Abstract
The historical reliance of biological research on the use of animal models has sometimes made it challenging to address questions that are specific to the understanding of human biology and disease. But with the advent of human organoids — which are stem cell-derived 3D culture systems — it is now possible to re-create the architecture and physiology of human organs in remarkable detail. Human organoids provide unique opportunities for the study of human disease and complement animal models. Human organoids have been used to study infectious diseases, genetic disorders and cancers through the genetic engineering of human stem cells, as well as directly when organoids are generated from patient biopsy samples. This Review discusses the applications, advantages and disadvantages of human organoids as models of development and disease and outlines the challenges that have to be overcome for organoids to be able to substantially reduce the need for animal experiments.
Subject terms: Stem cells, Biological techniques, Stem cells
Human organoids are valuable models for the study of development and disease and for drug discovery, thus complementing traditional animal models. The generation of organoids from patient biopsy samples has enabled researchers to study, for example, infectious diseases, genetic disorders and cancers. This Review discusses the advantages, disadvantages and future challenges of the use of organoids as models for human biology.
Introduction
The use of classical cell line and animal model systems in biomedical research during the late twentieth and early twenty-first centuries has been successful in many areas, such as improving our understanding of cellular signalling pathways, identifying potential drug targets and guiding the design of candidate drugs for pathologies including cancer and infectious disease. The value of the achievements these model systems have made possible is proved by the fact that their use has become near universal in biomedical research today. Historically, the investigation of disease mechanisms in animal models has progressed along a common discovery pipeline, whereby biological processes were initially investigated by genetic screens in invertebrates, followed by an analysis of evolutionary conservation in mammalian model systems, eventually leading to clinical translation to humans. The common principles of animal development and organ physiology derived from this approach have led to a detailed mechanistic understanding of many human diseases. Nevertheless, extrapolating results from model systems to humans has become a major bottleneck in the drug discovery process. Furthermore, recent studies have identified biological processes that are specific to the human body and cannot be modelled in other animals. These include, for example, brain development, metabolism and the testing of drug efficacy. The emergence of human in vitro 3D cell culture approaches using stem cells from different organs has therefore received widespread attention as having the potential to overcome these limitations.
Attempts to model the biology of human organs — including the differentiation of human stem cells in 2D, in either the presence or absence of a 3D matrix; bio-printing of human cells; and the culture of cells in a microfluidic device (‘organ-on-a-chip’)1 — were made prior to the emergence of organoids and have shown some potential for drug screening or human disease research. However, organoids are unique, in that they are self-organizing, 3D culture systems that are highly similar to — and in some cases, histologically indistinguishable from — actual human organs2–8. One feature that is common to all organoids is that they are generated from pluripotent stem cells (PSCs) or adult stem cells (AdSCs; also known as tissue stem cells) by mimicking human development or organ regeneration in vitro9. The analysis of organoid formation can thus provide valuable information about the mechanisms underlying human development and organ regeneration, highlighting their value for basic biological research in addition to their potential application in pharmaceutical drug testing and molecular medicine. The potential of organoids to complement existing model systems and extend basic biological research, medical research and drug discovery into a more physiologically relevant human setting is becoming ever more widely appreciated. However, the development of organoid technology is still in its infancy in comparison to established cell line and animal models, with challenges still to overcome.
In this Review, we describe examples of PSC-derived and AdSC-derived organoid models that have shown great potential and promise in modelling human disease for a broad spectrum of life stages, from early development through to adulthood. Many excellent reviews have already described the various organoid model systems9–14, highlighted their relevance for disease modelling15,16 and outlined organoid-associated protocols17. This Review will instead discuss organoids as a novel model system for understanding human development and genetics that complements, and in the future might reduce, the use of animal models. We emphasize the differences between the mouse and human systems, evaluating the advantages and disadvantages of animal models and human organoid models. We also highlight the tools and methodologies that are available for human organoid studies, in the hope that this information will lead basic biologists to consider using this emerging platform for the study of human pathophysiology.
The diversity of biological model systems
Building on the fundamental idea that biological mechanisms are conserved throughout evolution, biomedical research has traditionally focused on only a handful of model organisms. These models are typically robust, fast-growing species that generate a large number of offspring in a short time and can be propagated at low cost in the laboratory. Researchers benefit greatly from the use of an established model organism, as there are already detailed descriptions of its development and physiology, a large body of experimental techniques and many community resources, including reagents and species-specific databases. While some model organisms, such as yeast (Saccharomyces cerevisiae), fruit flies (Drosophila melanogaster) and mice (Mus musculus), have a long tradition of use in research, other model systems have been established more recently but nevertheless have helped broaden our knowledge by presenting us with a new set of experimental tools. These include the worm, Caenorhabditis elegans, whose reproducible cell lineage allows unparalleled analysis of cell fate decisions18, and the zebrafish, Danio rerio, the first vertebrate system in which saturation loss-of-function mutagenesis screening was performed19,20. C. elegans became a widely used experimental model from the mid 1970s, whereas the first large-scale genetic screens in D. rerio were conducted in the 1990s21,22. However, although it is extensive, the information obtained from animal models does not completely reflect human physiology. In this respect, human organoids (which emerged in the early 2010s)3,5,23,24 can be seen as a novel experimental model that bridges the gap between animal models and human beings.
Model organisms
The most commonly used model organisms currently are S. cerevisiae, C. elegans, D. melanogaster, zebrafish and the common house mouse. In addition, for cancer studies, patient-derived xenografts (PDX) and cancer cell lines are also used. Each model has its own unique advantages and limitations (Fig. 1). For example, 60–80% of genes in C. elegans have human orthologues, and about 42% of human disease genes have orthologues in C. elegans25–27. Moreover, many signalling pathways have been evolutionarily conserved from this species. The largely deterministic nature of C. elegans development makes it an ideal model for cell lineage analysis and has led to the discovery of key cell death pathways, yet pattern formation in C. elegans is different from that in other organisms, and mechanistic principles of its development are often not fully conserved. In addition, other aspects of the physiology of C. elegans, such as its lifespan, metabolism and immune system, are very different from human physiology.
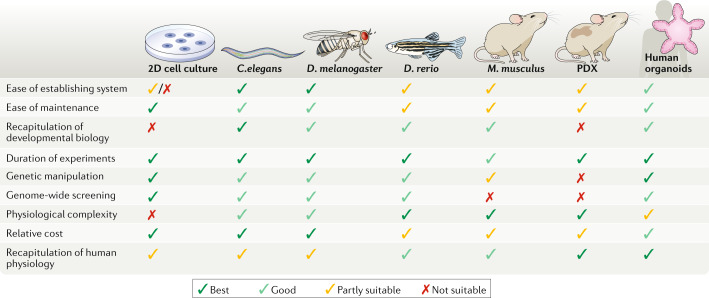
Fig. 1. Comparison of organoids with other model systems.
The most common model organisms that are used in biomedical research are Caenorhabditis elegans, Drosophila melanogaster, Danio rerio and Mus musculus, along with patient-derived xenografts (PDX). These models, as well as 2D cell cultures and human organoids, are assessed here for their relative benefits and limitations. Relative scores are represented as being the best (dark green tick), good (light green tick), partly suitable (yellow tick) and not suitable (red cross).
Among mammalian model systems, the availability of highly advanced genetic tools and germline-competent mouse embryonic stem cells (ESCs) that enable the generation of genetically engineered mouse models makes the mouse the most preferred system. Nevertheless, limitations remain with regard to the physical inaccessibility of early development, as compared with other, egg-laying models with visible development, the requirement for expensive animal facilities in which to maintain the mice and some important differences between mouse and human physiology (Box 1). The zebrafish was developed as a vertebrate model for large-scale genetic studies in the 1990s, to overcome some of the limitations of the mouse model: the transparent zebrafish embryo and this model’s potential to reproduce in huge numbers with a manageable price tag allowed zebrafish researchers to uncover fundamental principles of early embryonic development. However, the zebrafish model suffers from a hugely complex genome, a lack of comprehensive in vitro culture systems and the fact that it shares only a limited degree of biological similarity with humans. Over the past 30 years, the use of these various, equally valuable animal model systems, as wells as 2D human cancer cell lines, has led to an explosion of knowledge about human development and mechanisms of disease, but it has also revealed the limitations of these same model systems in emulating human pathophysiology.
Box 1 Differences between human and mouse stem cells.
The difference between mice and humans is evident in the culturing of their stem cells. Pluripotent mouse embryonic stem cells (ESCs) were established in the early 1980s45,46. Leukaemia inhibitory factor (Lif) was the first pluripotent stem cell (PSC) factor identified, with the later discovery that 2i (the kinase inhibitors PD0325901 and CHIR99021) and Lif (together termed 2iLif) are necessary as well as sufficient for the culture of ESCs from all mouse strains176–179. The first human PSCs to be successfully cultured were human ESCs, equivalent to mouse epi-stem cells that have lost germ-cell potential44,180,181. Initial cultures were propagated in media that were based on basic fibroblast growth factor, but for decades researchers continued to search for appropriate culture conditions in which to maintain human PSCs in the naive, pluripotent state182–188. Universally agreed-upon culture conditions for the maintenance of naive human PSC cultures remain elusive, with various reports suggesting different medium compositions including the use of a PKC inhibitor (Gö6983), ROCK inhibitor (Y-27632), MEK inhibitor (PD98059), GSK inhibitor (IM12), BRAF inhibitor (SB590885), LCK/SRC inhibitor (WH-4-023) or Wnt5A182,184,189,190. A similar fundamental discrepancy between mouse and human cultures, in their requirements for additional media components, has also been observed in a number of adult stem cell (AdSC)-derived organoid culture systems. However, one commonality appears to be that inhibitors of TGFβ and p38 signalling must be added to the growth factors used in murine AdSC cultures in order to enable the long-term culture of human AdSCs9. For example, mouse colonic organoids require epidermal growth factor, Noggin, R-spondin and Wnt as growth factors, while human colonic organoids additionally require two inhibitors, A83-01 and SB202190 (which block TGFβ and p38 signalling, respectively)3, in order to attain a similar level of longevity and robustness. The differences between mouse and human systems in both PSC-derived and AdSC-derived organoid cultures suggest rather surprising fundamental differences between these species in the intrinsic signalling pathways that are activated in their stem cells and that are required for stem cell identity, and not just differences during development or during an immune response. This confirms the need for human-based laboratory model systems in order to fully understand human development and pathophysiology.
The need for human-cell-based models
A number of biological phenomena that are specific to humans are not amenable to being reproduced in animal models. The human brain, for example, is far more complex than its mouse counterpart, owing partly to human-specific developmental events and mechanisms28. Neurons in the human cortex, for example, arise from a cell type, outer radial glia, that is not present — or is present only in minute numbers — in rodents28. Human physiology is also profoundly different from the mouse model system: it is perhaps unsurprising that there are huge differences in metabolism between humans and laboratory models, given that humans develop far more slowly than the models29. Common drugs such as ibuprofen30 and warfarin31 are metabolized in the liver in such different manners in humans and rodents that, in humans, ibuprofen is prescribed for pain, fever and inflammation, and warfarin is prescribed as an anticoagulation drug, whereas both drugs are toxic to rats. Finally, and in contrast to all animal models, humans are not inbred. Understanding human genetic diversity and its influence on disease onset and progression and drug responses is a prerequisite for developing personalized medical treatments and requires the establishment of human-specific model systems.
The advent of human induced pluripotent stem cell (iPSC) technology and diverse human AdSC culture methods has made it possible, for the first time, to generate laboratory models specific to an individual32. Reprogramming other cells into iPSCs has become a routine laboratory procedure, but generating disease models from those cell lines remains challenging. Early approaches focused on the differentiation of iPSCs into one type of cell (for example, neurons and cardiomyocytes) and their culture in 2D33,34. More recently, culture methods have been developed to mimic in vivo organ development in 3D, allowing more complex tissue structures and diverse cell types to be modelled simultaneously1,9,35–39. In this methodology, human iPSCs are sequentially exposed to a course of differentiation cues in order to simulate the stages of a human developmental process. During this process, differentiated iPSCs aggregate to form first an organ bud, and later organoids, that faithfully mimic the mature organ structure, including multiple cell types and the interactions between them.
Human AdSC-derived organoids have also emerged as an alternative organoid system that consists of a simpler structure composed mainly of cell types found in the epithelium3. In contrast to the complicated process of iPSC reprogramming followed by differentiation to the required organ type, these organoids can be generated from biopsies isolated directly from the organ of interest or from diseased patient tissue. However, the establishment of human AdSC-derived organoids is limited by accessibility to the tissue and prior knowledge of the culture conditions for that tissue, while an iPSC line, once established from a patient, can be used to repeatedly generate different tissue models without any time limit (that is, beyond the patient’s lifespan)40.
In summary, organoids constitute an improvement in the generation of model systems, with cell types that more closely resemble, and are in similar conditions to, those found in the human body7,41. Diverse organoid systems provide a useful variety in their complexity and modelling capability, ranging from the simple epithelial structures derived from AdSCs3 to more complex cultures generated from multiple germ layers6 obtained via multistep PSC differentiation protocols42,43. It is expected that, in the near future, the variety of organoid models available, coupled with advances in the technology, will provide a series of powerful and efficient platforms for studying human development, physiology and disease.
Human model systems for human biology
The past years have seen substantial progress in the development and use of human-cell-based model systems for the study of human biology, including both pluripotent and AdSC-based systems.
The introduction of human iPSC technology
The first human PSCs, known as human ESCs, were reported in 1998 (ref.44), nearly two decades after the initial discovery of murine pluripotent cell lines in 1981 (refs45,46). The human stem cell lines were predicted to be useful for the study of human developmental biology, drug discovery and cell therapy — in spite of the technical limitations imposed by the lack of knowledge of appropriate differentiation protocols at that time. As the generation of human PSC lines required the sacrifice of human embryos at the blastocyst stage, strong ethical concerns were raised47.
In 2007 the debate around human PSCs was largely circumvented by the introduction of human iPSC technology, which converts an ordinary differentiated fibroblast from an adult human into a pluripotent cell33. Reprogramming to a pluripotent state is usually achieved by forced expression of a specific set of transcription factors33, and the pluripotent cells can then be differentiated into specific cell types. iPSC technology not only allowed the generation of patient-specific stem cells but also enabled researchers to work with an unlimited supply of human stem cells and stem cell-derived tissues. Furthermore, iPSC technology allows the banking of patient-derived stem cells48. There are still technical limitations when using iPSCs — for example, the use of oncogenes for reprogramming, the genetically unstable nature of the reprogramming and the low efficiency of reprogramming. All these technical hurdles made it difficult to obtain error-free iPSC lines from patients49. But these limitations have been at least partly overcome by the use of non-integrating vectors and standardized quality control protocols to avoid or screen out unwanted genetic alterations49. Line-to-line variability caused by the genetic heterogeneity of humans has also been resolved by the use of isogenic controls generated by genetic engineering using CRISPR–Cas9 technology50. These improvements have enabled researchers to use iPSC-derived specialized cell types — for example, neurons, cardiomyocytes, haematopoietic progenitor cells and pancreatic β-cells — in disease modelling and drug screening51. However, just over a decade after their introduction, 3D organ culture methods have greatly increased the utility of human iPSCs.
iPSC-derived organoid models
Human PSC-derived organoids are generated by guided differentiation protocols that mimic developmental processes identified through previous work, both in vitro and in vivo (Fig. 2). As our knowledge of human development is very limited, most early studies that aimed at producing organoids with properties similar to those of human tissues were based on parallels drawn from mouse development. In principle, to generate an organoid, the entire process of organ development from PSCs should be faithfully mimicked. In reality, it is nearly impossible in vitro to provide all biochemical cues that drive cell differentiation and 3D tissue assembly and organization at precisely the right times, places and concentrations at which they would occur during embryonic development. Fortunately, cells in vitro tend to follow a semi-autonomous differentiation trajectory, as they do in vivo, and three main types of protocols are utilized to generate functional organoids.
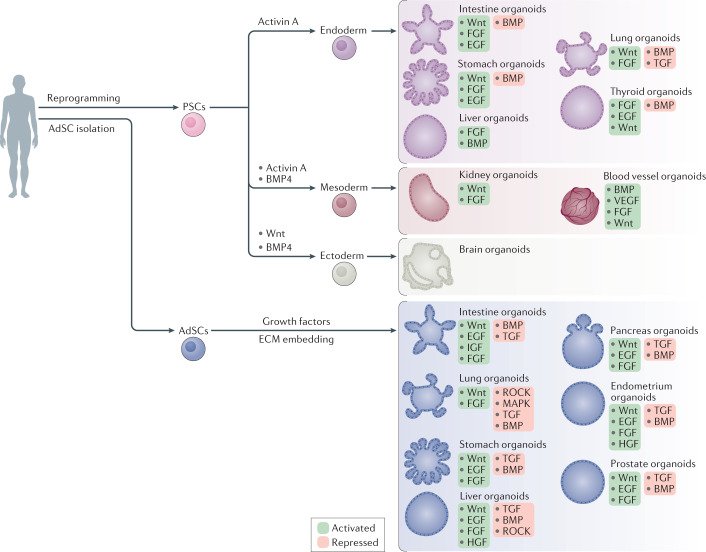
Fig. 2. Process for the establishment of human PSC-derived and AdSC-derived organoids.
Pluripotent stem cell (PSC)-derived organoids are established following directed differentiation of PSCs, which requires a first step that involves germ-layer specification (endoderm, mesoderm or ectoderm), followed by induction and maturation, by culturing with specific growth and signalling factors to obtain the specific cell types that form the desired organ. Some PSC-derived organoids may contain cells from multiple germ layers to closely mimic the in vivo counterpart. Adult stem cell (AdSC)-derived organoid cultures require isolation of the tissue-specific stem cell population, which can then be embedded into an extracellular matrix (ECM) with defined, tissue-specific combinations of growth factors to allow propagation. AdSC-derived organoids, as shown here, are of epithelial origin and lack a mesenchymal or immune component unless it is added separately. Signalling components that are important for guided differentiation and niche function are shown; activated signalling pathways are shown in green, and inhibited ones in red. BMP, bone morphogenetic protein; EGF, epidermal growth factor; FGF, fibroblast growth factors; HGF, hepatocyte growth factor; IGF, insulin-like growth factor; ROCK, RHO-associated protein kinase; TGF, transforming growth factor; VEGF, vascular endothelial growth factor.
In the case of brain organoids, human PSCs are initially guided to differentiate into embryoid bodies before further differentiation towards the neuroectodermal lineage. Once the cell aggregates contain the developmental precursors for brain tissue patterning, the rest of the developmental steps occur spontaneously in a spinning bioreactor5,52. For the development of liver primordia, guided differentiation to hepatoblasts (hepatocyte precursors) was not sufficient to form a complete set of organ precursors, which requires cells from different lineages. It was known from studies of mouse hepatogenesis that cell-to-cell communication between endothelial, mesenchymal and hepatic endoderm cells was important, so based on this knowledge the first human liver primordia were created by mixing human PSC-derived hepatoblasts, mesenchymal cells and endothelial cells. Through self-condensation and organization, these three cell types assemble in vitro to make an aggregate that mimics the architecture of developing human liver primordia23,53. Other organoids require more precise, lengthy protocols in order to acquire the appropriate progenitor cell types for the target epithelium. Many organoids that model endoderm-derived organs (such as oesophagus, stomach, colon, intestine and lung) undergo stepwise differentiation protocols, in which the timing, concentration and combination of specific growth factors and chemical inhibitors for modulating key developmental signalling pathways are crucial to developing the desired epithelial tissue in a manner analogous to fetal development24,43,54–57.
In all cases, the process of organoid formation involves three crucial steps. First, key signalling pathways regulating developmental patterning are activated or inhibited (using commercially available morphogens and signalling inhibitors) in order to establish a correct regional identity during stem cell differentiation. Usually this is achieved through induction of the signalling events that have been identified in the mouse as establishing cell fates in vivo. Second, media formulations that allow proper terminal differentiation of the desired cell types within the organoid are developed, generally following established methods of 2D culture or inspired by the murine developmental process. Finally, cultures are grown in a way that allows their expansion in three dimensions, which is achieved either by aggregating cells into 3D structures or by embedding the cultures into a 3D matrix.
Derivation of the first mouse and human intestinal organoids
AdSCs can be cultured in the presence of niche factors that function to maintain the stem cells in an undifferentiated state while allowing stem cell differentiation. As a result, the cultured stem cells can generate AdSC-derived organoids, which are composed of an epithelial monolayer that mimics the 3D architecture and contains the cell types of the desired organ to be modelled58. The first steps in the development of AdSC-derived organoids, however, are to identify and isolate the appropriate population of adult stem cells and to understand their niche requirements. Obtaining the first mouse intestinal organoid cultures required not only the identification of the intestinal stem cell population expressing the selective marker for these cells, the leucine-rich repeat-containing G protein-coupled receptor 5 (Lgr5), but also the essential niche factors to support stem cell activity59.
The re-creation of the intestinal stem cell niche in vitro was inspired by mouse genetic studies that had shown that epithelial proliferation and stem cell self-renewal are dependent on epidermal growth factor (EGF) and WNT activity, while differentiation is controlled by bone morphogenetic protein (BMP) signalling. Hence, the first culture medium that was successfully used to obtain mouse intestinal organoids contained EGF as a mitogen, R-spondin to activate the WNT signalling pathway and Noggin to block BMP activity2. However, the conditions identified as the minimum niche environment for the maintenance of mouse intestinal stem cells were not directly applicable to human intestinal or colonic organoid cultures. Unlike the Paneth cells that secrete some of the niche factors found in mouse intestinal organoid cultures, human gut organoid cultures are devoid of fully functional Wnt-secreting niche cells (for example, Paneth cells), and so require that exogenous Wnt3a be added to the medium. Moreover, long-term maintenance of human gut organoids through multiple passages requires both inhibition of the TGFβ pathway, by addition of the chemical inhibitor A83-01, and inhibition of the p38 MAPK pathway, by addition of SB202190 (ref.3). After it was discovered that SB202190 prevents secretory lineage differentiation, this inhibitor was replaced by the addition of insulin-like growth factor 1 (IGF1) and fibroblast growth factor 2 (FGF2)4; this modification enables concurrent multilineage differentiation and self-renewal in human gut organoids. All these changes and refinements to culture conditions, which are the results of another decade of research, improved the quality of human gut organoid cultures, enabling researchers to produce organoids that more closely resemble tissue in vivo. Nevertheless, mouse organoids remain more similar to the in vivo tissue of origin than do human organoids.
Generation of other AdSC-derived organoids
Using the procedure described above, multiple types of organoids from various tissues have been generated by modifying the basic medium that was initially developed for intestinal organoid culture (Fig. 2; Fig. 3). In most cases, a mouse organoid culture system was first established and then adapted to human cells. Mouse colonic organoids can be grown through the addition of Wnt3a3. Mouse stomach pyloric organoids and corpus organoids were grown with only slight modifications to the original protocol for mouse intestinal organoids60,61. Mouse liver and pancreas ductal organoids were also obtained similarly from injury-activated Lgr5-positive progenitors62,63.
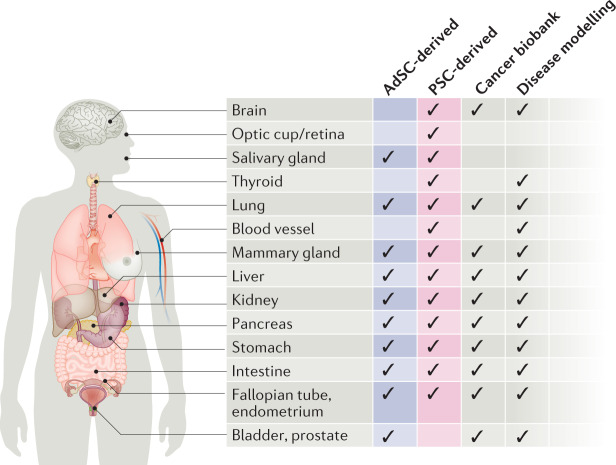
Fig. 3. Reported human organoid systems by tissue of origin.
The chart provides information on the type (either pluripotent stem cell (PSC)-derived or adult stem cell (AdSC)-derived), biobanking status and use in disease modelling of the human organoid systems reported to date, summarized by organ.
The number of organoids generated from diverse mouse tissues and organs is growing, and for each, previous knowledge of the signalling processes that comprise the organ-specific or tissue-specific stem cell niche environment has been key to developing the appropriate protocols. AdSC-derived human organoids have also become widely available (Fig. 3). They have been generated from almost all endoderm-derived tissues (intestine, colon, stomach, liver, pancreas, lung, bladder and so forth)3,7,41,64–70 and from gender-specific tissues (prostate, endometrium, fallopian tube and mammary gland)8,71–75. Additional components frequently required for the growth of human cultures, in comparison to the mouse cultures, are a TGFβ pathway inhibitor (such as A83-01) and a p38 MAPK inhibitor (such as SB202190)9. As for mouse organoids, human organoids can be derived from minimal amounts of tissue biopsies and can be cultured indefinitely, thus forming the basis for the building of living biobanks, which are an important resource in biomedical research.
Biomedical applications
Following the establishment of human stem cell-based organoids, various human diseases have been modelled using these systems. For example, human organoids have been used to study infectious diseases, inheritable genetic disorders and cancer. Moreover, with the advent of various genetic-engineering tools, pathogenic genes and mutations can be directly tested in organoids derived from cells isolated from healthy donors, which enables us to perform human genetic studies in controlled genetic backgrounds.
Human brain development and Zika virus
The human brain is the most highly developed brain in the animal kingdom and the most complex organ in our body. Because of its distinct complexity, it has been very difficult to model human brain development using animal systems. Human brain organoids, however, have now been successfully used to model human brain development and disease. The first disease studied in this model was microcephaly; unlike rodent models, human brain organoids with a mutation in CDK5RAP2 display a significantly reduced size, with a smaller number of progenitors that undergo premature neurogenesis5. CDK5RAP2 is localized at the centrosome and is required for correct orientation of the mitotic spindle, so that a mis-orientation of the cleavage plane prevents the expansion of the progenitor pool by symmetric division in the patient-derived organoids.
This human experimental system was also crucial in determining the relationship between the Zika virus (ZIKV) and microcephaly. During the recent ZIKV outbreak, a link was initially suggested between neurological disorders, including microcephaly, and ZIKV76–79. Nevertheless, even though ZIKV could be detected in microcephalic fetal tissues, the causal relationship and the mechanism by which ZIKV might compromise brain development were not clear. A number of studies have used 3D human stem cell-derived systems, including neurosphere culture and brain organoid models, to reveal the effect of ZIKV infection on human brain development80,81. Thus, the human organoid model provides a physiologically relevant platform for studying ZIKV pathogenesis. Further studies with human brain organoids have determined that Brazilian ZIKV strains possess increased virulence as compared to an African strain82, and that the NS2A protein of ZIKV causes decreased neural stem cell proliferation83. Various chemical compounds that alleviate the hypomorphic effect of ZIKV have also been identified using the human brain organoid system84.
These studies highlight the importance of human brain organoids as a model system in understanding the pathology and mechanistic basis of genetic and infectious diseases, as well as in discovering potential therapeutic reagents.
Human AdSC-derived organoids in the study of infection biology
As exemplified by the ZIKV studies above, a human model system is preferable to animal models when studying infectious disease pathogenesis, because pathogens often have a narrow species or tissue tropism; that is, they infect only certain species, and sometimes only specific cell types. For decades there were not any methods to culture human noroviruses, which cause acute gastroenteritis and can be fatal in young children or in old or immunocompromised individuals85. It recently became possible to successfully grow norovirus in culture by using a human intestinal organoid-derived epithelial monolayer, which led to the finding that certain viral strains specifically require both certain bile components and the activity of the host galactoside 2-alpha-l-fucosyltransferase 2 (FUT2) enzyme for infection86, which mirrors the patterns of epidemiological studies. Rotaviruses, which also cause diarrhoeal disease87, are another example of human intestinal organoids being used to cultivate patient-derived viral strains. The rapid replication of the virus was detected within one day of inoculation. An antiviral cytokine, a VP7-neutralizing antibody and ribavirin were demonstrated to suppress viral replication in the organoid system88,89.
Human airway organoids are suitable models for the study of respiratory viruses. Respiratory syncytial virus (RSV) is a single-stranded RNA virus that is a significant threat to young infants and the elderly41. It was shown to successfully infect human airway organoids and to cause dramatic changes to epithelial morphology and function: the NS2 protein of RSV was shown to cause increased motility of airway epithelial cells41. The human airway organoid system has also been used to assess the infectivity of animal-borne influenza viruses90–92. The diversity of two viral surface molecules — haemagglutinin and neuraminidase — has previously made it difficult to predict the infectivity of these viruses in humans93. The human airway organoids faithfully simulate human airway epithelium, and so provide a platform for rapidly testing the infectivity of newly emerging influenza viruses.
Organoids have also been used to co-cultivate human epithelia with bacteria (for example, Helicobacter pylori) and with protozoan parasites (for example, Cryptosporidium)64,94–96. The list of pathogens that can be grown in the surrogate human organ system is growing rapidly. We expect that, in the near future, numerous microbiome strains from the human gut or glands will be analysed by using organoids to study infectious diseases in a human cell context, and that organoid infection models will be used as screening platforms to identify novel drug candidates (Box 2).
Box 2 Human organoids and SARS-coronavirus 2.
Numerous epidemics and pandemics have threatened human health in history (of which a few examples are plague (Yersinia pestis), Spanish flu, smallpox, AIDS/HIV, Ebola virus, severe acute respiratory syndrome (SARS)-coronavirus (CoV) and Zika virus (ZIKV); WHO, Disease outbreaks), heavily disrupting human society. The emergence of the novel human coronavirus SARS-CoV-2 was first reported in Wuhan, China, at the end of 2019 (ref.191). SARS-CoV-2, reported to have crossed the species barrier, causes coronavirus disease-19 (COVID-19) in humans192. Owing to the lack of targeted antiviral medicines and a vaccine against COVID-19, the virus has spread globally and the WHO has declared COVID-19 a pandemic (WHO, Coronavirus disease (COVID-19) pandemic). Developing a vaccine and therapeutics against COVID-19 has become a priority, and human organoids can play a crucial role in advancing research, by providing human cells from different organs that are susceptible to SARS-CoV-2 infection. Several research groups have successfully shown that SARS-CoV-2 can infect and propagate in primary human organoids derived from liver and gut, as well as in pluripotent stem cell-derived organoids modelling blood vessels and the kidney193–195. SARS-CoV-2 uses the same protein, viral receptor angiotensin-converting enzyme 2 (ACE2), as SARS-CoV to enter the host cells. ACE2 is expressed in multiple tissues and cell types, enabling the virus to infect other tissues beyond the lung196–199. The ability of SARS-CoV-2 to infect different organs was modelled with human organoids193–195. Moreover, it has been shown that the viral infection of human blood vessel and kidney organoids can be inhibited by human recombinant soluble ACE2, a drug candidate initially developed for SARS-CoV194. Structural studies of the spike proteins of SARS-CoV-2 and SARS-CoV bound to ACE2 predicted a similar result200–202. These studies have provided additional evidence that human organoids can serve as an effective research platform for the study of human diseases, including the outbreak of new viruses such as SARS-CoV-2 and ZIKV.
Organoids for cystic fibrosis and the living cancer atlas
Human organoids are important resources for precision medicine. For example, they can be useful for selecting an appropriate drug for patients with genetic diseases or cancer. Cystic fibrosis (CF) is a relatively common genetic disease with approximately 90,000 patients worldwide. Although loss of function of the CFTR gene, encoding a chloride channel regulating the mucosal environment, is the main cause, nearly 2,000 mutations have been reported that affect the function or expression of CFTR, making this a genetically heterogeneous disease97. While two drugs — VX-770 (a CFTR potentiator that improves chloride-channel activity) and VX-809 (a CFTR corrector that helps mutant protein folding) — are already available for patients with CF who carry specific CFTR mutations in combination with the common mutation F508del, evaluating the efficacy of these drugs or discovering new drugs for patients with different mutations remains challenging. Rectal organoids isolated from small endoscopic biopsies have been used in forskolin-induced swelling (FIS) assays98–100 to assess forskolin–cAMP signalling-induced CFTR channel activity, and it was found that the assay can faithfully predict patient responses to individual drugs and to combined treatments99,100. It was also found that with this relatively simple in vitro assay it is possible to identify which patients with rare mutations would respond to, and could therefore be treated with, existing therapies99,100, which could be life-changing for those individuals. The rectal-organoid-based FIS assay screen is currently being performed by Hubrecht Organoid Technology for patients in the Netherlands with CF.
Human organoids also play a prominent role in enhancing our understanding of human cancers. Traditionally, human cancer cell lines, mouse cancer models or PDX have been the main experimental platforms for studies of human cancer. However, with the advent of AdSC-derived organoid technology, transformed cancer tissues have been grown in vitro as cancer organoids (also known as tumouroids or canceroids), demonstrating that this technology is applicable to diseased tissue as well as to normal epithelia. Patient samples from colon101–105, brain106,107, prostate108, pancreas109–111, liver112, breast113, bladder68, stomach114–116, oesophageal117, endometrial118 and lung41,119 cancers have readily been cultured as cancer organoid models. Recently, an analysis of drug responses in patients and in their matched cancer organoids led to the conclusion that responses to the drugs are highly similar in the two settings: a drug with no antitumour activity in the organoids did not demonstrate efficacy in the matched patient, and drugs that showed an effect in the organoid cultures were matched by a patient response in close to 90% of cases120. This initial study has been corroborated by several studies with larger cohorts105,121,122. The sample size and cancer types analysed remain limited, and more rigorous investigation will be needed before cancer organoids can be routinely adopted as in vitro patient ‘avatars’. Nevertheless, the rapidly increasing number of patient-derived cancer organoids and their use in xenograft formation and molecular profiling have already accelerated cancer research, and patient-derived organoid models in the future may provide an in vitro screening platform to predict the best therapeutic options for individual patients.
Genetic engineering in human cells
In 1987 Mario Capecchi reported highly frequent homologous recombination events in mouse ESCs123. By contrast, homologous recombination was found to be an extremely rare event in human PSCs124. Later studies reported that double-strand breaks (DSBs) facilitate homologous recombination events as part of the DNA repair mechanism in human cell lines125. Much effort has been directed at establishing an efficient method for generating DSBs at a specific target locus in order to harness this repair machinery in introducing desired genetic changes such as repair sequence or pathogenic mutation into the target locus. This was initially accomplished by the use of meganucleases (endonucleases with a long recognition site (12–40 bps)), zinc finger nucleases126 and transcription activator-like effector nucleases127, but with varying levels of target specificity and activity. The development of the CRISPR−Cas9 endonuclease technology has made diverse methods of genetic engineering readily available to all researchers128–131. Unlike the previous technologies, the Cas9 endonuclease is guided to the genomic sequence of interest as a means to generate a DSB by a guide RNA sequence (gRNA), making the system highly versatile and easy to apply132.
There are numerous examples of genetic engineering of human PSCs using this system, including the generation of isogenic cell lines with specific mutations. Such isogenic cell lines serve as an important control for genetic analysis in human PSCs, due to the strong phenotypical variation among different cell lines. Furthermore, large-scale loss-of-function analyses have been performed in many human cancer cell lines and human PSCs through the multiplexing of gRNAs in retroviral or lentiviral vectors, to provide genome-wide targeting coverage. Combining organoid systems with CRISPR–Cas9 genome editing broadens the applications of the organoid system in many ways. For example, this system has been used to model monogenic disorders such as CF. Human gut organoids with F508del, causing misfolded CFTR channel protein that leads to rapid degradation, were precisely corrected into the normal sequence by CRISPR–Cas9; the engineered organoids with the corrected CFTR amino acid sequence showed restored channel activity of CFTR in vitro133, showing the causal relationship between the mutation and disease phenotype, as well as the possibility of using a similar strategy to generate autologous organoids for transplantation. CRISPR–Cas9 technology has also been employed to identify and introduce oncogenic driver mutations to normal epithelial organoids. Two groups have independently reported the minimum set of mutations that can closely model a metastatic human colon cancer134,135. Simultaneous knockout of multiple genes or conditional gene knockout strategies have also been developed for AdSC-derived organoids and for PSCs that can be used for PSC-derived organoids136–138. CRISPR gRNA library screening analysis has been performed in multiple labs to maximize the utility of human organoids in genetic studies139,140. These developments in genetic engineering, combined with human PSC and human organoid technologies, have opened new opportunities for studies of human genetics, as it is possible to perform genetic experiments in small human organ models that closely reflect human physiology.
Opportunities and challenges
Organoids should largely be considered as a model system currently under development: much of the current excitement around the technology is based on their enormous potential rather than on what has already been achieved. Nevertheless, organoids could potentially revolutionize disease research in a profound manner (Fig. 4).
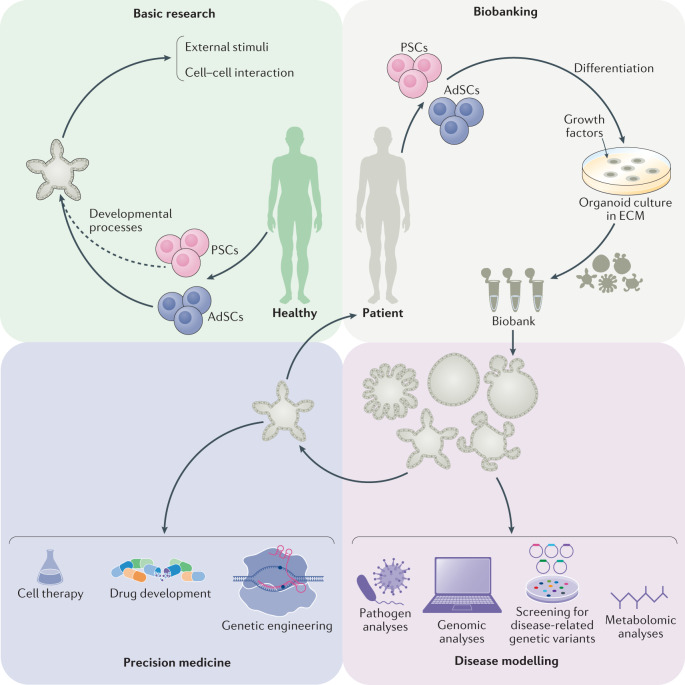
Fig. 4. Potential applications of human organoids.
Two types of organoids are widely used, derived from either pluripotent stem cells (PSCs) or adult stem cells (AdSCs). Organoids can be used for (1) basic research, including studies of human biology aiming to understand developmental processes, responses to external stimuli and stress signals, cell-to-cell interactions and mechanisms of stem cell homeostasis; (2) biobanking, whereby samples obtained from patients can be used to generate patient-derived organoids and stored as a resource for future research; (3) disease modelling, to understand the mechanisms of human diseases such as infectious diseases, inheritable genetic disorders and cancer using various laboratory techniques, including omics and drug-screening analyses; and (4) precision medicine, in which patient-derived organoids can be used to predict response to drugs and as resources for regenerative medicine coupled with genetic engineering. ECM, extracellular matrix.
Bridging the gap between animal models and humans
Generating animal models for a specific disease requires pre-existing insight into either the causative conditions or the genes involved. Animal models are typically created by applying harmful conditions to animals or by manipulating the genes responsible for the disorder, while organoid models can be directly generated from affected patients without prior knowledge of the specific genes responsible. This is particularly relevant for multigenic disorders such as inflammatory bowel disease, provided that the pathology is caused by the affected epithelium, and for cancers, where cancer organoids can be directly isolated from the patient141–146.
Human organoid cultures have a number of potential benefits over animal models (Box 3): organoids provide faster and more robust outcomes, are more readily accessible and provide both a more accurate representation of human tissue and a larger quantity of material to work with than animal models do. The mouse is one of the animal model systems most frequently used to explore human biology and disease, owing to its similarity to humans, in comparison with other animal models, and to the ability to generate transgenic and knockout mouse strains. However, the generation of a conventional transgenic mouse model to address questions regarding human disease generally takes more than a year, even with the technological advance of CRISPR–Cas9-mediated precision genome editing147–149. Furthermore, differences in microbiota and pathogen composition between animal models and humans, as well as the failure of certain phenomena observed in mice to translate directly to humans, limit the utility of animal models in human disease research150. Human stem cell-derived organoid culture is widely expected to bridge the remaining gaps between animal models and humans, primarily because the source material for organoid culture is a stem cell derived from a human35,40. The length of time required to establish the research platform is also faster for the organoid model than for animal models: a human organoid culture can be established within a few weeks or months with a high success rate, thus supporting the use of patient-derived organoids in personalized medicine to provide robust personalized data, including individual mutation profiles and drug responses. Organoid handling is also relatively easy, with researchers being able to handle a large number of organoid lines at the same time, although the initial determination of culture conditions for a new tissue type is rather complicated.
Box 3 Benefits and limitations of human organoids.
Benefits
Human-derived: human organoids represent human physiology, rather than being a ‘human-like’ or ‘similar’ system.
Rapid: adult stem cell-derived and pluripotent stem cell-derived organoids can be established rapidly and easily.
Robustness: once established, scale-up is usually possible for large-scale genomic screening and drug screening.
Genetic manipulation: most modern genetic engineering tools can be applied to induced pluripotent stem cells or directly to organoid systems.
Personalization: induced pluripotent stem cells and organoids can be obtained from individuals.
Limitations
Cellular components: the microenvironment is sometimes lacking, particularly in adult stem cell-derived organoids. Co-culture systems with other cell types are not firmly established.
Standardization: protocols for organoid establishment and quality control are not globally standardized.
Relatively costly: organoids cost less than mouse or fish models, but they are relatively expensive compared to traditional cell lines, fly, yeast or worm models.
Scale: studies at the level of whole organs are difficult.
Heterogeneity: owing to diversity between individuals and protocols, outcomes may vary from group to group.
Variability and single-cell profiling
In spite of the many advantages detailed above, human organoid culture remains under development, and numerous efforts to advance the technology are still in progress. One of the most pressing issues in organoid technology is the variability of the system: individual reports have described contrasting methods to generate organoids from stem cells, but a widely accepted, standardized protocol is still missing. For instance, human gut organoids derived in different laboratories, using human AdSCs and human PSCs, are different, even though both are termed ‘gut organoids’2,3,24. The possibility for further improvement of an already well-defined method was recently reported by showing that more refined culture conditions provide better cellular diversity and culture efficiency4. The lack of widely accepted, standardized protocols and guidance is an important issue to overcome in order to reduce the variability of the system from research group to group. A collective effort should be made to set clear guidelines and ways to assess the quality and validity of each system. Single-cell profiling technologies for transcriptome and epigenome analysis may be key in terms of highly accurate assays suitable for this purpose. With these assays, it would be possible to compare every cell type present in the organoids with their in vivo counterpart at the level of transcriptome and epigenome. In addition, individual differences such as age and genetic background might introduce further variability to the human organoid system, which, while potentially challenging, could also present an opportunity to assess the role of person-to-person variability in human biology.
Pros and cons of increasing cellular complexity
Additional considerations to be addressed include the modelling of cell-to-cell communication with stromal cell populations and the development of vasculature in organoid systems, although vascularization has been observed in some cases upon transplantation23,151. Significant progress has recently been achieved with blood vessel organoids152, which have a great potential to impact research into vascular diseases such as diabetes. However, despite steady progress, vascularization still remains a difficult obstacle to overcome.
Several reports already exist of organoid co-culture systems containing pathogens such as bacteria, viruses and parasites153,154, and moreover, organoid co-cultures with mesenchymal and/or immune cell populations have shown exciting progress in recent years and are of great interest as means to better model human disease23,155–162. Nevertheless, organoid systems already contain a large degree of complexity, and it is important to acknowledge the challenge of adding additional components to an already complex system. Both simple and complex organoid systems have their pros and cons, and therefore it is most important to use the most appropriate level of complexity for a given study. For example, cancer organoids containing only the epithelial component can be sufficient to test the efficacy of most cancer drugs. However, for immuno-oncology therapies or for an assessment of drug metabolism and availability to the target organ, more complex systems containing immune cells, mesenchymal cells and/or endothelial cells together with the organoids will be necessary. Unfortunately, a standardized protocol or guidelines concerning these matters are still lacking, so individual researchers are left to determine the most appropriate system for themselves. We foresee that full standardization of many of these protocols will be achieved in the near future, in a manner similar to increases in both the quality and reproducibility of human iPSC technology.
Assessing interactions with other organs and the environment
One clear drawback of organoid systems is the lack of interorgan communication. Human organoid systems fundamentally mimic a part of the human body, not the entire body. Therefore, human organoids are limited to the reproduction of organ-specific or tissue-specific microphysiology, a limitation to bear in mind prior to entering this exciting new field. However, efforts are already in progress to overcome this limitation. For example, multiple organoids have been connected in order to study communication between the liver, pancreas and gastrointestinal tract163; cell migration between the developing forebrain and hindbrain164–166; and the interaction between the brain and hormone-producing organs167. The development of tools to help us model organ-level communication will progress, although this capacity is likely to lag in comparison to progress in other areas of the field, owing to the complex nature of the studies undertaken. Of note, efforts to bring together the fields of organoid research and organ-on-a-chip research are particularly exciting, potentially resulting in an ‘organoid-on-a-chip’ technology168,169. We foresee, for example, the potential generation of a chamber device that enables the separate culture of distinct organoid types, thus preventing the uncontrolled fusion of organoids while permitting organoid–organoid communication.
Finally, the effect of extracellular matrix composition on organoid culture is yet to be defined; uncertainty in the composition of the extracellular matrix can heavily influence the outcomes in chemical screening or genetic screening of human organoids. Diverse efforts have been made, with some impressive successes in recent years170–175. This obstacle should be overcome not only as a means to produce more robust human model systems, but also to allow the translation of human organoid technology to regenerative medicine, where ‘good manufacturing practice’ requires all raw materials, including matrix materials, to be fully defined. Furthermore, work is ongoing towards the development of organoid culture platforms for large-scale production, organoid-based high-content screening platforms and micro-organoids-on-a-chip as miniature, finely controlled systems. For all these systems, it will be extremely important to know how to manufacture a synthetic, versatile extracellular matrix.
Conclusions
Despite the remaining challenges, human organoids hold great potential in clinical translational research, owing to the advantages outlined above and to rapid, ongoing technology development. From the initial full ‘laboratory life cycle’, which started with the isolation of patient samples, to the establishment of organoids and their cryopreservation, organoid technology has expanded to embrace genetic manipulation, various omics and drug-screening analyses and diverse co-culture system with viruses, bacteria and parasites (Fig. 4). Thus, technologies and experimental procedures that were developed in other model systems can now be applied to human organoid systems, which will accelerate our understanding of human biology and enable us to validate hypotheses and models generated from studies in animal model systems. Given the rapid technical advances in the field, we believe that human organoid systems will provide unprecedented opportunities to improve human health.
Acknowledgements
The authors thank A. Kavirayani and C. Hindley for proofreading, and S. Bae for initial design of the artwork. Work in J.A.K.’s laboratory is supported by the Austrian Academy of Sciences, the Austrian Science Fund (grant Z_153_B09) and an advanced grant from the European Research Council. B.-K.K.’s laboratory is supported by the Austrian Academy of Sciences, the European Research Council, the Human Frontier Science Program and the Interpark Bio-Convergence Center Grant Program.
Glossary
- Bio-printing
A printing technique to generate tissue-like structures using biomaterials such as cells.
- Adult stem cells
(AdSCs). Stem cells in the adult body that are important for maintaining organ homeostasis.
- Xenografts
Tissues or organs transplanted between different species.
- Outer radial glia
Part of neocortex where neuronal progenitors reside.
- Inbred
A breed with genetically closely related individuals.
- Induced pluripotent stem cell
(iPSC). Pluripotent stem cells reprogrammed from somatic cells.
- Isogenic controls
Control cell lines with the same genetic background as experimental cells.
- Embryoid bodies
Three-dimensional pluripotent stem cell aggregates.
- Spinning bioreactor
Culture system in which nutrients are supplied while cells are agitated.
- Niche factors
Growth factors provided by the surrounding environment of stem cells.
- Paneth cells
Epithelial cells that reside at the base of the intestinal crypt with secretory granules.
- Corpus
Middle part of the stomach, connected with cardia, fundus and pylorus.
- Living biobanks
Collection of biomaterials, such as cancer organoids, used for research.
- Neurosphere
Free-floating aggregates of neuronal stem cells.
- NS2A protein
Nonstructural protein 2A, encoded in a virus genome.
- Hypomorphic effect
An effect that causes small size of a body part.
- Tropism
The ability of a virus to infect various species and cells.
- Precision medicine
An approach to treating individual patients with the best possible drug, based on the individual’s genetic background and disease characteristics.
- Isogenic cell lines
Genetically identical cell lines that share a parental line.
- Multigenic disorders
Diseases caused by more than a single genetic factor.
Author contributions
The authors contributed equally to the writing and revisions of the article.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information
Nature Reviews Molecular Cell Biology thanks H. Clevers, P. Liberali and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
WHO, Disease outbreaks: www.who.int/emergencies/diseases/en/
WHO, Coronavirus disease 19 (Covid-19) pandemic: https://www.who.int/emergencies/diseases/novel-coronavirus-2019
Contributor Information
Bon-Kyoung Koo, Email: bonkyoung.koo@imba.oeaw.ac.at.
Juergen A. Knoblich, Email: juergen.knoblich@imba.oeaw.ac.at
References
- 1.Rossi G, Manfrin A, Lutolf MP. Progress and potential in organoid research. Nat. Rev. Genet. 2018;19:671–687. doi: 10.1038/s41576-018-0051-9. [DOI] [PubMed] [Google Scholar]
- 2.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 3.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 4.Fujii M, et al. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell. 2018;23:787–793. doi: 10.1016/j.stem.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 5.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takasato M, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564–568. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- 7.Hu H, et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell. 2018;175:1591–1606. doi: 10.1016/j.cell.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Turco MY, et al. Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat. Cell Biol. 2017;19:568–577. doi: 10.1038/ncb3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huch M, Koo BK. Modeling mouse and human development using organoid cultures. Development. 2015;142:3113–3125. doi: 10.1242/dev.118570. [DOI] [PubMed] [Google Scholar]
- 10.Simian M, Bissell MJ. Organoids: a historical perspective of thinking in three dimensions. J. Cell Biol. 2017;216:31–40. doi: 10.1083/jcb.201610056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 12.Kelava I, Lancaster MA. Dishing out mini-brains: current progress and future prospects in brain organoid research. Dev. Biol. 2016;420:199–209. doi: 10.1016/j.ydbio.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kretzschmar K, Clevers H. Organoids: modeling development and the stem cell niche in a dish. Dev. Cell. 2016;38:590–600. doi: 10.1016/j.devcel.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 15.Tiriac H, Plenker D, Baker LA, Tuveson DA. Organoid models for translational pancreatic cancer research. Curr. Opin. Genet. Dev. 2019;54:7–11. doi: 10.1016/j.gde.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 17.Kelava I, Lancaster MA. Stem cell models of human brain development. Cell Stem Cell. 2016;18:736–748. doi: 10.1016/j.stem.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 19.Mullins MC, Hammerschmidt M, Haffter P, Nusslein-Volhard C. Large-scale mutagenesis in the zebrafish: in search of genes controlling development in a vertebrate. Curr. Biol. 1994;4:189–202. doi: 10.1016/S0960-9822(00)00048-8. [DOI] [PubMed] [Google Scholar]
- 20.Haffter P, Nusslein-Volhard C. Large scale genetics in a small vertebrate, the zebrafish. Int. J. Dev. Biol. 1996;40:221–227. [PubMed] [Google Scholar]
- 21.Nusslein-Volhard C. The zebrafish issue of development. Development. 2012;139:4099–4103. doi: 10.1242/dev.085217. [DOI] [PubMed] [Google Scholar]
- 22.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takebe T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 24.Spence JR, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris TW, et al. WormBase: a multi-species resource for nematode biology and genomics. Nucleic Acids Res. 2004;32:D411–D417. doi: 10.1093/nar/gkh066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baumeister R, Ge L. The worm in us — Caenorhabditis elegans as a model of human disease. Trends Biotechnol. 2002;20:147–148. doi: 10.1016/S0167-7799(01)01925-4. [DOI] [PubMed] [Google Scholar]
- 27.Poulin G, Nandakumar R, Ahringer J. Genome-wide RNAi screens in Caenorhabditis elegans: impact on cancer research. Oncogene. 2004;23:8340–8345. doi: 10.1038/sj.onc.1208010. [DOI] [PubMed] [Google Scholar]
- 28.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuzawa CW, et al. Metabolic costs and evolutionary implications of human brain development. Proc. Natl Acad. Sci. USA. 2014;111:13010–13015. doi: 10.1073/pnas.1323099111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanoh S, et al. Predictability of metabolism of ibuprofen and naproxen using chimeric mice with human hepatocytes. Drug. Metab. Dispos. 2012;40:2267–2272. doi: 10.1124/dmd.112.047555. [DOI] [PubMed] [Google Scholar]
- 31.Inoue T, et al. CYP2C9-catalyzed metabolism of S-warfarin to 7-hydroxywarfarin in vivo and in vitro in chimeric mice with humanized liver. Drug. Metab. Dispos. 2008;36:2429–2433. doi: 10.1124/dmd.108.022830. [DOI] [PubMed] [Google Scholar]
- 32.McCauley HA, Wells JM. Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development. 2017;144:958–962. doi: 10.1242/dev.140731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 34.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 35.Fowler JL, Ang LT, Loh KM. A critical look: Challenges in differentiating human pluripotent stem cells into desired cell types and organoids. Wiley Interdiscip. Rev. Dev. Biol. 2019;9:e368. doi: 10.1002/wdev.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dutta D, Heo I, Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol. Med. 2017;23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 37.Marton RM, Pașca SP. Organoid and assembloid technologies for investigating cellular crosstalk in human brain development and disease. Trends Cell Biol. 2020;15:133–143. doi: 10.1016/j.tcb.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Nishinakamura R. Human kidney organoids: progress and remaining challenges. Nat. Rev. Nephrol. 2019;15:613–624. doi: 10.1038/s41581-019-0176-x. [DOI] [PubMed] [Google Scholar]
- 39.Prior N, Inacio P, Huch M. Liver organoids: from basic research to therapeutic applications. Gut. 2019;68:2228–2237. doi: 10.1136/gutjnl-2019-319256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lancaster MA, Huch M. Disease modelling in human organoids. Dis. Model Mech. 2019;12:dmm039347. doi: 10.1242/dmm.039347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sachs N, et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019;38:e100300. doi: 10.15252/embj.2018100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osakada F, Ikeda H, Sasai Y, Takahashi M. Stepwise differentiation of pluripotent stem cells into retinal cells. Nat. Protoc. 2009;4:811–824. doi: 10.1038/nprot.2009.51. [DOI] [PubMed] [Google Scholar]
- 43.McCracken KW, et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 45.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 46.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wert G, Mummery C. Human embryonic stem cells: research, ethics and policy. Hum. Reprod. 2003;18:672–682. doi: 10.1093/humrep/deg143. [DOI] [PubMed] [Google Scholar]
- 48.Huang CY, et al. Human iPSC banking: barriers and opportunities. J. Biomed. Sci. 2019;26:87. doi: 10.1186/s12929-019-0578-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu G, David BT, Trawczynski M, Fessler RG. Advances in pluripotent stem cells: history, mechanisms, technologies, and applications. Stem Cell Rev. Rep. 2020;16:3–32. doi: 10.1007/s12015-019-09935-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soldner F, Jaenisch R. Stem cells, genome editing, and the path to translational medicine. Cell. 2018;175:615–632. doi: 10.1016/j.cell.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Avior Y, Sagi I, Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Mol. Cell Biol. 2016;17:170–182. doi: 10.1038/nrm.2015.27. [DOI] [PubMed] [Google Scholar]
- 52.Lancaster MA, et al. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017;35:659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takebe T, et al. Generation of a vascularized and functional human liver from an iPSC-derived organ bud transplant. Nat. Protoc. 2014;9:396–409. doi: 10.1038/nprot.2014.020. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, et al. 3D modeling of esophageal development using human PSC-derived basal progenitors reveals a critical role for notch signaling. Cell Stem Cell. 2018;23:516–529. doi: 10.1016/j.stem.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trisno SL, et al. Esophageal organoids from human pluripotent stem cells delineate Sox2 functions during esophageal specification. Cell Stem Cell. 2018;23:501–515. doi: 10.1016/j.stem.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCracken KW, et al. Wnt/beta-catenin promotes gastric fundus specification in mice and humans. Nature. 2017;541:182–187. doi: 10.1038/nature21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dye BR, et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife. 2015;4:e05098. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barker N, van de Wetering M, Clevers H. The intestinal stem cell. Genes Dev. 2008;22:1856–1864. doi: 10.1101/gad.1674008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 60.Stange DE, et al. Differentiated Troy+chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 2013;155:357–368. doi: 10.1016/j.cell.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barker N, et al. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 62.Huch M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huch M, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bartfeld S, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–136. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schlaermann P, et al. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut. 2016;65:202–213. doi: 10.1136/gutjnl-2014-307949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huch M, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loomans CJM, et al. Expansion of adult human pancreatic tissue yields organoids harboring progenitor cells with endocrine differentiation potential. Stem Cell Rep. 2018;10:712–724. doi: 10.1016/j.stemcr.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee SH, et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell. 2018;173:515–528. doi: 10.1016/j.cell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rock JR, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl Acad. Sci. USA. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sampaziotis F, et al. Reconstruction of the mouse extrahepatic biliary tree using primary human extrahepatic cholangiocyte organoids. Nat. Med. 2017;23:954–963. doi: 10.1038/nm.4360. [DOI] [PubMed] [Google Scholar]
- 71.Boretto M, et al. Development of organoids from mouse and human endometrium showing endometrial epithelium physiology and long-term expandability. Development. 2017;144:1775–1786. doi: 10.1242/dev.148478. [DOI] [PubMed] [Google Scholar]
- 72.Linnemann JR, et al. Quantification of regenerative potential in primary human mammary epithelial cells. Development. 2015;142:3239–3251. doi: 10.1242/dev.123554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karthaus WR, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159:163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chua CW, et al. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat. Cell Biol. 2014;16:951–961. doi: 10.1038/ncb3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kessler M, et al. The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat. Commun. 2015;6:8989. doi: 10.1038/ncomms9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barkovich AJ, Guerrini R, Kuzniecky RI, Jackson GD, Dobyns WB. A developmental and genetic classification for malformations of cortical development: update 2012. Brain. 2012;135:1348–1369. doi: 10.1093/brain/aws019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heymann DL, et al. Zika virus and microcephaly: why is this situation a PHEIC? Lancet. 2016;387:719–721. doi: 10.1016/S0140-6736(16)00320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Calvet G, et al. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect. Dis. 2016;16:653–660. doi: 10.1016/S1473-3099(16)00095-5. [DOI] [PubMed] [Google Scholar]
- 79.Mlakar J, et al. Zika virus associated with microcephaly. N. Engl. J. Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 80.Dang J, et al. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell. 2016;19:258–265. doi: 10.1016/j.stem.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garcez PP, et al. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352:816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- 82.Cugola FR, et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534:267–271. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoon KJ, et al. Zika-virus-encoded NS2A disrupts mammalian cortical neurogenesis by degrading adherens junction proteins. Cell Stem Cell. 2017;21:349–358. doi: 10.1016/j.stem.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu M, et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med. 2016;22:1101–1107. doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ramani S, Atmar RL, Estes MK. Epidemiology of human noroviruses and updates on vaccine development. Curr. Opin. Gastroenterol. 2014;30:25–33. doi: 10.1097/MOG.0000000000000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ettayebi K, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rotavirus vaccines WHO position paper—January 2013. Wkly. Epidemiol. Rec. 2013;88:49–64. [PubMed] [Google Scholar]
- 88.Saxena K, et al. Human intestinal enteroids: a new model to study human Rotavirus infection, host restriction, and pathophysiology. J. Virol. 2016;90:43–56. doi: 10.1128/JVI.01930-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yin Y, et al. Modeling rotavirus infection and antiviral therapy using primary intestinal organoids. Antivir. Res. 2015;123:120–131. doi: 10.1016/j.antiviral.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 90.To KK, Chan JF, Chen H, Li L, Yuen KY. The emergence of influenza A H7N9 in human beings 16 years after influenza A H5N1: a tale of two cities. Lancet Infect. Dis. 2013;13:809–821. doi: 10.1016/S1473-3099(13)70167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou J, et al. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl Acad. Sci. USA. 2018;115:6822–6827. doi: 10.1073/pnas.1806308115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Klenk HD. Influenza viruses en route from birds to man. Cell Host Microbe. 2014;15:653–654. doi: 10.1016/j.chom.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 93.McAuley JL, Gilbertson BP, Trifkovic S, Brown LE, McKimm-Breschkin JL. Influenza virus neuraminidase structure and functions. Front. Microbiol. 2019;10:39. doi: 10.3389/fmicb.2019.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bartfeld S. Modeling infectious diseases and host–microbe interactions in gastrointestinal organoids. Dev. Biol. 2016;420:262–270. doi: 10.1016/j.ydbio.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 95.Leslie JL, et al. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect. Immun. 2015;83:138–145. doi: 10.1128/IAI.02561-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Heo I, et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 2018;3:814–823. doi: 10.1038/s41564-018-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rusnati M, et al. Recent strategic advances in CFTR drug discovery: an overview. Int. J. Mol. Sci. 2020;21:2407. doi: 10.3390/ijms21072407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dekkers JF, et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 99.Dekkers JF, et al. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci. Transl. Med. 2016;8:344ra384. doi: 10.1126/scitranslmed.aad8278. [DOI] [PubMed] [Google Scholar]
- 100.Berkers G, et al. Rectal organoids enable personalized treatment of cystic fibrosis. Cell Rep. 2019;26:1701–1708. doi: 10.1016/j.celrep.2019.01.068. [DOI] [PubMed] [Google Scholar]
- 101.van de Wetering M, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fujii M, et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18:827–838. doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 103.Weeber F, et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc. Natl Acad. Sci. USA. 2015;112:13308–13311. doi: 10.1073/pnas.1516689112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Engel RM, et al. Patient-derived colorectal cancer organoids upregulate revival stem cell marker genes following chemotherapeutic treatment. J. Clin. Med. 2020;9:128. doi: 10.3390/jcm9010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ooft SN, et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 2019;11:eaay2574. doi: 10.1126/scitranslmed.aay2574. [DOI] [PubMed] [Google Scholar]
- 106.Jacob F, et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180:188–204. doi: 10.1016/j.cell.2019.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fusco P, et al. Patient-derived organoids (PDOs) as a novel in vitro model for neuroblastoma tumours. BMC Cancer. 2019;19:970. doi: 10.1186/s12885-019-6149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gao D, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Boj SF, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Driehuis E, et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc. Natl. Acad. Sci. USA. 2019;116:26580–26590. doi: 10.1073/pnas.1911273116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Seino T, et al. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell. 2018;22:454–467. doi: 10.1016/j.stem.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 112.Broutier L, et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017;23:1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sachs N, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373–386. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 114.Seidlitz T, et al. Human gastric cancer modelling using organoids. Gut. 2019;68:207–217. doi: 10.1136/gutjnl-2017-314549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yan HHN, et al. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell. 2018;23:882–897. doi: 10.1016/j.stem.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 116.Nanki K, et al. Divergent routes toward Wnt and R-spondin niche independency during human gastric carcinogenesis. Cell. 2018;174:856–869. doi: 10.1016/j.cell.2018.07.027. [DOI] [PubMed] [Google Scholar]
- 117.Li X, et al. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nat. Commun. 2018;9:2983. doi: 10.1038/s41467-018-05190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Boretto M, et al. Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat. Cell Biol. 2019;21:1041–1051. doi: 10.1038/s41556-019-0360-z. [DOI] [PubMed] [Google Scholar]
- 119.Kim M, et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 2019;10:3991. doi: 10.1038/s41467-019-11867-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vlachogiannis G, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ganesh K, et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat. Med. 2019;25:1607–1614. doi: 10.1038/s41591-019-0584-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yao Y, et al. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell. 2020;26:17–26. doi: 10.1016/j.stem.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 123.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 124.Hockemeyer D, Jaenisch R. Gene targeting in human pluripotent cells. Cold Spring Harb. Symp. Quant. Biol. 2010;75:201–209. doi: 10.1101/sqb.2010.75.021. [DOI] [PubMed] [Google Scholar]
- 125.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 126.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 127.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 128.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 129.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 132.Pickar-Oliver A, Gersbach CA. The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019;20:490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schwank G, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 134.Drost J, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 135.Matano M, et al. Modeling colorectal cancer using CRISPR–Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 136.Andersson-Rolf A, et al. One-step generation of conditional and reversible gene knockouts. Nat. Methods. 2017;14:287–289. doi: 10.1038/nmeth.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Merenda A, et al. A protocol for multiple gene knockout in mouse small intestinal organoids using a CRISPR-concatemer. J. Vis. Exp. 2017;125:e55916. doi: 10.3791/55916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Andersson-Rolf A, et al. Simultaneous paralogue knockout using a CRISPR–concatemer in mouse small intestinal organoids. Dev. Biol. 2016;420:271–277. doi: 10.1016/j.ydbio.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Michels BE, et al. Pooled in vitro and in vivo CRISPR–Cas9 screening identifies tumor suppressors in human colon organoids. Cell Stem Cell. 2020;26:782–792. doi: 10.1016/j.stem.2020.04.003. [DOI] [PubMed] [Google Scholar]
- 140.Ringel T, et al. Genome-scale CRISPR screening in human intestinal organoids identifies drivers of TGF-beta resistance. Cell Stem Cell. 2020;26:e438. doi: 10.1016/j.stem.2020.02.007. [DOI] [PubMed] [Google Scholar]
- 141.Dotti I, et al. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut. 2017;66:2069–2079. doi: 10.1136/gutjnl-2016-312609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kraiczy J, et al. DNA methylation defines regional identity of human intestinal epithelial organoids and undergoes dynamic changes during development. Gut. 2019;68:49–61. doi: 10.1136/gutjnl-2017-314817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Suzuki K, et al. Single cell analysis of Crohn’s disease patient-derived small intestinal organoids reveals disease activity-dependent modification of stem cell properties. J. Gastroenterol. 2018;53:1035–1047. doi: 10.1007/s00535-018-1437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Howell KJ, et al. DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology. 2018;154:585–598. doi: 10.1053/j.gastro.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Drost J, Clevers H. Organoids in cancer research. Nat. Rev. Cancer. 2018;18:407–418. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- 146.Nanki K, et al. Somatic inflammatory gene mutations in human ulcerative colitis epithelium. Nature. 2020;577:254–259. doi: 10.1038/s41586-019-1844-5. [DOI] [PubMed] [Google Scholar]
- 147.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yang H, Wang H, Jaenisch R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat. Protoc. 2014;9:1956–1968. doi: 10.1038/nprot.2014.134. [DOI] [PubMed] [Google Scholar]
- 149.Yang H, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kostic AD, Howitt MR, Garrett WS. Exploring host-microbiota interactions in animal models and humans. Genes. Dev. 2013;27:701–718. doi: 10.1101/gad.212522.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Takebe T, et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell. 2015;16:556–565. doi: 10.1016/j.stem.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 152.Wimmer RA, et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature. 2019;565:505–510. doi: 10.1038/s41586-018-0858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bar-Ephraim, Y. E., Kretzschmar, K. & Clevers, H. Organoids in immunological research. Nat. Rev. Immunol. (2019). [DOI] [PubMed]
- 154.Kim J, Koo BK, Yoon KJ. Modeling host-virus interactions in viral infectious diseases using stem-cell-derived systems and CRISPR/Cas9 technology. Viruses. 2019;11:124. doi: 10.3390/v11020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Schreurs R, et al. Human fetal TNF-alpha-cytokine-producing CD4+ effector memory T cells promote intestinal development and mediate inflammation early in life. Immunity. 2019;50:462–476. doi: 10.1016/j.immuni.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 156.Neal JT, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–1988. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dijkstra KK, et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell. 2018;174:1586–1598. doi: 10.1016/j.cell.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Schnalzger TE, et al. 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J. 2019;38:e100928. doi: 10.15252/embj.2018100928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nozaki K, et al. Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes. J. Gastroenterol. 2016;51:206–213. doi: 10.1007/s00535-016-1170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Noel G, et al. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep. 2017;7:45270. doi: 10.1038/srep45270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Leeman KT, Pessina P, Lee JH, Kim CF. Mesenchymal stem cells increase alveolar differentiation in lung progenitor organoid cultures. Sci. Rep. 2019;9:6479. doi: 10.1038/s41598-019-42819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lee JH, et al. Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell. 2017;170:1149–1163. doi: 10.1016/j.cell.2017.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Koike H, et al. Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary. Nature. 2019;574:112–116. doi: 10.1038/s41586-019-1598-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bagley JA, Reumann D, Bian S, Levi-Strauss J, Knoblich JA. Fused cerebral organoids model interactions between brain regions. Nat. Methods. 2017;14:743–751. doi: 10.1038/nmeth.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Xiang Y, et al. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell. 2017;21:383–398. doi: 10.1016/j.stem.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Birey F, et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Adhya D, et al. Understanding the role of steroids in typical and atypical brain development: advantages of using a “brain in a dish” approach. J. Neuroendocrinol. 2018;30:e12547. doi: 10.1111/jne.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang C, Zhao Z, Abdul Rahim NA, van Noort D, Yu H. Towards a human-on-chip: culturing multiple cell types on a chip with compartmentalized microenvironments. Lab. Chip. 2009;9:3185–3192. doi: 10.1039/b915147h. [DOI] [PubMed] [Google Scholar]
- 169.Zhang YS, et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl Acad. Sci. USA. 2017;114:E2293–E2302. doi: 10.1073/pnas.1612906114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Giobbe GG, et al. Extracellular matrix hydrogel derived from decellularized tissues enables endodermal organoid culture. Nat. Commun. 2019;10:5658. doi: 10.1038/s41467-019-13605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jee JH, et al. Development of collagen-based 3D matrix for gastrointestinal tract-derived organoid culture. Stem Cell Int. 2019;2019:8472712. doi: 10.1155/2019/8472712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cruz-Acuna R, et al. PEG-4MAL hydrogels for human organoid generation, culture, and in vivo delivery. Nat. Protoc. 2018;13:2102–2119. doi: 10.1038/s41596-018-0036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ng S, Tan WJ, Pek MMX, Tan MH, Kurisawa M. Mechanically and chemically defined hydrogel matrices for patient-derived colorectal tumor organoid culture. Biomaterials. 2019;219:119400. doi: 10.1016/j.biomaterials.2019.119400. [DOI] [PubMed] [Google Scholar]
- 174.Gjorevski N, et al. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- 175.Broguiere N, et al. Growth of epithelial organoids in a defined hydrogel. Adv. Mater. 2018;30:e1801621. doi: 10.1002/adma.201801621. [DOI] [PubMed] [Google Scholar]
- 176.Smith AG, et al. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 177.Williams RL, et al. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 178.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nichols J, et al. Validated germline-competent embryonic stem cell lines from nonobese diabetic mice. Nat. Med. 2009;15:814–818. doi: 10.1038/nm.1996. [DOI] [PubMed] [Google Scholar]
- 180.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 181.Brons IG, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 182.Gafni O, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 183.Chan YS, et al. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell. 2013;13:663–675. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 184.Ware CB, et al. Derivation of naive human embryonic stem cells. Proc. Natl Acad. Sci. USA. 2014;111:4484–4489. doi: 10.1073/pnas.1319738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Theunissen TW, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:524–526. doi: 10.1016/j.stem.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Guo G, et al. Epigenetic resetting of human pluripotency. Development. 2017;144:2748–2763. doi: 10.1242/dev.146811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Guo G, et al. Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Rep. 2016;6:437–446. doi: 10.1016/j.stemcr.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Takashima Y, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Theunissen TW, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Van der Jeught M, et al. Application of small molecules favoring naive pluripotency during human embryonic stem cell derivation. Cell Reprogram. 2015;17:170–180. doi: 10.1089/cell.2014.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhao, B. et al. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell10.1007/s13238-020-00718-6 (2020). [DOI] [PMC free article] [PubMed]
- 194.Monteil V, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lamers MM, et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020 doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sungnak W, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Li W, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Walls AC, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yan R, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Shang J, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]