Abstract
Purpose
The standard treatment of patients with locally advanced rectal cancers comprises preoperative 5-fluorouracil-based chemoradiotherapy followed by standardized surgery. However, tumor response to multimodal treatment has varied greatly, ranging from complete resistance to complete pathologic regression. The prediction of the response is, therefore, an important clinical need.
Methods and Materials
To establish in vitro models for studying the molecular basis of this heterogeneous tumor response, we exposed 12 colorectal cancer cell lines to 3 μM of 5-fluorouracil and 2 Gy of radiation. The differences in treatment sensitivity were then correlated with the pretherapeutic gene expression profiles of these cell lines.
Results
We observed a heterogeneous response, with surviving fractions ranging from 0.28 to 0.81, closely recapitulating clinical reality. Using a linear model analysis, we identified 4,796 features whose expression levels correlated significantly with the sensitivity to chemoradiotherapy (Q <.05), including many genes involved in the mitogen-activated protein kinase signaling pathway or cell cycle genes. These data have suggested a potential relevance of the insulin and Wnt signaling pathways for treatment response, and we identified STAT3, RASSF1, DOK3, and ERBB2 as potential therapeutic targets. The microarray measurements were independently validated for a subset of these genes using real-time polymerase chain reactions.
Conclusion
We are the first to report a gene expression signature for the in vitro chemoradiosensitivity of colorectal cancer cells. We anticipate that this analysis will unveil molecular biomarkers predictive of the response of rectal cancers to chemoradiotherapy and enable the identification of genes that could serve as targets to sensitize a priori resistant primary tumors.
Keywords: Colorectal cancer, chemoradiotherapy, response, resistance, gene expression profiling
INTRODUCTION
Many features of primary tumors are recapitulated in derived cell lines. For instance, the genomic and transcriptomic aberration profiles that so dominantly define solid tumors are usually highly conserved (1, 2). This validates cell lines as in vitro models for functional analyses, and, accordingly, they have been widely used as model systems for target screening, drug discovery, and the determination of drug efficacy. The drug screening panel of the National Cancer Institute, for instance, has been used to correlate gene expression signatures and drug activity patterns (3) and to generate gene expression-based classifiers of drug sensitivity (4). More recent studies have demonstrated the value of cell culture systems for the prediction of radiosensitivity (5, 6) or the response to 5-fluorouracil (5-FU) (7).
Such attempts to predict in vitro responses are prompted by the thorny clinical problem of profoundly heterogeneous responses of primary tumors to therapeutic interventions. For locally advanced rectal cancer, preoperative 5-FU–based chemoradiotherapy represents the standard treatment; however, some tumors will have a complete pathologic response and others will be completely resistant (8, 9). The identification of biomarkers to predict the response to chemoradiotherapy is therefore urgently needed in clinical practice (10).
In the present study, we exposed colorectal cancer (CRC) cell lines to doses of both 5-FU and radiation similar to those used clinically. We hypothesized that identifying differentially expressed genes between the resistant and sensitive cell lines would point to gene expression signatures that could unveil relevant pathways involved in the heterogeneous treatment response. We also anticipated that the present analysis will enable the identification of novel target genes whose modification could be harnessed to sensitize a priori resistant primary tumors.
METHODS AND MATERIALS
Cell lines and cell culture
Twelve human CRC cell lines (Table 1) were obtained from the American Type Culture Collection (Manassas, VA), and cultured in their recommended medium (Invitrogen, Karlsruhe, Germany), supplemented with fetal bovine serum (Pan, Aidenbach, Germany) and 2 mM L-glutamine (BioWhittaker, Verviers, Belgium). Periodically, mycoplasma contamination was excluded by polymerase chain reaction (PCR) (11), and cell line cross-contamination was excluded by short tandem repeat profiling (12).
Table 1.
Characteristics of 12 colorectal cancer cell lines
| Cell Line | TP53 mutation | KRAS mutation | Doubling time (h) | SF2 |
|---|---|---|---|---|
| HT-29 | + | − | 22 | 0.81 |
| SW403 | + | + | 51 | 0.76 |
| SW837 | + | + | 64 | 0.71 |
| SW1116 | + | + | 54 | 0.68 |
| LS513 | − | + | 24 | 0.59 |
| LS1034 | + | + | 25 | 0.59 |
| Caco-2 | + | - | 38 | 0.58 |
| SW1463 | + | + | 60 | 0.56 |
| SW480 | + | + | 32 | 0.55 |
| SW620 | + | + | 28 | 0.46 |
| WiDr | + | − | 16 | 0.40 |
| LS411N | + | − | 27 | 0.28 |
Abbreviation: SF2 = surviving fraction after treatment with 3 μM of 5-fluorouracil and 2 Gy of radiation.
No correlation was found between chemoradiosensitivity and TP53 and KRAS mutation status or doubling time (data not shown).
5-FU–based chemoradiotherapy
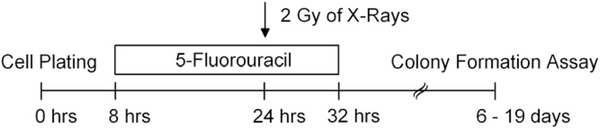
To estimate the sensitivity to chemoradiotherapy, tumor cells growing in log phase were seeded as single-cell suspensions into 6-well plates and allowed to adhere (Table e1). Before irradiation, the cells were exposed to 3 μM of 5-FU (Sigma, Steinheim, Germany). Sixteen hours later, the cells were irradiated with a single dose of 2 Gy of X-rays (200 kV, 15 mA, 0.5 mm Cu filter, Gulmay Medical, Camberley, United Kingdom). Drug treatment was stopped by a medium exchange, and colony formation was monitored as described in the next section. The experimental setup is shown in Fig. 1. In addition to chemoradiotherapy, we also tested the sensitivity of all cell lines to radiation alone (2 Gy).
Fig. 1.
Experimental setup. Twelve colorectal cell lines were treated with 3 μM of 5-fluorouracil, followed by radiation with 2 Gy.
No correlation was found between chemoradiosensitivity and TP53 and KRAS mutation status or doubling time (data not shown).
Determination of cell survival
To calculate the respective surviving fractions (SF) after treatment with 5-FU and radiation and with radiation alone, a standard colony-forming assay was performed, as previously described (13). The cells were treated for defined periods (Table e1), fixed with 70% ethanol, and stained with either crystal violet or Mayer’s hemalaun (Merck, Darmstadt, Germany). Nonirradiated cultures that were not exposed to 5-FU were used for normalization. Colonies with >50 cells were scored as survivors. The experiments were performed in triplicate, independently repeated three times, and calculated as the median.
RNA isolation
For each cell line, total RNA was isolated at three different passages (passages four to six after thawing), when the cells were approximately 60–70% confluent, using the RNeasy Mini kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. Using a 2100 Bioanalyzer analysis (Agilent Technologies, Palo Alto, CA), only samples with an RNA integrity number >9.5 were considered for additional experiments.
Gene expression profiling
Gene expression profiling was performed as previously described (14). In brief, 800 ng of total RNA was reverse transcribed, amplified, and labeled with Cy3 using the Low RNA Input Linear Amplification Kit PLUS (Agilent Technologies). Subsequently, 1.65 μg of labeled cRNA was fragmentized and hybridized overnight to a 4 × 44k gene expression microarray (Agilent Technologies). After a washing step, the arrays were scanned on an Agilent DNA microarray scanner G2505B (Agilent Technologies) at 5 micron resolution. The respective gene expression data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GSE20298).
Statistical analysis of gene expression data
The expression levels were analyzed using log2 transformation and quantile normalization (15). Except for control spots, all 43,376 features were used without any a priori filtering. Linear models, fitted on a gene-by-gene basis, were used to assess a linear correlation between the gene expression levels and the SFs at 2 Gy (SF2). We applied an empiric Bayes estimator (16) to compute the linear models for thousands of genes in parallel and assess their significance. To not exceed a false-discovery rate (Q value) of 5%, the p values were adjusted for multiple testing using the Benjamini-Hochberg method (17). Leave-one-out cross-validation was used to estimate the performance of selected genes for predicting the sensitivity to chemoradiotherapy. All analyses were performed using the free statistical software R (version 2.9.2; available from: www.r-project.org). Linear models were computed using the “limma” package (18).
Functional annotation and biologic pathway analysis
Gene lists were assessed for functional annotations and involvement in canonical pathways using Ingenuity Pathway Analysis (IPA) software (Ingenuity, Mountain View, CA). The significance of enriched pathways from the Kyoto Encyclopedia of Genes and Genomes database (available from: www.genome.jp/kegg/pathway.html) was established using a Wilcoxon rank test.
Real-time PCR
Total RNA (1 μg) was reverse transcribed in a 25-μL reaction volume into first-strand cDNA using Superscript III and random hexamers (Invitrogen). Next, 3 μL of cDNA mix was added to 12.5 μL of iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) and 375 ng primer solution to atotal of 25 μL/reaction. The amplification efficiency was assessed using LinRegPCR (19). The corresponding primer sequences can be found in Table e2. Real-time PCR analysis was performed in a Bio-Rad iCycler iQ5 (Bio-Rad, Munich, Germany) using the following cycling parameters: 10 min at 95°C, 40 cycles of 15 s at 95°C, and 1 min at 60°C. The resulting cycle threshold (Ct) values were normalized according to the mean of three housekeeping genes (i.e., FBXL12, HPRT1, and OTUB1). These genes were selected because their expression levels showed minimal variance within the 12 cell lines.
RESULTS
Sensitivity of CRC cell lines to chemoradiotherapy
To establish in vitro models to study the molecular basis of a heterogeneous tumor response to multimodal treatment, we exposed 12 CRC cell lines to 5-FU and radiation (Fig. 1). We chose a 5-FU concentration of 3 μM because it is similar to the serum concentration of patients treated with 5-FU–based chemoradiotherapy (unpublished data). However, to ensure that the tumor cells were still viable and proliferating at the point at which they were irradiated (Fig. 1), we first measured the cellular viability for all cell lines 16 hours after incubation with 3 μM of 5-FU, and cellular viability had not been impaired (data not shown).
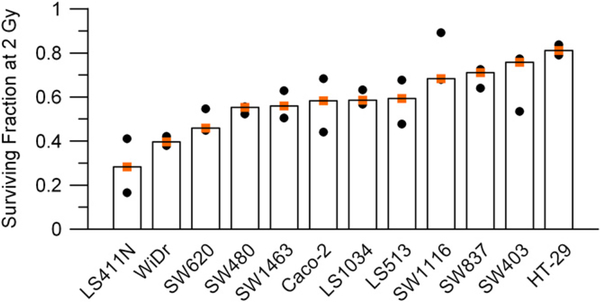
To establish the sensitivity to chemoradiotherapy, all cell lines were treated with 3 μM 5-FU and subsequently irradiated with 2 Gy, close to the clinically applied single dose of 1.8 Gy. We observed a heterogeneous response, recapitulating clinical reality. The most sensitive cell line was LS411N, with a SF2 of 0.28. The most resistant cell line, HT-29, had a SF2 of 0.81. The results for 5-FU–based chemoradiotherapy are displayed in Fig. 2. As shown in Fig. e1, the addition of 5-FU to radiation clearly changed the sensitivity of most cell lines compared to treatment with radiation alone. Hence, the sensitivity of the CRC cell lines is a combination of the sensitivity to both 5-FU and radiation.
Fig. 2.
Colorectal cancer cell lines showed a heterogeneous sensitivity to chemoradiotherapy. Respective surviving fractions are plotted after treatment with 3 μM of 5-fluorouracil and radiation at 2 Gy (SF2). Orange squares represent median values of three independent experiments.
A Gene expression signature for chemoradiosensitivity
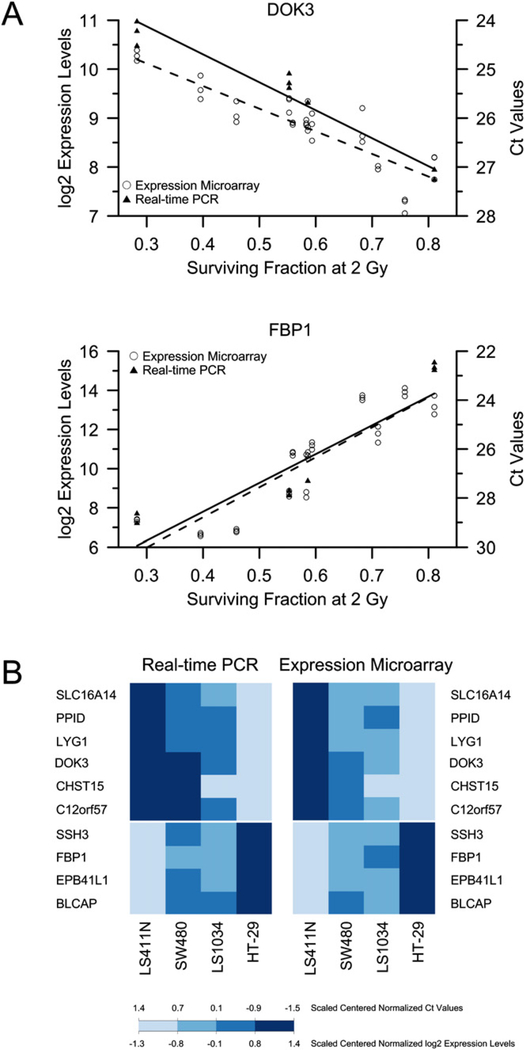
Next, we used genome-wide gene expression profiling to generate signatures of chemoradiosensitivity. The visualization of the gene expression levels using a correlation matrix showed that all biologic replicates clustered together, with the exception of one array (Caco-2), demonstrating the reliability of the experimental conditions (Fig. e2). To identify the genes whose expression levels correlated significantly with the sensitivity to chemoradiotherapy, we used a linear model analysis and identified 4,796 features that showed a correlation with chemoradiosensitivity at a significance level of Q <.05, corresponding to 2,770 genes (Table e3). A total of 2,065 features showed expression levels that increased with resistance to chemoradiotherapy, and 2,731 showed expression levels that decreased with resistance. The top 99 features are listed in Table 2, and their corressponding linear models are displayed in Fig. e3. Two examples of the linear model are provided in Fig. 3 a. Because the observed correlations between gene expression and chemoradiosensitivity were very high, only a very few genes would actually be required to accurately predict the respective SF2 (Fig. e4).
Table 2.
List of top 99 features*
| Rank | Gene Symbol | Slope | Q value | Rank | Gene symbol | Slope | Q value | Rank | Gene symbol | Slope | Q value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | FBP1 | 15.4 | 3.45E-09 | 34 | STK33 | −9.1 | 1.05E-06 | 67 | RHBDL2 | 10.7 | 7.53E-06 |
| 2 | DOK3 | −4.8 | 4.67E-09 | 35 | BCL2 | −4.0 | 1.31E-06 | 68 | MGC12935 | 5.1 | 7.62E-06 |
| 3 | LYG1 | −5.5 | 4.67E-09 | 36 | NFS1 | 3.0 | 1.43E-06 | 69 | OGFRL1 | −8.7 | 8.36E-06 |
| 4 | BLCAP | 4.3 | 4.67E-09 | 37 | C6orf204 | −2.1 | 1.43E-06 | 70 | POPDC2 | −2.1 | 8.39E-06 |
| 5 | EBP41L1 | 9.4 | 1.35E-08 | 38 | TUBE1 | −4.3 | 1.57E-06 | 71 | FAM111A | 3.5 | 8.39E-06 |
| 6 | SLC16A14 | −7.8 | 1.95E-08 | 39 | SAT1 | −5.8 | 1.59E-06 | 72 | STAT3 | 5.0 | 8.39E-06 |
| 7 | SSH3 | 3.7 | 2.02E-08 | 40 | MGLL | 12.1 | 1.61E-06 | 73 | STAT3 | 4.8 | 8.99E-06 |
| 8 | SLC16A14 | −7.5 | 2.02E-08 | 41 | DHX30 | −2.8 | 1.91E-06 | 74 | LONRF1 | −3.7 | 9.21E-06 |
| 9 | PPID | −4.3 | 9.91E-08 | 42 | BCL2 | −4.0 | 1.94E-06 | 75 | LRRC39 | −2.8 | 9.21E-06 |
| 10 | CHST15 | −7.7 | 1.07E-07 | 43 | TUSC1 | 11.9 | 1.96E-06 | 76 | ANKRD57 | 6.0 | 9.21E-06 |
| 11 | C12orf57 | −3.5 | 2.03E-07 | 44 | GBAS | −3.4 | 2.43E-06 | 77 | TTC33 | −3.5 | 9.21E-06 |
| 12 | EPB41L1 | 6.7 | 2.86E-07 | 45 | TGOLN2 | 3.8 | 2.54E-06 | 78 | RHBDL2 | 13.3 | 1.03E-05 |
| 13 | SAT1 | −6.4 | 3.77E-07 | 46 | AF074986 | 5.6 | 2.54E-06 | 79 | TCTN1 | −3.7 | 1.08E-05 |
| 14 | BCL2 | −4.1 | 3.91E-07 | 47 | MAN2A1 | 7.4 | 2.54E-06 | 80 | THC2611974 | −6.7 | 1.10E-05 |
| 15 | FARP2 | 3.4 | 3.91E-07 | 48 | CHMP4B | 3.4 | 2.59E-06 | 81 | ZNF586 | 3.5 | 1.10E-05 |
| 16 | SOBP | −8.6 | 3.96E-07 | 49 | HNRNPH1 | −3.5 | 2.90E-06 | 82 | MTL5 | −6.5 | 1.14E-05 |
| 17 | BCL2 | −3.8 | 4.83E-07 | 50 | THC2657554 | 3.5 | 2.90E-06 | 83 | THC2493066 | −3.4 | 1.15E-05 |
| 18 | NT5M | −5.6 | 4.83E-07 | 51 | ERLIN2 | 7.0 | 3.28E-06 | 84 | TUSC1 | 8.6 | 1.16E-05 |
| 19 | BCL2 | −4.0 | 4.83E-07 | 52 | PRDM16 | −8.8 | 3.35E-06 | 85 | DENND3 | 3.7 | 1.16E-05 |
| 20 | FARP2 | 3.0 | 4.83E-07 | 53 | PPP1R1A | −5.6 | 3.75E-06 | 86 | EPS8L2 | 4.5 | 1.16E-05 |
| 21 | TXNDC9 | 5.6 | 5.73E-07 | 54 | ALDH3B1 | 5.5 | 4.08E-06 | 87 | UBR4 | 3.3 | 1.16E-05 |
| 22 | RASSF1 | −3.2 | 6.20E-07 | 55 | DHX30 | −2.6 | 4.08E-06 | 88 | RHBDL2 | 11.9 | 1.21E-05 |
| 23 | THC2713710 | −7.4 | 6.90E-07 | 56 | POU6F1 | −4.3 | 4.08E-06 | 89 | BC027922 | 1.4 | 1.21E-05 |
| 24 | AK092508 | −2.8 | 7.08E-07 | 57 | CHAF1A | −2.8 | 4.21E-06 | 90 | BC035247 | 3.0 | 1.21E-05 |
| 25 | H2AFY2 | −10.3 | 7.08E-07 | 58 | TUG1 | 8.7 | 4.97E-06 | 91 | AK123861 | 3.3 | 1.21E-05 |
| 26 | BCL2 | −4.0 | 7.13E-07 | 59 | A_32_P108592 | 3.6 | 6.55E-06 | 92 | AV741130 | 11.2 | 1.22E-05 |
| 27 | BCL2 | −4.0 | 7.25E-07 | 60 | PARP14 | 4.7 | 6.55E-06 | 93 | WRN | −2.4 | 1.22E-05 |
| 28 | CASP10 | 3.3 | 7.86E-07 | 61 | RPAIN | −4.2 | 6.61E-06 | 94 | ZC3HAV1 | 4.9 | 1.28E-05 |
| 29 | BCL2 | −4.1 | 8.45E-07 | 62 | SH2B3 | −5.2 | 6.61E-06 | 95 | THC2656106 | −4.6 | 1.28E-05 |
| 30 | FBXL16 | −6.7 | 9.02E-07 | 63 | BCL2 | −3.5 | 6.61E-06 | 96 | STAT3 | 4.8 | 1.31E-05 |
| 31 | BCL2 | −4.0 | 9.38E-07 | 64 | ATP9A | 1.9 | 6.66E-06 | 97 | WRN | −2.4 | 1.32E-05 |
| 32 | ARNT2 | −12.4 | 1.00E-06 | 65 | PYCARD | 14.5 | 7.53E-06 | 98 | EOMES | −3.0 | 1.35E-05 |
| 33 | ATN1 | −2.4 | 1.05E-06 | 66 | MAN2A1 | 6.0 | 7.53E-06 | 99 | PPP1R3D | 3.8 | 1.43E-05 |
Top 99 features that showed significant linear correlation between their respective expression levels and sensitivity to chemoradiotherapy; a positive slope (correlation) indicates greater gene expression levels in resistant cell lines; a negative slope (correlation), lower gene expression levels.
Fig. 3.
(a) Linear model analysis of DOK3, which showed negative correlation between its expression levels and resistance to chemoradiotherapy, and FBP1, which showed positive correlation. For each cell line, microarray-based expression levels were plotted against respective surviving fractions at 2 Gy (SF2). Corresponding polymerase chain reaction (PCR)-based expression levels, measured in four cell lines, are also plotted. (b) Real-time PCR measurements nicely confirmed microarray-based gene expression measurements. Color-coded medians of gene expression levels of 10 genes in four cell lines are shown.
The cell lines were cultured in their recommended media (Table e1). To ensure that the sensitivity signatures were independent of the tissue culture medium, we grew three cell lines (i.e., SW480, SW837, and Caco-2) in their recommended medium and in Roswell Park Memorial Institute medium, and established gene expression signatures for each cell line specific for each culture medium. None of the genes of the “sensitivity signature” was differentially expressed at Q <.05 as a consequence of different media for Caco-2 and SW480. For SW837, only 171 features were differentially expressed (data not shown). Therefore, the effect of the tissue culture medium on the expression levels of genes from our “chemoradiosensitivity signature” was negligible.
Functional annotation of signature genes and pathway involvement
To interrogate the functional annotation of our signature genes, we used the IPA software. Focusing on the top 10 net-sssworks (as defined by the IPA), prominent networks centered around ERBB2 (Fig. 4a, IPA score 28), CCL5 and CDK2 (Fig. e5), STAT3 and ERK (Fig. 4b, IPA score 19), the NFkB complex and PI3K (Fig. 4c, IPA score 16), and MAPK14 (TP38) and WRN (Fig. 4d, IPA score 16). One network reflected TP53 signaling (Fig. e5). Other networks sigssssssnaled through CTNNB1, CASP1, and IgG (Fig. e5), FSH and LH (Fig. e5), RNA polymerase II, Histone H3, and Histone H4 (Fig. e5), and the E2F family and HNRNPA1 (Fig. e5). We then performed a pathway overrepresentation analysis to identify biologically annotated pathways using the Kyoto Encyclopedia of Genes and Genomes database. Potentially relevant pathways are listed in Table 3 and included the insulin, Wnt, and mitogen-activated protein kinase (MAPK) signaling pathways, cell cycle, apoptosis, and the spliceosome.
Fig. 4.
Ingenuity pathway analysis of genes correlated with chemoradiosensitivity. Red indicates genes with expression levels increasing with resistance to chemoradiotherapy; green, genes with expression levels decreasing with resistance.
Table 3.
Pathway analysis using Kyoto Encyclopedia of Genes and Genomes database
| Pathway | Pathway description | Pathway genes | Significant genes | Q value |
|---|---|---|---|---|
| PATH:hsa03040 | Spliceosome | 124 | 34 | 0.0002 |
| PATH:hsa04520 | Adherens junction | 75 | 16 | 0.0116 |
| PATH:hsa03018 | RNA degradation | 57 | 14 | 0.0116 |
| PATH:hsa05200 | Pathways in cancer | 325 | 60 | 0.0208 |
| PATH:hsa04310 | Wnt signaling | 148 | 28 | 0.0359 |
| PATH:hsa04110 | Cell cycle | 123 | 28 | 0.0388 |
| PATH:hsa00563 | GPI-anchor biosynthesis | 25 | 9 | 0.0578 |
| PATH:hsa05210 | Colorectal cancer | 82 | 13 | 0.0824 |
| PATH:hsa04622 | RIG-I-like receptor signaling | 66 | 18 | 0.0888 |
| PATH:hsa04530 | Tight junction | 127 | 26 | 0.0942 |
| PATH:hsa04910 | Insulin signaling | 133 | 25 | 0.0942 |
| PATH:hsa04144 | Endocytosis | 183 | 36 | 0.0942 |
| PATH:hsa00310 | Lysine degradation | 43 | 11 | 0.0942 |
| PATH:hsa00750 | Vitamin B6 metabolism | 6 | 3 | 0.0942 |
| PATH:hsa05416 | Viral myocarditis | 69 | 10 | 0.1101 |
| PATH:hsa05130 | Pathogenic Escherichia coli infection | 53 | 12 | 0.1101 |
| PATH:hsa04210 | Apoptosis | 85 | 16 | 0.1319 |
| PATH:hsa04010 | MAPK signaling | 262 | 54 | 0.1360 |
| PATH:hsa04912 | GnRH signaling | 97 | 19 | 0.1395 |
| PATH:hsa05220 | Chronic myeloid leukemia | 73 | 12 | 0.1395 |
| PATH:hsa00100 | Steroid biosynthesis | 17 | 5 | 0.1395 |
| PATH:hsa04662 | B cell receptor signaling | 74 | 14 | 0.1395 |
| PATH:hsa05322 | Systemic lupus erythematosus | 124 | 18 | 0.1487 |
Abbreviations: GPI = glycosylphosphatidylinositol; MAPK = mitogen-activated protein kinase; GnRH = gonadotropin-releasing hormone.
Real-time PCR validation
To independently validate the results obtained from our microarray platform, we selected the first 10 genes from our linear model analysis and measured their respective expressions levels in four cell lines using real-time PCR. As shown in Fig. 3b, the PCR results accurately confirmed the array-based measurements. The respective correlation coefficients for these genes ranged from 0.88 to 0.99 (Fig. e6), again demonstrating very robust and reproducible experimental conditions.
DISCUSSION
Patients with locally advanced rectal carcinomas benefit from preoperative combined modality therapy because local tumor control is significantly improved. Therefore, the standard treatment now includes 5-FU–based chemoradiotherapy followed by standardized surgery (8, 9). However, the heterogeneous tumor response and the lack of molecular markers to predict it remain major clinical problems.
Chemoradiosensitivity of CRC cell lines
To understand the molecular basis of the heterogeneous tumor response, we used cancer cell lines as a model system to identify the genes that contribute to this phenomenon. Toward this goal, we established the sensitivity of 12 CRC cell lines to chemoradiotherapy. Because of the limited number of immortalized cell lines from rectal cancers (SW837, SW1463), we included cell lines established from colon cancers, which exhibit analogous genomic and transcriptomic aberrations (20, 21). To recapitulate the clinical conditions, we chose doses of both chemotherapy and radiation that are close to those used for the treatment of cancer patients (i.e., 3 μM of 5-FU and 2 Gy of radiation). The sensitivity to chemoradiotherapy was measured using a colony formation assay, and the respective SFs ranged from 0.28 to 0.81 (Fig. 2). Accordingly, we observed a heterogeneous response, similar to the clinical situation. We then correlated the respective chemoradiosensitivities with the pretreatment gene expression profiles obtained from these cell lines. Because we observed a gradient in the sensitivity, the analysis of the differential gene expression levels was done using a linear regression model.
Signature for chemoradiosensitivity and involvement of pathways and networks
When we examined our list of sensitivity-correlated genes, we were reassured to find genes with a reported role for tumor progression and, occasionally, response to therapy. One of those genes that negatively correlated with chemoradioresistance was DOK3. DOK3 blocks the complex formation of GRB2 and SOS and, therefore, represents an inhibitor of the RAS signaling pathway (22). Although the relevance of KRAS mutations on the response of primary rectal cancers to preoperative chemoradiotherapy remains to be ultimately resolved (23, 24), our analysis has corroborated the interpretation that the deregulation of this pathway plays a prominent role for chemoradioresistance. This is supported by the manifold appearance of members of the RAS-MAPK cascade as signature genes (Table 2 and Table e3), through our pathway and network analyses that also showed enrichment (Fig. 4 and Fig. e3), and by recent publications that have provided functional evidence (25–27).
The RAS association domain family member 1 (RASSF1) also correlated negatively with chemoradioresistance. The loss of RASSF1 protein expression was recently demonstrated to correlate with a decreased responsiveness to DNA damaging therapy, and clonogenic survival of siRNA-transfected HeLa cells increased after radiation with 2 Gy compared with control cells (28).
One of the genes that correlated positively with chemoradioresistance was the signal transducer and activator of transcription 3 (STAT3). Through its cooperation with other transcription factors, STAT3 regulates the expression of a plethora of genes that mediate cellular proliferation and survival, including survivin, FOS, MYC, and Cyclin D1 (29). STAT3 mediates the survival of CRC cells in response to topoisomerase inhibition (30) but has not yet been associated with resistance to chemoradiotherapy. However, RNAi-mediated silencing of STAT3 resulted in radiosensitization of squamous carcinoma cells (31). Also, one of the top IPA networks was centered around STAT3 and ERK (Fig. 4b).
Another IPA network revealed ERBB2 as a focus gene (Fig. 4a). Although overexpression of ERBB2 has been correlated with chemoradioresistance of esophageal cancers (32), the potential involvement of ERBB2 in the resistance of rectal cancers to chemoradiotherapy is a novel finding.
We also identified the insulin signaling pathway as significantly deregulated (Table 3). The binding of insulin to its receptor induces receptor autophosphorylation and activation of PI3K and, subsequently, AKT (33). The insulin receptor was also capable of forming a hybrid receptor with the insulin-like growth factor 1 receptor. Preliminary evidence has suggested that RNAi-mediated silencing of insulin-like growth factor 1 receptor sensitizes colon cancer cells to radiation (34).
Surprisingly, the expression levels of many components of the spliceosomal complex correlated significantly with chemoradiosensitivity (Table 3). The spliceosome is involved in RNA splicing and has been very recently discovered as a novel anticancer target (35). Although, to the best of our knowledge, we are the first to report this novel association between the spliceosome and sensitivity to chemoradiotherapy, it remains to be elucidated whether this link is of functional relevance.
Finally, we observed a significant overrepresentation of genes involved in the Wnt signaling pathway in our “sensitivity signature” (Table 3). The Wnt signaling pathway plays a central role in colorectal tumorigenesis and interacts with several regulatory pathways for cell cycle, cell proliferation, and cell survival (36). Despite the notoriety of this pathway, its involvement in the resistance to chemoradiotherapy is a novel finding. Preliminary evidence has suggested, however, that activated Wnt/beta-catenin signaling mediates radiation resistance of mammary progenitor cells in mice (37).
Comparison with other signatures of sensitivity
We then compared our gene expression signature with the gene lists from other publications that sought to identify signatures of response to therapy (5, 6, 38, 39). Our own group had previously identified a set of 54 genes that were differentially expressed between primary rectal cancers that were either responsive or resistant to preoperative 5-FU–based chemoradiotherapy (38). Of these 54 genes, 35 were represented on our array platform; 12 of these 35 genes were also present in our gene list, and 6 genes were deregulated in the same direction, comparing primary tumors and cancer cell lines (Table e4). The incomplete overlap of both signatures could have been because of several reasons. In our previous analysis, we analyzed primary rectal cancers (38). It might, therefore, be possible that this signature was enriched by expression changes caused by the tumor stroma or the interaction of the tumor with its microenvironment. Additionally, we previously defined a response as downsizing of the T category of the primary tumor after preoperative chemoradiotherapy (38). In the present study, we analyzed the clonogenic survival of the irradiated tumor cells.
Although the present study is the first to establish a gene expression signature of in vitro sensitivity of CRC cell lines to a combination of 5-FU and radiation, two recent reports described the development of a gene expression-based model for the sensitivity to radiation alone. Eschrich et al. (5, 6) analyzed 48 cancer cell lines. They correlated the respective SF2 with gene expression profiles, KRAS and TP53 mutation status, and tissue of origin and extracted a network of 10 signature genes. One of those nine genes that were also represented on our array platform, STAT1, was also significantly correlated in our data set (Table e5).
From our viewpoint, three potential reasons exist for why our results might be more representative of the clinical situation. First, the panel of cell lines used by Eschrich et al. (5, 6) represented a mixture of tumor entities, not only cell lines derived from CRCs. Second, the seven CRC cell lines they analyzed included mismatch-repair proficient and mismatch-repair deficient cell lines. However, the tumors from which these cell lines were derived develop through pathways that are fundamentally different (40). Third, they correlated the gene expression signatures with sensitivity to radiation alone; however, the clinical treatment of rectal cancer patients comprises a combination of 5-FU and radiation.
These limitations also apply to the study performed by Amundson et al. (39), who correlated the gene expression profiles of the entire NCI-60 panel with their respective SFs at 2 Gy and 8 Gy and identified a set of 30 radiation-responsive genes. Twenty-seven of these genes were also represented on our array platform, but only Cathepsin D correlated with treatment sensitivity in our data set (Table e6).
Finally, it should be noted that survivin was not a part of our list. The expression of survivin has been previously shown by Rodel et al. (41–43) to correlate inversely with spontaneous and radiation-induced apoptosis, and they recently demonstrated that inhibition of survivin sensitizes CRC cells to radiotherapy.
CONCLUSION
We are the first to report a gene expression signature for in vitro chemoradiosensitivity of CRC cell lines. This signature now requires validation in primary rectal carcinomas. Our analysis has confirmed the prominent role of the MAPK signaling pathway and was enriched for cell cycle genes, and, importantly, revealed the potential relevance of the insulin and Wnt signaling pathways and the spliceosome for treatment response. Furthermore, we identified STAT3, RASSF1, DOK3, and ERBB2 as potential novel targets to sensitize resistant tumor cells.
Supplementary Material
Acknowledgments
The authors are grateful to Antje Schneeberg, Jessica Eggert, and Chan-Rong Lai for their excellent technical support. We also thank Reinhard Ebner for helpful discussions and critically reading our manuscript, Markus Schirmer for discussions on 5-fluorouracil, Gabriela Salinas-Riester and Lennart Opitz for help with microarray hybridizations, and Michael Klintschar for the short tandem repeat profiling analysis.
Supported by the Deutsche Forschungsgemeinschaft (KFO 179).
Footnotes
Conflict of interest: none.
REFERENCES
- 1.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006;10:515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solomon DA, Kim JS, Ressom HW, et al. Sample type bias in the analysis of cancer genomes. Cancer Res 2009;69:5630–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scherf U, Ross DT, Waltham M, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet 2000;24:236–244. [DOI] [PubMed] [Google Scholar]
- 4.Staunton JE, Slonim DK, Coller HA, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci USA 2001;98:10787–10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eschrich SA, Pramana J, Zhang H, et al. A gene expression model of intrinsic tumor radiosensitivity: Prediction of response and prognosis after chemoradiation. Int J Radiat Oncol Biol Phys 2009;75:489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eschrich S, Zhang H, Zhao H, et al. Systems biology modeling of the radiation sensitivity network: A biomarker discovery plat-form. Int J Radiat Oncol Biol Phys 2009;75:497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mariadason JM, Arango D, Shi Q, et al. Gene expression profiling-based prediction of response of colon carcinoma cells to 5-fluorouracil and camptothecin. Cancer Res 2003;63:8791–8812. [PubMed] [Google Scholar]
- 8.Sauer R, Becker H, Hohenberger W, et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med 2004;351:1731–1740. [DOI] [PubMed] [Google Scholar]
- 9.Minsky BD. Is preoperative chemoradiotherapy still the treatment of choice for rectal cancer? J Clin Oncol 2009;27:5115–5116. [DOI] [PubMed] [Google Scholar]
- 10.Kuremsky JG, Tepper JE, McLeod HL. Biomarkers for response to neoadjuvant chemoradiation for rectal cancer. Int J Radiat Oncol Biol Phys 2009;74:673–688. [DOI] [PubMed] [Google Scholar]
- 11.Uphoff CC, Drexler HG. Detection of mycoplasma contaminations in cell cultures by PCR analysis. Hum Cell 1999;12:229–236. [PubMed] [Google Scholar]
- 12.Masters JR, Thomson JA, Daly-Burns B, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc Natl Acad Sci USA 2001;98:8012–8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rave-Frank M, Schmidberger H, Christiansen H, et al. Comparison of the combined action of oxaliplatin or cisplatin and radiation in cervical and lung cancer cells. Int J Radiat Biol 2007; 83:41–47. [DOI] [PubMed] [Google Scholar]
- 14.Grade M, Hummon AB, Camps J, et al. A genomic strategy for the functional validation of colorectal cancer genes identifies potential therapeutic targets. Int J Cancer Epub 2010. May 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolstad BM, Irizarry RA, Astrand M, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003;19:185–193. [DOI] [PubMed] [Google Scholar]
- 16.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:Article3. [DOI] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc, Series B (Methodol) 1995;57:289–300. [Google Scholar]
- 18.Smyth GK. Limma: Linear Models for Microarray Data. New York: Springer; 2005. [Google Scholar]
- 19.Ramakers C, Ruijter JM, Deprez RH, et al. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 2003;339:62–66. [DOI] [PubMed] [Google Scholar]
- 20.Grade M, Ghadimi BM, Varma S, et al. Aneuploidy-dependent massive deregulation of the cellular transcriptome and apparent divergence of the Wnt/beta-catenin signaling pathway in human rectal carcinomas. Cancer Res 2006;66:267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grade M, Hormann P, Becker S, et al. Gene expression profiling reveals a massive, aneuploidy-dependent transcriptional deregulation and distinct differences between lymph node-negative and lymph node-positive colon carcinomas. Cancer Res 2007; 67:41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honma M, Higuchi O, Shirakata M, et al. Dok-3 sequesters Grb2 and inhibits the Ras-Erk pathway downstream of protein-tyrosine kinases. Genes Cells 2006;11:143–151. [DOI] [PubMed] [Google Scholar]
- 23.Zauber NP, Marotta SP, Berman E, et al. Molecular genetic changes associated with colorectal carcinogenesis are not prognostic for tumor regression following preoperative chemoradiation of rectal carcinoma. Int J Radiat Oncol Biol Phys 2009;74: 472–476. [DOI] [PubMed] [Google Scholar]
- 24.Gaedcke J, Grade M, Jung K, et al. KRAS and BRAF mutations in patients with rectal cancer treated with preoperative chemoradiotherapy. Radiother Oncol 2010;94:76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim IA, Bae SS, Fernandes A, et al. Selective inhibition of Ras, phosphoinositide 3 kinase, and Akt isoforms increases the radiosensitivity of human carcinoma cell lines. Cancer Res 2005;65:7902–7910. [DOI] [PubMed] [Google Scholar]
- 26.Oh JS, Kim JJ, Byun JY, et al. Lin28-let7 modulates radiosensitivity of human cancer cells with activation of K-Ras. Int J Radiat Oncol Biol Phys 2010;76:5–8. [DOI] [PubMed] [Google Scholar]
- 27.Dent P, Yacoub A, Fisher PB, et al. MAPK pathways in radiation responses. Oncogene 2003;22:5885–5896. [DOI] [PubMed] [Google Scholar]
- 28.Hamilton G, Yee KS, Scrace S, et al. ATM regulates a RASSF1A-dependent DNA damage response. Curr Biol 2009;19:2020–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aggarwal BB, Kunnumakkara AB, Harikumar KB, et al. Signal transducer and activator of transcription-3, inflammation, and cancer: how intimate is the relationship? Ann NY Acad Sci 2009;1171:59–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vigneron A, Gamelin E, Coqueret O. The EGFR-STAT3 oncogenic pathway up-regulates the Eme1 endonuclease to reduce DNA damage after topoisomerase I inhibition. Cancer Res 2008;68:815–825. [DOI] [PubMed] [Google Scholar]
- 31.Bonner JA, Trummell HQ, Willey CD, et al. Inhibition of STAT-3 results in radiosensitization of human squamous cell carcinoma. Radiother Oncol 2009;92:339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akamatsu M, Matsumoto T, Oka K, et al. c-erbB-2 oncoprotein expression related to chemoradioresistance in esophageal squamous cell carcinoma. Int J Radiat Oncol Biol Phys 2003;57: 1323–1327. [DOI] [PubMed] [Google Scholar]
- 33.Bevan P Insulin signaling. J Cell Sci 2001;114:1429–1430. [DOI] [PubMed] [Google Scholar]
- 34.Yavari K, Taghikhani M, Maragheh MG, et al. SiRNA-mediated IGF-1R inhibition sensitizes human colon cancer SW480 cells to radiation. Acta Oncol 2010;49:70–75. [DOI] [PubMed] [Google Scholar]
- 35.van Alphen RJ, Wiemer EA, Burger H, et al. The spliceosome as target for anticancer treatment. Br J Cancer 2009;100:228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moon RT, Kohn AD, De Ferrari GV, et al. WNT and betacatenin signaling: Diseases and therapies. Nat Rev Genet 2004;5:691–701. [DOI] [PubMed] [Google Scholar]
- 37.Woodward WA, Chen MS, Behbod F, et al. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci USA 2007;104:618–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghadimi BM, Grade M, Difilippantonio MJ, et al. Effectiveness of gene expression profiling for response prediction of rectal adenocarcinomas to preoperative chemoradiotherapy. J Clin Oncol 2005;23:1826–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amundson SA, Do KT, Vinikoor LC, et al. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res 2008;68:415–424. [DOI] [PubMed] [Google Scholar]
- 40.Walther A, Johnstone E, Swanton C, et al. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer 2009;9:489–499. [DOI] [PubMed] [Google Scholar]
- 41.Rodel C, Haas J, Groth A, et al. Spontaneous and radiation-induced apoptosis in colorectal carcinoma cells with different intrinsic radiosensitivities: Survivin as a radioresistance factor. Int J Radiat Oncol Biol Phys 2003;55:1341–1347. [DOI] [PubMed] [Google Scholar]
- 42.Rodel F, Hoffmann J, Distel L, et al. Survivin as a radioresistance factor; and prognostic and therapeutic target for radiotherapy in rectal cancer. Cancer Res 2005;65:4881–4887. [DOI] [PubMed] [Google Scholar]
- 43.Rodel F, Frey B, Leitmann W, et al. Survivin antisense oligonucleotides effectively radiosensitize colorectal cancer cells in both tissue culture and murine xenograft models. Int J Radiat Oncol Biol Phys 2008;71:247–255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.