Abstract
Colon cancer is one of the most frequent malignant tumors, and microRNA (miR)-205 is involved in the tumor progression. The present study aimed to explore the effects of miR-205 on human colon cancer and its targeting mechanism. The levels of miR-205 and mouse double minute 4 (MDM4) were determined via reverse transcription-quantitative (RT-q)PCR and western blot analysis. A luciferase activity assay was performed to analyze the association between miR-205 and MDM4. Cell viability, migration and invasion were determined via Cell Counting Kit-8, wound healing and Transwell assays, respectively. The levels of epithelial-mesenchymal transition (EMT)-associated factors were determined by RT-qPCR and western blot analysis. It was identified that MDM4 was overexpressed in colon cancer tissues and cells, and that there was a negative correlation between miR-205 and MDM4 expression in colon cancer. Similarly, miR-205 inhibited MDM4 expression by binding to its 3′untranslated region. in addition, miR-205 directly targeted MDM4, accompanied by suppressed proliferation, migration and invasion of HCT116 cells. EMT processes were suppressed in miR-205-overexpressed cells; upregulation of E-cadherin, and downregulation of N-cadherin, vimentin, matrix metalloproteinase (MMP)2 and MMP9 were observed. Collectively, miR-205 conspicuously depressed the viability, migration, invasion and EMT process of human colon cancer cells via targeting MDM4. miR-205 could be potentially used in the treatment of human colon cancer.
Keywords: microRNA-205, mouse double minute 4, cell migration, cell invasion, epithelial-mesenchymal transition
Introduction
Colon cancer is one of the most frequent malignant tumors and the second leading cause of tumor-related mortality in the United States (1,2). The incidence of colon cancer is increasing annually, and the disease seriously threatens the physical and mental health of patients (3). In recent years, a large number of clinical studies have reported that metastatic recurrence is the primary cause of the prognosis of colon cancer (4–6). In the early stages, the 5-year survival rate of colon cancer is >90%; however, when colon cancer is diagnosed with local lymph node metastasis, the 5-year survival rate decreases to 65% (7). Hence, it is of great clinical significance to explore the inhibition of tumor metastasis.
Tumor metastasis is a sequential process of interaction among tumor cells, host cells and the tissue microenvironment (8). Epithelial-mesenchymal transition (EMT), which plays a key role in tumor metastasis (9,10), is characterized by the deficiency of epithelial phenotypes, loss of cell polarity, reduced contact with surrounding cells and matrix, and enhanced cell migration and invasion in the presence of interstitial phenotype (11). Furthermore, EMT participates in nearly all physiological and pathological processes, such as the differentiation of various tissues and organs, repair of tissue damage, tissue fibrosis, tumor occurrence and metastasis (12–14).
MicroRNAs (miRNAs/miRs), a class of small, 18- to 28-nucleotide-long, noncoding RNA molecules, are involved in the progression of tumors (15,16). To date, the human genome contains ~1,000 miRNAs, and each miRNA is expected to interact with dozens or even hundreds of genes via matching 5′ sequences and 3′ untranslated regions (3′UTRs) of target mRNAs (17–19). miR-205 is a highly conserved miRNA, and its homologous chromosomes can be found in different species (20,21). Homo sapiens (hsa)-miR-205 is located in the second intron of the LOC642587 site of the first chromosome (22). Whether miR-205 is an oncogene or a tumor suppressor still remains controversial (23), though studies have reported that miR-205 participates in the EMT process of tumor cells (24–26).
Mouse double minute 4 (MDM4), which was isolated and identified in 1996, is also known as MDMX, and is an important upstream regulator of p53 (27). Following MDM4 phosphorylation, the p53 binding domain of MDM4 can combine with the transcriptional activation domain of the wild and mutant p53 proteins to form a MDM4/p53 complex to inhibit the transcriptional activity of p53 (28,29). Increasing evidence has shown that MDM4 is abnormally expressed in a number of tumor tissues such as breast cancer, retinoblastoma, lung cancer, colon cancer and gastric cancer (30–33). Accordingly, the inhibition of abnormal expression of MDM4 has attracted increasing attention of researchers in the field of anti-tumor mechanisms.
The current study determined the expression of miR-205 and MDM4 in colon cancer tissues, adjacent normal tissues, and colon and colorectal cancer cell lines. In addition, the correlation between the expression of miR-205 and MDM4 colon cancer tissue was studied. A binding site for miR-205 in the 3′UTR of MDM4 was identified by TargetScan prediction software, and the role of miR-205 in the migration, invasion and EMT process of tumor cells gene was explored by targeting MDM4.
Materials and methods
Tissue source
The colon cancer tissues and adjacent normal tissues (distance from tumor margin, 2 cm) were obtained from 47 patients with colon cancer who were diagnosed at Beijing Jishuitan Hospital between March 2011 and March 2016. All patients signed informed consent and agreed that their tissues would be used for clinical research. The relationship between miR-205 expression and clinical characteristics of colon cancer are presented in Table I. Patients were divided into high- and low-expression groups for analysis of associations with clinical characteristics based on the mean miR-205 expression value in tumor tissues. The study was reviewed and approved by the Ethics Committee of Beijing Jishuitan Hospital (permit no. J20110104015).
Table I.
Relationship between miR-205 expression and clinical characteristics of colon cancer.
| miR-205 expression | |||||
|---|---|---|---|---|---|
| Clinicopathological variable | n | Low | High | χ2 | P-value |
| All cases | 47 | 24 | 23 | ||
| Age | 0.180 | 0.671 | |||
| ≤65 | 21 | 10 | 11 | ||
| >65 | 26 | 14 | 12 | ||
| Sex | 0.216 | 0.642 | |||
| Female | 20 | 11 | 9 | ||
| Male | 27 | 13 | 14 | ||
| Pathological grade | 4.381 | 0.036 | |||
| I–II | 35 | 21 | 14 | ||
| III | 12 | 3 | 9 | ||
| Stage | 7.817 | 0.005 | |||
| I–II | 28 | 19 | 9 | ||
| III–IV | 19 | 5 | 14 | ||
| Lymph node metastasis | 4.846 | 0.028 | |||
| Positive | 19 | 6 | 13 | ||
| Negative | 28 | 18 | 10 | ||
miR, microRNA.
Cell culture and transfection
The human colorectal cancer cell line (HT29), colon cancer cell lines (HCT116, HCT8, LS174T and SW480) and 293T cells were purchased from Shanghai Gaining Biotechnology Co., Ltd. HT29, HCT116, HCT8, LS174T and SW480 cells were maintained in RPMI-1640 Medium (Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 100X penicillin-streptomycin mixed solution (Beijing Leagene Biotech Co., Ltd.) in an incubator (cat. no. DH-160I; Shanghai SANTN Instrument Co., Ltd.) with 5% CO2 at 37°C and 95% humidity. 293T cells were cultured in DMEM (Thermo Fisher Scientific, Inc.) containing 10% FBS and 100X penicillin-streptomycin mixed solution and used for luciferase activity assays.
Human wild-type (WT) and mutant (MT) MDM4 3′UTRs were cloned downstream of Renilla luciferase in a psiCHECK-2 vector (Hangzhou Hibio Technology Co., Ltd.). Subsequently, miR-205 mimic/control mimic (mimic NC; 30 µmol/l) was co-transfected into 293T cells (3×103 cells/well) using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) for 24 h at 37°C. HCT116 cells (5×105 cells/well) were transfected with 50 nmol/l miR-205 mimic (cat. no. HmiR0026; GeneCopoeia, Inc.), mimic NC (cat. no. CmiR0001; GeneCopoeia, Inc.), miR-205 inhibitor (cat. no. HmiR-AN0307; GeneCopoeia, Inc.), negative control for inhibitor (IC; cat. no. CmiR-AN0001; GeneCopoeia, Inc.), control small interfering (si)RNA (siNC; cat. no. AM4641; Thermo Fisher Scientific, Inc.), MDM4-siRNA (siMDM4; cat. no. AM16708; Thermo Fisher Scientific, Inc.), miR-205 inhibitor + siMDM4, control + siNC or miR-205 inhibitor + siNC using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) for 24 h at 37°C. The transfection efficiency was assessed by western blotting analysis. Subsequent experiments were conducted at 24 h post-transfection.
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from tissues and cells (1.3×105 cells/well) using RNAiso Plus (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. A total of 1 µg of RNA was used to synthesize cDNA using a RevertAid™ cDNA Synthesis kit (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. Subsequently, qPCR was performed using the SYBR Premix Ex Taq™ II kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The following thermocycling conditions were used for qPCR: Initial denaturation at 95°C for 10 min; followed by 40 cycles of 94°C for 2 min, 60°C for 50 sec; a final extension at 60°C for 1 min; and storage at 4°C. The primer sequences are listed in Table II. U6 and GAPDH were used as internal references. The formula 2−ΔΔCq was used to calculate relative gene expression (34).
Table II.
Primer sequences.
| Sequence (5′→3′) | ||
|---|---|---|
| Primer | Forward | Reverse |
| MDM4 | GAAAGACCCAAGCCCTCTCT | GCAGTGTGGGGATATCGTCT |
| miR-205 | CTCGAGCAGGTGCAAGGACGTGTTG | GGATCCGTGGCTTAGAAGGCCGGG |
| E-cadherin | ACGCATTGCCACATACACTC | GGTGTTCACATCATCGTCCG |
| N-cadherin | CTTGCCAGAAAACTCCAGGG | TGTGCCCTCAAATGAAACCG |
| MMP2 | CAGCCCTGCAAGTTTCCATT | GTTGCCCAGGAAAGTGAAGG |
| MMP9 | GAGACTCTACACCCAGGACG | GAAAGTGAAGGGGAAGACGC |
| Vimentin | AATAAGATCCTGCTGGCCGA | GGTGTTTTCGGCTTCCTCTC |
| U6 | ACACCAAGCAGTCCGAAGAG | ACAAAATTTCTCACGCCGGT |
| GAPDH | CCATCTTCCAGGAGCGAGAT | TGCTGATGATCTTGAGGCTG |
MDM4, mouse double minute 4; miR, microRNA; MMP, matrix metalloproteinase.
Western blot analysis
Total protein in tissues and cells were extracted using RIPA lysis buffer (Beyotime Institute of Biotechnology). Protein quantification was performed using a BCA protein assay kit (Bio-Rad Laboratories, Inc.). Subsequently, proteins (40 µg) were separated via 10% SDS-PAGE and transferred to a PVDF membrane. The membrane was blocked in 5% non-fat milk at room temperature for 2 h. Subsequently, the membrane was incubated overnight at 4°C with the following primary antibodies: Anti-MDM4 (1:1,200; cat. no. ab154324; Abcam), anti-E-cadherin (1:800; cat. no. MAB1838; R&D Systems, Inc.), anti-N-cadherin (1:1,200; cat. no. ab18203; Abcam), anti-vimentin (1:700; cat. no. AF2105; R&D Systems, Inc.), anti-matrix metalloproteinase (MMP)2 (1:1,000; cat. no. MA1-772; Invitrogen; Thermo Fisher Scientific, Inc.), anti-MMP9 (1:800; cat. no. AF911; R&D Systems, Inc.) and anti-GAPDH (1:800; cat. no. AF5718; R&D Systems, Inc.). Following primary incubation, the membranes were incubated with corresponding horseradish peroxidase-conjugated secondary antibodies for 90 min at room temperature [rabbit anti-mouse IgG (1:5,000; cat. no. 58802; Cell Signaling Technology, Inc.); goat anti-mouse IgG (1:8,000; cat. no. 31430; Invitrogen; Thermo Fisher Scientific, Inc.); mouse anti-rabbit IgG (1:10,000; cat. no. 31464; Invitrogen; Thermo Fisher Scientific, Inc.)]. Finally, the protein was exposed using an ECL chemiluminescence kit [Yeasen Biotechnology (Shanghai) Co., Ltd.]. Protein expression was quantified using ImageJ software (version 5.0; Bio-Rad Laboratories, Inc.) with GAPDH as the loading control.
Bioinformatics prediction
According to the computational analysis performed using TargetScan software (version 7.2; www.targetscan.org/vert_72) (35), the 3′UTR of MDM4 contained a predicted binding site for miR-205.
Luciferase activity analysis
At 24 h post-transfection, the 293T cells were lysed using RIPA lysis buffer. The cell suspension was centrifuged at 500 × g for 5 min at room temperature, the supernatant was placed in 96-well plates and luciferase detection reagent was added (Promega Corporation). Luciferase activity was detected with a Nano-Glo® Dual-Luciferase Reporter Assay System (Promega Corporation). Firefly luciferase activity was normalized to Renilla luciferase activity.
Cell Counting Kit-8 (CCK-8) analysis
CCK-8 (Beyotime Institute of Biotechnology) was carried out to determine the cell viability of HCT116 cells following the manufacturer's protocol. Cells were transfected for 24 h, re-seeded into 96-well plates (6×103 cells/well) and incubated for 0, 24, 48 and 72 h at 37°C. Subsequently, CCK-8 reagent (10 µl) was added to the cells and incubated for 4 h at 37°C. The absorbance was then analyzed at 450 nm using a microplate reader (FilterMax F3/F5; Molecular Devices, LLC).
Transwell analysis
BD Matrigel (Qcbio Science & Technologes Co., Ltd.) was added into the upper chambers of Transwell inserts (96-well inserts; pore size, 0.4 µm; diameter, 4.26 mm) at room temperature for 25 min, and RPMI-1640 medium was added into the upper chambers. Subsequently, the Transwell inserts were placed in the culture plate. RPMI 1640 medium with 15% FBS was placed in the lower chamber to attract cells. HCT116 cell suspensions (4×105 cells/well) were cultured in RPMI-1640 medium in the upper chambers at 37°C for 24 h. The cells were fixed with 4% paraformaldehyde for 15 min at room temperature, stained with 0.05% crystal violet (Beijing Solarbio Science & Technology Co., Ltd.) for 20 min at room temperature, and washed with PBS three times. Finally, the cells were observed and photographed under a fluorescence microscope (magnification, ×200; MF53; Guangzhou Micro-shot Technology Co., Ltd.).
Wound healing assay
Cells were transfected with 50 nM PBS, miR-205 mimic, mimic NC, siNC or siMDM4 for 0 and 12 h. During the wound healing assay, cells were serum-starved (0.2% FBS). Following transfection, HCT116 cells were seeded in 6-well plates (2×104 cell/well) and cultured in an incubator for 24 h at 37°C. Following culturing, a 6-µm width scratch was created in the cells using a pipette tip, and the cells were washed by the medium 3 times. Cells were observed and photographed under an inverted microscope (magnification, ×200).
Statistical analysis
All experiments were conducted in triplicate. The data were shown as the mean ± standard deviation using SPSS software (version 20; IBM, Corp.). Associations between miR-205 expression and clinicopathological characteristics were analyzed using χ2 tests. One-way analysis of variance and Bonferroni's post hoc test were used to evaluate the differences among groups. The correlation between the miR-205 and MDM4 mRNA expression was analyzed by Pearson correlation analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Negative correlation between miR-205 and MDM4 expression in colon cancer tissue
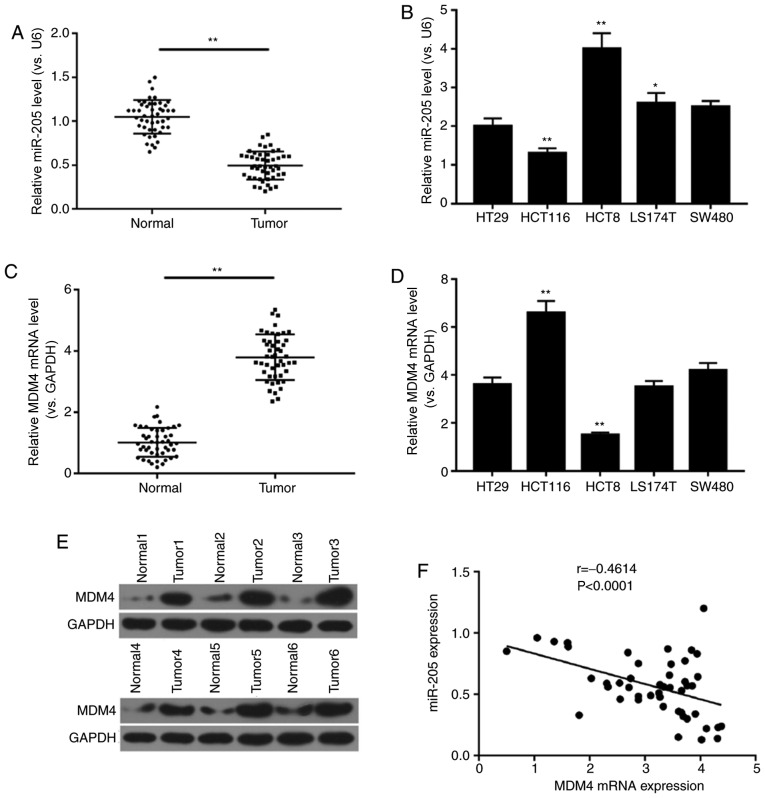
In order to determine the association between miR-205 and MDM4 in colon cancer tissues and cells, RT-qPCR and western blot analyses were performed. The results demonstrated that the expression level of miR-205 in normal tissue was higher compared with tumor tissue. In addition, miR-205 expression was determined in different colon tumor cells, and it was found that miR-205 had the lowest expression in HCT116 cells. However, mRNA and protein levels of MDM4 in normal tissue were lower compared with tumor tissue, and mRNA expression of MDM4 was the highest in HCT116 cells (P<0.01; Fig. 1A-E). Thus, HCT116 cells were selected for subsequent experiments. In addition, the data revealed a negative correlation between miR-205 and MDM4 mRNA expression in colon cancer tissue (r=−0.4614, P<0.0001; Fig. 1F). As Table I demonstrates, miR-205 expression was closely associated with pathological grade, stage and lymph node metastasis (P<0.05), and age and sex had no significant effect.
Figure 1.
Negative correlation between miR-205 and MDM4 expression in colon cancer tissue. Reverse transcription-quantitative PCR was performed to determine the mRNA levels of (A and B) miR-205 and (C and D) MDM4 in the colon cancer tissues, adjacent normal tissues and human colon cancer cell lines (HT29, HCT116, HCT8, LS174T and SW480). U6 and GAPDH were internal references. *P<0.05 and **P<0.01 vs. normal or HT29. (E) Western blot analysis was used to determine the protein expression of MDM4 in the colon cancer tissues and adjacent normal tissues. (F) Correlation between the miR-205 and MDM4 mRNA expression was quantified. miR, microRNA; MDM4, mouse double minute 4.
miR-205 silences MDM4 by binding with its 3′UTR
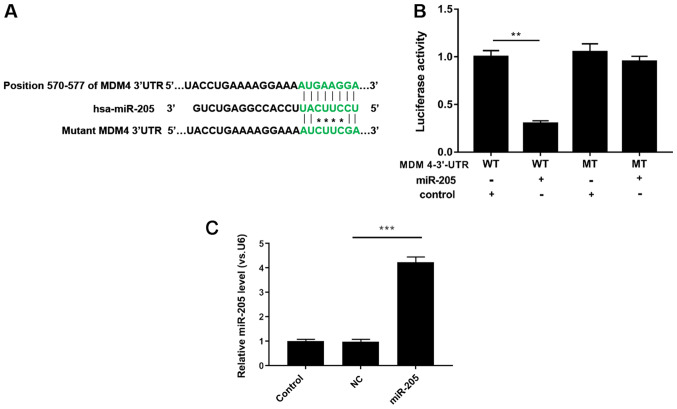
TargetScan prediction software was used to examine whether MDM4 is a potential target gene for miR-205, and the results indicated that there was a single eight-nucleotide complementary sequence at position 570–577 of the MDM4 3′UTR (Fig. 2A). Subsequently, luciferase activity assays demonstrated that when 293T cells were co-transfected with miR-205 and WT MDM4 3′UTR, the luciferase activity was significantly reduced. In addition, MT MDM4 3′UTR had no effect on luciferase activity (P<0.05; Fig. 2B). In addition, RT-qPCR analysis demonstrated that miR-205 was significantly upregulated in 293T cells transfected with miR-205 mimic compared with mimic NC (P<0.001; Fig. 2C). Thus, it was demonstrated that miR-205 inhibited MDM4 expression by interacting with its 3′UTR.
Figure 2.
miR-205 silences MDM4 by binding with its 3′UTR. (A) TargetScan prediction software was used to scan 3′UTR of MDM4 gene to detect the miR-205 combining site. (B) WT and MT MDM4 3′UTR were cloned to the downstream of firefly luciferase in the psiCHECK-2 vector. Then, plasmids with miR-205 mimic/NC were co-transfected into 293T cells. Luciferase activity was analyzed using a Nano-Glo Dual-Luciferase Reporter Assay System. (C) miR-205 expression was determined via reverse transcription-quantitative PCR. **P<0.01 and ***P<0.001. miR, microRNA; MDM4, mouse double minute 4; WT, human wild-type; MT, mutant; NC, mimic negative control; UTR, untranslated region.
miR-205 suppresses cell viability
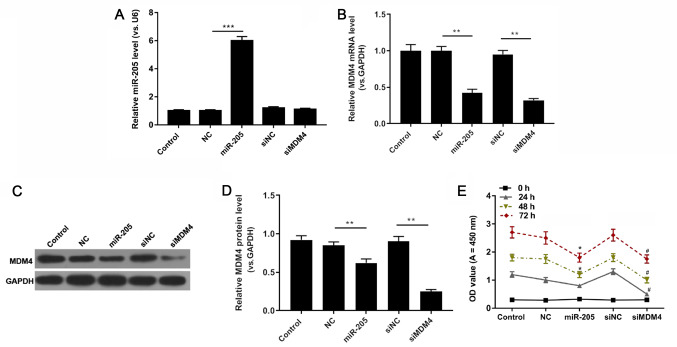
RT-qPCR, western blot and CCK-8 analyses were performed to evaluate the expression levels of miR-205 and MDM4, and the viability of HCT116 cells exposed to PBS, miR-205 mimic, mimic NC, siNC and siMDM4. miR-205 was markedly upregulated in the miR-205 mimic group compared with the mimic NC group (P<0.001; Fig. 3A). As RT-qPCR and western blot assay results revealed, siMDM4 significantly downregulated the mRNA and protein level of MDM4 compared with the siNC group. In comparison with the mimic NC group, miR-205 mimic transfection also significantly reduced the expression of MDM4 (P<0.01; Fig. 3B-D). In addition, the CCK-8 data demonstrated that compared with the mimic NC group, the OD value significantly decreased in the miR-205 mimic group at 48 and 72 h, and MDM4 silencing in cells reduced the value of OD compared with the siNC group at 24, 48 and 72 h (P<0.05; Fig. 3E).
Figure 3.
miR-205 suppresses cell viability. HCT116 cells were treated with PBS, miR-205, NC, siNC and siMDM4. (A) miR-205 and (B) MDM4 expression were determined using reverse transcription-quantitative PCR. **P<0.01 and ***P<0.001, as indicated. MDM4 protein expression was (C) determined by western blotting and (D) quantified. **P<0.01, as indicated. (E) Cell Counting Kit-8 was used to investigate cell viability. *P<0.05 vs. NC and #P<0.05 vs. siNC. miR, microRNA; control, PBS; miR-205, miR-205 mimic; NC, (mimic) negative control; si, small interfering RNA; MDM4, mouse double minute 4; OD, optical density.
miR-205 suppresses cell invasion and migration
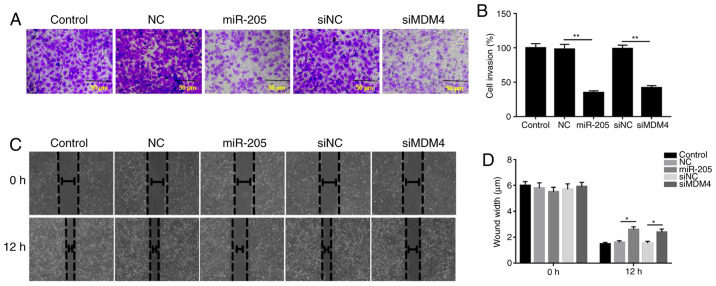
The invasive and migratory abilities of cells were explored via Transwell and wound healing assays. The data from Transwell assays demonstrated that the invasion of cells was inhibited by siMDM4 compared with siNC group; similarly, miR-205 overexpression significantly decreased cell invasion compared with the mimic NC group (P<0.05; Fig. 4A). In addition, the wound healing assay results revealed that the wound width was increased in the miR-205 mimic and siMDM4 groups compared with the mimic NC and siNC groups, respectively. The wound healing results demonstrated that miR-205 suppressed cell migration (P<0.05; Fig. 4B).
Figure 4.
miR-205 suppresses cell invasion and migration. (A and B) Cell invasion was assessed by Transwell analysis (magnification, ×200). (C and D) Cell migration was determined via a wound healing assay (magnification, ×100). *P<0.05, **P<0.01. miR, microRNA; control, PBS; miR-205, miR-205 mimic; NC, (mimic) negative control; si, small interfering RNA; MDM4, mouse double minute 4.
miR-205 mediates the expression of EMT-associated factors
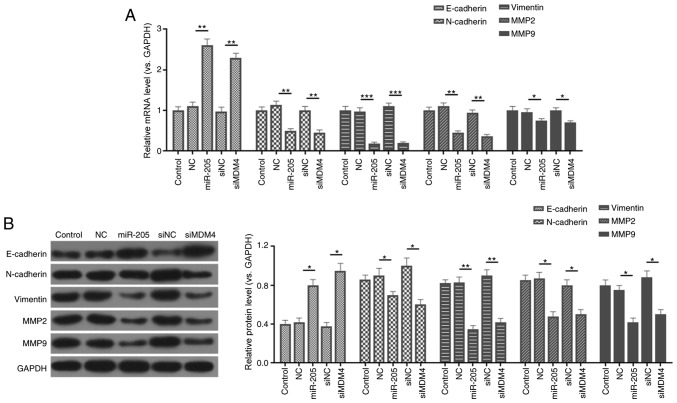
In order to further study the molecular mechanism of miR-205 inhibiting cell invasion and migration, the expression levels of E-cadherin, N-cadherin, vimentin, MMP-2 and MMP-9 were measured by RT-qPCR and western blotting. As RT-qPCR and western blot assays revealed, siMDM4 downregulated the mRNA and protein levels of MDM4, N-cadherin, vimentin, MMP2 and MMP9 compared with the siNC group. In comparison with the mimic NC group, miR-205 overexpression also significantly reduced the expression of N-cadherin, vimentin, MMP2 and MMP9. However, the expression levels of E-cadherin were upregulated in the siMDM4 and miR-205 mimic groups compared with the siNC and mimic NC groups (P<0.05; Fig. 5).
Figure 5.
miR-205 mediates the expressions of epithelial-mesenchymal transition-associated factors. (A) Reverse transcription-quantitative PCR was carried out to determine the mRNA levels of E-cadherin, N-cadherin, vimentin, MMP2, and MMP9. (B) Protein expressions of E-cadherin, N-cadherin, vimentin, MMP2, and MMP9 were detected by western blot assay. *P<0.05, **P<0.01 and ***P<0.001. miR, microRNA; control, PBS; miR-205, miR-205 mimic; NC, (mimic) negative control; si, small interfering RNA; MDM4, mouse double minute 4; MMP, matrix metalloproteinase.
Silencing MDM4 partially reverses the regulatory effects of miR-205 inhibition on invasion, migration and EMT
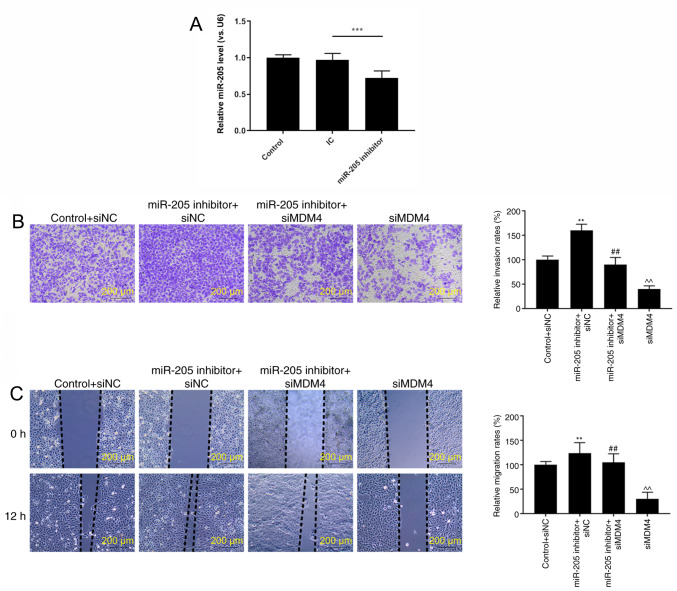
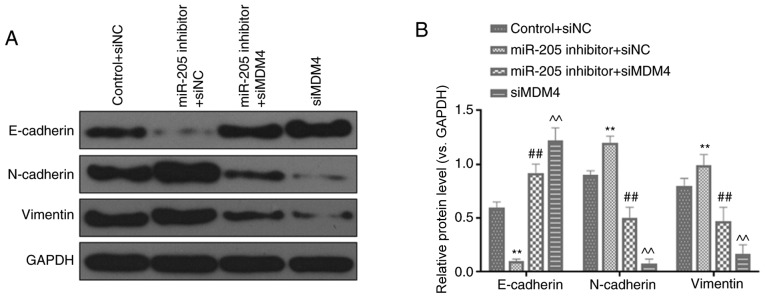
RT-qPCR analysis demonstrated that miR-205 was significantly downregulated in cells transfected with miR-205 inhibitor compared with the inhibitor control (P<0.001; Fig. 6A). In order to further verify the effects of miR-205 through MDM4, rescue experiments were performed. It was identified that miR-205 inhibitor significantly promoted cell invasion (P<0.01; Fig. 6B) and migration (P<0.01; Fig. 6C). In addition, silencing MDM4 could partially reversed the increase effects of miR-205 inhibition on invasion and migration (P<0.01). Furthermore, the expression of EMT-related proteins was also observed. miR-205 inhibition significantly inhibited E-cadherin expression, while increasing N-cadherin and vimentin levels. Silencing MDM4 could also partially reversed the regulatory effects of miR-205 inhibition on EMT (P<0.01; Fig. 7A and B).
Figure 6.
Silencing MDM4 partially reverses the enhancing effects of miR-205 inhibition on invasion and migration. (A) Downregulation of miR-205 following transfection with miR-205 inhibitor. ***P<0.001, as indicated. (B) Cell invasion was assessed by Transwell analysis. (C) Cell migration was determined via a wound healing assay. **P<0.01 vs. control + siNC; ##P<0.01 vs. miR-205 inhibitor + siNC; ^^P<0.01 vs. miR-205 inhibitor + siMDM4. MDM4, mouse double minute 4; miR, microRNA; control, PBS; NC, negative control; si, small interfering RNA.
Figure 7.
Silencing MDM4 partially reverses the regulatory effects of miR-205 inhibition on epithelial-mesenchymal transition-associated factors. (A and B) Protein expression of E-cadherin, N-cadherin and vimentin were detected via western blot analysis. **P<0.01 vs. control + siNC; ##P<0.01 vs. miR-205 inhibitor + siNC; ^^P<0.01 vs. miR-205 inhibitor + siMDM4. MDM4, mouse double minute 4; miR, microRNA; control, PBS; NC, negative control; si, small interfering RNA.
Discussion
MDM4, which mediates p53-independent activities, is abnormally expressed in various cancer cells and contributes to the development of cancer (36–38). Gilkes et al (39) noted that MDM4 is overexpressed in human colon tumors; consistent with these findings, the present results also revealed overexpressed MDM4 in human colon cancer tissues and cells. In addition, it has been found that inhibition of the expression of MDM4 can impede the proliferation and metastasis of tumor cells (40). To some extent, the present study demonstrated that MDM4 silencing in human colon tumor HCT116 cells significantly suppressed proliferation, migration and invasion.
A number of miRNAs can regulate cancer cell progress by targeting MDM4. For example, Jiang et al (41) reported that overexpressed miR-33a can suppress renal cell cancer growth by inhibiting the expression of MDM4. miR-766 can increase human colon cancer cell apoptosis through MDM4 (42). Previous studies suggested that miR-205 possibly has distinct functions in different cancers. It is reported that miR-205 is downregulated in colon, breast and prostate cancers (43–45), but upregulated in lung, bladder and ovarian cancers (46,47). The present study also demonstrated the downregulation of miR-205 in human colon tumor tissues and cells. miR-205 also can interact with the 3′UTR of certain genes, and then mediate the translation of genes and regulate tumor processes (48–50). Zhuang et al (51) demonstrated that miR-205 suppresses human pancreatic cancer progression by targeting runt-related transcription factor 2. A previous study indicated that miR-205 downregulates Prospero homeobox 1 by binding to its 3′UTR, thus further suppressing the viability and metastasis of human colon cancer cells (44). Thus, the present study investigated the relationship between miR-205 and MDM4 in colon cancer, and the data demonstrated that the expression levels of miR-205 and MDM4 were negatively correlated. In addition, the prediction results indicated that there was a single 8-nucleotide complementary sequence between hsa-miR-205 and the position 570–577 of MDM4 3′UTR.
miR-205 plays a vital role in the growth, migration and invasion of tumors (52,53). Previous studies have confirmed that miR-205 has anti-proliferation and anti-invasion effects on gastric and cervical tumors (54,55). As expected, the upregulation of miR-205 in HCT116 cells notably attenuated cell proliferation, migration and invasion by silencing the MDM4 gene. Furthermore, EMT-related proteins, including E-cadherin, N-cadherin, vimentin, MMP2 and MMP9, were detected. E-cadherin is an important adhesion molecule for maintaining epithelial cell characteristics. N-cadherin, which plays a key role in promoting cell movement, is considered as one of the characteristic molecular markers of mesenchymal cells (56,57). A recent study observed that the overexpression of miR-205 in anaplastic thyroid carcinoma predominantly blocks the process of EMT by targeting zinc finger E-box-binding homeobox 1 gene, which upregulates E-cadherin expression, and downregulates N-cadherin, vimentin, MMP2 and MMP9 expression levels (26). Similarly, the results of the present study demonstrated that miR-205 targeted the MDM4 gene to suppress EMT, followed by downregulation of N-cadherin, vimentin, MMP2 and MMP9, and upregulation of E-cadherin.
In conclusion, the present study characterized the miR-205-MDM4 mechanism in human colon cancer. It found that miR-205 and MDM4 expressions are negatively correlated in human colon cancer. In addition, miR-205 significantly suppressed the proliferation, migration, invasion and EMT of human colon cancer cells by silencing MDM4 gene. Thus, miR-205 could be employed in the treatment of human colon cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YF made substantial contributions to the study conception and design. KW performed data acquisition, data analysis and interpretation. YF drafted the manuscript and critically revised it for important intellectual content. Both authors gave final approval to the published version of the study and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of the work are appropriately investigated and resolved. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
The current study was reviewed and approved by the Ethics Committee of Beijing Jishuitan Hospital (approval no. J20110104015). All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and the Declaration of Helsinki. All patients signed informed consent and agreed that their tissues would be used for clinical research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 4.Muto Y, Chochi K, Morita D, Oka A, Rikiyama T. An elderly patient with metastatic colon cancer achieved long-term survival following single-agent chemotherapy with S-1. Gan To Kagaku Ryoho. 2018;45:55–57. (In Japanese) [PubMed] [Google Scholar]
- 5.Sun LY. Essential and interpretation of Japanese society for cancer of the colon and rectum (JSCCR) guidelines 2019 for the treatment of colorectal cancer. Zhonghua Wei Chang Wai Ke Za Zhi. 2019;22:1088–1094. doi: 10.3760/cma.j.issn.1671-0274.2019.11.016. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 6.Taieb J, Shi Q, Pederson L, Alberts S, Wolmark N, Van Cutsem E, de Gramont A, Kerr R, Grothey A, Lonardi S, et al. Prognosis of microsatellite instability and/or mismatch repair deficiency stage III colon cancer patients after disease recurrence following adjuvant treatment: Results of an ACCENT pooled analysis of seven studies. Ann Oncol. 2019;30:1466–1471. doi: 10.1093/annonc/mdz208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American joint committee on cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 8.Fang WG, Tian XX. Identification of a new pro-invasion factor in tumor microenvironment: Progress in function and mechanism of extracellular ATP. Beijing Da Xue Xue Bao Yi Xue Ban. 2017;49:188–195. (In Chinese) [PubMed] [Google Scholar]
- 9.Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20:1531–1541. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: A clinical and in vitro investigation. Oncol Rep. 2014;31:358–364. doi: 10.3892/or.2013.2850. [DOI] [PubMed] [Google Scholar]
- 11.Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: Yes, no, maybe? Curr Opin Cell Biol. 2016;43:7–13. doi: 10.1016/j.ceb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Lou Y, Diao L, Cuentas ER, Denning WL, Chen L, Fan YH, Byers LA, Wang J, Papadimitrakopoulou VA, Behrens C, et al. Epithelial-mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin Cancer Res. 2016;22:3630–3642. doi: 10.1158/1078-0432.CCR-15-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan TZ, Miow QH, Miki Y, Noda T, Mori S, Huang RY, Thiery JP. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med. 2014;6:1279–1293. doi: 10.15252/emmm.201404208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu BB, Wang H, Chi YF, Wang YM, Yao XM, Liu S, Qiu H, Fang J, Yin PH, Zhang XM, Peng W. Protective effects of probucol on Ox-LDL-induced epithelial-mesenchymal transition in human renal proximal tubular epithelial cells via LOX1/ROS/MAPK signaling. Mol Med Rep. 2018;17:1289–1296. doi: 10.3892/mmr.2017.7935. [DOI] [PubMed] [Google Scholar]
- 15.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 16.Ma S, Deng X, Yang Y, Zhang Q, Zhou T, Liu Z. The lncRNA LINC00675 regulates cell proliferation, migration, and invasion by affecting Wnt/β-catenin signaling in cervical cancer. Biomed Pharmacother. 2018;108:1686–1693. doi: 10.1016/j.biopha.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 18.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 19.Stojkovic S, Jurisic M, Kopp CW, Koppensteiner R, Huber K, Wojta J, Gremmel T. Circulating microRNAs identify patients at increased risk of in-stent restenosis after peripheral angioplasty with stent implantation. Atherosclerosis. 2018;269:197–203. doi: 10.1016/j.atherosclerosis.2018.01.020. [DOI] [PubMed] [Google Scholar]
- 20.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 21.Zhang G, Hou X, Li Y, Zhao M. MiR-205 inhibits cell apoptosis by targeting phosphatase and tensin homolog deleted on chromosome ten in endometrial cancer Ishikawa cells. BMC Cancer. 2014;14:440. doi: 10.1186/1471-2407-14-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lebanony D, Benjamin H, Gilad S, Ezagouri M, Dov A, Ashkenazi K, Gefen N, Izraeli S, Rechavi G, Pass H, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27:2030–2037. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 23.Qin AY, Zhang XW, Liu L, Yu JP, Li H, Wang SZ, Ren XB, Cao S. MiR-205 in cancer: An angel or a devil? Eur J Cell Biol. 2013;92:54–60. doi: 10.1016/j.ejcb.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Grelet S, Link LA, Howley B, Obellianne C, Palanisamy V, Gangaraju VK, Diehl JA, Howe PH. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol. 2017;19:1105–1115. doi: 10.1038/ncb3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tellez CS, Juri DE, Do K, Bernauer AM, Thomas CL, Damiani LA, Tessema M, Leng S, Belinsky SA. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer Res. 2011;71:3087–3097. doi: 10.1158/0008-5472.CAN-10-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vosgha H, Ariana A, Smith RA, Lam AK. miR-205 targets angiogenesis and EMT concurrently in anaplastic thyroid carcinoma. Endocr Relat Cancer. 2018;25:323–337. doi: 10.1530/ERC-17-0497. [DOI] [PubMed] [Google Scholar]
- 27.Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, Hateboer G, van der Eb AJ, Jochemsen AG. MDMX: A novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–5357. doi: 10.1002/j.1460-2075.1996.tb00919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golestanian S, Sharifi A, Popowicz GM, Azizian H, Foroumadi A, Szwagierczak A, Holak TA, Amanlou M. Discovery of novel dual inhibitors against Mdm2 and Mdmx proteins by in silico approaches and binding assay. Life Sci. 2016;145:240–246. doi: 10.1016/j.lfs.2015.12.047. [DOI] [PubMed] [Google Scholar]
- 29.Popowicz GM, Czarna A, Holak TA. Structure of the human Mdmx protein bound to the p53 tumor suppressor transactivation domain. Cell Cycle. 2008;7:2441–2443. doi: 10.4161/cc.6365. [DOI] [PubMed] [Google Scholar]
- 30.Gansmo LB, Romundstad P, Birkeland E, Hveem K, Vatten L, Knappskog S, Lonning PE. MDM4 SNP34091 (rs4245739) and its effect on breast-, colon-, lung-, and prostate cancer risk. Cancer Med. 2015;4:1901–1907. doi: 10.1002/cam4.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu L, Zhang H, Bergholz J, Sun S, Xiao ZX. MDM2/MDMX: Master negative regulators for p53 and RB. Mol Cell Oncol. 2016;3:e1106635. doi: 10.1080/23723556.2015.1106635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pedram N, Pouladi N, Feizi MA, Montazeri V, Sakhinia E, Estiar MA. Analysis of the association between MDM4 rs4245739 single nucleotide polymorphism and breast cancer susceptibility. Clin Lab. 2016;62:1303–1308. doi: 10.7754/Clin.Lab.2016.151128. [DOI] [PubMed] [Google Scholar]
- 33.Siebring-van Olst E, Blijlevens M, de Menezes RX, van der Meulen-Muileman IH, Smit EF, van Beusechem VW. A genome-wide siRNA screen for regulators of tumor suppressor p53 activity in human non-small cell lung cancer cells identifies components of the RNA splicing machinery as targets for anticancer treatment. Mol Oncol. 2017;11:534–551. doi: 10.1002/1878-0261.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chee SMQ, Wongsantichon J, Siau J, Thean D, Ferrer F, Robinson RC, Lane DP, Brown CJ, Ghadessy FJ. Structure-activity studies of Mdm2/Mdm4-binding stapled peptides comprising non-natural amino acids. PLoS One. 2017;12:e0189379. doi: 10.1371/journal.pone.0189379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu Z, Zhang H, Tao Y, Li X, Li G. MDM4 genetic variants predict HPV16-positive tumors of patients with squamous cell carcinoma of the oropharynx. Oncotarget. 2017;8:86710–86717. doi: 10.18632/oncotarget.21414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ten Kate FJ, Suzuki L, Dorssers LC, Dinjens WN, Jones DT, Nieboer D, Doukas M, Van Lanschot JJ, Wijnhoven BP, Looijenga LH, Biermann K. Pattern of p53 protein expression is predictive for survival in chemoradiotherapy-naive esophageal adenocarcinoma. Oncotarget. 2017;8:104123–104135. doi: 10.18632/oncotarget.22021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilkes DM, Pan Y, Coppola D, Yeatman T, Reuther GW, Chen J. Regulation of MDMX expression by mitogenic signaling. Mol Cell Biol. 2008;28:1999–2010. doi: 10.1128/MCB.01633-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCubrey JA, Lertpiriyapong K, Fitzgerald TL, Martelli AM, Cocco L, Rakus D, Gizak A, Libra M, Cervello M, Montalto G, et al. Roles of TP53 in determining therapeutic sensitivity, growth, cellular senescence, invasion and metastasis. Adv Biol Regul. 2017;63:32–48. doi: 10.1016/j.jbior.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Jiang K, Sun F, Zhu J, Luo G, Ban Y, Zhang P. miR-33a inhibits cell growth in renal cancer by downregulation of MDM4 expression. Mol Genet Genomic Med. 2019;7:e833. doi: 10.1002/mgg3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen W, Cai G, Liao Z, Lin K, Li G, Li Y. miRNA-766 induces apoptosis of human colon cancer cells through the p53/Bax signaling pathway by MDM4. Exp Ther Med. 2019;17:4100–4108. doi: 10.3892/etm.2019.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gulei D, Magdo L, Jurj A, Raduly L, Cojocneanu-Petric R, Moldovan A, Moldovan C, Florea A, Pasca S, Pop LA, et al. The silent healer: miR-205-5p up-regulation inhibits epithelial to mesenchymal transition in colon cancer cells by indirectly up-regulating E-cadherin expression. Cell Death Dis. 2018;9:66. doi: 10.1038/s41419-017-0102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen-Vu T, Wang J, Mesmar F, Mukhopadhyay S, Saxena A, McCollum CW, Gustafsson JÅ, Bondesson M, Williams C. Estrogen receptor beta reduces colon cancer metastasis through a novel miR-205-PROX1 mechanism. Oncotarget. 2016;7:42159–42171. doi: 10.18632/oncotarget.9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamada Y, Nishikawa R, Kato M, Okato A, Arai T, Kojima S, Yamazaki K, Naya Y, Ichikawa T, Seki N. Regulation of HMGB3 by antitumor miR-205-5p inhibits cancer cell aggressiveness and is involved in prostate cancer pathogenesis. J Hum Genet. 2018;63:195–205. doi: 10.1038/s10038-017-0371-1. [DOI] [PubMed] [Google Scholar]
- 46.Braicu OL, Budisan L, Buiga R, Jurj A, Achimas-Cadariu P, Pop LA, Braicu C, Irimie A, Berindan-Neagoe I. miRNA expression profiling in formalin-fixed paraffin-embedded endometriosis and ovarian cancer samples. Onco Targets Ther. 2017;10:4225–4238. doi: 10.2147/OTT.S137107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li JH, Sun SS, Li N, Lv P, Xie SY, Wang PY. MiR-205 as a promising biomarker in the diagnosis and prognosis of lung cancer. Oncotarget. 2017;8:91938–91949. doi: 10.18632/oncotarget.20262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei J, Zhang L, Li J, Zhu S, Tai M, Mason CW, Chapman JA, Reynolds EA, Weiner CP, Zhou HH. MicroRNA-205 promotes cell invasion by repressing TCF21 in human ovarian cancer. J Ovarian Res. 2017;10:33. doi: 10.1186/s13048-017-0328-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu CG, Yang MF, Fan JX, Wang W. MiR-30a and miR-205 are downregulated in hypoxia and modulate radiosensitivity of prostate cancer cells by inhibiting autophagy via TP53INP1. Eur Rev Med Pharmacol Sci. 2016;20:1501–1508. [PubMed] [Google Scholar]
- 50.Yang G, Zhang P, Lv A, Liu Y, Wang G. MiR-205 functions as a tumor suppressor via targeting TGF-α in osteosarcoma. Exp Mol Pathol. 2016;100:160–166. doi: 10.1016/j.yexmp.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 51.Zhuang L, Guo J, Yao Y, Li Z. miR-205 targets runt-related transcription factor 2 to inhibit human pancreatic cancer progression. Oncol Lett. 2019;17:843–848. doi: 10.3892/ol.2018.9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S, Jin L, Nie S, Han L, Lu N, Zhou Y. MiR-205 inhibits growth and invasion of neuroblastoma by targeting cAMP responsive element binding protein 1. Oncol Res. 2018;26:445–455. doi: 10.3727/096504017X14974834436195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu C, Li M, Zhang L, Bi Y, Wang P, Li J, Jiang X. MicroRNA-205 suppresses the invasion and epithelial-mesenchymal transition of human gastric cancer cells. Mol Med Rep. 2016;13:4767–4773. doi: 10.3892/mmr.2016.5849. [DOI] [PubMed] [Google Scholar]
- 54.Pang H, Yue X. MiR-205 serves as a prognostic factor and suppresses proliferation and invasion by targeting insulin-like growth factor receptor 1 in human cervical cancer. Tumour Biol. 2017;39:1010428317701308. doi: 10.1177/1010428317701308. [DOI] [PubMed] [Google Scholar]
- 55.Yin WZ, Li F, Zhang L, Ren XP, Zhang N, Wen JF. Down-regulation of microRNA-205 promotes gastric cancer cell proliferation. Eur Rev Med Pharmacol Sci. 2014;18:1027–1032. [PubMed] [Google Scholar]
- 56.Chen T, You Y, Jiang H, Wang ZZ. Epithelial-mesenchymal transition (EMT): A biological process in the development, stem cell differentiation, and tumorigenesis. J Cell Physiol. 2017;232:3261–3272. doi: 10.1002/jcp.25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodriguez-Monterrosas C, Diaz-Aragon R, Leal-Orta E, Cortes-Reynosa P, Perez Salazar E. Insulin induces an EMT-like process in mammary epithelial cells MCF10A. J Cell Biochem. 2018;119:4061–4071. doi: 10.1002/jcb.26582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.