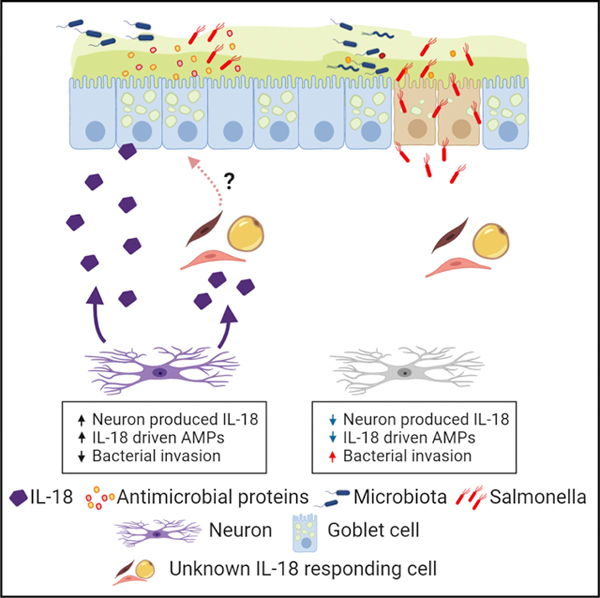
SUMMARY
Mucosal barrier immunity is essential for the maintenance of the commensal microflora and combating invasive bacterial infection. Although immune and epithelial cells are thought to be the canonical orchestrators of this complex equilibrium, here, we show that the enteric nervous system (ENS) plays an essential and non-redundant role in governing the antimicrobial protein (AMP) response. Using confocal microscopy and single-molecule fluorescence in situ mRNA hybridization (smFISH) studies, we observed that intestinal neurons produce the pleiotropic cytokine IL-18. Strikingly, deletion of IL-18 from the enteric neurons alone, but not immune or epithelial cells, rendered mice susceptible to invasive Salmonella typhimurium (S.t.) infection. Mechanistically, unbiased RNA sequencing and single-cell sequencing revealed that enteric neuronal IL-18 is specifically required for homeostatic goblet cell AMP production. Together, we show that neuron-derived IL-18 signaling controls tissue-wide intestinal immunity and has profound consequences on the mucosal barrier and invasive bacterial killing.
In Brief
IL-18 produced by the enteric nervous system (ENS) is crucial for host protection against Salmonella infection via goblet cell antimicrobial peptide production, highlighting the ENS as a crucial immune mediator in bacterial defense.
Graphical Abstract
INTRODUCTION
The intestinal mucosal barrier provides essential protection from pathogenic microbial invasion while also facilitating a non-inflammatory, symbiotic relationship with the commensal microflora (Hooper and Macpherson, 2010). Although it is well understood how immune and epithelial cell crosstalk arbitrates this balance (Clark and Coopersmith, 2007), little is known of how intestinal neurons influence this critical axis during homeostasis or upon bacterial infection. Intriguingly, although the enteric nervous system (ENS) permeates the entirety of the intestinal tissue and has the capacity to rapidly relay sensory information to a vast network of both autologous and non-autologous cell types (Furness et al., 2014), its ability to act as a major immunological sensory platform is only beginning to emerge (Lai et al., 2019; Matheis et al., 2020; Talbot et al., 2019; Yoo and Mazmanian, 2017).
Patients born with Hirschsprung disease are characterized by an absence of distal bowel ganglia resulting in loss of colonic motility. These patients are at risk of developing Hirschsprung-associated enterocolitis (HAEC), a serious and life-threatening complication (Austin, 2012; Hirschsprung, 1887; Okamoto and Ueda, 1967). Although the pathogenesis of HAEC is currently unknown, it correlates with major mucus dysregulation and microbial dysbiosis (Thiagarajah et al., 2014), suggesting an intimate connection between the ENS and the mucosal barrier. Concordantly, exogenous administration of neuro-modulatory compounds targeting neurotransmitter receptors have been shown to trigger widespread effects on mucus and antimicrobial protein (AMP) secretion by intestinal epithelial cells (Birchenough et al., 2016; Satoh et al., 1992). Numerous observational studies have concluded that the cells of the ENS have the capacity to respond to pathogen-associated molecular patterns inferred by their expression of classical pattern recognition receptors (Barajon et al., 2009). This work was followed by studies showing that the ENS can physiologically contribute to the inflammatory response through intrinsic expression of not only immunomodulatory neuropeptides and neurotransmitters, but also known inflammatory cytokines (Coquenlorge et al., 2014). Taken together, these observations are highly suggestive that the ENS may play a key role in regulating mucosal barrier immunity.
Intense research efforts using reverse genetics have greatly aided our collective mechanistic understanding of how the immune and epithelial cell compartments control the mucosal barrier. Interleukin 18 (IL-18) has emerged as a key pleiotropic cytokine in intestinal homeostasis and host defense (Nowarski et al., 2015). In immune cells, IL-18 is a proinflammatory cytokine and required to combat invasive bacterial infections such as Salmonella typhimurium (S.t.) (Broz et al., 2012; Lara-Tejero et al., 2006; Raupach et al., 2006). Under homeostatic conditions, IL-18-deficent mice display dysbiosis (Elinav et al., 2011) and a major defect in epithelial AMP production, a process thought to occur through an autocrine epithelial IL-18 signaling axis (Levy et al., 2015). Indeed, the individual importance of both immune and epithelial IL-18 in the intestine was recently demonstrated in models of autoinflammatory colitis using conditional IL-18- and IL-18R1-deficient mice (Nowarski et al., 2015). Specific ablation of IL-18 from either immune or epithelial compartments was sufficient to protect mice from chemically induced colitis, indicating that both sources can act in concert to drive pathogenic inflammation (Nowarski et al., 2015). Although IL-18 is clearly critical to immunity and maintenance of the mucosal barrier, our understanding of this cytokine outside of immune and epithelial cells remains largely unknown. In this present study, we discovered that canonical sources of IL-18 were redundant in combating invasive S.t. infection. We identified enteric neurons as the essential source of IL-18 that drives goblet cell AMP production and S.t. protection. This study demonstrates that the ENS is a critical arm of the innate immune response and is essential for coordinating not only mucosal barrier homeostasis but also combatting invasive bacterial infection.
RESULTS
Epithelial Cell- and Immune Cell-Derived IL-18 Does Not Protect against Enteric S.t. Infection
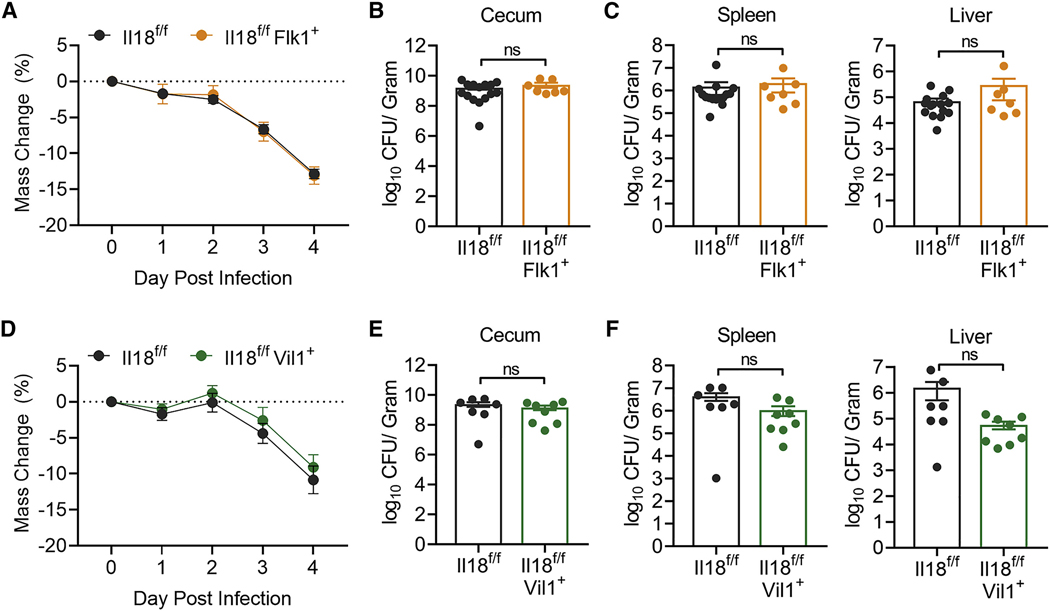
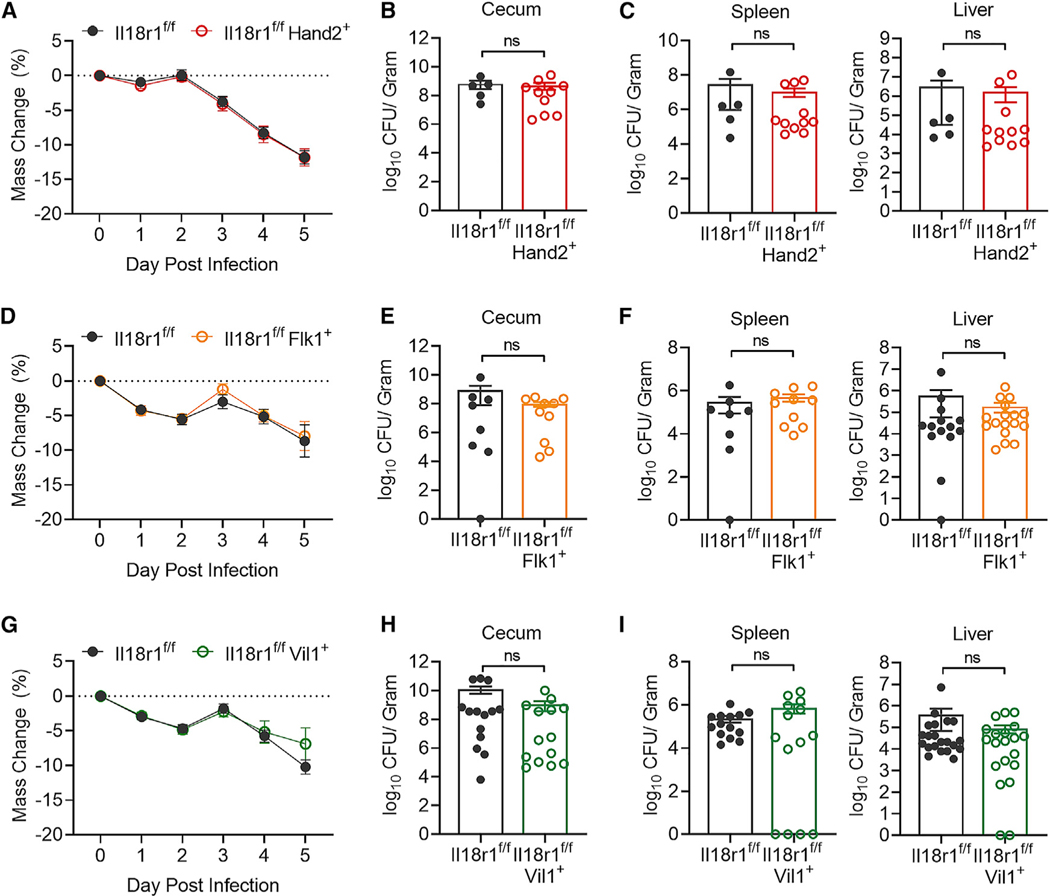
Becasue epithelial cells produce the majority of IL-18 in the intestine at homeostasis and myeloid cells rapidly upregulate IL-18 during inflammation, both cell types are thought to be the major contributors to IL-18-mediated host defense. To test this, we crossed Il18f/f mice with Flk1-Cre (Il18f/fFlk1+), which enables genetic IL-18 deletion in hematopoietic and endothelial cell lineages (Motoike et al., 2003; Nowarski et al., 2015). Il18f/f and Il18f/fFlk1+ mice were subjected to the streptomycin pretreatment S.t. model of colonic infection (Barthel et al., 2003). Interestingly, contrary to the phenotype observed in IL-18-deficient animals, absence of IL-18 in hematopoietic cells did not result in increased susceptibly to S.t. induced weight loss (Figure 1A), cecal bacterial abundance (Figure 1B), or dissemination to the spleen or liver (Figure 1C). We next crossed Il18f/f mice with mice expressing Villin1-Cre (Il18f/fVil1+) for targeted deletion of IL-18 in intestinal epithelial cells (Madison et al., 2002). Similarly, loss of epithelial-derived IL-18 had no effect on S.t.-induced weight loss (Figure 1D), cecal colonization (Figure 1E), or peripheral tissue infection (Figure 1F). Thus, neither immune- nor epithelial-derived IL-18 is required to drive anti-bacterial defense, leaving open the possibility that there is an unidentified source of IL-18 that confers protection against S.t. infection.
Figure 1. Epithelial Cell- and Immune Cell-Derived IL-18 Does Not Protect against Enteric S.t. Infection.
(A) Weight loss of Il18f/f (n = 15) or Il18f/fFlk1+ (n = 7) mice infected with S.t. Data represent mean ± SEM; unpaired t test was used for statistical analysis. Data represent two independent experiments combined.
(B and C) S.t. colony-forming unit (CFU)/g of (B) cecum, (C) spleen, and liver from Il18f/f or Il18f/fFlk1+ mice 4 days post-infection. Data represent mean ± SEM; Mann-Whitney test was used for statistical analysis. Each dot represents one mouse. Data represent two independent experiments combined.
(D) Weight loss of Il18f/f (n = 7) or Il18f/fVil1+ (n = 8) mice infected with S.t. Data represent mean ± SEM; unpaired t test was used for statistical analysis. Data represent two independent experiments combined from three total independent experiments.
(E and F) S.t. CFU/g of (E) cecum, (F) spleen, and liver from Il18f/f or Il18f/fVil1+ mice 4 days post-infection. Data represent mean ± SEM; Mann-Whitney test was used for statistical analysis. Each dot represents one mouse. Data represent two independent experiments combined from three total independent experiments.
Enteric Neurons Express IL-18
We were particularly interested in the possibility that enteric neurons could produce IL-18, because neurons in the brain are reported to produce the cytokine (Kuwahara-Otani et al., 2017; Sugama et al., 2002). To investigate this, we conducted whole-mount confocal microscopy of the myenteric plexus isolated from rat tissue for IL-18 reactivity. Strikingly, we uncovered a large subset of neuronal ganglia, assessed by the pan-neuronal marker beta tubulin 3 class III (Tubb3) (Caccamo et al., 1989; Drokhlyansky et al., 2019), that were expressing IL-18 in the myenteric plexus (Figure 2A). Orthogonal x,y,z plane analysis confirmed IL-18 immunoreactivity was within the neuronal ganglia itself (Figure S1A). In addition, we observed IL-18-positive staining in a population of neurons located near the base of crypts and innervating villi (Figures 2B and 2C), suggesting that neuronal IL-18 may function in both the myenteric plexus and submucosa. To determine if IL-18+ neurons were nitrergic or cholinergic in nature, we co-stained for neuronal nitric oxide synthase (nNOS) and choline acetyltransferase (ChAT) (Qu et al., 2008). We observed a population of IL-18+ nNOS+ neurons but did not observe IL-18+ ChAT+ neurons (Figures 2D and S1F; data not shown). In order to confirm this observation, we performed single-molecule fluorescence in situ mRNA hybridization (smFISH) using Il18 mRNA probes in mice (Table S1). We verified the specificity of our Il18 mRNA probes using colon sections from Il18−/− mice (Figure S1B). We observed co-localization of Il18 mRNA probes with neuron-specific Tubb3 mRNA probes (Figure 2E). Together, these data demonstrate that enteric neurons are novel producers of IL-18 in the colon.
Figure 2. Enteric Neurons Express IL-18.
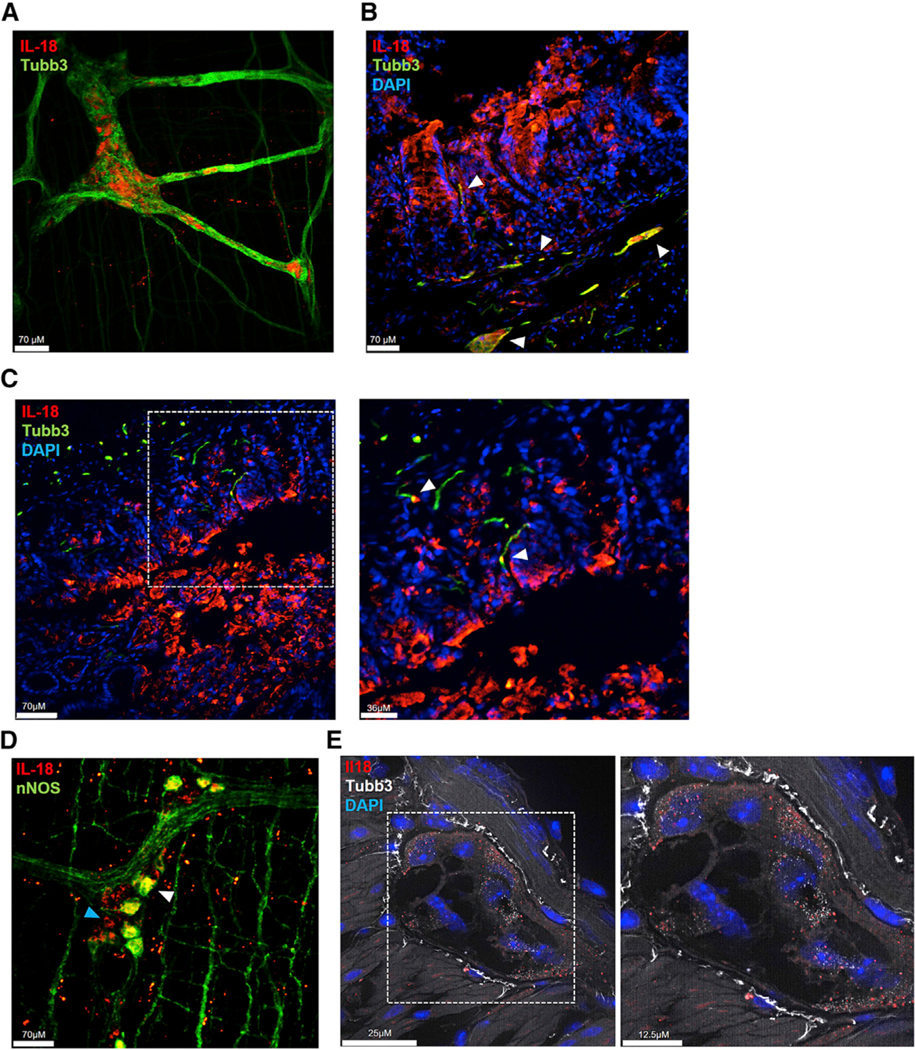
(A) Confocal immunofluorescence (IF) image of the myenteric plexus (MP) isolated from rat colon stained for IL-18 (red), the neuronal marker Tubulin beta 3 (Tubb3; green), and DAPI (blue).
(B and C) Confocal IF images of rat colon cross-sections stained for IL-18 (red), Tubb3 (green), and DAPI (blue). Arrows highlight IL-18+ Tubb3+ neurons, which can be seen near the base of crypts and in villi.
(D) Confocal IF image of the MP isolated from rat colon stained for IL-18 (red), nNOS (green), and DAPI (blue). White arrow highlights an IL-18+ nNOS+ cell body, blue arrow highlights an IL-18+ nNOS− cell body.
(E) Visualization of Il18 (red) and Tubb3 (white) transcripts and DAPI (blue) in mouse colon cross-sections detected by single-molecule fluorescence in situ mRNA hybridization.
We next mined two published single-cell RNA sequencing (scRNA-seq) datasets for expression of IL-18 in neurons. scRNA-seq conducted on enteric sensory neurons showed high expression of IL-18 in all neuron subtypes (Figure S1C) (Hockley et al., 2019). Expression of IL-18 in these cells was comparable with neuronal marker genes (Figure S1D). We next investigated a scRNA-seq dataset that examined central, peripheral, and enteric neurons (Zeisel et al., 2018). IL-18 is highly expressed in several neuron populations, including enteric neurons (Figure S1G). Interestingly, we did not observe expression of the closely related cytokine IL-1β in any neuron population (Figures S1E and S1G). The distinct presence of IL-18 and lack of IL-1β expression in neurons suggests there is a potential specific, yet unknown role for enteric neuronal IL-18.
Enteric Neuronal IL-18 Is Protective against S.t. Infection
To investigate the impact of ENS-derived IL-18 on intestinal immunity, we initially interbred Il18f/f mice to Nestin-Ert2-Cre (Il18f/f NesERT+) mice (Lagace et al., 2007). This allows Tamoxifen (TMX)-inducible deletion of IL-18 in a broad array of Nestin-expressing cells that include ENS cells, neuronal and glial precursor cells, mesenchymal stem cells, and vascular and lymphatic endothelial cells (Belkind-Gerson et al., 2013; Joseph et al., 2011; Koning et al., 2016; Kulkarni et al., 2017; Mendez-Ferrer et al., 2010; Zeisel et al., 2018). Because of coprophagy behavior, Il18f/fNesERT+ littermate mice were separated and administered TMX or vehicle daily for 5 days following 7 days of rest. We conducted 16S bacterial sequencing of feces and observed no significant difference between groups, indicating that loss of IL-18 did not introduce detectable confounding alterations to the luminal microbiota (Figure S2A). We next infected Il18f/fNesERT+ mice with S.t. and observed that TMX-treated mice had exaggerated weight loss and became moribund significantly quicker than vehicle-treated mice (Figures 3A and 3B). TMX-treated Il18f/fNesERT+ mice also exhibited increased bacterial abundance in the cecum (Figure 3C), liver, and spleen (Figure 3D) compared with vehicle control mice. To determine whether TMX administration itself could be a contributing factor to the observed S.t. susceptibility, we treated wild-type mice with TMX or vehicle and subjected them to S.t. infection. No differences were observed in terms of infection-induced weight loss or survival (Figures S2B and S2C). Therefore, we concluded that IL-18 from Nestin-expressing cells protects mice from invasive bacterial infection. As Nestin is highly expressed in ENS glial precursor cells (Shah et al., 1994; Wiese et al., 2004), we crossed Plp1-Ert-Cre mice with Il18f/f mice to conditionally delete IL-18 in enteric glial cells (Il18f/fPlp1+) (Doerflinger et al., 2003; Rao et al., 2015). Using TMX or vehicle treatment followed by S.t. infection, we demonstrated that deficiency of enteric glial IL-18 does not contribute to bacterial-induced weight loss (Figure 3E), cecal colonization (Figure 3F), or dissemination to the spleen and liver (Figure 3G). Due to Nestin’s broad expression pattern, to address if IL-18 specifically from enteric neurons provides protection against S.t. infection, we interbred Il18f/f mice with an animal harboring an enteric-neuron-specific Cre recombinase. The Hand2 transcription factor is essential for enteric neuron differentiation (D’Autréaux et al., 2007; Hendershot et al., 2007), and Hand2-Cre mice have proven to be an invaluable tool for conditional ablation and labeling of enteric neurons (Gabanyi et al., 2016; Ruest et al., 2003). Interbreeding Hand2-Cre mice with ROSA26loxStoploxTdTomato reporter mice (Madisen et al., 2010) confirmed highly specific and widespread enteric neuron recombination in the colon myenteric plexus (Figure S2D). Furthermore, scRNA-seq studies have recently confirmed that Hand2 is highly expressed in intestinal neurons (Zeisel et al., 2018) (Figure S1G). Therefore, we next generated Il18f/fHand2+ mice to conditionally delete IL-18 from enteric neurons. When infected with S.t., Il18f/fHand2+ mice displayed more severe weight loss and became moribund significantly quicker than their Il18f/f littermates (Figures 3H and 3I). Il18f/fHand2+ mice also had higher bacterial burden in their cecum (Figure 3J) and peripheral tissues (Figure 3K) than wild-type mice. Finally, we tested if enteric neuronal lL-18 is protective against oral S.t. infection in the absence of antibiotic pretreatment. We observed that Il18f/f Hand2+ mice were again more susceptible than their wild-type littermates in this model of infection (Figure S2E). Taken together, these results indicate that neuron-derived IL-18 is essential for host immunity to invasive bacterial infection.
Figure 3. Enteric Neuronal IL-18 Is Protective against S.t. Infection.
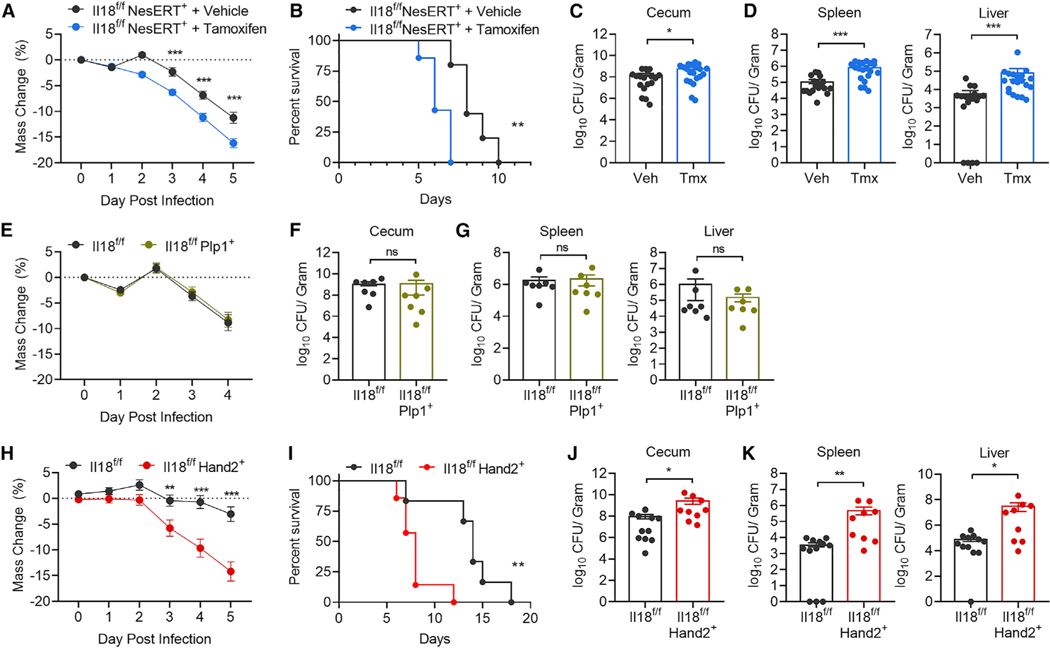
(A) Weight loss of Il18f/fNesERT+ mice pretreated with tamoxifen (TMX) (n = 20) or vehicle (n = 18) then infected with S.t. Data represent mean ± SEM; unpaired t test was used for statistical analysis. Data represent two independent experiments combined from three total independent experiments.
(B) Survival curve for S.t.-infected Il18f/fNesERT+ mice pretreated with TMX (n = 5) or vehicle (n = 7). Log rank test was used for analysis. Data represent two independent experiments combined.
(C and D) S.t. CFU/g of (C) cecum, (D) spleen, and liver from Il18f/fNesERT+ mice, pretreated with vehicle or TMX, 5 days post-infection. Data represent mean ± SEM; Mann-Whitney test was used for statistical analysis. Each dot represents one mouse. Data represent two independent experiments combined from three total independent experiments.
(E) Weight loss of Il18f/f (n = 7) or Il18f/fPlp1+ (n = 7) mice pretreated with TMX then infected with S.t. Data represent mean ± SEM; unpaired t test was used for statistical analysis. Data represent two independent experiments combined.
(F and G) S.t. CFU/g of (F) cecum, (G) spleen, and liver from Il18f/f (n = 7) or Il18f/fPlp1+ (n = 7) pretreated with TMX, 4 days post-infection. Data represent mean ± SEM; Mann-Whitney test was used for statistical analysis. Each dot represents one mouse. Data represent two independent experiments combined.
(H) Weight loss of Il18f/f (n = 20) or Il18f/fHand2+ (n = 17) mice infected S.t. Data represent mean ± SEM; unpaired t test was used for statistical analysis. Data represent two independent experiments combined from three total independent experiments.
(I) Survival curve for S.t.-infected Il18f/f (n = 6) or Il18f/fHand2+ (n = 7) mice. Log rank test was used for analysis. Data represent two independent experiments combined.
(J and K) S.t. CFU/g of (J) cecum, (K) spleen, and liver from Il18f/f or Il18f/fHand2+ mice 5 days post-infection. Data represent mean ± SEM; Mann-Whitney test was used for statistical analysis. Each dot represents one mouse. Data represent two independent experiments combined from three total independent experiments.
*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Intrinsic IL-18 Signaling to Epithelial, Immune, or Neuronal Cells Does Not Mediate Protection against S.t. Infection
To address mechanistically how enteric neuronal IL-18 facilitates defense against S.t. infection, we first investigated whether deficiency in IL-18 affected intestinal motility as nNOS+ neurons are important regulators of peristalsis and bacterial clearance from the intestinal lumen. We measured both intestinal transit time and colonic expulsion time but observed no differences in motility between 1118f/f and Il18f/fHand2+ mice (Figures S3A and S3B). Therefore, we next sought to identify if a single cell type expressing the IL-18 receptor was required to mediate protection. Because of the surprising finding that immune and epithelial IL-18 production was dispensable for anti-S.t. defense, we hypothesized that neuron-derived IL-18 may function through an intrinsic ENS signaling mechanism. To test this, we interbred Hand2+ mice with Il18r1f/f mice (Il18r1f/fHand2+). Maintaining Il18r1f/f and Il18r1f/fHand2+ animals as cohoused littermates prevented alterations to the luminal microbiota between groups (Figure S3C). When challenged with S.t., Il18r1f/f and Il18r1f/fHand2+ mice displayed similar weight loss (Figure 4A) and bacterial colonization in their cecum (Figure 4B) and peripheral tissues (Figure 4C). Because Il18r1f/fNesERT+ mice treated with TMX were also no more susceptible to S.t. induced weight loss, bacterial colonization, or dissemination than vehicle control mice (Figures S3D–S3F), we conclude that IL-18 signaling to neurons does not mediate an intrinsic mechanism of protection to infection.
Figure 4. Intrinsic IL-18 Signaling to Epithelial, Immune, or Neuronal Cells Does Not Mediate Protection against S.t. Infection.
(A) Weight loss of II18r1f/f (n = 14)or Il18r1 f/fHand2+ (n = 25) infected with S.t. Data represent mean ± SEM; unpaired t test was used for statistical analysis. Data represent two independent experiments combined.
(B and C) S.t. CFU/g of (B) cecum, (C) spleen, and liverfrom Il18r1f/f (n = 5) or Il18r1f/fHand2+ (n = 11) mice 5 days post-infection. Data represent mean ± SEM; Mann-Whitney test was used for statistical analysis. Each dot represents one mouse. Data represent one independent experiment of two total independent experiments.
(D) Weight lossof Il18r1f/f(n = 8) or Il18r1f/fFlk1+ (n = 10) mice infected with S.t. Data represent mean ± SEM; unpaired t testwas used forstatistical analysis. Data represent two independent experiments combined.
(E and F)S.t. CFU/g of(E) cecum, (F) spleen, and liverfrom Il18r1f/f(n = 8) or Il18r1f/fFlk1+ (n = 10) mice 5 days post-infection. Data represent mean ± SEM; Mann-Whitney test was used for statistical analysis. Each dot represents one mouse. Data represent two independent experiments combined.
(G) Weight loss of Il18r1f/f (n = 14) or Il18r1f/fVil1+ (n = 14) mice infected S.t. Data represent mean ± SEM; unpaired t test was used for statistical analysis. Data represent two independent experiments combined.
(H and I) S.t. CFU/g of(H) cecum, (I) spleen, and liverfrom Il18r1f/f(n = 14)or Il18r1f/fVil1+ (n = 14) mice 5 days post-infection. Data represent mean ± SEM; Mann-Whitney test was used for statistical analysis. Each dot represents one mouse. Data represent two independent experiments combined.
These findings prompted us to investigate whether IL-18 signaling to immune cells was required for neuronal IL-18 protection from S.t. because IL-18 is a potent driver of T cell- and natural killer (NK) cell-mediated anti-bacterial immunity (Dinarello, 2009). To address whether IL-18 mediated immune or endothelial cell activation was responsible for the protective effect of neuronal IL-18, we crossed Il18r1f/f to Flk1-Cre mice (Il18r1f/f Flk1+). We infected Il18r1f/f and Il18r1f/fFlk1+ mice with S.t. and observed that both genotypes displayed comparable weight loss (Figure 4D), cecal colonization (Figure 4E), and dissemination into peripheral tissues (Figure 4F), suggesting that IL-18 signaling to immune cells is also dispensable for neuronal IL-18-mediated protection. Epithelial cells also express the IL-18R1 and contribute to mucosal barrier immunity through their production of mucus and AMPs (Hansson, 2012; Levy et al., 2015; McDonald et al., 2006). To address whether epithelial IL-18 signaling conferred protection to S.t., we interbred Il18r1f/f with Vil1 Cre mice (Il18r1f/fVil1+) and infected the littermate progeny with S.t. Interestingly, epithelial cell deficiency in IL-18R1 did not alter host susceptibility to S.t. infection (Figures 4G–4I). Taken together, these data indicate that neuronal-derived IL-18 does not facilitate its protective effects during S.t. infection by signaling directly to neuronal, glial, immune, endothelial, or epithelial cells in the intestine.
Enteric Neuronal IL-18 Is a Specific Driver of Goblet Cell Antimicrobial Protein Expression
Because it was not obvious from our IL-18R1 loss-of-function studies how neuronal IL-18 signaling protects against S.t., we next sought to understand whether unique signaling events occur in neuronal IL-18-deficient mice compared with loss of the cytokine in immune and epithelial cell compartments. We conducted RNA-seq on proximal colon tissue from Il18f/f and Il18f/fHand2+ cohoused littermates and compared it with RNA-seq of Il18f/fFlk1+ and Il18f/fVil1+ mice and their wild-type littermates. Surprisingly, RNA-seq analysis revealed distinct and predominantly non-overlapping gene signatures specifically regulated by neuronal-, epithelial-, or hematopoietic-derived IL-18. By comparing genes reduced in Il18f/fHand2+ mice with those reduced in Il18f/fVil1+ and Il18f/fFlk1+, we identified that bactericidal and antimicrobial genes were exclusively reduced in neuronal but not hematopoietic or epithelial IL-18-deficient animals (Figures 5A and 5B). This was the major dysregulated pathway in ll18f/fHand2+ mice with no major effects on other known immunologically important pathways. Conversely, in Il18f/fVil1+ colons, major alterations in genes associated with hematopoietic cells known to be IL-18 responsive were observed (Figure 5A; Table S2). Therefore, we conclude that epithelial-derived IL-18 is essential in maintaining the immunologic tone of the intestine during homeostasis. Finally, transcriptomic analysis of Il18f/fFlk1+ colons revealed no major defects in immune or antimicrobial gene signatures, suggesting that this source of IL-18 may be more important during an inflammatory response (Figure 5B; Table S2). Together, these data reveal that the cellular source of IL-18 has profound and diverse effects on the expression landscape of the intestine and indicates each compartment has unique regulatory roles that we are only now beginning to understand.
Figure 5. Enteric Neuronal IL-18 Is a Specific Driver of Goblet Cell Derived-AMP Expression.
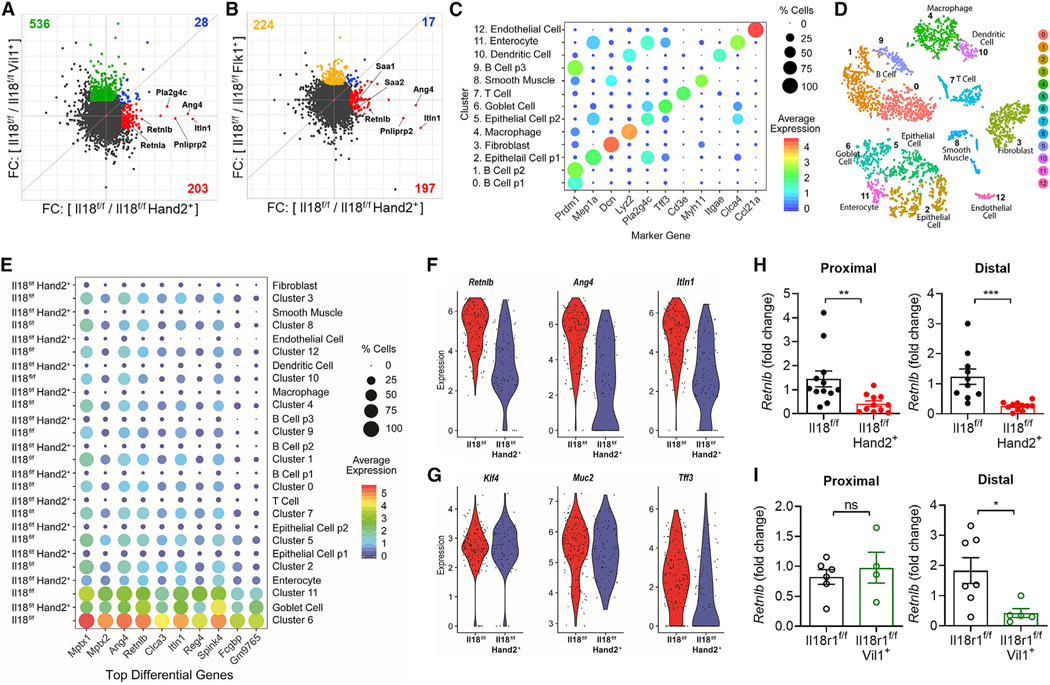
(A and B) Matched proximal colons (n = 2 per genotype) from cohoused llttermatesot the Indicated genotypes were collected, RNA Isolated, and RNA sequencing analysis conducted.
(A) Comparison of wild-type/knockout fold-change between Il18f/f and Il18f/fHand2+ or Il18f/f and Il18f/fVil1+. Gene sets with fold-change ≥ 2 that are unique to Il18f/fHand2+ (red) or Il18f/fVil1+ (green) or shared by both genotypes (blue) are highlighted. AMPs are labeled.
(B) Comparison of wild-type/knockout fold-change between Il18f/f and Il18f/fHand2+ or Il18f/f and Il18f/fFlk1+. Gene sets with fold-change ≥ 2 that are unique to Il18f/fHand2+ (red) or Il18f/fFlk1+ (yellow) or shared by both genotypes (blue) are highlighted. AMPs are labeled.
(C–G). Single-cell RNA sequencing was performed on colonic cells isolated from Il18f/f (n = 2) and Il18f/fHand2+ (n = 2) mice then analyzed by ALRA (Adaptively-thresholded Low-Rank Approximation).
(C) Integrative analysis reveals marker genes for each cluster.
(D) Clustering and labeling of these cells visualized on a t-SNE plot.
(E) Analysis of the top 10 differentially regulated genes between Il18f/f and Il18f/fHand2+ in all clusters.
(F and G) Violin plots illustrate expression distribution of (F) the AMPs Retnlb, Ang4, and Itln1 or (G) goblet cell genes Klf4, Muc2, and Tff3 in the annotated goblet cell cluster (cluster 6) of Il18f/f or Il18f/fHand2+ samples.
(H) Gene expression of Retnlb in tissue from the proximal or distal colon of Il18f/f and Il18f/fHand2+ mice, results are presented as relative to an Il18f/f sample. Data represent mean ± SEM; each dot represents one mouse; unpaired t test was used for statistical analysis.
(I) Gene expression of Retnlb in tissue from the proximal or distal colon of Il18r1f/f and Il18r1f/fVil1+ mice, results are presented as relative to an Il18f/f sample. Data represent mean ± SEM; each dot represents one mouse; unpaired t test was used for statistical analysis.
*p < 0.05, **p < 0.01, ****p < 0.0001
Comparative RNA-seq analysis revealed that neuronal IL-18 was specifically promoting AMP production in the colon. To investigate how this was occurring at a single-cell level of resolution, we conducted scRNA-seq of colonocytes from Il18f/f and Il18f/fHand2+ mice. Using microfluidic scRNA Drop-seq and employing Adaptively-thresholded Low-Rank Approximation (ALRA) analysis (Linderman et al., 2018; Macosko et al., 2015), we identified 13 individual clusters of cell types that we confirmed with expression of cell lineage marker genes (Figures 5C and 5D). We next analyzed our data to identify the most differentially regulated genes from all clusters. In line with our RNA-seq data, the AMPs Ang4, Retnlb, and Itln1 were among the most significantly reduced genes in Il18f/fHand2+ mice compared with wild-type littermates (Figure 5E). In order to ascertain which cell types were most affected by loss of neuronal IL-18 in an unbiased manner, we analyzed the correlation of average gene expression between genotypes for each cluster. Epithelial cell population 1 and goblet cells exhibited the lowest correlation scores, indicating the largest divergence in gene expression between genotypes compared with all other clusters (Figures S4A–S4D). Indeed, these clusters showed the greatest number of genes differentially regulated between Il18f/f and Il18f/fHand2+ groups with the most significant genes again being antimicrobial in nature. This indicates that the major defect in colons lacking neuronal IL-18 are epithelial and goblet cells transcription of important antimicrobial genes. As goblet cells were the highest expressing cell type for AMPs identified, we visualized the expression of known goblet cell developmental markers to identify if there was a general dysregulation of goblet cell maturation. While the AMPs Ang4, Retnlb, and Itln1 displayed significant reduction in both average expression and number of expressing cells per cluster (Figure 5F), known transcriptional regulators of goblet cell differentiation Klf4 and mucus producing genes such as Muc2 were unaffected (Figure 5G). These findings suggest that deficiency of neuronal-IL-18 leads to a specific loss of AMP production rather than a global dysregulation of goblet cell development. We next sought to investigate if neuronal IL-18 directly signals to epithelial cells to drive AMP expression. To address this, we measured AMP expression in the distal and proximal colons of Il18f/fHand2+ and Il18r1f/fVil1+ mice and their wild-type counterparts by qPCR. We identified a significant loss of Retnlb in the distal and proximal colons of Il18f/fHand2+ mice, recapitulating the findings from the RNA-seq experiments (Figure 5H). AMP levels could also be rescued in Il18f/fHand2+ mice by injection with recombinant IL-18 (Figure S4E). Interestingly, although we also observed a significant loss of Retnlb in the distal colon of Il18r1f/fVil1+ mice, we observed no robust difference in AMPs in the proximal colon (Figure 5I). This discordant defect in AMP production in the proximal and distal colon of Il18r1f/fVil1+ strongly indicates that both direct neuronal IL-18 to epithelial cell signaling and indirect signaling mechanisms must contribute to AMP expression patterns in the intestine during homeostasis.
We next endeavored to understand whether canonical inflammasome processing of IL-18 through caspase-1 was required for neuronal IL-18 driven AMP production. To test this, we interbred Casp1f/f mice with Hand2+ mice (Casp1f/fHand2+). Strikingly, neuronal-intrinsic expression of caspase-1 was not required for goblet cell expression of Retlnb, Ang4, or Itln1 (Figure S4F). Taken together, our results indicate that neuronal IL-18 is a major driver of the intestinal AMP landscape by a process that does not require neuronal intrinsic classical inflammasome processing.
Neuronal IL-18-Driven AMP Expression Prevents Bacterial Infiltration and Infection
To investigate whether deficiency of neuronal IL-18 leads to targeted loss of AMPs or whether more fundamental perturbations of the intestine that may not score in transcriptional analysis were present, we conducted histological examination of wild-type and Il18f/fHand2+ colons. H/E, AB/Pas staining, and confocal immunofluorescence studies revealed no major defects in intestinal architecture or goblet cell number, development, maturation, and mucus production (Figures 6A, S6A, and S6B). To characterize the immunological tone of the intestine, we conducted flow cytometry of all major immune cell populations in the intestine and the capacity of lymphocytes to produce IL-18-dependent molecules, including interferon-γ and granzyme B. We observed no difference in cell number, frequency, or activation capacity of cells from Il18f/f or Il18f/fHand2+ mice at steady state or 48 h post S.t. infection (Figure S5). Recent studies have also highlighted that the neurotransmitter norepinephrine is essential to intestinal immunity (Gabanyi et al., 2016; Straub et al., 2006). To address if norepinephrine was altered by IL-18, we measured norepinephrine in the cecal contents from wild-type and Il18f/f Hand2+ mice and observed no difference between littermates (Figure S6C). Taken together, these data demonstrated that loss of neuronal IL-18 had no effect on goblet cell development, immunological tone, intestinal motility, or catecholamine production. These findings prompted us to further investigate whether the differences we observed in AMPs were the operative mechanism governing neuronal IL-18 protection against bacterial infection.
Figure 6. Neuronal IL-18-Driven AMP Expression Prevents Bacterial Infiltration and Infection.
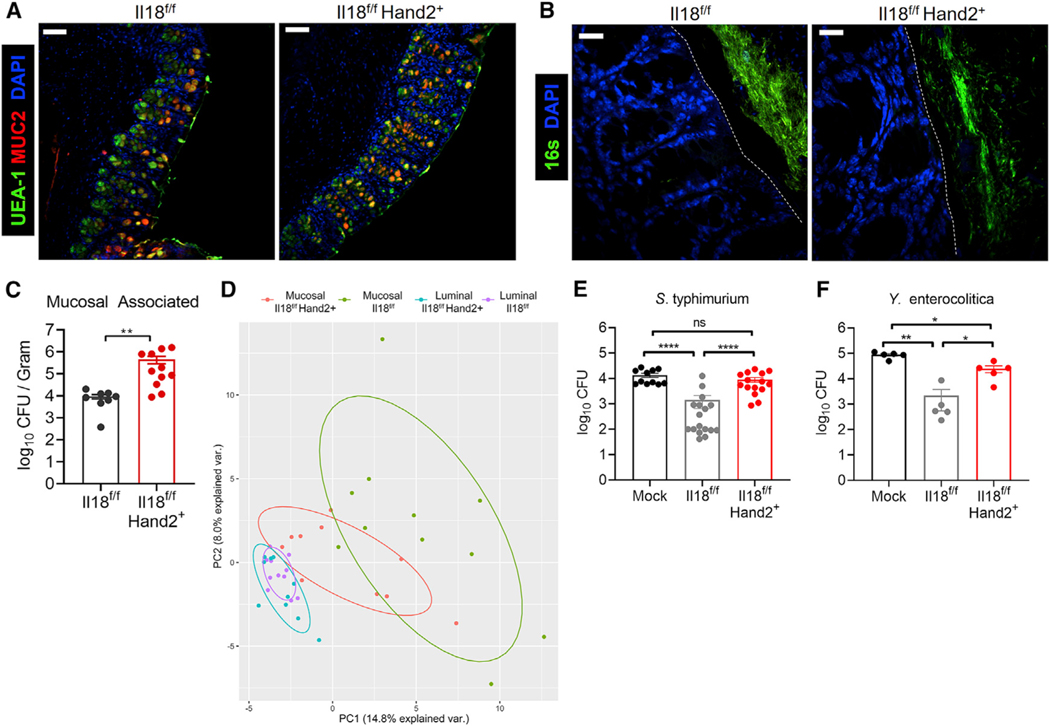
(A) Confocal IF images of colon cross-sections from Il18f/f or Il18f/fHand2+ stained for Muc2 (red) and with UEA I (green) and DAPI (blue). Scale bar represents 70 μm.
(B) Visualization of bacteria in relation to the epithelial brush border by 16S rRNA fluorescence in situ hybridization (green) and DAPI (blue). The brush boarder is demarked by a dotted line. Scale bar represents 18μm.
(C) Quantification of viable mucosal-associated anaerobic bacteria in proximal colon biopsies from Il18f/f or Il18f/fHand2+ mice. Each dot represents a biopsy from an individual mouse colon. Data represent two independent experiments combined from three total independent experiments. Data represent mean ± SEM; Mann-Whitney test was used for statistical analysis.
(D) Principal component analysis based on the relative abundance (>0.001) of bacterial operational taxonomic units from mucosa-associated or luminal microbiota. Luminal and mucosa-associated samples were collected from the same animals housed in Yale animal facilities.
(E and F) Media or colonic explant supernatants from Il18f/f or Il18,f/fHand2+ mice were generated and incubated with (E) 104 CFU S.t. or (F) 105 CFU Y. enterocolitica for 1 h. Surviving bacterial CFUs were enumerated. Each dot represents a control media sample or explant supernatant from an individual mouse colon. Data represent mean ± SEM; Mann-Whitney test was used for statistical analysis.
(E) Data represent two independent experiments combined from three total independent experiments.
(F) Data represent one independent experiment.
*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
AMPs are secreted into the mucus layer of the intestine to limit bacterial association with the apical surface of epithelial cells. Studies using whole-body knockout mice of AMPs have demonstrated that these proteins shape the commensal mucosal bacterial communities in the intestine. To investigate the impact of neuronal IL-18 on the microbiota, we ran 16S sequencing on fecal samples from Il18f/f and Il18f/fHand2+ mice. We observed no significant alteration to the luminal bacterial microbiota (Figure S6D). We next investigated whether loss of AMP expression in Il18f/fHand2+ resulted in increased bacterial infiltration into the inner mucus layer as previously reported in Retn/b-deficient mice (Propheter et al., 2017). Analysis of bacterial localization by 16S FISH revealed widespread infiltration of bacteria into the inner mucus layer of Il18f/fHand2+ intestine but not of their wild-type littermates (Figure 6B). To confirm this, we quantified the abundance and viability of mucosal-associated bacteria using anaerobic culturing conditions and observed that Il18f/fHand2+ mice had significantly more viable mucosal-associated bacteria than their wild-type littermates (Figure 6C). Furthermore, 16S sequencing of mucosal-associated and luminal bacteria from the same animal showed that in the absence of neuronal IL-18, the composition of mucosal-associated bacteria changed to a composition more similar to the luminal microbiota than in wild-type animals (Figure 6D; Table S3). Together, these data reveal that neuronal IL-18-driven AMP production is necessary to exclude microbes from the epithelium and maintain a quasisterile inner mucus layer.
To test if dysregulated AMP production could explain the increased susceptibility to S.t. colonization exhibited in neuronal IL-18-deficient mice, we used a reductionist ex vivo colon explant assay that allowed us to measure the effectiveness of antimicrobial killing (Udden et al., 2017). AMP-containing supernatants were collected from Il18f/f and Il18f/fHand2+ colon explants and incubated with S.t. or Yersinia enteroco/itica for 1 h to allow for bacterial killing. Surviving bacteria was plated and enumerated. These experiments revealed that bacterial killing was significantly reduced for both pathogens in Il18f/f Hand2+-derived explants compared with wild-type counterparts (Figures 6E and 6F). These data demonstrate that the reduction in AMPs observed in Il18f/fHand2+ mice results in an inability to kill pathogenic bacteria and limit pathogenesis. Taken together, we have identified that IL-18 produced from enteric neurons promotes goblet cell AMP production, mediates direct bactericidal activity against the indigenous microbiota, and is required for protection against invasive microbial species to maintain mucosal barrier immunity.
DISCUSSION
In this study, we discovered that IL-18 produced by enteric neurons is necessary for protection against invasive bacterial infection. Unlike immune cell- and epithelial cell-derived sources, neuronal expression of IL-18 non-redundantly directs a program of AMP production in goblet cells, which, during homeostasis, enforces a sterile inner-mucus barrier and, during infection, facilitates killing of enteric pathogens. Previously, bone marrow chimera studies have identified non-hematopoietic IL-18 sources to be required for epithelial cell AMP production, and because epithelial cells are the major producers of the cytokine, the epithelium was attributed as the source (Levy et al., 2015; Nowarski et al., 2015). However, by pairing conditional genetic studies with transcriptomic analysis, we uncover that epithelial IL-18 is completely dispensable for AMP expression, which is facilitated by neuronal IL-18. IL-18 has also been reported to promote proliferation of intraepithelial T lymphocytes and NK cells (Kanai et al., 2000). RNA-seq of whole colon revealed genes such as Gzmb, Cd8a, Klrc2, and Cd103 are specifically reduced in epithelial IL-18-deficient mice, supporting a specific non-redundant role for epithelial IL-18 in maintaining intraepithelial lymphocytes. Furthermore, RNA-seq of hematopoietic IL-18-deficient colons revealed that there is no dramatic change in the immunological tone of the tissue, suggesting that immune-derived IL-18 may function primarily during an inflammatory response. Thus, our data demonstrate that the cellular source and likely location of IL-18 greatly influences its downstream effects and help explain the previously puzzling divergent observations that IL-18 drives colonic inflammation and maintains microbial homeostasis.
Although we have identified that neuronal IL-18 expression is essential for mucosal barrier homeostasis and response to infection, unresolved questions remain and are the focus of our ongoing research efforts pertaining to the direct and indirect mechanism of signaling and neuronal IL-18 activation and release.
Expression of IL-18 in neurons in the submucosal layer is suggestive of a direct ability of these cells to signal to epithelial cells and drive AMP production. In support of this model, we observed that in the distal colon of mice, IL-18R1 on the epithelium is essential for expression of goblet cell-expressed AMPs. Interestingly, in the proximal colon, there was no requirement for direct IL-18 to epithelial cell signaling, indicating that indirect neuronal IL-18 signaling mechanisms exist to regulate AMP production. This seemingly incongruent finding could be because of the diversity among IL-18-expressing neurons throughout the intestine. Additionally, factors such as pH, neuronal pattering, and composition of the microbiota differ dramatically in the proximal and distal regions of the colon and may influence the effects of neuronal IL-18. While further studies are necessary to address this observation specifically, our data indicate that IL-18 is essential for colonic AMP production and S.t. defense and that direct IL-18 signaling to the epithelium of the proximal colon is insufficient in providing robust protection against infection and indirect signaling mechanisms are also required.
In our studies, we identified that a population of IL-18+ neurons in the myenteric plexus also express nNOS. Nitrergic neurons are a heterogenous population of cells comprised mainly of inhibitory interneurons and inhibitory motor neurons, which regulate intestinal motility and fluid secretion (Bódi et al., 2019). In our studies, we did not observe that neuronal IL-18 regulates intestinal motility, suggesting that the effect of neuronal IL-18 in nNOS+ neurons is independent of their functions in regulating gastric motility. A unique feature of nNOS+ neurons is their production of nitric oxide. In macrophages, nitric oxide production promotes NF-κB signaling, which is known to drive production of pro-IL-18 (Baig et al., 2015). Thus, it is interesting to consider whether IL-18 expression in nNOS+ neurons is tied to their unique ability to produce nitric oxide. In addition to IL-18+ nNOS+ neurons, we also observe IL-18 expression in nNOS-cells, suggesting that multiple populations of neurons may express IL-18; therefore, we cannot rule out the expression of IL-18 in neurons that express other important immune-modulatory factors such as vasoactive intestinal peptide and substance P. Further studies will address what population of neurons express IL-18 and whether the unique properties or location of these neurons influence the expression and function of the cytokine.
Our findings also raise the intriguing question of how IL-18 is activated and released from enteric neurons. In epithelial and immune cells, mature IL-18 release is understood to occur via a NOD-like receptor (NLR) inflammasome that triggers caspase-1 activity to cleave cytoplasmic IL-18 into a bioactive cytokine (Gu et al., 1997; Martinon et al., 2002; Strowig et al., 2012). In turn, caspase-1 and/or caspase-11 cleaves and activates the pore-forming protein, gasdermin D, to facilitate IL-18 release (He et al., 2015; Kayagaki et al., 2015; Shi et al., 2015). However, here, we observe that loss of caspase-1 in enteric neurons has no effect on intestinal epithelial AMP production, indicating that IL-18 release and activation must occur through a distinct mechanism in neurons. Indeed, release of IL-18 from neurons could be achieved through a variety of mechanisms, which will require new tools and genetic models to interrogate. For instance, caspase-11 can also cause pore formation and release of IL-1 family members during pyroptosis, potentially facilitating IL-18 release in a caspase-1-independent manner (Kayagaki et al., 2011). However, as steady-state neuronal IL-18 is required for AMP expression, it seems unlikely that a pyroptotic mechanism of neuronal cell death is required for steady-state IL-18 release. Instead, it may be possible that caspase-11 contributes to neuronal IL-18 release specifically during invasive bacterial infection to directly limit bacterial invasion. In support of this, as demonstrated by Matheis and colleagues in this issue of Cell, caspase-11 is essential for neural cell death after S.t. infection (Matheis et al., 2020). Recent studies have uncovered other examples and mechanisms for IL-1 family member release independent of cell death and pro-inflammatory caspases. In the CNS, IL-33 is released through an elusive mechanism thought to occur independent of cell death in order to regulate neuronal synapse formation (Vainchtein et al., 2018). Mechanistically, mature IL-1 b can be released from immune cells independently of gasdermin D pore formation by interaction with PIP-2 membrane microdomains and subsequent exocytosis (Monteleone et al., 2018). In neurons, it is exciting to speculate that IL-18 could be released from neuronal synaptic vesicles. While we have not seen evidence for this in our preliminary studies (data not shown), IL-18 is a similar size as some neuropeptides, suggesting that it could be exported via neuronal synaptic vesicles. Identifying whether IL-18 can be exported this route and what signals mediate its release are of the upmost interest. In the intestinal epithelium, NLRP6-mediated release of IL-18 is dependent on microbial-derived polyamine metabolites that activate the inflammasome and trigger IL-18 release (Levy et al., 2015). It is possible that these or other unidentified molecules can trigger release of IL-18 from neurons. Regardless of how IL-18 is released from neurons, the finding that its function in regulating AMPs is independent of intrinsic neuronal caspase-1 is both surprising and thought provoking. It is possible that neuronal IL-18 can be processed by non-autologous sources of caspase-1 either through extracellular interaction or by neuronal uptake and internalization of active caspase-1 inflammasome complexes (Franklin et al., 2014). Alternatively, although caspase-1 is the archetypical protease for IL-18 processing, other enzymes can cleave the N-terminal domain of IL-18 to trigger its bioreactivity. For example, CD8 T cell and NK cell release of granzyme B has been demonstrated to cleave pro-IL-18 into its active form (Omoto et al., 2010). In addition, Mast cells, which are known to form neuro-immunological like synapses, produce chymases with the capacity to cleave and activate IL-18 (Omoto et al., 2006; Schemann and Camilleri, 2013). Taken together, these studies highlight a multitude of possible mechanisms that may facilitate neuron IL-18 activation and signaling potential. Therefore, further investigations are focused on identifying the mechanism and signals that regulate IL-18 processing and release.
Our understanding of how the nervous system influences intestinal immunity has grown rapidly in the last half decade. Here, we show for the first time that neurons can produce an immunomodulatory cytokine, IL-18, that drives an antimicrobial program necessary to prevent pathogenic bacterial infection. This axis is mediated exclusively by neuron-produced IL-18, not IL-18 from epithelial or immune cell sources. Based on these unexpected findings, we propose that targeting neurons to enhance epithelial AMP production could be used to limit pathogenicity in patients suffering from dysregulated barrier immunity.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Richard A. Flavell (richard.flavell@yale.edu). All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
For conditional deletion of Caspase-1, IL-18 and IL-18R1, Casp1f/f, Il18f/f and Il18r1f/f animals previously generated in our laboratory (Case et al., 2013; Nowarski et al., 2015), were crossed to Cre expressing lines obtained from Jackson Laboratories: Nestin-Cre-ERT2 (Jax #016261) to target nestin-expressing cells, PLP-Cre-ER (Jax #005975) to target glial cells, oligodendrocytes and Schwann cells, Flk1 Cre (Jax #018976) to target hematopoietic, endothelial and some muscle cells, and Villin1 Cre (Jax #021504) to target intestinal epithelial cells. For conditional deletion in enteric neurons mice with loxP-floxed alleles were crossed to Hand2-Cre mice generated and generously gifted to us by Dr. David E. Clouthier (Ruest et al., 2003). To validate Cre recombinase expression in Hand2-Cre animals, Cre expressing mice were crossed to ROSA26loxStoploxTdTomato mice (Jax #007914). All experiments were performed using cohoused littermate control mice. Both male and female mice 7–12 weeks old were used for all experiments. Animal experimentation was conducted in compliance with Yale Institutional Animal Care and Use Committee protocols, and Harvard Institutional Animal Care and Use Committee protocols.
Rats
Female naive adult rat colon tissue was used for confocal immunofluorescence studies investigating IL-18 localization. Rat tissue was a generous gift from Dr. Justus Verhagen who obtained Sprague Dawley animals from Charles Rivers. All procedures were performed in accordance with protocols approved by the Pierce Animal Care and Use Committee (PACUC). These procedures are in agreement with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (8th edition).
METHOD DETAILS
Tamoxifen treatment
Mice were administered 75 mg tamoxifen / kg body weight via intraperitoneal injection daily for five days, then rested for 7 days before experimental use. Tamoxifen treated mice were separated from vehicle (sunflower oil) treated animals during and following tamoxifen treatment to inhibit tamoxifen ingestion due to coprophagic behavior.
In vivo S. typhimurium infection
In vivo infections were performed as previously described (Jackson et al., 2018). Briefly, mice were gavaged with streptomycin (20mg), then 24 h later infected with streptomycin-resistant S. enterica subsp. enterica serovar typhimurium (SL1344 strain, generously provided by Dr. Jorge E. Galán). Before infection, bacteria were propagated overnight in LB containing streptomycin (100 μg ml−1), then subcultured the next day in antibiotic-free LB containing 0.3 M NaCl (Chen et al., 1996). Mice were infected with 1 × 103 CFUs. To calculate bacterial CFU from the cecum, liver and spleen of infected mice, organs were isolated and added to 2 mL of PBS. Tissue was then dissociated with GentleMacs C Tubes (Miltenyi Biotech) as per manufacturer’s instructions, and tissue slurries were clarified at 50 g for 10 min, and bacteria-containing supernatants serially diluted and plated in triplicate on LB agar streptomycin plates. CFU counts were performed blinded. In experiments without streptomycin pretreated (Figure S2E), bacteria were cultured as above. Mice were infected with 1 × 106 CFUs by oral gavage and monitored twice daily.
Ex vivo S. typhimurium killing assay
Ex vivo bacterial killing assays were performed as previously described (Udden et al., 2017), with minor modifications pertaining to S. Typhimurium conditions. Briefly, colons were removed and washed thoroughly with ice-cold PBS. Colons were then cut into 1cm pieces, weighed, and incubated in DMEM/F12 medium containing 5% FBS, penicillin-streptomycin, and gentamycin (20ug/mL) for 2 h at 37°C with 5% CO2. After 2 h, tissue segments were washed 3 times in antibiotic-free DMEM/F12 medium containing 5% FBS to remove all antibiotics. Tissue was then incubated for 12 h at 37°C with 5% CO2 in DMEM/F12 medium containing 5% FBS (without any antibiotics) at 100mg tissue / 1 mL medium. After 12 h incubation, supernatants were collected, clarified, and diluted 1:10 with antibiotic free DMEM/F12 with 5% serum. For S. typhimurium, bacteria was propagated and subcultured as described above then 1×104 CFU S. typhimurium was added to 250ul diluted supernatant and mixtures were incubated at 37 degrees for 1 h. After 1 h samples were serially diluted, plated on LB agar streptomycin plates and grown overnight at 37 degrees. CFU counts were performed blinded. For Y. enterocolitica the same protocol was followed with the minor adjustment that the bacteria were grown and subcultured in antibiotic-free LB, then plated on antibiotic-free LB agar plates.
Intestinal motility and expulsion assay
To measure intestinal motility mice were fasted for 4 h then gavaged with 200ul of 6% (w/v) carmine red dye in methylcellulose. Mice were observed until the dye could be seen in the feces.
To measure colonic motility, a 2mm glass bead was inserted 2cm into the colon of mice using a glass rod and bead expulsion time was recorded.
Colon immune cell isolation and flow cytometry
Colons were removed and luminal contents flushed with ice cold HBSS. The tissue was then cut into 0.5 cm pieces and incubated in HBSS with 5mM EDTA and 1mM DTT in a bacterial shaker at 200 RPM, 37°C for 15 min. Tissue was then removed and placed into new HBSS/EDTA/DTT solution and this cycle was repeated for a total of three washes. Each epithelial wash was collected, spun down and then intraepithelial immune cells were purified using a 40% / 80% percoll gradient. The remaining tissue was washed in PBS then digested in DMEM with 10% FBS, 300 U/mL Collagenase XI (Sigma C7657), 0.1 mg/mL Dispase and 50 u/mL DNase II type V (Sigma, 1100 u/mg) for 1 h at 37 degrees with gentle horizontal shaking. After 1 h of gentle shaking tissue was then shaken quickly to release the majority of cells. Following this, the tissue slury was strained to remove any undigested material and washed in DMEM with 10% FBS before processing for flow cytometry.
Single-cell suspensions were stained with a cocktail of antibodies (See Star Methods) to identify specific subsets of immune cells. For intracellular staining of transcription factors, cells were stained for surface markers then fixed and permeablized using the Foxp3 Transcription Factor Staining Buffer Set (eBioscience), followed by intracellular antibody staining overnight (Sefik et al., 2015). For intracellular cytokine staining, cells were stimulated with PMA (20ng/mL) and ionomycin (20ng/mL) for 4 h, with the addition of brefeldin A1 h into the stimulation. Cells were then fixed using Fixation/Permeabilization Solution kit (BD) and stained with antibodies overnight.
Isolation and growth of mucosal-associated bacteria
To isolate mucosal-associated bacteria, 1cm of proximal colon was removed of fat, opened longitudinally and gently washed through 4 Petri dishes with 20 mL of PBS per dish. A 5mm punch biopsy was collected from this piece of tissue then weighed and put into a Lysing Matrix D tube (MP Biomedicals) with PBS at 50mg/mL. Tissue pieces were homogenized using a bead beater and spun for 1 min at 50 g in a 4 degree centrifuge. A fraction of the sample was then serially diluted, plated on 5% Sheep Blood Agar plates (BD 221734), and grown in anaerobic conditions at 37 degrees for 24 h.
Whole mount microscopy
Rat or mouse tissue segments from the colon were harvested and the lumen was flushed of debris with ice cold HBSS plus magnesium and calcium. Longitudinal muscle and the adjoining myenteric plexus (LMPP) was removed by placing the tissue on a glass rod and peeling the LMPP from the circular muscle and adjoining submucosal plexus (SMP) as previously described (Smith et al., 2013). The LMPP was then pinned to sylagard discs and fixed in 2% PFAin PBS for 20 min at room temperature (RT). Samples were washed 3x in PBS and permeabilized in 0.3% Triton, 5% BSA and 5% donkey serum for 2 h at RT. Samples were washed 2x in PBS and incubated with unconjugated primary antibodies diluted in 1% BSA, 3% donkey serum and 0.1% Tween overnight at 4°C. The next day samples were washed 3x with PBS + 0.1% tween and incubated with secondary antibodies and directly conjugated antibodies for 2 h at RT. Samples were then washed 3x with PBS + 0.1% tween, incubated for 10 min with PBS+DAPI, then washed again in PBS. Samples were mounted on slides using ProLong Diamond Antifade (ThermoFisher Scientific), cured overnight, and sealed. Confocal imaging was conducted with a Nikon-Ti microscope combined with UltraVox spinning disk (PerkinElmer) and data was analyzed using the Volocity software (PerkinElmer).
Immunohistochemistry
Rat or mouse intestinal tissue segments were harvested, and the lumen was flushed of debris with ice cold HBSS containing magnesium and calcium. Samples were fixed in 2% PFA diluted in PBS for 20 min at room temperature, then dehydrated for 1 h each in 10%, 20% and 30% sucrose containing PBS. Dehydrated tissue was frozen in OCT and cut 8–10uM thick using a Leica cryostat. For staining, tissue sections were permeabilized with 0.1% triton in 2% BSA diluted in PBS for 15 min at RT then washed 2x in PBS and incubated for 5 min with Serum-Free Protein Block (Agilent Dako). Primary antibody was diluted in 1% BSA and 0.1% tween and incubated overnight at 4°C. The next day slides were washed 3x with PBS, incubated with secondary antibodies and directly conjugated antibodies for 1 h at RT, then washed 3x with PBS and mounted with ProLong Diamond Antifade (ThermoFisher Scientific).
Histology
Tissue was fixed in Carnoy’s solution for 2 h on ice. Post-fixation samples were moved to 70% ethanol then sent to HistoWiz Inc. (Histowiz.com) for processing. Samples were embedded in paraffin and sectioned at 5 mm prior to AB/PAS staining. Whole slide scanning (40x) was performed on an Aperio AT2 (Leica Biosystems).
16S Fluorescence In situ hybridization
16S rRNA FISH was performed as previously described (Canny et al., 2006) with minor adjustments. Briefly, colon tissue sections were dissected and immediately placed in ice-cold Carnoy’s solution (Ethanol:Chloroform:Glacial acetic acid at 6:3:1) for 1–2 h. The tissue was then washed twice in 100% ethanol for 15 min followed by two washes in xylenes for 15 min then embedded in paraffin and cut to 5 mm. Slides were allowed to sit in sunlight for at least 24 h to reduce background autofluorescence. Following this, slides were deparaffinized by heating to 50°C for 1 h followed by 5 min incubations with xylene (2x) then 100% ethanol (2x) at room temperature. Slides were next incubated with a universal bacterial 16S-DNA probe (EUB-338: [Cy3]-5’-GCTGCCTCCCGTAGGAGT-3’-[Cy5]) at 0.5ng/uL in hybridization buffer (0.9M NaCl, 0.02M Tris pH 7.5, 20% Formamide, 0.05% SDS) for 2 h at 46°C. Following this incubation slides were washed in post-hybridization buffer (0.215M NaCl, 0.02 Tris pH 7.5, 0.025M EDTA, 0.05% SDS) 2x for 5 min at 46°C then rinsed with deionized water and allowed to air dry before coverslips were mounted using Antifade Mounting media with DAPI (ThermoFisher Scientific).
Single-molecule fluorescence in situ hybridization (smFISH)
Mice were sacrificed and the colon was removed and flushed with cold PBS. Colonic tissue was opened longitudinally and spread on whatman filter paper. Flat tissue was then fixed in 4% paraformaldehyde (PFA, Santa Cruz Biotechnology, sc-281692) in PBS for 3 h and subsequently incubated in a 30% sucrose, 4% PFA in PBS solution at 4°C overnight with gentle agitation. Fixed tissues were then embedded in Tissue-Tek OCT Compund (Sakura, 4583). 7mm thick sections of fixed colon were sectioned onto poly L-lysine coated coverslips and used for smFISH staining. Probe libraries were designed using the Stellaris FISH Probe Design Software (Biosearch Technologies) (Table S1). Probe libraries were coupled to Cy5 (Il-18) orTMR (Tubb3). Colonic sections were hybridized with smFISH probe sets based on a previously published protocol (Mooret al., 2018). Before mounting the samples on slides, colonic tissue samples were treated with 1XTrueBlack Lipofuscin Autofluorescence Quencher (Biotium, 23007) to remove signal from autofluorescent granules within mesenteric neurons. smFISH imaging was performed on a Leica THUNDER 3D Live Cell Imaging system using the following THUNDER Computational Clearing Settings, Feature Scale (nm): 383, Strength (%): 97.75, Deconvolution settings: Auto and Optimization: High.
Norepinephrine ELISA
Cecal contents from Il18f/f or Il18f/fHand2+ mice were resuspended in norepinephrine stabilization buffer (10 mM HCl, 1 mM EDTA, 4mM Na2S2O5) to 200mg/mL and vortexed for 1 min to homogenize the sample. Homogenized samples were spun for 5 min at 2300 RCF at 4°C and 100ul of clarified supernatant was used as input for a norepinephrine ELISA which was conducted according to the manufacturer’s protocol (Labor Diagnostika Nord: BA E-5200 Noradrenaline Research ELISA).
RNA sequencing
Proximal tissue was flushed of fecal debris, snap frozen and then homogenized in TRIzol reagent (Invitrogen) before being purified following the manufacturer’s instructions. RNA was further purified using a RNAeasy Plus Mini Kit as per manufactures instructions. Purified RNA was processed by the Yale Center for Genome Analysis using standard methodology and sequenced on a HiSeq2000 with 75-bp paired-ended reads. Raw RNA sequencing reads were aligned to the mouse mm10, GRCm38 genome with Tophat. Gene expression levels and differential analysis were performed with Cufflinks and Cuffdiff respectively. Genes with a Log2FoldChange of ± 1 and < 0.05 p value were considered significantly differentially expressed. Volcano plots were constructed using Origin lab graphical software.
Intestinal cell isolation for single-cell RNA sequencing
Whole colon was dissected from euthanized mice and luminal contents were removed by vigorous flushing with ice cold HBSS containing 2% FBS and 1x HEPES. Clean colons were cut into 0.5cm pieces and incubated shaking for 15 min at 37 degrees in 10mLof HBSS, 5mM EDTA, 1mM DTT. After 15 min of shaking, HBSS wash was collected, diluted with 10% FBS containing DMEM and placed on ice. New HBSS/EDTA/DTT solution was added to the tissue sections and this step was repeated for a total of 3 wash, with the epithelial fraction collected at the end of each wash. In order to retain intraepithelial lymphocytes, the epithelial wash containing cells were subjected to Percol separation using an 80% and 40% gradient. Immune cells were isolated and later combined with tissue digestion liberated cells from the remaining colon tissue. The remaining colon tissue was then washed in PBS without calcium or magnesium and digested in 10mL of DMEM containing 10% FBS, Collagenase XI (Sigma C7657), Dispase, and DNase 1 type V (Sigma) for approximately 60 min at 37 degrees and shaking. When tissue appeared “stringy,” it was shaken vigorously, and the digested tissue was passed through a 40 mm strainer and spun at 200 g for 5 min at 4°C. Samples are then washed 3–5 times with 10mL DMEM containing 2% sorbitol in 15 mL tube to remove digestion associated cellular debris. Cells were counted and resuspended in PBS containing 0.01% BSA(Ambion #AM2616) at a concentration of 100,000 cells per ml for further downstream single cell sequencing.
Single-cell RNA sequencing and data analysis
Intestinal cells from Il18f/f and Il18f/fHand2+ mice were diluted to a concentration of 100 cells/ml and 1 μL aliquots were used as input to the Drop-seq protocol which was performed as described previously (Macosko et al., 2015; Shekhar et al., 2016) with minor modifications. The beads were purchased from ChemGenes Corporation, Wilmington MA (catalog number Macosko201110) and the PDMS co-flow microfluidic droplet generation device was generated by Nanoshift, Emeryville CA. For both conditions the 1ml Drop-Seq collection was performed. Samples were processed for cDNA amplification within 15 min of collection. Populations of 5000 beads (150 cells) were separately amplified for 15 cycles of PCR (conditions identical to those previously described) and pairs of PCR products were co-purified by the addition of 0.6x AMPure XP beads (Agencourt). The cDNA from an estimated 1500 cells was prepared and fragmented by Nextera XT using 1000 pg of cDNA input, and the custom primers P5_TSO_Hybrid (Macosko et al., 2015) and Nextera XT primers - N701 and N702 (Illumina). Both libraries were sequenced on the Illumina NextSeq 500 using 2.0 pM in a volume of 1.3 mL HT1, and 2 mL of 0.3 μM Read1CustomSeqB (Macosko et al., 2015) for priming of read 1. Read 1 was 20 bp; read 2 (paired end) was 60 bp. Single cell RNA-seq data was processed as described in in (Linderman et al., 2018; Macosko et al., 2015). After the generation of the digital expression matrix, data was first filtered to include only cells with more than 100 genes and less than 10% mitochondria transcripts, and genes expressed in more than 10 cells; then normalized in terms of log (CPM/100 + 1), where CPM stands for counts per million (meaning that the sum of all gene-levels is equal 1,000,000). After normalization, we used ALRA (Adaptively-thresholded Low Rank Approximation) (Linderman et al., 2018) to impute the matrix and fill in the technical drop-ped-out values. Subsequently, to visualize the cell subpopulations in two dimensions, principal component analysis (PCA) followed by t-Distributed Stochastic Neighbor Embedding (t-SNE) (Maaten and Hinton, 2008), a non-linear dimensional reduction method, were applied to the log transformed data. Graph-based clustering (Butler et al., 2018) were then used to generate clusters that were overlaid on the t-SNE coordinates to investigate cell subpopulations. Marker genes for each cluster of cells were identified using the Wilcoxon test with Seurat. To investigate the differences between Il18f/f and Il18f/fHand2 then conducted differential expression analysis on each cluster.
4-way plot analysis
Cre+ genotypes were compared to their wild type (Cre−) controls. Analysis focused on genes with a coefficient of variation ≤ 0.5 to generate lists of significantly induced or repressed genes (Log2FC ± 2, P Value ≤ 0.05). These lists were compared between genotypes (Il18f/f/Il18f/fHand2+ versus Il18f/f/Il18f/fFlk1+ or Il18f/f/Il18f/fHand2+ versus Il18f/f/Il18f/fVil1+) to identify genes that were preferentially over-represented in each Cre+ genotype or in both Cre+ genotypes compared with wild type counterparts. Scatter analysis of foldchanges across two different genotypes each compared to wild type was presented as 4-way plots generated using Multiplot Studio (Scott P. Davis and Christophe Benoist).
Production of recombinant IL-18
The coding sequence of the mature form of murine IL-18 (amino acids 37–193) was subcloned into the pE-SUMO vector (LifeSensors) with an N-terminal SUMO tag and C-terminal hexa-histidine tag. The resulting plasmid was transformed into BL21(DE3) cells and protein expression was induced by addition of 0.5 mM IPTG once the culture reached an optical density of 1.2. The induced culture was incubated at 16°C and the cells were harvested by centrifugation after 20 h. Harvested cells were resuspended in lysis buffer (10 mM Tris pH 7.5,150 mM NaCl, 1 mM TCEP) and disrupted with a french press. The soluble fraction was clarified by centrifugation at 24,000 RCF for 20 min and the supernatant applied to a Ni-NTA column. The SUMO tag was removed by addition of the SUMO protease Ulp1 (produced in-house) to a 1:1000 dilution at 4°C for 16 h, then buffered exchanged with a spin concentrator into lysis buffer and re-applied to a second Ni-NTA column to remove the free SUMO tag. The cleaved IL-18 protein was then applied to an ENrich SEC70 gel-filtration column (Bio-Rad) to remove aggregates. The mono-disperse fractions were pooled and applied to a final Ni-NTA column for endotoxin removal by washing with 0.2% Triton X-114 at 4°C. The eluted protein was buffered exchanged into endotoxin-free phosphate buffered saline using a sterile PD-10 column (GE Healthcare). Endotoxin levels were quantified using an EndoSafe nexgen-PTS device (Charles River) and found to be < 5 endotoxin units/mg protein.
Bacterial DNA isolation
For fecal DNA isolation, fecal pellets were collected in 2mL screw cap cryo-storage vials with 250ul of 20% SDS. 500ul of PB Buffer (QIAGEN PCR Purifcation Kit #28104) and 100ul of 0.1mm diameter Zirconia/silica beads (Biospec# 11079101z) were then added to each sample followed by 550ul of phenol:chloroform:IAA. Samples were bead beated for 4 min then centrifuged at 8000 RCF for min at room temperature. 500ul of the clarified aqueous phase was then run through a QIAGEN PCR purification column and processed following the manufacture’s protocol. Isolated DNA was used for 16 s sequencing.
For mucosal associated DNA isolation, samples were prepared as previously reported (Zoetendal et al., 2006). Briefly, colons were dissected, removed of fat and opened longitudinally. Tissue was then washed through 4 dishes full of PBS until no fecal matter was observed. The colon was then flayed open and a 5mm biopsy was collected from the proximal colon. This tissue was either processed immediately or flash frozen until processing. To process the tissue, 1 mL of TE buffer was added to each tissue biopsy followed by 50ul of 10% SDS and 10ul of Protease K (20 mg ml-1). Samples were then incubated at 55°C for 1 h. After incubation samples were pipetted to resuspend the tissue and then 0.5g of 0.1 mm zirconia beads and 4 glass beads were added to each tube. Samples were then vortexed until the sample was homogenized followed by 2 min bead beatings at room temperature. Following homogenization samples were heated to 95°C for 15 min and DNA was isolated by proceeding to Step 4 of the protocol “Isolation of DNA from Stool and Pathogen Detection (Qiagen).”
16 s sequencing
16 s rRNA sequencing was performed on an Illumina miSeq (Paired-end 2×250) using barcoded primers(Kozich et al., 2013). Microbial diversity and tactical analysis were performed with QIIME as previously described (Geva-Zatorsky et al., 2017; Nowarski et al., 2015; Palm etal., 2014)
RNA isolation and RT-PCR
For whole tissue RNA extraction, intestinal tissue was washed of fecal debris then snap frozen. RNA processing and cDNA synthesis was conducted as previously described Jarret et al., 2016, with minor modifications. TRIzol reagent (Invitrogen) was added to frozen tissue segments, homogenized, and then processed as per manufacturer’s instructions. RNA was further purified using the QIAGEN RNeasy Plus Mini kit. Maxima H Minus (ThermoFisher Scientific) was used for cDNA synthesis and real time PCR was carried out using the Biorad CFX384 system with Universal SYBR Green Supermix (Biorad).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis details of each individual experiments, including number of replicates is reported either in the figure legends or relevant method section. Data are presented as means with error bars indicating SE unless otherwise stated. The calculations were performed either with Microsoft Excel or with GraphPad Prism.
DATA AND CODE AVAILABILITY
The transcriptome datasets generated during this study are available at Gene Expression Omnibus (GEO) under the superseries number GSE141810. The bulk RNaseq is available under the accession number: GSE141808. The DropSeq data is available under the accession number: GSE141809. The 16S sequencing datasets generated during this study are available at NCBI BioProject under the number: PRJNA594999.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3e | Biolegend | RRID: AB_312670 |
| CD4 | Biolegend | RRID: AB_312690 |
| CD8a | Biolegend | RRID: AB_312750 |
| CD11b | Biolegend | RRID: AB_10897942 |
| CD11c | Biolegend | RRID: AB_313778 |
| CD45 | Biolegend | RRID: AB_312976 |
| CD103 | Biolegend | RRID: AB_10709438 |
| CD326 | Biolegend | RRID: AB_1501158 |
| F4/80 | Biolegend | RRID: AB_893500 |
| Ly6c | Biolegend | RRID: AB_1186134 |
| IA/IE | Biolegend | RRID: AB_313328 |
| Gzmb | Biolegend | RRID: AB_2294995 |
| TCRgd | Biolegend | RRID: AB_313829 |
| TCRB | Biolegend | RRID: AB_313428 |
| NK1.1 | Biolegend | RRID: AB_313396 |
| Nkp46 | Biolegend | RRID: AB_10612749 |
| Gata3 | Biolegend | RRID: AB_2562722 |
| Foxp3 | eBioscience | RRID: AB_465936 |
| tbet | eBioscience | RRID: AB_925761 |
| Ifng | Biolegend | RRID: AB_315403 |
| Rorg | eBioscience | RRID: AB_10807092 |
| Rat IL-18/IL-1F4 | R&D | RRID: AB_2248910 |
| nNOS | Genetex | GTX133403 |
| Choline Acetyltransferase/ChAT | Novus Biologicals | NBP2-46620 |
| Muc2 | Novus Biologicals | RRID: AB_10003763 |
| Alexa Fluor 647 anti-Tubulin β 3 (TUBB3) | Biolegend | RRID: AB_2686930 |
| Alexa Fluor 488 anti-Tubulin β 3 (TUBB3) Antibody | Biolegend | RRID: AB_2564757 |
| Alexa Fluor 594 anti-Tubulin β 3 (TUBB3) Antibody | Biolegend | RRID: AB_2650635 |
| RFP | Rockland | RRID: AB_2209751 |
| Bacterial and Virus Strains | ||
| Yersinia enterocolitica subsp. enterocolitica (Schleifstein and Coleman) | ATCC | 9610 |
| Salmonella enterica subsp. enterica serovar Typhimurium | Jorge Galan (Yale) | SL1344 strain |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase XI | Sigma | C7657 |
| Dispase II | Sigma | D4693 |
| Dnase II type V | Sigma | D8764 |
| Lectin from Ulex europaeus | Sigma | L9006 |
| 1XTrueBlack Lipofuscin Autofluorescence Quencher | Biotium | 23007 |
| Critical Commercial Assays | ||
| Noradrenaline ELISA | LDN | BA E-5200 |
| Foxp3 Transcription Factor Staining Buffer Set | eBioscience | 00-5523-00 |
| Fixation/Permeabilization Solution kit with BD GolgiPlug | BD | 555028 |
| QIAamp DNA Stool Mini Kit | QIAGEN | 51504 |
| Deposited Data | ||
| Bulk RNaSeq Data | Gene Expression Omnibus (Geo) | GSE141808 |
| Single-cell DropSeq Data | Gene Expression Omnibus (Geo) | GSE141809 |
| 16S Sequencing | BioProject | PRJNA594999 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6-Tg(Nes-cre/ERT2)KEisc/J | Jackson Laboratory | 016261 |
| B6.Cg-Tg(Plp1-cre/ERT)3Pop/J | Jackson Laboratory | 005975 |
| Kdrtm1(cre)Sato/J | Jackson Laboratory | 018976 |
| B6.Cg-Tg(Vil1-cre)1000Gum/J | Jackson Laboratory | 021504 |
| B6.Cg-Gt(ROSA)26Sortm14(CAG -tdTomato)Hze/J | Jackson Laboratory | 007914 |
| dHAND-Cre (Hand2Cre C57BL/6 x SJL backcrossed to Il18f/f C57BL/6) | Dr. David E. Clouthier | N/A |
| SAS Sprague Dawley Rat | Charles River | 400 |
| Oligonucleotides | ||
| Universal 16S-DNA Probe [Cy3]-5’-GCTGCCTCCCGTAGGAGT-3’-[Cy5]) | IDT | EUB-338 |
| Software and Algorithms | ||
| Adaptively-thresholded Low Rank Approximation | Linderman et al., 2018 | N/A |
Highlights.
Epithelial and immune cell IL-18 are not required to combat S. typhimurium
Enteric neurons express IL-18
Enteric neuronal IL-18 controls goblet cell antimicrobial protein expression
Neuronal IL-18 directs killing of enteric bacterial pathogens
ACKNOWLEDGMENTS
We would like to thank J. Alderman, C. Lieber, J. Stein, L. Evangelisti, C. Hughes, and E. Hughes-Picard for technical and administrative assistance. We would like to thank Dr. David E. Clouthier for providing the Hand2-cre mice and Dr. Justus Verhagen for providing rat tissue. We would like to thank Dr. I. Chiu and Dr. A. York for helpful discussion and R. Brecht and H. Steach for comments on the manuscript. R.J. is supported by the Crohn’s & Colitis Foundation of America Research Fellowship under award number 569103. A.J. is supported by National Science Foundation Graduate Research Fellowship Program under program grant number Fall 2007 DGE1752134. K.N.S. was supported by Australian National Health and Medical Research Council CJ Martin Overseas Biomedical Early Career Fellowship. S.H. was supported by the Austrian Marshall Plan Foundation. M.R.d.Z. was supported by a VIDI grant from the Netherlands Organization for Scientific Research (NWO, 91715377) and the Utrecht Exposome Hub. A.W. was supported by a grant from the Swiss National Science Foundation (SNF 320030_182764). This work was also supported by NIH grants R01HG008383 (Y.K. and J.Z.), R01GM131642A (Y.K., J.Z., and R.A.F), and R01GM135928 (Y.K., J.Z., and R.A.F.) This work was supported by the Howard Hughes Medical Institute and the Blavatnik Family Foundation (R.A.F.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2019.12.016.
DECLARATION OF INTERESTS
R.A.F. is a scientific advisor to GlaxoSmithKline. A.M.R. is the founder of Simcha Therapeutics, an immuno-oncology company developing IL-18 based therapies. All other authors declare no competing interests.
REFERENCES
- Austin KM (2012). The pathogenesis of Hirschsprung’s disease-associated enterocolitis. Semin. Pediatr. Surg. 21, 319–327. [DOI] [PubMed] [Google Scholar]
- Baig MS, Zaichick SV, Mao M, de Abreu AL, Bakhshi FR, Hart PC, Saqib U, Deng J, Chatterjee S, Block ML, et al. (2015). NOS1-derived nitric oxide promotes NF-κB transcriptional activity through inhibition of suppressor of cytokine signaling-1. J. Exp. Med. 212, 1725–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajon I, Serrao G, Arnaboldi F, Opizzi E, Ripamonti G, Balsari A, and Rumio C (2009). Toll-like receptors 3, 4, and 7 are expressed in the enteric nervous system and dorsal root ganglia. J. Histochem. Cytochem. 57, 1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, and Hardt WD (2003). Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71, 2839–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkind-Gerson J, Carreon-Rodriguez A, Benedict LA, Steiger C, Pieretti A, Nagy N, Dietrich J, and Goldstein AM (2013). Nestin-expressing cells in the gut give rise to enteric neurons and glial cells. Neurogastroenterol. Motil. 25, 61–69.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchenough GMH, Nystrom EEL, Johansson MEV, and Hansson GC (2016). A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 352, 1535–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bódi N, Szalai Z, and Bagyanszki M (2019). Nitrergic Enteric Neurons in Health and Disease-Focus on Animal Models. Int J Mol Sci. 20, E2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, Ruby T, Belhocine K, Bouley DM, Kayagaki N, Dixit VM, and Monack DM (2012). Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature 490, 288–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccamo DV, Herman MM, Frankfurter A, Katsetos CD, Collins VP, and Rubinstein LJ (1989). An immunohistochemical study of neuropeptides and neuronal cytoskeletal proteins in the neuroepithelial component of a spontaneous murine ovarian teratoma. Primitive neuroepithelium displays immunoreactivity for neuropeptides and neuron-associated beta-tubulin isotype. Am. J. Pathol. 135, 801–813. [PMC free article] [PubMed] [Google Scholar]
- Canny G, Swidsinski A, and McCormick BA (2006). Interactions of intestinal epithelial cells with bacteria and immune cells: methods to characterize microflora and functional consequences. Methods Mol. Biol. 341, 17–35. [DOI] [PubMed] [Google Scholar]
- Case CL, Kohler LJ, Lima JB, Strowig T, Zoete M.R.d., Flavell RA, Zamboni DS, and Roy CR (2013). Caspase-11 stimulates rapid flagellin-independent pyroptosis in response to Legionella pneumophila. Proc Natl Acad Sci USA 110, 1851–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LM, Kaniga K, and Galan JE (1996). Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol. 21, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Clark JA, and Coopersmith CM (2007). Intestinal crosstalk: a new paradigm for understanding the gut as the “motor” of critical illness. Shock 28, 384–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquenlorge S, Duchalais E, Chevalier J, Cossais F, Rolli-Derkinderen M, and Neunlist M. (2014). Modulation of lipopolysaccharide-induced neuronal response by activation of the enteric nervous system. J. Neuroinflammation 11, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Autréaux F, Morikawa Y, Cserjesi P, and Gershon MD (2007). Hand2 is necessary for terminal differentiation of enteric neurons from crest-derived precursors but not for their migration into the gut or for formation of glia. Development 134, 2237–2249. [DOI] [PubMed] [Google Scholar]
- Dinarello CA (2009). Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 27, 519–550. [DOI] [PubMed] [Google Scholar]
- Doerflinger NH, Macklin WB, and Popko B. (2003). Inducible site-specific recombination in myelinating cells. Genesis 35, 63–72. [DOI] [PubMed] [Google Scholar]
- Drokhlyansky E, Smillie CS, Van Wittenberghe N, Ericsson M, Griffin GK, Dionne D, Cuoco MS, Goder-Reiser MN, Sharova T, Aguirre AJ, et al. (2019). The enteric nervous system of the human and mouse colon at a single-cell resolution. bioRxiv. 10.1101/746743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, and Flavell RA (2011). NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin BS, Bossaller L, De Nardo D, Ratter JM, Stutz A, Engels G, Brenker C, Nordhoff M, Mirandola SR, Al-Amoudi A, et al. (2014). The adaptor ASC has extracellular and ‘prionoid’ activities that propagate inflammation. Nat. Immunol. 15, 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness JB, Callaghan BP, Rivera LR, and Cho HJ (2014). The enteric nervous system and gastrointestinal innervation: integrated local and central control. Adv. Exp. Med. Biol. 817, 39–71. [DOI] [PubMed] [Google Scholar]
- Gabanyi I, Muller PA, Feighery L, Oliveira TY, Costa-Pinto FA, and Mucida D. (2016). Neuro-immune Interactions Drive Tissue Programming in Intestinal Macrophages. Cell 164, 378–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A , Yanortsang TB, Yang L, Jupp R, Mathis D, et al. (2017). Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 168, 928–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Kuida K, Tsutsui H, Ku G, Hsiao K, Fleming MA, Hayashi N, Hi-gashino K, Okamura H, Nakanishi K, et al. (1997). Activation of Interferon-γ Inducing Factor Mediated by Interleukin-1β Converting Enzyme. Science 275, 206–209. [DOI] [PubMed] [Google Scholar]
- Hansson GC (2012). Role of mucus layers in gut infection and inflammation. Curr. Opin. Microbiol. 15, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He WT, Wan H, Hu L, Chen P, Wang X, Huang Z, Yang ZH, Zhong CQ, and Han J. (2015). Gasdermin D is an executor of pyroptosis and required for interleukin-1 b secretion. Cell Res. 25, 1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot TJ, Liu H, Sarkar AA, Giovannucci DR, Clouthier DE, Abe M, and Howard MJ (2007). Expression of Hand2 is sufficient for neurogenesis and cell type-specific gene expression in the enteric nervous system. Dev. Dyn. 236, 93–105. [DOI] [PubMed] [Google Scholar]
- Hirschsprung H. (1887). Stuhtragheit Neugeborener infolge Dilatationen und hypertrophie des Colons. Jahruch Kinderheikunde 27, 1. [Google Scholar]
- Hockley JRF, Taylor TS, Callejo G, Wilbrey AL, Gutteridge A, Bach K, Winchester WJ, Bulmer DC, McMurray G, and Smith ESJ (2019). Single-cell RNAseq reveals seven classes of colonic sensory neuron. Gut 68, 633–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, and Macpherson AJ (2010). Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10, 159–169. [DOI] [PubMed] [Google Scholar]
- Jackson R, Kroehling L, Khitun A, Bailis W, Jarret A, York AG, Khan OM, Brewer JR, Skadow MH, Duizer C, et al. (2018). The translation of non-canonical open reading frames controls mucosal immunity. Nature 564, 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarret A, McFarland AP, Horner SM, Kell A, Schwerk J, Hong M, Badil S, Joslyn RC, Baker DP, Carrington M, et al. (2016). Hepatitis-C-virus-induced microRNAs dampen interferon-mediated antiviral signaling. Nat Med. 22, 1475–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph NM, He S, Quintana E, Kim Y-G, Nunez G, and Morrison SJ (2011). Enteric glia are multipotent in culture but primarily form glia in the adult rodent gut. J. Clin. Invest. 121, 3398–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai T, Watanabe M, Okazawa A, Nakamaru K, Okamoto M, Naganuma M, Ishii H, Ikeda M, Kurimoto M, and Hibi T. (2000). Interleukin 18 isa potent proliferative factor for intestinal mucosal lymphocytes in Crohn’s disease. Gastroenterology 119, 1514–1523. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, et al. (2011). Non-canonical inflammasome activation targets caspase-11. Nature 479, 117–121. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, et al. (2015). Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671. [DOI] [PubMed] [Google Scholar]
- Koning JJ, Konijn T, Lakeman KA, O’Toole T, Kenswil KJG, Raaij-makers MHGP, Michurina TV, Enikolopov G, and Mebius RE (2016). Nestin-Expressing Precursors Give Rise to Both Endothelial as well as Nonendothelial Lymph Node Stromal Cells. J Immunol. 197, 2686–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, and Schloss PD (2013). Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl Environ Microbiol. 79, 5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Micci M-A, Leser J, Shin C, Tang S-C, Fu Y-Y, Liu L, Li Q, Saha M, Li C, et al. (2017). Adult enteric nervous system in health is maintained by a dynamic balance between neuronal apoptosis and neurogenesis. Proc. Natl. Acad. Sci. USA 114, E3709–E3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara-Otani S, Maeda S, Kobayashi K, Minato Y, Tanaka K, Yamanishi K, Hata M, Li W, Hayakawa T, Noguchi K, et al. (2017). Interleukin-18 and its receptor are expressed in gonadotropin-releasing hormone neurons of mouse and rat forebrain. Neurosci. Lett. 650, 33–37. [DOI] [PubMed] [Google Scholar]
- Lagace DC, Whitman MC, Noonan MA, Ables JL, DeCarolis NA, Arguello AA, Donovan MH, Fischer SJ, Farnbauch LA, Beech RD, et al. (2007). Dynamic contribution of nestin-expressing stem cells to adult neurogenesis. J. Neurosci. 27, 12623–12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai NY, Musser MA, Pinho-Ribeiro FA, Baral P, Ma P, Potts DE, Chen Z, Paik D, Soualhi S, Shi H, et al. (2019). Gut-innervating nociceptor neurons protect against enteric infection by modulating the microbiota and Peyer’s patch microfold cells. bioRxiv. 10.1101/580555. [DOI] [Google Scholar]
- Lara-Tejero M, Sutterwala FS, Ogura Y, Grant EP, Bertin J, Coyle AJ, Flavell RA, and Galan JE (2006). Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J. Exp. Med. 203, 1407–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Thaiss CA, Zeevi D, Dohnalova L, Zilberman-Schapira G, Mahdi JA, David E, Savidor A, Korem T, Herzig Y, et al. (2015). Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 163, 1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderman GC, Zhao J, and Kluger Y. (2018). Zero-preserving imputation of scRNA-seq data using low-rank approximation. bioRxiv. 10.1101/397588. [DOI] [Google Scholar]
- Maaten LVD, and Hinton G. (2008). Visualizing Data using t-SNE. J Mach Learn Res. 9, 2579–2605. [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, and Gumucio DL (2002). Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 277, 33275–33283. [DOI] [PubMed] [Google Scholar]
- Martinon F, Burns K, and Tschopp J. (2002). The inflammasome: a molecular platform triggering activation of inflammatory caspasesand processing of proIL-beta. Mol. Cell 10, 417–26. [DOI] [PubMed] [Google Scholar]
- Matheis F, Muller PA, Graves CL, Gabanyi I, Kerner ZJ, Costa-Borges D , Ahrends T, Rosenstiel P, and Mucida D. (2020). Adrenergic Signaling in Muscularis Macrophages Limits Infection-Induced Neuronal Loss. Cell 180. Published online January 9, 2020. https://doi.org/10.1016Zj.cell.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald V, Pollok RC, Dhaliwal W, Naik S, Farthing MJ, and Bajaj-Elliott M. (2006). A potential role for interleukin-18 in inhibition of the development of Cryptosporidium parvum. Clin. Exp. Immunol. 145, 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur AD, Lira SA, Scadden DT, Ma’ayan A, Enikolopov GN, and Frenette PS (2010). Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466, 829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone M, Stanley AC, Chen KW, Brown DL, Bezbradica JS, von Pein JB, Holley CL, Boucher D, Shakespear MR, Kapetanovic R, et al. (2018). Interleukin-1 ß Maturation Triggers Its Relocation to the Plasma Membrane for Gasdermin-D-Dependent and -Independent Secretion. Cell Rep. 24, 1425–1433. [DOI] [PubMed] [Google Scholar]
- Moor AE, Harnik Y, Ben-Moshe S, Massasa EE, Rozenberg M, Eilam R, Bahar Halpern K, and Itzkovitz S. (2018). Spatial Reconstruction of Single Enterocytes Uncovers Broad Zonation along the Intestinal Villus Axis. Cell 175, 1156–1167. [DOI] [PubMed] [Google Scholar]
- Motoike T, Markham DW, Rossant J, and Sato TN (2003). Evidence for novel fate of Flk1 + progenitor: contribution to muscle lineage. Genesis 35, 153–159. [DOI] [PubMed] [Google Scholar]
- Nowarski R, Jackson R, Gagliani N, de Zoete MR, Palm NW, Bailis W, Low JS, Harman CC, Graham M, Elinav E, and Flavell RA (2015). Epithelial IL-18 Equilibrium Controls Barrier Function in Colitis. Cell 163, 1444–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto E, and Ueda T. (1967). Embryogenesis of intramural ganglia of the gut and its relation to Hirschsprung’s disease. J. Pediatr. Surg. 2, 437–443. [Google Scholar]
- Omoto Y, Tokime K, Yamanaka K, Habe K, Morioka T, Kurokawa I, Tsutsui H, Yamanishi K, Nakanishi K, and Mizutani H. (2006). Human mast cell chymase cleaves pro-IL-18 and generates a novel and biologically active IL-18 fragment. J. Immunol. 177, 8315–8319. [DOI] [PubMed] [Google Scholar]
- Omoto Y, Yamanaka K, Tokime K, Kitano S, Kakeda M, Akeda T, Kurokawa I, Gabazza EC, Tsutsui H, Katayama N, et al. (2010). Granzyme B is a novel interleukin-18 converting enzyme. J. Dermatol. Sci. 59, 129–135. [DOI] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al. (2014). Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Propheter DC, Chara AL, Harris TA, Ruhn KA, and Hooper LV (2017). Resistin-like molecule b is a bactericidal protein that promotes spatial segregation of the microbiota and the colonic epithelium. Proc Natl Acad Sci USA. 114, 11027–11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu ZD, Thacker M, Castelucci P, Bagyanszki M, Epstein ML, and Furness JB (2008). Immunohistochemical analysis of neuron types in the mouse small intestine. Cell Tissue Res. 334, 147–161. [DOI] [PubMed] [Google Scholar]
- Rao M, Nelms BD, Dong L, Salinas-Rios V, Rutlin M, Gershon MD, and Corfas G. (2015). Enteric glia express proteolipid protein 1 and are a transcriptionally unique population of glia in the mammalian nervous system. Glia 63, 2040–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raupach B, Peuschel S-K, Monack DM, and Zychlinsky A. (2006). Caspase-1-mediated activation of interleukin-1 β (IL-1 β) and IL-18 contributes to innate immune defenses against Salmonella enterica serovar Typhimurium infection. Infect. Immun. 74, 4922–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest LB, Dager M, Yanagisawa H, Charite J, Hammer RE, Olson DN, Yanagisawa M, and Clouthier DE (2003). dHAND-Cre transgenic mice reveal specific potential functions of dHAND during craniofacial development. Dev. Biol. 257, 263–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh Y, Ishikawa K, Oomori Y, Takeda S, and Ono K. (1992). Bethanechol and a G-protein activator, NaF/AlCl3, induce secretory response in Paneth cells of mouse intestine. Cell Tissue Res. 269, 213–220. [DOI] [PubMed] [Google Scholar]
- Schemann M, and Camilleri M. (2013). Functions and imaging of mast cell and neural axis of the gut. Gastroenterology 144, 698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, et al. (2015). Individual intestinal symbionts induce a distinct population of RORg+ regulatory T cells. Science 349, 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah NM, Marchionni MA, Isaacs I, Stroobant P, and Anderson DJ (1994). Glial growth factor restricts mammalian neural crest stem cells to a glial fate. Cell 77, 349–360. [DOI] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. (2016). Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, and Shao F. (2015). Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665. [DOI] [PubMed] [Google Scholar]
- Smith TH, Ngwainmbi J, Grider JR, Dewey WL, and Akbarali HI (2013). An in-vitro preparation of isolated enteric neurons and glia from the myenteric plexus of the adult mouse. J. Vis. Exp. 10.3791/50688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub RH, Wiest R, Strauch UG, Harle P, and Scholmerich J. (2006). The role of the sympathetic nervous system in intestinal inflammation. Gut. 55, 1640–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strowig T, Henao-Mejia J, Elinav E, and Flavell R. (2012). Inflammasomes in health and disease. Nature 481, 278–286. [DOI] [PubMed] [Google Scholar]
- Sugama S, Cho BP, Baker H, Joh TH, Lucero J, and Conti B. (2002). Neurons of the superior nucleus of the medial habenula and ependymal cells express IL-18 in rat CNS. Brain Res. 958, 1–9. [DOI] [PubMed] [Google Scholar]
- Talbot J, Hahn P, Kroehling L, Nguyen H, Li D, and Littman DR (2019). VIP-producing enteric neurons interact with innate lymphoid cells to regulate feeding-dependent intestinal epithelial barrier functions. bioRxiv. 10.1101/721464. [DOI] [Google Scholar]
- Thiagarajah JR, Yildiz H, Carlson T, Thomas AR, Steiger C, Pieretti A, Zukerberg LR, Carrier RL, and Goldstein AM (2014). Altered goblet cell differentiation and surface mucus properties in Hirschsprung disease. PLoS ONE 9, e99944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udden SM, Waliullah S, Harris M, and Zaki H. (2017). The Ex Vivo Colon Organ Culture and Its Use in Antimicrobial Host Defense Studies. J. Vis. Exp. 10.3791/55347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainchtein ID, Chin G, Cho FS, Kelley KW, Miller JG, Chien EC, Liddelow SA, Nguyen PT, Nakao-Inoue H, Dorman LC, et al. (2018). Astrocyte-derived interleukin-33 promotes microglial synapse engulfment and neural circuit development. Science 359, 1269–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Rolletschek A, Kania G, Blyszczuk P, Tarasov KV, Tarasova Y, Wersto RP, Boheler KR, and Wobus AM (2004). Nestin expression-a property of multi-lineage progenitor cells? Cell. Mol. Life Sci. 61, 2510–2522. [DOI] [PubMed] [Google Scholar]
- Yoo BB, and Mazmanian SK (2017). The Enteric Network: Interactions between the Immune and Nervous Systems of the Gut. Immunity 46, 910–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lonnerberg P, Johnsson A, Memic F, van der Zwan J, Haring M, Braun E, Borm LE, La Manno G, et al. (2018). Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Heilig HG, Klaassens ES, Booijink CC, Kleerebezem M, Smidt H, and de Vos WM (2006). Isolation of DNAfrom bacterial samples of the human gastrointestinal tract. Nat. Protoc. 1, 870–873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptome datasets generated during this study are available at Gene Expression Omnibus (GEO) under the superseries number GSE141810. The bulk RNaseq is available under the accession number: GSE141808. The DropSeq data is available under the accession number: GSE141809. The 16S sequencing datasets generated during this study are available at NCBI BioProject under the number: PRJNA594999.