Abstract
Increasing evidence suggests that long non-coding RNAs (lncRNAs) serve a crucial role in predicting prognosis for hepatocellular carcinoma (HCC). However, prognostic performance may not be the same for alcohol-related HCC. The aim of the present study was to screen prognosis-associated lncRNAs and construct a risk scoring system for alcohol-related HCC. The expression profiles of lncRNAs in 113 patients with alcohol-related HCC and 224 with non-alcohol-related HCC were obtained from The Cancer Genome Atlas (TCGA) database and screened for differentially expressed lncRNAs. Cox regression analysis was performed to identify prognosis-associated lncRNAs and select the optimal lncRNA model. A risk scoring system was established to calculate the risk score for each patient. The prognostic ability of this system was tested. Functional enrichment analysis was performed for genes that were highly associated with lncRNA expression. A total of 102 differentially expressed lncRNAs were identified between alcohol-related and non-alcohol-related HCC. Four lncRNAs (AC012640.1, AC013451.2, AC062004.1 and LINC02334) were used to construct the risk assessment model to predict overall survival (OS), and five lncRNAs (ERVH48-1, LINC02043, LINC01605, AC062004.1 and AL139385) were used to predict recurrence-free survival (RFS). Patients were assigned to high- or low-risk groups according to the risk score. OS in the high-risk group was significantly shorter than that of the low-risk group. The area under the receiver operating characteristic (ROC) curve of risk scoring systems was >0.7. The risk score was an independent prognostic factor for alcohol-related HCC. Functional enrichment analysis demonstrated that lncRNA-related genes found in this system were mainly involved in chemical carcinogenesis, drug metabolism, and the cell cycle. In conclusion, this study developed and validated a prognostic scoring system for alcohol-related HCC based on lncRNAs.
Keywords: hepatocellular carcinoma, long noncoding RNA, risk scoring system, survival
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common malignancy and the fourth leading cause of cancer-related death globally, and worldwide incidence increases by 3–4% per year (1,2). Hepatitis B virus (HBV) or hepatitis C virus (HCV) infection, alcohol consumption, diabetes, nonalcoholic fatty liver disease and smoking are known to be major risk factors for HCC (3,4). HCC is highly heterogeneous, and the pathogenesis is extremely complex. The progression of HCC involves multiple processes such as mutation and signaling pathway maladjustment, reflecting the interaction among multiple genes (5,6). Despite the development of various drugs and breakthroughs in diagnosis, the prognosis of HCC remains poor, with a 5-year survival rate of only 5% for patients with advanced HCC (7). Timely and effective assessment of prognosis is of great significance to guide the treatment. At present, there are no biomarkers that effectively predict the survival of patients with HCC, and thus, finding effective prognostic biomarkers for patients with HCC is crucial.
Long non-coding RNAs (lncRNAs), non-coding transcripts >200 nucleotides, serve important cellular functions such as in chromatin modification as well as transcriptional and post-transcriptional regulation (8,9). Increasing evidence demonstrates that aberrant expression of lncRNAs is associated with the occurrence and development of various human diseases, especially cancer (10–12). For example, the overexpression of lncRNA HOX Transcript Antisense Intergenic RNA (HOTAIR) was demonstrated to predict tumor recurrence after liver transplantation (13). There was also a significant association between HOTAIR expression and tumor progression in patients with HCC (14–16). Increased biallelic expression of H19 and IGF2 may participate in an epigenetic mechanism of HCC development and progression (17). The lncRNA GPC3-AS1 promotes HCC progression by epigenetic GPC3 activation (18). However, the role of lncRNAs in alcohol-related HCC remains unclear.
Alcohol is a dose-related risk factor known to be associated with more than 200 diseases, including HCC (19,20). Heavy drinkers are at 3- to 10-fold higher risk of hepatocellular carcinoma than non-drinkers (2). In addition, the overall survival rate is lower for patients with alcohol-related HCC than for those with non-alcohol-related HCC, suggesting that there may be a link between alcohol and prognosis (21). Thus, the present study aimed to examine whether lncRNAs are differentially expressed in the presence of alcohol consumption that may be used as prognostic markers in HCC, and whether these differences might influence the risk of HCC recurrence or death. Using data from The Cancer Genome Atlas (TCGA), the present study developed a risk-scoring system based on lncRNA levels that may be valuable for predicting the prognosis of patients with alcohol-related HCC.
Materials and methods
Patient selection and data collection
Profiles of lncRNA and mRNA expression in HCC patients were downloaded from the University of California Santa Cruz (UCSC) Xena server (https://xenabrowser.net/datapages/). Corresponding clinical information was obtained from TCGA (version 09–14–2017 for HCC). The patients in this dataset had histologically confirmed HCC. Patient data included a complete lncRNA expression profile, alcohol consumption status and survival data for determining OS and RFS. A total of 113 patients with HCC and alcohol consumption, and 224 without alcohol consumption were selected (Table I). This study complied with TCGA publication guidelines and policies (http://cancergenome.nih.gov/publications/publicationguidelines). No ethics approval was required for this study since data were obtained from TCGA.
Table I.
Clinicopathological characteristics of 337 patients with alcohol- or non-alcohol-related hepatocellular carcinoma.
| Patients (n=337) | ||
|---|---|---|
| Clinicopathological characteristics | n | % |
| Age, years | ||
| ≤60 | 162 | 48.07 |
| >60 | 175 | 51.93 |
| BMI | ||
| <25 | 161 | 47.77 |
| ≥25 | 150 | 44.51 |
| Not reported | 26 | 7.72 |
| Race | ||
| Non-Asian | 186 | 55.19 |
| Asian | 141 | 41.84 |
| Not reported | 10 | 2.97 |
| Sex | ||
| Female | 107 | 31.75 |
| Male | 230 | 68.25 |
| Hepatitisa | ||
| No | 189 | 56.08 |
| Yes | 148 | 43.92 |
| Cirrhosis | ||
| Non-cirrhosis | 124 | 36.80 |
| Cirrhosis | 74 | 21.96 |
| Not reported | 139 | 41.25 |
| Alcohol consumption | ||
| No | 224 | 66.47 |
| Yes | 113 | 33.53 |
| Histologic grade | ||
| G1-2 | 209 | 62.02 |
| G3-4 | 123 | 36.50 |
| Not reported | 5 | 1.418 |
| New tumor event | ||
| No | 167 | 49.55 |
| Yes | 154 | 45.70 |
| Not reported | 16 | 4.75 |
| Pathologic stage | ||
| Stage I+II | 231 | 68.55 |
| Stage III+IV | 82 | 24.33 |
| Not reported | 24 | 7.12 |
| Cancer status | ||
| Tumor free | 181 | 53.71 |
| With tumor | 142 | 42.14 |
| Not reported | 14 | 4.15 |
| Family cancer history | ||
| No | 188 | 55.79 |
| Yes | 108 | 32.05 |
| Not reported | 41 | 12.17 |
| Residual tumor | ||
| R0 | 301 | 89.32 |
| Non-R0 | 29 | 8.61 |
| Not reported | 7 | 2.08 |
| Vascular invasion | ||
| Negative | 188 | 55.79 |
| Positive | 95 | 28.19 |
| Not reported | 54 | 16.02 |
Hepatitis B or C. BMI, Body mass index; AFP, alpha fetoprotein.
Identification of lncRNAs differentially expressed between alcohol-related or non-alcohol-related HCC
After eliminating lncRNAs showing zero expression in >50% of all patients, the edgeR package in R (https://www.r-project.org/) was used to identify lncRNAs differentially expressed between patients with or without alcohol consumption (22). Differential expression was defined as log2fold change (log2FC) >1 and false discovery rate (FDR) <0.05. Differentially expressed lncRNAs were then presented in cluster heat maps and volcano maps generated using the packages gplots and heatmap in R.
Construction of lncRNA-based risk scoring systems
Standardized expression of lncRNAs in multiple tissues of the same patient were averaged. Univariate Cox analysis was then performed to screen differentially expressed lncRNAs to determine their significant relationship with OS or RFS, with the threshold set at P=0.05. Selected lncRNAs were included in subsequent multivariate Cox regression by the backward stepwise method in order to identify the best model. The expression level of each lncRNA was multiplied by the corresponding regression coefficient β and linearly combined to generate a risk scoring system:
Risk score=(β1 × expression level of lncRNA1) + (β2 × expression level of lncRNA2) + (β3 × expression level of lncRNA3) + (βn × expression level of lncRNAn).
This formula was used to calculate a risk score for each patient. The prognosis prediction performance of this risk score was assessed using time-dependent receiver operating characteristic (ROC) curves within three years (23). Patients with HCC were divided into a high- or a low-risk group according to the cut-off value of the median risk score, as demonstrated in non-cluster heat maps. Kaplan-Meier survival curves were generated and compared between high- and low-risk groups. All these analyses were conducted using R/Bioconductor (version 3.4.4, http://www.r-project.org/).
Prognostic performance of the risk scoring systems
To validate the prognostic performance of the risk scoring systems, univariate and multivariate Cox regression analyses were performed to determine whether the risk score was an independent factor for survival. This regression was performed in SPSS 16.0 (SPSS, Inc.), and a significance threshold of P=0.05 (two-sided).
Co-expression and functional enrichment analysis of related mRNAs
Pearson correlation was performed to screen for relationships between lncRNAs in the risk scoring systems and mRNAs based on data of 337 patients with HCC. Relationships were considered significant if the mRNA expression co-varied with that of lncRNAs with a two-sided absolute value of the Pearson correlation coefficient >0.30 and a z-test P<0.01. To obtain a deeper understanding of these mRNAs, enrichment analyses were performed using the Genomes pathway in the Kyoto Encyclopedia of Genes and Genomes by the package clusterProfiler in R (24). P<0.05 was considered to indicate a statistically significant difference.
Results
lncRNAs are differentially expressed in alcohol-related HCC or non-alcohol-related HCC
A total of 102 differentially expressed lncRNAs were identified, 47 (46.08%) of which were upregulated and 55 (53.92%) downregulated (Figs. 1 and 2); the first 20 up- and downregulated lncRNAs, together with the corresponding values for log2FC, P and FDR are demonstrated in Table II.
Figure 1.
Volcano map of the differentially expressed lncRNAs between alcohol-related and non-alcohol-related HCC. Red spots represent upregulated genes, and green spots represent downregulated genes. lncRNA, long non-coding RNA; HCC, hepatocellular carcinoma; FC, fold change; FDR, false discovery rate.
Figure 2.
Heat map based on the differentially expressed lncRNA between alcohol-related and non-alcohol-related HCC. lncRNA, long non-coding RNA; HCC, hepatocellular carcinoma.
Table II.
Differentially expressed lncRNAs in patients with alcohol-related or non-alcohol-related hepatocellular carcinoma.
| Top 20 upregulated lncRNAs | Top 20 downregulated lncRNAs | ||||||
|---|---|---|---|---|---|---|---|
| lncRNA | logFC | P-value | FDR | lncRNA | logFC | P-value | FDR |
| AC090921.1 | 5.26 | 1.35×10−50 | 7.79×10−47 | ERVH48-1 | −3.77 | 1.25×10−21 | 1.45×10−18 |
| AL451074.6 | 3.15 | 2.17×10−35 | 6.29×10−32 | AP000439.3 | −4.73 | 3.49×10−17 | 2.89×10−14 |
| AC007938.3 | 1.68 | 3.78×10−22 | 7.29×10−19 | AC139749.1 | −3.34 | 5.15×10−17 | 3.72×10−14 |
| AC105446.1 | 2.58 | 6.40×10−22 | 9.26×10−19 | LINC00473 | −4.00 | 1.51×10−16 | 9.73×10−14 |
| AC012640.1 | 1.94 | 2.45×10−18 | 2.36×10−15 | LINC01480 | −2.26 | 4.15×10−12 | 2.00×10−9 |
| AC025627.1 | 1.26 | 6.31×10−13 | 3.65×10−10 | AP000757.1 | −1.61 | 2.31×10−10 | 7.86×10−8 |
| AL445483.1 | 1.73 | 1.36×10−12 | 7.14×10−10 | UCA1 | −2.54 | 7.54×10−9 | 1.90×10−6 |
| MYOSLID | 1.59 | 1.35×10−11 | 6.03×10−9 | AC064807.2 | −1.85 | 9.35×10−9 | 2.16×10−6 |
| EMX2OS | 1.63 | 5.63×10−11 | 2.33×10−8 | AL359853.1 | −1.98 | 1.29×10−8 | 2.82×10−6 |
| AC156455.1 | 1.18 | 1.15×10−10 | 4.17×10−8 | PLCE1-AS1 | −2.03 | 2.86×10−8 | 5.71×10−6 |
| AP003354.2 | 1.27 | 6.76×10−10 | 2.17×10−7 | AC002398.2 | −2.03 | 9.19×10−8 | 1.77×10−5 |
| AC007220.1 | 1.14 | 1.70×10−9 | 5.18×10−7 | AC079305.1 | −1.12 | 2.12×10−7 | 3.71×10−5 |
| U62317.1 | 1.63 | 1.93×10−9 | 5.58×10−7 | AC007099.1 | −2.10 | 4.35×10−7 | 6.80×10−5 |
| LINC02043 | 1.22 | 2.25×10−9 | 6.19×10−7 | LINC01229 | −1.32 | 7.07×10−7 | 1.02×10−4 |
| AC068473.3 | 1.41 | 3.83×10−9 | 1.01×10−6 | AC098869.2 | −1.26 | 7.52×10−7 | 1.06×10−4 |
| AC069431.1 | 1.42 | 8.70×10−9 | 2×10−6 | LINC00624 | −1.28 | 1.16×10−6 | 1.51×10−4 |
| LINC01615 | 1.40 | 1.32×10−8 | 2.82×10−6 | AC005674.1 | −1.06 | 1.83×10−6 | 2.11×10−4 |
| AC079779.2 | 1.50 | 2.35×10−8 | 4.86×10−6 | AL161772.1 | −1.24 | 2.26×10−6 | 2.52×10−4 |
| AC062004.1 | 1.40 | 1.11×10−7 | 2.07×10−5 | BX640514.2 | −1.79 | 2.93×10−6 | 3.08×10−4 |
| AL391056.1 | 1.574 | 1.21×10−7 | 2.18×10−5 | AC023154.1 | −1.44 | 3.44×10−6 | 3.46×10−4 |
lncRNAs, long noncoding RNAs; logFC, log2fold change; FDR, false discovery rate.
Risk-scoring system based on lncRNA expression and OS
Univariate Cox analysis identified six lncRNAs that were significantly associated with OS: AC012640.1, AC013451.2, AC062004.1, LINC02334, AC090921.1 and LINC01605. The first four were independent prognostic indicators of OS based on multivariate Cox regression (Table III). The resulting risk scoring system was: Risk score=(0.186 × AC012640.1) + (0.363 × AC013451.2) + (−0.243 × AC062004.1) + (−0.275 × LINC02334).
Table III.
Four lncRNAs were correlated with overall survival in the best statistical model.
| lncRNA | β | HR | z | P-value |
|---|---|---|---|---|
| AC012640.1 | 0.186 | 1.205 | 1.71 | 0.088 |
| AC062004.1 | 0.243 | 0.784 | −1.98 | 0.047a |
| LINC02334 | −0.275 | 0.759 | −2.23 | 0.026a |
| AC013451.2 | 0.363 | 1.437 | 2.42 | 0.016a |
P<0.05. lncRNAs, long noncoding RNAs; HR, Hazard ratio.
In this scoring system, increased expression of AC012640.1 and AC013451.2 predicted worse OS (β>0), whereas increased expression of AC062004.1 and LINC02334 predicted better OS (β<0).
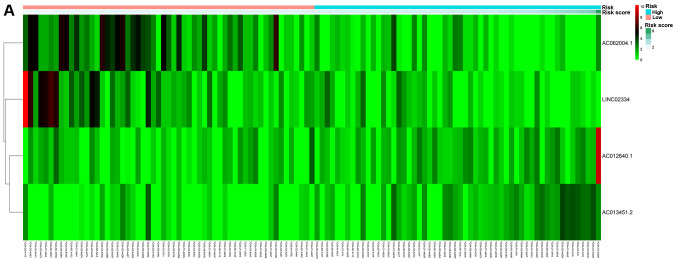
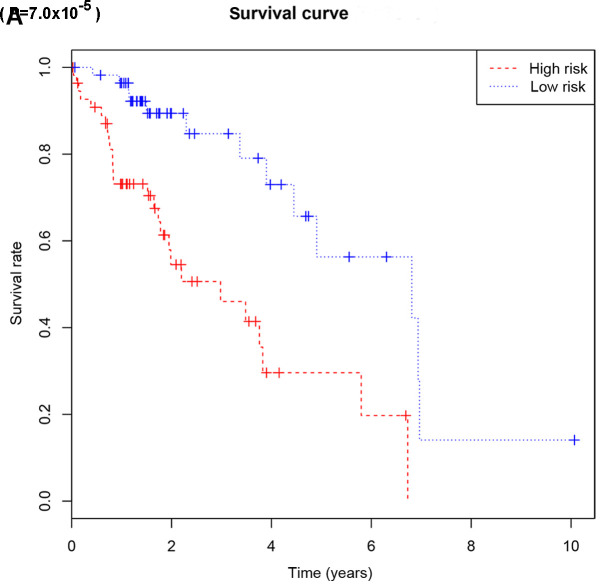
Based on their risk scores, patients were classified as at low or high risk of poor OS using the median risk score as cutoff (Fig. 3A). Kaplan-Meier curves demonstrated that patients with high risk had significantly lower OS at 3 and 5 years compared with that of patients with low risk (Fig. 4A). The area under the ROC curve (AUC) for the risk scoring system was 0.721 (Fig. 5A).
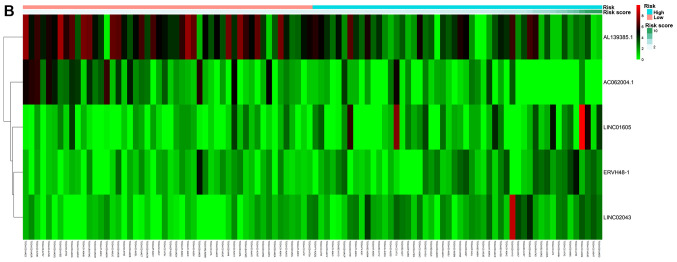
Figure 3.
Non-cluster risk heat map of long non-coding RNA-based risk scoring system for overall survival (A) or recurrence-free survival (B). The risk value gradually increases from left to right.
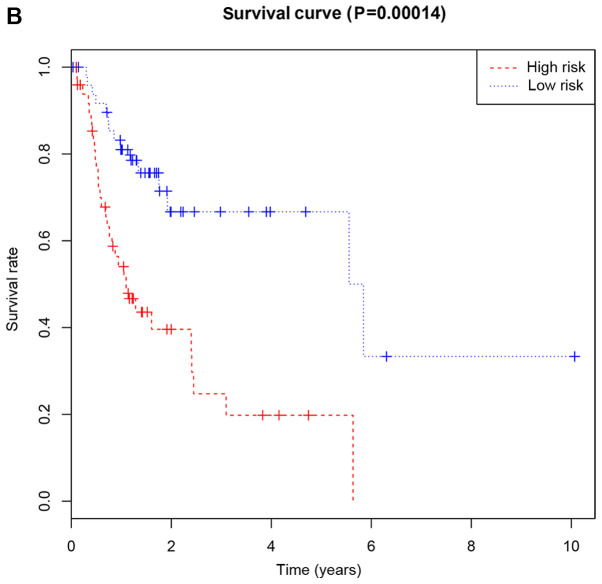
Figure 4.
Kaplan-Meier survival curves of overall survival (A) or recurrence-free survival (B) according to the risk cutoff point. Horizontal axis represents the survival period and the vertical axis represents the frequency.
Figure 5.
ROC curves analysis of the risk scoring system for overall survival (A) or recurrence-free survival (B). ROC, receiver operating characteristic; AUC, area under the curve.
The risk scoring system was used to predict OS of patients with different clinicodemographic characteristics. This is an important test of the scoring system because of the heterogeneity of HCC and the large number of factors that influence prognosis.
Univariate analysis identified risk score, family cancer history and vascular invasion as significantly associated with OS, but not age, body mass index (BMI), ethnicity, sex, hepatitis, cirrhosis, histological grade of cancer, new tumor event, pathology stage, cancer status or residual tumor. Multivariate Cox regression identified the following as independent predictors of poor OS: Risk score [hazard ratio (HR) 3.393, 95% confidence interval (CI) 1.597–7.210] and vascular invasion (HR 2.146, 95% CI 0.903–5.104, Table V).
Table V.
Univariate and multivariate Cox regression analysis for overall survival.
| Univariate Cox regression | Multivariate Cox regression | |||||
|---|---|---|---|---|---|---|
| Variables | P-value | HR | 95% CI | P-value | HR | 95% CI |
| Risk score (high/low) | 0.003a | 4.949 | 1.735–14.117 | 0.001a | 3.393 | 1.597–7.210 |
| Age (>60/≤60) | 0.202 | 1.816 | 0.726–4.543 | |||
| BMI | 0.060 | |||||
| <25 | Reference | |||||
| ≥25 | 0.245 | 0.073–0.818 | ||||
| Not reported | 0.779 | 0.096–6.340 | ||||
| Race | 0.423 | |||||
| Non-Asian | Reference | |||||
| Asian | 0.352 | 0.053–2.322 | ||||
| Not reported | 1.915 | 0.115–31.769 | ||||
| Sex (Male/Female) | 0.143 | 2.590 | 0.725–9.251 | |||
| Hepatitis B or C | 0.542 | |||||
| No | Reference | |||||
| Yes | 1.489 | 0.415–5.346 | ||||
| Cirrhosis | 0.216 | |||||
| Non-cirrhosis | Reference | |||||
| Cirrhosis | 3.183 | 0.524–19.324 | ||||
| Not reported | 0.572 | 0.178–1.843 | ||||
| Histologic grade | 0.586 | |||||
| G1-2 | Reference | |||||
| G3-4 | 1.326 | 0.481–3.649 | ||||
| New tumor event | 0.406 | |||||
| No | Reference | |||||
| Yes | 1.126 | 0.216–5.883 | ||||
| Not reported | 16.771 | 0.260–1080.40 | ||||
| Pathologic stage | 0.093 | |||||
| Stage I+II | Reference | |||||
| Stage III+IV | 2.857 | 0.935–8.730 | ||||
| Not reported | 3.173 | 0.740–13.617 | ||||
| Cancer status | 0.998 | |||||
| Tumor free | Reference | |||||
| With tumor | 1.052 | 0.196–5.635 | ||||
| Not reported | 0.000 | 0.000 | ||||
| Family cancer history | 0.031 | 0.126 | ||||
| No | Reference | Reference | ||||
| Yes | 0.667 | 0.234–1.899 | 0.868 | 0.419–1.799 | ||
| Not reported | 0.076 | 0.011–0.528 | 0.206 | 0.045–0.953 | ||
| Residual tumor | 0.611 | |||||
| R0 | Reference | |||||
| Non-R0 | 0.469 | 0.105–2.092 | ||||
| Not reported | 0.000 | 0.000 | ||||
| Vascular invasion | 0.006a | 0.005a | ||||
| Negative | Reference | Reference | ||||
| Positive | 3.019 | 0.798–11.416 | 2.146 | 0.903–5.104 | ||
| Not reported | 8.383 | 2.264–31.038 | 3.577 | 1.652–7.748 | ||
P<0.05. BMI, Body mass index; AFP, α fetoprotein; HR, Hazard ratio; CI, Confidence interval.
Risk-scoring system based on lncRNA expression and RFS
Univariate analysis identified 11 lncRNAs that were significantly correlated with RFS: ERVH48-1, LINC02043, LINC01605, AC062004.1, AL139385, AC007938.3, AC090921.1, AC025580.1, AC012640.1, C10orf91, and LINC01589. Multivariate analysis demonstrated the first five to be independent prognostic indicators of RFS (Table IV). The resulting risk scoring system was: Risk score=(0.3529 × ERVH48-1) + (0.3499 × LINC02043) + (0.1701 × LINC01605) + (−0.3531 × AC062004.1) + (−0.1924 × AL139385).
Table IV.
Five lncRNAs were correlated with recurrence-free survival in the best statistical model.
| lncRNA | β | HR | z | P-value |
|---|---|---|---|---|
| ERVH48-1 | 0.3529 | 1.4232 | 2.59 | 0.0096a |
| LINC02043 | 0.3499 | 1.4189 | 3.13 | 0.0017a |
| AC062004.1 | −0.3531 | 0.7025 | −3.12 | 0.0018a |
| LINC01605 | 0.1701 | 1.1855 | 2.19 | 0.0285a |
| AL139385 | −0.1924 | 0.8249 | −2.23 | 0.0259a |
P<0.05. lncRNAs, long noncoding RNAs; HR, Hazard ratio
In this scoring system, increased expression of ERVH48-1, LINC02043, and LINC01605 predicted worse RFS (β>0), whereas increased expression of AC062004.1 and AL139385 predicted better RFS (β<0).
Patients were classified as at low or high risk of poor RFS (Fig. 3B). Kaplan-Meier curves demonstrated that patients with high risk had significantly lower RFS at 3 and 5 years compared with that of patients with low risk (Fig. 4B). AUC for the risk scoring system was 0.777 (Fig. 5B).
Univariate analysis identified that risk score and vascular invasion were significantly correlated with RFS, but not age, BMI, ethnicity, sex, hepatitis, cirrhosis, histology grade, new tumor event, pathology stage, cancer status, family cancer history or residual tumor. Multivariate analysis identified the independent predictors to be risk score (HR 2.895, 95% CI 1.491–5.621) and vascular invasion (HR 2.398, 95% CI 1.104–5.210, Table VI).
Table VI.
Univariate and multivariate Cox regression analysis for recurrence-free survival.
| Univariate Cox regression | Multivariate Cox regression | |||||
|---|---|---|---|---|---|---|
| Variables | P-value | HR | 95% CI | P-value | HR | 95% CI |
| Risk score (high/low) | 0.009a | 4.883 | 1.485–16.051 | 0.002a | 2.895 | 1.491–5.621 |
| Age (>60/≤60) | 0.064 | 2.343 | 0.952–5.767 | |||
| BMI | 0.255 | |||||
| <25 | Reference | |||||
| ≥25 | 0.533 | 0.092–3.074 | ||||
| Not reported | 0.087 | 0.004–1.772 | ||||
| Race | 0.403 | |||||
| Non-Asian | Reference | |||||
| Asian | 4.064 | 0.514–32.105 | ||||
| Not reported | 1.065 | 0.034–33.341 | ||||
| Sex (Male/Female) | 0.680 | 1.523 | 0.207–11.220 | |||
| Hepatitis B or C | 0.210 | |||||
| No | Reference | |||||
| Yes | 3.102 | 0.528–18.223 | ||||
| Cirrhosis | 0.340 | |||||
| Non-cirrhosis | Reference | |||||
| Cirrhosis | 2.228 | 0.297–16.737 | ||||
| Not reported | 0.291 | 0.037–2.284 | ||||
| Histologic grade | 0.489 | |||||
| G1-2 | Reference | |||||
| G3-4 | 0.705 | 0.262–1.899 | ||||
| New tumor event | 0.860 | |||||
| No | Reference | |||||
| Yes | 316078 | 0.000–3.495×1066 | ||||
| Pathologic stage | 0.464 | |||||
| Stage I+II | Reference | |||||
| Stage III+IV | 1.896 | 0.638–5.632 | ||||
| Not reported | 0.997 | 0.127–7.803 | ||||
| Cancer status | 0.286 | |||||
| Tumor free | Reference | |||||
| With tumor | 2.632 | 0.323–21.431 | ||||
| Not reported | 11.747 | 0.399–345.797 | ||||
| Family cancer history | 0.242 | |||||
| No | Reference | |||||
| Yes | 0.726 | 0.147–3.588 | ||||
| Not reported | 0.129 | 0.010–1.703 | ||||
| Residual tumor | 0.749 | |||||
| R0 | Reference | |||||
| Non-R0 | 0.930 | 0.090–9.614 | ||||
| Not reported | 0.321 | 0.015–6.981 | ||||
| Vascular invasion | 0.039a | 0.001a | ||||
| Negative | Reference | Reference | ||||
| Positive | 10.023 | 1.408–71.326 | 2.398 | 1.104–5.210 | ||
| Not reported | 3.990 | 0.369–43.169 | 4.732 | 2.235–10.019 | ||
P<0.05. BMI, Body mass index; AFP, α fetoprotein; HR, Hazard ratio; CI, Confidence interval.
Functional analysis of co-expressed lncRNA and mRNAs
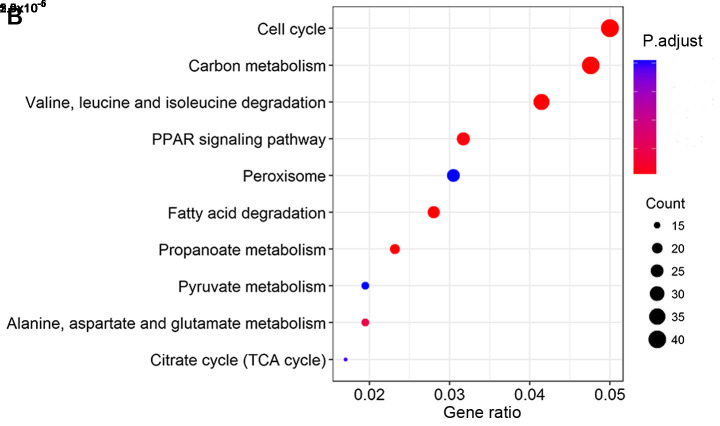
KEGG pathway analysis revealed that co-expressed lncRNA and mRNAs that correlated with OS were involved mainly in chemical carcinogenesis, cytochrome P450-mediated drug metabolism and retinol metabolism (Fig. 6A). Co-expressed lncRNAs and mRNAs that correlated with RFS were involved mainly in cell cycle and carbon metabolism (Fig. 6B).
Figure 6.
KEGG pathway analyses. Top 10 pathways of mRNAs that co-expressed with lncRNAs in the risk scoring system for (A) overall survival or (B) recurrence-free survival. KEGG, Kyoto Encyclopedia of Genes and Genomes; lncRNA, long non-coding RNA.
Discussion
HCC is a major health problem worldwide with poor overall prognosis (25,26). Most patients with HCC are diagnosed at advanced stages (III–IV) (27). Earlier diagnosis and more reliable prognosis, based on suitable biomarkers, are crucial for improving the management and therefore outcomes of patients with HCC. Accumulating evidence has suggested that the abnormal expression of lncRNAs is associated with the recurrence, metastasis and prognosis of HCC (28–30). Since the prognosis in HCC may differ depending on whether it is alcohol-related or not, the present study developed a risk scoring system based on lncRNA expression to evaluate the risk of poor OS or RFS in alcohol-related patients with HCC. The results of the present study may suggest good potential for lncRNAs to be prognostic biomarkers in alcohol-related HCC.
The results of the present study demonstrated that the risk-scoring system and vascular invasion were important independent predictors of prognosis in the sample of patients with HCC. The AUCs for OS and RFS risk scoring systems were high, suggesting good predictive power. Thus, an lncRNA-based risk scoring system may be used to estimate the risk scores of different alcohol-related patients with HCC, predict survival and determine treatment.
Previous studies have identified lncRNAs as prognostic biomarkers for HCC using the TCGA database (31,32). To the best of our knowledge, the present study is the first to analyze alcohol-related HCC. The present study identified eight lncRNAs as potential prognostic biomarkers for alcohol-related HCC. Among them, LINC01605 has been demonstrated to be upregulated in bladder cancer tissues and may be associated with poor prognosis (33), whereas ERVH48-1 has been identified as a prognostic biomarker for tongue squamous cell carcinoma (34). The remaining potential biomarkers from the present study (AC012640.1, AC013451.2, AC062004.1, LINC02334, LINC02043, and AL139385) do not appear to have been analyzed in detail. The eight lncRNAs in this model appear to be involved in chemical carcinogenesis, metabolism and the cell cycle. Investigating these lncRNA-mediated pathways may provide new insights into the development of alcohol-related HCC.
There are some limitations in this study. First, HCC treatment types were not included in the multivariate Cox regression due to lack of data. Second, Cox analyses may be less accurate because some clinical data were missing for some patients. Third, the sample was relatively small, and as a result the present study could not divide the samples into training and test dataset for determining and validating the model. Thus the findings of the present study should be verified and extended in larger studies.
Despite these limitations, the results of the present study suggested that an lncRNA-based risk scoring system may predict the risk of poor prognosis in patients with alcohol-related HCC. Eight lncRNAs are independent clinicopathological variables for alcohol-related HCC.
Acknowledgements
Not applicable.
Funding
This study was supported by the Guangxi Key Research and Development Program (grant. no. GuiKeAB16380215), Guangxi First-class Discipline Project for Basic Medicine Sciences (grant. no. GXFCDP-BMS-2018) and Guangxi Medical University Training Program for Distinguished Young Scholars.
Availability of data and material
The datasets used during the present study are included in this published article.
Authors' contributions
YUL. and JY designed and performed the research, analyzed and interpreted the data, and drafted the manuscript. JW collected and analyzed the data. YOL and JZ conceived the study, designed the methodology and reviewed the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380:1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 2.Matsushita H, Takaki A. Alcohol and hepatocellular carcinoma. BMJ Open Gastroenterol. 2019;6:e000260. doi: 10.1136/bmjgast-2018-000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrick JL, Kelly SP, Altekruse SF, McGlynn KA, Rosenberg PS. Future of hepatocellular carcinoma incidence in the United States forecast through 2030. J Clin Oncol. 2016;34:1787–1794. doi: 10.1200/JCO.2015.64.7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 5.Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149:1226–1239 e1224. doi: 10.1053/j.gastro.2015.05.061. [DOI] [PubMed] [Google Scholar]
- 6.Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol. 2016;64(1 Suppl):S84–S101. doi: 10.1016/j.jhep.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 7.Louafi S, Boige V, Ducreux M, Bonyhay L, Mansourbakht T, de Baere T, Asnacios A, Hannoun L, Poynard T, Taïeb J. Gemcitabine plus oxaliplatin (GEMOX) in patients with advanced hepatocellular carcinoma (HCC): Results of a phase II study. Cancer. 2007;109:1384–1390. doi: 10.1002/cncr.22532. [DOI] [PubMed] [Google Scholar]
- 8.Hung T, Chang HY. Long noncoding RNA in genome regulation: Prospects and mechanisms. RNA Biol. 2010;7:582–585. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, Li J. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 10.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauptman N, Glavac D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14:4655–4669. doi: 10.3390/ijms14034655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 14.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39:2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 15.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang L, Peng X, Li Y, Zhang X, Ma Y, Wu C, Fan Q, Wei S, Li H, Liu J. Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol Cancer. 2019;18:78. doi: 10.1186/s12943-019-0990-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim KS, Lee YI. Biallelic expression of the HI9 and ZGF2 genes in hepatocellular carcinoma. Cancer Lett. 1997;119:143–148. doi: 10.1016/S0304-3835(97)00264-4. [DOI] [PubMed] [Google Scholar]
- 18.Zhu XT, Yuan JH, Zhu TT, Li YY, Cheng XY. Long noncoding RNA glypican 3 (GPC3) antisense transcript 1 promotes hepatocellular carcinoma progression via epigenetically activating GPC3. FEBS J. 2016;283:3739–3754. doi: 10.1111/febs.13839. [DOI] [PubMed] [Google Scholar]
- 19.Ganne-Carrie N, Nahon P. Hepatocellular carcinoma in the setting of alcohol-related liver disease. J Hepatol. 2019;70:284–293. doi: 10.1016/j.jhep.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Pimpin L, Cortez-Pinto H, Negro F, Corbould E, Lazarus JV, Webber L, Sheron N, EASL HEPAHEALTH Steering Committee Burden of liver disease in Europe: Epidemiology and analysis of risk factors to identify prevention policies. J Hepatol. 2018;69:718–735. doi: 10.1016/j.jhep.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Bucci L, Garuti F, Camelli V, Lenzi B, Farinati F, Giannini EG, Ciccarese F, Piscaglia F, Rapaccini GL, Di Marco M, et al. Comparison between alcohol- and hepatitis C virus-related hepatocellular carcinoma: Clinical presentation, treatment and outcome. Aliment Pharmacol Ther. 2016;43:385–399. doi: 10.1111/apt.13485. [DOI] [PubMed] [Google Scholar]
- 22.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341X.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 24.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Budhu A, Forgues M, Ye QH, Jia HL, He P, Zanetti KA, Kammula US, Chen Y, Qin LX, Tang ZY, Wang XW. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99–111. doi: 10.1016/j.ccr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 26.Lin Y, Liang R, Qiu Y, Lv Y, Zhang J, Qin G, Yuan C, Liu Z, Li Y, Zou D, Mao Y. Expression and gene regulation network of RBM8A in hepatocellular carcinoma based on data mining. Aging (Albany NY) 2019;11:423–447. doi: 10.18632/aging.101749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y, Zheng S, Yao J, Li M, Yang G, Zhang N, Zhang S, Zhong B. Decreased expression of protocadherin 20 is associated with poor prognosis in hepatocellular carcinoma. Oncotarget. 2017;8:3018–3028. doi: 10.18632/oncotarget.13822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Xie X, Xiao YF, Xie R, Hu CJ, Tang B, Li BS, Yang SM. The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett. 2015;360:119–124. doi: 10.1016/j.canlet.2015.02.035. [DOI] [PubMed] [Google Scholar]
- 29.Xiong H, Li B, He J, Zeng Y, Zhang Y, He F. lncRNA HULC promotes the growth of hepatocellular carcinoma cells via stabilizing COX-2 protein. Biochem Biophys Res Commun. 2017;490:693–699. doi: 10.1016/j.bbrc.2017.06.103. [DOI] [PubMed] [Google Scholar]
- 30.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z, Li J, He T, Ouyang Y, Huang Y, Liu Q, Wang P, Ding J. The competitive endogenous RNA regulatory network reveals potential prognostic biomarkers for overall survival in hepatocellular carcinoma. Cancer Sci. 2019;110:2905–2923. doi: 10.1111/cas.14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin P, Wen DY, Li Q, He Y, Yang H, Chen G. Genome-wide analysis of prognostic lncRNAs, miRNAs, and mRNAs forming a competing endogenous RNA network in hepatocellular carcinoma. Cell Physiol Biochem. 2018;48:1953–1967. doi: 10.1159/000492519. [DOI] [PubMed] [Google Scholar]
- 33.Qin Z, Wang Y, Tang J, Zhang L, Li R, Xue J, Han P, Wang W, Qin C, Xing Q, et al. High LINC01605 expression predicts poor prognosis and promotes tumor progression via up-regulation of MMP9 in bladder cancer. Biosci Rep. 2018;38:BSR20180562. doi: 10.1042/BSR20180562. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Zhang S, Cao R, Li Q, Yao M, Chen Y, Zhou H. Comprehensive analysis of lncRNA-associated competing endogenous RNA network in tongue squamous cell carcinoma. PeerJ. 2019;7:e6397. doi: 10.7717/peerj.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are included in this published article.