Abstract
Prenatal clinical detection of thalassemia involves gap-PCR and reverse dot blot (RDB) analysis of fetal DNA acquired through invasive methods. The present study aimed to develop a non-invasive prenatal diagnostic method for thalassemia based on next-generation sequencing (NGS). A total of eight families with proband children with thalassemia were recruited for the study during a subsequent pregnancy. The sequence of the thalassemia genes of the parents and proband were determined using NGS, based on a thalassemia AmpliSeq panel. Cell-free plasma DNA from pregnant women related to the aforementioned proband was analyzed using an NGS panel, based on thalassemia-associated capture probes. Heterozygous single nucleotide polymorphisms within the 10 kb regions flanking exons of the targeted thalassemia genes were acquired using probes or AmpliSeq and employed for parental haplotype construction using Trio-based panel sequencing. The fetal haplotype was deduced from the parental haplotypes and relative haplotype dosage, and subsequently validated using gap-PCR and RDB, based on invasively sampled amniotic fluid. A non-invasive prenatal diagnosis procedure from maternal plasma fetal DNA was successfully developed based on haplotype analysis. The deduced haplotypes of eight fetuses were identical to the results of invasive prenatal diagnosis procedures, with an accuracy rate of 100%. Taken together, the present study demonstrated the potential for non-invasive prenatal diagnosis of α- and β-thalassemia using NGS and haplotype-assisted analysis.
Keywords: thalassemia, non-invasive prenatal diagnosis, multiplex PCR, target capture, next-generation sequencing
Introduction
Thalassemia is one of the most common monogenetic diseases in Southern China, Southeast Asia and the Mediterranean region (1). The frequency of carriers in Guangxi, China is 26.9% for α-thalassemia (OMIM 604131) (omim.org) and 19.9% for β-thalassemia (OMIM 603902) (2). Thalassemia is a form of hemolytic anemia caused by an imbalance in the rate of synthesis of α- and β-globin peptide chains, due to mutation or deletion of the human α- or β-globin gene (3). The two most common types of thalassemia are α-thalassemia and β-thalassemia (1). Genes associated with α-thalassemia include hemoglobin α locus 2 (HBA2; OMIM 141850) and HBA1 (OMIM 141800). β-thalassemia is associated with the hemoglobin subunit β (HBB) gene (OMIM 141900) (4). Moreover, ~7% of the global population is estimated to carry the genes associated with thalassemia and the birth rate of children with hemoglobin disorders is >2.4% per year (5). As thalassemia has no effective treatment (6), prenatal diagnosis is an important medical requirement for thalassemia prevention strategies (7). Amniocentesis and chorionic villus sampling are two commonly used invasive prenatal diagnostic procedures. However, these traumatic operations may cause injury to the fetus, miscarriage or intrauterine infection (8). Furthermore, anxiety associated with these invasive procedures has been reported by numerous pregnant women (9). Therefore, non-invasive prenatal diagnostic techniques for thalassemia detection are urgently required.
Since the presence of cell-free fetal DNA (cffDNA) in maternal plasma during pregnancy was first reported in 1997 (10), great efforts have been made to use this source of fetal material for non-invasive prenatal diagnosis (11). Non-invasive prenatal testing (NIPT) has been established for the detection of fetal chromosomal abnormalities, such as chromosome 21, 18 or 13 aneuploidy, in the plasma of pregnant women, and major variations in copy number can be rapidly detected for clinical prenatal screening and diagnosis (12,13). In addition to NIPT for fetal aneuploidies, non-invasive testing techniques for monogenic diseases are also being developed (14). A challenge facing this field is the need to target low concentrations of fetal mutations that differ by only one or a few nucleotides from the overwhelming background of maternal DNA in the mother's plasma (15,16). At present, assessing the relative mutation dosage and the relative haplotype dosage (RHDO) are the main analytical approaches used for NIPT of monogenic diseases (17–20). Early attempts to diagnose monogenic diseases focused on approaches targeting paternally inherited fetal mutations absent from the maternal genome. This strategy has proven to be successful for the detection of achondroplasia (21), myotonic dystrophy (22) and Huntington's disease (23). However, to assess fetal mutations in maternal plasma that share the same genetic identity between the mother and the fetus, more sophisticated strategies are required. These approaches have been facilitated by technological advances, including massively parallel sequencing (24) and digital PCR (25), which enable the sensitive and precise measurement of circulating plasma DNA. However, the large target region used for these methods substantially increase the cost of sequencing (26), a factor which may impede their clinical application. Furthermore, these methods have not yet been applied for the detection of large deletion mutations.
In the present study, a technique was described for non-invasive prenatal detection of paternal and maternal mutations associated with α- and β-thalassemia using multiplex PCR or target capture combined with next-generation sequencing (NGS). Furthermore, the present study demonstrated the feasibility of using NGS data to detect targeted copy number variations (CNVs) and single nucleotide variations (SNVs) (3).
Materials and methods
Sample collection and DNA extraction
A total of eight families were recruited at The Third Affiliated Hospital of Sun Yatsen University. A total of 24 samples of peripheral blood (~10 ml per sample) of known thalassemia genotypes were collected. There were 13 female and 11 male patients with thalassemia, with an age range of 2–38 years (mean age, 15 years). The maternal plasma of pregnant women was collected at an average gestation period of 20 weeks for non-invasive prenatal diagnostic assays. A total of ~10 ml amniotic fluid was also collected for direct molecular diagnosis. In addition, ~5 ml peripheral blood from the parents and their first child was collected into EDTA tubes. For each sample, genomic DNA (gDNA) was extracted using the DNeasy Blood and Tissue kit (Qiagen Sciences, Inc.). genomic DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen Sciences, Inc.), according to the manufacturer's instructions.
Moreover, ~2 ml plasma was separated in two steps from each sample obtained from a pregnant woman. First, 5 ml peripheral blood from the pregnant women was centrifuged at 1,600 × g for 15 min at 4°C. Subsequently, ~2 ml supernatant was centrifuged at 16,000 × g for 10 min at 4°C. The plasma cffDNA was extracted using the QIAamp Circulating Nucleic Acid kit (Qiagen Sciences, Inc.), according to the manufacturer's instructions., then quantified using a Qubit® fluorometer (Thermo Fisher Scientific, Inc.) and stored at −20°C until further use.
Molecular genetic diagnosis of thalassemia
DNA extracted from amniotic cells of pregnant women was amplified with the α-thalassemia Genetic Deletion diagnostic kit using the gap-PCR method (Daan Gene Co., Ltd.). The thermocycling conditions were as follows: i) 96°C for 5 min, 98°C for 45 sec; ii) 10 cycles at 65°C for 1.5 min, 72°C for 3 min; iii) 25 cycles at 98°C for 30 sec, 65°C for 45 sec and 72°C for 3 min; and iv) 13 min at 72°C in a reaction volume of 25 µl with 50 ng genomic DNA (primer sequences not commercially available). The target products detected by agarose gel electrophoresis are shown in Table SI.
For samples in which HBA2 levels were below the normal range (2.5–3.5%) a β-thalassemia Mutation Genetic diagnostic kit (DaAN Gene Co., Ltd.) was used. The thermocycling conditions were: i) 50°C for 15 min; ii) 95°C for 10 min; iii) 94°C for 1 min; iv) 35 cycles at 55° for 30 sec, 72°C for 30 sec; and v) 5 min at 72°C in a reaction volume of 25 µl with 50 ng genomic DNA (primer sequences not commercially available). The PCR products were hybridized with the membrane strip at 43°C for 2 h, and subsequently the membranes were washed (washing reagent supplied as part of the kit) for 15 min, incubated at 25°C for 30 min, shaken and washed twice for 10 min. The film was subsequently washed for 2 min, then according to the instructions of the present solution and displayed at 25°C for 15 min. The results were determined by observing the presence of blue spots over the whole membrane, with blue spots at the mutation site representing the presence of the mutation.
Primer and probe design
For α-thalassemia, primers and probes were designed using the reference sequence for chromosome 16 (GenBank accession no. NC_000016.9; range, 222,846-223,709). For β-thalassemia, the reference sequence for chromosome 11 was used (GenBank accession no. NC_000011.9; range, 5,246,696-5,248,301). Reference sequences were obtained from the GenBank repository (ncbi.nlm.nih.gov/genbank). The up- and downstream 10 kb regions of HBA and HBB, with 195 single nucleotide polymorphisms (SNPs) associated with α-thalassemia and 275 SNPs with β-thalassemia, were screened from the dbSNP database (ncbi.nlm.nih.gov/SNP/) using Ion AmpliSeq Designer (ampliseq.com/) and the Agilent Sure Design website (earray.chem.agilent.com/suredesign/home.htm). The SNP site selection criteria used in the database were: i) SNP loci in the 1,000 Genomes Project database (gloshospitals.nhs.uk/about-us/research-our-hospitals/100000-genomes-project/) from North and South China; ii) minor allele frequency (MAF) ≥20%; iii) location of the SNP 100 bp sequence within a specific region; iv) no homology identified in the genome; v) guanine-cytosine content >45 and <70%; and vi) the SNP did not result in three consecutive identical bases. The Ion AmpliSeq Thalassemia panel was manufactured by Thermo Fisher Scientific, Inc., which consisted of four primer pools comprising 1,354 pairs of primers for α-thalassemia and 361 pairs of primers for β-thalassemia. The thalassemia probes were constructed by Agilent Technologies, Inc., with a target region size of ~274 kb
Library construction
gDNA from the parents was used for library construction using the Life Technologies™ Ion AmpliSeq Library kit v. 2.0 (Thermo Fisher Scientific, Inc.), as follows: 10 ng gDNA, 4 µl 5X Ion AmpliSeq HiFi mix, 10 µl 2X Ion AmpliSeq primer pool and 4 µl nuclease-free water were mixed per reaction to amplify the target regions. Subsequently, 2 µl FuPa reagent was added to each amplified sample to partially digest the primer sequences, and each library was ligated to a unique barcode and universal adapter in the form of Life Technologies™ Ion Xpress™ barcode adapters (Thermo Fisher Scientific, Inc.). Each library was purified using AMPure XP beads (Beckman Coulter, Inc.) and quantified using a Life Technologies™ Qubit® 3.0 fluorometer (Thermo Fisher Scientific, Inc.). The size distributions of the libraries were verified using a High Sensitivity DNA kit with a 2100 Bioanalyzer (both Agilent Technologies, Inc.).
Libraries were constructed from cffDNA extracted from the plasma of pregnant women using the Life Technologies™ Ion Plus Fragments Library kit (Thermo Fisher Scientific, Inc.) with the SureSelect Target Enrichment kit PTN Hyb Module Box2 and SureSelect TE Reagent kit (both Agilent Technologies, Inc.). First, 30 µl cffDNA, 9.5 µl 5X End Repair buffer, 0.5 µl End Repair enzyme and 10 µl nuclease-free water were mixed per reaction for end repair of the cffDNA. Subsequently, the library was ligated to a unique barcode and universal adapter using Life Technologies™ Ion Xpress™ barcode adapters (Thermo Fisher Scientific, Inc.). Each library was concentrated to 3.4 µl, and then 13 µl Hybridization Buffer mix, 5.6 µl SureSelect Block mix and 2 µl SureSelect Library were added, followed by incubation for 16 or 24 h at 65°C to capture the target regions. Each positive library was enriched using Life Technologies™ Dynabeads MyOne Streptavidin T1 (Thermo Fisher Scientific, Inc.). The purified libraries were quantified using a Qubit® 3.0 fluorometer. The size distributions of the libraries were verified using a High Sensitivity DNA kit with a 2100 Bioanalyzer (Agilent Technologies, Inc.).
Template preparation and enrichment and NGS
Each library was diluted to 100 pM based on its concentration quantified with the Qubit® 3.0 fluorometer. Subsequently, ten libraries (for blood samples) or four libraries (for plasma) were mixed equally and diluted to 100 pM, respectively. The diluted sample was amplified through emulsion PCR on Ion Proton™ HiQ™ ion sphere particles (ISPs) using a Life Technologies™ Ion OneTouch™ 2 Instrument according to the manufacturer's instructions. Template-positive ISPs were enriched using a Life Technologies™ Ion OneTouch™ ES Instrument, according to the manufacturer's instructions.
The enriched templates were loaded onto a Life Technologies™ Ion PI™ chip kit V2 and sequenced on the Life Technologies™ Ion Torrent Proton semiconductor sequencing platform. All instruments and reagents were from Thermo Fisher Scientific, Inc.
Haplotype
Sequences with a high quality of Q≥20 were mapped to the human reference sequence hgl9 (Genome Reference Consortium, GRCh37) (sanger.ac.uk/science/data/genome-reference-consortium). The generated BAM files were then subjected to the quality control process using the analysis system of Proton semiconductor sequencing platform. The significance index, which must be met for continued analysis, is achieved when the fraction of the target region with coverage in excess of 30X >85% (27). Variants associated to Ion AmpliSeq Thalassemia panel were screened using the Genome Analysis Toolkit (GATK; version 3.4) (broadinstitute.org/gatk/) (28). Variants were annotated using the bioinformatics software tool Annovar (docopenbio.readthedocs.io/projects/annovar/en/latest/) as well as in-house ad hoc bioinformatics tools (28). The detected variants were subjected to a rigorous manual curation process that included querying variant databases [e.g., dbSNP (ncbi.nlm.nih.gov/SNP/), Exome Aggregation Consortium (exac.broadinstitute.org), 1,000 Genomes (gloshospitals.nhs.uk/about-us/research-our-hospitals/100000-genomes-project/) and Clinvar databases (ncbi.nlm.nih.gov/clinvar/)] and a literature review. Common variation loci were screened in samples from the parents and their first child, according to the exclusion criterion MAF >0.1%. Subsequently, genotype information was used to determine the parental haplotype and its linkage to the pathogenic allele, following the rules of Mendelian inheritance.
RHDO sequential probability ratio test (SPRT)
High quality plasma sequence data were mapped to the human reference sequence hgl9 (Genome Reference Consortium GRCh37), and variants were called using GATK (version 3.4) (29) with parameters optimized for thalassemia. First, gDNA samples from the mother, father and proband (‘trio’) were subjected to analysis of the target region. For paternal inheritance, SNPs that were homozygous in the mother and heterozygous in the father were used. Paternal haplotypes that were inherited by the proband or absent in the proband were thus identified. For maternal inheritance, SNPs that were heterozygous in the mother and homozygous in the father were used. Maternal haplotypes (linked or not linked to the proband's mutation) were determined. Secondly, the target region of the pregnant mother was determined from the plasma cffDNA. Detection of paternal specific alleles in the maternal plasma revealed the inheritance of the paternal haplotype, which could be either linked or not linked to the proband's mutation, from the father. Maternal inheritance was determined through RHDO analysis as previously described (24). Using the fractional fetal DNA concentration, SPRT classification was used to determine the statistical significance of the allelic imbalance within a haplotype block for RHDO analysis. Cumulative sequencing counts of SNP alleles from plasma cffDNA were inputted to the SPRT in order of chromosomal position (30), until a classification was possible. The SPRT curve was calculated according to the following formula:
Where
and
‘Upper boundary’ and ‘lower boundary’ refer to the upper and lower bounds, respectively (3). N is the number of samples for all sites used for classification. q0 represents fetal inheritance of maternal haplotype 2, based on statistics from the haplotype 1 ratio. q1 is the case of fetal inheritance of maternal haplotype 1, from statistics of the haplotype 1 ratio. Haplotype 1 was defined as the upper bound in this assay, and haplotype 2 as the lower bound. Using the SPRT algorithm, the filtered points were used to calculate the cumulative depth and cumulative frequency of variations.
Analysis of amniotic fluid
All amniotic fluid samples were specifically amplified as aforementioned using the α-thalassemia genetic diagnostic kit by gap-PCR and the thalassemia RDB genotyping assay kit (both DaAN Gene Co., Ltd.), according to the manufacturer's instructions. Subsequently, the target products of gap-PCR were detected by agarose gel electrophoresis.
Results
Hematological parameters and genotypes of eight recruited families with thalassemia
Thalassemia screening generally includes the detection of mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), HBA (α2β2) and HBA2 (α2δ2). The MCV reference value usually falls within the range 80–100 fl while the MCH reference value is in the 27–34 pg range. In addition, the HBA reference value is within 96.5–97.5%, and the HBA2 reference value 2.5–3.5%.
The results for hypochromic anemia in all families are presented in Table I. The MCV and MCH values of all the subjects were found to be lower than normal values. The HBA2 values of four individuals were lower than normal values. The HBA2 values of another two individuals were normal and the HBA2 values of ten individuals were higher than normal values.
Table I.
Results of hypochromic anemia analysis for all families.
| Family number | Family member | MCV, fl | MCH, pg | HBA, % | HBA2, % | Type |
|---|---|---|---|---|---|---|
| 1 | Mother | 62.2 | 19.7 | 97.9 | 2.5 | αQSα/αα |
| Father | 69.4 | 20.2 | 97.8 | 2.2 | --SEA/αα | |
| 2 | Mother | 64.6 | 19.6 | 97.7 | 2.3 | --SEA/αα |
| Father | 63.4 | 18.9 | 95.9 | 2.2 | -α3.7/αα | |
| 3 | Mother | 72.5 | 22.4 | 94.2 | 5.2 | βN/β+ (−28A>G) |
| Father | 76.9 | 23.9 | 92.0 | 5.7 | βN/β° (IVS-2-654) | |
| 4 | Mother | 58.5 | 20.2 | 93.6 | 6.0 | βN/β° (CD41-42_CTTT) |
| Father | 63.7 | 22.2 | 95.5 | 4.5 | βN/β+ (−28A>G) | |
| 5 | Mother | 79.9 | 25.4 | 97.1 | 2.9 | αCSα/αα |
| Father | 63.2 | 19.9 | 97.9 | 2.1 | --SEA/αα | |
| 6 | Mother | 65.0 | 21.0 | 93.2 | 6.5 | --SEA/αα& βN/β° (CD41-42_CTTT) |
| Father | 61.8 | 19.7 | 92.5 | 5.9 | βN/β° (CD41-42_CTTT) | |
| 7 | Mother | 66.8 | 21.1 | 93.2 | 6.0 | βN/β0N(CD71-72+A) |
| Father | 72.4 | 22.2 | 95.1 | 4.9 | βN/β+ (−28A>G) | |
| 8 | Mother | 60.4 | 18.5 | 94.0 | 6.0 | βN/β° (CD41-42_CTTT) |
| Father | 59.9 | 19.1 | 92.1 | 5.9 | βN/β° (CD17A>T) |
MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; HBA, adult hemoglobin α2β2; HBA2, adult hemoglobin, α2δ2.
The results for microcytic anemia in all families are shown in Table I. Moreover, in total, three families exhibited α-thalassemia, four families had β-thalassemia and one family had both α- and β-thalassemia (Figs. S1–S3).
Sequencing statistics
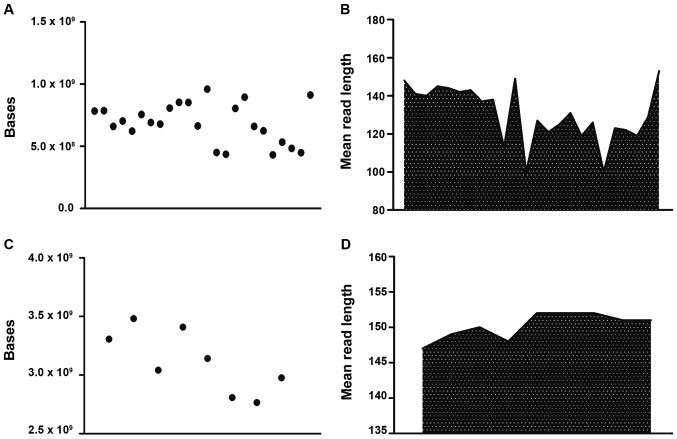
For blood DNA samples, the average number of total raw bases was 728,011,603 (range, 431,538,785-1,100,881,706). The average read length was 137 bp. The mean percentage of sequencing reads mapped to the reference hg19 genome was 98%. After the filtering of low-quality reads, polyclonal reads and primer dimer reads, the number of sequenced bases with Q≥20 ranged between 345,306,892-921,298,977 (Fig. 1A and B). After mapping to the hg19 genome and the removal of Q<20 reads, polyclonal reads and primer dimer reads, the remaining sequences were mapped to the target regions, containing 195 SNPs for α-thalassemia and 275 SNPs for β-thalassemia (data not shown). Furthermore, an average depth of coverage of 118X (range, 70-178X) was obtained for all 470 SNPs across the 24 blood samples (data not shown).
Figure 1.
Sequenced bases and mean read lengths of 24 samples. (A) Bases sequenced from genomic DNA samples. (B) Mean read length of genomic DNA samples. (C) Bases sequenced from plasma samples. (D) Mean read length of plasma samples.
For plasma DNA samples, the average number of total raw bases was 2,986,588,135 (range, 2,509,364,780-3,482,168,649). The average read length was 151 bp. The mean percentage of sequencing reads mapped to the reference hg19 genome was 98%. After filtering of low-quality reads, polyclonal reads and primer dimer reads, the number of sequenced bases with Q-values ≥20 ranged between 2,157,982,851-2,975,281,429 (Fig. 1C and D). Finally, an average depth of coverage of 836X (range, 702–975X) was obtained for all 470 SNPs across the eight plasma samples.
Haplotypes
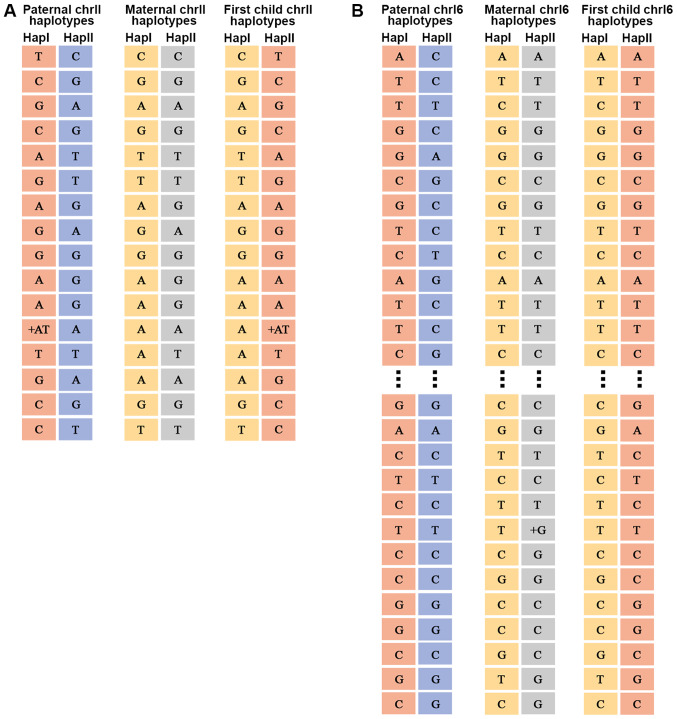
The fractional fetal DNA concentration in the maternal plasma, f, can be calculated from sequencing data as f=2p/(p + q), where p is the number of sequenced reads of the fetal specific allele and q is the read count of the other allele, which is shared by the maternal and fetal genomes (31). The average percentage of fetal DNA in maternal plasma was found to be 17.3% (range, 15–19%) (data not shown). For the father, mother and child, the chromosome 11 and 16 haplotypes were sorted into 16 and 87 different SNP categories, respectively (data not shown). An example of chromosome 11 and 16 haplotypes for a family is illustrated in Fig. 2. The genotype information of the parents was used to determine the parental haplotype and its linkage to the pathogenic allele according to Mendelian inheritance laws (32). Detailed information regarding the haplotypes associated with the pathogenic allele is provided in Table II.
Figure 2.
Haplotype results. (A) Chromosome 11 haplotype of family 1. (B) Chromosome 16 haplotype of family 1. Hap, haplotype; chr, chromosome.
Table II.
Haplotypes associated with the pathogenic allele.
| Family number | Maternal haplotype | Paternal haplotype |
|---|---|---|
| 1 | Hap II | Hap II |
| 2 | Hap I | Hap I |
| 3 | Hap II | Hap II |
| 4 | Hap II | Hap I |
| 5 | Hap II | Hap II |
| 6 | α: Hap I; β: Hap II | Hap II |
| 7 | Hap I | Hap II |
| 8 | Hap II | Hap I |
Hap, haplotype.
Genetic analysis of paternal and maternal genotypes
The genetic results for the father were based on an analysis of SNPs that were heterozygous in the plasma, while being homozygous in the mother and heterozygous in the father, or homozygous in both, but with different genotypes. Overall, the eight fetuses inherited haplotype I of their father (Table III). For maternal inheritance, SNPs were analyzed where the mother was heterozygous and the father was homozygous, allowing the detection of slight allelic imbalances in the maternal plasma using RHDO and SPRT.
Table III.
Parental genetic source results for family 1.
| Chr | Position | Hap I of father | Hap II of father | Hap I of mother | Hap II of mother | Fetal | Inherited |
|---|---|---|---|---|---|---|---|
| Chr11 | 4178706 | T | C | C | C | C:T | Hap I |
| Chr11 | 4186666 | T | C | C | C | C:T | Hap I |
| Chr11 | 4415266 | C | A | A | A | A:C | Hap I |
| Chr11 | 4415319 | A | G | G | G | G:A | Hap I |
| Chr11 | 5452526 | G | A | A | A | A:G | Hap I |
| Chr11 | 5546041 | C | G | G | G | G:C | Hap I |
| Chr11 | 5550754 | C | G | G | G | G:C | Hap I |
| Chr11 | 5554559 | C | G | G | G | G:C | Hap I |
| Chr11 | 5573193 | T | A | A | A | A:T | Hap I |
| Chr16 | 142200 | A | G | G | G | G:A | Hap I |
| Chr16 | 218532 | C | SEA | T | T | T:C | Hap I |
| Chr16 | 219497 | G | SEA | G | G | G:A | Hap I |
| Chr16 | 248418 | C | T | T | T | T:C | Hap I |
| Chr16 | 306062 | G | C | C | C | C:G | Hap I |
| Chr16 | 326525 | C | A | A | A | A:C | Hap I |
| Chr16 | 754288 | T | C | C | C | C:T | Hap I |
| Chr16 | 893160 | A | G | G | G | G:A | Hap I |
Chr, chromosome; hap, haplotype; SEA, Southeast Asian deletion.
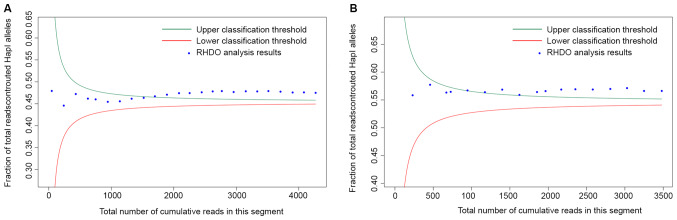
In total, eight fetuses inherited haplotype I from their mother. The results of RHDO SPRT for family 1 are illustrated in Fig. 3 and Tables IV and V. A summary of the results for all eight fetal genotypes is shown in Table VI. It was found that three fetuses (from family 1, family 3 and family 5) had no inheritance of their parents' pathogenic sites; three fetuses (from family 4, family 7 and family 8) only inherited the pathogenic site of one parent; and one fetus (family 2) inherited the pathogenic sites of both parents.
Figure 3.
SPRT classification. SPRT classification process for the RHDO analysis of (A) type-α and (B) type-β single nucleotide polymorphisms in a region near the p-ter of chromosome 16 in family 1. Classification was carried out from the telomeric end to the centromere. RHDO, relative haplotype dosage; SPRT, sequential probability ratio test; hap, haplotype.
Table IV.
SPRT classification process for relative haplotype dosage analysis of type α single nucleotide polymorphisms near the p-ter of chromosome 16 in family 1.
| Position | Hap I count | Hap II count | Hap I cum. | Hap II cum. | Total reads | Hap I fraction | Upper boundary | Lower boundary | SPRT |
|---|---|---|---|---|---|---|---|---|---|
| 234632 | 23 | 25 | 23 | 25 | 48 | 0.4792 | 0.8506 | 0.0569 | Unclassified |
| 256278 | 84 | 108 | 107 | 133 | 240 | 0.4458 | 0.5331 | 0.3744 | Unclassified |
| 281299 | 97 | 95 | 204 | 228 | 432 | 0.4722 | 0.4978 | 0.4096 | Unclassified |
| 281885 | 85 | 109 | 289 | 337 | 626 | 0.4617 | 0.4842 | 0.4233 | Unclassified |
| 306243 | 56 | 68 | 345 | 405 | 750 | 0.4600 | 0.4791 | 0.4283 | Unclassified |
| 311853 | 84 | 110 | 429 | 515 | 944 | 0.4544 | 0.4739 | 0.4336 | Unclassified |
| 312253 | 89 | 104 | 518 | 619 | 1,137 | 0.4556 | 0.4705 | 0.4370 | Unclassified |
| 312635 | 94 | 95 | 612 | 714 | 1,326 | 0.4615 | 0.4681 | 0.4394 | Unclassified |
| 315557 | 92 | 101 | 704 | 815 | 1,519 | 0.4635 | 0.4663 | 0.4412 | Unclassified |
| 317393 | 89 | 90 | 793 | 905 | 1,698 | 0.4670 | 0.4650 | 0.4425 | Hap I |
| 319761 | 97 | 96 | 890 | 1,001 | 1,891 | 0.4707 | 0.4638 | 0.4437 | Hap I |
| 336660 | 98 | 95 | 988 | 1,096 | 2,084 | 0.4741 | 0.4629 | 0.4446 | Hap I |
| 377617 | 81 | 89 | 1,069 | 1,185 | 2,254 | 0.4743 | 0.4622 | 0.4453 | Hap I |
| 390780 | 96 | 97 | 1,165 | 1,282 | 2,447 | 0.4761 | 0.4615 | 0.4460 | Hap I |
| 423420 | 89 | 87 | 1,254 | 1,369 | 2,623 | 0.4781 | 0.4610 | 0.4465 | Hap I |
| 427516 | 72 | 74 | 1,326 | 1,443 | 2,769 | 0.4789 | 0.4606 | 0.4469 | Hap I |
| 641445 | 84 | 105 | 1,410 | 1,548 | 2,958 | 0.4767 | 0.4602 | 0.4473 | Hap I |
| 651517 | 98 | 96 | 1,508 | 1,644 | 3,152 | 0.4784 | 0.4598 | 0.4477 | Hap I |
| 674029 | 87 | 97 | 1,595 | 1,741 | 3,336 | 0.4781 | 0.4594 | 0.4480 | Hap I |
| 678843 | 88 | 90 | 1,683 | 1,831 | 3,514 | 0.4789 | 0.4592 | 0.4483 | Hap I |
| 878161 | 88 | 106 | 1,771 | 1,937 | 3,708 | 0.4776 | 0.4589 | 0.4486 | Hap I |
| 880431 | 85 | 109 | 1,856 | 2,046 | 3,902 | 0.4757 | 0.4586 | 0.4489 | Hap I |
| 928867 | 87 | 94 | 1,943 | 2,140 | 4,083 | 0.4759 | 0.4584 | 0.4491 | Hap I |
| 940706 | 85 | 102 | 2,028 | 2,242 | 4,270 | 0.4749 | 0.4582 | 0.4493 | Hap I |
SPRT, sequential probability ratio test; hap, haplotype.
Table V.
SPRT classification process for relative haplotype dosage analysis of type β single nucleotide polymorphisms near the p-ter of chromosome 16 in family 1.
| Position | Hap I count | Hap II count | Hap I cum. | Hap II cum. | Total reads | Hap I fraction | Upper boundary | Lower boundary | SPRT |
|---|---|---|---|---|---|---|---|---|---|
| 151136 | 129 | 102 | 129 | 102 | 231 | 0.5584 | 0.6287 | 0.4638 | Unclassified |
| 192314 | 136 | 92 | 265 | 194 | 459 | 0.5773 | 0.5878 | 0.5048 | Unclassified |
| 225159 | 113 | 99 | 378 | 293 | 671 | 0.5633 | 0.5746 | 0.5179 | Unclassified |
| 231541 | 33 | 24 | 411 | 317 | 728 | 0.5646 | 0.5724 | 0.5201 | Unclassified |
| 235660 | 126 | 93 | 537 | 410 | 947 | 0.5671 | 0.5664 | 0.5262 | Hap I |
| 241210 | 125 | 102 | 662 | 512 | 1,174 | 0.5639 | 0.5625 | 0.5300 | Hap I |
| 258741 | 134 | 92 | 796 | 604 | 1,400 | 0.5686 | 0.5599 | 0.5327 | Hap I |
| 326826 | 112 | 112 | 908 | 716 | 1,624 | 0.5591 | 0.5580 | 0.5345 | Hap I |
| 534954 | 138 | 92 | 1,046 | 808 | 1,854 | 0.5642 | 0.5565 | 0.5360 | Hap I |
| 902172 | 66 | 45 | 1,112 | 853 | 1,965 | 0.5659 | 0.5560 | 0.5366 | Hap I |
| 903426 | 122 | 83 | 1,234 | 936 | 2,170 | 0.5687 | 0.5550 | 0.5375 | Hap I |
| 905559 | 106 | 78 | 1,340 | 1,014 | 2,354 | 0.5692 | 0.5544 | 0.5382 | Hap I |
| 922134 | 128 | 99 | 1,468 | 1,113 | 2,581 | 0.5688 | 0.5536 | 0.5389 | Hap I |
| 935560 | 133 | 96 | 1,601 | 1,209 | 2,810 | 0.5698 | 0.5530 | 0.5395 | Hap I |
| 937413 | 129 | 89 | 1,730 | 1,298 | 3,028 | 0.5713 | 0.5526 | 0.5400 | Hap I |
| 945728 | 110 | 112 | 1,840 | 1,410 | 3,250 | 0.5662 | 0.5521 | 0.5404 | Hap I |
| 947817 | 131 | 100 | 1,971 | 1,510 | 3,481 | 0.5662 | 0.5517 | 0.5408 | Hap I |
SPRT, sequential probability ratio test; hap, haplotype; Cum, cumulative count for alleles on Hap I/Hap II.
Table VI.
Results for the eight fetal genotypes.
| Fetus number | Haplotype inherited from father | Haplotype inherited from mother | Type |
|---|---|---|---|
| 1 | Hap I | Hap I | αα/αα |
| 2 | Hap I | Hap I | --SEA/-α3.7 |
| 3 | Hap I | Hap I | βN/βN |
| 4 | Hap I | Hap I | β+[-28(A>G)]/βN |
| 5 | Hap I | Hap I | αα/αα |
| 6 | Hap I | Hap I | --SEA/αα, βN/βN |
| 7 | Hap I | Hap I | β°[CD71-72(+A)]/βN |
| 8 | Hap I | Hap I | β°[CD17(AAG>TAG)]/βN |
Hap, haplotype; SEA, Southeast Asian deletion.
Accuracy of fetus haplotype determination
To further verify the accuracy of fetal haplotype inference for NIPT, the results obtained from maternal plasma DNA sequencing were validated using gap-PCR and RDB of fetal gDNA collected from amniotic fluid cells. The non-invasive results were 100% consistent with the gap-PCR and RDB results from amniotic fluid for these eight samples (Figs. 1 and 2).
Discussion
The discovery of cffDNA in maternal plasma has allowed for the possibility of non-invasive prenatal diagnosis of genetic disorders. Applications of cffDNA include screening for aneuploidies, prenatal diagnosis of mutation based monogenic diseases, and fetal Rhesus D status determination (33). These techniques are based on qualitative and quantitative comparisons of cffDNA with the background of maternal cffDNA in maternal plasma (10). Non-invasive prenatal screening for aneuploidies based on NGS was rapidly incorporated into routine clinical practice (30). Accordingly, guidelines for clinical application and committee guidance regarding NIPT have been issued by numerous medical societies, including the Society of Obstetricians and Gynaecologists of Canada (34), the Italian College of Fetal Maternal Medicine (35), the American College of Medical Genetics and Genomics (36), the National Society of Genetic Counselors and the International Society for Prenatal Diagnosis (37). Furthermore, genome-wide micro deletion or micro duplication syndromes have been screened for in an expanded NIPT protocol.
Maternal plasma cffDNA includes cffDNA and maternal cffDNA, and half of fetal alleles are inherited paternally. Xiong et al (38) reported one case of this type of non-invasive prenatal diagnosis using PCR and NGS for the detection of a paternal pathogenic mutation. Yan et al (39) and Ho et al (40) reported exclusion of fetal homozygous α-thalassemia based on the presence of the father's SNP within the break point sequence. However, these methods can only be used to exclude fetal risk and do not provide the exact genotype of the fetus. Quantification of specific alleles can provide accurate fetal genotype results. In this manner, several monogenic diseases have been successfully detected non-invasively using cffDNA and PCR based methods (41,42). NGS-based approaches have also been applied to non-invasive prenatal diagnosis of β-thalassemia through deduction of the fetal inheritance of maternally transmitted mutations via the quantification of the relative levels of haplotypes with SNP alleles in and around the targeted gene. Recently, Yang et al (43) reported a robust and versatile NGS-based cffDNA allelic molecule counting system termed the cffDNA barcode enabled single molecule test, which was developed for the non-invasive prenatal diagnosis of β-thalassemia. However, these methods mainly aim to identify SNV-associated thalassemia and have not been applied to CNV-associated thalassemia (44).
The present study aimed to develop a system for non-invasive prenatal detection of paternal and maternal mutations associated with α- and β-thalassemia. It was demonstrated that multiplex PCR or target capture combined with NGS of highly heterozygous SNPs within the 10 kb flanking region of the gene of interest effectively reduced the target region size for detection of paternal and maternal mutations. Furthermore, this technique may be used to detect both α- and β-thalassemia, as well as SNPs and large deletion mutations. gDNA of the parents and their children were amplified via multiplex PCR using a thalassemia panel, and cffDNA was captured with a set of thalassemia probes. The products were sequenced using NGS. Subsequently, the parental haplotype was determined using a trio-based strategy. The genetic results for the father were based on analysis of SNPs that were heterozygous in the plasma and were also either homozygous in the mother and heterozygous in the father, or homozygous in both, but of different genotypes. For maternal inheritance, SNPs were analyzed where the mother was heterozygous and the father was homozygous, and whether a slight allelic imbalance was present in maternal plasma was investigated using RHDO SPRT. In total, three cases of α-thalassemia, four of β-thalassemia and one case of combined α- and β-thalassemia were successfully diagnosed using this non-invasive prenatal diagnosis method, and the results exhibited high consistency with the traditional invasive method. Fetal genotypes were successfully deduced non-invasively for eight families. The non-invasively determined haplotypes of the eight fetuses were identical with those obtained through invasive prenatal diagnosis procedures, with an accuracy rate of 100% in the target region.
In the present study, the average percentage of fetal DNA was 17.3%. As accuracy is affected by the fetal fraction, in follow-up studies, additional cases should be tested, especially using samples collected in early gestation, in order to thoroughly evaluate the method prior to clinical application. This study, however, has provided an example of non-invasive prenatal diagnosis of single gene diseases that can be exploited for other single gene diseases, such as congenital adrenal hyperplasia, Ellis van Creveld syndrome, hemophilia and Hunter's syndrome. Overall, multiplex PCR and target capture combined with NGS of highly heterozygous SNPs flanking the gene of interest is an effective method to reduce sequencing costs.
In conclusion, the routine prenatal diagnosis method for thalassemia currently used clinically is gap-PCR and RDB of fetal DNA, which can be acquired from ultrasound mediated abdominal biopsy, amniocentesis or cordocentesis. However, these invasive procedures may increase the risk of miscarriage and cause anxiety for pregnant women. The present study demonstrated the feasibility of non-invasive prenatal diagnosis of thalassemia using cffDNA from the plasma of pregnant women through target capture and NGS combined with RHDO analysis. Furthermore, this method can be used to detect deletion and mutation-based thalassemia at a relatively lower cost, as well as to investigate other diseases caused by CNVs and SNVs.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was funded by the Science and Technology Program of Guangzhou (grant nos. 201604020104 and 201704020114), the Key Program for Health Care Collaborative Innovation of Guangzhou (grant no. 201803040009) and the Natural Scientific Research Foundation of Guangdong Province (grant no. 2018A030313286).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XY, YCY, XXY and JZ conceived and designed the study. YCY and DMF performed the experiments. XY, YCY and DMF wrote the manuscript. XXY and JZ improved the manuscript. DMF, XY, SL ML and HYH analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study protocol was approved by the Medical Ethics Committee of The Third Affiliated Hospital of Sun Yatsen University, and the Committee on Human Research, Publications and Ethics of School of Laboratory Medicine and Biotechnology, Southern Medical University. Prior to recruitment and sample collection, meetings were held to explain in detail the purpose and procedures of the study. The inconveniences involved, including blood sampling, were also explained to the participants. Written informed consent was obtained from each participant or the participant's guardian. The study was undertaken according to the principles of the Helsinki Declaration of 1975 (as revised 2008).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Higgs DR, Engel JD, Stamatoyannopoulos G. Thalassaemia. Lancet. 2012;379:373–383. doi: 10.1016/S0140-6736(11)60283-3. [DOI] [PubMed] [Google Scholar]
- 2.Pan HF, Long GF, Li Q, Feng YN, Lei ZY, Wei HW, Huang YY, Huang JH, Lin N, Xu QQ, et al. Current status of thalassemia in minority populations in Guangxi, China. Clin Genet. 2007;71:419–426. doi: 10.1111/j.1399-0004.2007.00791.x. [DOI] [PubMed] [Google Scholar]
- 3.Lo YM, Chan KC, Sun H, Chen EZ, Jiang P, Lun FM, Zheng YW, Leung TY, Lau TK, Cantor CR, Chiu RW. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2:61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 4.Yin A, Li B, Luo M, Xu L, Wu L, Zhang L, Ma Y, Chen T, Gao S, Liang J, et al. The prevalence and molecular spectrum of α- and β-globin gene mutations in 14,332 families of Guangdong province, China. PLoS One. 2014;9:e89855. doi: 10.1371/journal.pone.0089855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao A, Kan YW. The prevention of thalassemia. Cold Spring Harb Perspect Med. 2013;3:a11775. doi: 10.1101/cshperspect.a011775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mourad FH, Hoffbrand AV, Sheikh-Taha M, Koussa S, Khoriaty AI, Taher A. Comparison between desferrioxamine and combined therapy with desferrioxamine and deferiprone in iron overloaded thalassaemia patients. Br J Haematol. 2003;121:187–189. doi: 10.1046/j.1365-2141.2003.04240.x. [DOI] [PubMed] [Google Scholar]
- 7.Lidonnici MR, Ferrari G. Gene therapy and gene editing strategies for hemoglobinopathies. Blood Cells Mol Dis. 2018;70:87–101. doi: 10.1016/j.bcmd.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Allyse M, Minear M, Rote M, Hung A, Chandrasekharan S, Berson E, Sridhar S. Non-invasive prenatal testing: A review of international implementation and challenges. Int J Women's Health. 2015;7:113–126. doi: 10.2147/IJWH.S67124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mujezinovic F, Alfirevic Z. Procedure-relatedcomplications of amniocentesis and chorionic villous sampling: A systematic review. Obstet Gynecol. 2007;110:687–694. doi: 10.1097/01.AOG.0000278820.54029.e3. [DOI] [PubMed] [Google Scholar]
- 10.Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 11.Babkina N, Graham JM. New genetic testing in prenatal diagnosis. Semin Fetal Neonatal Med. 2014;19:214–219. doi: 10.1016/j.siny.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Chiu RW, Akolekar R, Zheng YW, Leung TY, Sun H, Chan KC, Lun FM, Go AT, Lau ET, To WW, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: Large scale validity study. BMJ. 2011;342:c7401. doi: 10.1136/bmj.c7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.You Y, Sun Y, Li X, Li Y, Wei X, Chen F, Ge H, Lan Z, Zhu Q, Tang Y, et al. Integration of targeted sequencing and NIPT into clinical practice in a Chinese family with maple syrup urine disease. Genet Med. 2014;16:594–600. doi: 10.1038/gim.2013.197. [DOI] [PubMed] [Google Scholar]
- 14.McCullough RM, Almasri EA, Guan X, Geis JA, Hicks SC, Mazloom AR, Deciu C, Oeth P, Bombard AT, Paxton B, et al. Non-invasive prenatal chromosomal aneuploidy testing-clinical experience: 100,000 clinical samples. PLoS One. 2014;9:e109173. doi: 10.1371/journal.pone.0109173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Zimmermann B, Rusterholz C, Kang A, Holzgreve W, Hahn S. Size separation of circulatory DNA in maternal plasma permits ready detection of fetal DNA polymorphisms. Clin Chem. 2004;50:1002–1011. doi: 10.1373/clinchem.2003.029835. [DOI] [PubMed] [Google Scholar]
- 16.Chan KC, Zhang J, Hui AB, Wong N, Lau TK, Leung TN, Lo KW, Huang DW, Lo YM. Size distributions of maternal and fetal DNA in maternal plasma. Clin Chem. 2004;50:88–92. doi: 10.1373/clinchem.2003.024893. [DOI] [PubMed] [Google Scholar]
- 17.Hui WW, Jiang P, Tong YK, Lee WS, Cheng YK, New MI, Kadir RA, Chan KC, Leung TY, Lo YM, Chiu RW. Universal haplotype-based noninvasive prenatal testing for single gene diseases. Clin Chem. 2017;63:513–524. doi: 10.1373/clinchem.2016.268375. [DOI] [PubMed] [Google Scholar]
- 18.Xu Y, Li X, Ge H, Xiao B, Zhang Y, Ying X, Pan X, Wang L, Xie W, Ni L, et al. Haplotype-based approach for noninvasive prenatal tests of Duchenne muscular dystrophy using cell-free fetal DNA in maternal plasma. Genet Med. 2015;17:889–896. doi: 10.1038/gim.2014.207. [DOI] [PubMed] [Google Scholar]
- 19.Ma D, Ge H, Li X, Jiang T, Chen F, Zhang Y, Hu P, Chen S, Zhang J, Ji X, et al. Haplotype-based approach for noninvasive prenatal diagnosis of congenital adrenal hyperplasia by maternal plasma DNA sequencing. Gene. 2014;544:252–258. doi: 10.1016/j.gene.2014.04.055. [DOI] [PubMed] [Google Scholar]
- 20.Lo YM, Hjelm NM, Fidler C, Sargent IL, Murphy MF, Chamberlain PF, Poon PM, Redman CW, Wainscoat JS. Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma. N Engl J Med. 1998;339:1734–1738. doi: 10.1056/NEJM199812103392402. [DOI] [PubMed] [Google Scholar]
- 21.Saito H, Sekizawa A, Morimoto T, Suzuki M, Yanaihara T. Prenatal DNA diagnosis of a single-gene disorder from maternal plasma. Lancet. 2000;356:1170. doi: 10.1016/S0140-6736(00)02767-7. [DOI] [PubMed] [Google Scholar]
- 22.Amicucci P, Gennarelli M, Novelli G, Dallapiccola B. Prenatal diagnosis of myotonic dystrophy using fetal DNA obtained from maternal plasma. Clin Chem. 2000;46:301–302. doi: 10.1093/clinchem/46.2.301. [DOI] [PubMed] [Google Scholar]
- 23.González-González MC, Trujillo MJ, Rodríguez de Alba M, García-Hoyos M, Lorda-Sánchez I, Díaz-Recasens J, Ayuso C, Ramos C. Huntington disease-unaffected fetus diagnosed from maternal plasma using QF-PCR. Prenatal Diag. 2003;23:232–234. doi: 10.1002/pd.570. [DOI] [PubMed] [Google Scholar]
- 24.Lam KW, Jiang P, Liao GJ, Chan KC, Leung TY, Chiu RW, Lo YM. Noninvasive prenatal diagnosis of monogenic diseases by targeted massively parallel sequencing of maternal plasma: Application to β-Thalassemia. Clin Chem. 2012;58:1467–1475. doi: 10.1373/clinchem.2012.189589. [DOI] [PubMed] [Google Scholar]
- 25.Camunas-Soler J, Lee H, Hudgins L, Hintz SR, Blumenfeld YJ, El-Sayed YY, Quake SR. Noninvasive prenatal diagnosis of single-gene disorders by use of droplet digital PCR. Clin Chem. 2018;64:336–345. doi: 10.1373/clinchem.2017.278101. [DOI] [PubMed] [Google Scholar]
- 26.Hudecova I, Chiu RW. Non-invasive prenatal diagnosis of thalassemias using maternal plasma cell free DNA. Best Pract Res Clin Obstet Gynaecol. 2017;39:63–73. doi: 10.1016/j.bpobgyn.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 27.Wenke L, Fengyu L, Siyao Z, Bin C, Na Z, Yu N, Dao Z, Qian Z. Automatic analysis pipeline of next-generation sequencing data. Yi Chuan. 2014;36:618–624. doi: 10.3724/SP.J.1005.2014.0618. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 28.De Summa S, Malerba G, Pinto R, Mori A, Mijatovic V, Tommasi S. GATK hard filtering: Tunable parameters to improve variant calling for next generation sequencing targeted gene panel data. BMC Bioinformatics. 2017;18(Suppl 5):S119. doi: 10.1186/s12859-017-1537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bauer DC. Variant calling comparison CASAVA1.8 and GATK. Nat Prec. 2011 Jul 18; doi: 10.1038/npre.2011.6107.1. doi: 10.1038/npre.2011.6107.1. [DOI] [Google Scholar]
- 30.Lo YM, Lun FM, Chan KC, Tsui NB, Chong KC, Lau TK, Leung TY, Zee BC, Cantor CR, Chiu RW. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc Natl Acad Sci USA. 2007;104:13116–13121. doi: 10.1073/pnas.0705765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng M, Li X, Ge H, Chen F, Han M, Zhang Y, Kang D, Xie W, Gao Z, Pan X, et al. Noninvasive prenatal testing for autosomal recessive conditions by maternal plasma sequencing in a case of congenital deafness. Genet Med. 2014;16:972–976. doi: 10.1038/gim.2014.51. [DOI] [PubMed] [Google Scholar]
- 32.Allen S, Young E, Bowns B. Noninvasive prenatal diagnosis for single gene disorders. Curr Opin Obstet Gynecol. 2017;29:73–79. doi: 10.1097/GCO.0000000000000347. [DOI] [PubMed] [Google Scholar]
- 33.Walsh JM, Goldberg JD. Fetal aneuploidy detection by maternal plasma DNA sequencing: A technology assessment. Prenatal Diag. 2013;33:514–520. doi: 10.1002/pd.4109. [DOI] [PubMed] [Google Scholar]
- 34.Galbiati S, Monguzzi A, Damin F, Soriani N, Passiu M, Castellani C, Natacci F, Curcio C, Seia M, Lalatta F, et al. COLD-PCR and microarray: Two independent highly sensitive approaches allowing the identification of fetal paternally inherited mutations in maternal plasma. J Med Genet. 2016;53:481–487. doi: 10.1136/jmedgenet-2015-103229. [DOI] [PubMed] [Google Scholar]
- 35.Langlois S, Brock J, Genetics Committee Current status in non-invasive prenatal detection of down syndrome, trisomy 18, and trisomy 13 using cell-Free DNA in Maternal Plasma. J Obstetr Gynaecol Can. 2013;35:177–181. doi: 10.1016/S1701-2163(15)31025-2. (In English, French) [DOI] [PubMed] [Google Scholar]
- 36.Benn P, Borrell A, Chiu RW, Cuckle H, Dugoff L, Faas B, Gross S, Huang T, Johnson J, Maymon R, et al. Position statement from the chromosome abnormality screening committee on behalf of the board of the international society for prenatal diagnosis. Prenatal Diag. 2015;35:725–734. doi: 10.1002/pd.4608. [DOI] [PubMed] [Google Scholar]
- 37.Gregg AR, Gross SJ, Best RG, Monaghan KG, Bajaj K, Skotko BG, Thompson BH, Watson MS. ACMG statement on noninvasive prenatal screening for fetal aneuploidy. Genet Med. 2013;15:395–398. doi: 10.1038/gim.2013.29. [DOI] [PubMed] [Google Scholar]
- 38.Xiong L, Barrett AN, Hua R, Tan TZ, Ho SS, Chan JK, Zhong M, Choolani M. Non-invasive prenatal diagnostic testing for β-thalassaemia using cell-free fetal DNA and next-generation sequencing. Prenat Diagn. 2014;35:258–265. doi: 10.1002/pd.4536. [DOI] [PubMed] [Google Scholar]
- 39.Yan TZ, Mo QH, Cai R, Chen X, Zhang CM, Liu YH, Chen YJ, Zhou WJ, Xiong F, Xu XM. Reliable detection of paternal SNPs within deletion breakpoints for non-invasive prenatal exclusion of homozygous α-thalassemia in maternal plasma. PLoS One. 2011;6:e24779. doi: 10.1371/journal.pone.0024779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho SS, Chong SS, Koay ES, Ponnusamy S, Chiu L, Chan YH, Rauff M, Baig S, Chan J, Su LL, et al. Noninvasive prenatal exclusion of haemoglobin Bart's using foetal DNA from maternal plasma. Prenat Diagn. 2010;30:65–73. doi: 10.1002/pd.2413. [DOI] [PubMed] [Google Scholar]
- 41.Devers PL, Cronister A, Ormond KE, Facio F, Brasington CK, Flodman P. Noninvasive prenatal Testing/Noninvasive prenatal diagnosis: The Position of the National Society of Genetic Counselors. J Genet Couns. 2013;22:291–295. doi: 10.1007/s10897-012-9564-0. [DOI] [PubMed] [Google Scholar]
- 42.Lee ST, Weykamp CW, Lee YW, Kim JW, Ki CS. Effects of 7 hemoglobin variants on the measurement of glycohemoglobin by 14 analytical methods. Clin Chem. 2007;53:2202–2205. doi: 10.1373/clinchem.2007.093963. [DOI] [PubMed] [Google Scholar]
- 43.Yang X, Zhou Q, Zhou W, Zhong M, Guo X, Wang X, Fan X. A Cell-free DNA Barcode-enabled single-molecule test for noninvasive prenatal diagnosis of monogenic disorders: Application to β-thalassemia. Adv Sci. 2019;6:1802332. doi: 10.1002/advs.201802332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan DM, Yang X, Huang LM, Ouyang GJ, Yang XX, Li M. Simultaneous detection of target CNVs and SNVs of thalassemia by multiplex PCR and nextgeneration sequencing. Mol Med Rep. 2019;19:2837–2848. doi: 10.3892/mmr.2019.9896. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.