Abstract
T cell receptor (TCR) stimulation and cytokine cues drive the differentiation of CD4+ naïve T cells into effector T cell populations with distinct pro-inflammatory or regulatory functions. Unlike adult naïve T cells, human fetal naïve CD4+ T cells preferentially differentiate into FOXP3+ regulatory T (Treg) cells upon TCR activation independent of exogenous cytokine signalling. This cell-intrinsic predisposition for Treg differentiation is implicated in the generation of tolerance in utero; however, the underlying mechanisms remain largely unknown. Here, we identify epigenetic and transcriptional programs shared between fetal naive T and committed Treg cells that are inactive in adult naive T cells, and show that fetal-derived induced Treg (iTreg) cells retain this transcriptional program. We show that a subset of Treg-specific enhancers is accessible in fetal naive T cells, including two active super-enhancers at Helios. Helios is expressed in fetal naïve T cells but not in adult naïve T cells, and fetal iTreg cells maintain Helios expression. CRISPR-Cas9 ablation of Helios in fetal naïve T cells impaired their differentiation into iTreg cells upon TCR stimulation, reduced expression of immunosuppressive genes in fetal iTreg cells such as IL10, and increased expression of pro-inflammatory genes including IFNG. Consequently, Helios knockout fetal iTreg cells had reduced IL-10 and increased IFN-γ cytokine production. Together, our results reveal important roles for Helios in enhancing preferential fetal Treg differentiation and fine-tuning eventual Treg function. The Treg-biased programs identified within fetal naive T cells could potentially be utilized to engineer enhanced iTreg populations for adoptive cellular therapies.
One Sentence Summary:
Helios contributes to a transcriptional and epigenetic program in human fetal naïve T cells promoting Treg differentiation.
Introduction
The adaptive immune system must generate immunotolerance in order to prevent or resolve pro-inflammatory responses that can cause host damage (1–3), while still permitting functional effector responses for host defense against pathogens (4, 5). A primary mechanism that achieves this flexibility is the capacity of CD4+ naive T cells to differentiate into multiple specialized T helper (Th) subsets with either pro-inflammatory or immunosuppressive functions. The presence of polarizing cytokines within their immediate environment determines the eventual Th cell fate by triggering the expression and/or activation of master transcription factors that enact lineage specific transcriptional programs (6). For example, signaling by transforming growth factor-β (TGF-β) promotes the induction of FOXP3 (7–9), which is the master transcription factor required for the differentiation of naïve T cells into immunosuppressive regulatory T (Treg) cells.
Mutations of the FOXP3 gene leading to the absence or dysfunction of Treg cells result in the loss of Treg-mediated immunotolerance, and trigger fatal, early onset multi-organ autoimmunity in both mice and humans (10–15). Autoimmunity resulting from the loss of FOXP3+ Treg cell-mediated tolerance in humans, defined as the IPEX (Immune dysregulation, polyendocrinopathy, enteropathy, X-linked) syndrome, can manifest within the fetus in utero and result in miscarriage, preterm birth, or death in childhood without hematopoietic stem cell transplant (16–20). The initiation of autoimmunity in IPEX coincides with the emergence of T cells in the second trimester of human development, suggesting that Treg cell-mediated peripheral tolerance is required during fetal development (21, 22). This is supported by the presence of an abundant population of fetal Treg cells in the secondary lymphoid tissues, which comprise a larger percentage of the total CD4+ T cell population compared to adults (23–25). However, thymic Treg cell frequencies in fetal and infant thymus are not significantly different (24), indicating that increased thymic output is not responsible for the increased frequency of fetal Treg cells. Fetal naive T cells, unlike their adult counterparts, preferentially differentiate into functional Treg cells upon antigen stimulation, which include non-inherited maternal alloantigens (i.e. NIMAs) on maternal antigen-presenting cells (24). These findings imply that the abundance of fetal Treg cells observed in fetal lymphoid tissues is due to peripheral conversion from naïve T cells. This propensity for Treg differentiation is retained in vitro, as a high frequency of fetal naïve T cells differentiate into FOXP3+ Treg cells upon TCR activation even in the absence of exogenous TGF-β (26). The unique capability of fetal naïve T cells to initiate Treg differentiation in the absence of exogenous TGF-β suggests that this ability is cell-intrinsic; however, the molecular mechanisms that underlie this predisposition are largely unknown.
Chromatin changes are also implicated in driving the final effector phenotype and function of differentiated T cells, defined by increases in chromatin accessibility of active lineage-specific genes, and the silencing of genes associated with other effector lineages (27, 28). In thymic and peripheral Treg cells, permissive/active histone marks and DNA demethylation at Treg-associated genes such as IL2RA (i.e., CD25), CTLA4, IKZF2 (i.e., Helios), and IKZF4 (i.e., Eos) (29, 30) must be acquired for commitment to and maintenance of the Treg phenotype (29–32). This Treg-chromatin landscape is acquired within developing thymic Treg precursors before FOXP3 protein expression (30), indicating that a Treg-specific epigenome may be responsible for initiating and promoting the expression of FOXP3. Additionally, other key genes associated with the Treg epigenome, such as Helios, are expressed independently of FOXP3 expression (29, 30, 33), and can direct the partial acquisition of the Treg-specific transcriptional signature when over-expressed in FOXP3-CD4+ T cells (34). We therefore hypothesized that fetal naïve T cells might already possess a partial Treg-specific epigenetic and transcriptional signature that predisposes them for differentiation towards the Treg cell fate even without exogenous TGF-β signaling.
Here, we interrogated the transcriptional and chromatin landscape of fetal and adult naïve and Treg cells, and discovered that components of the Treg gene regulatory program are activated only in fetal naïve T cells. We then show that the partial Treg-specific gene signature detected at steady state in fetal naïve T cells is retained only in fetal-derived, but not adult-derived induced Treg (iTreg) cells. We next identify two Treg-specific super-enhancers (SEs) associated with the Helios locus that are active in fetal naïve T cells, in which we subsequently demonstrate the expression of Helios protein. Only iTreg cells generated from fetal naïve T cells in vitro retained Helios expression and were characterized by repression of interleukin-2 (IL-2) production; neither of which were observed in adult iTreg cells. CRISPR (clustered regular interspaced short palindromic repeats)-Cas9 (CRISPR-associated protein 9) mediated ablation of Helios in fetal naive T cells impaired their cell-intrinsic ability to differentiate into iTreg cells in the absence of exogenous TGF-β. Analysis of the transcriptome in Helios knockout iTreg cells revealed that Helios enhanced the upregulation of Treg-specific genes (e.g., IL10) and mediated the repression of pro-inflammatory genes involved in T effector differentiation and function (e.g., IFNG). Helios ablation in fetal iTreg cells resulted in decreased IL-10 production concurrent with increased IFN-γ and IL-2. Given that Helios has been previously characterized to be a gene specific to thymic Treg cells, our data reveal a new role for Helios as part of a pre-existing epigenetic and transcriptional program within human fetal naïve T cells that lowers the threshold for Treg differentiation and functional commitment. Taken together, we thus identify a TGF-β-independent mechanism unique to fetal naïve T cells that favors their differentiation into Treg cells, which may contribute insights into better engineering Treg cells in vitro from naïve T cells for use in immunotherapy.
Results
Human fetal naïve T cells express a partial Treg transcriptome.
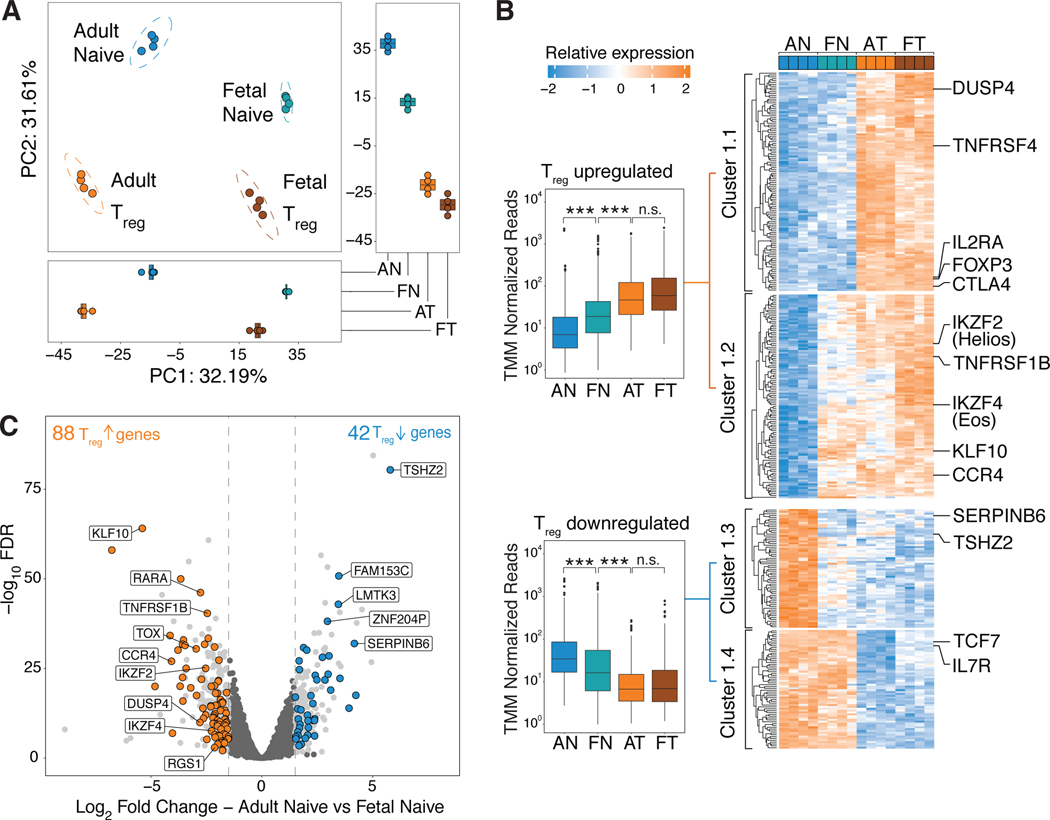
Given that fetal naïve T cells preferentially differentiate into Treg cells upon TCR stimulation alone (26), we first asked if fetal naïve T cells shared elements of their transcriptome with Treg cells that could predispose them towards the Treg lineage. We performed RNA sequencing (RNAseq) on sorted fetal and adult CD4+ naïve and Treg cells (see fig. S1 for sort gating strategy and purity confirmation). Principal component analysis (PCA) revealed that fetal and adult populations first segregated by cell origin (PC1, fetal or adult) before cell phenotype (PC2, naïve versus Treg, Fig. 1A). PC2 scores for fetal naïve samples were intermediate to adult naïve and Treg samples (Fig. 1A), suggesting intermediate expression of Treg-specific genes in fetal naïve T cells. In order to test this hypothesis, we first defined a Treg-specific transcriptional signature by identifying genes differentially expressed in both fetal and adult Treg cells relative to adult naïve T cells based on a false discovery rate cut-off of FDR<0.05 and log 2 fold change (log2FC) increase in expression of 1.5 (fig. S2, table S1). Fetal naïve T cells possessed intermediate upregulation/downregulation (Fig. 1B) across genes upregulated/downregulated in our Treg-specific transcriptional signature. More specifically, relative to adult naïve T cells, fetal naïve T cells had 88 Treg-upregulated and 42 Treg-downregulated genes (Fig. 1C). We defined four different clusters within the Treg-specific transcriptome (Fig. 1B, table S1) – two of which corresponded to all Treg-upregulated genes (Cluster 1.1, 1.2), while the other two clusters contained all Treg-downregulated genes (Cluster 1.3, 1.4). Within Treg-upregulated genes, fetal naïve T cells did not express canonical Treg genes such as FOXP3, IL2RA and CTLA-4 (Cluster 1.1, Fig. 1B). Instead, fetal naïve T cells had increased expression of Treg-upregulated genes previously associated with Treg function such as CCR4 (35, 36) and KLF10 (37–39). Additionally, fetal naïve T cells had increased expression of the transcription factors IKZF2 (Helios) (40–42) and IKZF4 (Eos) (34, 43) (Cluster 1.2, Fig. 1B,1C) which are transcribed independently of FOXP3 expression in mice (29, 33, 44). Fetal naïve T cells and both the Treg cell populations shared similar downregulation of a subset of genes previously characterized as being downregulated in Treg cells such as TSHZ2 and SERPINB6 (45–47) (Cluster 1.3, Fig. 1B). In contrast, genes shared between fetal and adult naïve T cells included known genes contributing to the naïve T cell phenotype such as TCF7 and IL7R (Cluster 1.4, Fig. 1B). Taken together, the presence of a partial Treg-specific signature in fetal naïve T cells could thus potentiate Treg differentiation upon the receipt of TCR signaling, consistent with the lowered threshold and greater propensity for these cells towards Treg differentiation (24, 26).
Figure 1. Fetal naïve T cells have expression of a partial Treg transcriptome.
(A) Principal Component Analysis (PCA) plot of RNAseq data comparing adult naïve (AN), fetal naïve (FN), adult Treg (AT) and fetal Treg (FT) cells. Boxplots show scores for PC1 (bottom) and PC2 (right).
(B) Heatmap shows relative expression levels of Treg-specific differentially expressed (DE) genes in AN, FN, AT and FT cells. Clusters are labeled and defined by k-means clustering. Genes associated with Treg and naïve T cell function are labeled. Boxplots (left) show the averaged trimmed mean of M values (TMM) normalized reads across all upregulated (top, Cluster 1&2) and downregulated (bottom, Cluster 3&4) genes. Kruskal-Wallis test and Dunn’s multiple comparison test with Bonferroni correction for multiple testing, *** p<0.001, n.s., p>0.05 (n = 4 biological replicates per group).
(C) Volcano plot of DE (log2 fold change >1.5, p.adj<0.05) genes in fetal and adult naïve T cells, all DE genes in light grey. Dotted lines (grey) denote fold change cut-offs. 88 and 42 Treg-specific genes are upregulated (orange) and downregulated (blue) in fetal naïve T cells. Among Treg-upregulated genes, genes previously associated with Treg function are labeled. Among Treg-downregulated genes, the top five genes with the lowest p.adj values are labeled.
All boxplots in this figure show median (centre line), interquartile range (box) and tenth and ninetieth percentiles (whiskers).
Fetal-derived iTreg cells maintain differential expression of the partial Treg-specific transcriptome present in fetal naïve T cells.
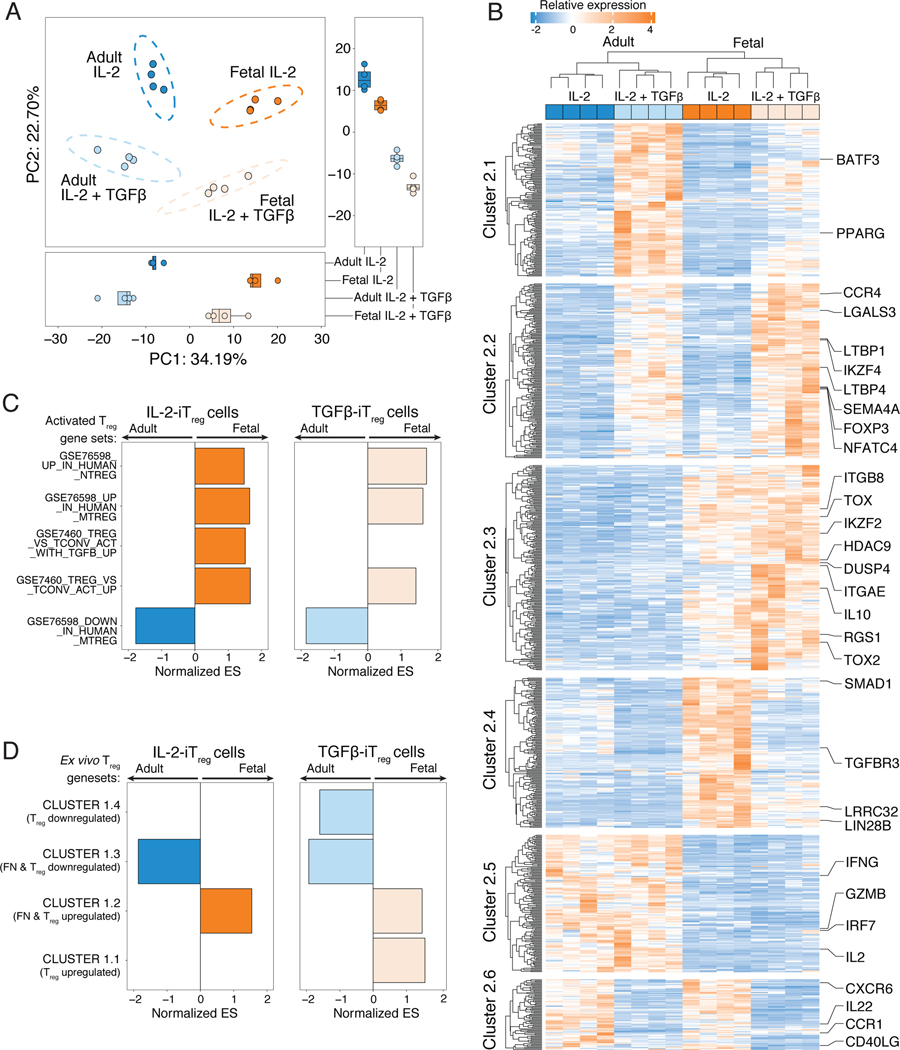
We next asked if the partial Treg-specific transcriptome detected in fetal naïve T cells would remain differentially expressed in fetal iTreg cells, and whether adult iTreg cells also acquire the Treg-specific transcriptome after in vitro differentiation. To assess this, we performed RNAseq on fetal and adult iTreg cells that underwent differentiation with TCR stimulation in media supplemented with IL-2 alone (IL-2-iTreg) or additionally supplemented with TGF-β (TGF-β-iTreg). PCA revealed that PC1 still segregated iTreg populations by cell origin (fetal or adult), after which populations segregated by the stimulus received during differentiation (absence/presence of TGF-β, PC2, Figure 2A). This indicated that fetal-derived iTreg cells were still transcriptionally different from adult-derived iTreg cells, although the addition of TGF-β was sufficient to drive differential expression of a shared set of genes. We then evaluated all differentially expressed genes (FDR<0.05 and log2FC>1.5) across all iTreg populations (Fig. 2B), and defined six clusters (table S2). Both fetal and adult TGF-β-iTreg cells upregulated two gene clusters (Cluster 2.1 & 2.2, Fig. 2B, fig. S3A), and downregulated one gene cluster (Cluster 2.6, Fig. 2B, fig. S3A). Treg-specific genes within Cluster 2.1 and 2.2 included genes known to be upregulated with TGF-β signaling such as FOXP3 and IKZF4 (48), as well genes potentially implicated in Treg differentiation and function such as SEMA4A (49), LTBP1, LTBP4 (50) and LGALS3 (51, 52). However, only fetal IL-2- and TGF-β-iTreg cells had increased expression of Treg-specific genes that were upregulated in ex vivo fetal naïve T cells (Fig. 1C) such as IKZF2, DUSP4, TOX and RGS1 (Cluster 2.3, Fig. 2B, fig. S3A). Decreased transcription of pro-inflammatory transcripts such as IFNG, IL2, GZMB and IRF7 (Cluster 2.5, Fig. 2B, fig. S3A), and increased expression of genes involved in Treg cell suppressive function such as IL10 (Cluster 2.3, Fig. 2B) were only observed in fetal, but not adult, iTreg cells. This suggested that, qualitatively, fetal iTreg cells may have a transcriptome more reflective of ex vivo Treg populations relative to adult iTreg cells. We thus utilized Gene Set Enrichment Analysis (GSEA) to independently evaluate the transcriptomes of fetal or adult-derived iTreg cells against published gene sets comparing Treg and conventional T cell populations that also underwent TCR- and cytokine-stimulated activation in vitro (fig. S3B, table S3). In comparison to adult iTreg cells, fetal iTreg cells differentiated in both stimulation conditions had enrichment in genes upregulated in activated Treg populations (Fig. 2B), and had enrichment of the partial Treg-specific signature as defined by Cluster 1.2 and 1.3 (Fig. 1B, 2C, left, fig. S3C). Exogenous TGF-β signaling during Treg differentiation resulted in the enrichment of Cluster 1.1 and depletion of Cluster 1.4 genes within fetal TGF-β-iTreg but not adult TGF-β-iTreg cells (Fig. 2C, right, fig. S3D). Our data thus show that differentiating fetal naïve T cells, independently of exogenous TGF-β, retain increased expression of the partial Treg-specific transcriptional signature detected in ex vivo naïve T cells. Furthermore, the expression of these genes does not reach the same levels in adult iTreg cells, suggesting that upstream mechanisms might be responsible for driving the transcriptional differences that favor Treg differentiation in fetal naïve T cells.
Figure 2. Fetal induced Treg cells retain expression of the partial Treg-specific transcriptome detected in fetal naïve T cells in steady state.
(A) Principal Component Analysis (PCA) of RNAseq data comparing fetal and adult induced Treg cells generated in IL-2 alone (IL-2-iTreg) or with added exogenous TGF-β (TGF-β-iTreg). Boxplots show scores for PC1 (bottom) and PC2 (right).
(B) Heatmap shows relative expression levels of differentially expressed genes (log2 fold change >1.5, false discovery rate, FDR<0.05) in adult and fetal IL-2- and TGF-β-iTreg Clusters are labeled and defined by k-means clustering. Genes associated with Treg or pro-inflammatory/effector T cell function are labeled.
(C) Pre-ranked Gene Set Enrichment Analysis (GSEA) was used to assess overrepresentation of pre-defined activated Treg-associated gene sets (see fig. S3B for details of all gene sets) in fetal (orange) or adult iTreg cells (blue) differentiated with IL-2 (left) or IL-2 and TGF-β (right), n=4 for all conditions. Barplot shows normalized Enrichment Scores (ES) for gene sets with FDR<0.05, arrows denote direction of enrichment in adult or fetal iTreg cells.
(D) Pre-ranked GSEA was used to assess overrepresentation of each of the gene clusters identified in Figure 1B. Barplot shows normalized Enrichment Scores (ES) for all clusters with FDR <0.05 for fetal (orange) or adult (blue) iTreg cells differentiated with IL-2 (left) or IL-2 and TGF-β (right).
Fetal iTreg cells have increased sensitivity to TGF-β signaling
In addition to increased expression of genes associated with Treg function, fetal IL-2-iTreg cells strongly upregulated a gene cluster which contained key genes associated with TGF-β sequestration and downstream signaling – including SMAD1, TGFBR3, LRRC32 (54, 55) and LIN28B (26) (Cluster 2.4, Fig. 2B, fig. S4A). Expression of Lin28b protein in fetal naïve T cells contributes to increased expression of TGFBR1, TGFBR3, SMAD2, as well as increased phosphorylation of SMAD2/3 (26). GARP (LRRC32) is expressed highly on the cell surface of activated Treg cells and captures inactive TGF-β bound to the latency-associated peptide (LAP) (54, 56). This reservoir of cell-surface associated TGF-β is implicated in the maintenance of oral tolerance in mice (55), and in the induction of FOXP3 in naïve T cells co-cultured with GARP+Treg cells (54). Here we demonstrate that fetal but not adult iTreg cells highly expressed GARP (fig. S4B) and LAP in a linear fashion (fig. S4C). Furthermore, fetal iTreg cells have increased transcription of ITGB8 (Cluster 3, fig. 2B), the beta chain for the integrin αvβ8 which processes and releases bioactive TGF-β1 from LAP (55). Since fetal iTreg cells have increased cell surface-associated TGF-β and the machinery to mediate its potential release, we tested whether blockade of TGF-β1 with TGF-β-neutralizing antibodies resulted in decreased fetal Treg differentiation in response to TCR stimulation alone. As hypothesized, fetal Treg induction was blunted in the setting of TGF-β blockade (fig. S4D, E). However, fetal naïve T cells still retained an increased ability for Treg differentiation over adult naïve T cells, even when exogenous bioactive TGF-β was added (fig. S4D,E). Hence, although active TGF-β1 biogenesis may contribute to fetal iTreg differentiation in the absence of exogenous TGF-β, additional upstream mechanisms are responsible for driving enhanced fetal Treg differentiation in vitro.
Fetal naïve T cells share a partial epigenetic landscape with adult Treg cells
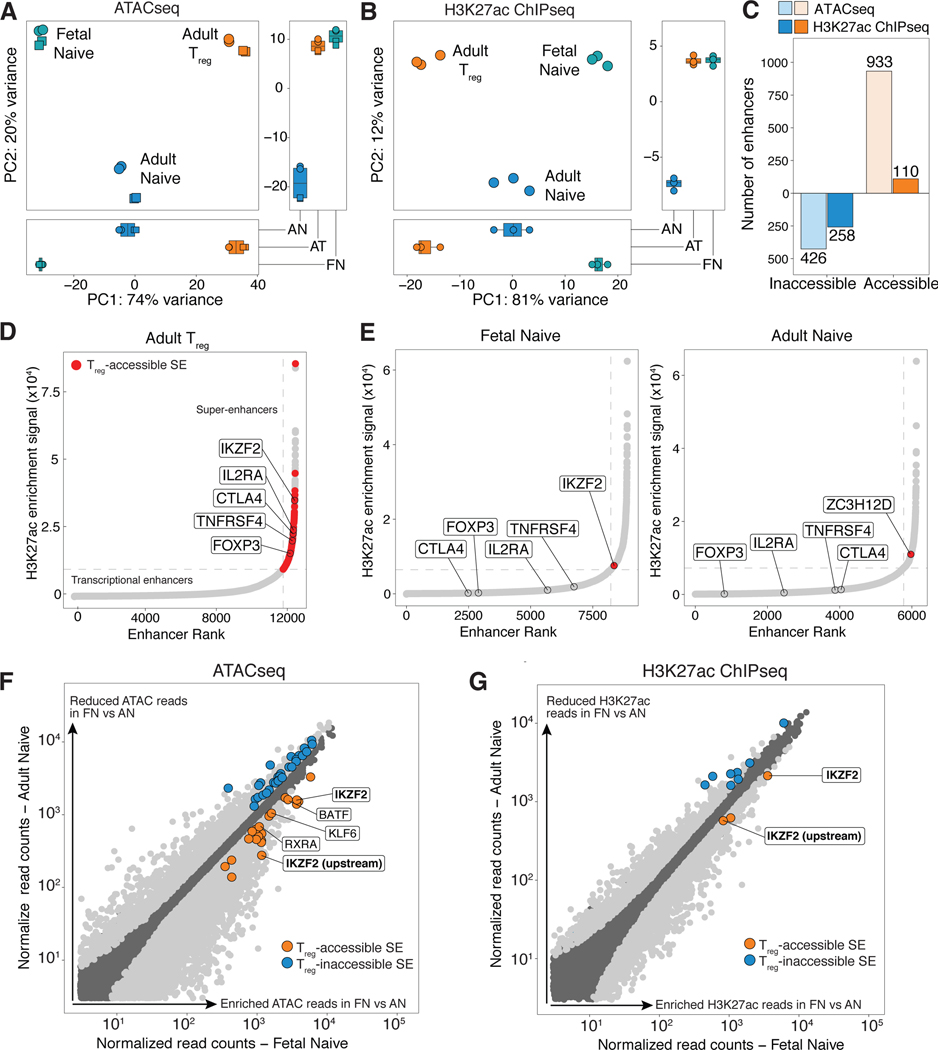
Given that fetal naïve T cells already express a partial Treg-specific signature, we next assessed if we could also detect the presence of permissive epigenetic marks in fetal naïve T cells that could further drive the predisposition towards more robust Treg cell differentiation. In order to identify these chromatin features, we used Assay for Transposase-Accessible Chromatin followed by sequencing (ATACseq) and H3K27ac chromatin immunoprecipitation sequencing (ChIPseq) to compare regions of open and active chromatin in adult Treg cells relative to fetal and adult naïve T cells. Super-enhancers (SEs) and typical transcriptional enhancers (TEs) were classified using the ROSE algorithm (57, 58), and PCA was performed across enhancers identified in all samples. Cell origin (fetal versus adult) was the primary source of variance (PC1) in both ATACseq (Fig. 3A) and H3K27ac ChIPseq (Fig. 3B), and together with cell phenotype (naïve or Treg), largely accounted for differences in the epigenome across all three populations. However, fetal naïve and adult Treg cell samples clustered together across the second source of variance (PC2) in both datasets (Fig. 3A,B). This suggested that, in addition to expression of a partial Treg-specific transcriptome, fetal naïve T cells share a small subset of open and active Treg-specific enhancers with adult Treg cells.
Figure 3. Fetal naïve T cells have increased ATAC and H3K27ac enrichment at two Treg-accessible superenhancers associated with Helios.
(A,B) Principal component analysis performed on all super- (SE) and transcriptional enhancers (TE) with mapped (A) ATAC or (B) H3K27ac reads. Boxplots show scores for PC1 (bottom) and PC2 (right).
(C) Barplots show number of Treg-accessible/inaccessible enhancers with increased (orange) or decreased (blue) ATAC and H3K27ac signal in fetal naïve T cells relative to adult naïve T cells (Treg-accessible/inaccessible enhancers defined in Supplementary Materials and Methods).
(D) Plots show cumulative H3K27ac signal at stitched enhancers within 12.5kb against enhancer rank, and SEs were defined where the tangent of the plotted curve is 1. Dotted lines (grey) show cutoff for SEs for one representative sample each for adult Treg (n = 3). SEs defined as Treg-accessible and meet fold change (FC) > 1.5 and false discovery rate (FDR) <0.05 cut-offs are colored in red, and SEs associated with key Treg genes are labeled.
(E) Plots as with (D) show one representative sample for fetal (left) and adult (right) naïve T cells (n = 3). Treg-accessible SEs identified in (D) was assessed for differential enrichment in fetal naïve against adult naïve T cells and vice versa. Differentially enriched Treg-SEs in each sample with FC>1.5 and FDR<0.05 are colored red and labeled. TEs associated with key Treg genes in (D) are also labeled.
(F,G) Scatterplots of normalized (F) ATACseq reads and (G) H3K27ac reads at all stitched enhancer regions of fetal naïve T against adult naïve T cells. Differentially enriched SEs and TEs with FC>1.5 and FDR<0.05 are shown in light grey, with Treg-accessible SEs (orange) and Treg-inaccessible SEs (blue). Treg-accessible SEs associated with transcription factors labeled; Helios (IKZF2) labeled in bold.
To address this possibility, we first independently defined enhancers differentially enriched for ATAC (fig. S5A) or H3K27ac (fig. S5B) signal in adult Treg cells relative to adult naïve T cells (FDR<0.05; expression fold change>1.5). Differentially enriched enhancers classified as having both increased H3K27ac and ATAC signals in adult Treg cells were termed Treg-accessible enhancers (fig. S5C, table S4), while common enhancers were defined as having no difference in enrichment of both signals (fig. S5C). Treg-inaccessible enhancers with decreased signals were similarly defined (table S5). We then assessed if any Treg-accessible enhancers were enriched in fetal naïve T cells relative to adult naïve T cells. We found that fetal naive T cells had increased ATAC and H3K27ac signal at 38.8% (933/2405) and 4.6% (110/2405) of Treg-accessible enhancers respectively (Fig. 3C, fig. S5D,E). These Treg-accessible enhancers (table S4) were annotated to genes previously described to be part of the Treg-specific epigenome, such as IKZF2 (i.e., Helios), IKZF4 (i.e., Eos) and RXRA (i.e., retinoic receptor RXR-alpha) (29, 30, 32). Similarly, 23.1% (426/1837) and 14% (258/1837) of Treg-inaccessible enhancers also had decreased ATAC and H3K27ac signal, respectively, in fetal naive T cells (Fig. 3C, fig. S5D,E). Taken together, these data suggest that fetal naïve T cells at steady state are poised for Treg differentiation by the acquisition of a partial Treg epigenomic landscape characterized by increased chromatin accessibility at more than a third of all Treg-accessible enhancers. Given that chromatin accessibility may precede H3K27ac deposition (59), acquisition of the Treg epigenetic signature within fetal naïve T cells could occur in a stepwise fashion where full enhancer activation via H3K27 acetylation is acquired with the triggering of Treg cell differentiation.
In light of this hypothesis, we evaluated transcription factor motif enrichment within all Treg-accessible peaks shared between fetal naïve and committed adult Treg cells (defined in a similar manner as TEs/SEs). Peak calls were utilized to minimize false positives stemming from the broadness of SE regions. Fetal naïve T cells had minimal enrichment of Treg-accessible H3K27ac peaks, but had increased chromatin accessibility at a third of all Treg-accessible ATAC peaks (fig. S6A). These shared Treg-accessible peaks were enriched in binding motifs for the AP-1 complex (fig. S6B) and RUNX1 (fig. S6C, table S6), which are downstream of TCR signaling and play critical roles as transcriptional regulators of the FOXP3 locus and as co-factors for FOXP3 (60). We also detected a smaller subset of peaks that had enrichment of binding motifs for STAT5 (fig. S6D) and SMAD2/3 (fig. S6E). As such, increased chromatin accessibility could potentially synergize with enhancer activation and faster transcription of genes underlying STAT5 and SMAD2/3 binding sites with IL-2 and TGF-β signaling during fetal Treg differentiation. Lastly, we examined differentially enriched Treg-accessible peaks in fetal naïve T cells for the presence of FOXP3 binding sites previously identified in human Treg cells(61). We show that only 5% (116/2213) of shared Treg-accessible peaks with increased ATACseq signal have FOXP3 binding sites (fig. S6F). Taken together, we further illustrate that increased chromatin accessibility within fetal naïve T cells is largely poised to synergize with TCR and cytokine signaling cues, and to a smaller extent, direct binding of FOXP3, to drive their preferential differentiation into Treg cells.
Fetal naïve T cells have increased open and active chromatin at two Treg-accessible SEs associated with Helios
SEs are defined by high density regions of H3K27ac modifications, and they nucleate the assembly of transcription factors to drive expression of genes associated with cell lineage commitment (57, 58, 62). We saw that highly ranked SEs shared across fetal naïve, adult naïve, and adult Treg samples corresponded to genes commonly associated with global T cell development and function such as BCL11B (63) and ETS1 (64) (fig. S7A, table S7). Because increased accessibility at Treg SEs in murine thymic Treg progenitors precedes FOXP3 upregulation and commitment to the Treg lineage (30), we asked if increased accessibility at Treg-accessible SEs in fetal naïve T cells could contribute to their priming towards Treg differentiation. We identified 121 SEs within all Treg-accessible enhancers, many of which were proximal to canonical Treg genes including FOXP3, IL2RA, CTLA4, TNFRSF4 (i.e., TNF receptor superfamily member 4) and IKZF2 (Fig. 3D) as previously described in mice (30). Globally, fetal naïve T cells did not have greater accessible chromatin or H3K27ac enrichment at all Treg-accessible SEs compared to adult naïve T cells (fig. S7B). This suggested that unlike thymic Treg progenitors, fetal naive T cells might acquire active enhancer marks at the full Treg-accessible SE signature only after Treg differentiation. We next wondered if any SEs independently classified by ROSE, and preferentially enriched within fetal naïve T cells, were proximal to genes associated with Treg accessible-SEs, since their presence would have been masked by the global analysis. Most genes associated with Treg accessible-SEs did not have enrichment of H3K27ac signal that met the SE cut-off in both fetal and adult naïve T cells (Fig. 3E). One exception was the transcription factor IKZF2 (i.e., Helios), which was unique to fetal naïve T cells (left, Fig. 3E), and previously identified to be one of the first Treg-SEs to acquire permissive epigenetic marks in murine thymic Treg progenitors (30). Adult naïve T cells had independent SE classification for one gene, ZC3H12D (right, Fig. 3E), which currently has no reported association with Treg cell function.
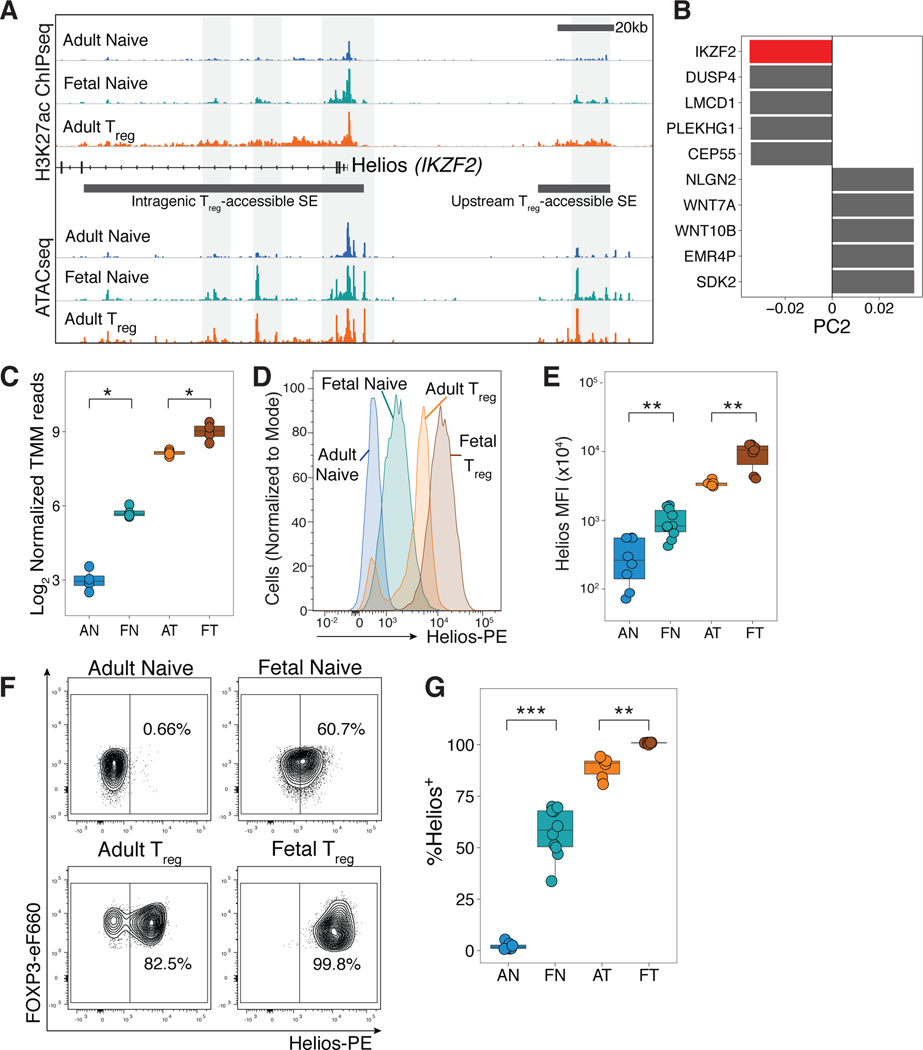
Cell-specific SE regions are typically found proximal to genes encoding transcription factors that play key roles in cell identity by controlling the transcription of lineage-specific transcriptional programs (30, 57). We next focused our analysis on evaluating if any Treg-accessible SEs defined within adult Treg cells had increased enrichment of either ATAC or H3K27ac signal in fetal naive T cells (Fig. 3C), and were also associated with known transcription factors. Five Treg-accessible SEs associated with four different transcription factors were identified to have increased ATAC signal (Fig. 3F), of which only two Treg-accessible SEs were also differentially enriched for H3K27ac (Figure 3G). These active, H3K27ac-marked Treg-accessible SEs were located in the intragenic and upstream regions within the Helios locus (Figure 4A), indicating that active expression of Helios might already be present in fetal naïve T cells. We further observed that Helios was a substantial contributor to the negative directionality of PC2 (Fig. 4B), which drove segregation of Treg cells away from naïve T cell populations in our RNAseq dataset (Fig. 1A). Helios was also among the significant Treg-upregulated genes with increased RNA transcription in fetal naïve T cells (Fig. 1C, Fig. 4C). Since enriched permissive epigenetic marks and transcription at the Helios gene locus regulate Treg phenotype and function independent of FOXP3 expression in mice (29, 33, 65, 66), we further investigated Helios as a candidate contributing to the program of fetal Treg differentiation.
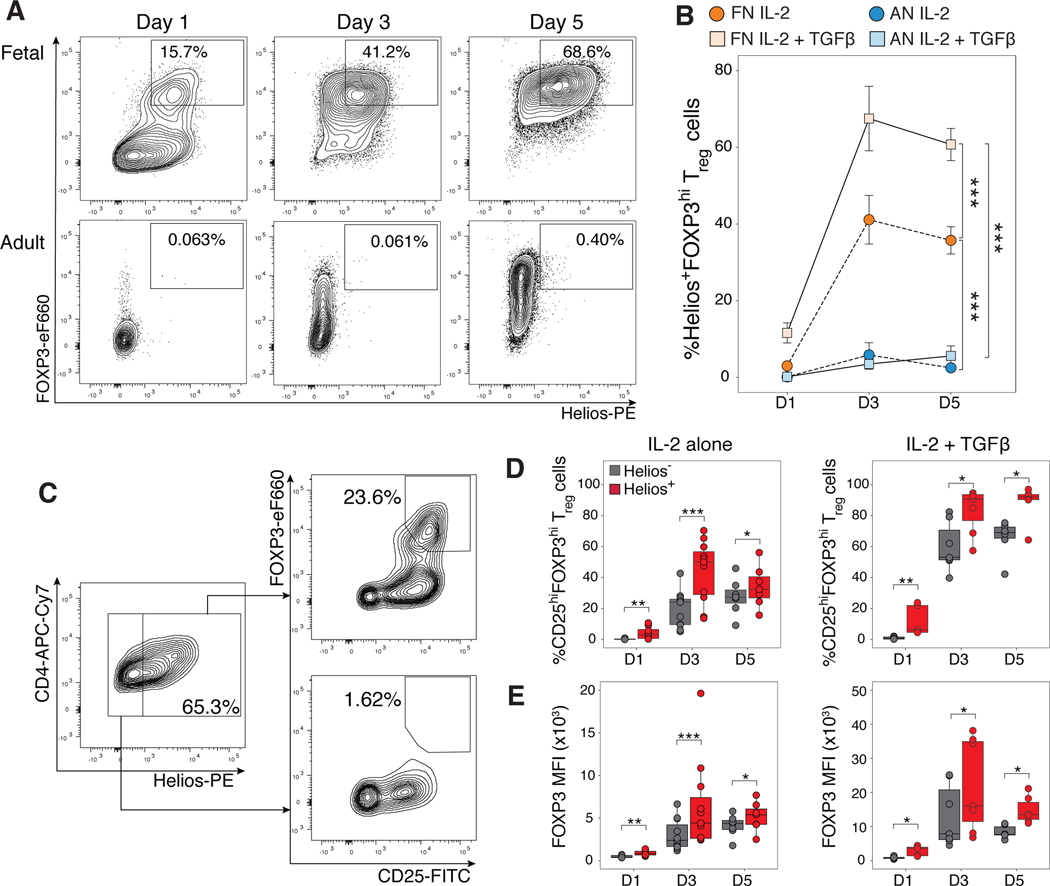
Figure 4. Helios expression is increased in fetal naïve T cells.
(A) Tracks show H3K27ac and ATAC signals at two Treg accessible super-enhancers (SEs) associated with the Helios (IKZF2) locus. Representative tracks of one replicate shown (H3K27ac ChIPseq, n=3, ATACseq, n=2).
(B) The top 5 genes contributing to PC2, which segregates cells by functional subtype (naïve versus Treg) are plotted for both directions; Helios (IKZF2) highlighted in red.
(C) Boxplot shows log2 trimmed mean of M values of normalized RNAseq reads for Helios in adult naïve (AN), fetal naïve (FN), adult Treg (AT) and fetal Treg (FT) cells (n=4).
(D) Helios staining intensity in sorted CD4+CD25-CD27+CD45RA+ adult (blue) and fetal naïve (green), and adult CD25hiCD127loFOXP3 (orange) and fetal Treg (brown).
(E) Boxplot shows quantification of mean fluorescence intensity of Helios for adult naïve (AN, n=8), fetal naïve (FN, n=10), adult Treg (AT, n=8), and fetal Treg (FT, n=10) cells.
(F) Flow cytometry analyses of sorted populations in (D). Helios+ and Helios- gates were set based on negative and positive populations in adult Treg samples (bottom left). Adult naïve T cells were universally Helios-.
(G) Quantification of Helios+ cells among sorted populations in (F).
All statistics were calculated by unpaired two-sided Mann-Whitney test. ***p<0.001, **p<0.01, *p<0.05. All boxplots show median (centre line), interquartile range (box) and tenth and ninetieth percentiles (whiskers)
Fetal naïve T cells have increased Helios protein expression at baseline.
Using flow cytometry staining, we show that fetal naïve T cells had higher Helios protein expression compared to adult naïve T cells (Fig. 4D,E). As previously described (40, 67), we identified Helios- and Helios+ FOXP3+ populations in adult Treg cells (Fig. 4F). Fetal Treg cells were all uniformly Helios+, which could indicate that retention of permissive epigenetic marks at Helios Treg-accessible SEs may drive high Helios expression (Fig. 4F). An average of 60% of fetal naïve T cells were Helios+, while adult naïve T cells did not express Helios (Fig. 4F&G). In comparison, we also examined the protein expression of two other Treg-specific genes with increased transcription in fetal naïve T cells with differentially enriched ATAC signal, (CCR4; fig. S8C) or H3K27ac signal (Eos; fig. S8D). Relative to FOXP3 expression, CCR4 and Eos expression did not demonstrate a similar shift in expression within fetal naïve T cells from adult naïve T cells when compared to Helios (fig. S8E), which led us to focus on investigating the role of Helios expression in fetal Treg cell differentiation.
The predisposition of fetal naïve T cells towards Treg differentiation is not explained by increased incidence of CD31+ cells in the naïve T cell population or increased proliferative ability.
Prior to further investigations into potential contributions of Helios to Treg differentiation, we sought to address potential confounding factors in our analysis. Previous studies have demonstrated that CD31+ population within the human naïve T cell population is enriched for recent thymic emigrants and have increased Treg differentiation potential (68), making them potential precursors of Treg cells in the periphery. We assessed if increased CD31+ cell frequency was a contributor to increased Treg differentiation in fetal naïve T cells, since the fraction of the CD31+ population is highest at birth and decline with age (69). Unexpectedly, CD31+ proportions were not different between adult and fetal naïve T cell populations (fig. S9A,B). Additionally, CD31+ naïve T cells isolated from human peripheral blood do not demonstrate increased differentiation in the absence of exogenous TGF-β (67). We thus concluded that the predisposition towards Treg differentiation that we observed within fetal naïve T cells was not attributed to differences in CD31+ proportions.
We further observed that mean CD31 expression levels were reduced within fetal CD31+ naïve T cells (fig. S9C). CD31 is downregulated with TCR signaling (69), and a subset of fetal CD4+ T cells are CD69+ and actively cycling (23). We therefore assessed the expression of CD69 and Ki67, a marker of active proliferation, relative to Helios expression in fetal naïve T cells. Neither fetal nor adult naïve T cells expressed CD69 (fig. S10A). However, as previously characterized (23), a subset of fetal naïve T cells are actively proliferating, while adult naïve T cells are mainly Ki67- (fig. S10B), thus possibly accounting for the reduced CD31 expression in fetal naïve T cells. The majority of the Ki67+ population in fetal naïve T cells was also Helios+ (fig. S10C), suggesting that Helios might regulate proliferation. However, with TCR stimulation, both adult and fetal naïve T cells upregulated Ki67 to a similar extent after 5 days (fig. S10D), indicating that Helios expression does not confer any selective proliferation advantage on fetal naïve T cells during Treg differentiation that may account for their increased Treg differentiation potential.
Fetal naïve T cells do not have increased demethylation at the FOXP3 Treg-specific demethylated region (TSDR)
As Helios was first identified as a marker of thymic Treg cells (40), we sought to rule out possible contamination of thymic Treg cells by assessing demethylation of the TSDR at the conserved non-coding sequence 2 (CNS2) region within the FOXP3 gene in our sorted naïve T cell populations (fig. S1). We saw that as expected, only fetal and adult Treg populations had complete TSDR demethylation, while both fetal and adult naïve T cells had a fully methylated TSDR (fig. S11A). This indicated that Helios expression within fetal naïve T cells was cell-intrinsic and not due to contamination with thymic Treg cells.
Fetal naïve T cells upregulate and maintain Helios expression during iTreg differentiation
Helios is expressed independently of FOXP3 expression (29, 33, 44) and can enhance the acquisition of a Treg-transcriptional signature with the co-expression of FOXP3 (34). As such, Helios expression in fetal naïve T cells might allow them to bypass the need for TGF-β to initiate FOXP3 upregulation and underlie their preferential differentiation into Treg cells. To assess this, we tracked Helios expression within fetal and adult naïve T cells during Treg differentiation with IL-2 alone or with TGF-β added at 1, 3, or 5 days. As previously observed, a higher frequency of fetal naïve T cells differentiated into CD25hiFOXP3hi iTreg cells relative to adult naïve T cells either in the presence or absence of exogenous TGF-β (24, 25) (fig. S12A,B). Fetal naïve T cells highly upregulated and maintained Helios protein expression during iTreg differentiation, whereas adult naïve T cells did not (fig. S12C,D). Concurrent upregulation of both FOXP3 and Helios was observed only in fetal iTreg cells differentiated in both stimulation conditions; even as FOXP3 expression increased with TGF-β stimulation, Helios expression was not upregulated in adult iTreg cells at any time point (Fig. 5A, B). Although Helios has been implicated as a marker of activation in proliferating cells (70), we show that both fetal and adult iTreg cells upregulated Ki67 to the same extent, but only fetal iTreg maintained upregulation of Helios (fig. S12E), thus excluding the probability that Helios upregulation in fetal iTreg cells resulted from increased cell proliferation. We also excluded the possibility of thymic Treg outgrowth leading to Helios expression within fetal iTreg cells. As previously shown (71–73), we did not detect any TSDR demethylation for adult TGF-β-iTreg populations (fig. S11A–C). Assessment of sorted fetal FOXP3-, FOXP3+Helios- and FOXP3+Helios+ IL-2- or TGF-β-iTreg populations (fig. S11D,E) did not reveal TSDR demethylation in any fetal iTreg populations (fig. S11A), thus suggesting that the increase in Helios expression happens de novo in fetal iTreg cells.
Figure 5. Helios+ fetal induced Treg cells have increased FOXP3 expression.
Sorted fetal and adult naïve T cells were stimulated with αCD3/αCD28/αCD2 tetramers and IL-2 in the absence or presence of TGF-β over 1, 3 or 5 days, and analyzed by flow cytometry. The number of biological replicates for each time point and stimulation condition are specified in table S8.
(A) Representative flow cytometry plots show Treg induction in the presence of IL-2 and TGF-β for fetal (top) and adult (bottom) naïve T cells respectively gated on live, CD4+ T cells.
(B) Quantification of percentage of Helios+FOXP3+ iTreg cells gated in (A) for adult (AN) and fetal naïve (FN) T cells stimulated in the presence or absence of TGFβ. Statistics calculated using 2-way ANOVA with Tukey’s honest significant difference post-test, ***p<0.001. Error bars denote mean ± s.d.
(C) Representative flow cytometry plots shown for one fetal sample stimulated with IL-2 and TGF-β at day 1. (D) Quantification of the proportion of fetal Treg cells in the Helios+ or Helios- population as gated in (C) over 1, 3 and 5 days in the absence (left) or presence (right) of TGF-β.
(E) Quantification of FOXP3 mean fluorescence intensity for Helios+ and Helios- iTreg cells as gated in (C) across all time points in the absence (left) or presence (right) of TGF-β. Statistics for (D,E) were calculated by two-sided Wilcoxon signed-rank test, ***p<0.001, **p<0.01, *p<0.05. All boxplots show median (center line), interquartile range (box) and tenth and ninetieth percentiles (whiskers)
The proportions of fetal IL-2-iTreg cells generated across time tracked closely with proportions of adult TGF-β-iTreg cells (fig. S12B), suggesting that Helios expression within fetal naïve T cells could enhance their preferential differentiation into Treg cells independently of exogenous TGF-β. We further examined fetal Helios+ and Helios- populations after Treg differentiation and found that the majority of fetal iTreg cells were within the Helios+ population 24 hours after the initiation of Treg induction (Fig. 5C). The increased frequency of iTreg cells present within the Helios+ over the Helios- population was maintained over time and in both stimulation conditions (Fig. 5D). Helios+ cells also consistently had higher FOXP3 expression (Fig. 5E) relative to Helios- cells, suggesting that Helios expression could potentially drive increased FOXP3 expression in differentiating fetal naive T cells.
Helios knockout in fetal naïve T cells impairs their ability to preferentially differentiate into Treg cells
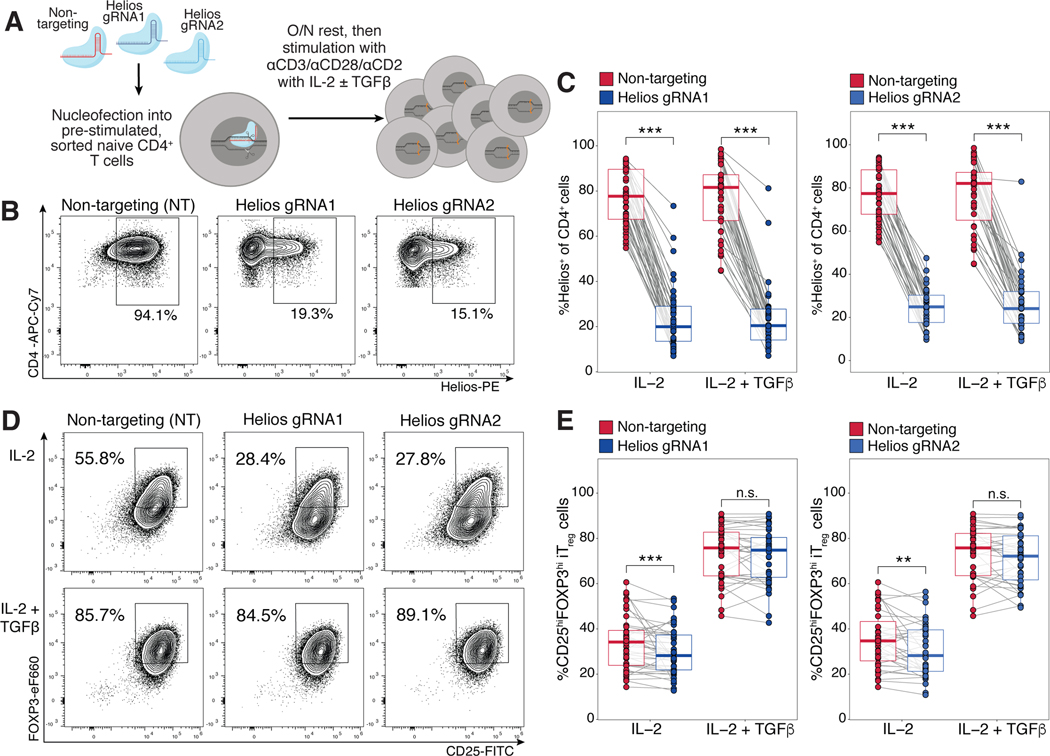
Given the hypothesized role of Helios in enhancing FOXP3 upregulation during fetal Treg cell differentiation, we predicted that reduced Helios expression in fetal naïve T cells would subsequently inhibit their cell-intrinsic propensity for Treg differentiation. Using CRISPR-Cas9 mediated editing, we knocked out Helios with two independent guide RNAs (gRNAs) targeting different exons of the gene. Fetal naïve T cells were then assessed for Treg induction post editing after differentiation in the presence or absence of exogenous TGF-β (Fig. 6A). We first confirmed that both gRNAs were able to successfully disrupt the Helios locus (fig. S13A–C), and observed specific reduction of Helios protein (Fig. 6B) in comparison to the non-targeting (NT) guide. Both gRNAs resulted in an average of 70% of fetal naïve T cells losing Helios expression (fig. S13D), and the reduction was maintained after 6 days of Treg induction in both stimulation conditions (Fig. 6B,C). Helios knockout in stimulated fetal naïve T cells reduced subsequent Treg differentiation in the absence of exogenous TGF-β compared to cells that received the NT guide (Fig. 6D,E), and the reduction in Treg percentage correlated with the extent of knockout generated (fig. S14A). Adult naïve T cells nucleofected with the same guides were used as Treg gating controls (fig. S13E). In contrast, Helios knockout had no effect on fetal iTreg differentiation with addition of exogenous TGF-β (Fig. 6D, E, fig. S14B). This indicated that signaling via TGF-β compensated for the loss of Helios-driven Treg differentiation, and that Helios and TGF-β may participate in shared signaling pathways. Our data show that Helios expression within fetal naive T cells plays a role in enhancing preferential Treg differentiation specifically in the absence of exogenous TGF-β. This mechanism present within fetal naïve T cells could lower the threshold required for Treg cell differentiation, thus potentially allowing for the default generation of peripheral Treg-mediated tolerance upon antigen encounter during fetal development.
Figure 6. CRISPR-Cas9 mediated knockout of Helios in fetal naïve T cells reduces their preferential differentiation into Treg cells.
A) Schematic showing experimental design. CRISPR-Cas9 editing at the Helios locus with two independent guide RNAs (gRNA1, gRNA2, see Supplementary Materials & Methods) was carried out in pre-stimulated fetal naïve T cells, with a non-targeting (NT) gRNA as a control. Edited cells were stimulated with αCD3/αCD28/αCD2 tetramers in the absence or presence of TGF-β. Treg induction was assessed at 6 days. The numbers of biological replicates for each guide and stimulation condition are specified in table S9.
(B) Flow cytometry plots showing Helios expression in fetal iTreg cells differentiated in IL-2 alone at day 6 after CRISPR-Cas9 editing with Helios gRNA1/2 or NT controls. Representative plots of one experiment are shown gated on live, CD4+ T cells.
(C) Quantification of Helios expression post-editing in fetal naïve T cells after Treg induction as in (B). Boxplots show paired samples for gRNA1 (left) and gRNA2 (right).
(D) Flow cytometry plots showing FOXP3 and CD25 staining in fetal iTreg cells at D6 post CRISPR-Cas9 editing with Helios gRNA1/2 or NT controls. Representative plots of one experiment are shown gated on live, CD4+ T cells.
(E) Quantification of induced Treg proportions in edited fetal naïve T cells as in (D). Boxplots show paired samples for gRNA1 (left) and gRNA2 (right). All statistics were calculated by two-sided Wilcoxon signed-rank test, ***p<0.001, **p<0.01, *p<0.05. All boxplots show median (center line), interquartile range (box) and tenth and ninetieth percentiles (whiskers).
Helios suppresses IL-2 secretion in fetal iTreg cells.
Helios maintains an anergic and non-proliferative state characteristic of the Treg phenotype (74) by mediating the epigenetic silencing of the IL2 locus in Treg cells (75). In contrast to conventional T cells, Treg cells have reduced IL-2 production upon TCR stimulation and depend heavily on paracrine IL-2 for their maintenance (76). Since Helios is highly expressed and maintained in fetal iTreg cells, we investigated whether this led to a corresponding suppression of IL-2 production. As hypothesized, fetal iTreg cells demonstrated less IL-2 production upon restimulation compared to adult iTreg cells (fig. S15A), and suppression of IL-2 was observed regardless of iTreg differentiation conditions (fig. S15B). When delineated on the basis of Helioshi and Helioslo expression (fig. S15C), Helioshi fetal iTreg cells consistently had lower IL-2 production across both stimulation conditions (fig. S15D), indicating that high Helios expression may be associated with greater repression of the IL2 locus. Helios knockout in fetal iTreg cells then resulted in increased IL-2 production in IL-2-iTreg cells (fig. S15E,F). Furthermore, Helios knockout TGF-β-iTreg cells also produced more IL-2 upon restimulation when compared to the NT control (fig. S15E,F). This demonstrates that continued Helios expression in fetal naïve T cells not only enhances preferential Treg differentiation, but also aids in the repression of IL-2 production in fetal iTreg cells.
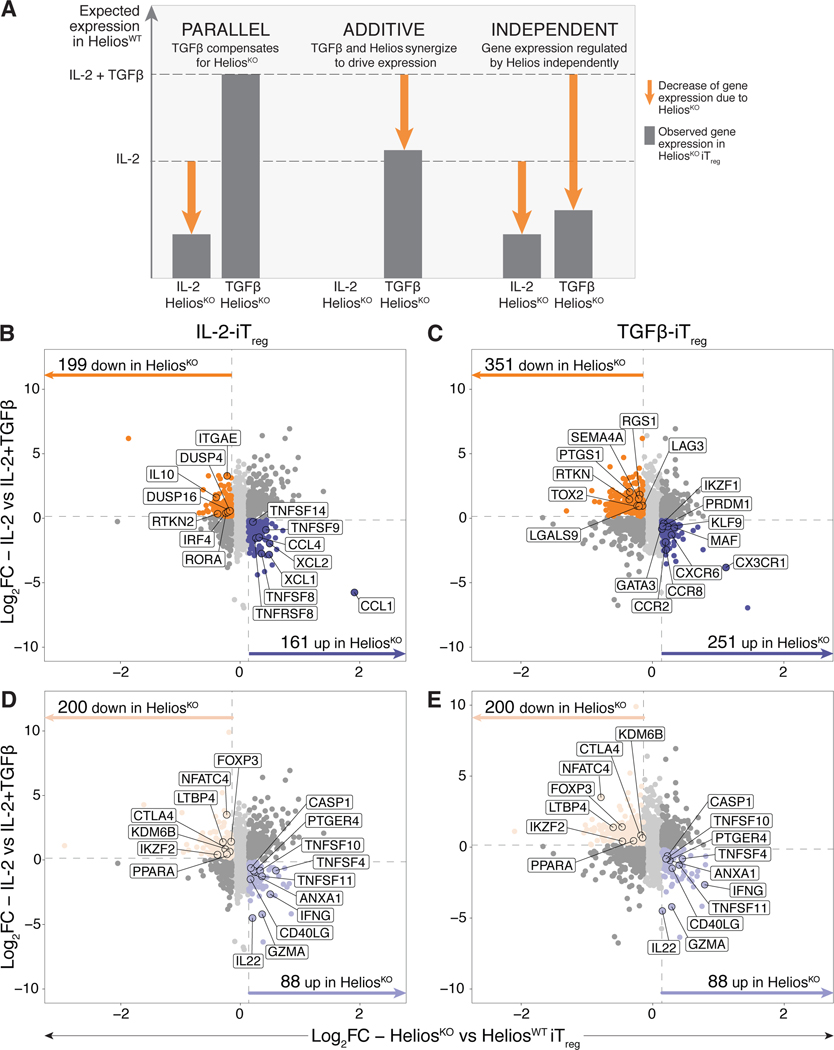
Helios knockout results in the downregulation of genes associated with Treg differentiation and function and the concurrent upregulation of pro-inflammatory genes
Helios controls the expression of several key genes involved in Treg suppressive function (77) including GARP. Helios knockout in fetal naïve T cells did not impact FOXP3 or CD25 expression (fig. S14C, D), but resulted in decreased CTLA-4 expression in fetal iTreg cells (fig. S14E). Helios knockout also resulted in a trend towards downregulation of GARP and LAP on fetal iTreg cells (fig. S14F,G). Since the impact of Helios knockout was variable across the conventional Treg markers surveyed, and fetal iTreg cells retain expression of a partial Treg-specific transcriptional signature, we wondered whether transcriptional control of other Treg genes by Helios could enhance the conversion of fetal naïve T cells into iTreg cells as well as influence their subsequent function. As such, we further assessed the impact of Helios ablation on the fetal iTreg transcriptome by RNA sequencing. CRISPR-Cas9 editing was carried out in fetal naïve T cells with Helios gRNA1 (HeliosKO) or the NT control (HeliosWT) before Treg differentiation was induced in the absence or presence of TGF-β (fig. S16A).
PCA revealed that HeliosKO and HeliosWT iTreg cells segregated largely according to whether differentiation occurred in the presence or absence of TGF-β (PC1, fig. S16B). This was not unexpected, since TGF-β signaling is responsible for the upregulation and repression of a significant subset of Treg-specific genes (Fig. 2B–D). We also detected a small but distinct segregation of HeliosKO from HeliosWT cells, with PC2 mainly segregating HeliosKO and HeliosWT IL-2-iTreg cells (fig. S15C), while PC3 mainly distinguished HeliosKO and HeliosWT TGF-β-iTreg cells (fig. S15D). As full knockout of Helios expression is not achieved within the total iTreg population with an average of 30% of all cells still retaining Helios expression (fig. S16A), we expected that this would result in a lowered signal-to-noise ratio. We thus utilized a more generous cutoff, where genes with at least a 10% change in expression (FC>1.1, FDR<0.05) were defined to be differentially expressed (fig. S15E,F).
Given that TGF-β signaling is able to compensate for the defect in Treg induction in HeliosKO iTreg cells (Fig. 6D,E), we decided to dissect possible pathways controlled by Helios and TGF-β signaling in parallel (Fig. 7A). We first identified differentially expressed genes within both stimulation conditions that would normally be upregulated with TGF-β signaling within HeliosWT iTreg cells but were downregulated in HeliosKO cells. This allowed us to detect genes whose expression was potentially enhanced by Helios in a complementary fashion – these genes would show decreased expression in HeliosKO IL-2-iTreg cells, but due to compensation with TGF-β signaling, would have no change in expression in HeliosKO TGF-β-iTreg cells compared to HeliosWT controls (Fig. 7B,C). Similar cutoffs were used to define genes potentially suppressed by Helios in parallel, which would be upregulated in HeliosKO IL-2-iTreg cells. We identified 199 downregulated and 161 upregulated genes within HeliosKO IL-2-iTreg cells that were not differentially expressed in TGF-β-iTreg cells (Fig. 7B). HeliosKO IL-2-iTreg cells had reduced expression of Treg-specific genes previously identified to be exclusively upregulated in fetal iTreg cells such as DUSP4, IL10 and ITGAE (CD103) (Fig. 7B, table S10). Concurrently, HeliosKO IL-2-iTreg cells upregulated several chemokine and tumor necrosis factor (TNF) superfamily genes (Fig. 7B, table S10), indicating that Helios may enhance the expression of a subset of Treg genes while simultaneously reducing expression of pro-inflammatory genes associated with effector function in the absence of TGF-β signaling.
Figure 7. Ablation of Helios in fetal iTreg results in downregulation of Treg-specific genes, and the concurrent upregulation of pro-inflammatory genes.
CRISPR-Cas9 editing was carried out in fetal naïve T cells (n=6) with Helios gRNA1 (HeliosKO) or the non-targeting control guide (HeliosWT). Edited cells stimulated with αCD3/αCD28/αCD2 tetramers in the absence or presence of TGF-β for 6 days, after which changes in their overall transcriptome was assessed by RNAseq.
(A) Schematic showing hypothesized expression levels (dotted lines) of a Treg-specific gene that is upregulated with IL-2 in the absence or presence of TGF-β signaling, with proposed changes in transcription level given one of the three proposed scenarios of transcriptional control by Helios and TGF-β. Gene expression that is driven by Helios is shown, with the arrows denoting the corresponding decrease in gene expression occurring with Helios knockout.
(B, C) Scatterplots show log 2 fold change (log2FC) values comparing the absence or presence of exogenous TGFβ during the differentiation process (y-axis) and HeliosKO against HeliosWT iTreg (x-axis) that underwent differentiation in IL-2 alone (B) or with exogenous TGF-β added (C). Dotted lines in grey denote log2FC cut-offs. Only genes upregulated (dark purple) or downregulated (dark orange) in HeliosKO relative to HeliosWT iTreg cells are colored and shown (fold change, FC>1.1, false discovery rate, FDR<0.05). Genes associated with Treg or pro-inflammatory immune functions are outlined and labeled.
(D, E) Same scatterplots as in (B,C) for IL-2 only iTreg cells (D) and IL-2 + TGF-β-iTreg (E) now colored to only show shared genes meeting the same cutoffs that were upregulated (light purple) or downregulated (light orange) in HeliosKO relative to HeliosWT iTreg cells. Genes associated with Treg or pro-inflammatory immune functions within the shared Helios-regulated transcriptome are outlined and labeled.
Additionally, synergy between Helios and TGF-β signaling might occur, thus amplifying the expression of Treg-specific genes in an additive manner (Fig.7A). We thus assessed genes that had decreased expression specifically in HeliosKO TGF-β-iTreg cells (Fig. 7C). Loss of Helios expression resulted in the downregulation of 351 genes in TGF-β-iTreg cells (Fig. 7C), including SEMA4, PTGS1 and RTKN, previously identified in the TGF-β-iTreg gene signature (Cluster 2.2, Fig. 2B), as well as TOX2 and RGS1, which are upregulated in fetal but not adult iTreg cells (Cluster 2.3, Fig. 2B, table S10). Conversely, HeliosKO TGF-β-iTreg cells had upregulation of 251 genes; these comprised chemokine receptor genes, as well as transcription factors involved with Th1, Th2 and Th17 cell differentiation and function such as PRDM1 (Blimp1)(78), GATA3 (79), IKZF1 (Ikaros) (80, 81), and MAF (c-MAF) (82)(Fig. 7C). These data suggest that Helios performs both parallel and additive roles in enhancing the upregulation of genes associated with the Treg transcriptional signature and repressing genes that might drive differentiation towards other effector T cell pathways during Treg differentiation.
Lastly, we identified genes that were either upregulated or downregulated in HeliosKO iTreg cells across both induction conditions, implicating possible transcriptional control by Helios independent of TGF-β signaling during Treg differentiation. 200 genes were downregulated in HeliosKO iTreg cells, including genes related to Treg phenotype and function such as CTLA4 and LTBP4 (Fig. 7D,E). Although we did not observe reduced protein expression of FOXP3 in HeliosKO iTreg cells at Day 6 of differentiation (Fig. S14C), we observed decreased FOXP3 transcription across both iTreg populations, suggesting that additional post-transcriptional mechanisms probably regulate FOXP3 expression downstream of Helios. HeliosKO iTreg cells also had reduced expression of NFATC4 (NFAT3) and PPARA (PPARα) transcription factors which regulate the repression of pro-inflammatory cytokines such as IL-2, IFN-γ and TNF-α (83, 84), as well as the histone H3K27 demethylase KDM6B (JMJD3), which suppresses Th2 and Th17 programs (85) (Fig. 7D,E). Loss of Helios expression also led to the upregulation of 88 genes, including genes attributed to pro-inflammatory effector T cell function such as IFNG (Fig. 7D,E). Taken together, we propose that Helios could potentially play a role in enhancing fetal Treg differentiation through the transcriptional regulation of a key subset of genes that restrict differentiation towards effector T cell helper phenotypes while favoring differentiation towards the Treg cell fate.
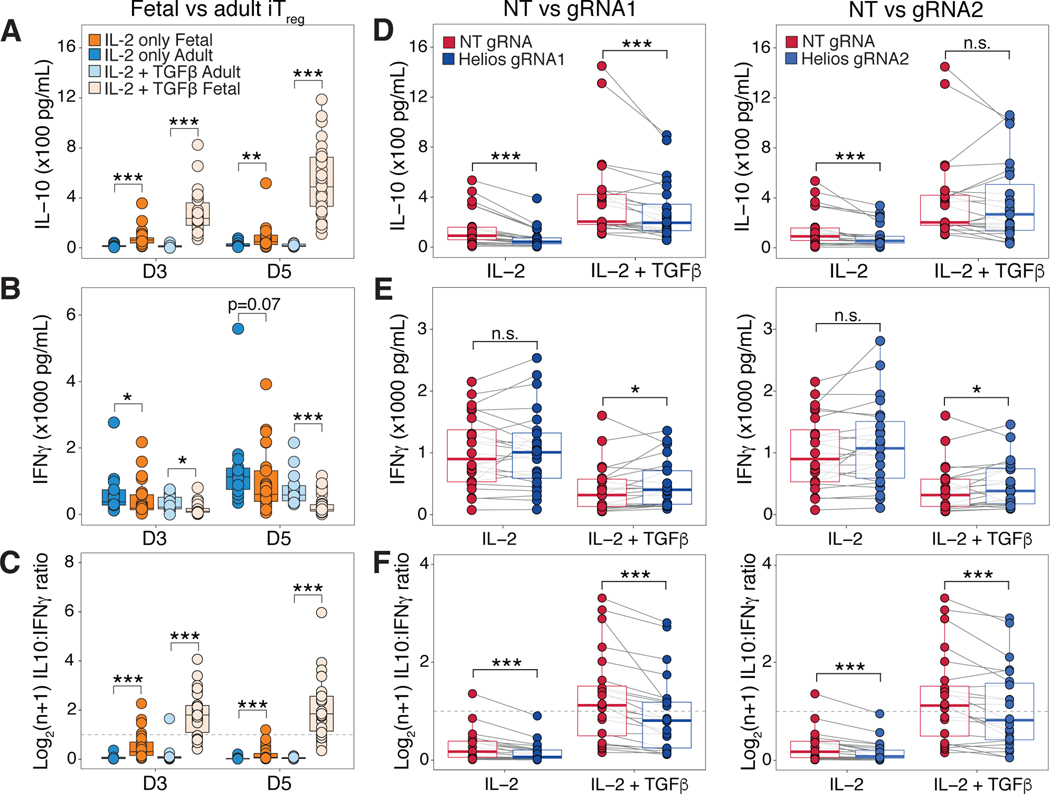
Helios knockout fetal iTreg cells have decreased IL-10 and increased IFN-γ production
The regulation of cytokine production in Treg cells is important for their suppressive ability; Treg cells must repress secretion of pro-inflammatory cytokines such as IFN-γ while maintaining production of immunosuppressive cytokines such as IL-10 (86). Given that we detected decreased IL-10 expression in HeliosKO IL-2-iTreg cells (Fig. 7B), with a corresponding increase in IFN-γ transcription in both HeliosKO iTreg populations (Fig. 7D,E), we next validated these observations by assessing IL-10 and IFN-γ in supernatant during iTreg differentiation. We first confirmed that only fetal iTreg cells produced IL-10 during iTreg differentiation in the absence of TGF-β (Figure. 8A), as observed in our transcriptomic analysis of fetal and adult iTreg cells (Fig. 2B). This is further augmented by exogenous TGF-β (Fig. 8A), which may be due to an increased frequency of Treg cell differentiation (Fig. S5B). Fetal iTreg cells also produced less IFN-γ than adult iTreg generated in both stimulation conditions (Fig. 8B). Lastly, both fetal iTreg cell populations had a greater ratio of IL-10 produced over IFN-γ compared to their adult counterparts, and this effect was enhanced in TGF-β-iTreg cells (Figure 8C). Ablation of Helios with either gRNA1 or 2 then resulted in a reduction of IL-10 produced in HeliosKO IL-2-iTreg cells (Fig. 8D). We observed a small but sustained decrease in IL-10 across HeliosKO TGF-β-iTreg cells that received gRNA1 but did not detect this decrease in gRNA2 (Fig. 8D), which reflected our RNA sequencing results showing a minimal decrease in IL-10 in gRNA1 treated TGF-β-iTreg that did not meet fold change cutoffs (fig. S16G). This is consistent with our prediction that TGF-β signaling can compensate for Helios knockout to upregulate IL10 transcription within HeliosKO TGF-β-iTreg cells (Figure 7A). Helios knockout resulted in increased IFN-γ production in HeliosKO TGF-β-iTreg cells, but the effect was less obvious in IL-2-iTreg cells (Fig. 8E). However, across all differentiation conditions, HeliosKO iTreg cells had a decrease in their IL-10 to IFN-γ ratio (Fig. 8F), especially in TGF-β-iTreg cells. Overall, this suggests an potential role for Helios in regulating the balance of cytokine output from fetal iTreg cells, which could help prevent pro-inflammatory responses that can be detrimental in utero.
Figure 8. Fetal Helios knockout iTreg cells have decreased IL-10 and increased IFN-γ cytokine production.
A) Boxplots quantify IL-10 cytokine concentration within culture supernatants collected at day 3 and 5 of differentiation for adult (blue, n=27) or fetal iTreg cells (orange, n=58) differentiated in IL-2 alone (IL-2 iTreg), as well as adult (light blue, n=27) or fetal (light orange, n=55) iTreg cells differentiated with added TGF-β (TGF-β-iTreg).
(B) Boxplots quantify IFN-γ cytokine concentrations for samples as in (A).
(C) Boxplots show log2(n+1) ratio of IL-10 to IFN-γ cytokine concentration for samples as in (A). Dotted line marks the ratio at which cells produce IL-10 and IFN-γ at 1:1. All statistics for (A,B,C) calculated by unpaired two-sided Mann-Whitney test,*** p<0.001, ** p<0.01, *p<0.05.
(D) Boxplots quantify IL-10 cytokine concentrations at day 5 of differentiation for iTreg cells that received the non-targeting guide (HeliosWT, red) or for Helios knockout (HeliosKO, blue) with gRNA1 (left, n=24) and gRNA2 (right, n=23).
(E) Boxplots quantify IFN-γ cytokine concentrations for samples in (D).
(F) Boxplots show log2(n+1) ratio of IL-10 to IFN-γ cytokine concentration for samples as in (D). Dotted line marks the ratio where cells produce IL-10 and IFN-γ at 1:1. All statistics for (D,E,F) calculated by two-sided Wilcoxon signed-rank test, *** p<0.001, * p<0.05, n.s. p>0.05. All boxplots show median (center line), interquartile range (box) and tenth and ninetieth percentiles (whiskers).
Discussion
The predisposition of human fetal naïve T cells towards Treg cell differentiation presents an opportunity to identify potential underlying cell-intrinsic factors that not only enhance our understanding of Treg differentiation and function, but also may ultimately be manipulated to improve in vitro iTreg cell generation for cell-based immunotherapies. We show here that fetal naïve T cells possess a partial Treg-specific transcriptome and epigenome, and that only fetal-derived iTreg cells retain expression of this transcriptome upon differentiation in vitro. Fetal naïve T cells had increased chromatin accessibility at approximately a third of all defined Treg-accessible enhancers, but only a small percentage were marked by H3K27ac, including two SEs associated with Helios. This suggests that many Treg-specific enhancers are held in a poised (i.e., accessible), but not active, state in quiescent fetal naïve T cells. As such, the full acquisition of the Treg-specific epigenetic, and subsequently, transcriptional signature might only occur upon TCR activation and/or cytokine signaling which triggers the final commitment to the Treg cell fate. These data thus implicate a broad landscape of Treg-poised chromatin in fetal naïve T cells that contribute to their propensity for Treg differentiation. It is likely that there is also a contribution of additional histone marks to the overall chromatin landscape. For example, since Helios interacts with other histone-modifying proteins such as the NuRD co-repressor complex (87, 88), sequencing of additional histone marks such as the repressive H3K27me3 mark would allow us to assess possible repression of genes associated with other effector lineages. Future experiments that identify Helios binding sites by Helios ChIPseq within fetal naïve or iTreg populations will be critical to determine if Helios is required for the acquisition and/or maintenance of active or suppressive epigenetic marks at key Treg-specific genes. These further analyses would reveal a more complete understanding of the contribution of Helios to the fetal epigenome and how it contributes to the overall Treg cell differentiation phenotype.
Our group and others have shown that the human fetal immune system is skewed towards tolerance by the presence of a large Treg cell population (24, 25) which is likely derived from preferential conversion of fetal naive T cells into Treg cells upon antigen encounter (26). Here, we show that Helios enhances the preferential differentiation and phenotypic commitment of fetal naïve T cells towards the Treg cell fate. Ablation of Helios expression in fetal naïve T cells impaired their cell-intrinsic predisposition for iTreg differentiation, and reduction in Treg proportions were correlated with the extent of Helios knockout. Although the addition of exogenous TGF-β compensates for loss of Helios during Treg generation, the resulting transcriptional landscape in HeliosKO fetal TGF-β-iTreg cells was still impacted. Our results suggest that Helios-mediated transcriptional regulation may potentially play a dual role in fetal Treg differentiation – first, by enhancing Treg differentiation through upregulation of a set of Treg-specific genes even in the absence of TGF-β, and second, by repressing pro-inflammatory genes and genes that mediate differentiation towards other effector T cell subsets. This is consistent with the established role of Helios in restraining effector Th1 and Th17 cell programs within committed Treg cells in mice (41, 42). While global transcriptional differences observed between HeliosKO and HeliosWT iTreg cells were modest, we propose that the observed enhancement of fetal iTreg differentiation is the result of cumulative effects of upregulation or downregulation of genes transcriptionally controlled by Helios. Validation of other gene candidates identified here would further clarify the extent of Helios transcriptional control on the human fetal iTreg differentiation. Lastly, we demonstrate that Helios expression in fetal iTreg cells has functional consequences, as the loss of Helios increased the ratio of pro-inflammatory (IFN-γ) over anti-inflammatory (IL-10) cytokines by these cells. This is consistent with previous observations that Helios deficiency in mouse Treg cells in vivo does not result in overt distortions in FOXP3 expression or Treg proportions at steady state (89), but rather in the loss of immunosuppressive ability and the manifestation of autoimmunity in later life or in response to inflammatory insults (41, 42). Regulation of cytokine and other pro-inflammatory genes by Helios could then potentially play a role in maintaining the functional stability of fetal iTreg cells in utero to prevent potentially harmful pro-inflammatory responses.
Given the nature of this study, and the exclusive use of primary human cells, there are important limitations to acknowledge in its interpretation. We primarily compared T cells isolated from fetal spleen and adult peripheral blood for our studies, due to practical difficulties in obtaining adult splenic tissue and fetal peripheral blood. While we cannot fully rule out immunological differences associated with tissue residence, we have taken steps to reduce confounders resulting from this factor by deriving the epigenetic Treg signature from comparisons of adult naïve and adult Treg cells, thus ensuring that only the Treg signature was the main point of comparison between adult and fetal naive T cells. We were able to additionally rule out contributions from age specific differences by selecting genes shared between adult and fetal Treg cells in our RNAseq analysis to ensure that only genes truly contributing to the Treg cell phenotype were included. Gestational age, and on some occasions when it could be determined, sex, were the only demographics available for fetal cells. Hence, we were unable to fully rule out any other potential confounders or stratify our samples accordingly. Furthermore, technical variation due to the kinetics of CRISPR-Cas9 editing, together with the inherent biological variability in primary human samples manifested in the high sample variability that we observed within our editing experiments. We also observed heterogeneity in Helios expression at steady state in fetal naïve T cells, as well as the extent of Helios upregulation from sample to sample, which could contribute to differences in sensitivity to Helios disruption and subsequent Treg differentiation. While we focused here on Helios, additional transcription factors unregulated in fetal naive T cells, such as Eos (IKZF4), could also play complementary or compensatory roles in fetal iTreg differentiation. We envision that future work will aim to utilize single cell RNA or ATAC sequencing techniques to separate and identify fetal naïve T cell populations that will explain the observed biological heterogeneity. This will allow us to potentially identify subsets that have greater predisposition towards Treg differentiation, and further characterized the underlying factors leading to this phenotype.
Our work here suggests a key role for Helios in establishing early life peripheral tolerance in humans, and there are some indications to support this hypothesis from studies in neonatal mice. Notably, when Helios−/− mice were first generated, the authors reported the presence of significant fatality in the first two weeks of neonatal life, and a 100% fatality was observed with subsequent crosses to achieve a full B6 background (89). This timing coincides with the emergence and migration of functional T cells into the secondary lymphoid organs within the neonatal mouse, and is developmentally equivalent to when T cells emerge during the second trimester in humans (21). Furthermore in mice, thymic Treg cell populations generated in an early window from birth to 10 days later are qualitatively different from those generated later in life (90), indicating that Treg cell populations generated in early life are indispensable to the maintenance of lifelong tolerance. However, these studies did not examine the potential contributions of peripheral Treg differentiation from naïve T cells, which is likely an important factor contributing to the dominant tendency toward tolerance observed in the human fetus. This is particularly significant since there is an increased frequency of rapidly proliferating CD4+ and CD8+CD25- human fetal T cell populations (23) potentially bearing autoreactive TCRs that have escaped thymic deletion as observed in neonatal mice (91). In light of our data presented here, we thus speculate that Helios plays a previously unappreciated role in the generation of peripheral Treg cells in this critical period in early human fetal development where the need for peripheral tolerance is perhaps the most acute.
In vitro human Treg cell differentiation from naïve T cells for therapeutic purposes has encountered significant roadblocks due to difficulties in generating pure populations of Treg cells that maintain a stable phenotype over time (71–73). Previous studies have show that Helios+, but not Helios- ex vivo Treg cells retain a more highly demethylated TSDR when expanded in vitro (92, 93). We did not observe TSDR demethylation in Helios+ fetal iTreg cells, likely reflecting the inability of in vitro differentiation to capture the conditions or environmental factors that trigger TSDR demethylation. Regardless, we demonstrate that fetal iTreg cells have greater phenotypic resemblance to ex vivo Treg cells in that they retain expression of Helios, have diminished IL-2 production upon restimulation, and produce IL-10 upon Treg differentiation. These attributes are commonly associated with the Treg cell phenotype, but are not acquired in adult human iTreg cell populations. Lastly, our data shows that Helios enhances the expression of genes in parallel with, in addition to, and independently of TGF-β signaling, which favor Treg over helper T effector commitment. This may represent a mechanism by which Helios maintains stable Treg function and identity in vivo (41, 42, 67, 75, 77). Additional studies are thus required to assess this possibility, particularly in pro-inflammatory environments, and whether manipulation of Helios expression within adult naïve T cells can recapture this effect. Further identification of upstream factors in addition to Helios that contribute to the acquisition of the permissive enhancer landscape in fetal naïve T cells will likely provide important deeper insight into their predisposition toward Treg differentiation. These findings may then ultimately inform strategies for the generation of stable iTreg cells for use in for immunotherapy to establish tolerance in autoimmunity and transplantation.
Materials and Methods
Study design
The objective of this study was to determine the molecular mechanisms that underlie preferential Treg cell differentiation in human fetal naïve T cells. As such, primary human CD4+ naïve T and Treg cells from fetal spleen and adult peripheral blood mononuclear cells (PBMCs) were the primary cell sources used for this study. Transcriptional and epigenetic profiling was carried out utilizing RNAseq, H3K27ac ChIPseq and ATACseq. Flow cytometry was utilized to confirm observations from sequencing datasets, to validate expression levels at baseline, and to assess the activation and subsequent differentiation of fetal and adult naïve T cells in vitro. CRISPR-Cas9 mediated knockout was used to confirm the results of the observational studies. The sample size (n = 2–6 per experiment) for the sequencing datasets was determined to be the optimal size for statistical analysis and to allow for independent replicates, given the scarcity of cells isolated from each fetal sample. The sample size and experimental replicates for in vitro experiments are subsequently indicated in all accompanying figure legends or supplementary tables. Sample size was determined to be adequate based on the magnitude and consistency of measurable differences between groups. Investigators were not blinded, and samples were equally divided between treatments, i.e. gRNA received. Further details on dataset analysis and experimental technique are detailed in Supplementary Materials and Methods.
Magnetic isolation of CD4+ T cells and fluorescence activated cell sorting (FACS) for sequencing and cell culture
Prior to FACS, fetal splenocytes and adult PBMCs were pre-enriched for CD4+ T cells using the EasySep Human CD4+ T cell isolation kit (#17952, STEMCELL Technologies). In order to obtain sufficient numbers of adult regulatory T (Treg) cells for epigenetic analyses, adult PBMCs were also pre-enriched for CD4+CD127lo T cells using the Human CD4+CD127lo T cell Pre-enrichment kit (#19231, STEMCELL Technologies). Enriched cell fractions were incubated in FACS staining buffer (phosphate-buffered saline, PBS, with 2% HI-FBS and 2mM EDTA) with fluorochrome-conjugated anti-human surface monoclonal antibodies (mAbs): CD25 FITC (2A3, BD Biosciences), CD127 PE/BV421 (hIL-7R-M21, BD Pharmingen), CD45RA PE-CF594 (HI100, BD Horizon), CD4 PE-Cy7 (SK3, BD Pharmingen), CD27 eFluor780 (O323, ThermoFisher Scientific). All cells were stained with the live-dead marker (GhostDye Violet 510; Tonbo Biosciences) to exclude dead cells. Cells were sorted into supplemented RPMI (10% HI-FBS, 300 mg/L L-glutamine, 10 U/mL penicillin, and 10 μg/mL streptomycin) based on gating in Fig. S1. All sorts were carried out on a BD FACS Aria III.
RNAseq for ex vivo sorted cells
RNAseq was carried out for 2.5×105 cells for 4 biological replicates of adult and fetal naïve and Treg cells. RNA was extracted and purified with the Nucleospin RNA kit (Machery-Nagel) and assessed for quality by RNA Pico (Agilent Technologies). Library preparation and sequencing on a HiSeq300 were carried out by the Technology Center for Genomics and Bioinformatics (TCGB) core at University of California, Los Angeles.
RNAseq for adult and fetal induced Treg populations
4 biological replicates of sorted adult or fetal naïve T cells were stimulated in U-bottomed 96 wells with 5 μl of Immunocult Human T cell Activator (CD3/CD28/CD2 tetramers, STEMCELL Technologies) in 200 μl of culture media (RPMI-1640 supplemented with 10% HI-FBS (heat inactivated fetal bovine serum), 300 mg/L L-glutamine, 10 U/mL penicillin, and 10 μg/mL streptomycin, 10 mM HEPES, 1X MEM-NEAA, β-mercaptoethanol). Exogenous IL-2 (10 ng/ml, Peprotech) and TGF-β (50 ng/ml, Peprotech) were added according to the relevant experimental setup. 100 μl media changes were performed every two days starting from the first day of stimulation. At day 6 post-induction, RNA from 2.5×104 iTreg cells was extracted and purified with the Nucleospin XS RNA kit (Machery-Nagel) and assessed for quality by RNA Pico (Agilent Technologies). Ribosomal RNA was removed using the NEBNext® Poly(A) mRNA Magnetic Isolation Module (New England Biolabs), and library preparation was carried out using the NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina® (New England Biolabs). Sequencing on a HiSeq4000 was carried out by the Center for Advanced Technology core at UCSF.
Flow cytometry staining in sorted naïve and Treg cells
Sorted naïve and Treg cells from fetal and adult samples were incubated in FACS buffer with fluorochome-conjugated, anti-human surface mAbs. Fixation and permeabilization was performed using the Foxp3/Transcription Factor Staining Buffer set (Tonbo Biosciences). All cells were stained with a live/dead marker (GhostDye Violet 510; Tonbo Biosciences) to exclude dead cells from analysis. All data were acquired with an LSRII flow cytometer (BD Biosciences) and analyzed with FlowJo (BD Biosciences) software. Helios+ and Helios- gates were set based on the clearly positive and negative populations seen in adult Treg samples; all adult naïve T cell samples were observed to be fully within the Helios- population, and were subsequently used to define the Helios- gate for subsequent experiments as a biological negative control.
Treg induction assays
Sorted adult or fetal naïve T cells were differentiated into Treg cells as detailed above according to the relevant experimental setup. For Treg induction with TGF-β blockade, 2.5 μg/ml or 0.5 μg/ml of neutralizing anti-TGF-β (1D11, R&D Systems) antibody was added to the culture medium where specified. Cells were cultured and harvested after 1, 3 and 5 days for analysis by flow cytometry. 100 μl media changes were performed every two days starting from the first day of stimulation. Cells were stained, fixed and permeabilized as detailed in flow cytometry staining. All data were acquired with an LSRII flow cytometer (BD Biosciences) and analyzed with FlowJo (BD Biosciences) software.
CRISPR-Cas9 editing of the Helios locus
CRISPR-Cas9 editing was carried out as previously detailed (94, 95). Cas9 gRNA ribonucleoproteins (RNPs) were assembled before experiments by assembling the gRNAs through incubating 160 μM of crRNA with 160 μM of tracrRNA (Dharmacon) at a 1:1 ratio for 30 minutes at 37oC to a final concentration of 80 μM. Assembled gRNAs are then subsequently incubated with 40 μM of Cas9-NLS (QB3 MacroLab, UC Berkeley) at a 1:1 ratio for 15 minutes at 37oC for a final concentration of 20 μM RNPs. Nucleofection was performed using the Amaxa P3 Primary Cell 96-well Nucleofector kit and 4D-Nucleofector (Lonza). 1×105 sorted fetal naïve T cells per well were stimulated overnight in 96-well plates pre-coated with anti-CD3 antibodies (1 μg/mL, HIT3a, BD Biosciences), and supplemented with soluble anti-CD28 antibodies (2 μg/mL, CD28.2, BD Biosciences) and IL-2 (10 ng/ml, Peprotech). Stimulated fetal naïve T cells were washed with PBS, and resuspended in 20 μl of P3 solution. 5 μl of the final 20 μM RNP solution was added, along with 1 μl of 100 μM HDRT solution. Cells were gently mixed, and transferred to the 96-well shuttle device. Cells were electroporated using program EH-115 on the Amaxa 4D-Nucleofector (Lonza). 80 μl of prewarmed culture media was added immediately post-nucleofection, and cells were allowed to recover for 15 minutes at 37oC. Nucleofected cells were then transferred into a 96-well U-bottom plate, and additional culture media added to a final volume of 200 μl. After 5 hours, cells were spun down and 150 μl of the culture media changed to increase cell viability and left to incubate overnight. Cells were stimulated the next day for Treg induction assays as detailed above, and harvested at Day 6 for analysis.
RNAseq for HeliosKO and HeliosWT Treg populations
CRISPR-Cas9 mediated Helios knockout in 6 biological replicates of fetal naïve T cells was performed as described above using Helios gRNA1 or the NT control guide for (all samples paired across nucleofection conditions). Cells were stimulated for Treg induction assays as detailed above, and 2.5×104 iTreg cells harvested at Day 6 for RNA extraction and analysis. RNA extraction, library preparation and sequencing were carried out as for fetal and adult iTreg populations.
Cytokine bead assays
100ul of culture supernatant were collected at D3 and D5 of Treg differentiation with IL-2 only or in the presence of added TGF-β. Supernatants collected at D5 of Treg differentiation after CRISPR-Cas9 mediated Helios knockout were also analyzed. Supernatants were harvested and stored at −80oC prior to analysis. IL-10 (#558274) and IFN-γ (#558269) concentrations were measured using the Cytokine Bead Assay (CBA) Flex kits (BD Biosciences).
Statistical analyses
All statistical analyses were performed using R (cran.r-project.org, v3.5.2) and Bioconductor (www.bioconductor.org). Tests were specified with each experiment in each figure legend, and p<0.05 level of confidence was accepted for statistical significance. All statistical tests are non-parametric and two-sided unless mentioned. Paired testing was used when comparisons were made within the same sample in order to increase resistance towards outlier effects, and are specified in the figure legends. Kruskal-Wallis test and Dunn’s post-test with Bonferroni correction were performed using the PMCMRplus (v1.4.1) package. Graphs were made using the ggplot2 (v3.1.0), ggrepel (v0.8.0) and cowplot (v0.9.4) R packages. Heatmaps were generated with the ComplexHeatmap (v1.12.0) R package.
Supplementary Material
Table S7. Top 25 ranked super-enhancers in adult naive, fetal naive and adult Treg cells (Excel spreadsheet).
Table S6. De novo motif enrichment in Treg -accessible ATAC peaks shared between fetal naive and adult Treg cells (Excel spreadsheet).
Table S10. Upregulated and downregulated differentially expressed genes defined in Helios KO versus Helios WT fetal induced Treg cells (Excel spreadsheet).
Table S11. Raw data file (Excel spreadsheet).
Table S2. Genes upregulated/downregulated within iTreg populations with cluster assignment (Excel spreadsheet).
Table S3. Statistics from pre-ranked gene set enrichment analysis for fetal versus adult iTreg populations (Excel spreadsheet).
Figure S1. Gating strategy and purity assessment for sorted naïve and Treg cells.
Figure S2. Definition of the Treg transcriptional signature.
Figure S3. Assessing the enrichment of Treg upregulated or downregulated genes in fetal and adult induced Treg (iTreg) populations.
Figure S4. Fetal induced Treg cells have increased sensitivity to TGF-β signaling.
Figure S5. Identification of Treg-accessible and inaccessible enhancers.
Figure S6. Binding motifs for downstream effectors of Treg differentiation are enriched within shared Treg-accessible peaks in fetal naïve T cells.
Figure S7. The highest ranked super-enhancers shared across all cell populations are associated with T cell development and function.
Figure S8. Chromatin accessibility and H3K27ac enrichment at the Helios locus in fetal naïve T cells correlate with increased RNA and protein expression.
Figure S9. Fetal naïve T cells do not have an increased proportion of CD31+ cells relative to adult naïve T cells.
Figure S10. A fraction of fetal naïve T cells are highly proliferative.
Figure S11. Fetal naïve T cells do not have demethylation at the FOXP3 CNS2 (conserved non-coding sequence 2) Treg-specific demethylated region (TSDR).
Figure S12. Fetal naïve T cells upregulate Helios during Treg induction.
Figure S13. Validation of CRISPR-Cas9 editing at the Helios locus.
Figure S14. – The effect of CRISPR-Cas9 knockout of Helios on protein expression of Treg functional markers is variable.
Figure S15. Fetal, but not adult, induced Treg cells have suppressed IL-2 production after restimulation.
Figure S16. Helios knockout in fetal iTreg cells result in a subtle shift in the underlying transcriptome.
Table S8. Experimental setup for Treg induction time course carried out for adult and fetal naïve T cells.
Table S9. Experimental setup for Helios CRISPR-Cas9 mediated editing for subsequent Treg induction carried out for fetal naïve T cells.
Table S4. Treg accessible enhancers (Excel spreadsheet).
Table S5. Treg inaccessible enhancers (Excel spreadsheet).
Table S1. RNAseq Treg upregulated and downregulated signature genes (Excel spreadsheet).
Acknowledgements:
We would like to thank S. Fisher, P. Odorizzi, E. Rackaityte, J. Halkias, D. Bunis, J. Mold, J.M, McCune, and J.Y. Lim for helpful discussion. We thank D. Simeonov and K. Schumann for help with CRISPR-Cas9 editing. We thank M. Nguyen for advice and help with cell fixation and DNA extraction for TSDR analysis. We thank R. Krishnakumar, A. Chen, S. Thomas and R. Thomas for their technical help with ChIPseq and advice on bioinformatics analyses. Bioinformatics analyses for genome mapping and bigwig file generation for ATACseq was carried out by the Gladstone Bioinformatics Core at the Gladstone Institutes. Bioinformatic analyses for RNAseq in ex vivo naïve and Treg cells were carried out by the Technology Center for Genomics and Bioinformatics (TCGB) at University of California, Los Angeles. Sequencing for ChIPseq and RNAseq in iTreg cells were carried out at the Center for Advanced Technologies (CAT) Core at UCSF.
Funding: This work was supported by an Agency for Science, Technology and Research (A*STAR, Singapore) National Science Scholarship (PhD) awarded to M.N, and research grants NIH K08HD067295 and R21AI120032 to T.D.B., who was also supported by an award from the Preterm Birth Initiative of the Burroughs Wellcome Fund. T.L.R. was supported by the UCSF Medical Scientist Training Program (T32GM007618) and the UCSF Endocrinology Training Grant (T32DK007418). A.M. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund, has received funding from the Innovative Genomics Insitute (IGI) and the Parker Institute for Cancer Immunotherapy (PICI), and is an investigator at the Chan Zuckerberg Biohub. We also thank the Flow Cytometry Core at UCSF, supported by the Diabetes Research Center grant NIH P30 DK063720, and Shared Instrument Grant 1S10OD021822–01.
Footnotes
Competing Interests: A.M. is a co-founder of Spotlight Therapeutics. A.M. and T.L.R. are co-founders of Arsenal Biosciences. A.M. serves on the scientific advisory board of PACT Pharma, is an advisor to Trizell, and previously served as an advisor to Juno Therapeutics. A.M. has had speaking engagements with or done consulting for Amgen, ThermoFisher, Health Advances, Lonza, Bernstein, AbbVie, Genentech, Merck, Illumina, Arcus, Jackson Laboratories, Nanostring Technologies, GLG, RCM and Soteria. The Marson laboratory has received sponsored research support from Juno Therapeutics, Epinomics and Sanofi, and a gift from Gilead. The other authors declare that they have no competing interests.
Data and materials availability: ATACseq, H3K27ac ChIPseq, and all RNAseq raw and processed files are deposited in the Gene Expression Omnibus database under a superseries entry with the accession number GSE110472. The transcriptional and epigenetic gene lists defined in this manuscript are supplied in Supplementary Tables as described. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References
- 1.Mucida D, Kutchukhidze N, Erazo A, Russo M, Lafaille JJ, de Lafaille M. A. Curotto, Oral tolerance in the absence of naturally occurring Tregs, J. Clin. Invest. 115, 1923–1933 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coombes JL, Siddiqui KRR, Arancibia-Cárcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F, A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism, J. Exp. Med. 204, 1757–1764 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haribhai D, Williams JB, Jia S, Nickerson D, Schmitt EG, Edwards B, Ziegelbauer J, Yassai M, Li S-H, Relland LM, Wise PM, Chen A, Zheng Y-Q, Simpson PM, Gorski J, Salzman NH, Hessner MJ, Chatila TA, Williams CB, A requisite role for induced regulatory T cells in tolerance based on expanding antigen receptor diversity, Immunity 35, 109–122 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J, Paul WE, CD4 T cells: fates, functions, and faults, Blood 112, 1557–1569 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J, Yamane H, Paul WE, Differentiation of effector CD4 T cell populations (*), Annu. Rev. Immunol. 28, 445–489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sundrud MS, Nolan MA, Synergistic and combinatorial control of T cell activation and differentiation by transcription factors, Curr. Opin. Immunol. 22, 286–292 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Chen W, Jin W, Hardegen N, Lei K-J, Li L, Marinos N, McGrady G, Wahl SM, Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3, J. Exp. Med. 198, 1875–1886 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fantini MC, Becker C, Monteleone G, Pallone F, Galle PR, Neurath MF, Cutting edge: TGF-beta induces a regulatory phenotype in CD4+CD25- T cells through Foxp3 induction and down-regulation of Smad7, J. Immunol. 172, 5149–5153 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Kim JK, Hirning AJ, Josi K, Bennett MR, Emergent genetic oscillations in a synthetic microbial consortium, Science 349, 986–989 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F, Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse, Nat. Genet. 27, 68–73 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, Levy-Lahad E, Mazzella M, Goulet O, Perroni L, Bricarelli FD, Byrne G, McEuen M, Proll S, Appleby M, Brunkow ME, X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy, Nat. Genet. 27, 18–20 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD, The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3, Nat. Genet. 27, 20–21 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Khattri R, Cox T, Yasayko S-A, Ramsdell F, An essential role for Scurfin in CD4+CD25+ T regulatory cells, Nat Immunol 4, 337–342 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Fontenot JD, Gavin MA, Rudensky AY, Foxp3 programs the development and function of CD4+CD25+ regulatory T cells, Nat Immunol 4, 330–336 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Sakaguchi S, Naturally arising Foxp3-expressing CD25+ CD4+ regulatory T cells in immunological tolerance to self and non-self, Nat Immunol (2005). [DOI] [PubMed] [Google Scholar]
- 16.Xavier-da-Silva MM, Moreira-Filho CA, Suzuki E, Patricio F, Coutinho A, Carneiro-Sampaio M, Fetal-onset IPEX: report of two families and review of literature, Clin. Immunol. 156, 131–140 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Vasiljevic A, Poreau B, Bouvier R, Lachaux A, Arnoult C, Fauré J, Cordier MP, Ray PF, Immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome and recurrent intrauterine fetal death, Lancet 385, 2120 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Rae W, Gao Y, Bunyan D, Holden S, Gilmour K, Patel S, Wellesley D, Williams A, A novel FOXP3 mutation causing fetal akinesia and recurrent male miscarriages, Clin. Immunol. 161, 284–285 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Reichert SL, McKay EM, Moldenhauer JS, Identification of a novel nonsense mutation in the FOXP3 gene in a fetus with hydrops--Expanding the phenotype of IPEX syndrome, Am. J. Med. Genet. A 170A, 226–232 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Shehab O, Tester DJ, Ackerman NC, Cowchock FS, Ackerman MJ, Whole genome sequencing identifies etiology of recurrent male intrauterine fetal death, Prenat. Diagn. 37, 1040–1045 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Burt TD, Fetal Regulatory T Cells and Peripheral Immune Tolerance In Utero: Implications for Development and Disease, Am J Reprod Immunol 69, 346–358 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bacchetta R, Barzaghi F, Roncarolo MG, Rose NR, Ed. From IPEX syndrome to FOXP3 mutation: a lesson on immune dysregulation, Annals of the New York Academy of Sciences 1417, 5–22 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Michaelsson J, Mold JE, McCune JM, Nixon DF, Regulation of T Cell Responses in the Developing Human Fetus, J. Immunol. 176, 5741–5748 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Mold JE, Michaelsson J, Burt TD, Muench MO, Beckerman KP, Busch MP, Lee TH, Nixon DF, McCune JM, Maternal Alloantigens Promote the Development of Tolerogenic Fetal Regulatory T Cells in Utero, Science 322, 1562–1565 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mold JE, Venkatasubrahmanyam S, Burt TD, Michaelsson J, Rivera JM, Galkina SA, Weinberg K, Stoddart CA, McCune JM, Fetal and Adult Hematopoietic Stem Cells Give Rise to Distinct T Cell Lineages in Humans, Science 330, 1695–1699 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bronevetsky Y, Burt TD, McCune JM, Lin28b Regulates Fetal Regulatory T Cell Differentiation through Modulation of TGF-β Signaling, The Journal of Immunology 197, 4344–4350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tripathi SK, Lahesmaa R, Transcriptional and epigenetic regulation of T-helper lineage specification, Immunological Reviews 261, 62–83 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen MLT, Jones SA, Prier JE, Russ BE, Transcriptional Enhancers in the Regulation of T Cell Differentiation, Front Immunol 6, 462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y, Osaki M, Tanaka Y, Yamashita R, Nakano N, Huehn J, Fehling HJ, Sparwasser T, Nakai K, Sakaguchi S, T Cell Receptor Stimulation-Induced Epigenetic Changes and Foxp3 Expression Are Independent and Complementary Events Required for Treg Cell Development, Immunity 37, 785–799 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Kitagawa Y, Ohkura N, Kidani Y, Vandenbon A, Hirota K, Kawakami R, Yasuda K, Motooka D, Nakamura S, Kondo M, Taniuchi I, Kohwi-Shigematsu T, Sakaguchi S, Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment, Nat Immunol 18, 173–183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY, Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate, Nature 463, 808–812 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arvey A, van der Veeken J, Plitas G, Rich SS, Concannon P, Rudensky AY, Genetic and epigenetic variation in the lineage specification of regulatory T cells, Elife 4, e07571 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, Mathis D, Benoist C, Foxp3 Transcription-Factor-Dependent and -Independent Regulation of the Regulatory T Cell Transcriptional Signature, Immunity 27, 786–800 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Fu W, Ergun A, Lu T, Hill JA, Haxhinasto S, Fassett MS, Gazit R, Adoro S, Glimcher L, Chan S, Kastner P, Rossi D, Collins JJ, Mathis D, Benoist C, A multiply redundant genetic switch “locks in” the transcriptional signature of regulatory T cells, Nat Immunol 13, 972–980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, Zhu Y, Wei S, Kryczek I, Daniel B, Gordon A, Myers L, Lackner A, Disis ML, Knutson KL, Chen L, Zou W, Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival, Nature Medicine 10, 942–949 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Miyara M, Sakaguchi S, Natural regulatory T cells: mechanisms of suppression, Trends Mol Med 13, 108–116 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Cao Z, Wara AK, Icli B, Sun X, Packard RRS, Esen F, Stapleton CJ, Subramaniam M, Kretschmer K, Apostolou I, von Boehmer H, Hansson GK, Spelsberg TC, Libby P, Feinberg MW, Kruppel-like Factor KLF10 Targets Transforming Growth Factor- 1 to Regulate CD4+CD25- T Cells and T Regulatory Cells, Journal of Biological Chemistry 284, 24914–24924 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong Y, Khanna S, Grzenda AL, Sarmento OF, Svingen PA, Lomberk GA, Urrutia RA, Faubion WA, Polycomb Antagonizes p300/CREB-binding Protein-associated Factor to Silence FOXP3 in a Kruppel-like Factor-dependent Manner, Journal of Biological Chemistry 287, 34372–34385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong Y, Svingen PA, Sarmento OO, Smyrk TC, Dave M, Khanna S, Lomberk GA, Urrutia RA, Faubion WA, Differential coupling of KLF10 to Sin3-HDAC and PCAF regulates the inducibility of the FOXP3 gene, AJP: Regulatory, Integrative and Comparative Physiology 307, R608–R620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, Shevach EM, Expression of Helios, an Ikaros Transcription Factor Family Member, Differentiates Thymic-Derived from Peripherally Induced Foxp3+ T Regulatory Cells, The Journal of Immunology 184, 3433–3441 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim H-J, Barnitz RA, Kreslavsky T, Brown FD, Moffett H, Lemieux ME, Kaygusuz Y, Meissner T, Holderried TAW, Chan S, Kastner P, Haining WN, Cantor H, Stable inhibitory activity of regulatory T cells requires the transcription factor Helios, Science 350, 334–339 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sebastian M, Lopez-Ocasio M, Metidji A, Rieder SA, Shevach EM, Thornton AM, Helios Controls a Limited Subset of Regulatory T Cell Functions, The Journal of Immunology 196, 144–155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan F, Yu H, Dang EV, Barbi J, Pan X, Grosso JF, Jinasena D, Sharma SM, McCadden EM, Getnet D, Drake CG, Liu JO, Ostrowski MC, Pardoll DM, Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells, Science 325, 1142–1146 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morikawa H, Ohkura N, Vandenbon A, Itoh M, Nagao-Sato S, Kawaji H, Lassmann T, Carninci P, Hayashizaki Y, Forrest ARR, Standley DM, Date H, Sakaguchi S, FANTOM Consortium, Itoh M, Sakaguchi S, Differential roles of epigenetic changes and Foxp3 expression in regulatory T cell-specific transcriptional regulation, Proc. Natl. Acad. Sci. U.S.A. 111, 5289–5294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu A, Zhu L, Altman NH, Malek TR, A low interleukin-2 receptor signaling threshold supports the development and homeostasis of T regulatory cells, Immunity 30, 204–217 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joller N, Lozano E, Burkett PR, Patel B, Xiao S, Zhu C, Xia J, Tan TG, Sefik E, Yajnik V, Sharpe AH, Quintana FJ, Mathis D, Benoist C, Hafler DA, Kuchroo VK, Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses, Immunity 40, 569–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furlan SN, Watkins B, Tkachev V, Cooley S, Panoskaltsis-Mortari A, Betz K, Brown M, Hunt DJ, Schell JB, Zeleski K, Yu A, Giver CR, Waller EK, Miller JS, Blazar BR, Kean LS, Systems analysis uncovers inflammatory Th/Tc17-driven modules during acute GVHD in monkey and human T cells, Blood 128, 2568–2579 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takimoto T, Wakabayashi Y, Sekiya T, Inoue N, Morita R, Ichiyama K, Takahashi R, Asakawa M, Muto G, Mori T, Hasegawa E, Saika S, Shizuya S, Hara T, Nomura M, Yoshimura A, Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development, The Journal of Immunology 185, 842–855 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Delgoffe GM, Woo S-R, Turnis ME, Gravano DM, Guy C, Overacre AE, Bettini ML, Vogel P, Finkelstein D, Bonnevier J, Workman CJ, Vignali DAA, Stability and function of regulatory T cells is maintained by a neuropilin-1-semaphorin-4a axis, Nature 501, 252–256 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Travis MA, Sheppard D, TGF-β activation and function in immunity, Annu. Rev. Immunol. 32, 51–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ocklenburg F, Moharregh-Khiabani D, Geffers R, Janke V, Pfoertner S, Garritsen H, Groebe L, Klempnauer J, Dittmar KEJ, Weiss S, Buer J, Probst-Kepper M, UBD, a downstream element of FOXP3, allows the identification of LGALS3, a new marker of human regulatory T cells, Lab. Invest. 86, 724–737 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Probst-Kepper M, Geffers R, Kröger A, Viegas N, Erck C, Hecht H-J, Lünsdorf H, Roubin R, Moharregh-Khiabani D, Wagner K, Ocklenburg F, Jeron A, Garritsen H, Arstila TP, Kekäläinen E, Balling R, Hauser H, Buer J, Weiss S, GARP: a key receptor controlling FOXP3 in human regulatory T cells, J. Cell. Mol. Med. 13, 3343–3357 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pesenacker AM, Wang AY, Singh A, Gillies J, Kim Y, Piccirillo CA, Nguyen D, Haining WN, Tebbutt SJ, Panagiotopoulos C, Levings MK, A Regulatory T-Cell Gene Signature Is a Specific and Sensitive Biomarker to Identify Children With New-Onset Type 1 Diabetes, Diabetes 65, 1031–1039 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Tran DQ, Ramsey H, Shevach EM, Induction of FOXP3 expression in naive human CD4+FOXP3 T cells by T-cell receptor stimulation is transforming growth factor-beta dependent but does not confer a regulatory phenotype, Blood 110, 2983–2990 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edwards JP, Thornton AM, Shevach EM, Release of active TGF-β1 from the latent TGF-β1/GARP complex on T regulatory cells is mediated by integrin β8, The Journal of Immunology 193, 2843–2849 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang R, Kozhaya L, Mercer F, Khaitan A, Fujii H, Unutmaz D, Expression of GARP selectively identifies activated human FOXP3+ regulatory T cells, Proc. Natl. Acad. Sci. U.S.A. 106, 13439–13444 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA, Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes, Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA, Selective inhibition of tumor oncogenes by disruption of super-enhancers, Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shlyueva D, Stampfel G, Stark A, Transcriptional enhancers: from properties to genome-wide predictions, Nature Publishing Group 15, 272–286 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, Neph S, Sabo P, Kim JM, Liao W, Li MO, Leslie C, Stamatoyannopoulos JA, Rudensky AY, Foxp3 Exploits a Pre-Existent Enhancer Landscape for Regulatory T Cell Lineage Specification, Cell 151, 153–166 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidl C, Hansmann L, Lassmann T, Balwierz PJ, Kawaji H, Itoh M, Kawai J, Nagao-Sato S, Suzuki H, Andreesen R, Hayashizaki Y, Forrest ARR, Carninci P, Hoffmann P, Edinger M, Rehli M, FANTOM Consortium, The enhancer and promoter landscape of human regulatory and conventional T-cell subpopulations, Blood 123, e68–78 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, Polak L, Kadaja M, Asare A, Zheng D, Fuchs E, Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice, Nature 521, 366–370 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, Hitomi J, Yamamoto T, Utsuyama M, Niwa O, Aizawa S, Kominami R, Bcl11b is required for differentiation and survival of alphabeta T lymphocytes, Nat Immunol 4, 533–539 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Muthusamy N, Barton K, Leiden JM, Defective activation and survival of T cells lacking the Ets-1 transcription factor, Nature 377, 639–642 (1995). [DOI] [PubMed] [Google Scholar]
- 65.Sugimoto N, Oida T, Hirota K, Nakamura K, Nomura T, Uchiyama T, Sakaguchi S, Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis, International Immunology 18, 1197–1209 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Gottschalk RA, Corse E, Allison JP, Expression of Helios in peripherally induced Foxp3+ regulatory T cells, The Journal of Immunology 188, 976–980 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Getnet D, Grosso JF, Goldberg MV, Harris TJ, Yen H-R, Bruno TC, Durham NM, Hipkiss EL, Pyle KJ, Wada S, Pan F, Pardoll DM, Drake CG, A role for the transcription factor Helios in human CD4(+)CD25(+) regulatory T cells, Molecular Immunology 47, 1595–1600 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paiva RS, Lino AC, Bergman M-L, Caramalho I, Sousa AE, Zelenay S, Demengeot J, Recent thymic emigrants are the preferential precursors of regulatory T cells differentiated in the periphery, Proc. Natl. Acad. Sci. U.S.A. 110, 6494–6499 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kimmig S, Przybylski GK, Schmidt CA, Laurisch K, Möwes B, Radbruch A, Thiel A, Two subsets of naive T helper cells with distinct T cell receptor excision circle content in human adult peripheral blood, J. Exp. Med. 195, 789–794 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Akimova T, Beier UH, Wang L, Levine MH, Hancock WW, Molina-Paris C, Ed. Helios expression is a marker of T cell activation and proliferation, PLoS ONE 6, e24226 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang H-D, Bopp T, Schmitt E, Klein-Hessling S, Serfling E, Hamann A, Huehn J, Marrack P, Ed. Epigenetic control of the foxp3 locus in regulatory T cells, PLoS Biol. 5, e38 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koenecke C, Czeloth N, Bubke A, Schmitz S, Kissenpfennig A, Malissen B, Huehn J, Ganser A, Förster R, Prinz I, Alloantigen-specific de novo-induced Foxp3+ Treg revert in vivo and do not protect from experimental GVHD, Eur. J. Immunol. 39, 3091–3096 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Beres A, Komorowski R, Mihara M, Drobyski WR, Instability of Foxp3 Expression Limits the Ability of Induced Regulatory T Cells to Mitigate Graft versus Host Disease, Clinical Cancer Research 17, 3969–3983 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jonuleit H, Schmitt E, Stassen M, Tuettenberg A, Knop J, Enk AH, Identification and functional characterization of human CD4(+)CD25(+) T cells with regulatory properties isolated from peripheral blood, J. Exp. Med. 193, 1285–1294 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baine I, Basu S, Ames R, Sellers RS, Macian F, Helios Induces Epigenetic Silencing of Il2 Gene Expression in Regulatory T Cells, J. Immunol. 190, 1008–1016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gasteiger G, Kastenmuller W, Foxp3+ Regulatory T-cells and IL-2: The Moirai of T-cell Fates? Front Immunol 3, 179 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takatori H, Kawashima H, Matsuki A, Meguro K, Tanaka S, Iwamoto T, Sanayama Y, Nishikawa N, Tamachi T, Ikeda K, Suto A, Suzuki K, Kagami S-I, Hirose K, Kubo M, Hori S, Nakajima H, Helios Enhances Treg Cell Function in Cooperation With FoxP3, Arthritis Rheumatol 67, 1491–1502 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Fu S-H, Yeh L-T, Chu C-C, Yen BL-J, Sytwu H-K, New insights into Blimp-1 in T lymphocytes: a divergent regulator of cell destiny and effector function, J. Biomed. Sci. 24, 49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Onodera A, Kokubo K, Nakayama T, Epigenetic and Transcriptional Regulation in the Induction, Maintenance, Heterogeneity, and Recall-Response of Effector and Memory Th2 Cells, Front Immunol 9, 2929 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Quirion MR, Gregory GD, Umetsu SE, Winandy S, Brown MA, Cutting edge: Ikaros is a regulator of Th2 cell differentiation, The Journal of Immunology 182, 741–745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wong LY, Hatfield JK, Brown MA, Ikaros sets the potential for Th17 lineage gene expression through effects on chromatin state in early T cell development, J. Biol. Chem. 288, 35170–35179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wheaton JD, Yeh C-H, Ciofani M, Cutting Edge: c-Maf Is Required for Regulatory T Cells To Adopt RORγt+ and Follicular Phenotypes, The Journal of Immunology 199, 3931–3936 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Choi J-M, Bothwell ALM, The nuclear receptor PPARs as important regulators of T-cell functions and autoimmune diseases, Mol. Cells 33, 217–222 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kaminuma O, Kitamura N, Nishito Y, Nemoto S, Tatsumi H, Mori A, Hiroi T, Downregulation of NFAT3 Due to Lack of T-Box Transcription Factor TBX5 Is Crucial for Cytokine Expression in T Cells, The Journal of Immunology 200, 92–100 (2018). [DOI] [PubMed] [Google Scholar]
- 85.Li Q, Zou J, Wang M, Ding X, Chepelev I, Zhou X, Zhao W, Wei G, Cui J, Zhao K, Wang HY, Wang R-F, Critical role of histone demethylase Jmjd3 in the regulation of CD4+ T-cell differentiation, Nat Commun 5, 5780 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shevach EM, Mechanisms of foxp3+ T regulatory cell-mediated suppression, Immunity 30, 636–645 (2009). [DOI] [PubMed] [Google Scholar]
- 87.Sridharan R, Smale ST, Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes, Journal of Biological Chemistry 282, 30227–30238 (2007). [DOI] [PubMed] [Google Scholar]
- 88.Zhao S, Liu W, Li Y, Liu P, Li S, Dou D, Wang Y, Yang R, Xiang R, Liu F, Song C, Ed. Alternative Splice Variants Modulates Dominant-Negative Function of Helios in T-Cell Leukemia, PLoS ONE 11, e0163328 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cai Q, Dierich A, Oulad-Abdelghani M, Chan S, Kastner P, Helios deficiency has minimal impact on T cell development and function, The Journal of Immunology 183, 2303–2311 (2009). [DOI] [PubMed] [Google Scholar]
- 90.Yang S, Fujikado N, Kolodin D, Benoist C, Mathis D, Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance, Science, 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smith H, Chen IM, Kubo R, Tung KS, Neonatal thymectomy results in a repertoire enriched in T cells deleted in adult thymus, Science 245, 749–752 (1989). [DOI] [PubMed] [Google Scholar]
- 92.Kim YC, Bhairavabhotla R, Yoon J, Golding A, Thornton AM, Tran DQ, Shevach EM, Oligodeoxynucleotides stabilize Helios-expressing Foxp3+ human T regulatory cells during in vitro expansion, Blood 119, 2810–2818 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thornton AM, Lu J, Korty PE, Kim YC, Martens C, Sun PD, Shevach EM, Helios+ and Helios- Treg subpopulations are phenotypically and functionally distinct and express dissimilar TCR repertoires, Eur. J. Immunol. 112, 137 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schumann K, Lin S, Boyer E, Simeonov DR, Subramaniam M, Gate RE, Haliburton GE, Ye CJ, Bluestone JA, Doudna JA, Marson A, Generation of knock-in primary human T cells using Cas9 ribonucleoproteins, Proc. Natl. Acad. Sci. U.S.A. 112, 10437–10442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hultquist JF, Schumann K, Woo JM, Manganaro L, McGregor MJ, Doudna J, Simon V, Krogan NJ, Marson A, A Cas9 Ribonucleoprotein Platform for Functional Genetic Studies of HIV-Host Interactions in Primary Human T Cells, Cell Rep 17, 1438–1452 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McCarthy DJ, Chen Y, Smyth GK, Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation, Nucleic Acids Research 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1:417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ, Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position, Nat Meth 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ, ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide, Curr Protoc Mol Biol 109, 21.29.1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Collas P, A chromatin immunoprecipitation protocol for small cell numbers, Methods Mol. Biol. 791, 179–193 (2011). [DOI] [PubMed] [Google Scholar]
- 102.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK, Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities, Molecular Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E, CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering, Nucleic Acids Research 44, W272–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Richardson CD, Ray GJ, Bray NL, Corn JE, Non-homologous DNA increases gene disruption efficiency by altering DNA repair outcomes, Nat Commun 7, 12463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S7. Top 25 ranked super-enhancers in adult naive, fetal naive and adult Treg cells (Excel spreadsheet).
Table S6. De novo motif enrichment in Treg -accessible ATAC peaks shared between fetal naive and adult Treg cells (Excel spreadsheet).
Table S10. Upregulated and downregulated differentially expressed genes defined in Helios KO versus Helios WT fetal induced Treg cells (Excel spreadsheet).
Table S11. Raw data file (Excel spreadsheet).
Table S2. Genes upregulated/downregulated within iTreg populations with cluster assignment (Excel spreadsheet).
Table S3. Statistics from pre-ranked gene set enrichment analysis for fetal versus adult iTreg populations (Excel spreadsheet).
Figure S1. Gating strategy and purity assessment for sorted naïve and Treg cells.
Figure S2. Definition of the Treg transcriptional signature.
Figure S3. Assessing the enrichment of Treg upregulated or downregulated genes in fetal and adult induced Treg (iTreg) populations.
Figure S4. Fetal induced Treg cells have increased sensitivity to TGF-β signaling.
Figure S5. Identification of Treg-accessible and inaccessible enhancers.
Figure S6. Binding motifs for downstream effectors of Treg differentiation are enriched within shared Treg-accessible peaks in fetal naïve T cells.
Figure S7. The highest ranked super-enhancers shared across all cell populations are associated with T cell development and function.
Figure S8. Chromatin accessibility and H3K27ac enrichment at the Helios locus in fetal naïve T cells correlate with increased RNA and protein expression.
Figure S9. Fetal naïve T cells do not have an increased proportion of CD31+ cells relative to adult naïve T cells.
Figure S10. A fraction of fetal naïve T cells are highly proliferative.
Figure S11. Fetal naïve T cells do not have demethylation at the FOXP3 CNS2 (conserved non-coding sequence 2) Treg-specific demethylated region (TSDR).
Figure S12. Fetal naïve T cells upregulate Helios during Treg induction.
Figure S13. Validation of CRISPR-Cas9 editing at the Helios locus.
Figure S14. – The effect of CRISPR-Cas9 knockout of Helios on protein expression of Treg functional markers is variable.
Figure S15. Fetal, but not adult, induced Treg cells have suppressed IL-2 production after restimulation.
Figure S16. Helios knockout in fetal iTreg cells result in a subtle shift in the underlying transcriptome.
Table S8. Experimental setup for Treg induction time course carried out for adult and fetal naïve T cells.
Table S9. Experimental setup for Helios CRISPR-Cas9 mediated editing for subsequent Treg induction carried out for fetal naïve T cells.
Table S4. Treg accessible enhancers (Excel spreadsheet).
Table S5. Treg inaccessible enhancers (Excel spreadsheet).
Table S1. RNAseq Treg upregulated and downregulated signature genes (Excel spreadsheet).