Abstract
BACKGROUND.
The arachidonic acid (AA) pathway is suspected to be involved in the development of various cancers, including prostate cancer. However, the role of single nucleotide polymorphisms (SNPs) of AA pathway genes remains unclear. The purpose of this case-control study was to evaluate the association between prostate cancer risk and 14 such SNPs in the PTGS2, PTGES2, ALOX5, ALOX5AP, and LTA4H genes.
METHODS.
Genotyping was conducted on 585 white prostate cancer cases and 585 healthy, age-matched controls. The best genetic model for each SNP was determined using Akaike’s information criterion. Odds ratios for the association between each SNP and prostate cancer risk were calculated, both overall and stratified by obesity (BMI≥30). Haplotype analysis was conducted for the PTGES2 SNPs.
RESULTS.
LTA4H rs1978331 was inversely associated with prostate cancer risk overall (overdominant model OR=0.68, 95% CI: 0.51-0.91 for TC vs TT/CC). Among non-obese individuals, the GG genotype of PTGES2 rs10987883 was associated with an increased risk for prostate cancer (recessive model OR=3.23, 95% CI: 1.27-8.23).
CONCLUSION.
Our results indicate that SNPS in certain AA metabolism genes may influence prostate cancer susceptibility. Furthermore, it is possible that obesity, which induces a chronic state of low-level inflammation in addition to several metabolic sequelae, may modify the impact of these SNPs. These findings should be confirmed in a larger study with power to detect differential effects by obesity. It would also be interesting to see if these effects differed by racial/ethnic group, especially since African-American men are at higher risk of prostate cancer.
Introduction
Over the course of the last 30 years, an enormous body of research on prostate cancer etiology has attempted to clarify the role of lipid metabolism, including that of the arachidonic acid (AA) pathway, in carcinogenesis (1-3). Arachidonic acid is an ω-6 polyunsaturated fatty acid that is obtained from dietary sources and stored as phospholipids in the bilayer of plasma membranes (4). The pathway regulating the release and metabolism of this essential fatty acid has been well-characterized, and some of the eicosanoid products of this pathway, such as prostaglandins, leukotrienes, and hydroxyeicosatetraenoic acids (HETEs), have been previously shown to be involved in cancer cell proliferation, cell motility, and angiogenesis (1,2,5-12). However, the impact of single-nucleotide polymorphisms (SNPs) of AA metabolism genes on prostate cancer risk is a newer area of research that merits further investigation.
The majority of recent studies that have examined SNPs of AA metabolism genes have only focused on a few selected genes, rather than taking a broader pathway-based approach (13-18). The results of these studies have been somewhat inconsistent, and the SNPs of several AA pathway genes have not yet been examined. The benefit of a pathway-based, exploratory analysis is that such an approach provides greater coverage of the genetic variation present in genes representing different parts of the pathway and thus, could direct future studies to the most influential candidate genes in the pathway. To date, a disproportionate number of studies on AA metabolism genes have examined SNPs in PTGS2/COX2; whereas analyses on SNPs in other genes of biosynthetic enzymes involved in the AA pathway, such as ALOX5 and PTGES2, are uncommon in the literature.
Related to lipid metabolism, the effects of obesity on prostate cancer risk have been explored in several studies (19-23). While obesity appears to be associated with worse prostate cancer prognosis, some studies suggest that it may be inversely associated with overall prostate cancer risk (20,23). However, this hypothesis remains controversial, and few studies have addressed it directly. Nevertheless, it is clear that obesity and its related cascade of physiological effects (i.e., higher levels of lipids, adipokines, and other pro-inflammatory factors) may be relevant to the way in which SNPs in lipid metabolism genes influence prostate cancer risk.
The purpose of this pathway-based study was to examine the effects, both overall and stratified by obesity, of 14 SNPs in five AA metabolism pathway genes (PTGS2, PTGES2, ALOX5, ALOX5AP, and LTA4H) on prostate cancer risk in a population of 585 non-Hispanic white cases and 585 age-matched, non-Hispanic white controls.
Methods
Study Population.
DNA collected from prostate cancer cases and healthy control subjects was purchased from BioServe Biotechnologies, Ltd. (Beltsville, MD). BioServe’s Global Biorepository contains DNA, RNA, and tissue samples from over 120,000 participants that are available to researchers. Tissue and blood samples of cases were obtained from patients recruited from hospitals and urology clinics. Controls were recruited from primary care and specialists’ offices. We purchased DNA, collected in the United States, from 585 non-Hispanic white prostate cancer cases and 585 age-matched, white cancer-free controls. We also obtained self-reported information on demographics and other characteristics from the standard questionnaires used by the biorepository. All subjects consented to have their specimens become part of BioServe’s biorepository to be used for research.
Candidate Gene & SNP Selection.
Fourteen SNPs in five AA metabolism pathway genes, PTGS2, PTGES2, ALOX5, ALOX5AP, and LTA4H, were chosen for this analysis based on the previous literature (Table I). The gene products of these five candidate genes are representative of two different branches of the AA metabolism pathway (1,3,10). One branch of the pathway leads to the synthesis of prostaglandins and thromboxanes. PTGS2 codes for cyclooxygenase-2 (COX2), also called prostaglandin-endoperoxide synthase 2, which is an inducible enzyme that has pro-inflammatory effects and is overexpressed in several cancers (11,24). COX2 catalyzes the conversion of prostaglandin G2 to prostaglandin H2; whereas prostaglandin E synthase-2, the product of the PTGES2 gene, facilitates the next step in the pathway, the conversion of prostaglandin H2 to prostaglandin E2 (PGE2). Like COX2, PGE2 has also been shown to be dramatically overexpressed in malignant prostate tissue compared to benign surrounding tissue (2,9,25,26). As a result, COX2, PTGES2, PGE2, and related proteins that act further downstream have been considered to be possible therapeutic targets for cancer (9).
Table I.
Information about 14 genotyped SNPs in five arachidonic acid metabolism pathway genes
| Genes | SNP ID | SNP region | Base change |
P, HWEa | Basis of selection |
|||
|---|---|---|---|---|---|---|---|---|
| Details | Referenceb | Epidemiologic Associations | Reference | |||||
| Prostaglandin Synthesis Subpathway | ||||||||
| PTGS2/ COX2 | rs2745557 | Intronic | G>A | 0.40 | (15) | Prostate cancer | (16) | |
| PTGES2 | rs884115 | Intronic | C>T | 0.72 | Haplotype tagging SNP | (36) | ||
| rs10987883 | Intronic | A>G | 0.85 | Haplotype tagging SNP | (36) | |||
| rs13283456 | Intronic | C>T | 0.25 | Missense; Arg298His | Body mass index; Type 2 diabetes | (36,37) | ||
| rs4837240 | 3’ UTR | G>A | 0.18 | Haplotype tagging SNP | (36) | |||
| Leukotriene Synthesis Subpathway | ||||||||
| ALOX5 | rs2029253 | Intronic | A>G | <.001 | Acute coronary syndrome | (38) | ||
| rs12762303 | Promoter | T>C | 0.41 | Coronary artery disease | (39) | |||
| rs2228065 | Exon 6 | G>T | 1.0 | Missense; Glu254Lys | (40) | Tuberculosis | (41) | |
| rs6593482 | VNTR | G>T | <.001 | |||||
| ALOX5AP | rs12721458 | Intronic | C>G | 0.84 | ||||
| rs4076128 | 5’UTR | A>G | <.001 | Breast cancer | (10) | |||
| rs4073259 | 5’UTR | G>A | 0.13 | Myocardial infarction | (42) | |||
| LTA4H | rs2660845 | 5’UTR | A>G | 0.17 | Asthma | (43) | ||
| rs1978331 | Intronic | T>C | 0.65 | Higher level of LB4c associated with T allele | (33,42) | Asthma; leprosy; tuberculosis | (30,33) | |
P-values from Chi-squared test for Hardy-Weinberg equilibrium in controls. SNPs with significant p-values were excluded from further analysis.
Information from HapMap where not otherwise indicated.
LB4= leukotriene B4
ALOX5, ALOX5AP, and LTA4H represent a different subpathway of AA metabolism. These genes encode arachidonate 5-lipoxygenase and arachidonate 5-lipoxygenase-activating protein (aka, FLAP), respectively, which are involved in synthesizing leukotrienes from arachidonic acid. Leukotrienes are important eicosanoid immunomodulators that are overexpressed in malignant cells of several cancers (8). Previous research has indicated that leukotrienes may have a role in increasing cell proliferation and decreasing apoptosis among cancer cells (8,27). LTA4H is the leukotriene A4 hydrolase gene. LTA4H acts downstream of ALOX5 and converts leukotriene A4 into leukotriene B4, which is a chemotactic agent for neutrophils (28,29). Thus, these five genes were selected both on the basis of biological plausibility and the availability of previous research implying that polymorphisms in these genes may be relevant to prostate cancer susceptibility and development.
The 14 SNPs of the five candidate genes were chosen based on the following criteria: 1) commonality (>5% minor allele frequency in non-Hispanic whites); 2) potential biological significance (nonsynonymous SNPs or those shown to be associated with other cancers or inflammatory diseases); and 3) for the purpose of capturing more of the genetic variation within the candidate genes. More information about these SNPs is provided in Table I.
Genotyping.
DNA was originally extracted from peripheral blood lymphocytes using the QIAmp DNA Blood Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol and stored in the Global Biorepository. DNA quality was assessed by measuring the A260/A280 ratio on a spectrometer. Genotyping was done using the Sequenom MassARRAY iPLEX platform according to the manufacturer’s instructions (http://www.sequenom.com/sEq.genotyping.html). The quality control analysis included the genotyping of internal positive control samples, the use of no template controls, and the use of replicates for 10% of the samples. Genotyping plates were reviewed for results from positive / negative / DNA control wells that were organized in specific patterns to assist in the quality control process and to ensure correct plate orientations during processing and data review. Call rates ranged from 83-94% with only two being <90%.
Statistical Analysis.
For each SNP, deviations from Hardy-Weinberg equilibrium (HWE) were assessed among controls using the Χ2-test. Allele and genotype frequency distributions were compared between the cases and controls, and the best genetic model for each SNP was determined using Akaike’s information criterion (AIC). Crude and age-adjusted odds ratios and 95% confidence intervals (CIs) for the association between each SNP and prostate cancer risk were calculated using logistic regression, both in the overall study population and stratified by obesity (BMI≥30). As previously discussed, analyses were stratified by obesity because we hypothesized that the chronic low-level inflammation induced by obesity could potentially modify the effect of the polymorphisms on prostate cancer risk.
Pairwise linkage disequilibrium (LD) between the SNPs was examined, using LD coefficient r2 and Lewontin’s standardized coefficient D’. Haplo.stats (http://www.mayo.edu/stagen) was used to conduct haplotype analyses on the four PTGES2 SNPs.
Results
Information on selected characteristics of prostate cancer cases and cancer-free controls is provided in Table II. Although controls were slightly younger and less likely to smoke, these differences were not statistically significant.
Table II.
Characteristics of study population by case-control status
| Characteristics | Cases (n=585) No. (%) |
Controls (n=585) No. (%) |
P-value |
|---|---|---|---|
| Mean age in years ± SD | 70.6 ± 9.1 | 66.3 ± 8.1 | 0.99 |
| Smokinga | 0.05 | ||
| Never | 221 (37.8) | 254 (43.4) | |
| Ever | 364 (62.2) | 331 (56.6) | |
| Obesityb | 0.63 | ||
| BMI <30 | 448 (76.6) | 455 (77.8) | |
| BMI ≥30 | 137 (23.4) | 130 (22.2) | |
| Gleason Score | |||
| ≤4 | 48 (8.3) | ||
| 5-7 | 468 (80.6) | ||
| ≥8 | 65 (11.2) | ||
Includes cigarettes, cigars, and/or pipe use.
Obesity status at study recruitment. Cases and controls were matched on body mass index.
The genotype distributions of three (ALOX5 rs2029253, ALOX5 rs6593482, and ALOX5AP rs4076128) of the 14 SNPs of interest demonstrated deviations from HWE in the controls (Table I); these SNPs were excluded from further analysis. Logistic regression analyses of the remaining 11 SNPs revealed that two, PTGES2 rs10987883 and LTA4H rs1978331, were associated with prostate cancer risk (Table III). An increased risk, of borderline statistical significance, was associated with the minor allele (G) of PTGES2 rs10987883 among non-obese individuals, using a recessive model as determined by AIC (age-adjusted OR=2.53, 95% CI: 0.99-6.55 for GG vs. AA + AG). Among the overall study population, the heterozygous genotype (TC) of LTA4H rs1978331 was significantly protective against prostate cancer, compared to the homozygous genotypes (TT + CC) [age-adjusted OR=0.66, 95% CI: 0.50-0.86]. This inverse association remained present among both obese and non-obese subgroups. None of the other SNPs of interest were found to be significantly associated with prostate cancer risk in the overall study population or stratified by obesity.
Table III.
Genotype distributions and age-adjusted odds ratios for significant associations between arachidonic acid metabolism pathway SNPs and prostate cancer risk
| Arachidonic Acid Metabolism Subpathway |
Gene & SNP ID |
Modela | Genotype | No. of Cases/Controls |
Overall Study Population (n= 585 cases; 585 controls) |
Stratified by Obesity | |
|---|---|---|---|---|---|---|---|
| Non-Obese | Obese | ||||||
| OR (95% C.I.) | OR (95% C.I.) | OR (95% C.I.) | |||||
| Leukotriene Synthesis Subpathway | |||||||
| LTA4H rs1978331 | Overdominant | TT + CC | 284/267 | 1.00 | 1.00 | 1.00 | |
| TC | 174/244 | 0.66 (0.50-0.86) | 0.70 (0.52-0.95) | 0.51 (0.29-0.90) | |||
| Unrestricted | TT | 183/175 | 1.00 | ||||
| TC | 174/244 | 0.67 (0.50-0.90) | |||||
| CC | 101/193 | 1.05 (0.73-1.51) | |||||
| Prostaglandin Synthesis Subpathway | |||||||
| PTGES2 rs10987883 | Recessive | AA + AG | 503/526 | 1.00 | 1.00 | 1.00 | |
| GG | 19/10 | 1.52 (0.69-3.35) | 2.53 (0.99-6.55) | 0.17 (0.02-1.57) | |||
| Unrestricted | AA | 384/404 | 1.00 | ||||
| AG | 119/122 | 1.09 (0.81-1.46) | |||||
| GG | 19/10 | 1.55 (0.70-3.42) | |||||
The best genetic model for each SNP (bold), as determined by AIC, is provided along with the unrestricted model. Stratified analysis results are only provided for the best genetic models.
Strong LD (r2 > 0.8) was observed between the four SNPs in the PTGES2 gene. Haplotype analyses were conducted on these SNPs, but no significant associations were found (Table IV). The global score test indicated that haplotype distributions were not significantly different between cases and controls (p=0.12).
Table IV.
Age-adjusted associations between PTGES2 haplotypes and prostate cancer risk
| Gene | Haplotype | Frequencya |
OR (95% CI) | Global score test p-valueb |
||
|---|---|---|---|---|---|---|
| Total | Case | Control | ||||
| PTGES2 | 0.12 | |||||
| C A C G | 0.633 | 0.645 | 0.620 | 1.00 | ||
| C A T G | 0.187 | 0.185 | 0.188 | 1.05 (0.83-1.32) | ||
| T G C A | 0.118 | 0.119 | 0.117 | 0.99 (0.76-1.30) | ||
| C A C A | 0.032 | 0.033 | 0.032 | 1.11 (0.67-1.84) | ||
Only haplotypes with frequencies>0.01 were examined; PTGES2 rs884115, rs10987883, rs13283456, and rs4837240.
Generated by permutation test (10,000x).
Discussion
In this study, we conducted a pathway-based analysis to investigate the role of putative functional arachidonic acid metabolism gene polymorphisms in prostate cancer susceptibility. Our findings from the single-locus analyses indicate that one SNP from each of the two major branches of AA metabolism, the prostaglandin synthesis subpathway (PTGES2 rs10987883) and the leukotriene synthesis subpathway (LTA4H rs1978331), may be relevant to prostate cancer risk. While the TC genotype of LTA4H rs1978331 was significantly protective against prostate cancer overall, the GG genotype of PTGES2 rs10987883 was associated with an increased prostate cancer risk only among non-obese individuals. Interestingly, the divergent point estimates obtained from the stratified analyses imply that obesity, and its associated inflammatory and metabolic sequelae, may potentially be effect modifiers of the relationship between this PTGES2 SNP and prostate cancer risk.
Previous studies examining the relationship between SNPs in AA metabolism genes and prostate cancer risk have largely focused on the genetic variation in the PTGS2/COX2 gene, rather than taking a more inclusive pathway-based approach (13-16,18). Although these studies have provided some evidence that polymorphisms in the PTGS2 gene may influence prostate cancer risk, their findings have been relatively inconsistent. For example, Cheng et al. reported that the heterozygous genotype of PTGS2 rs2745557 (GA) was associated with a 36% lower odds of prostate cancer development in a hospital-based case-control study of 506 advanced prostate cancer cases and 506 healthy controls (GA vs. GG, OR=0.64, 95% CI: 0.49-0.84) (16). By contrast, an analysis conducted in a large, population-based Swedish case-control study of 1378 cases and 782 controls did not find an association between this SNP and localized or advanced disease (15). Like the latter study, we were also unable to detect an association between PTGS2 rs274557 and prostate cancer risk. Because our study is one of the first to take a pathway-based approach to identifying AA metabolism SNPs relevant to prostate cancer risk, we cannot corroborate the two significant associations reported here (PTGES2 rs10987883 and LTA4H rs1978331) with the previous literature.
One reason why the previous literature on AA metabolism genes and prostate cancer has focused so heavily on PTGS2 is the plethora of data suggesting that overexpression of COX2, which is considered to be the rate-limiting enzyme in prostaglandin synthesis, is involved in prostate carcinogenesis (11). However, several lines of evidence also support the potential importance of PTGES2 and LTA4H expression in the carcinogenic process (2,9,25,26). The product of the PTGES2 gene, prostaglandin E synthase-2, facilitates the conversion of prostaglandin H2 to PGE2, which is also significantly overexpressed in cancer tissues (2,25,26). Overproduction of PGE2 may play a role in immunosuppression, angiogenesis, and prostate cancer progression (2,9,25,26). In fact, it has been suggested that PGE2 should be a target for anti-angiogenic cancer therapy to provide an alternative to treatment with COX2 inhibitors, which have been associated with numerous side effects (9).
There is also strong biological plausibility behind a potential relationship between SNPs of the LTA4H gene and prostate cancer susceptibility. LTA4H codes for leukotriene A4 hydrolase, which facilitates the synthesis of leukotriene B4 (LB4) from leukotriene A4. LB4 is a strong chemotactic factor for mast cells and neutrophils and has been implicated in the pathogenesis of several chronic inflammatory diseases, including asthma, rheumatoid arthritis, and inflammatory bowel disease (29,30). Leukotrienes, including LB4, are involved in a multitude of functions that are relevant to carcinogenesis, including increasing transcription of oncogenes, encouraging tumor cell proliferation, and improving tumor survival by decreasing apoptosis (8). It has, therefore, been suggested that disruption of leukotriene synthesis may prevent cancer growth (8,31). Here, we found that the TC genotype of LTA4H rs1978331 is associated with a significantly reduced risk of prostate cancer, compared to either homozygous genotype. Because the exact functional relevance of the heterozygous genotype of this intronic SNP is unknown, we cannot fully explain this seeming heterozygote advantage. However, given that the T allele of this SNP is associated with higher LB4 level (32,33), the TC genotype could be associated with somewhat lower LB4 levels than the TT genotype, but higher levels than the CC genotype, thus potentially reducing the tumorigenic activities of this leukotriene without completely diminishing its inflammatory effects. In fact, Tobin et al. arrived at a similar hypothesis with regard to the function of the heterozygous genotype of this SNP when they found that the TC genotype conferred protection against both tuberculosis and leprosy (33). They postulated that if the variant allele of this SNP resulted in lower LB4 expression levels, then the heterozygous genotype may confer an ideal balance of eicosanoids with pro- and anti-inflammatory effects during infection. Alternatively, we cannot discount the possibility that our finding may be either an artifact of our data or due to chance alone. Therefore, replication of this association in other larger study populations, as well as additional functional studies on this SNP, are warranted.
Perhaps the most interesting implication of our study is that obesity may be modifying the relationship between PTGES2 rs10987883 and prostate cancer risk. We found that among the non-obese subgroup of our population, the GG genotype of PTGES2 rs10987883 was associated with an increased prostate cancer risk, although this association was only of borderline statistical significance. Conversely, this genotype was associated with a non-significant protective effect among obese individuals. Obesity induces numerous metabolic, hormonal, and inflammatory consequences, including lower testosterone levels and increased secretion of certain cytokines (e.g. IL-12, IL-6, and VEGF), C-reactive protein, and other proinflammatory agents (20,22,23,34). Transcriptional activation of AA metabolism genes can be regulated by various cytokines. Thus, it is feasible that in the context of the chronic state of inflammation induced by obesity, the impact of this SNP on prostate cancer risk could differ, even by a mechanism as simple as a differential level of expression among obese individuals. Nevertheless, the vast inconsistencies in the results of studies that have attempted to clarify the relationship between obesity and prostate cancer point to the involvement of an extremely complicated network of physiological processes that cannot be easily discerned (20,21,23). We acknowledge that our stratified analysis on the effect of PTGES2 rs10987883 was dependent on a very small number of individuals with the GG genotype and that the width of the confidence intervals reflects this fact. However, the possibility of an interaction between PTGES2 rs10987883 and obesity in prostate cancer susceptibility poses an interesting hypothesis to explore in future research.
A limitation of our study is that body mass index was measured at study recruitment, which was after cancer diagnosis for the cases. However, the inability to assess exposures at the etiologic time period of interest is a common problem in case-control studies, and the stratified analyses that we conducted were intended for the purposes of hypothesis generation. In addition, information on cancer stage and grade was unavailable for a substantial proportion of the cases (60.5% and 39.8%, respectively), precluding us from being able to examine associations specifically with advanced prostate cancer. Despite these limitations, our findings have provided some evidence that SNPs in the PTGES2 and LTA4H genes may be relevant to prostate cancer etiology. It will be essential to validate these associations in future studies that include information on advanced disease and have greater statistical power for the detection of potential interactions. Furthermore, because our current study was restricted to non-Hispanic white participants, the effects of these SNPs should also be examined in African-American and Hispanic men, who tend to have higher risk and worse prognosis for prostate cancer, to see if these associations hold across racial and ethnic groups (35).
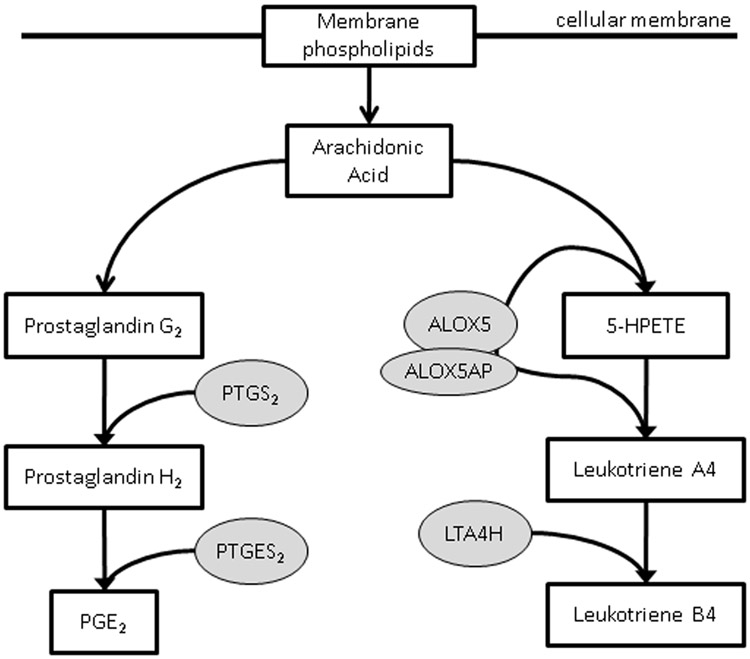
Figure 1.
Simplified depiction of the arachadonic acid metabolism pathway, illustrating the roles of the protein products of genes of interest in the prostaglandin synthesis and leukotriene synthesis branches of the pathway.
Footnotes
Disclosure Statement:
The authors have no relationships to disclose.
References
- 1.Patel MI, Kurek C, Dong Q. The arachidonic acid pathway and its role in prostate cancer development and progression. J Urol 2008;179(5):1668–1675. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh J, Myers CE Jr.. Arachidonic acid metabolism and cancer of the prostate. Nutrition 1998;14(1):48–49. [DOI] [PubMed] [Google Scholar]
- 3.Harizi H, Corcuff JB, Gualde N. Arachidonic-acid-derived eicosanoids: roles in biology and immunopathology. Trends Mol Med 2008;14(10):461–469. [DOI] [PubMed] [Google Scholar]
- 4.Taber L, Chiu CH, Whelan J. Assessment of the arachidonic acid content in foods commonly consumed in the American diet. Lipids 1998;33(12):1151–1157. [DOI] [PubMed] [Google Scholar]
- 5.Rose DP, Connolly JM. Regulation of tumor angiogenesis by dietary fatty acids and eicosanoids. Nutr Cancer 2000;37(2):119–127. [DOI] [PubMed] [Google Scholar]
- 6.Rose DP, Connolly JM. Effects of fatty acids and eicosanoid synthesis inhibitors on the growth of two human prostate cancer cell lines. Prostate 1991;18(3):243–254. [DOI] [PubMed] [Google Scholar]
- 7.Liu B, Maher RJ, De Jonckheere JP, Popat RU, Stojakovic S, Hannun YA, Porter AT, Honn KV. 12(S)-HETE increases the motility of prostate tumor cells through selective activation of PKC alpha. Adv Exp Med Biol 1997;400B:707–718. [PubMed] [Google Scholar]
- 8.Peters-Golden M, Henderson WR Jr. Leukotrienes. N Engl J Med 2007;357(18):1841–1854. [DOI] [PubMed] [Google Scholar]
- 9.Jain S, Chakraborty G, Raja R, Kale S, Kundu GC. Prostaglandin E2 regulates tumor angiogenesis in prostate cancer. Cancer Res 2008;68(19):7750–7759. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, John EM, Ingles SA. 5-lipoxygenase and 5-lipoxygenase-activating protein gene polymorphisms, dietary linoleic acid, and risk for breast cancer. Cancer Epidemiol Biomarkers Prev 2008;17(10):2748–2754. [DOI] [PubMed] [Google Scholar]
- 11.Hussain T, Gupta S, Mukhtar H. Cyclooxygenase-2 and prostate carcinogenesis. Cancer Lett 2003;191(2):125–135. [DOI] [PubMed] [Google Scholar]
- 12.Hyde CA, Missailidis S. Inhibition of arachidonic acid metabolism and its implication on cell proliferation and tumour-angiogenesis. Int Immunopharmacol 2009;9(6):701–715. [DOI] [PubMed] [Google Scholar]
- 13.Panguluri RC, Long LO, Chen W, Wang S, Coulibaly A, Ukoli F, Jackson A, Weinrich S, Ahaghotu C, Isaacs W, Kittles RA. COX-2 gene promoter haplotypes and prostate cancer risk. Carcinogenesis 2004;25(6):961–966. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez P, de Beer PM, van der Merwe L, Heyns CF. COX-2 promoter polymorphisms and the association with prostate cancer risk in South African men. Carcinogenesis 2008;29(12):2347–2350. [DOI] [PubMed] [Google Scholar]
- 15.Shahedi K, Lindstrom S, Zheng SL, Wiklund F, Adolfsson J, Sun J, Augustsson-Balter K, Chang BL, Adami HO, Liu W, Gronberg H, Xu J. Genetic variation in the COX-2 gene and the association with prostate cancer risk. Int J Cancer 2006;119(3):668–672. [DOI] [PubMed] [Google Scholar]
- 16.Cheng I, Liu X, Plummer SJ, Krumroy LM, Casey G, Witte JS. COX2 genetic variation, NSAIDs, and advanced prostate cancer risk. Br J Cancer 2007;97(4):557–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu W, Wei BB, Shan X, Liu P. −765G>C and 8473T>C polymorphisms of COX-2 and cancer risk: a meta-analysis based on 33 case-control studies. Mol Biol Rep 2010;37(1):277–288. [DOI] [PubMed] [Google Scholar]
- 18.Danforth KN, Hayes RB, Rodriguez C, Yu K, Sakoda LC, Huang WY, Chen BE, Chen J, Andriole GL, Calle EE, Jacobs EJ, Chu LW, Figueroa JD, Yeager M, Platz EA, Michaud DS, Chanock SJ, Thun MJ, Hsing AW. Polymorphic variants in PTGS2 and prostate cancer risk: results from two large nested case-control studies. Carcinogenesis 2008;29(3):568–572. [DOI] [PubMed] [Google Scholar]
- 19.Hsing AW, Sakoda LC, Chua S Jr. Obesity, metabolic syndrome, and prostate cancer. Am J Clin Nutr 2007;86(3):s843–857. [DOI] [PubMed] [Google Scholar]
- 20.Mistry T, Digby JE, Desai KM, Randeva HS. Obesity and prostate cancer: a role for adipokines. Eur Urol 2007;52(1):46–53. [DOI] [PubMed] [Google Scholar]
- 21.Clarke NW, Brown MD. The influence of lipid metabolism on prostate cancer development and progression: is it time for a closer look? Eur Urol 2007;52(1):3–4. [DOI] [PubMed] [Google Scholar]
- 22.Wang MH, Helzlsouer KJ, Smith MW, Hoffman-Bolton JA, Clipp SL, Grinberg V, De Marzo AM, Isaacs WB, Drake CG, Shugart YY, Platz EA. Association of IL10 and other immune response- and obesity-related genes with prostate cancer in CLUE II. Prostate 2009;69(8):874–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freedland SJ, Platz EA. Obesity and prostate cancer: making sense out of apparently conflicting data. Epidemiol Rev 2007;29:88–97. [DOI] [PubMed] [Google Scholar]
- 24.Wang W, Bergh A, Damber JE. Cyclooxygenase-2 expression correlates with local chronic inflammation and tumor neovascularization in human prostate cancer. Clin Cancer Res 2005;11(9):3250–3256. [DOI] [PubMed] [Google Scholar]
- 25.Chaudry A, McClinton S, Moffat LE, Wahle KW. Essential fatty acid distribution in the plasma and tissue phospholipids of patients with benign and malignant prostatic disease. Br J Cancer 1991;64(6):1157–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaudry AA, Wahle KW, McClinton S, Moffat LE. Arachidonic acid metabolism in benign and malignant prostatic tissue in vitro: effects of fatty acids and cyclooxygenase inhibitors. Int J Cancer 1994;57(2):176–180. [DOI] [PubMed] [Google Scholar]
- 27.Ihara A, Wada K, Yoneda M, Fujisawa N, Takahashi H, Nakajima A. Blockade of leukotriene B4 signaling pathway induces apoptosis and suppresses cell proliferation in colon cancer. J Pharmacol Sci 2007;103(1):24–32. [DOI] [PubMed] [Google Scholar]
- 28.Bray MA. Leukotriene B4: an inflammatory mediator with vascular actions in vivo. Agents Actions Suppl 1982;11:51–61. [PubMed] [Google Scholar]
- 29.Griffiths RJ, Pettipher ER, Koch K, Farrell CA, Breslow R, Conklyn MJ, Smith MA, Hackman BC, Wimberly DJ, Milici AJ, et al. Leukotriene B4 plays a critical role in the progression of collagen-induced arthritis. Proc Natl Acad Sci U S A 1995;92(2):517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holloway JW, Barton SJ, Holgate ST, Rose-Zerilli MJ, Sayers I. The role of LTA4H and ALOX5AP polymorphism in asthma and allergy susceptibility. Allergy 2008;63(8):1046–1053. [DOI] [PubMed] [Google Scholar]
- 31.Rioux N, Castonguay A. Inhibitors of lipoxygenase: a new class of cancer chemopreventive agents. Carcinogenesis 1998;19(8):1393–1400. [DOI] [PubMed] [Google Scholar]
- 32.Helgadottir A, Manolescu A, Helgason A, Thorleifsson G, Thorsteinsdottir U, Gudbjartsson DF, Gretarsdottir S, Magnusson KP, Gudmundsson G, Hicks A, Jonsson T, Grant SF, Sainz J, O'Brien SJ, Sveinbjornsdottir S, Valdimarsson EM, Matthiasson SE, Levey AI, Abramson JL, Reilly MP, Vaccarino V, Wolfe ML, Gudnason V, Quyyumi AA, Topol EJ, Rader DJ, Thorgeirsson G, Gulcher JR, Hakonarson H, Kong A, Stefansson K. A variant of the gene encoding leukotriene A4 hydrolase confers ethnicity-specific risk of myocardial infarction. Nat Genet 2006;38(1):68–74. [DOI] [PubMed] [Google Scholar]
- 33.Tobin DM, Vary JC Jr., Ray JP, Walsh GS, Dunstan SJ, Bang ND, Hagge DA, Khadge S, King MC, Hawn TR, Moens CB, Ramakrishnan L. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell 2010;140(5):717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahima RS, Osei SY. Adipokines in obesity. Front Horm Res 2008;36:182–197. [DOI] [PubMed] [Google Scholar]
- 35.Altekruse SF, Huang L, Cucinelli JE, McNeel TS, Wells KM, Oliver MN. Spatial patterns of localized-stage prostate cancer incidence among white and black men in the southeastern United States, 1999-2001. Cancer Epidemiol Biomarkers Prev 2010;19(6):1460–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nitz I, Fisher E, Grallert H, Li Y, Gieger C, Rubin D, Boeing H, Spranger J, Lindner I, Schreiber S, Rathmann W, Gohlke H, Doring A, Wichmann HE, Schrezenmeir J, Doring F, Illig T. Association of prostaglandin E synthase 2 (PTGES2) Arg298His polymorphism with type 2 diabetes in two German study populations. J Clin Endocrinol Metab 2007;92(8):3183–3188. [DOI] [PubMed] [Google Scholar]
- 37.Fischer A, Grallert H, Bohme M, Gieger C, Boomgaarden I, Heid I, Wichmann HE, Doring F, Illig T. Association analysis between the prostaglandin E synthase 2 R298H polymorphism and body mass index in 8079 participants of the KORA study cohort. Genet Test Mol Biomarkers 2009;13(2):223–226. [DOI] [PubMed] [Google Scholar]
- 38.Wung SF, Aouizerat BE. Candidate genes of the 5-lipoxygenase pathway in acute coronary syndrome: a pilot study. Biol Res Nurs 2008;9(4):280–292. [DOI] [PubMed] [Google Scholar]
- 39.Assimes TL, Knowles JW, Priest JR, Basu A, Volcik KA, Southwick A, Tabor HK, Hartiala J, Allayee H, Grove ML, Tabibiazar R, Sidney S, Fortmann SP, Go A, Hlatky M, Iribarren C, Boerwinkle E, Myers R, Risch N, Quertermous T. Common polymorphisms of ALOX5 and ALOX5AP and risk of coronary artery disease. Hum Genet 2008;123(4):399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crosslin DR, Shah SH, Nelson SC, Haynes CS, Connelly JJ, Gadson S, Goldschmidt-Clermont PJ, Vance JM, Rose J, Granger CB, Seo D, Gregory SG, Kraus WE, Hauser ER. Genetic effects in the leukotriene biosynthesis pathway and association with atherosclerosis. Hum Genet 2009;125(2):217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herb F, Thye T, Niemann S, Browne EN, Chinbuah MA, Gyapong J, Osei I, Owusu-Dabo E, Werz O, Rusch-Gerdes S, Horstmann RD, Meyer CG. ALOX5 variants associated with susceptibility to human pulmonary tuberculosis. Hum Mol Genet 2008;17(7):1052–1060. [DOI] [PubMed] [Google Scholar]
- 42.Helgadottir A, Manolescu A, Thorleifsson G, Gretarsdottir S, Jonsdottir H, Thorsteinsdottir U, Samani NJ, Gudmundsson G, Grant SF, Thorgeirsson G, Sveinbjornsdottir S, Valdimarsson EM, Matthiasson SE, Johannsson H, Gudmundsdottir O, Gurney ME, Sainz J, Thorhallsdottir M, Andresdottir M, Frigge ML, Topol EJ, Kong A, Gudnason V, Hakonarson H, Gulcher JR, Stefansson K. The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat Genet 2004;36(3):233–239. [DOI] [PubMed] [Google Scholar]
- 43.Lima JJ, Zhang S, Grant A, Shao L, Tantisira KG, Allayee H, Wang J, Sylvester J, Holbrook J, Wise R, Weiss ST, Barnes K. Influence of leukotriene pathway polymorphisms on response to montelukast in asthma. Am J Respir Crit Care Med 2006;173(4):379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]