Abstract
Maternal antibodies provide short-term protection to infants against many infections. However, they can inhibit de novo antibody responses in infants elicited by infections or vaccination, leading to increased long-term susceptibility to infectious diseases. Thus, there is a need to develop vaccines that are able to elicit protective immune responses in the presence of antigen-specific maternal antibodies. Here, we used a mouse model to demonstrate that influenza virus–specific maternal antibodies inhibited de novo antibody responses in mouse pups elicited by influenza virus infection or administration of conventional influenza vaccines. We found that a recently developed influenza vaccine, nucleoside-modified mRNA encapsulated in lipid nanoparticles (mRNA-LNP), partially overcame this inhibition by maternal antibodies. The mRNA-LNP influenza vaccine established long-lived germinal centers in the mouse pups and elicited stronger antibody responses than did a conventional influenza vaccine approved for use in humans. Vaccination with mRNA-LNP vaccines may offer a promising strategy for generating robust immune responses in infants in the presence of maternal antibodies.
INTRODUCTION
Infections account for more than 2 million infant deaths each year; however, we lack effective vaccines against many pathogens for this population (1). In particular, infants have the highest rate of hospitalization for severe lower respiratory tract infections (2), yet there are no licensed influenza vaccines for children under 6 months old. Maternal antibodies (matAbs) can effectively protect infants against pathogens early in life (3–7). For example, vaccination of mothers during pregnancy with inactivated influenza vaccine confers a 50% reduction in laboratory-confirmed influenza virus infections in their infants (8). However, matAbs can also inhibit de novo antibody responses in infants elicited by infections or vaccinations. Such inhibition is well established for measles and pertussis vaccines (9, 10), and some evidence suggests the same effect for influenza vaccines in mice (11–13) and humans (14). A recent meta-analysis of serological data from randomized clinical trials suggested that preexisting maternal antibodies inhibited infants’ vaccine responses for 20 of the 21 antigens studied (9). Many vaccination strategies rely on delayed vaccination or multiple booster doses to elicit protective responses in vaccinees who have high titers of matAbs. For example, the measles vaccine is administered at 12 to 15 months of age, in part, because of interference by matAbs (15).
Better vaccine strategies need to be developed to overcome matAb inhibition of infant immune responses. Vaccines with viral vector delivery of antigens have successfully elicited de novo antibody responses in poultry in the presence of matAbs (16–19) but have been less successful in mammals (20–22). DNA vaccines have also shown varying degrees of success in the presence of matAbs in some studies (20, 23–27) but were unsuccessful in others (12, 13, 28–31). Thus, eliciting protective antibody responses in the presence of matAbs remains a challenge. To address this, we established an influenza virus matAb mouse model, and we used this mouse model to identify a vaccine platform that elicited de novo influenza virus antibodies in mouse pups in the presence of influenza virus–specific matAbs.
RESULTS
Establishment of a mouse model of matAbs against influenza virus
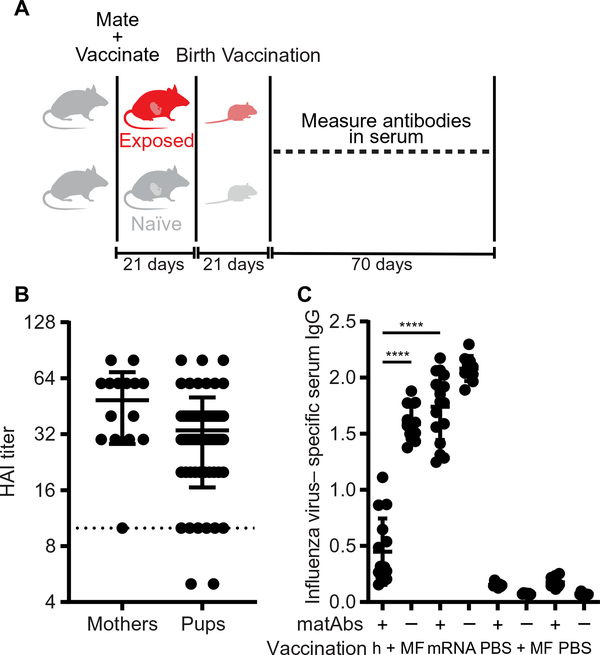
To examine the effect of matAbs on infant responses to influenza virus infection, we established a mouse model in which we intranasally infected adult female BALB/c mice with a subclinical dose of the A/Puerto Rico/8/1934 (PR8) H1N1 influenza virus strain. These mice cleared the infection and mounted an antibody response, which was confirmed using a hemagglutination inhibition assay (HAI). We then mated these mice along with unexposed control female mice to males and let them deliver pups (Fig. 1A). In mouse dams, matAbs are transferred to offspring both in utero and in milk (4, 32). To verify that matAbs were transferred to the pups, we collected serum from the mothers and pups at weaning (~21 days old) and measured influenza virus–specific serum IgG titers by enzyme-linked immunosorbent assay (ELISA) (Fig. 1B) and HAI (Fig. 1C). Female mice efficiently transferred influenza virus–specific antibodies to their pups as most offspring:mother pairs had a ratio of ~1 (mean, 1.03; range, 0.65 to 1.81) (fig. S1A). Efficiency of antibody transfer was not related to the number of pups in the litter (fig. S1B), the mother’s age (fig. S1C), or pregnancy history because all mothers had only a single litter. After weaning, matAbs in the pups waned over time. Consistent with previous reports (33, 34), serum influenza virus–specific IgG decreased over time by exponential decay with a half-life of 12 ± 2 days (Fig. 1D).
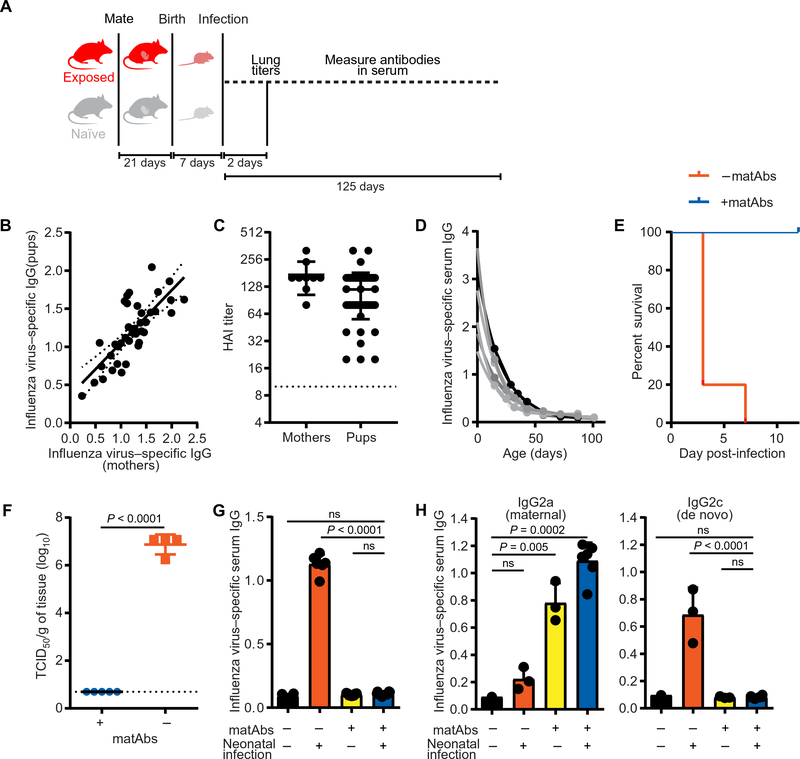
Fig. 1. matAbs protect mouse pups from influenza disease but inhibit de novo antibody responses.
(A) The experimental design is shown. (B and C) Serum was collected from mothers and mouse pups on the day of weaning, and influenza virus–specific antibody titers were measured by ELISA (B) or hemagglutination inhibition assay (C). Each point represents one litter (1 to 10 pups per litter; mean, 5.3) (B) or one mouse (C). In (B), maternal antibody titers are shown on the x axis, and pup titers are shown on the y axis; dotted line, 95% confidence interval (CI) (R2 = 0.58). (D) Serum was collected at the indicated time points from pups with influenza virus–specific matAbs, and influenza virus–specific ELISA titers were measured. One-phase decay (R2 > 0.97 for each mouse) was fitted to titer data; each line represents one mouse (n = 6 mice). (E and F) Seven-day-old mice with or without influenza virus–specific matAbs were intranasally inoculated with a 30 tissue culture infectious dose (TCID)50 of PR8 influenza virus. (E) Survival was measured over 14 days post-inoculation. Mouse groups were n = 3 (+matAbs) or n = 5 (−matAbs). P = 0.008, log-rank test. (F) Influenza virus titers in the lungs of pups were measured 2 days post-inoculation. Each point represents one mouse; n = 4 (+matAbs) or n = 5 (−matAbs) mice per group; P < 0.0001, two-tailed Welch’s t test. (G) Seven-day-old mice were intranasally inoculated with influenza virus or PBS in the presence or absence of matAbs. Serum influenza virus–specific IgG was measured by ELISA 125 days post-inoculation. n = 6 (black and orange), n = 8 (blue), or n = 9 (yellow) mice per group. Groups were compared using one-way ANOVA with Tukey’s post hoc test. (H) C57BL/6 mouse pups born to mothers not exposed to influenza virus were fostered by virus-exposed BALB/c mothers and then inoculated with 3 TCID50 of PR8 influenza virus or PBS intranasally at 7 days old. Serum influenza virus–specific IgG2a (maternal antibody) or IgG2c (de novo antibody) was measured by ELISA 14 days post-inoculation. n = 3 (orange and yellow) or n = 6 (blue) mice per group. Groups were compared using one-way ANOVA with Tukey’s post hoc test. Data in (C), (F), (G), and (H) are shown as means ±SD. Panels (D) to (G) show the results of one experiment that is representative of three independent biological replicates. Panel (H) shows results of one experiment that is representative of two independent biological replicates. ns, not significant.
Next, we tested whether influenza virus–specific matAbs could protect infant mice from infection with influenza virus. We intranasally infected 7-day-old pups with or without influenza virus–specific matAbs with a 30 tissue culture infectious dose (TCID)50 of PR8 influenza virus. This dose of virus caused 80 to 100% mortality in pups born to naïve mothers (Fig. 1E). However, pups inoculated in the presence of influenza virus–specific matAbs were protected (Fig. 1E). We also quantified viral titers in the lungs of pups 2 days after infection. We found high titers of virus (~107 TCID50/g of tissue) in pups without influenza virus–specific matAbs, but we were unable to detect virus in the lungs of pups with influenza virus–specific matAbs (Fig 1F; P < 0.0001). Together, these results show that influenza virus–specific matAbs were efficiently transferred to infant mice and protected them from influenza disease.
We next sought to determine the effect of matAbs on infant antibody responses elicited by influenza virus infection. We measured influenza virus–specific IgG titers in the sera of pups that were intranasally infected with PR8 influenza virus at 7 days of age in the presence or absence of influenza virus–specific matAbs. We used a dose of virus (3 TCID50 PR8 influenza virus) that was sublethal for pups in the presence or absence of influenza virus–specific matAbs for these experiments. At 125 days post-infection (dpi), a time point at which matAbs have waned below the limit of detection (Fig. 1G, yellow bar), influenza virus–specific serum IgG titers of mice inoculated with PR8 influenza virus in the presence of influenza virus–specific matAbs were equivalent to those of unexposed mice (Fig. 1G, blue and black bars; P = 0.8). In contrast, high titers of influenza virus–specific serum IgG antibodies were present in mice that were infected in the absence of matAbs (Fig. 1G, orange bar; P < 0.0001). Because maternally derived and de novo antibodies are indistinguishable when the mother and pup are of the same strain, we confirmed that matAbs suppressed de novo antibody responses to influenza virus in mouse offspring using a cross-fostering system in which pups received matAbs only from a foster mother. Murine matAbs are efficiently transferred in milk (4, 32), and different strains of mice have differences in IgG subclasses (BALB/c, IgG2a; C57BL/6, IgG2c). C57BL/6 pups born to unexposed mothers were nursed by BALB/c foster mothers who had been infected with influenza virus. These pups thus acquired influenza virus–specific IgG2a matAbs only through milk (Fig. 1H, left; P < 0.005). These C57BL/6 pups with IgG2a influenza virus–specific matAbs failed to mount de novo IgG2c antibodies after influenza virus infection (Fig. 1H, right; P = 0.99), confirming that matAbs inhibited de novo antibody responses in mouse pups elicited by influenza virus infection.
matAbs inhibit de novo antibody responses in mouse pups elicited by influenza virus infections or conventional influenza vaccines
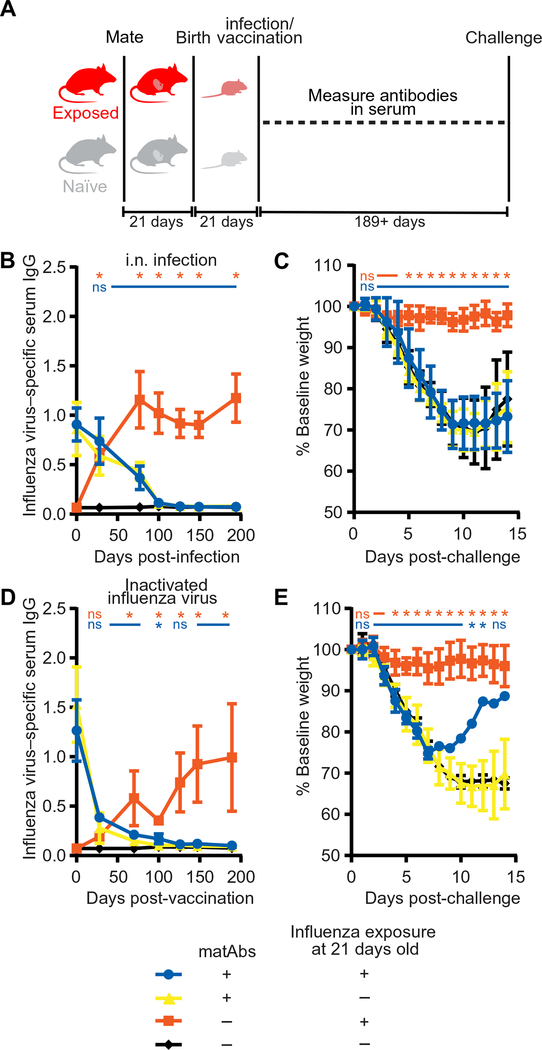
It was possible that antigen-specific matAbs inhibited de novo antibody responses in mouse pups elicited by live virus infections by limiting virus replication and antigen production (Fig. 1F). It is unknown whether matAbs similarly could inhibit live attenuated influenza vaccines (LAIVs) that require viral replication and inactivated vaccines that do not require viral replication. We intranasally infected 21-day-old juvenile mice with subclinical doses of PR8 influenza virus to model LAIVs, and we intramuscularly injected beta propiolactone–inactivated purified PR8 virus to model inactivated vaccines (Fig. 2A). Because of their size, we were not able to obtain sufficient amounts of prevaccination sera from 7-day-old pups, and therefore, all further experiments were conducted on 21-day-old juvenile mice. Consistent with experiments with 7-dayold mice, 21-day-old mice receiving subclinical doses of live PR8 influenza virus in the absence of influenza virus–specific matAbs generated high serum antibody titers, whereas mice exposed to live virus in the presence of influenza virus–specific matAbs did not (Fig. 2B; P < 0.05). To determine whether intranasal vaccination early in life protects from influenza virus infection in adulthood in our model, we then challenged the same mice with 300 TCID50 PR8 influenza virus at 189 days post-vaccination (dpv), at which time residual matAbs had waned below the protective threshold. Adult mice that were previously exposed to influenza virus in the absence of influenza virus–specific matAbs were protected from infection, whereas adult mice that were previously exposed to virus in the presence of influenza virus–specific matAbs were susceptible to infection (Fig 2C; P < 0.05). We found similar results after intramuscular vaccination of 21-day-old mice with inactivated influenza virus. Mice vaccinated with inactivated PR8 influenza virus in the presence of matAbs did not generate a de novo antibody response (Fig. 2D; P < 0.05) and were not protected during virus challenge (Fig. 2E; P < 0.05) compared to mice vaccinated in the absence of matAbs. Thus, antigen-specific matAbs in mouse pups inhibited de novo antibody responses after intranasal inoculation with live influenza virus or intramuscular vaccination with inactivated virus in our mouse model.
Fig. 2. matAbs inhibit antibody responses to conventional influenza vaccines.
(A) The experimental design is shown. (B and D) Mice (21 days old) with or without influenza virus–specific matAbs were inoculated with 10 TCID50 of PR8 influenza virus intranasally (B) or 1000 hemagglutination units of purified inactivated PR8 influenza virus intramuscularly (D) or PBS as a vehicle control. Serum influenza virus–specific antibody responses were measured over time. *P < 0.05 after comparison of serum titers from mice exposed to influenza virus versus PBS in the presence (blue *) or absence (orange *) of influenza virus–specific matAbs; one-way ANOVA with Tukey’s post hoc test at each time point. ns, not significant. (C and E) Mice inoculated in (B) and (D) were challenged at 189 or 194 days post-vaccination with 300 TCID50 of PR8 influenza virus intranasally, and weight loss was measured over 14 days. Data are shown as percentage of baseline weight (current weight divided by prechallenge weight). *P < 0.05 after comparison of percentage of baseline weight on each day for mice exposed to influenza virus versus PBS as infants in the presence (blue *) or absence (orange *) of influenza virus–specific matAbs; one-way ANOVA with Tukey’s post hoc test at each day. ns, not significant. In (E), one mouse survived in the blue group. n = 5 (blue), n = 6 (yellow), or n = 7 (orange and black) mice per group (B and C); n = 3 (black), n = 4 (blue and orange), or n = 11 (yellow) mice per group (D and E). Data in (B) to (E) are shown as means ± SD. Panels (B) and (C) show the results of one experiment that is representative of three independent biological replicates. Panels (D) and (E) show the results of one experiment that is representative of two independent biological replicates.
An mRNA-LNP influenza vaccine elicited antibody responses in mouse pups in the presence of matAbs
We and others recently demonstrated that vaccines composed of nucleoside-modified mRNA encapsulated in lipid nanoparticles (mRNA-LNPs) expressing the surface glycoprotein hemagglutinin of influenza viruses elicit robust antibody responses in animal models and humans (35–40). We found that mRNA-LNPs are efficiently taken up by cells that then endogenously express large amounts of antigen for more than 1 week (35, 41). mRNA-LNP vaccination has features that suggest that it may be able to circumvent inhibition by matAbs. First, the vaccine itself does not include antigen, precluding binding of antigen-specific matAbs to the vaccine, and second, the vaccine expresses antigens in cells for long periods of time, which could be beneficial as matAbs decline.
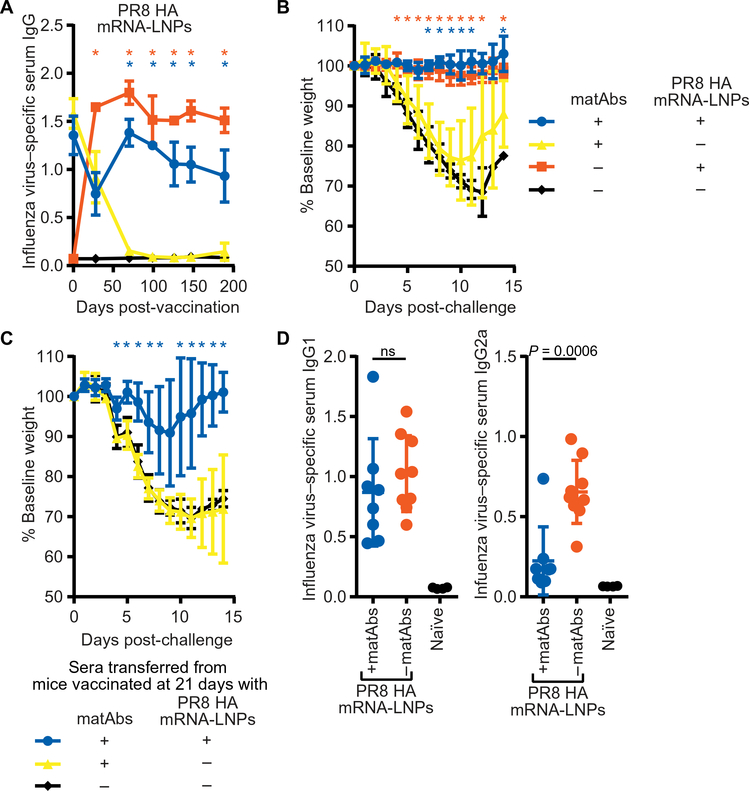
To determine whether nucleoside-modified mRNA-LNP influenza vaccines could circumvent antigen-specific matAbs, we intramuscularly injected 21-day-old mice with or without influenza virus–specific matAbs with 1 μg of mRNA-LNP vaccine encoding the immunodominant surface glycoprotein hemagglutinin from the PR8 influenza virus. The PR8 hemagglutinin mRNA-LNP vaccine elicited high titers of de novo influenza virus–specific antibodies in the presence of influenza virus–specific matAbs (Fig. 3A; P < 0.05). Mice vaccinated with the PR8 hemagglutinin mRNA-LNP vaccine in the presence or absence of matAbs were protected when they were subsequently challenged with PR8 influenza virus in adulthood 189 dpv (Fig. 3B; P < 0.05). To test whether this protection was antibody mediated, we passively transferred serum from vaccinated mice into naïve adult mice and then challenged them with 300 TCID50 of PR8 influenza virus. Mice that were administered serum from the control groups of animals that received PBS instead of vaccine displayed severe disease after virus challenge (Fig. 3C). In contrast, mice that received serum from animals vaccinated with PR8 hemagglutinin mRNA-LNP vaccine in the presence of matAbs were protected (Fig. 3C; P < 0.05). The mRNA-LNP vaccine elicited equivalent influenza virus–specific IgG1 antibodies in the presence or absence of influenza virus–specific matAbs (Fig. 3D, left) but elicited fewer IgG2a antibodies in the presence compared to the absence of matAbs (Fig. 3D, right). This apparent shift toward a T helper 2 (TH2) immune response in the presence of matAbs is interesting because previous studies suggested that TH2 responses are favored after immunization with immune complexes (42, 43).
Fig. 3. PR8 hemagglutinin mRNA-LNP vaccine elicits protective antibody responses in the presence of matAbs.
(A) Mice (21 days old) were vaccinated intramuscularly with 1 μg of nucleoside-modified PR8 hemagglutinin (HA) mRNA-LNP vaccine, and influenza virus–specific serum antibody responses were measured by ELISA. *P < 0.05 after comparison of serum titers from mice vaccinated with PR8 HA mRNA-LNP vaccine versus PBS in the presence (blue *) or absence (orange *) of influenza virus–specific matAbs; one-way ANOVA with Tukey’s post hoc test at each time point. (B) Mice in (A) were challenged at 189 days after vaccination with 300 TCID50 of PR8 influenza virus intranasally, and weight loss was measured over 14 days. Data are shown as percentage of baseline weight (current weight divided by prechallenge weight). *P < 0.05 after comparison of percentage of baseline weight on each day for mice vaccinated as infants with PR8 HA mRNA-LNP vaccine versus PBS in the presence (blue *) or absence (orange *) of influenza virus–specific matAbs; one-way ANOVA with Tukey’s post hoc test at each day. (A and B) n = 3 (+matAbs) or n = 4 (−matAbs) mice per group. (C) Serum was collected at 100+ days after vaccination with 1 μg of PR8 HA mRNA-LNP vaccine or PBS in the presence or absence of influenza virus–specific matAbs and was pooled. Pooled serum (500 μl) was intraperitoneally transferred to 6- to 8-week-old naïve mice, and 4 to 5 hours later, mice were intranasally challenged with 300 TCID50 of PR8 influenza virus. Weight loss was measured over 14 days. n = 4 mice per group. *P < 0.05 after comparison of percentage of baseline weight on each day for mice that received sera from mice vaccinated with PR8 HA mRNA-LNP vaccine or PBS as infants in the presence of influenza virus–specific matAbs; one-way ANOVA with Tukey’s post hoc test at each day. (D) Sera from mice vaccinated with 1 μg of PR8 HA mRNA-LNP vaccine in the presence or absence of influenza virus–specific matAbs and from naïve mice were collected 189 days post-vaccination. Sera were analyzed for influenza virus–specific IgG1 (left) or IgG2a (right). n = 8 (blue), n = 9 (orange), or n = 4 (black) mice per group. Serum titers of mice vaccinated with PR8 HA mRNA-LNP vaccine in the presence or absence of influenza virus–specific matAbs were compared using an unpaired two-tailed t test. In (D), each point represents one mouse. Data are shown as means ± SD. Panels (A) to (D) show data from one experiment that is representative of two independent biological replicates.
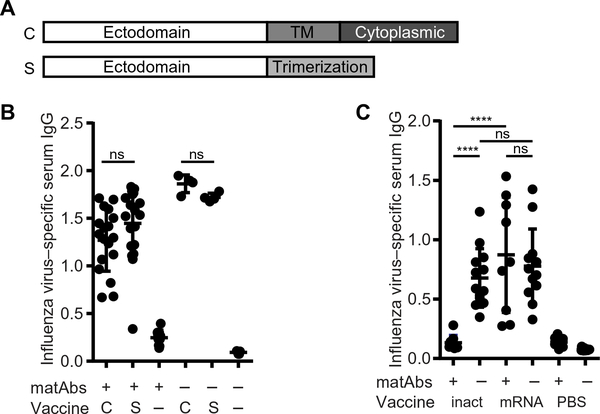
The hemagglutinin produced by our mRNA-LNP vaccine was cell associated because it was produced in host cells and had an intact transmembrane domain (Fig. 4A). In contrast, hemagglutinin in conventional vaccines is in particulate form. It was possible that matAbs preferentially formed immune complexes and sequestered particulate antigens rather than cell-associated antigens. To address this, we designed a secreted form of the PR8 hemagglutinin mRNA-LNP vaccine by removing the transmembrane and cytoplasmic domains of hemagglutinin and introducing a trimerization domain (Fig. 4A). We found that the mRNA-LNP vaccine encoding either cell-associated or secreted hemagglutinin elicited similarly high titers of hemagglutinin-specific antibodies in the presence of influenza virus–specific matAbs (Fig. 4B). This indicated that mRNA-LNP vaccines could circumvent matAb inhibition regardless of whether the mRNA-expressed antigen was cell associated or in particulate form.
Fig. 4. Cell-associated and secreted PR8 HA mRNA-LNP vaccines elicit similar antibody responses, and a low dose of mRNA-LNP vaccine overcomes matAb inhibition.
(A) The schematic shows PR8 influenza virus hemagglutinin (HA) mRNA constructs. mRNA expressing full-length PR8 HA produced cell-associated (C) HA. For some experiments, we used mRNA expressing secreted (S) HA. For this construct, the transmembrane (TM) and cytoplasmic domains were removed, and a trimerization domain was introduced. (B) Mice (21 days old) were vaccinated with cell-associated (C) or secreted (S) PR8 influenza virus HA mRNA-LNP vaccine or PBS (−) in the presence or absence of influenza virus–specific matAbs. Serum was collected 70 days post-vaccination, and influenza virus–specific IgG was measured by ELISA. n = 4 (−matAbs/C vaccine and −matAbs/S vaccine), n = 5 (naïve mice), n = 10 (+matAbs/PBS), n = 18 (+matAbs/C vaccine), or n = 19 (+matAbs/S vaccine) mice per group. (C) Mice (21 days old) were intramuscularly vaccinated with 1000 hemagglutination units of purified inactivated PR8 influenza virus (inact), 0.3 μg of PR8 influenza virus HA mRNA-LNP vaccine (mRNA), or PBS as control in the presence or absence of PR8 influenza virus–specific matAbs. Serum was collected 70 days post-vaccination, and influenza virus–specific serum IgG was measured by ELISA. Data are pooled from two independent experiments (n = 9 to 15 mice per group). Each point represents one mouse. Data are shown as means ± SD. Titers were compared by one-way ANOVA with Sidak’s (B) or Tukey’s (C) post hoc test. ****P < 0.0001. ns, not significant. Panel (B) shows results of two independent experiments. ns, not significant.
Previous studies have shown that the ratio of antigen to matAbs at the time of vaccination is an important consideration in the formation of immune responses (44–46). To determine whether the mRNA-LNP vaccine could overcome matAb inhibition simply by eliciting stronger antibody responses than inactivated virus, we repeated experiments using a lower dose of PR8 hemagglutinin mRNA-LNP vaccine (0.3 μg) that elicited similar antibody titers compared to inactivated PR8 virus in the absence of matAbs (Fig. 4C). We found that the lower dose of mRNA-LNP vaccine elicited equivalent antibody titers in the presence or absence of matAbs (Fig. 4C). This indicated that mRNA-LNP vaccines could circumvent the inhibitory effects of matAbs even at vaccine doses that elicited similar antibody responses to those elicited by inactivated influenza vaccines in the absence of matAbs.
mRNA-LNP vaccines establish long-lived germinal centers in the presence of matAbs
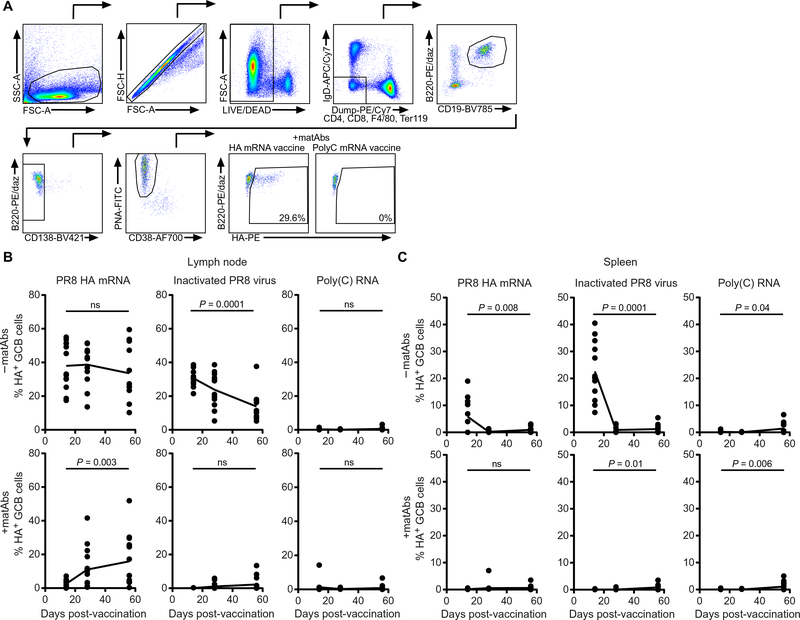
We previously found that nucleoside-modified mRNA-LNP vaccines elicited robust germinal centers (35), which are lymphoid structures that promote antibody class switching, somatic hypermutation, and affinity maturation of antibodies (47, 48). We hypothesized that mRNA-LNP vaccines may elicit protective responses in the presence of antigen-specific matAbs by establishing robust long-lived germinal center reactions. To address this, we measured hemagglutinin-specific germinal center B cells by flow cytometry (gating strategy in Fig. 5A) in the spleen and draining (popliteal) lymph nodes of mice vaccinated intramuscularly with PR8 hemagglutinin mRNA-LNP vaccine, poly(C) RNA-LNP as a negative control, or inactivated PR8 virus. As expected, mice vaccinated with poly(C) RNA-LNP in the presence or absence of influenza virus–specific matAbs did not generate hemagglutinin-specific germinal center B cells in the lymph nodes (Fig. 5B) or spleen (Fig. 5C) of vaccinated mice. In the absence of matAbs, inactivated influenza virus elicited a transient germinal center B cell response in the spleen (Fig. 5C) and a germinal center B cell response in the lymph nodes that was high at 14 dpv but declined over time (Fig. 5B). Consistent with the serum influenza virus–specific IgG titers (Fig. 2C), matAbs inhibited germinal center B cell responses in mice vaccinated with inactivated influenza virus in both the spleen and lymph nodes. In contrast, the PR8 hemagglutinin mRNA-LNP vaccine elicited germinal center B cell responses in the presence and absence of influenza virus–specific matAbs. In the absence of influenza virus–specific matAbs, PR8 hemagglutinin mRNA-LNP vaccine elicited a transient hemagglutinin-specific germinal center B cell response in the spleen (Fig. 5C) and persistent germinal center responses in lymph nodes that remained high at 56 dpv (Fig. 5B). In the presence of influenza virus–specific matAbs, PR8 hemagglutinin mRNA-LNP vaccine failed to elicit germinal center B cells in the spleen (Fig. 5C) and lymph nodes (Fig. 5B) at 14 dpv. However, hemagglutinin-specific germinal center B cells became elevated in the lymph nodes of these mice at 28 and 56 dpv (Fig. 5B). This suggested that the mRNA-LNP vaccine might circumvent the inhibitory effects of matAbs by establishing sustained germinal center reactions that were required for the induction of robust de novo antibody responses.
Fig. 5. PR8 influenza virus HA mRNA-LNP vaccine elicits prolonged germinal center responses in the presence of matAbs.
(A) Flow cytometry gating strategy for hemagglutinin (HA)–positive germinal center (GC) B cells. (B and C) Mice were intramuscularly vaccinated with 1 μg of nucleoside-modified PR8 influenza virus HA mRNA-LNP vaccine, 1000 hemagglutination units of inactivated PR8 influenza virus, or 1 μg of poly(C) RNA-LNP at 21 days of age. Draining (popliteal) lymph nodes (B) and spleens (C) were collected, and HA-specific germinal center B cells (HA probe+ CD19+ B220+ CD138− PNA+ CD38−) were analyzed by flow cytometry. Data are pooled from three independent experiments. n = 4 (−matAbs) or n = 5 (+matAbs) mice per group per experiment at each time point. Each point represents one mouse. The line represents mean. Four- and 8-week post-vaccination time points within each condition were compared with the 2-week post-vaccination time point by one-way ANOVA with Dunnett’s post hoc test.
mRNA-LNP vaccines elicit better responses in mice compared to an adjuvanted human influenza vaccine in the presence of matAbs
We completed additional experiments to directly compare our mRNA-LNP vaccine to a human influenza vaccine. For these experiments, we vaccinated mice early in pregnancy with an A/California/07/2009 (Cal09) monovalent hemagglutinin-based vaccine approved for human use (Fig. 6A). We used an MF59-like adjuvant for these experiments because we found that the unadjuvanted human vaccine did not elicit robust antibody responses in mice. Similar to our experiments using the mouse-adapted PR8 virus, we found that Cal09 hemagglutinin–specific matAbs were transferred efficiently from mothers to pups (Fig. 6B). We then vaccinated pups (in the presence or absence of Cal09 hemagglutinin–specific matAbs) at 21 days of age with human monovalent Cal09 vaccine with MF59-like adjuvant or Cal09 hemagglutinin mRNA-LNP vaccine. Then, we measured serum anti-Cal09 hemagglutinin IgG responses over time. The adjuvanted Cal09 human vaccine elicited robust antibody responses in the absence of matAbs, but these responses were decreased in the presence of matAbs (Fig. 6C). Conversely, the Cal09 hemagglutinin mRNA-LNP vaccine elicited high antibody responses in the presence and absence of matAbs (Fig. 6C). These experiments demonstrate that the mRNA-LNP vaccine elicits stronger antibody responses in the presence of matAbs compared to a vaccine licensed for use in humans, even when the human vaccine was delivered with an adjuvant.
Fig. 6. mRNA-LNP vaccine expressing Cal09 influenza virus strain hemagglutinin overcomes matAb inhibition.
(A) The experimental design is shown. (B) Serum was collected from mothers and pups on the day of weaning and A/California/07/2009 (Cal09) hemagglutinin (HA)–specific antibody titers were measured by hemagglutination inhibition. Each point represents one mouse. (C) Mice (21 days old) were vaccinated with human monovalent Cal09 influenza virus vaccine with MF59-like adjuvant (h + MF), 10 μg of mRNA-LNP vaccine expressing Cal09 HA (mRNA), PBS with MF59-like adjuvant (PBS + MF), or PBS alone (PBS) in the presence or absence of Cal09 HA-specific matAbs. Serum was collected 70 days post-vaccination, and Cal09 HA-specific serum IgG was measured by ELISA. Data are pooled from two independent experiments (n = 7 to 17 mice per group). Each point represents one mouse. Groups were compared by one-way ANOVA with Tukey’s post hoc test. ****P < 0.0001.
DISCUSSION
In this study, we developed a mouse model that demonstrated that influenza virus–specific matAbs inhibited de novo antibody responses in mouse pups elicited by influenza virus infections or conventional influenza vaccines. The mechanisms by which matAbs inhibit de novo antibody responses remain only partially understood. The leading hypotheses include neutralization of live virus by matAbs, epitope masking, clearance of antigen through Fc-mediated uptake of immune complexes by phagocytic cells, and engagement of the inhibitory FcγRIIB receptor on B cells by matAb-vaccine antigen immune complexes (46, 49). In the case of influenza virus respiratory infections, neutralization by matAbs likely occurs via maternal IgG that passes onto the respiratory epithelium, given that maternal IgA remains in the gut (33). Inhibition of antibody responses to inactivated vaccines likely involves a combination of epitope masking, Fc-mediated clearance, and FcγRIIB engagement.
We found that a nucleoside-modified mRNA-LNP vaccine established prolonged germinal center reactions and was able to partially overcome matAb inhibition. It is unclear why mRNA-LNP vaccines are efficient at establishing germinal centers in adult (35) and young mice, but this may be related to prolonged antigen expression by these vaccines (35, 41). Several studies have demonstrated that prolonged antigen availability leads to stronger germinal center responses [reviewed in (50)]. A recent study (51) showed that naïve B cells do not enter germinal centers as efficiently as B cells preloaded with antigen. It is possible that matAb-antigen immune complexes may facilitate the entry of B cells into germinal centers, thus expanding the pool of responding B cells. Whereas the percentage of germinal center B cells decreased over time in mice vaccinated with inactivated influenza virus in our study, the percentage of germinal center B cells remained steady or increased in mice vaccinated with the hemagglutinin mRNA-LNP vaccine. Germinal centers are key sites for affinity maturation, class switching, and differentiation into memory and plasma cell subsets (48). A prolonged germinal center life span may increase all of these important processes.
We observed that our mRNA-LNP vaccine elicited germinal center responses in the draining lymph nodes and spleen in the absence of matAbs but only in the draining lymph nodes in the presence of matAbs. Multiple studies using varied mouse models have reported differences in responses in lymph nodes versus the spleen (52–58). Many factors are likely involved, including type of antigen, dose, type of adjuvant, and local inflammatory signals. There are also differences in ontogeny between secondary lymphoid organs, leading to distinctive cell populations in the developed organ (59–61). We speculate that matAbs interact with specialized cell populations in the lymph nodes to restrict antigen distribution.
We identified several potential limitations that will need to be addressed to optimize mRNA-LNP vaccination in the presence of matAbs. Although we found that mRNA-LNP vaccines partially overcame matAb inhibition at later time points after vaccination, we found that de novo antibody responses elicited by these vaccines were reduced at earlier time points. Future studies will need to determine whether this apparent delay in de novo antibody production in the presence of matAbs is related to different anatomical priming of germinal centers in the presence and absence of matAbs. Future studies will also need to address why mRNA-LNP vaccine–elicited de novo antibody responses were skewed toward TH2 in the presence of antigen-specific matAbs. This may be due to immune complex formation (42, 43), lower overall amounts of antigen (62, 63), or priming in distinct immunological sites. TH2-skewed responses to an inactivated split influenza vaccine preparation used in the 2000–2001 influenza season in Canada were associated with type 2–like adverse events in human vaccine recipients (64). A link between TH2 bias and waning immunity to the acellular pertussis vaccine has been suggested (65, 66). For these reasons, caution should be taken as mRNA-LNP vaccines are considered for human use. However, it is worth noting that the pups vaccinated with the mRNA-LNP vaccine in the presence of matAbs in our study were fully protected from subsequent viral challenge despite having antibody isotypes associated with TH2 responses.
Future studies should directly compare how matAbs differentially affect mRNA-LNP vaccines and other genetic vaccines, such as DNA vaccines, which have had variable success in the presence of matAbs (12, 13, 23, 27, 30, 67). It is unclear why nucleoside-modified mRNA-LNP vaccines expressing hemagglutinin, but not DNA vaccines expressing hemagglutinin (12, 13), are able to elicit protective antibody responses in the presence of influenza virus–specific matAbs. This might be due to differences in magnitude and duration of antigen expression or due to unique properties of mRNA-LNP vaccines. Our studies suggest that nucleoside-modified mRNA-LNP vaccines are an attractive candidate for use in young children. mRNA vaccines have entered clinical trials in adults and seem to be well tolerated (36, 40, 68–74). These vaccines do not require live pathogens; there is no concern about incomplete inactivation, and there is no possibility of reversion to virulence. Insertional mutagenesis is also not possible because mRNA cannot integrate into host DNA. In addition, mRNA is easily catabolized by host machinery and thus essentially has a built-in “off switch.” However, the safety and efficacy of these vaccines must still be carefully evaluated in children and infants, especially because of issues related to TH2-associated immunity.
Children under 6 months of age remain at increased risk of severe disease from viral and bacterial infections, yet many currently available vaccines are not licensed for this age group (75). Whereas maternal vaccination can provide passive protection, it also negatively affects the infant’s own active immunity. Nucleoside-modified mRNA-LNP vaccines offer a promising strategy to vaccinate this vulnerable population because they can elicit antibody responses in the presence of antigen-specific matAbs. Because the mRNA-LNP platform is easily adaptable to different antigens (76), this system could offer a general solution to matAb inhibition of immune responses to current vaccines.
MATERIAL AND METHODS
Study design
The study objectives were to determine the effect of matAbs on mouse pup immune responses and to assess the ability of an mRNA-LNP vaccine to elicit de novo immune responses in the presence of matAbs. Mouse pups with or without influenza virus–specific matAbs were infected with influenza virus or vaccinated, and immune responses and protection against influenza virus challenge were assessed. The number of pups in each experiment varied because of litter size. Both male and female pups were used in experiments. In experiments involving infection of 7-day-old pups, all pups within each litter received the same treatment to prevent the transmission of virus from infected to uninfected pups. In experiments involving infection or vaccination of 21-day-old pups, pups of each sex and litter were randomized to treatment groups. No pups were excluded from experiments. Outliers were included in the analyses. Investigators were not blinded. Two to four biological replicates were performed for each experiment, as specified in each figure legend.
Experiments involving mice complied with all relevant ethical regulations. All protocols involving mice were approved by the Institutional Animal Care and Use Committee of the Wistar Institute and the University of Pennsylvania.
Mouse model
BALB/c and C57BL/6 mice were purchased from Charles River Laboratories or bred in-house. For most experiments, 6- to 8-week-old female mice were intranasally infected with 20 TCID50 of PR8 influenza virus under isoflurane anesthesia. After 3 weeks, serum was collected, and antibody titers were measured by HAI. At least 3 weeks after infection, exposed female mice and unexposed controls were mated with males of the same strain and allowed to have pups. For experiments with A/California/07/2009, 6- to 8-week-old female mice were mated with male mice for 3 days. On the third day, male mice were removed, and female mice were intramuscularly injected with 1.5 μg of Influenza A (H1N1) 2009 Monovalent Vaccine (Sanofi Pasteur; NR-20347 from BEI Resources) or phosphate-buffered saline (PBS) mixed 1:1 with MF59-like adjuvant (InvivoGen AddaVax) in a total volume of 100 μl. Pups were either infected at ~7 days old (range, 6 to 8 days) or infected or vaccinated at ~21 days old (range, 19 to 23 days). Pups were weaned at ~21 days old (range, 19 to 23 days). For cross-fostering experiments, 1-day-old C57BL/6 pups were removed from their naïve mothers and fostered with lactating influenza virus–exposed BALB/c females.
Serum collection
Blood was collected at the indicated time points by submandibular puncture into 1.1-ml Z-Gel tubes (Sarstedt) using a 5-mm lancet (MEDIpoint). Sera were heat-treated at 55°C for 30 min and stored at 4°C. For some experiments, sera were treated with receptor-destroying enzyme (RDE; Seiken). RDE and serum were combined 3:1 and incubated at 37°C for 2 hours and then at 55°C for 30 min and stored at 4°C.
Infections and vaccinations
All intranasal infections were performed under isoflurane anesthesia. Virus was diluted in PBS and instilled into the nostrils in 50 μl (adults), 25 μl (21-day-old mice), or 5 μl (neonates). Seven-day-old mice were infected with 3 or 30 TCID50 PR8 virus. Twenty-one–day–old mice were infected with 10 TCID50. Adult mice were challenged with 300 TCID50. After challenge, mice were monitored at least once daily and were euthanized when they became lethargic, cachexic, or unresponsive to stimuli. For intramuscular (i.m.) injections, virus or vaccine was diluted in PBS and injected into the lower or upper hind leg in 50 μl per leg.
Viruses
A/Puerto Rico/8/1934 (H1N1) (PR8) virus and a 2 × 6 recombinant virus with A/California/07/2009 (H1N1) hemagglutinin (HA) and neuraminidase (NA) segments and internal genes from PR8 were propagated in 10-day-old fertilized chicken eggs. Allantoic fluid was clarified and aliquoted, and titer was determined by TCID50 on Madin-Darby canine kidney (MDCK) cells. For inactivated virus vaccine, allantoic fluid was purified by sucrose gradient ultracentrifugation and inactivated with 0.1% beta propiolactone (BPL) with 0.1 M Hepes. Titer for inactivated virus was determined by hemagglutination unit (HAU) assay. Influenza A (H1N1) 2009 Monovalent Vaccine (Sanofi Pasteur) was obtained from BEI Resources (NR-20347).
mRNA production
mRNAs were produced as previously described (77) using T7 RNA polymerase (MEGAscript, Ambion) on linearized plasmids encoding codon-optimized (78) PR8 influenza virus hemagglutinin (HA) (pTEV-PR8 HA-A101 and pTEV-sPR8 HA-A101) or A/California/07/2009 HA (pTEV-A/Cal09 HA-A101). For some experiments, we used mRNA constructs producing secreted HA. For mRNA producing secreted HA, the transmembrane and cytoplasmic domains of HA were removed and replaced with a codon-optimized sequence for the trimerization domain (foldon; amino acid sequence GSGYIPEAPRDGQAYVRKDGEWVLLSTFL) (79–81). mRNAs were transcribed to contain 101-nucleotide-long poly(A) tails. One-methylpseudouridine (m1Ψ)–5′-triphosphate (TriLink) instead of UTP was used to generate modified nucleoside-containing mRNA. RNAs were capped using the m7G capping kit with 2′-O-methyltransferase (ScriptCap, CELLSCRIPT) to obtain cap1. mRNA was purified by fast protein liquid chromatography (FPLC) (ÄKTA purifier, GE Healthcare), as described (82). All mRNAs were analyzed by denaturing or native agarose gel electrophoresis and were stored frozen at −20°C.
LNP formulation of the mRNA
FPLC-purified m1Ψ-containing RNAs were encapsulated in LNP using a self-assembly process, as previously described (41), wherein an ethanolic lipid mixture of ionizable cationic lipid, phosphatidylcholine, cholesterol, and polyethylene glycol (PEG)–lipid was rapidly mixed with an aqueous solution containing mRNA at acidic pH. The RNA-loaded particles were characterized and subsequently stored at −80°C at a concentration of 1 μg/μl. The mean hydrodynamic diameter of these mRNA-LNPs was ~80 nm with a polydispersity index of 0.02 to 0.06 and an encapsulation efficiency of ~95%.
Viral titer measurements
Pups were intranasally inoculated at 7 days old with 30 TCID50 PR8 virus. Two days later, mice were euthanized by decapitation with a sharp blade under isoflurane anesthesia, and the lungs were removed. Viral titers in lung homogenates were quantified by TCID50 assay using MDCK cells (using the Reed and Muench calculator).
Passive transfer
Sera were pooled, and 500 μl was injected intraperitoneally (i.p.) into naïve 6- to 8-week-old female BALB/c mice. Four to 5 hours later, sera were collected to assure efficient passive transfer, and mice were intranasally challenged with 300 TCID50 PR8 virus under anesthesia. After challenge, mice were monitored at least once daily and were euthanized when they became lethargic, cachexic, or unresponsive to stimuli. Transfer of antibodies was verified by ELISA.
Cells
Madin-Darby canine kidney (MDCK) cells were maintained in modified Eagle’s medium (MEM; Mediatech) with 10% fetal bovine serum (FBS; Sigma-Aldrich). 293T cells were maintained in Dulbecco’s MEM (DMEM; Mediatech) with 10% FBS.
ELISA
Immulon 4HBX plates (Thermo Fisher Scientific) were coated with beta propiolactone–inactivated PR8 allantoic fluid (0.4 HAU/μl) or A/California/07/2009 HA (2 μg/ml) (NR-44074, BEI Resources) diluted in PBS overnight at 4°C. Plates were blocked with 3% bovine serum albumin (BSA; Sigma-Aldrich) in PBS for 2 hours at room temperature and then washed with distilled water. Primary antibodies or sera were diluted in 1% BSA in PBS and incubated in the plates for 2 hours at room temperature. Plates were washed, and secondary antibody (goat anti-mouse IgG, IgG1, or IgG2a, human-adsorbed and conjugated to alkaline phosphatase; SouthernBiotech) was diluted 1:1000 in 1% BSA in PBS and incubated in the plates for 1 hour at room temperature. Plates were washed and developed with p-nitrophenyl phosphate (PNPP) (1 mg/ml) for 1 hour at room temperature and read on a SpectraMAX 190 (Molecular Devices). Data are reported as absorbance (OD450).
HAU
Virus was twofold serially diluted in a volume of 50 μl across a 96-well polystyrene U-bottom plate (Falcon) and incubated for 45 min at room temperature with 12.5 μl of 2% (v/v) washed turkey erythrocytes (LAMPIRE) in PBS.
HAI
Sera were twofold serially diluted in PBS in a 96-well polystyrene U-bottom plate (Falcon) and mixed with four agglutinating doses of virus in a total volume of 100 μl. 12.5 μl of 2% (v/v) washed turkey erythrocytes (LAMPIRE) in PBS was added. Agglutination was read out after 45 min at room temperature. HAI titers are expressed as the highest dilution of serum that inhibited four agglutinating doses of virus. Titers <10 were not detectable.
Flow cytometry
At the specified time after vaccination, mice were euthanized by carbon dioxide inhalation, and spleens and draining lymph nodes were collected and stored on ice. Organs were quickly processed into single-cell suspensions in 5% RPMI. Spleen samples were treated with ACK lysing buffer (Thermo Fisher Scientific). Dead cells were stained with the Zombie Aqua Fixable Viability Kit (BioLegend). Fc receptors were blocked with anti-mouse CD16/CD32 (Mouse Fc Block, BD). Cells were stained with the following antibodies/reagents: anti–CD19-BV785 (clone 6D5, BioLegend), anti–B220-PE-dazzle (clone RA3–6B2, BioLegend), anti–CD38-AF700 (clone 90, eBioscience), PNA-FITC (Vector Laboratories), anti–CD138-BV421 (clone 281–2, BioLegend), anti–IgD-APC-Cy7 (clone 11–26c.2a, BioLegend), anti–CD4-PE-Cy7 (clone RM4–5, eBioscience), anti–CD8-PE-Cy7 (clone 53–6.7, eBioscience), anti–F4/80-PE-Cy7 (clone BM8, eBioscience), and anti–Ter119-PE-Cy7 (clone TER-119, eBioscience). To identify HA-specific B cells, cells were stained with PR8 HA protein with a Y98F mutation in the receptor binding site, which prevents nonspecific binding to sialic acid (81), conjugated to PE. Singly stained Igκ compensation microparticles (BD) or cells were used to determine appropriate compensation settings for each experiment. Events were acquired on an LSRII flow cytometer (BD). Data were analyzed with FlowJo software (FlowJo LLC).
Statistics
Data were graphed and analyzed using GraphPad Prism 7. Survival data were analyzed by log-rank test. Time course data at each time point and antibody titer data were analyzed by one-way analysis of variance (ANOVA) with Tukey’s post hoc test. Viral titers in the lungs were analyzed by two-tailed Welch’s t test. IgG subtype titers were analyzed by two-tailed t test. Germinal center B cell responses were analyzed by one-way ANOVA with Dunnett’s post hoc test, comparing 4 and 8 weeks to 2 weeks after vaccination. Titers against cell-associated and secreted PR8 influenza virus hemagglutinin mRNA-LNP vaccine were compared by one-way ANOVA with Sidak’s post hoc test. Data are reported as means ± standard deviation (SD) unless otherwise noted.
Supplementary Material
Fig. S1. Efficiency of matAb transfer does not depend on litter size or mother’s age.
Data file S1. Individual subject-level data for
Acknowledgments
The Vaccine Research Center of the National Institutes of Health provided the hemagglutinin probe used in flow cytometry experiments.
Funding: This work was supported by the Burroughs Wellcome Fund and the National Institute of Allergy and Infectious Diseases (1R01AI113047 and 1R01AI108686 to S.E.H.; R01-AI050484, R01-AI124429, and R01-AI084860 to D.W.; and T32-AI070077 and T32-AI055428 to E.W.). S.E.H. holds an Investigators in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Fund.
Footnotes
Competing interests: S.E.H. has received consulting fees from Sanofi Pasteur, Lumen, Novavax, and Merck. B.L.M. and Y.K.T. are employed by and own shares of Acuitas Therapeutics, a company involved in the development of mRNA-LNP therapeutics. D.W. is a coinventor on the following patents that describe the use of nucleoside-modified mRNA as a platform to deliver therapeutic proteins: patent no. 11/990,646 “RNA containing modified nucleosides and methods of use thereof,” patent no.14/776,525 “Purification and purity assessment of RNA molecules synthesized with modified nucleosides,” and patent no. 13/585,517 “RNA containing modified nucleosides and methods of use thereof.” D.W. and N.P. are coinventors on patents describing the use of modified mRNA in lipid nanoparticles as a vaccine platform: patent no. WO/2016/176330 “Nucleoside-modified RNA for inducing an adaptive immune response” and patent no. WO/2018/081638 “Nucleoside-modified RNA for inducing an adaptive immune response.” B.L.M. and Y.K.T. are coinventors on patents that describe lipid nanoparticles for delivery of nucleic acid therapeutics including mRNA: patent no. WO/2016/176330 “Nucleoside-modified RNA for inducing an adaptive immune response,” patent no. WO/2018/081638 “Nucleoside-modified RNA for inducing an adaptive immune response,” patent no. WO/2018/191719 “Lipid delivery of therapeutic agents to adipose tissue,” patent no. WO/2018/081480 “Lipid nanoparticle formulations,” patent no. WO/2018/078053 “Lipid nanoparticle mRNA vaccines,” and patent no. WO/2019/089828 “Lamellar lipid nanoparticles.”
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. An MTA between the University of Pennsylvania and Acuitas Therapeutics exists for mRNAs produced by the laboratory of D.W. Requests for mRNAs should be directed to D.W.
REFERENCES AND NOTES
- 1.Sanchez-Schmitz G, Levy O, Development of newborn and infant vaccines. Sci. Transl. Med. 3, 90ps27 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenbaum AH, Chen J, Reed C, Beavers S, Callahan D, Christensen D, Finelli L, Fry AM, Hospitalizations for severe lower respiratory tract infections. Pediatrics 134, 546–554 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Nunes MC, Madhi SA, Influenza vaccination during pregnancy for prevention of influenza confirmed illness in the infants: A systematic review and meta-analysis. Hum. Vaccin. Immunother. 14, 758–766 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jennewein MF, Abu-Raya B, Jiang Y, Alter G, Marchant A, Transfer of maternal immunity and programming of the newborn immune system. Semin. Immunopathol. 39, 605–613 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Baxter R, Bartlett J, Fireman B, Lewis E, Klein NP, Effectiveness of vaccination during pregnancy to prevent infant pertussis. Pediatrics 139, (2017). [DOI] [PubMed] [Google Scholar]
- 6.Marchant A, Sadarangani M, Garand M, Dauby N, Verhasselt V, Pereira L, Bjornson G, Jones CE, Halperin SA, Edwards KM, Heath P, Openshaw PJ, Scheifele DW, Kollmann TR, Maternal immunisation: Collaborating with mother nature. Lancet Infect. Dis. 17, e197–e208 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Omer SB, Maternal immunization. N. Engl. J. Med. 376, 1256–1267 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Madhi SA, Cutland CL, Kuwanda L, Weinberg A, Hugo A, Jones S, Adrian PV, van Niekerk N, Treurnicht F, Ortiz JR, Venter M, Violari A, Neuzil KM, Simoes EA, Klugman KP, Nunes MC; Maternal Flu Trial (Matflu) Team, Influenza vaccination of pregnant women and protection of their infants. N. Engl. J. Med. 371, 918–931 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Voysey M, Kelly DF, Fanshawe TR, Sadarangani M, O’Brien KL, Perera R, Pollard AJ, The influence of maternally derived antibody and infant age at vaccination on infant vaccine responses: An individual participant meta-analysis. JAMA Pediatr. 171, 637–646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mooi FR, de Greeff SC, The case for maternal vaccination against pertussis. Lancet Infect. Dis. 7, 614–624 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Reuman PD, Paganini CM, Ayoub EM, Small PA Jr., Maternal-infant transfer of influenza-specific immunity in the mouse. J. Immunol. 130, 932–936 (1983). [PubMed] [Google Scholar]
- 12.Chen J, Zhang F, Fang F, Chang H, Chen Z, Vaccination with hemagglutinin or neuraminidase DNA protects BALB/c mice against influenza virus infection in presence of maternal antibody. BMC Infect. Dis. 7, 118 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pertmer TM, Oran AE, Moser JM, Madorin CA, Robinson HL, DNA vaccines for influenza virus: Differential effects of maternal antibody on immune responses to hemagglutinin and nucleoprotein. J. Virol. 74, 7787–7793 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halasa NB, Gerber MA, Chen Q, Wright PF, Edwards KM, Safety and immunogenicity of trivalent inactivated influenza vaccine in infants. J Infect Dis 197, 1448–1454 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards KM, Maternal antibodies and infant immune responses to vaccines. Vaccine 33, 6469–6472 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Bublot M, Pritchard N, Le Gros FX, Goutebroze S, Use of a vectored vaccine against infectious bursal disease of chickens in the face of high-titred maternally derived antibody. J. Comp. Pathol. 137 (Suppl. 1), S81–S84 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Kilany WH, Hassan MK, Safwat M, Mohammed S, Selim A, VonDobschuetz S, Dauphin G, Lubroth J, Jobre Y, Comparison of the effectiveness of rHVT-H5, inactivated H5 and rHVT-H5 with inactivated H5 prime/boost vaccination regimes in commercial broiler chickens carrying MDAs against HPAI H5N1 clade 2.2.1 virus. Avian Pathol. 44, 333–341 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Sonoda K, Sakaguchi M, Okamura H, Yokogawa K, Tokunaga E, Tokiyoshi S, Kawaguchi Y, Hirai K, Development of an effective polyvalent vaccine against both Marek’s and Newcastle diseases based on recombinant Marek’s disease virus type 1 in commercial chickens with maternal antibodies. J. Virol. 74, 3217–3226 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertran K, Lee DH, Criado MF, Balzli CL, Killmaster LF, Kapczynski DR, Swayne DE, Maternal antibody inhibition of recombinant Newcastle disease virus vectored vaccine in a primary or booster avian influenza vaccination program of broiler chickens. Vaccine 36, 6361–6372 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Capozzo AV, Cuberos L, Levine MM, Pasetti MF, Mucosally delivered Salmonella live vector vaccines elicit potent immune responses against a foreign antigen in neonatal mice born to naive and immune mothers. Infect. Immun. 72, 4637–4646 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramirez K, Ditamo Y, Galen JE, Baillie LW, Pasetti MF, Mucosal priming of newborn mice with S. Typhi Ty21a expressing anthrax protective antigen (PA) followed by parenteral PA-boost induces B and T cell-mediated immunity that protects against infection bypassing maternal antibodies. Vaccine 28, 6065–6075 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welter J, Taylor J, Tartaglia J, Paoletti E, Stephensen CB, Vaccination against canine distemper virus infection in infant ferrets with and without maternal antibody protection, using recombinant attenuated poxvirus vaccines. J. Virol. 74, 6358–6367 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manickan E, Yu Z, Rouse BT, DNA immunization of neonates induces immunity despite the presence of maternal antibody. J. Clin. Invest. 100, 2371–2375 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Rooij EM, Moonen-Leusen HW, de Visser YE, Middel WG, Boersma WJ, Bianchi AT, A DNA vaccine coding for gB and gD of pseudorabies virus (suid herpes type 1) primes the immune system in the presence of maternal immunity more efficiently than conventional vaccines. Vaccine 24, 1264–1273 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Hsieh MK, Wu CC, Lin TL, DNA-mediated vaccination conferring protection against infectious bursal disease in broiler chickens in the presence of maternal antibody. Vaccine 28, 3936–3943 (2010). [DOI] [PubMed] [Google Scholar]
- 26.van Drunen Littel-van den Hurk S, Lawman Z, Wilson D, Luxembourg A, Ellefsen B, van den Hurk JV, Hannaman D, Electroporation enhances immune responses and protection induced by a bovine viral diarrhea virus DNA vaccine in newborn calves with maternal antibodies. Vaccine 28, 6445–6454 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Hassett DE, Zhang J, Whitton JL, Neonatal DNA immunization with a plasmid encoding an internal viral protein is effective in the presence of maternal antibodies and protects against subsequent viral challenge. J. Virol. 71, 7881–7888 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radu DL, Antohi S, Bot A, Miller A, Mirarchi A, Bona C, Effect of maternal antibodies on influenza virus-specific immune response elicited by inactivated virus and naked DNA. Scand. J. Immunol. 53, 475–482 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Monteil M, Le Potier MF, Cariolet R, Houdayer C, Eloit M, Effective priming of neonates born to immune dams against the immunogenic pseudorabies virus glycoprotein gD by replication-incompetent adenovirus-mediated gene transfer at birth. J. Gen. Virol. 78, 3303–3310 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Siegrist C-A, Barrios C, Martinez X, Brandt C, Berney M, Cordova M, Kovarik J, Lambert P-H, Influence of maternal antibodies on vaccine responses: Inhibition of antibody but not T cell responses allows successful early prime-boost strategies in mice. Eur. J. Immunol. 28, 4138–4148 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Bot A, Bona C, Genetic immunization of neonates. Microbes Infect. 4, 511–520 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Hemmings WA, Morris IG, An attempt to affect the selective absorption of antibodies from the gut in young mice. Proc. R. Soc. Lond. B Biol. Sci. 150, 403–409 (1959). [DOI] [PubMed] [Google Scholar]
- 33.Malanchere E, Huetz F, Coutinho A, Maternal IgG stimulates B lineage cell development in the progeny. Eur. J. Immunol. 27, 788–793 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Lee PX, Ong LC, Libau EA, Alonso S, Relative contribution of dengue IgG antibodies acquired during gestation or breastfeeding in mediating dengue disease enhancement and protection in type I interferon receptor-deficient mice. PLOS Negl. Trop. Dis. 10, e0004805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pardi N, Hogan MJ, Naradikian MS, Parkhouse K, Cain DW, Jones L, Moody MA, Verkerke HP, Myles A, Willis E, LaBranche CC, Montefiori DC, Lobby JL, Saunders KO, Liao HX, Korber BT, Sutherland LL, Scearce RM, Hraber PT, Tombacz I, Muramatsu H, Ni H, Balikov DA, Li C, Mui BL, Tam YK, Krammer F, Kariko K, Polacino P, Eisenlohr LC, Madden TD, Hope MJ, Lewis MG, Lee KK, Hu SL, Hensley SE, Cancro MP, Haynes BF, Weissman D, Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 215, 1571–1588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bahl K, Senn JJ, Yuzhakov O, Bulychev A, Brito LA, Hassett KJ, Laska ME, Smith M, Almarsson O, Thompson J, Ribeiro AM, Watson M, Zaks T, Ciaramella G, Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol. Ther. 25, 1316–1327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang F, Lindgren G, Lin A, Thompson EA, Ols S, Röhss J, John S, Hassett K, Yuzhakov O, Bahl K, Brito LA, Salter H, Ciaramella G, Loré K, Efficient targeting and activation of antigen-presenting cells in vivo after modified mRNA vaccine administration in rhesus macaques. Mol. Ther. 25, 2635–2647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindgren G, Ols S, Liang F, Thompson EA, Lin A, Hellgren F, Bahl K, John S, Yuzhakov O, Hassett KJ, Brito LA, Salter H, Ciaramella G, Lore K, Induction of robust B cell responses after influenza mRNA vaccination is accompanied by circulating hemagglutinin-specific ICOS+ PD-1+ CXCR3+ T follicular helper cells. Front. Immunol 8, 1539 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pardi N, Parkhouse K, Kirkpatrick E, McMahon M, Zost SJ, Mui BL, Tam YK, Karikó K, Barbosa CJ, Madden TD, Hope MJ, Krammer F, Hensley SE, Weissman D, Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun 9, 3361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feldman RA, Fuhr R, Smolenov I, Mick Ribeiro A, Panther L, Watson M, Senn JJ, Smith M, Almarsson Ö, Pujar HS, Laska ME, Thompson J, Zaks T, Ciaramella G, mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine 37, 3326–3334 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Pardi N, Tuyishime S, Muramatsu H, Kariko K, Mui BL, Tam YK, Madden TD, Hope MJ, Weissman D, Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release 217, 345–351 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson CF, Gerber JS, Mosser DM, Modulating macrophage function with IgG immune complexes. J. Endotoxin Res. 8, 477–481 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Anderson CF, Mosser DM, Cutting edge: Biasing immune responses by directing antigen to macrophage Fc gamma receptors. J. Immunol. 168, 3697–3701 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Albrecht P, Ennis FA, Saltzman EJ, Krugman S, Persistence of maternal antibody in infants beyond 12 months: Mechanism of measles vaccine failure. J. Pediatr. 91, 715–718 (1977). [DOI] [PubMed] [Google Scholar]
- 45.Appaiahgari MB, Glass R, Singh S, Taneja S, Rongsen-Chandola T, Bhandari N, Mishra S, Vrati S, Transplacental rotavirus IgG interferes with immune response to live oral rotavirus vaccine ORV-116E in Indian infants. Vaccine 32, 651–656 (2014). [DOI] [PubMed] [Google Scholar]
- 46.C. A. Siegrist, Mechanisms by which maternal antibodies influence infant vaccine responses: Review of hypotheses and definition of main determinants. Vaccine 21, 3406–3412 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Dufaud CR, McHeyzer-Williams LJ, McHeyzer-Williams MG, Deconstructing the germinal center, one cell at a time. Curr. Opin. Immunol. 45, 112–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mesin L, Ersching J, Victora GD, Germinal center B cell dynamics. Immunity 45, 471–482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim D, Huey D, Oglesbee M, Niewiesk S, Insights into the regulatory mechanism controlling the inhibition of vaccine-induced seroconversion by maternal antibodies. Blood 117, 6143–6151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cirelli KM, Crotty S, Germinal center enhancement by extended antigen availability. Curr. Opin. Immunol. 47, 64–69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turner JS, Benet ZL, Grigorova IL, Antigen acquisition enables newly arriving B cells to enter ongoing immunization-induced germinal centers. J. Immunol. 199, 1301–1307 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krueger JG, Segal RA, Moyer RC, Differences in the responsiveness of splenic, lymph node, and peripheral blood lymphoid cells to tumor membrane extracts. Cancer Res. 37, 320–322 (1977). [PubMed] [Google Scholar]
- 53.Gonzalez SF, Lukacs-Kornek V, Kuligowski MP, Pitcher LA, Degn SE, Kim Y-A, Cloninger MJ, Martinez-Pomares L, Gordon S, Turley SJ, Carroll MC, Capture of influenza by medullary dendritic cells via SIGN-R1 is essential for humoral immunity in draining lymph nodes. Nat. Immunol. 11, 427–434 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yokochi T, Fuji Y, Inoue Y, Ito H, Kimura Y, Kato N, Differential response of B cells in the lymph node and the spleen to bacterial lipopolysaccharide. Cell. Immunol. 128, 79–88 (1990). [DOI] [PubMed] [Google Scholar]
- 55.Lofano G, Mancini F, Salvatore G, Cantisani R, Monaci E, Carrisi C, Tavarini S, Sammicheli C, Rossi Paccani S, Soldaini E, Laera D, Finco O, Nuti S, Rappuoli R, De Gregorio E, Bagnoli F, Bertholet S, Oil-in-water emulsion MF59 increases germinal center B cell differentiation and persistence in response to vaccination. J. Immunol. 195, 1617–1627 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Frank GM, Angeletti D, Ince WL, Gibbs JS, Khurana S, Wheatley AK, Max EE, McDermott AB, Golding H, Stevens J, Bennink JR, Yewdell JW, A simple flow-cytometric method measuring B cell surface immunoglobulin avidity enables characterization of affinity maturation to influenza A virus. MBio 6, e01156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boyden AW, Legge KL, Waldschmidt TJ, Pulmonary infection with influenza A virus induces site-specific germinal center and T follicular helper cell responses. PLOS ONE 7, e40733 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Junt T, Moseman EA, Iannacone M, Massberg S, Lang PA, Boes M, Fink K, Henrickson SE, Shayakhmetov DM, Di Paolo NC, van Rooijen N, Mempel TR, Whelan SP, von Andrian UH, Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature 450, 110–114 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Anjuère F, Martin P, Ferrero I, Fraga ML, del Hoyo GM, Wright N, Ardavin C, Definition of dendritic cell subpopulations present in the spleen, Peyer’s patches, lymph nodes, and skin of the mouse. Blood 93, 590–598 (1999). [PubMed] [Google Scholar]
- 60.Ettinger R, Browning JL, Michie SA, van Ewijk W, McDevitt HO, Disrupted splenic architecture, but normal lymph node development in mice expressing a soluble lymphotoxin-β receptor–IgG1 fusion protein. Proc. Natl. Acad. Sci. U.S.A. 93, 13102–13107 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moriyama A, Fujishima J, Furukawa T, Yoshikawa T, Kodama R, Sasaki Y, Nagaoka T, Kamimura Y, Maeda H, Hirai T, Yamaguchi R, Quantitative analyses of lymphoid tissue in the spleen, lymph nodes and Peyer’s patches in cynomolgus monkeys. J. Vet. Med. Sci. 73, 1459–1464 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Tubo NJ, Jenkins MK, TCR signal quantity and quality in CD4+ T cell differentiation. Trends Immunol. 35, 591–596 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Panhuys N, TCR signal strength alters T–DC activation and interaction times and directs the outcome of differentiation. Front. Immunol. 7, 6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Babiuk S, Skowronski DM, De Serres G, HayGlass K, Brunham RC, Babiuk L, Aggregate content influences the Th1/Th2 immune response to influenza vaccine: Evidence from a mouse model. J. Med. Virol. 72, 138–142 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Burdin N, Handy LK, Plotkin SA, What is wrong with pertussis vaccine immunity? The problem of waning effectiveness of pertussis vaccines. Cold Spring Harb. Perspect. Biol 9, a029454 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Domenech de Cellès M, Riolo MA, Magpantay FMG, Rohani P, King AA, Epidemiological evidence for herd immunity induced by acellular pertussis vaccines. Proc. Natl. Acad. Sci. U.S.A. 111, E716–E717 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Monteil M, Le Potier MF, Guillotin J, Cariolet R, Houdayer C, Eloit M, Genetic immunization of seronegative one-day-old piglets against pseudorabies induces neutralizing antibodies but not protection and is ineffective in piglets from immune dams. Vet. Res. 27, 443–452 (1996). [PubMed] [Google Scholar]
- 68.Kübler H, Scheel B, Gnad-Vogt U, Miller K, Schultze-Seemann W, Vom Dorp F, Parmiani G, Hampel C, Wedel S, Trojan L, Jocham D, Maurer T, Rippin G, Fotin-Mleczek M, von der Mulbe F, Probst J, Hoerr I, Kallen KJ, Lander T, Stenzl A, Self-adjuvanted mRNA vaccination in advanced prostate cancer patients: A first-in-man phase I/IIa study. J. Immunother. Cancer 3, 26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sebastian M, Boehmer L. v., Zippelius A, Mayer F, Reck M, Atanackovic D, Thomas M, Schneller F, Stoehlmacher J, Goekkurt E, Bernhard H, Groeschel A, Bals R, Schmidt S, Scheel B, Koch SD, Lander T, Kallen K, Knuth A, Messenger RNA vaccination in NSCLC: Findings from a phase I/IIa clinical trial. 29, 2584–2584 (2011). [Google Scholar]
- 70.Rittig SM, Haentschel M, Weimer KJ, Heine A, Muller MR, Brugger W, Horger MS, Maksimovic O, Stenzl A, Hoerr I, Rammensee HG, Holderried TA, Kanz L, Pascolo S, Brossart P, Intradermal vaccinations with RNA coding for TAA generate CD8+ and CD4+ immune responses and induce clinical benefit in vaccinated patients. Mol. Ther. 19, 990–999 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rittig SM, Haentschel M, Weimer KJ, Heine A, Müller MR, Brugger W, Horger MS, Maksimovic O, Stenzl A, Hoerr I, Rammensee H-G, Holderried TA, Kanz L, Pascolo S, Brossart P, Long-term survival correlates with immunological responses in renal cell carcinoma patients treated with mRNA-based immunotherapy. Oncoimmunology 5, e1108511 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kranz LM, Diken M, Haas H, Kreiter S, Loquai C, Reuter KC, Meng M, Fritz D, Vascotto F, Hefesha H, Grunwitz C, Vormehr M, Hüsemann Y, Selmi A, Kuhn AN, Buck J, Derhovanessian E, Rae R, Attig S, Diekmann J, Jabulowsky RA, Heesch S, Hassel J, Langguth P, Grabbe S, Huber C, Türeci Ö, Sahin U, Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534, 396–401 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Weide B, Carralot J-P, Reese A, Scheel B, Eigentler TK, Hoerr I, Rammensee HG, Garbe C, Pascolo S, Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J. Immunother. 31, 180–188 (2008). [DOI] [PubMed] [Google Scholar]
- 74.Weide B, Pascolo S, Scheel B, Derhovanessian E, Pflugfelder A, Eigentler TK, Pawelec G, Hoerr I, Rammensee HG, Garbe C, Direct injection of protamine-protected mRNA: Results of a phase 1/2 vaccination trial in metastatic melanoma patients. J. Immunother. 32, 498–507 (2009). [DOI] [PubMed] [Google Scholar]
- 75.Hodgins DC, Shewen PE, Vaccination of neonates: Problem and issues. Vaccine 30, 1541–1559 (2012). [DOI] [PubMed] [Google Scholar]
- 76.Pardi N, Hogan MJ, Porter FW, Weissman D, mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pardi N, Muramatsu H, Weissman D, Karikó K, In vitro transcription of long RNA containing modified nucleosides. Methods Mol. Biol. 969, 29–42 (2013). [DOI] [PubMed] [Google Scholar]
- 78.Thess A, Grund S, Mui BL, Hope MJ, Baumhof P, Fotin-Mleczek M, Schlake T, Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 23, 1456–1464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Frank S, Kammerer RA, Mechling D, Schulthess T, Landwehr R, Bann J, Guo Y, Lustig A, Bächinger HP, Engel J, Stabilization of short collagen-like triple helices by protein engineering. J. Mol. Biol. 308, 1081–1089 (2001). [DOI] [PubMed] [Google Scholar]
- 80.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA, Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312, 404–410 (2006). [DOI] [PubMed] [Google Scholar]
- 81.Whittle JRR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, Narpala SR, Yassine HM, Frank GM, Yewdell JW, Ledgerwood JE, Wei C-J, McDermott AB, Graham BS, Koup RA, Nabel GJ, Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J. Virol. 88, 4047–4057 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weissman D, Pardi N, Muramatsu H, Karikó K, HPLC purification of in vitro transcribed long RNA. Methods Mol. Biol. 969, 43–54 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Efficiency of matAb transfer does not depend on litter size or mother’s age.
Data file S1. Individual subject-level data for