass spectrum of the tetramer precursor (Figure S5, Supporting Information).
These observations reveal that the avidin tetramer is best described as a heterogeneous ensemble composed of a multitude of tetramer species with different ion mobilities and masses that do not interconvert on the ~100 ms time scale of the tandem-TIMS measurement. The structural complexity of the avidin tetramer potentially arises from four different aspects. First, solvent or salts may remain attached to the protein complex. Second, several sequence variants are reported for avidin in UniProt. And third, a number of distinct glycoforms were reported for monomeric avidin.38–40 Finally, each species may also adopt different conformations with similar mobilities that are unresolved by the experiment.
Native-like avidin tetramers are composed of various glycoforms with buried solvent adducts
To probe the presence of solvent adducts and/or different glycoforms in the avidin tetramer, we first select two mobility-regions within tetramer charge state 18+ and then collisionally-activate the selected ions in the interface between TIMS-1 and TIMS-2 (see Figures S2, S3 and S6, Supporting Information). The corresponding mass spectra and ion mobility spectra for the two mobility-windows obtained upon collisional-activation of up to 100 V are found in Figures S8 and S9 (Supporting Information).
Figure 3A plots the relative abundances of avidin tetramers and monomers as a function of the activation voltage. The data show that collisionally-activated tetramers begin to dissociate (CID) at activation voltages around 80 V. The data further show that the abundance of the avidin tetramer 18+ is negligible for activation voltages of ~100 V. Figure 3B plots the tetramer cross sections as a function of the activation voltage. The data reveal marginal changes of the cross section for activation voltages below 70 V, at which point the cross sections abruptly increase until about 80 V, indicating a collision-induced unfolding (CIU). Hence, in line with prior reports,57 our results show that avidin tetramers are unfolded before they dissociate.
Figure 3. Collisional-activation of mobility-selected avidin tetramers reveal stages of cleaning, unfolding, and dissociation.

(A) Relative abundances of avidin tetramers (solid) and monomers (open) as a function of activation voltage. Strong increase of monomer abundance above 80 V indicates collisional-induced dissociation of the tetramer. (B) Tetramer cross sections increase significantly between 70 to 80 V, indicating collision-induced unfolding. (C-D) The FWHM and center of the tetramer mass peaks decrease between 70 V and 80 V. Both observations suggest that solvent molecules are released during unfolding. (K0 indicated in 3A).
Figures 3C and 3D show the full-width-at-half-maximum (FWHM) and the center of the tetramer mass peaks as a function of the activation voltage. The figures reveal that both the center and the width of the peaks decrease as the ions are collisionally-activated. We stress that our data in Figure 3 cannot be rationalized by fragmentation of the glycans or the protein backbone (see Figure S8A, Supporting Information). Thus, our data imply that non-covalently bound solvent particles detach from the avidin tetramer upon collisional-activation, in line with “collisional cleaning” reported for similar systems.58
Our tandem-TIMS data, however, reveal that loss of solvent particles occurs in two distinct stages. Some solvent particles are lost by collisional activation of up to 70 V, i.e. before the CIU and CID processes have begun. This means that loss of these solvent particles is unrelated to structural changes of the avidin tetramer. Hence, these solvent particles were most likely non-specifically bound to the surface of avidin, but then detached upon collisional-activation. By contrast, other solvent particles appear to be lost only during the unfolding process. The FWHM and center of the tetramer peak decrease during the CIU process (see Figure 3B–D and Table S1, Supporting Information). These solvent particles thus dissociate from the tetramer as it unfolds, implying that these particles were initially buried in the native-like avidin tetramer, i.e. within pockets of the monomer chains or, alternatively, in the binding interfaces of the monomers. We stress that water molecules buried inside the avidin tetramer are reported in x-ray structures59 (see Fig. S4C, Supporting Information).
Finally, for activation voltages lower than 80 V (onset of CID), we observe similar solvent-loss behavior for the two mobility fractions. Thus, the broadness of the mass and ion mobility peaks observed for native-like avidin tetramers (Figures 1C and 1D) cannot arise from solvent-adducts alone. If this were the case, then ions with larger cross sections and greater masses (i.e. more solvent adducts) would have shown a more significant loss of solvent upon collisional activation. Further, a slight increase of tetramer mass peaks at activation voltages >80 V is apparent for the high mobility fraction (K0,II = 0.88-0.90 cm2/Vs, Fig. 3D), which we rationalize by different dissociation energy thresholds for distinct avidin glycoforms (see Figure S10, Supporting Information). This observation further supports the notion that distinct glycoform combinations contribute to the broadness of the native avidin peak.
Tandem-TIMS reveals subunit architecture of avidin tetramers
As discussed above, activation voltages of ~80 to 100 V in the interface of tandem-TIMS lead to dissociation of avidin tetramers into monomers and trimers. These monomers are highly unfolded with cross sections (CCSN2) ranging from 2104 Å2 to 2740 Å2 (see Table S2, Supporting Information) and approximately half of the charge (7+ to 11+) of the precursor tetramers. By contrast, the trimers (charge states 7+ to 10+) are compact with cross sections (CCSN2) between 3161 Å2 to 3274 Å2. These observations are consistent with a “typical” CID mechanism of protein complexes,60–62 which starts with charge migration and unfolding of one avidin monomer chain. Subsequently, the unfolded avidin monomer detaches while taking up approximately half of the total charges. Hence, the product ions do not reflect the subunit architecture of the native protein complex.
Figure 4A and 4B show nested ion mobility – mass spectra, in which avidin tetramer 18+ ions are mobility-selected, collisionally-activated, and then mobility-analyzed in TIMS-2. The data shown in Figure 4A (140 V activation voltage) are consistent with Figure 3 and reflect the typical CID behavior of protein complexes mentioned above. By contrast, the data plotted in Figure 4B (260 V activation voltage) reflect an “atypical” CID behavior. While extended monomers still predominate, the plot shows significant abundance of species that were not abundant upon collisional-activation at 140 V. These additional species are avidin monomers with low charge states (3+ to 6+) as well as avidin dimers (charge states 5+ to 7+). The cross sections of these additional monomers (CCSN2 = 1568-1671 Å2) and dimers (CCSN2 = 2439-2499 Å2, see Table S2, Supporting Information) indicate that their structures are compact. Indeed, these avidin dimers are only slightly larger than neutravidin dimers55 produced by surface-induced dissociation (SID). Considering that neutravidin is a deglycosylated form of avidin, this means that the structure of avidin dimers in Figure 4B potentially resemble those generated for neutravidin by SID.
Figure 4. Collision-induced dissociation (CID) of native-like avidin tetramers in the interface of tandem-TIMS.
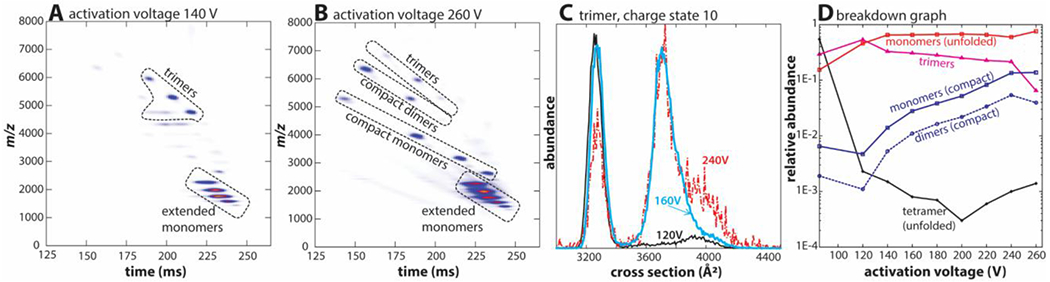
(A) At an activation voltage of 140 V, avidin dissociates mainly into extended monomers and compact trimers indicating a “typical” CID mechanism. (B) At an activation voltage of 260 V, compact monomers and dimers emerge, which indicates the presence of an “atypical” CID mechanism at higher activation voltages. (C) Cross section distributions for avidin trimers 10+ generated at 120 V (black), 160 V (blue), and 240 V (red) reveal that the trimers unfold at higher activation voltages. (D) The breakdown graph reveals presence of an “atypical” CID mechanism at activation voltages above ~150 V.
Figure 4C plots the ion mobility spectrum of avidin trimers 10+ produced by CID of the tetramer 18+ at activation voltages ranging from 120 V to 240 V. The cross section of the trimers (CCSN2 ~3250 Å2) at low activation voltages indicates a folded structure. As the activation voltage is increased, an extended structure (~3650 Å2) emerges and predominates above 240 V. These observations imply that the compact trimer ions produced at low activation voltages cannot be refolded gas-phase structures. Further, compact monomers, dimers, and trimers agree with the native-like tetramers in terms of their mass density (Figure S11, Supporting Information). Thus, our results imply that the compact subunits produced by CID in tandem-TIMS at high activation voltage might reflect their topology within the native, folded avidin tetramer. A conclusive structural interpretation of our data, however, would require a detailed computational analysis by our Structure Relaxation Approximation (SRA) method63, as carried out for the protein ubiquitin. Due to space constraints, we defer this discussion to a separate publication.
Figure 4D plots the abundances of the avidin tetramer and its product ions as we increase the activation voltage from 85 V to 260 V (the corresponding mass spectra are found in Figure S12, Supporting Information). The plot reveals that compact monomers and dimers become significant at activation voltages above ~150 V and that their abundances increase with activation voltage. Finally, at 260 V, the compact monomers and dimers account for about ~15% of the total ion count. Thus, our data suggest that a gradual transition occurs from a “typical” CID process predominating at low activation voltages to an “atypical” CID process at higher activation voltages. Such “atypical” dissociation pathways were previously reported for CID at high drift gas temperatures,49 and for charge reduced protein complexes.57,64
We rationalize the significant abundance of “atypical” CID product ions in our tandem-TIMS system by the combination of high electric field strengths (~105 V/m, see Figure S3, Supporting Information) and a short distance for activation (1.6 mm). In other words: ions are collisionally-activated in the interface of tandem-TIMS by highly energetic ion-neutral collisions over a short time scale. Another explanation would be that activated ions might closely approach the metallic wire-mesh grid located at deflector-2, thereby colliding with the “surface” of the wire. Hence, both the compact nature of the product ions and also the activation mechanism of this “atypical” CID mechanism appear to resemble surface induced dissociation (SID).29
Tandem-TIMS reveals the glycoform combinations in avidin assemblies
Variations of the glycan and amino acid sequence strongly influence the biological activity of a glycoprotein (complex), as shown by the env glycoproteins gp41 and gp120 in HIV65 and the plasma proteins α1-acid glycoprotein and haptoglobin.66 Therefore, identification and quantification of individual glycoproteoforms both in monomeric and oligomeric states is important. Here, we examine the mass spectra of subunits generated upon CID of avidin tetramers in order to reveal the relationships between monomeric and oligomeric avidin glycoproteoforms.
Figure 5A–D show charge-deconvolved mass spectra for avidin monomers, dimers, trimers and tetramers obtained upon collisional-activation. All spectra exhibit a broad envelope. A closer inspection reveals a distinct fine structure with a 40.5 Da repeat pattern superimposed on each envelope (Figures 5A–D). The 40.5 Da interval can be linked to the mass differences between an N-acetyl glucosamine (203.20 Da) and a mannose residue (162.10 Da) of the glycans. Thus, the peak pattern observed for avidin monomers and oligomers indicates the presence of distinct glycoforms.
Figure 5. Charge-deconvolved mass spectra for avidin monomers, dimers, trimers, and tetramers.

(A) Charge-deconvolved mass spectrum obtained for avidin monomers at activation voltage of 260 V. The repetitive pattern with a 40.5 Da spacing arises from the presence of avidin glycoforms. (B-D) Charge-deconvolved mass spectra recorded for avidin dimers (B), trimers (C), and tetramers (D) reveal repetitive patterns with 40.5 Da spacing. (E-F) Comparison of experimental (black, box-filtered) and expected (red) spectra obtained by convolving two monomer spectra (E) and a dimer with a monomer spectra (F). The experimental and expected spectra are consistent with each other in term of peak position, however differs slightly in peak intensity (insets). (G) Random association of two dimer spectra (red, shifted by +200 Da) compared to the experimental tetramer spectrum (black, box-filtered). (H) Proposed structure for avidin tetramer detected by tandem-TIMS/MS with solvent molecules (red) and glycans with mannose- (green) and glycosaminoglycan (blue) residues indicated.
Figure S13 (Supporting Information) shows the charge-deconvolved mass spectra acquired for avidin monomers at activation voltages of 140 V (extended, charge state 8+ to 11+), and 260 V (compact, charge states 3+ to 6+). We notice that the positions and intensities of the peaks do not vary with activation voltage. Hence, the data are inconsistent with neutral-losses (NH3, H2O) or other fragmentation during collisional-activation. Indeed, we can assign individual peaks to the same monomeric avidin glycoforms reported by Oliver et al.40 (Table S3, Supporting Information).
Further, we investigate how monomeric glycoforms of avidin combine to form the dimers. If avidin dimers were composed of random combinations of monomeric glycoforms, then the dimer mass spectrum would be given by a convolution of the monomer spectrum (Figure 5A) with itself. We superimpose this convoluted, “random dimer” spectrum on the experimental dimer spectrum in Figure 5E. The plot shows overall a strong agreement (Pearson correlation coefficient ρ≈0.98) with respect to both the position and the width of the mass spectral envelope, and with respect to the fine structure comprising the 40.5 Da spacing between the peaks. Hence, the broad envelope visible in the dimer spectrum is consistent with the presence of many glycoform combinations that our time-of-flight mass detector is unable to resolve. Furthermore, we notice that the relative intensities for a number of peaks in the experimental spectrum are lower than expected for a random combination of glycoforms (Figure 5E, inset). This implies that some glycoform combinations are less favored than others.
Figure 5F compares the experimental mass spectrum of the avidin trimer to the spectrum obtained by convolving a monomer spectrum with that of a dimer. As observed for the dimers, the position and width of the broad envelope as well as the fine structures in the spectra agree well with each other (Figure 5F, inset; ρ≈0.99). By contrast, the mass spectrum recorded for the avidin tetramer at an activation voltage of 85 V is shifted by ~200 Da with respect to the mass spectrum expected for a random combination of a monomer and trimer (Figure 5G, ρ≈0.98). We rationalize this observation by solvent particles that remain bound to the extended avidin tetramer, likely buried within cavities or the interfaces between the subunits.
Figure 5H graphically summarizes our observations that (1) the broad avidin oligomer peaks arise from the presence of a plethora of glycoform combinations that differ in their masses but remain unresolved by our mass analyzer; (2) avidin assemblies are composed of (almost) random glycoforms combinations; (3) native-like avidin tetramers contain deeply buried solvent particles, presumably the water molecules observed in biotin-binding pockets or interfacial water observed by x-ray crystallography (Figure S4C, Supporting Information).
Tandem-TIMS enables native top-down sequence analysis
In order to link conformational heterogeneity to proteoform variations of protein complexes, tandem-TIMS/MS must be capable to conduct native top-down protein sequence analysis. We previously demonstrated the ability of tandem-TIMS to dissociate ubiquitin (8.6 kDa) by CID in the interface between TIMS-1 and TIMS-2.36 Here, we want to assess the utility of tandem-TIMS to perform native top-down sequencing on avidin, a 64 kDa glycoproteins with strongly bonded subunits.
We mobility-select avidin charge state 18+ and collisionally-activate the selected ions by an activation voltage of 270 V, followed by mobility-analysis in TIMS-2. The resulting nested ion mobility-mass spectrum is shown in Figure 6A. In addition to extended monomer peaks (8+ to 11+), the spectrum shows many fragment ions produced from cleavage of the avidin backbone. These fragment ions separate into several bands, as commonly observed in bottom-up proteomics using ion mobility spectrometry.67,68 These bands correspond mainly to fragment ions with charge states 1+ to 4+, of which the band with predominantly doubly-charged ions is shown in Figure 6B.
Figure 6. Native top-down sequence analysis of avidin by tandem-TIMS/MS.
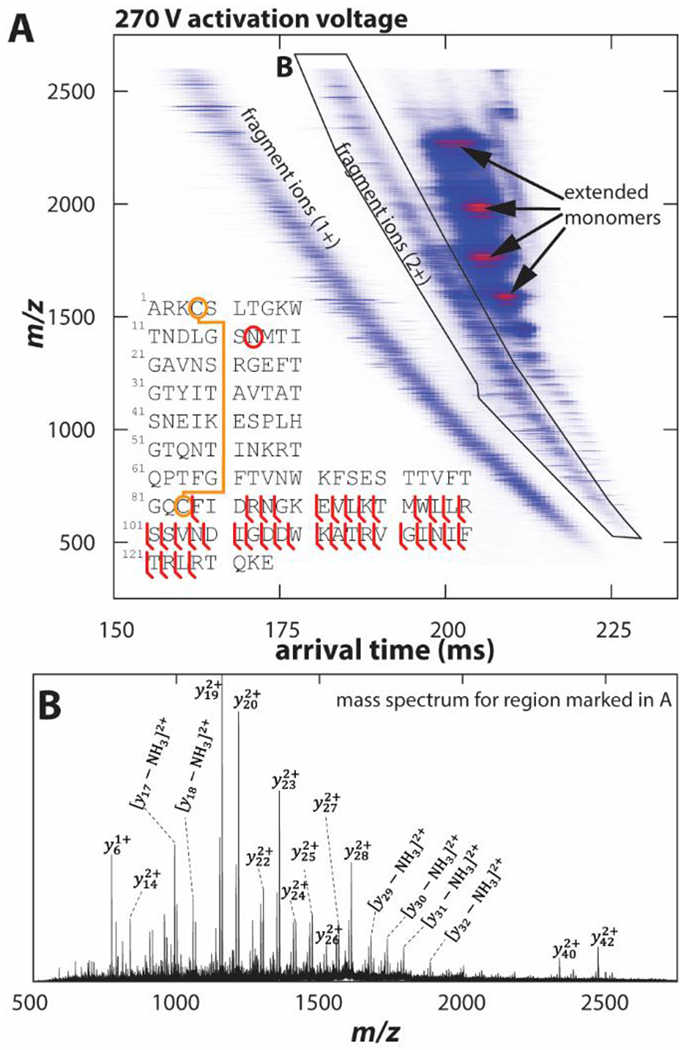
(A) Nested ion mobility–mass spectrum recorded by mobility-selecting avidin charge state 18+, followed by collisional-activation at 270 V and mobility-analysis in TIMS-2. A plethora of fragment ions, mostly y-ions, are observed, with a sequence coverage of ~29 %. (B) Mass spectrum for the region marked in (A) with y-ions annotated.
We manually assign the fragment ion spectrum by comparing the isotopic patterns observed in Figure 6A to those expected for a, b, and y fragment ions for avidin, including their neutral loss fragment ions (see Figure S14, Supporting Information). We achieve an overall sequence coverage of 29 %. All identified ions correspond to cleavages C-terminal of the disulfide bond (Cys4-Cys83, see fragmentation map in Fig. 6A). Overall, the sequence coverage of avidin tetramer recorded in our tandem-TIMS is similar to the one reported for a travelling wave ion mobility instrument.35
One additional feature of the tandem-TIMS/MS instrument is the ability to mass-select generated fragment ions in the quadrupole, followed by CID in the collision cell (Figure S15A, Supporting Information). This allows us to perform TIMS2-MS2 experiments, which enables the sequencing of fragment ions, and increases the sequence coverage. Here, we mass-selected fragment ions generated by CID of avidin tetramer 18+ with m/z 1159 ± 5 in the quadrupole, which corresponds to y192+ and an internal ion (Figure S15C, Supporting Information). We then collisionally-activated the m/z-selected ions by 90 V bias in the collision cell. The resulting MS/MS spectrum confirms the presence of y192+ (see Figure S15D–F, Supporting Information).
CONCLUSIONS
Using the glycoprotein complex avidin as a reference system, we demonstrated the versatility of tandem-trapped ion mobility spectrometry – mass spectrometry (tandem-TIMS/MS). Utilizing various operational modes in tandem-TIMS/MS, we conclude from our data:
Tandem-TIMS preserves intact, native-like avidin tetramers with buried solvent particles attached.
The native-like avidin tetramer is a heterogeneous ensemble of non-interconverting species.
Avidin tetramers are composed of (almost) random glycoform combinations whereas some combinations appear favored over others in the dimer subunits.
Collision-induced dissociation in the tandem-TIMS interface at low activation voltages produces compact trimers that likely resemble the structure of the trimer within the folded tetramer.
At high activation voltages in the tandem-TIMS interface, native avidin tetramers dissociate into compact monomers and dimers with cross sections consistent with x-ray structures and reports from surface-induced dissociation (SID).
Native top-down sequence analysis of the avidin tetramer is possible by CID in the interface between two TIMS analyzers. In addition, MS2 experiments using the quadrupole/collision cell allow the sequence identification of fragment ions produced in tandem-TIMS.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the National Science Foundation under grant CHE-1654608 and the National Institutes of Health under award R01GM135682 (C.B.). Support from the Office of Postdoctoral Affairs of the Florida State University is acknowledged (F.C.L.).
Footnotes
ASSOCIATED CONTENT
Supporting Information
Expanded experimental details, cross section calculations, cross section calibration, 16 Figures and 3 Tables. The Supporting Information is available free of charge on the ACS Publications website.
REFERENCES
- (1).Robinson CV; Sali A; Baumeister W The Molecular Sociology of the Cell. Nature 2007, 450 (7172), 973–982. [DOI] [PubMed] [Google Scholar]
- (2).Frauenfelder H; Sligar S; Wolynes P The Energy Landscapes and Motions of Proteins. Science 1991, 254 (5038), 1598–1603. [DOI] [PubMed] [Google Scholar]
- (3).Wand AJ Dynamic Activation of Protein Function: A View Emerging from NMR Spectroscopy. Nat. Struct. Biol 2001, 8 (11), 926–931. [DOI] [PubMed] [Google Scholar]
- (4).Lange OF; Lakomek N-A; Farès C; Schröder GF; Walter KF; Becker S; Meiler J; Grubmüller H; Griesinger C; De Groot BL Recognition Dynamics up to Microseconds Revealed from an RDC-Derived Ubiquitin Ensemble in Solution. Science 2008, 320 (5882), 1471–1475. [DOI] [PubMed] [Google Scholar]
- (5).Smith LM; Kelleher NL Proteoforms as the next Proteomics Currency. Science 2018, 359 (6380), 1106–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Koike R; Kidera A; Ota M Alteration of Oligomeric State and Domain Architecture Is Essential for Functional Transformation between Transferase and Hydrolase with the Same Scaffold. Protein Sci. 2009, 18 (10), 2060–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Marcos E; Crehuet R; Bahar I Changes in Dynamics upon Oligomerization Regulate Substrate Binding and Allostery in Amino Acid Kinase Family Members. PLoS Comput. Biol 2011, 7 (9), e1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Gristick HB; Wang H; Bjorkman PJ X-Ray and EM Structures of a Natively Glycosylated HIV-1 Envelope Trimer. Acta Crystallogr. Sect. Struct. Biol 2017, 73 (10), 822–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Crispin M; Ward AB; Wilson IA Structure and Immune Recognition of the HIV Glycan Shield. Annu. Rev. Biophys 2018, 47 (1), 499–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Alley WR; Novotny MV Structural Glycomic Analyses at High Sensitivity: A Decade of Progress. Annu. Rev. Anal. Chem 2013, 6 (1), 237–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Braakman I; Bulleid NJ Protein Folding and Modification in the Mammalian Endoplasmic Reticulum. Annu. Rev. Biochem 2011, 80 (1), 71–99. [DOI] [PubMed] [Google Scholar]
- (12).Chen I-H; Aguilar HA; Paez Paez JS; Wu X; Pan L; Wendt MK; Iliuk AB; Zhang Y; Tao WA Analytical Pipeline for Discovery and Verification of Glycoproteins from Plasma-Derived Extracellular Vesicles as Breast Cancer Biomarkers. Anal. Chem 2018, 90 (10), 6307–6313. [DOI] [PubMed] [Google Scholar]
- (13).Fuertes-Martín R; Taverner D; Vallvé J-C; Paredes S; Masana L; Correig Blanchar X; Amigó Grau N Characterization of 1 H NMR Plasma Glycoproteins as a New Strategy To Identify Inflammatory Patterns in Rheumatoid Arthritis. J. Proteome Res 2018, 17 (11), 3730–3739. [DOI] [PubMed] [Google Scholar]
- (14).Varki A Biological Roles of Glycans. Glycobiology 2017, 27 (1), 3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Moremen KW; Tiemeyer M; Nairn AV Vertebrate Protein Glycosylation: Diversity, Synthesis and Function. Nat. Rev. Mol. Cell Biol 2012, 13 (7), 448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Araki K; Nagata K Protein Folding and Quality Control in the ER. Cold Spring Harb. Perspect. Biol 2011, 3 (11), a007526–a007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Woods RJ Predicting the Structures of Glycans, Glycoproteins, and Their Complexes. Chem. Rev 2018, 118 (17), 8005–8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Meyer B; Moeller H Conformation of Glycopeptides and Glycoproteins In Glycopeptides and Glycoproteins; Topics in Current Chemistry; Springer, Berlin, Heidelberg, 2006; Vol. 267, pp 187–251. [Google Scholar]
- (19).Livnah O; Bayer EA; Wilchek M; Sussman JL Three-Dimensional Structures of Avidin and the Avidin-Biotin Complex. Proc Natl Acad Sci USA 1993, 90, 5076–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Pazy Y; Kulik T; Bayer EA; Wilchek M; Livnah O Ligand Exchange between Proteins: Exchange of Biotin and Biotin Derivatives between Avidin and Streptavidin. J. Biol. Chem 2002, 277 (34), 30892–30900. [DOI] [PubMed] [Google Scholar]
- (21).Ruotolo BT; Giles K; Campuzano I; Sandercock AM; Bateman RH; Robinson CV Evidence for Macromolecular Protein Rings in the Absence of Bulk Water. Science 2005, 310 (5754), 1658–1661. [DOI] [PubMed] [Google Scholar]
- (22).Koeniger SL; Merenbloom SI; Clemmer DE Evidence for Many Resolvable Structures within Conformation Types of Electrosprayed Ubiquitin Ions. J. Phys. Chem. B 2006, 110 (13), 7017–7021. [DOI] [PubMed] [Google Scholar]
- (23).Wyttenbach T; Bowers MT Structural Stability from Solution to the Gas Phase: Native Solution Structure of Ubiquitin Survives Analysis in a Solvent-Free Ion Mobility–Mass Spectrometry Environment. J. Phys. Chem. B 2011, 115 (42), 12266–12275. [DOI] [PubMed] [Google Scholar]
- (24).Zhou M; Politis A; Davies RB; Liko I; Wu K-J; Stewart AG; Stock D; Robinson CV Ion Mobility-Mass Spectrometry of a Rotary ATPase Reveals ATP-Induced Reduction in Conformational Flexibility. Nat. Chem 2014, 6 (3), 208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Lai AL; Clerico EM; Blackburn ME; Patel NA; Robinson CV; Borbat PP; Freed JH; Gierasch LM Key Features of an Hsp70 Chaperone Allosteric Landscape Revealed by Ion-Mobility Native Mass Spectrometry and Double Electron-Electron Resonance. J. Biol. Chem 2017, 292 (21), 8773–8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Katta Viswanatham.; Chowdhury SK; Chait BT. Use of a Single-Quadrupole Mass Spectrometer for Collision-Induced Dissociation Studies of Multiply Charged Peptide Ions Produced by Electrospray Ionization. Anal. Chem 1991, 63 (2), 174–178. [DOI] [PubMed] [Google Scholar]
- (27).Loo JA; Edmonds CG; Smith RD Tandem Mass Spectrometry of Very Large Molecules. 2. Dissociation of Multiply Charged Proline-Containing Proteins from Electrospray Ionization. Anal. Chem 1993, 65 (4), 425–438. [DOI] [PubMed] [Google Scholar]
- (28).Senko MW; Speir J. Paul.; McLafferty FW Collisional Activation of Large Multiply Charged Ions Using Fourier Transform Mass Spectrometry. Anal. Chem 1994, 66 (18), 2801–2808. [DOI] [PubMed] [Google Scholar]
- (29).Wysocki VH; Joyce KE; Jones CM; Beardsley RL Surface-Induced Dissociation of Small Molecules, Peptides, and Non-Covalent Protein Complexes. J. Am. Soc. Mass Spectrom 2008, 19 (2), 190–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Zinnel NF; Pai P-J; Russell DH Ion Mobility-Mass Spectrometry (IM-MS) for Top-Down Proteomics: Increased Dynamic Range Affords Increased Sequence Coverage. Anal. Chem 2012, 84 (7), 3390–3397. [DOI] [PubMed] [Google Scholar]
- (31).Belov ME; Damoc E; Denisov E; Compton PD; Horning S; Makarov AA; Kelleher NL From Protein Complexes to Subunit Backbone Fragments: A Multi-Stage Approach to Native Mass Spectrometry. Anal. Chem 2013, 85 (23), 11163–11173. [DOI] [PubMed] [Google Scholar]
- (32).Konijnenberg A; Bannwarth L; Yilmaz D; Koçer A; Venien-Bryan C; Sobott F Top-down Mass Spectrometry of Intact Membrane Protein Complexes Reveals Oligomeric State and Sequence Information in a Single Experiment: Top-down MS of Membrane Proteins. Protein Sci. 2015, 24 (8), 1292–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Yang Y; Wang G; Song T; Lebrilla CB; Heck AJR Resolving the Micro-Heterogeneity and Structural Integrity of Monoclonal Antibodies by Hybrid Mass Spectrometric Approaches. mAbs 2017, 9 (4), 638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Li H; Nguyen HH; Ogorzalek Loo RR; Campuzano IDG; Loo JA An Integrated Native Mass Spectrometry and Top-down Proteomics Method That Connects Sequence to Structure and Function of Macromolecular Complexes. Nat. Chem 2018, 10 (2), 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Polasky DA; Lermyte F; Nshanian M; Sobott F; Andrews PC; Loo JA; Ruotolo BT Fixed-Charge Trimethyl Pyrilium Modification for Enabling Enhanced Top-Down Mass Spectrometry Sequencing of Intact Protein Complexes. Anal. Chem 2018, 90 (4), 2756–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Liu FC; Ridgeway ME; Park MA; Bleiholder C Tandem Trapped Ion Mobility Spectrometry. Analyst 2018, 143 (10), 2249–2258. [DOI] [PubMed] [Google Scholar]
- (37).DeLange R Egg White Avidin. J. Biol. Chem 1970, 245 (5), 907–9016. [PubMed] [Google Scholar]
- (38).Bruch RC; White HB Compositional and Structural Heterogeneity of Avidin Glycopeptides. Biochemistry 1982, 21 (21), 5334–5341. [DOI] [PubMed] [Google Scholar]
- (39).Yang Y; Orlando R Identifying the Glycosylation Sites and Site-Specific Carbohydrate Heterogeneity of Glycoproteins by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. Rapid Commun. Mass Spectrom 1996, 10 (8), 932–936. [DOI] [PubMed] [Google Scholar]
- (40).Oliver RWA; Green BN; Harvey DJ The Use of Electrospray Ionization MS to Determine the Structure of Glycans in Intact Glycoproteins. Biochem. Soc. Trans 1996, 24, 917–927. [DOI] [PubMed] [Google Scholar]
- (41).Hernandez DR; DeBord JD; Ridgeway ME; Kaplan DA; Park MA; Fernandez-Lima F Ion Dynamics in a Trapped Ion Mobility Spectrometer. The Analyst 2014, 139 (8), 1913–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Ridgeway ME; Silveira JA; Meier JE; Park MA Microheterogeneity within Conformational States of Ubiquitin Revealed by High Resolution Trapped Ion Mobility Spectrometry. The Analyst 2015, 140, 6964–6972. [DOI] [PubMed] [Google Scholar]
- (43).Michelmann K; Silveira JA; Ridgeway ME; Park MA Fundamentals of Trapped Ion Mobility Spectrometry. J. Am. Soc. Mass Spectrom 2015, 26 (1), 14–24. [DOI] [PubMed] [Google Scholar]
- (44).Chai M; Young MN; Liu FC; Bleiholder C A Transferable, Sample-Independent Calibration Procedure for Trapped Ion Mobility Spectrometry (TIMS). Anal. Chem 2018, 90 (15), 9040–9047. [DOI] [PubMed] [Google Scholar]
- (45).Stow SM; Causon TJ; Zheng X; Kurulugama RT; Mairinger T; May JC; Rennie EE; Baker ES; Smith RD; McLean JA; et al. An Interlaboratory Evaluation of Drift Tube Ion Mobility–Mass Spectrometry Collision Cross Section Measurements. Anal. Chem 2017, 89 (17), 9048–9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Kirk SR; Liu FC; Cropley TC; Carlock HR; Bleiholder C On the Preservation of Non-Covalent Peptide Assemblies in a Tandem-Trapped Ion Mobility Spectrometer-Mass Spectrometer (TIMS-TIMS-MS). J. Am. Soc. Mass Spectrom 2019, 30 (7), 1204–1212. [DOI] [PubMed] [Google Scholar]
- (47).Liu FC; Kirk SR; Bleiholder C On the Structural Denaturation of Biological Analytes in Trapped Ion Mobility Spectrometry – Mass Spectrometry. Analyst 2016, 141 (12), 3722–3730. [DOI] [PubMed] [Google Scholar]
- (48).Schwartz BL; Light-Wahl KJ; Smith RD Observation of Noncovalent Complexes to the Avidin Tetramer by Electrospray Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom 1994, 5 (3), 201–204. [DOI] [PubMed] [Google Scholar]
- (49).Pacholarz KJ; Barran PE Distinguishing Loss of Structure from Subunit Dissociation for Protein Complexes with Variable Temperature Ion Mobility Mass Spectrometry. Anal. Chem 2015, 87 (12), 6271–6279. [DOI] [PubMed] [Google Scholar]
- (50).Bush MF; Hall Z; Giles K; Hoyes J; Robinson CV; Ruotolo BT Collision Cross Sections of Proteins and Their Complexes: A Calibration Framework and Database for Gas-Phase Structural Biology. Anal. Chem 2010, 82 (22), 9557–9565. [DOI] [PubMed] [Google Scholar]
- (51).Bleiholder C; Wyttenbach T; Bowers MT A Novel Projection Approximation Algorithm for the Fast and Accurate Computation of Molecular Collision Cross Sections (I). Method. Int. J. Mass Spectrom 2011, 308 (1), 1–10. [Google Scholar]
- (52).Anderson SE; Bleiholder C; Brocker ER; Stang PJ; Bowers MT A Novel Projection Approximation Algorithm for the Fast and Accurate Computation of Molecular Collision Cross Sections (III): Application to Supramolecular Coordination-Driven Assemblies with Complex Shapes. Int. J. Mass Spectrom 2012, 330–332, 78–84. [Google Scholar]
- (53).Bleiholder C; Contreras S; Do TD; Bowers MT A Novel Projection Approximation Algorithm for the Fast and Accurate Computation of Molecular Collision Cross Sections (II). Model Parameterization and Definition of Empirical Shape Factors for Proteins. Int. J. Mass Spectrom 2013, 345–347, 89–96. [Google Scholar]
- (54).Light-Wahl KJ; Schwartz BL; Smith RD Observation of the Noncovalent Quaternary Associations of Proteins by Electrospray Ionization Mass Spectrometry. J. Am. Chem. Soc 1994, 116 (12), 5271–5278. [Google Scholar]
- (55).Quintyn RS; Yan J; Wysocki VH Surface-Induced Dissociation of Homotetramers with D2 Symmetry Yields Their Assembly Pathways and Characterizes the Effect of Ligand Binding. Chem. Biol 2015, 22 (5), 583–592. [DOI] [PubMed] [Google Scholar]
- (56).McKay AR; Ruotolo BT; Ilag LL; Robinson CV Mass Measurements of Increased Accuracy Resolve Heterogeneous Populations of Intact Ribosomes. J. Am. Chem. Soc 2006, 128 (35), 11433–11442. [DOI] [PubMed] [Google Scholar]
- (57).Bornschein RE; Niu S; Eschweiler J; Ruotolo BT Ion Mobility-Mass Spectrometry Reveals Highly-Compact Intermediates in the Collision Induced Dissociation of Charge-Reduced Protein Complexes. J. Am. Soc. Mass Spectrom 2016, 27 (1), 41–49. [DOI] [PubMed] [Google Scholar]
- (58).Benesch JLP Collisional Activation of Protein Complexes: Picking up the Pieces. J. Am. Soc. Mass Spectrom 2009, 20 (3), 341–348. [DOI] [PubMed] [Google Scholar]
- (59).Repo S; Paldanius TA; Hytönen VP; Nyholm TKM; Halling KK; Huuskonen J; Pentikäinen OT; Rissanen K; Slotte JP; Airenne TT; et al. Binding Properties of HABA-Type Azo Derivatives to Avidin and Avidin-Related Protein 4. Chem. Biol 2006, 13 (10), 1029–1039. [DOI] [PubMed] [Google Scholar]
- (60).Jurchen JC; Williams ER Origin of Asymmetric Charge Partitioning in the Dissociation of Gas-Phase Protein Homodimers. J. Am. Chem. Soc 2003, 125 (9), 2817–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Benesch JLP; Aquilina JA; Ruotolo BT; Sobott F; Robinson CV Tandem Mass Spectrometry Reveals the Quaternary Organization of Macromolecular Assemblies. Chem. Biol 2006, 13 (6), 597–605. [DOI] [PubMed] [Google Scholar]
- (62).Ruotolo BT; Hyung S-J; Robinson PM; Giles K; Bateman RH; Robinson CV Ion Mobility–Mass Spectrometry Reveals Long-Lived, Unfolded Intermediates in the Dissociation of Protein Complexes. Angew. Chem. Int. Ed 2007, 46 (42), 8001–8004. [DOI] [PubMed] [Google Scholar]
- (63).Bleiholder C; Liu FC Structure Relaxation Approximation (SRA) for Elucidation of Protein Structures from Ion Mobility Measurements. J. Phys. Chem. B 2019, 123 (13), 2756–2769. [DOI] [PubMed] [Google Scholar]
- (64).Pagel K; Hyung S-J; Ruotolo BT; Robinson CV Alternate Dissociation Pathways Identified in Charge-Reduced Protein Complex Ions. Anal. Chem 2010, 82 (12), 5363–5372. [DOI] [PubMed] [Google Scholar]
- (65).Raska M; Takahashi K; Czernekova L; Zachova K; Hall S; Moldoveanu Z; Elliott MC; Wilson L; Brown R; Jancova D; et al. Glycosylation Patterns of HIV-1 Gp120 Depend on the Type of Expressing Cells and Affect Antibody Recognition. J. Biol. Chem 2010, 285 (27), 20860–20869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Wu D; Struwe WB; Harvey DJ; Ferguson MAJ; Robinson CV N-Glycan Microheterogeneity Regulates Interactions of Plasma Proteins. Proc. Natl. Acad. Sci 2018, 115 (35), 8763–8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Meier F; Beck S; Grassl N; Lubeck M; Park MA; Raether O; Mann M Parallel Accumulation–Serial Fragmentation (PASEF): Multiplying Sequencing Speed and Sensitivity by Synchronized Scans in a Trapped Ion Mobility Device. J. Proteome Res 2015, 14 (12), 5378–5387. [DOI] [PubMed] [Google Scholar]
- (68).Sproß J; Muck A; Gröger H Detection and Fragmentation of Doubly Charged Peptide Ions in MALDI-Q-TOF-MS by Ion Mobility Spectrometry for Improved Protein Identification. Anal. Bioanal. Chem 2019, 411 (24), 6275–6285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


