Abstract
The nuclear receptors REV-ERBα and REV-ERBβ have been demonstrated to play key roles in the regulation of numerous physiological functions, such as metabolism and the circadian rhythm. Recent studies have established the REV-ERBs’ roles in immunity, including macrophage and T cell responses. In contrast, their roles in dendritic cells have not been well defined. Dendritic cells are potent antigen presenting cells, connecting microbial sensing and innate immunity to adaptive immune responses. We demonstrate that both REV-ERBα and REV-ERBβ expression is upregulated during the course of bone marrow derived dendritic cell (BMDC) differentiation. BMDCs from REV-ERBα and REV-ERBβ deficient mice showed enhanced expression of maturation markers like CD86, MHCII, and proinflammatory cytokines. Conversely, treatment of BMDCs with a REV-ERB-specific agonist, SR9009, inhibited the expression of maturation markers and proinflammatory cytokines. Our study suggests the REV-ERBs act as negative regulators of dendritic cell development and activation. These results indicate that pharmacological modulation of REV-ERB activity could be an attractive strategy to modulate DC activation status and for DC-based therapies.
Keywords: REV-ERB, dendritic cell, nuclear receptors, NR1D1, NR1D2, SR9009
1. Introduction
Dendritic cells (DCs) operate at the interface between the innate and adaptive immune systems. One important function of DCs is to instruct T cells to elicit an immune response to pathogens or promote tolerance [1, 2]. Many factors influence how DCs develop, migrate, and function during immune responses, however, there has been limited success in exploiting DCs for immune-based therapies. Therefore, understanding the molecular mechanisms that control DCs is imperative in order to identify novel therapeutic targets for the development of DC-based therapies.
Nuclear receptors are a unique and evolutionarily conserved superfamily of ligand-regulated transcription factors that control almost all aspects of mammalian physiology. Because their activity can be modulated by ligand interaction, NRs represent attractive therapeutic targets for the treatment of a variety of diseases. The nuclear receptors REV-ERBα (NR1D1) and REV-ERBβ (NR1D2) have been shown to regulate proinflammatory responses in several immune cell types. Importantly, use of REV-ERB selective ligands have modulated proinflammatory responses in REV-ERB-expressing T cells and macrophages [3–8]. However, whether the REV-ERBs are expressed in and regulate other immune cell subsets is not well defined.
Both REV-ERBα and REV-ERBβ are best known for regulating genes involved in metabolism and the circadian rhythm [9]. Although considerably more is known about the function of REV-ERBα than REV-ERBβ, their similarities suggest they likely have overlapping functions, but this is still not well understood. The REV-ERBs are unique among nuclear receptors in that they lack the regulatory helix important for binding coactivator proteins. Thus, they are thought to interact exclusively with corepressor proteins to mediate transcriptional repression which can be enhanced in the presence of ligand, either endogenous or synthetic, to further augment transcriptional repression of target genes [9]. We and others have recently demonstrated that use of REV-ERB-selective agonist ligands are effective in the treatment of several autoimmune and chronic inflammatory diseases [6, 8, 10, 11].
Several years ago, REV-ERBα was shown to be expressed in splenic DCs in a circadian manner in vivo[12]. However, little work has been done since so the role of the REV-ERBs in DCs remains elusive. While the REV-ERBs have been shown to play a number of roles in macrophage proinflammatory responses, the role of the REV-ERBs in DC cytokine production is unknown despite both cell types sharing the same common progenitor during development. [3–5, 10, 11, 13, 14]. In this study we set out to assess whether the REV-ERBs played a role in the development and maturation of DCs, whether the REV-ERBs affected proinflammatory cytokine function in DCs, and whether ligand regulation of REV-ERB activity affected these roles. Understanding REV-ERB-mediated roles and whether REV-ERB-selective ligands affect these roles may prove beneficial in improving DC-based therapies.
2. Materials and Methods
2.1. Mice
All experimental procedures were approved by the Scripps Florida Institutional Animal Care and Use Committee (IACUC) and were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. C57BL/6J (B6) mice were either purchased from Jackson Laboratory (Bar Harbor, ME) or bred at Scripps, Florida. REV-ERBα−/− deficient mice [B6.Cg-Nr1d1tm1Ven/LazJ [15]] were purchased from Jackson Laboratory. REV-ERBβ−/− mice have been previously described [16]. Both female and male mice between 8–10 weeks of age were used. Mice were sacrificed between 8 and 11am. Mice were kept at controlled temperature (22–23°C), ~60% humidity with 12h light: 12h dark cycles. Mice had access to regular chow (Harlan 2920X, Harlan Laboratories, Indianapolis, Indiana) and water, ad libitum. All mice were kept under specific pathogen free conditions.
2.2. Chemicals
SR9009 [17] has been previously described.
2.3. In vitro generation of dendritic cells
Dendritic cells were generated from bone marrow isolated from femurs and tibia from indicated mice. 2×106 bone marrow cells were cultured in media containing GM-CSF (25 ng/ml; Stem Cell Technologies, Vancouver, Canada) and IL-4 (5 ng/ml; Stem Cell Technologies) for 7 days[18]. On day 3, cultures were replenished with media containing GM-CSF and IL-4. On Day 6, immature DCs were stimulated with LPS (1μg/ml; Sigma-Aldrich, St. Louis, MO, USA) for 24 hrs. DCs were cultured at 37°C, 5% CO2 in RPMI 1640 (Invitrogen, Waltham, MA, USA) supplemented with 10% FBS, 100IU/mL penicillin, 100 μg/ml streptomycin, 50uM β-mercaptoethanol, and 2mM L-glutamine.
2.4. Quantitative real-time PCR
Total RNA was extracted using RNeasy columns (Zymo Research, Irvine, CA) and reverse transcribed using qscript cDNA synthesis kit (Quanta bio, MA). Real-time PCR was performed using SYBR Green including passive reference dye (ROX) (Roche) on a HT7900 Fast Real Time PCR system (Life Technologies, Carlsbad, CA). All gene expression data were normalized to a housekeeping gene, using a ΔΔ cycle threshold-based algorithm followed by fold change comparison with the average of the control group. Primer sequences can be found in Table 1.
Table 1.
qRT-PCR primers
| Gene name | Forward primer (5’ - 3’) | Reverse Primer (5’ - 3’) |
|---|---|---|
| B2m | AGAATGGGAAGCCGAACATAC | GAAAGACCAGTCCTTGCTGAA |
| Cd86 | ATCAAGGACATGGGCTCGTA | GAAGTTGGCGATCACTGACA |
| Cxcl10 | TCCTTGTCCTCCCTAGCTCA | ATAACCCCTTGGGAAGATGG |
| Il1b | GTTTTCCTCCTTGCCTCTGA | GCTGCCTAATGTCCCCTT |
| Il2 | GCATGCAGCTCGCATCCTGT | GAGCTTGAAGTGGGTGCGCT |
| Il6 | CATGTTCTCTGGGAAATCGTG | TCCAGTTTGGTAGCATCCATC |
| Il10 | ACCAGCTGGACAACATACTGC | CAAATGCTCCTTGATTTCTGG |
| Il12b | GGAAGCACGGCAGCAGAATA | AACTTGAGGGAGAAGTAGGAATGG |
| MhcII | GAGCATCCCAGCCTGAAGA | CGATGCCGCTCAACATCTT |
| Nr1d1 | ACCTTTGAGGTGCTGATGGT | CTCGCTGAAGTCAAACATGG |
| Nr1d2 | TGTGAAAACAGGCAAAACCA | CCCTTACAGCCTTCACAAGC |
2.5. Qiagen Array
A Mouse Dendritic and Antigen Presenting Cell RT2 PCR array (PAMM-406Z, Qiagen, Frederick, MD., USA) was used to screen a panel of 84 genes focused on dendritic cell activation and maturation. Total RNA was isolated from BMDCs using a Qiagen RNeasy Mini Kit by following manufacturer’s protocol. BMDCs were treated with SR9009 for the duration of the DC culture. DMSO was used as the vehicle. The array was performed on an HT7900 Fast Real Time PCR system according to the RT2 Profiler PCR Array instructions. Microarray data was normalized against the house keeping genes included in the array by calculating the ΔCt for each gene of interest in the plate. Fold changes of gene expression, scatterplot and heatmap were analyzed and generated by using RT2 PCR array data analysis web portal version 3.5 (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php). Genes that had fold changes of more than 1.5 in expression against DMSO control were considered significant.
2.6. Flow cytometry
BMDCs were collected, washed with PBS and prepared as single cell suspensions. Cells were stained with fluorescence-conjugated antibodies for 30 minutes, washed, then resuspended in FACS buffer (0.5% BSA, 2mM EDTA in PBS). Flow cytometric analysis was performed on a BD LSRII (BD Biosciences, Franklin Lakes, NJ, USA) instrument and analyzed using FlowJo software (TreeStar, Ashland, OR, USA). All antibodies used in experiments are described Table 2.
Table 2.
FACS antibodies
| Antibody | Fluorophore | Species | Isotype | Clone | Company | Cat # |
|---|---|---|---|---|---|---|
| CD11b | FITC | Rat | IgG2b, κ | M1/70 | eBioscience | 11-0112-85 |
| CD11c | PE/Dazzle 594 | Hamster | IgG | N418 | Biolegend | 117347 |
| MHC II | APC | Rat | IgG2b, κ | M5/114.15.2 | Biolegend | 107614 |
| CD86 | PE/Cy7 | Rat | IgG2a, κ | GL1 | Biolegend | 105013 |
| Viability | eFluor 506 | eBioscience | 65-0866-14 |
2.7. Statistical analysis
Data are expressed as mean ± s.e.m. Statistical analyses were performed using GraphPad PRISM 6 (GraphPad Software, Inc., La Jolla, CA). A student’s two tailed t-test was used for comparison between two groups. A p value of < 0.05 was considered statistically significant.
3. Results
3.1. The REV-ERBs are upregulated during DC development
To establish whether both REV-ERBs, REV-ERBα and REV-ERBβ, were expressed in DCs, we isolated bone marrow from C57BL/6 mice and differentiated the cells into bone marrow derived DCs (BMDCs) [18]. BMDCs were collected at different time points during differentiation in order to assess the mRNA expression of the REV-ERBs during development. Both REV-ERBα (Nr1d1) and REV-ERBβ (Nr1d2) were upregulated during differentiation, with a significant increase in expression observed following LPS treatment (Day 6 to Day 7, Fig. 1A). The mRNA expression of Major Histocompatibility Class protein II (MhcII) and the co-stimulatory molecule CD86 (Cd86) were also assessed to verify the differentiation and maturation (Fig. 1A). These data indicate that both REV-ERBα and REV-ERBβ are upregulated during bone marrow derived DC differentiation, with a significant increase in expression occurring in response to LPS treatment.
Figure 1. Loss of the REV-ERBs leads to increased expression of MHCII and the co-stimulatory molecule CD86.
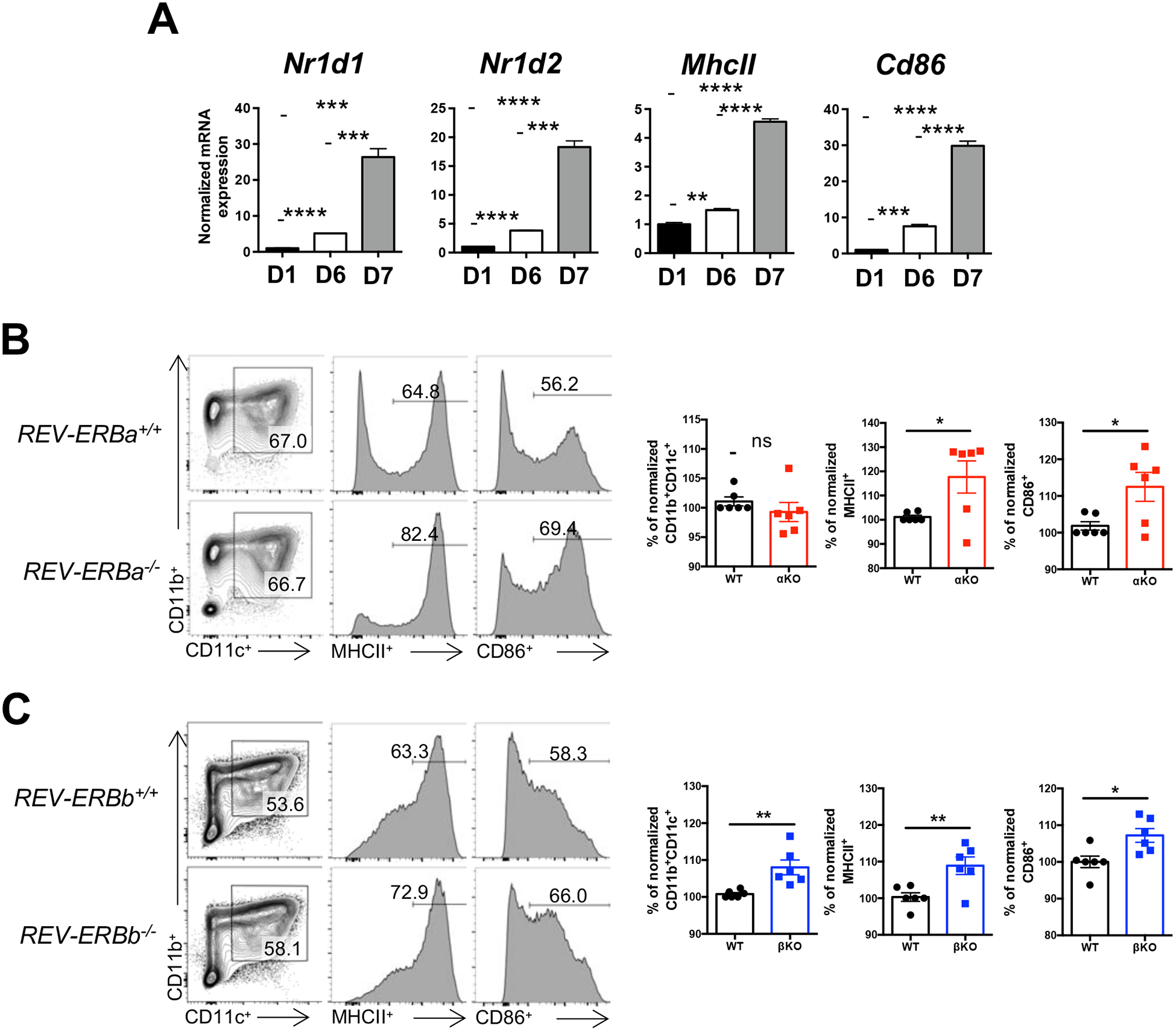
(A) Bone marrow cells from C57BL/6 mice were differentiated into bone marrow derived dendritic cells (BMDCs). Cells were collected during differentiation to assess the mRNA expression of REV-ERBα (Nr1d1), REV-ERBβ (Nr1d2), and mature dendritic cell markers, MhcII and Cd86 by quantitative real time PCR. B2M was used as the housekeeping control gene. Data are representative of three separate independent experiments generating similar results. (B) Bone marrow from WT or REV-ERBα KO mice were differentiated into BMDCs and analyzed for the expression of MHCII and CD86 from CD11b+CD11c+ cells by flow cytometry (left). Graphs (right) depict all of the experiments performed with data normalized to WT within each individual experiment. (n=6) (C) Similar experiments for bone marrow from WT and REV-ERBβ KO mice were performed as in B. Data represents mean ± SEM. (n=6). Student’s two-tailed t-tests were performed for statistical analysis. *p,0.05, **p<0.01. ***p<0.001, ****p<0.0001, ns, not significant.
3.2. Loss of the REV-ERBs affects the expression of MHCII and the co-stimulatory receptor CD86
When immature, DCs express low levels of MHC proteins and co-stimulatory molecules, including CD86, and are not equipped to activate naïve T cells. However, upon activation, DCs rapidly upregulate these proteins and are considered “mature”. To test whether the REV-ERBs are required for the development and/or maturation of DCs, we differentiated BMDCs from REV-ERBα-deficient (αKO), REV-ERBβ-deficient (βKO) and their respective wild-type (WT) littermate control mice. While we did not observe any differences between WT and αKO CD11b+CD11c+ populations, both of which are surface markers for DCs, we did observe that MHCII and CD86 expression were increased in αKO DCs relative to WT DCs (Fig. 1B). Similar to REV-ERBα KO DCs, REV-ERBβ-deficient DCs also demonstrated increased MHCII and CD86 expression relative to WT DCs (Fig. 1C). In contrast to αKO DCs, βKO DCs showed an increase in the percent of CD11b+CD11c+ cells relative to WT DCs. These data suggest that REV-ERBβ may play a unique role in CD11b+CD11c+ DC development, one in which REV-ERBα does not appear to be able to compensate for.
3.3. A REV-ERB-selective agonist suppresses DC development and maturation
To determine whether overexpression of REV-ERB activity affected DC development in vitro, we differentiated BMDCs from WT bone marrow in the presence of vehicle or the REV-ERBα/β agonist, SR9009 [17], for the duration of the experiment (Fig. 2). SR9009 has been demonstrated to be selective for the REV-ERBs in multiple past studies[6, 10, 17, 19, 20]. FACS analysis of DCs demonstrated that SR9009 dose dependently inhibited the percent of CD11b+CD11c+ cells, as well as MHCII and CD86 expression relative to vehicle treated cells (Fig. 2). Importantly, this inhibition was not a consequence of cell death as the viability was not affected in any of the culture conditions.
Figure 2. SR9009 inhibits the development of BMDCs.
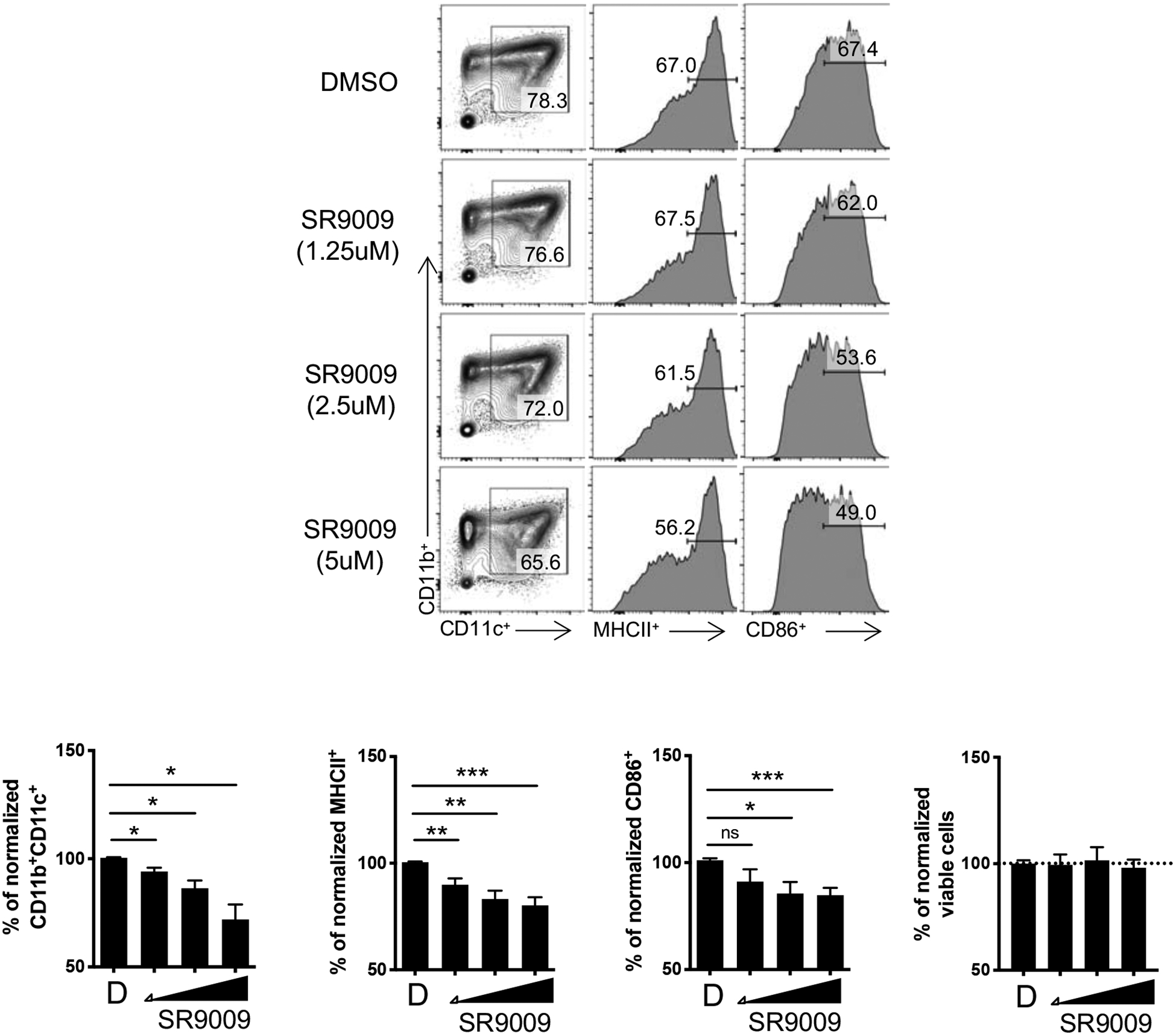
(A) Bone marrow cells from C57BL/6 mice were differentiated into BMDCs in the presence of vehicle (DMSO) and increasing doses of SR9009 starting at day 0. BMDCs were analyzed for the expression of MHCII and CD86 from CD11b+CD11c+ cells by flow cytometry (top). Graphs (bottom) depict all of the experiments performed with data normalized to DMSO within each individual experiment. Data represents mean ± SEM. (n=7) Student’s two-tailed t-tests were performed for statistical analysis. *p,0.05, **p<0.01, ***p<0.001, ns, not specific.
3.4. SR9009 suppress pro-inflammatory gene expression in DCs
To further explore the role of the REV-ERBs in DCs we utilized a Mouse Dendritic and Antigen Presenting Cell PCR array. This array contains genes important for DC activation and maturation, including cytokines, chemokines, and their respective receptors. Many of the genes in the array are highly expressed in mature DCs or show significant changes during differentiation. Therefore, we differentiated BMDCs from WT bone marrow in the presence of vehicle or SR9009 for the duration of the experiment after which we collected cells for the array. Array analysis of drug-treated samples revealed that ligand treatment lead to changes in a number of genes, (greater than 1.5 fold over DMSO), including repression of Cd86 and Cd80, with CD86 being consistent with our flow cytometry data (Fig. 3A). Both CD80 and CD86 are co-stimulatory receptors indicating that use of REV-ERB ligands could inhibit the ability of DCs to properly activate T cells. We also observed repression of a number of proinflammatory cytokines and chemokines, some of which have been demonstrated to be regulated by the REV-ERBs and/or de-repressed in REV-ERBα/β double knock-out macrophages, including Il6, Il1b, Ccr2, and Cxcl10 [4, 5, 13]. We also noticed a number of other cytokine genes that were differentially regulated by SR9009 treatment, including Il12b, Il10, and Il2. Expression of these cytokines, either individually or in combination, could impact TH17 and TH1 CD4+ T helper cell development, with IL-12p40 (Il12b) being a component of both the IL-12 heterodimeric complex and IL-23 [21]. We chose several of the genes from the array that had previously been described as REV-ERB-regulated, albeit largely in human macrophages, for verification and repeated the experiments in order to validate the PCR array expression data. We found that SR9009 efficiently repressed the expression of Il1b, Il10, Il6, Il12b, Cxcl10, and Il2 (Fig. 3B).
Figure 3. SR9009 has differential effects on gene expression in BMDCs.
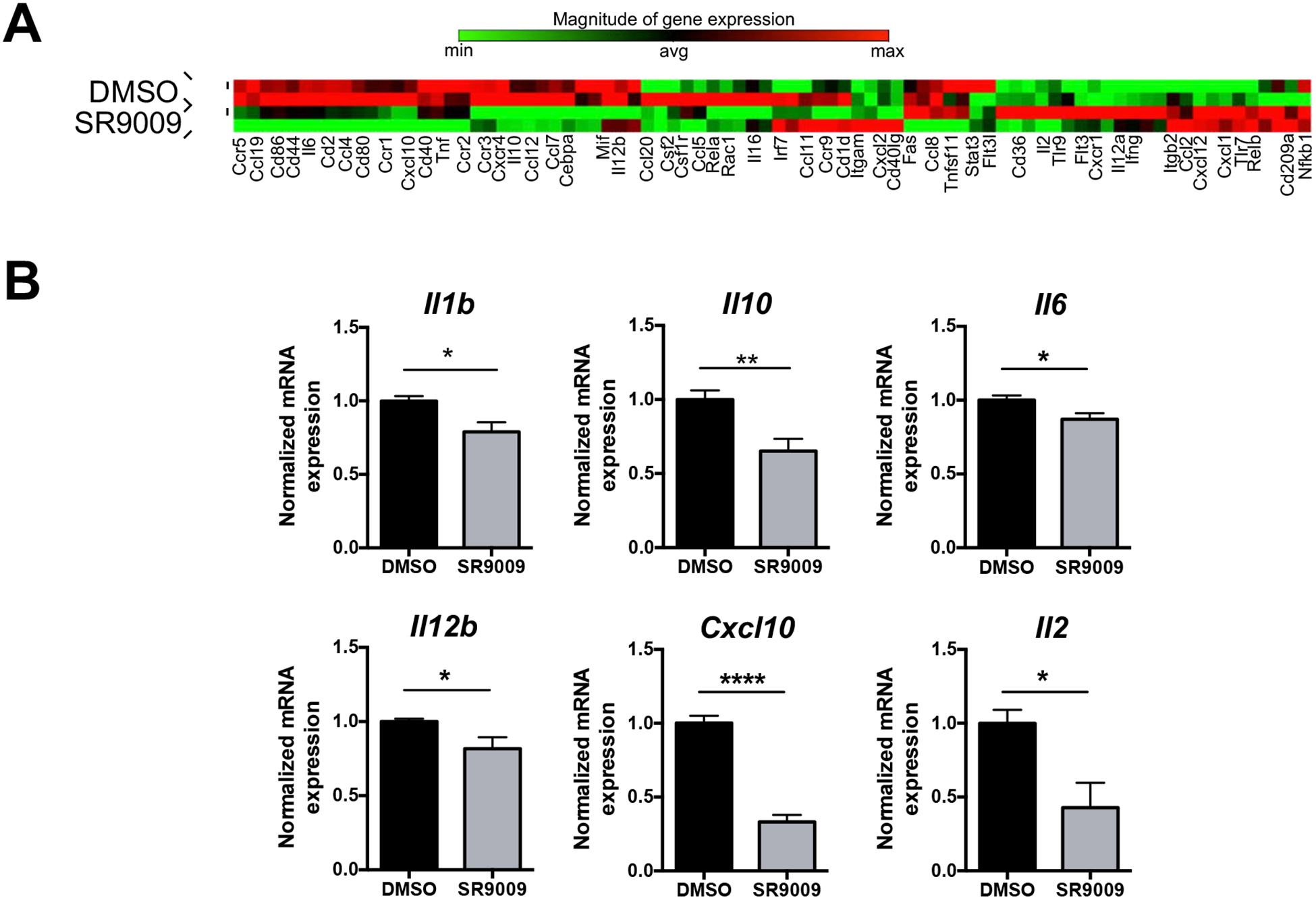
(A) Bone marrow cells from C57BL/6 mice were differentiated into BMDCs in the presence of vehicle (DMSO) or SR9009 (5μM) for the duration of the experiment. BMDCs were sorted on live CD11b+CD11c+ cells in order to perform the Qiagen array analysis and the mRNA expression of the depicted pro-inflammatory cytokines and chemokines. Data represents mean ± SEM. (n=3) B2M was used as the housekeeping control. Student’s two-tailed t-tests were performed for statistical analysis. *p,0.05, **p<0.01, ***p<0.001, ****p<0.0001.
3.5. REV-ERB-deficient DCs have altered proinflammatory gene expression
To assess whether deletion of the REV-ERBs would have any effect on genes identified in the PCR array, we isolated bone marrow from αKO, βKO, and their respective WT littermate controls and differentiated these cells into DCs. We hypothesized that if SR9009 repressed the genes in the array by increasing the REV-ERBs repressive activity, loss of the REV-ERBs should lead to de-repression of many of these target genes. Indeed, loss of REV-ERBα lead to de-repression of many of the genes, including Il1b, Il10, Il6, and Il12b, which was consistent with the drug-treated samples (Fig. 4A). There was some de-repression observed at Cxcl10, but the effect did not reach significance. Interestingly, Il2 was still repressed in αKO DCs relative to WT controls. Repression was likely not a function of increased REV-ERBβ (Nr1d2) as its expression was unchanged in αKO vs. WT DCs. In contrast to REV-ERBα-deficient DCs, REV-ERBβ-deficient DCs demonstrated a different gene expression pattern. While several genes were de-repressed similar to what was observed in αKO DCs, including Il10, Il6, and Il12b, other genes, such as Il1b, Cxcl10, and Il2 were actually repressed, contrary to what we had hypothesized (Fig. 4B). We noted that repression of Il1b, Cxcl10, and Il2 may be due to the increased expression of REV-ERBα (Nr1d1) observed in the βKO DCs, indicating that these genes may be exclusively regulated by REV-ERBα and effects at these genes by REV-ERBβ may be indirect. Increased REV-ERBα expression in βKO DCs also suggests that REV-ERBβ may regulate REV-ERBα but not vice-versa as there was no effect on REV-ERBβ in the αKO DCs. Collectively, these data indicate that the REV-ERBs coordinately regulate gene expression in DCs in vitro.
Figure 4. Loss of the REV-ERBs has differential effects on gene expression in BMDCs.
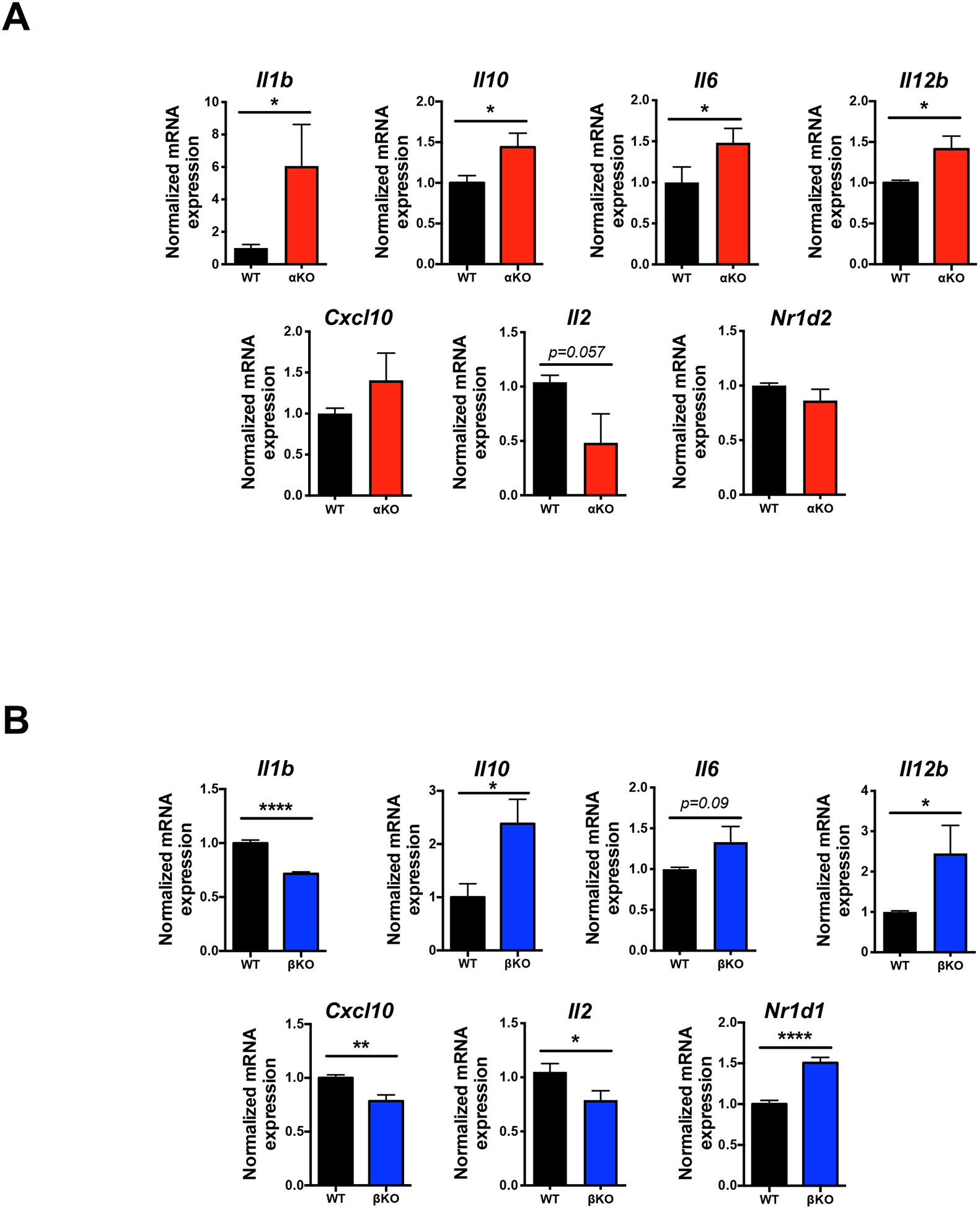
(A) mRNA analysis of BMDCs from WT or REV-ERBα KO mice to assess the expression of various pro-inflammatory cytokines, chemokines, and transcription factors. (n=3) (B) mRNA analysis of BMDCs from WT or REV-ERBβ KO mice to assess the expression of various pro-inflammatory cytokines, chemokines, and transcription factors. Data represents mean ± SEM. (n=3) B2M was used as the housekeeping control. Student’s two-tailed t-tests were performed for statistical analysis. *p<0.05, **p<0.01, ****p<0.0001.
4. Discussion
Dendritic cells (DCs) are potent, professional antigen presenting cells that can prime the adaptive immune response against pathogens. Alternatively, DCs play key roles in the induction and maintenance of tolerance. While much work has been done to understand the diverse functions of DCs, there are still many questions regarding the molecular mechanisms that DCs use to regulate immune function. The present study showed that the nuclear receptors, REV-ERBα and REV-ERBβ, are expressed in DCs in vitro and appear to regulate expression of surface molecules and genes important for DC maturation and regulation of immune responses. Furthermore, modulation of REV-ERB activity using a REV-ERB selective agonist lead to changes in these same surface molecules and target genes, indicating targeting of these receptors may be a viable therapeutic option for DC-based therapies.
A number of works have demonstrated that the REV-ERBs, specifically REV-ERBα, are important for innate immunity by regulating macrophage development and function, including cytokine secretion, wound repair, and disease development [3–5, 10, 11, 13, 14]. These works show, either through genetics or small molecule regulation, that the REV-ERBs regulate cytokines, chemokines, and chemokine receptors, including Il6, Il1b, Il12b, Cxcl10, and Il10, in human or mouse macrophages [3–5, 10, 11, 13, 14]. Repression of these genes are consistent with previously published reports demonstrating that REV-ERBα directly and indirectly regulates their expression, which indicates that the REV-ERBs roles are to restrain cytokine production in activated DCs [3–5, 13, 14]. It should be noted that the REV-ERB-mediated regulation of Il10 expression was identified in human macrophages, but this element did not translate to mouse Il10 [22]. However, the authors did not take into account that the REV-ERBs may tether to other transcription factors to regulate mouse Il10 expression [23]. Il2 was another cytokine that did not fit the previous paradigms. While small molecule modulation suppressed Il2 expression, so did genetic deletion. While this could be accounted for in the REV-ERBβ KO DCs due to upregulation of REV-ERBα, no reverse explanation was obvious in the REV-ERBα KO DCs. However, given the pleiotropic role for IL-2 in immune responses, its expression must be tightly regulated. Thus, it is possible that there are multiple feedback loops to regulate the expression of IL-2 in DCs[24]. Future studies are warranted to mechanistically determine how the REV-ERBs regulate cytokine expression in DCs.
The REV-ERBs have also been shown to be key factors dictating adaptive immunity, regulating TH17 cell development and function, with small molecule modulators of REV-ERB activity effective in the treatment of mouse models of autoimmunity and chronic inflammation [6–8]. However, a critical role for DCs is to present antigen and give appropriate signals to naïve T cells to influence T cell differentiation and function. Our results demonstrate that the REV-ERBs regulate the expression of MHCII and CD86, expression of which are important for proper activation of T cell responses. We recently identified a role for the REV-ERBs in TH17 cell differentiation [6]. Importantly, we and others established that small molecule modulation of REV-ERB activity was effective in the treatment of autoimmunity and chronic inflammation[6, 8]. Given the effects we observed with drug treatment of DCs in vitro, it is possible that some effects observed with SR9009 in vivo could be due to both dampened TH17 and DC responses. Indeed, if REV-ERB-selective small molecules inhibit DC maturation this could lead to weak antigen presentation and inefficient priming of T cells. Additionally, given the suppression of cytokine genes observed in DCs, including Il6, Il1b, and Il12b, T cell effector responses may also be suppressed in vivo, thus augmenting the development of TH17- and TH1-mediated responses. More work is needed to understand the extent of REV-ERB-selective small molecule regulation of DC responses in vivo.
Overall, our findings suggest that the REV-ERBs act to repress a combination of genes important for DC development and maturation. Given that many of these genes are important for antigen presentation, T cell activation, and inflammatory responses, the REV-ERBs may be a novel therapeutic target to either boost or inhibit the activation status of DCs, depending on whether an agonist or antagonist ligand is used. Thus, the REV-ERBs represent an attractive target for DC-based vaccination strategies for the treatment of cancer or autoimmunity, respectively.
Highlights:
The REV-ERBs are upregulated during BMDC development
The REV-ERBs regulate MHCII and CD86 surface expression in BMDCs
The REV-ERBs act as negative regulators of BMDC pro-inflammatory cytokine gene expression
Small molecule modulation of REV-ERB activity affects BMDC maturation and gene expression
Funding
This work was supported by the US National Institutes of Health (1R01AI116885 to L.A.S.). the American Association of Immunologists Careers in Immunology Fellowship Program (M.A.) and the Crohn’s and Colitis Foundation of America (#546172 to M.A.).
Abbreviations
- DC
dendritic cell
- BMDC
bone marrow derived dendritic cell
- MHCII
Major histocompatibility complex class II
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interest
The authors confirm that there are no conflicts of interest.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Worbs T, Hammerschmidt SI, Forster R, Dendritic cell migration in health and disease, Nat Rev Immunol, 17 (2017) 30–48. [DOI] [PubMed] [Google Scholar]
- [2].Macri C, Pang ES, Patton T, O’Keeffe M, Dendritic cell subsets, Semin Cell Dev Biol, 84 (2018) 11–21. [DOI] [PubMed] [Google Scholar]
- [3].Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, Lee CY, Watt A, Grossman TR, Rosenfeld MG, Evans RM, Glass CK, Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription, Nature, 498 (2013) 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, Farrow SN, Else KJ, Singh D, Ray DW, Loudon AS, The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines, Proceedings of the National Academy of Sciences of the United States of America, 109 (2012) 582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Eichenfield DZ, Troutman TD, Link VM, Lam MT, Cho H, Gosselin D, Spann NJ, Lesch HP, Tao J, Muto J, Gallo RL, Evans RM, Glass CK, Tissue damage drives co-localization of NF-kappaB, Smad3, and Nrf2 to direct Rev-erb sensitive wound repair in mouse macrophages, Elife, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Amir M, Chaudhari S, Wang R, Campbell S, Mosure SA, Chopp LB, Lu Q, Shang J, Pelletier OB, He Y, Doebelin C, Cameron MD, Kojetin DJ, Kamenecka TM, Solt LA, REV-ERBalpha Regulates TH17 Cell Development and Autoimmunity, Cell reports, 25 (2018) 3733–3749 e3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yu X, Rollins D, Ruhn KA, Stubblefield JJ, Green CB, Kashiwada M, Rothman PB, Takahashi JS, Hooper LV, TH17 cell differentiation is regulated by the circadian clock, Science, 342 (2013) 727–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chang C, Loo CS, Zhao X, Solt LA, Liang Y, Bapat SP, Cho H, Kamenecka TM, Leblanc M, Atkins AR, Yu RT, Downes M, Burris TP, Evans RM, Zheng Y, The nuclear receptor REV-ERBalpha modulates Th17 cell-mediated autoimmune disease, Proceedings of the National Academy of Sciences of the United States of America, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kojetin DJ, Burris TP, REV-ERB and ROR nuclear receptors as drug targets, Nat Rev Drug Discov, 13 (2014) 197–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang S, Lin Y, Yuan X, Li F, Guo L, Wu B, REV-ERBalpha integrates colon clock with experimental colitis through regulation of NF-kappaB/NLRP3 axis, Nature communications, 9 (2018) 4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sitaula S, Billon C, Kamenecka TM, Solt LA, Burris TP, Suppression of atherosclerosis by synthetic REV-ERB agonist, Biochem Biophys Res Commun, 460 (2015) 566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Silver AC, Arjona A, Hughes ME, Nitabach MN, Fikrig E, Circadian expression of clock genes in mouse macrophages, dendritic cells, and B cells, Brain Behav Immun, 26 (2012) 407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sato S, Sakurai T, Ogasawara J, Shirato K, Ishibashi Y, Oh-ishi S, Imaizumi K, Haga S, Hitomi Y, Izawa T, Ohira Y, Ohno H, Kizaki T, Direct and indirect suppression of interleukin-6 gene expression in murine macrophages by nuclear orphan receptor REV-ERBalpha, ScientificWorldJournal, 2014 (2014) 685854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sato S, Sakurai T, Ogasawara J, Takahashi M, Izawa T, Imaizumi K, Taniguchi N, Ohno H, Kizaki T, A circadian clock gene, Rev-erbalpha, modulates the inflammatory function of macrophages through the negative regulation of Ccl2 expression, J Immunol, 192 (2014) 407–417. [DOI] [PubMed] [Google Scholar]
- [15].Chomez P, Neveu I, Mansen A, Kiesler E, Larsson L, Vennstrom B, Arenas E, Increased cell death and delayed development in the cerebellum of mice lacking the rev-erbA(alpha) orphan receptor, Development, 127 (2000) 1489–1498. [DOI] [PubMed] [Google Scholar]
- [16].Banerjee S, Wang Y, Solt LA, Griffett K, Kazantzis M, Amador A, El-Gendy BM, Huitron-Resendiz S, Roberts AJ, Shin Y, Kamenecka TM, Burris TP, Pharmacological targeting of the mammalian clock regulates sleep architecture and emotional behaviour, Nat Commun, 5 (2014) 5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, Yoo SH, Takahashi JS, Butler AA, Kamenecka TM, Burris TP, Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists, Nature, 485 (2012) 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang J, Dai X, Hsu C, Ming C, He Y, Zhang J, Wei L, Zhou P, Wang CY, Yang J, Gong N, Discrimination of the heterogeneity of bone marrowderived dendritic cells, Mol Med Rep, 16 (2017) 6787–6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chang C, Loo CS, Zhao X, Solt LA, Liang Y, Bapat SP, Cho H, Kamenecka TM, Leblanc M, Atkins AR, Yu RT, Downes M, Burris TP, Evans RM, Zheng Y, The nuclear receptor REV-ERBalpha modulates Th17 cell-mediated autoimmune disease, Proceedings of the National Academy of Sciences of the United States of America, 116 (2019) 18528–18536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sulli G, Rommel A, Wang X, Kolar MJ, Puca F, Saghatelian A, Plikus MV, Verma IM, Panda S, Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence, Nature, 553 (2018) 351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Stockinger B, Omenetti S, The dichotomous nature of T helper 17 cells, Nat Rev Immunol, 17 (2017) 535–544. [DOI] [PubMed] [Google Scholar]
- [22].Chandra V, Mahajan S, Saini A, Dkhar HK, Nanduri R, Raj EB, Kumar A, Gupta P, Human IL10 gene repression by rev-erbalpha ameliorates Mycobacterium tuberculosis clearance, The Journal of biological chemistry, 288 (2013) 10692–10702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, Armour SM, Remsberg JR, Jager J, Soccio RE, Steger DJ, Lazar MA, GENE REGULATION. Discrete functions of nuclear receptor Rev-erbalpha couple metabolism to the clock, Science, 348 (2015) 1488–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zelante T, Fric J, Wong AY, Ricciardi-Castagnoli P, Interleukin-2 production by dendritic cells and its immuno-regulatory functions, Front Immunol, 3 (2012) 161. [DOI] [PMC free article] [PubMed] [Google Scholar]


