Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of the novel viral disease COVID-19. With no approved therapies, this pandemic illustrates the urgent need for broad-spectrum antiviral countermeasures against SARS-CoV-2 and future emerging CoVs. We report that remdesivir (RDV) potently inhibits SARS-CoV-2 replication in human lung cells and primary human airway epithelial cultures (EC50 = 0.01 μM). Weaker activity is observed in Vero E6 cells (EC50 = 1.65 μM) because of their low capacity to metabolize RDV. To rapidly evaluate in vivo efficacy, we engineered a chimeric SARS-CoV encoding the viral target of RDV, the RNA-dependent RNA polymerase of SARS-CoV-2. In mice infected with the chimeric virus, therapeutic RDV administration diminishes lung viral load and improves pulmonary function compared with vehicle-treated animals. These data demonstrate that RDV is potently active against SARS-CoV-2 in vitro and in vivo, supporting its further clinical testing for treatment of COVID-19.
Keywords: COVID-19, SARS-CoV-2, coronavirus, antiviral, remdesivir, RdRp, RNA-dependent RNA polymerase, therapeutic, mouse, GS-441524
Graphical Abstract
SARS-CoV-2 causes severe lung disease (COVID-19) in humans. Pruijssers et al. demonstrate that the antiviral drug remdesivir potently inhibits SARS-CoV-2 in human lung cell cultures. Therapeutic treatment of infected mice with remdesivir reduces viral loads and improves clinical outcomes, further supporting use of remdesivir for the treatment of COVID-19.
Introduction
Coronaviruses (CoVs) are genetically diverse positive-sense RNA viruses that circulate in animals and humans. In the past 20 years, three new human CoVs have emerged: severe acute respiratory syndrome CoV (SARS-CoV) in 2002, Middle East respiratory syndrome (MERS)-CoV in 2012, and SARS-CoV-2, the causative agent of the current CoV disease 2019 (COVID-19) pandemic (de Wit et al., 2016; Zhou et al., 2020b). Although four endemic human CoVs (HCoV-OC43, -229E, -NL63, and -HKU1) typically cause mild respiratory disease with common cold-like symptoms, SARS-CoV, MERS-CoV, and SARS-CoV-2 cause severe respiratory disease with respective mortality rates of 11% (Chan-Yeung and Xu, 2003), 35% (Arabi et al., 2017), and an estimated 3% (Chen, 2020). The development of effective broad-spectrum antiviral agents has been hampered by viral diversity, the capacity of CoVs to adaptively overcome negative selective pressure, and the ability to actively counteract drugs through the action of a proofreading exoribonuclease. We reported previously that remdesivir (RDV), a monophosphoramidate prodrug of the C-adenosine analog GS-441524, potently inhibits replication of a broad spectrum of pre-pandemic bat CoVs and human epidemic CoVs in primary human lung cell cultures (Agostini et al., 2018; Brown et al., 2019; Sheahan et al., 2017). Biochemical analysis of the mechanism of inhibition of the SARS-CoV-2, SARS-CoV, and MERS-CoV RNA-dependent RNA polymerase (RdRp) revealed that incorporation of the active metabolite RDV triphosphate (RDV-TP) was more efficient than the natural substrate ATP and led to delayed chain termination 3 nt downstream of incorporation (Gordon et al., 2020a, 2020b). Prolonged passaging of murine hepatitis virus (MHV), a group 2a CoV, in the presence of GS-441524 resulted in low-level resistance through mutations in the RdRp, further implicating this protein as the drug target (Agostini et al., 2018). RDV showed prophylactic and therapeutic efficacy in mouse models of SARS and MERS and against MERS-CoV challenge in a rhesus macaque model (Sheahan et al., 2017, 2020a; de Wit et al., 2020). Here we report that RDV potently inhibits SARS-CoV-2 replication in Calu3 human lung cells with sub-micromolar 50% maximal effective concentration (EC50) and in primary human airway epithelial (HAE) cultures with nanomolar EC50. Notably, we detected a comparably lower potency of RDV in established human and monkey cell lines because of their lower metabolic capacity to activate the compound. Mice infected with chimeric SARS-CoV encoding the SARS-CoV-2 RdRp and treated therapeutically with RDV show decreased viral loads in the lungs and increased pulmonary function. These data emphasize the potential of RDV as a promising countermeasure against the ongoing COVID-19 pandemic.
Results
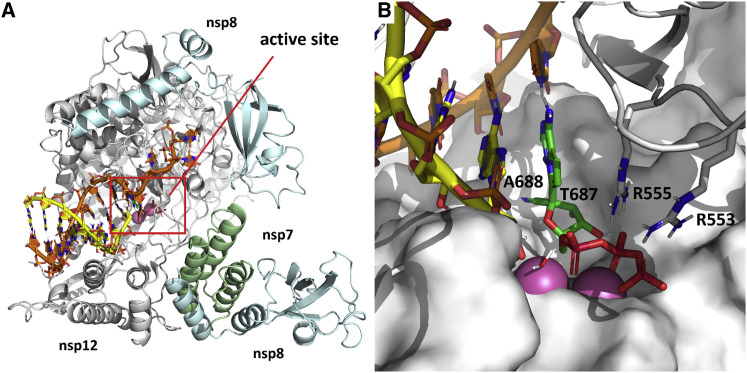
A Structural Model of RDV Incorporation by the SARS-CoV-2 Polymerase and Conservation of the Active Site across Human CoV
Drug function and performance are heavily influenced by microvariation in target genes across virus families, biodistribution in the organism, and, importantly, host cell and tissue expression patterns that influence drug stability and metabolism. We previously modeled RDV on a homology model of SARS-CoV-2 based on the cryo-EM structure of the SARS-CoV polymerase complex (Gordon et al., 2020b; Kirchdoerfer and Ward, 2019). Composed of nsp12, nsp7, and nsp8, the model was consistent with biochemical findings predicting efficient incorporation of RDV-TP into the growing RNA strand and provided an explanation for the observed delayed chain termination after incorporation of three additional nucleotides. We have since refined this model using the recently released cryoelectron microscopy (cryo-EM) structure of the SARS-CoV-2 polymerase complex (Gao et al., 2020). The major qualitative change is a more complete picture of the N-terminal NiRAN (nidovirus RdRp-associated nucleotidyltransferase) domain of nsp12, which was not resolved in the SARS-CoV structure. The current model of the pre-incorporation state, with RDV-TP, RNA primer and template strands, and catalytic metals was well-optimized with a series of constrained energy minimizations and conformational searches, as described previously (Figure 1 A). Bound to the two catalytic Mg2+ ions, RDV-TP is coordinated by two basic residues (R553 and R555). The ribose 2′OH forms hydrogen bonds to T680 and N691, and the 1′CN resides in a shallow pocket formed by T687 and A688 (Figure 1B). The interaction with T680 distinguishes CoVs from other structurally related families, including noroviruses, picornaviruses, and flaviviruses. Although key residues, including D623, S682, and N691, are conserved across all of these virus families and have been shown to govern positioning of the nucleoside triphosphate (NTP) into the active site, the role of T680 appears to be novel. While writing this manuscript, another model of RDV-TP in the SARS-CoV-2 active site (Shannon et al., 2020) was published that predicts a role for S682 as well. The position of T680 relative to N691 strongly implies that it will contribute to recognition of the ribose 2′OH, likely diminishing the role of S682 as a result, consistent with earlier predictions (Kirchdoerfer and Ward, 2019). Furthermore, while this manuscript was under review, a cryo-EM ternary structure of the complex with RDV was released (Yin et al., 2020). The new structure (PDB: 7BV2) aligned with the model with a root-mean-square deviation (RMSD) of 0.91 Å, with an RMSD of 1.66 Å for the double-stranded RNA (dsRNA) less the template overhang. Even though the new structure captures the post-incorporation/pre-translocation state, and our model depicts the pre-incorporation state, the new structure predicts no interaction between S682 and the incorporated RDV and, thus, confirms its diminished role. This contrasts with structures that capture this same post-incorporation state of the poliovirus polymerase, in which the homologous serine residue is seen to directly hydrogen-bond to the 2′OH of the incorporated nucleotide (Gong and Peersen, 2010). The new structure also reinforces our previous prediction (Gordon et al., 2020b) that S861 is the residue most likely responsible for the delayed chain termination caused by RDV incorporation.
Figure 1.
Modeling of RDV onto the SARS-CoV-2 RdRp Structure
(A) Model of the SARS-CoV-2 polymerization complex in its elongating state. The model was based on the cryo-EM apo structures of SARS-CoV (http://www.rcsb.org/structure/6NUR) and SARS-CoV-2 (http://www.rcsb.org/structure/6M71). The active site is boxed in red.
(B) Enlarged view of the active site, depicting RDV pre-incorporation. The 1′CN substituent sits in a shallow pocket formed by residues T687 and A688. Bound to the two catalytic Mg2+ ions (pink), the TP is coordinated by two basic residues (R553 and R555).
See also Figures S1–S3.
Modeling of the RDV resistance mutations identified in MHV (Agostini et al., 2018) onto the homologous residues V557 and F480 in the SARS-CoV-2 RdRp structure reveals that V557L shifts the position of the template base, which, in turn, shifts the positioning of the incoming NTP (Figures S1A and S1B). This will affect RDV activity in that it alters the position of the 1′CN in the pocket. Because the model predicts no direct interaction of F480 with the NTP, primer, or template, the effect of the F480L mutation is more difficult to discern. The F480L mutation could potentially induce a subtle change to the 1′CN binding pocket (Figures S1C and S1D). Alignment of nsp12 sequences from SARS-CoV-2 used in other studies of RDV shows complete conservation of nsp12 nucleotide sequences, predicting comparable antiviral activity of RDV against these isolates (Figure S2). We next modeled the active sites of the six other human CoVs SARS-CoV (Figure S3A), MERS-CoV (Figure S3B), HCoV-OC43 (Figure S3C), HCoV-229E (Figure S3D), HCoV-NL63 (Figure S3E), and HCoV-HKU1 (Figure S3F). The models show that SARS-CoV-2 is identical to SARS-CoV out to a radius of 18 Å from the active site. Differences detected on the periphery of the active site of MERS-CoV and HCoV-OC43, -229E, -NL63, and -HKU1 correspond to residues that do not directly interact with RDV-TP. Together, these data demonstrate high structural conservation of the RdRp active site interacting with RDV-TP across all seven known human CoV strains.
RDV and GS-441524 Potently Inhibit SARS-CoV-2 Replication
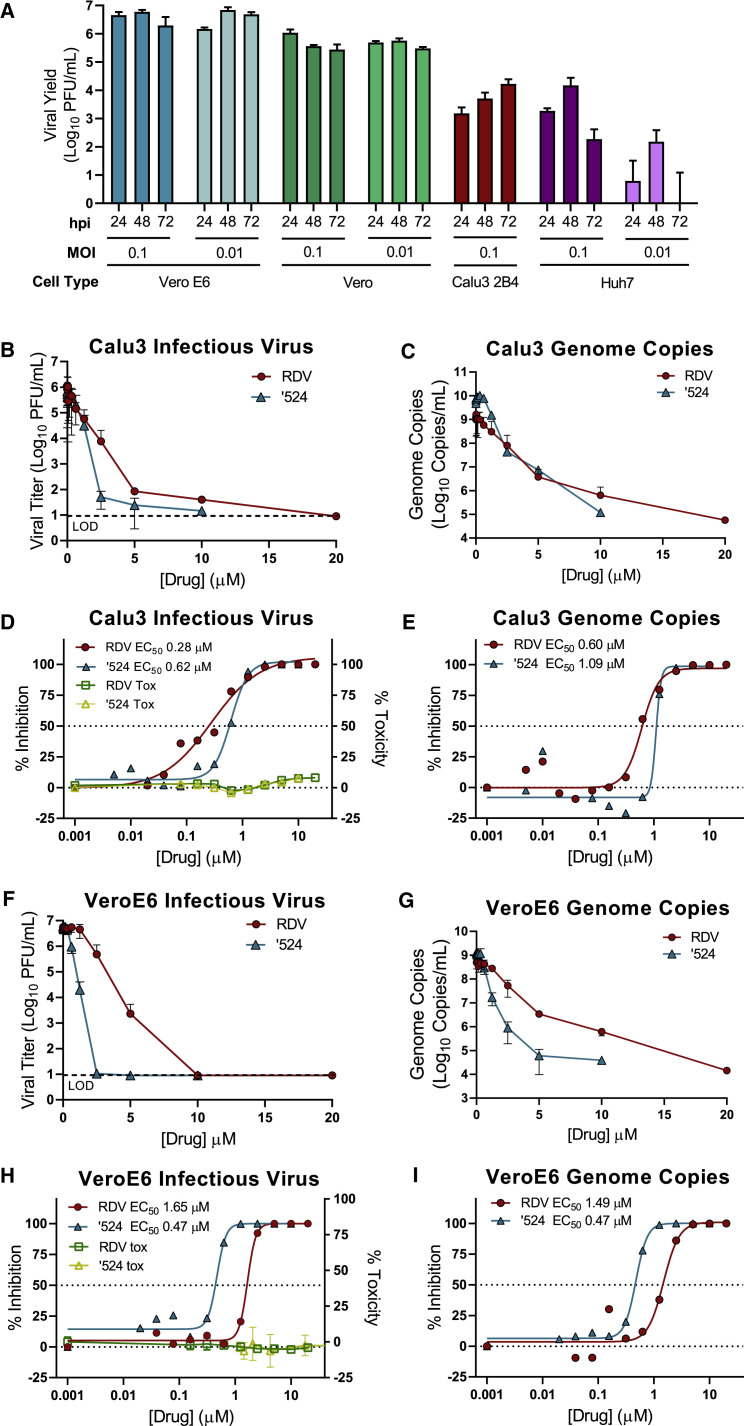
RDV and its parent nucleoside analog GS-441524 inhibit CoVs and multiple other viruses (Agostini et al., 2018; Cho et al., 2012; Lo et al., 2017; Sheahan et al., 2017; Warren et al., 2016). Previous reports (Choy et al., 2020; Wang et al., 2020; Runfeng et al., 2020) suggest that RDV inhibits SARS-CoV-2, but comparative studies of anti-SARS-CoV-2 activity using authentic compounds in physiologically relevant cell lines are lacking. We first compared SARS-CoV-2 replication in established cell lines to determine which cell types could potentially be suitable for studying RDV efficacy against SARS-CoV-2. Viral yields were determined 24, 48, and 72 h post-infection (hpi) in Vero E6, Vero CCL-81 (Vero), Huh7, and Calu3 2B4 cells (Yoshikawa et al., 2010; Figure 2 A). Vero E6 and Vero cells supported the highest levels of SARS-CoV-2 replication, consistent with a previous study (Harcourt et al., 2020). Maximum yields were detected at 48 hpi in Vero E6 cells (>6 logs at MOI = 0.1 and 0.01 PFU/cell), 24 hpi in Vero cells infected at MOI = 0.1 PFU (>5 logs), 48 hpi in Vero cells infected at MOI = 0.01 PFU/cell (>5 logs), 72 hpi in Calu3 2B4 cells (>4 logs at MOI = 0.1 PFU/cell), and 48 hpi in Huh7 cells (>4 logs at MOI = 0.1 PFU/cell and <2 logs at MOI = 0.01 PFU/cell). These results indicate that Vero E6, Vero, and Calu3 2B4 cells support varying levels of SARS-CoV-2 replication and that the cell type should be chosen for a given study depending on the study goals.
Figure 2.
The Prodrug Remdesivir (RDV) and Parent Nucleoside GS-441524 (’524) Potently Inhibit SARS-CoV-2 Replication
(A) Vero E6, Vero CCL-81 (Vero), Huh7, and Calu3 2B4 cells were infected with an MOI of 0.01 and/or 0.1 PFU/cell SARS-CoV-2 (2019-nCoV/USA-WA1/2020), and infectious viral titers were determined by plaque assay 0.5, 24, 48, and 72 h post-infection (hpi). Viral yields were calculated by subtracting the average 0.5 h (post-adsorption, pre-incubation) titer from each subsequent time point. Data represent the average of three replicates from one experiment. Error bars indicate SD. B.D., below detection. Calu3 2B4 cells were infected with 0.1 PFU/cell SARS-CoV-2, and Vero cells were infected with 0.01 PFU/cell SARS-CoV-2 and treated with RDV, GS-441524 (’524), or DMSO only (control) in cell culture medium. Supernatants were collected 48 hpi (Vero E6 cells) or 72 hpi (Calu3 2B4 cells).
(B and C) Reduction of SARS-CoV-2 replication by RDV in Calu3 2B4 cells, as determined by infectious viral titer (B) and qRT-PCR (C).
(D) Percent inhibition of SARS-CoV-2 replication by RDV and GS-441524 in Calu3 2B4 cells, as determined by infectious viral titer (RDV: EC50 = 0.28 μM, EC90 = 2.48 μM; GS-441524: EC50 = 0.62 μM, EC90 = 1.34 μM). No significant cytotoxicity of either compound was detected in Calu3 cells.
(E) Percent inhibition of SARS-CoV-2 replication by RDV and GS-441524 in Calu3 2B4 cells, as determined by qRT-qPCR (RDV: EC50 = 0.60 μM, EC90 = 1.28 μM; GS-441524: EC50 = 1.09 μM, EC90 = 1.37 μM).
(F and G) Reduction of SARS-CoV-2 replication by RDV in Vero E6 cells, as determined by infectious viral titer (F) and qRT-PCR (G).
(H) Percent inhibition of SARS-CoV-2 replication by RDV and GS-441524 in Vero E6 cells, as determined by infectious viral titer (RDV: EC50 = 1.65 μM, EC90 = 2.40 μM; GS-441524: EC50 = 0.47 μM, EC90 = 0.71 μM). No significant cytotoxicity of either compound was detected in Vero E6 cells.
(I) Percent inhibition of SARS-CoV-2 replication by RDV and GS-441524 in Vero E6 cells, as determined by qRT-PCR (RDV: EC50 = 1.49 μM, EC90 = 3.03 μM; GS-441524: EC50 = 0.47 μM, EC90 = 0.80 μM).
Data represent means of 2–4 independent experiments with 2–3 replicates each. Error bars represent SEM. See also Figure S5.
To determine whether RDV and GS-441524 inhibit SARS-CoV-2 replication in established cell lines, Calu3 2B4 human lung adenocarcinoma cells and Vero E6 African green monkey kidney cells were infected with the SARS-CoV-2 clinical isolate 2019-nCoV/USA-WA1/2020 and treated with a range of RDV or GS-441524 concentrations. Supernatants were harvested at time points corresponding to peak viral replication for each cell type, and infectious viral titers and viral genome copy numbers in the supernatant were quantified by plaque assay and qRT-PCR, respectively. RDV and GS-441524 potently inhibited SARS-CoV-2 replication in a dose-dependent manner in both cell types (Figure 2; Table 1 ). In Calu3 2B4 cells, both compounds displayed dose-dependent inhibition of viral replication, as determined by plaque assay (Figure 2B) and qRT-PCR (Figure 2C). RDV inhibited SARS-CoV-2 with an EC50 value of 0.28 μM and a 90% maximal effective concentration (EC90) value of 2.48 μM. The parent compound GS-441524 displayed similar potency: an EC50 value of 0.62 μM and EC90 value of 1.34 μM (Figure 2D; Table 1). EC50 values determined by quantification of viral genome copies showed a similar trend (Figure 2E; Table 1). Both compounds also displayed dose-dependent inhibition of viral replication in Vero E6 cells, as determined by infectious viral titers and genome copy numbers (Figure 2F). RDV inhibited SARS-CoV-2 with an EC50 value of 1.65 μM and EC90 value of 2.40 μM. However, in this cell type, GS-441524 was more potent (EC50 = 0.47 μM and EC90 = 0.71 μM) than RDV, as determined by infectious viral titers (Figure 2G; Table 1) and genome copy numbers (Figure 2H; Table 1). Thus, RDV inhibits SARS-CoV-2 more potently in Calu3 2B4 than in Vero E6 cells, and the relative potencies of the prodrug and parent nucleoside are cell type dependent.
Table 1.
Cell-Specific SARS-CoV-2 Potency and Metabolism
| Analysis | RDV |
GS-441524a |
|||
|---|---|---|---|---|---|
| Vero E6 | Calu3 2B4 | HAEb | Vero E6 | Calu3 2B4 | |
| Plaque assay EC50 (μM) | 1.65 | 0.28 | 0.010 | 0.47 | 0.62 |
| Genome copy EC50 (μM) | 1.49 | 0.60 | n.d. | 0.47 | 1.09 |
| RDV-TP at 24 hc (pmol/million cells) | 0.50 ± 0.15d | 2.17 ± 0.14e | 10.6 ± 5.3b | 1.78 ± 0.68d | 0.85 ± 0.16e |
GS-441524 was not tested for antiviral potency or RDV-TP levels in HAE cultures.
HAE antiviral potencies and RDV-TP levels were determined independently in differentiated cultures from two donors. RDV-TP levels in HAE cultures are presented as the mean ± SD of quadruplicate technical replicates from two donors.
Values represent mean ± SD from four independent experiments, each performed with duplicate samples.
Values represent mean ± SD from two independent experiments, each performed with duplicate samples.
RDV Is a Highly Potent Antiviral Inhibitor of SARS-CoV-2 in Primary HAE Cultures
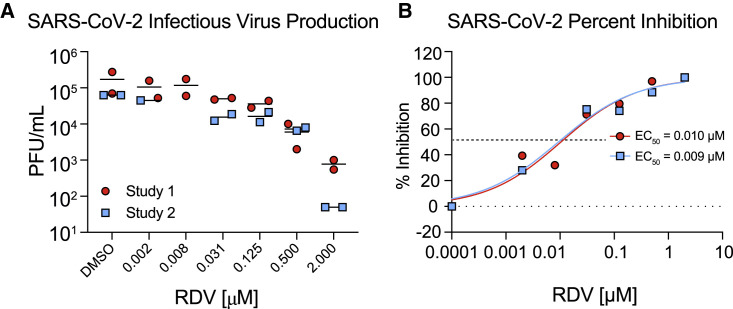
SARS-CoV-2 is known to replicate in the upper and lower airways in humans (Adachi et al., 2020; Wölfel et al., 2020). In addition, we have shown recently that SARS-CoV-2 replicates in human primary cells from the nasal to alveolar epithelium (Hou et al., 2020). To demonstrate the antiviral activity of RDV against SARS-CoV-2 in a human primary epithelial culture system, we performed antiviral assays in HAE cultures, which are grown on air-liquid interface and recapitulate the cellular complexity and physiology of the human conducting airway (Sims et al., 2005). HAE cultures are the model system in which we have amassed data for many other enzootic, emerging, and endemic CoVs, allowing comparison of SARS-CoV-2 data with previous reports (Sheahan et al., 2017, 2020a). In RDV-treated, SARS-CoV-2-infected HAE cultures, we observed a dose-dependent reduction in infectious virus production with more than 100-fold inhibition at the highest tested concentration (Figure 3 A). Importantly, RDV demonstrates potent antiviral activity with EC50 values of 0.0010 and 0.009 μM in two independent experiments (Figure 3B). Similar to previously reported studies, RDV did not cause cytotoxicity in HAE cultures across the dose range where we see potent antiviral effects (Figure S4; Sheahan et al., 2017). Together, these data demonstrate that RDV is potently antiviral against SARS-CoV-2 in primary human lung cultures with a selectivity index of more than 1,000.
Figure 3.
RDV Is Potently Antiviral against SARS-CoV-2 in Primary Human Airway Epithelial (HAE) Cultures
HAE cultures were infected with a SARS-CoV-2 clinical isolate (2019-nCoV/USA-WA1/2020) at an MOI of 0.5 PFU/cell for 2 h, and then the virus was removed and cultures were washed 3 times, followed by incubation at 37°C for 48 h.
(A) SARS-CoV-2 infectious virus production in two independent studies. The virus was titered via plaque assay in apical washes 48 hpi. Each symbol represents the titer from a single culture, and a line is drawn at the mean.
(B) Percent inhibition generated from the titer data in (A).
See also Figure S4.
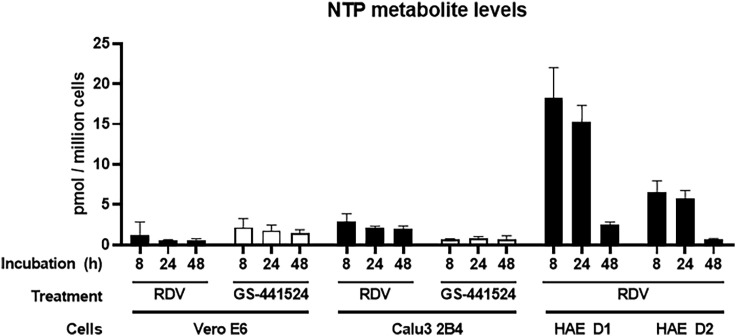
Antiviral Activities of RDV and GS-441524 Correlate with RDV-TP Metabolite Levels
Cell-type-specific expression of genes for enzymes that metabolize ribonucleoside analogs and their prodrugs can have a profound effect on activity (Eriksson, 2013; Koczor et al., 2012). Table 1 and prior studies (Bojkova et al., 2020; Choy et al., 2020; Jeon et al., 2020) demonstrate that the antiviral activity of RDV against SARS-CoV-2 is highly variable in different cell culture models. RDV and GS-441524 undergo intracellular conversion to the active metabolite RDV-TP, involving several metabolic steps (Figure S5), and the efficiency of each step might differ between cell types. Therefore, to reconcile the differences in antiviral activity of RDV and GS-441524 observed in our and other studies, we compared intracellular RDV-TP concentrations in Vero E6, Calu3 2B4, and HAE cultures following compound incubation. RDV-TP levels per million cells produced after 8- to 48-h treatment with RDV were substantially higher in primary HAE cultures than in Calu3 2B4 or Vero E6 cultures (Figure 4 ; Table 1; Tables S1 and S2). Given the primary nature of HAE cultures, we used cells from two independent donors with similar demographic profiles. RDV-TP was formed efficiently in both donor cultures following incubation with RDV, with a difference of less than 3-fold from each other. The lowest levels of RDV-TP were observed following RDV treatment of Vero E6 cells and were approximately 4- and 20-fold lower than those observed in Calu3 2B4 and HAE cultures, respectively. Incubation of Vero E6 cells with GS-441524 yielded 3.5-fold higher RDV-TP levels compared with incubation with RDV, corresponding to higher antiviral potency of GS-441524 relative to RDV (Table S1). In conclusion, the RDV-TP levels in the different cell types directly correlated with the antiviral potency of RDV against SARS-CoV-2. HAE cultures produced substantially higher levels of RDV-TP, which translated into markedly more potent antiviral activity of RDV (Table 1). Importantly, the metabolism of RDV in Vero E6 cells appeared to be less efficient, particularly in comparison with the HAE cultures, indicating that Vero E6 cells might not be an appropriate cell type to characterize the antiviral activity of RDV and, potentially, also other nucleotide prodrug-based antiviral agents.
Figure 4.
RDV-TP Levels in Vero E6, Calu3, and HAE Cultures
Vero E6 cells, Calu3 2B4 cells, and HAE cultures were incubated with RDV or GS-441524. At 8, 24, and 48 h of treatment, whole-cell extracts were prepared, and RDV-TP levels were quantified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) as described in STAR Methods. RDV-TP levels in Vero E6 and Calu3 2B4 cells represent mean ± SD from n = 2 or 4 independent experiments, respectively, each performed with duplicate samples. RDV-TP levels in HAEs represent the mean ± SD of four replicates for each individual donor (D1 and D2). See also Tables S1 and S2 and Figure S5.
RDV Is Active against the SARS-CoV-2 RdRp In Vivo
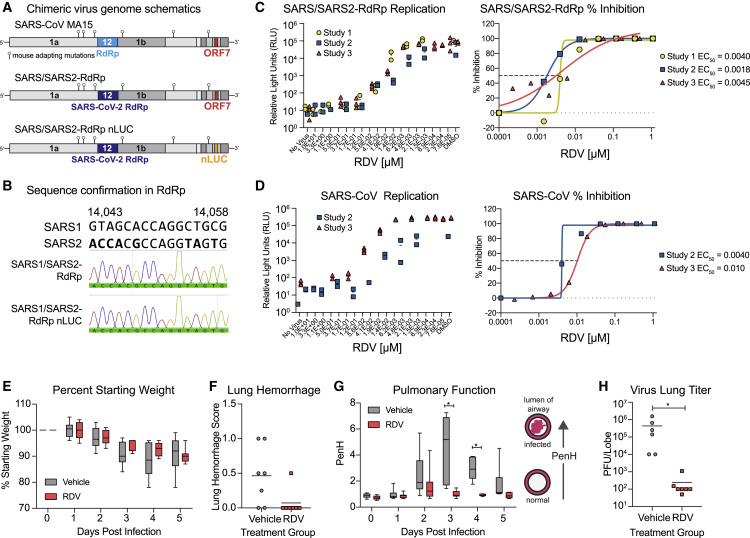
SARS-CoV-2 does not bind the murine ortholog of the human entry receptor (i.e., mouse angiotensin-converting enzyme-2 [mACE2]) to enter cells (Zhou et al., 2020b). To rapidly assess the therapeutic efficacy of RDV against SARS-CoV-2 in vivo, we constructed a chimeric mouse-adapted SARS-CoV variant encoding the target of RDV antiviral activity, the RdRp, of SARS-CoV-2 (SARS/SARS2-RdRp) (Figure 5 A). Although other chimeric replicase open reading frame (ORF) recombinant CoVs have been shown to be viable (Stobart et al., 2013), this is the first demonstration that the RdRp from a related but different CoV can support efficient replication of another. After recovery and sequence confirmation (Figure 5B) of recombinant chimeric viruses with and without a nanoluciferase reporter, we compared SARS-CoV and SARS/SARS2-RdRp replication and sensitivity to RDV in Huh7 cells. Replication of both viruses was inhibited similarly in a dose-dependent manner by RDV (SARS-CoV mean EC50 = 0.007 μM, SARS/SARS2-RdRp mean EC50 = 0.003 μM) (Figures 5C and 5D). We then determined the therapeutic efficacy of RDV against the SARS/SARS2-RdRp in mouse models employed for previous studies of RDV (Sheahan et al., 2017). Mice produce a serum esterase absent in humans, carboxyl esterase 1c (Ces1c), which dramatically reduces the half-life of RDV. Thus, to mirror pharmacokinetics observed in humans, mouse studies with RDV must be performed in transgenic C57BL/6 Ces1c −/− mice (Sheahan et al., 2017). We infected female C57BL/6 Ces1c −/− mice with 103 PFU SARS/SARS2-RdRp and initiated subcutaneous treatment with 25 mg/kg RDV BID 1 day post-infection (dpi). This regimen was continued until study termination. Although weight loss and lung hemorrhage did not differ significantly between vehicle- and RDV-treated animals (Figures 5E and 5F), we found differences in pulmonary function, as measured by whole-body plethysmography (WBP) between RDV- and vehicle-treated animals. The WBP metric, PenH, is a surrogate marker of pulmonary obstruction (Menachery et al., 2015a). Therapeutic RDV significantly ameliorated the loss of pulmonary function observed in the vehicle-treated group (Figure 5G). Importantly, RDV treatment dramatically reduced the lung viral load (Figure 5H). Taken together, these data demonstrate that therapeutically administered RDV can reduce virus replication and improve pulmonary function in an ongoing infection with a chimeric SARS-CoV/SARS-CoV-2 virus encoding the RdRp target of RDV.
Figure 5.
RDV Is Active against the SARS-CoV-2 RdRp In Vivo
The activity of RDV against the SARS-CoV-2 RdRp was evaluated using a chimeric SARS-CoV encoding the SARS-CoV-2 RdRp (SARS/SARS2-RdRp).
(A) Schematic of the recombinant SARS-CoV mouse-adapted MA15 strain chimeric virus genomes generated for these studies. SARS/SARS2-RdRp and SARS/SARS2-RdRp-nanoluciferase (nLUC) were constructed by exchanging the SARS-CoV MA15 RdRp with the SARS-CoV-2 RdRp. ORF7 is replaced by nLUC in SARS2-RdRp-nLUC.
(B) The presence of the SARS-CoV-2 RdRp was confirmed by Sanger sequencing in stocks of both recombinant chimeric viruses. Alignment of a stretch of nucleotides from the SARS-CoV and SARS-CoV-2 RdRp highlighting nucleotides that differ between the two strains is shown in boldface. These SARS-CoV-2 RdRp-specific nucleotides are present in both chimeric viruses used in this study, as determined by Sanger sequencing and shown in the histogram.
(C) SARS/SARS2-RdRp-nLUC replication in Huh7 cells in the presence of RDV (left) and associated percent inhibition (right).
(D) SARS-CoV replication in Huh7 cells in the presence of RDV (left) and associated percent inhibition (right).
(E) Percent starting weight of 17-week-old female Ces1c−/− mice infected intranasally with 1 × 103 PFU of SARS/SARS2-RdRp and treated subcutaneously with 25 mg/kg RDV or vehicle 1 day post-infection (dpi) and twice daily thereafter.
(F) Lung hemorrhage at 5 dpi.
(G) Pulmonary function by WBP. The PenH metric shown is a surrogate marker of pulmonary obstruction. p < 0.0001 as determined by two-way ANOVA with Sidak’s multiple comparisons test.
(H) Lung titer at 5 dpi as measured by plaque assay. p = 0.0012 by Mann-Whitney test.
In (E) and (G), boxes encompass the 25th–75th percentile, a line is drawn at the median, and whiskers represent the range.
Discussion
The COVID-19 pandemic has gravely illustrated the need for countermeasures against emerging epidemic and pandemic CoVs. Broad-spectrum antiviral drugs, antibodies, and vaccines are needed to combat the current pandemic and those that will emerge in the future. RDV shows potent activity against an array of genetically diverse CoVs as well as against unrelated emerging viruses like Ebola (Agostini et al., 2018; Brown et al., 2019; Sheahan et al., 2017, 2020a; Warren et al., 2016). In this study, we demonstrate that RDV and its parent nucleoside GS-441524 are active against SARS-CoV-2 in the physiologically relevant Calu3 2B4 cell line and that RDV has strong antiviral activity in primary human airway cultures. The potency of RDV was directly related to the intracellular concentration of the pharmacologically active TP metabolite, which was markedly higher in primary HAE cultures compared with human lung cells (Calu3 2B4) and monkey kidney cells (Vero E6). Our data are consistent with recent studies demonstrating important contributions of natural variation in host- and tissue-specific gene expression patterns and microbiome-specific contributions to drug metabolism, stability, and bioavailability in different tissues (Eriksson, 2013; Koczor et al., 2012). Modeling of RDV-TP onto the SARS-CoV-2 RdRp revealed that the positioning of RDV-TP into the active site closely resembled that of the cognate natural substrate ATP, consistent with efficient incorporation into RNA during replication of the viral genome. RDV decreased viral loads and improved lung function in mice infected with the SARS/SARS2-RdRp chimeric virus when treated at 1 dpi. This is the first rigorous demonstration of potent inhibition of SARS-CoV-2 in continuous and primary human lung cultures and the first study suggesting efficacy of RDV against SARS-CoV-2 in mice.
Previous studies of RDV anti-SARS-CoV-2 activity reported EC50 values of 0.77 μM as determined by quantification of genome copy number (Wang et al., 2020), 23.2 μM as determined by 50% tissue infectious dose (TCID50), 26.9 μM as determined by RNA copy number (Choy et al., 2020), and 0.65 μM as determined by cytopathic effect (CPE) (Runfeng et al., 2020), all in Vero E6 cells. The potency of RDV in Vero E6 cells (EC50, 1.65 μM) observed in our study is comparable with values reported by Wang et al. (2020) and Runfeng et al. (2020) but greater than reported by Choy et al. (2020). Sequence comparison of the nsp12 from the Seattle, Washington isolate used in this study versus SARS-CoV-2 isolates used in the previously mentioned studies assessing RDV potency did not reveal consensus changes in the nsp12 sequence, suggesting that any isolate-specific variation in RDV sensitivity is not likely to be due to differences in the RDV-TP interaction with the RdRp. Therefore, the differences in EC50 values might be partially explained by intrinsic differences in SARS-CoV-2 virus isolates, quantification methods, Vero cell lineages, and assay conditions, such as incubation period and virus input.
Although Vero E6 cells support robust replication of SARS-CoV-2, as illustrated here and elsewhere, our study emphasized that caution should be exercised when interpreting nucleoside prodrug potency experiments performed using Vero cell lineages. Nucleoside analog potency is dependent on metabolism into the active form. In contrast to the nucleoside GS-441524, RDV is a monophosphoramidate prodrug with moieties that mask the negative charges of its monophosphate group, which enhances its cellular uptake. Intracellularly, RDV is then rapidly metabolized to its monophosphate, which is efficiently converted to the active TP (Mehellou et al., 2018). Consistent with previous reports for SARS-CoV and MERS-CoV (Agostini et al., 2018), RDV showed a similar potency of inhibition against SARS-CoV-2 over GS-441524 in Calu3 2B4 cells. In contrast, RDV was 2-fold less potent than GS-441524 in Vero E6 cells. The relative potency of the two compounds was directly linked to intracellular concentration of the active TP metabolite, suggesting an altered uptake and/or intracellular metabolism of RDV, consistent with a previous report describing inefficient metabolism of the nucleotide prodrug sofosbuvir in Vero cells (Mumtaz et al., 2017). Drug potency in Vero E6 was similar whether quantified by infectious virus or genome copy number. In Calu3 2B4 cells, the potency determined by qRT-PCR was about 2-fold lower than when quantified by plaque assay. It is possible that qRT-PCR, which was developed to detect nucleocapsid (N) RNA, also detects packaged subgenomic RNAs and defective genomes in addition to full-length genomes, resulting in underestimation of the reduction in infectious titer. Notably, the potency of RDV against SARS-CoV encoding the SARS-CoV-2 RdRp in Huh7 cells was more than 100-fold higher than that of RDV against bona fide SARS-CoV-2 in Huh7 and Calu3 2B4 cells. This difference could be due to infectivity, which is driven by the SARS-CoV instead of the SARS-CoV-2 spike protein. In addition, SARS-CoV infects Huh7 cells at low frequency at the MOI used in this study and does not appear to spread throughout the culture over the course of the experiment. The number of Huh7 cells in which the virus replicates is relatively lower compared with Calu3 2B4 cells, possibly enhancing the overall antiviral effect of RDV in Huh7 compared with Calu3 2B4 cells. Interestingly, the antiviral potency of RDV against SARS-CoV-2 in HAE cultures was comparable with SARS-CoV and MERS-CoV (Sheahan et al., 2017), which is consistent with the high conservation of the RdRp active site across these different CoVs. Together, these results emphasize the need for careful selection and use of cell types and methods to study the potency and efficacy of nucleoside analogs.
The target of RDV antiviral activity is the viral RdRp. To mirror the pharmacokinetic exposures observed in humans, RDV studies in mice must be performed in Ces1c −/− animals (Sheahan et al., 2017). In addition, SARS-CoV-2 does not readily infect wild-type mice because of incompatibilities between the virus spike and the murine ACE2 ortholog, which serves as the SARS-CoV-2 entry receptor (Wan et al., 2020). Breeding of a double-transgenic (hACE2, Ces1c −/−) mouse for use in RDV efficacy studies is ongoing. To rapidly assess the therapeutic efficacy of RDV against SARS-CoV-2, we constructed SARS/SARS2-RdRp. Virus entry, tropism, and pathogenesis of this chimeric virus are driven by the parental mouse-adapted SARS-CoV virus. Similar to our previous studies with SARS-CoV (Sheahan et al., 2017), we now show that therapeutic administration of RDV 1 dpi can reduce the viral load and improve pulmonary function in mice. The kinetics of SARS-CoV replication and disease are notably compressed in mice compared with humans, where the virus titer peaks 10–15 days after onset of symptoms (Hung et al., 2004; Peiris et al., 2004). In comparison, initial reports suggest that SARS-CoV-2 replication peaks around 5–6 days after symptom onset, just prior to onset of dyspnea (Pan et al., 2020; Zhou et al., 2020a). Importantly, a recent preprint described the therapeutic efficacy of RDV against SARS-CoV-2 in rhesus macaques, where RDV treatment reduced respiratory pathology and viral loads in bronchoalveolar lavage fluid, consistent with our study with a chimeric virus in mice (Williamson et al., 2020). Prior to the emergence of pandemic SARS-CoV-2, RDV was evaluated in phase 1 clinical trials as well as phase 2 randomized controlled trials to treat acute Ebola virus disease in the Democratic Republic of Congo (DRC), and human safety data are available (Mulangu et al., 2019). Thus, our preclinical development of RDV supported immediate compassionate use of RDV for severely ill COVID-19 patients with promising early results (Grein et al., 2020). Furthermore, a preliminary report of a large-scale, double-blind, randomized, placebo-controlled trial in hospitalized adults suggests that RDV shortened the time to recovery (Beigel et al., 2020). This and other ongoing phase III randomized controlled trials for treatment of patients with COVID-19 will ultimately determine efficacy, safety, and optimal dosing of RDV in patients with different stages of COVID-19.
Despite worldwide drug discovery efforts and over 300 active clinical evaluations of potential treatments, no effective countermeasure currently exists to combat COVID-19 (Sanders et al., 2020) or likely future CoV pandemics. Large-scale deployment of antiviral monotherapies creates a high risk for emergence of drug resistance. Our previous work demonstrates that CoV resistance to RDV is generated slowly and conferred by two mutations in the RdRp. In addition, RDV-resistant CoVs exhibit reduced replication capacity and are also more sensitive to another potently active nucleoside analog inhibitor, β-D-N4-hydroxycytidine (NHC; EIDD-1931/2801) (Agostini et al., 2019; Sheahan et al., 2020b). Therapies combining direct-acting antivirals (DAAs), such as RDV and EIDD-2801, along with other DAAs, such as antibodies and protease inhibitors that target different stages of the viral replication cycle, could be considered for counteracting resistance if it emerges in patients treated with antiviral monotherapy. In addition, attention should be given to combining DAAs with anti-inflammatory drugs to potentially extend the treatment window during which DAAs can improve outcomes. With zoonotic relatives of SARS-CoV, SARS-CoV-2, and MERS-CoV continuing to circulate in bat species, more outbreaks of novel CoVs are expected (Menachery et al., 2015b, 2016). Identification and evaluation of broadly efficacious, robust anti-CoV therapies are thus urgently needed at present and in the future.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| SARS-CoV-2 2019-nCoV/USA-WA1/2020 clinical isolate | CDC Atlanta | Accession number MN985325 |
| SARS/SARS2-RdRp | This paper | N/A |
| SARS/SARS2-RdRp-nLUC | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Remdesivir | Gilead Sciences | GS-5734 (RDV) |
| GS-441524 | Gilead Sciences | GS-441524 |
| Critical Commercial Assays | ||
| Nano-Glo® Luciferase Assay System | Promega | N1110 |
| CellTiter-Glo® Luminescent Cell Viability Assay | Promega | G7570 |
| TaqMan Fast Virus 1-Step Master Mix | Life Technologies | 4444436 |
| mMESSAGE mMACHINE T7 Transcription Kit | ThermoFisher Scientific | AM1344 |
| Deposited Data | ||
| PDB SARS-CoV-2 RDV-TP | This manuscript | Mendeley data https://doi.org/10.17632/x3sw5z2bg9.1 |
| Experimental Models: Cell Lines | ||
| Vero | ATCC | CCL-81 |
| Vero C1008 [Vero 76, clone E6, Vero E6] | ATCC | CRL-1586 |
| Calu3 2B4 cells | Chien-Te (Kent) Tseng, UTMB | N/A |
| Huh7 | Dr. Mark Heise, UNC | N/A |
| Primary HAE cell cultures | Tissue Procurement and Cell Culture Core Laboratory in the Marsico Lung Institute/Cystic Fibrosis Research Center, UNC | N/A |
| Differentiated HAE cultures | MatTek Corporation; Ashland, MA | AIR-100 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6 Ces1c−/− mice | Jackson Labs | Stock 014096 |
| Oligonucleotides | ||
| N gene forward primer: 5′-GACCCCAAAATCAGCGAAAT | Integrated DNA Technologies | 10006606 |
| N gene reverse primer: 5′-TCTGGTTACTGCCAGTTGAATCTG | Integrated DNA Technologies | 10006606 |
| N gene probe: 5′- FAM-ACCCC GCATTACGTTTGGTGGACC-BHQ1 |
Integrated DNA Technologies | 10006606 |
| T7 forward primer to create in vitro transcription template 5′- TAATACG ACTCACTATAGGGATGTCTGATA |
This manuscript | N/A |
| ATGGACCCCA | ||
| T7 reverse primer to create in vitro transcription template: 5′- TTAGG CCTGAGTTGAG |
This manuscript |
N/A |
| TCAG | ||
| Recombinant DNA | ||
| SARS-CoV-2 N gene positive control plasmid | Integrated DNA Technologies | 10006625 |
| Software and Algorithms | ||
| Prime | Schrödinger, LLC, New York, NY, 2020 | https://www.schrodinger.com/prime |
| Macromodel | Schrödinger, LLC, New York, NY, 2020 | https://www.schrodinger.com/macromodel |
| CLC Main Workbench 20 | QIAGEN | https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-clc-genomics-workbench/ |
| Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Andrea Pruijssers (ardina.pruijssers@vumc.org).
Materials Availability
Plasmids generated in this study are available upon request.
Data and Code Availability
The pdb file containing SARS-CoV-2 model showing RDV-TP in the active site has been deposited to Mendeley Data: https://doi.org/10.17632/x3sw5z2bg9.1.
Experimental Model and Subject Details
Cells, viruses, and compounds
Vero (ATCC CCL-81) and Vero E6 (ATCC CRL-1586) cells were purchased from ATCC and cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (GIBCO, ThermoFisher Scientific) or 10% FCS fetal clonal serum (FCS)(HyClone, GE Life Sciences), 100 U/ml penicillin and streptomycin (GIBCO, ThermoFisher Scientific), and 0.25 μM amphotericin B (Corning). Human hepatoma (Huh7) cells were provided by Dr. Mark Heise at UNC Chapel Hill and grown in DMEM supplemented with 10% FBS (Hyclone) and 1 × antibiotic-antimycotic (GIBCO, ThermoFisher Scientific). Calu3 2B4 cells (Yoshikawa et al., 2010) were cultured in DMEM supplemented with 20% FBS, 100 U/mL penicillin and streptomycin, and 0.25 μM amphotericin B.
Primary HAE cell cultures used in antiviral activity assays were obtained from the Tissue Procurement and Cell Culture Core Laboratory in the Marsico Lung Institute/Cystic Fibrosis Research Center at UNC. All assays in this report were performed using a single HAE cell donor. Human tracheobronchial epithelial cells provided by Dr. Scott Randell were obtained from airway specimens resected from patients undergoing surgery under University of North Carolina Institutional Review Board-approved protocols (#03-1396) by the Cystic Fibrosis Center Tissue Culture Core. Primary cells were expanded to generate passage 1 cells and passage 2 cells were plated at a density of 250,000 cells per well on Transwell-COL (12mm diameter) supports (Corning). Human airway epithelium cultures (HAE) were generated by provision of an air-liquid interface for 6 to 8 weeks to form well-differentiated, polarized cultures that resembled in vivo pseudostratified mucociliary epithelium (Fulcher et al., 2005).
Clinical specimens of SARS-CoV-2 from a case-patient who acquired COVID-19 during travel to China and diagnosed in Washington State, USA upon return were collected as described (Holshue et al., 2020). Virus isolation from this patient’s specimens was performed as described in Harcourt et al. (2020). The sequence is available through GenBank (accession number MN985325.1). A passage 3 stock of the SARS-CoV-2 Seattle isolate was obtained from the CDC and passed twice in Vero E6 cells to generate high-titer passage 5 stock for experiments described in this manuscript.
SARS-CoV expressing GFP (GFP replaces ORF7) was created from molecular cDNA clones as described (Scobey et al., 2013; Sims et al., 2005). To create SARS-CoV expressing nanoluciferase (nLUC), the gene for GFP was replaced with nLUC and isolated using our existing mouse adapted SARS-CoV (MA15) SARS-CoV Urbani molecular clone (Yount et al., 2003). A synthetic cDNA encoding the SARS-CoV-2 RdRp (Integrated DNA Technologies) was cloned into SARS MA15 D fragment using StuI (5′) and BsaI (3′) via Gibson assembly. The resultant plasmids were sequence confirmed and then utilized to generate recombinant virus with or without nanoluciferase as described. Recombinant virus stocks were confirmed to harbor SARS-CoV-2 RdRp by Sanger sequencing.
Remdesivir (RDV; GS-5734) and GS-441524 were synthesized by the Department of Medicinal Chemistry, Gilead Sciences (Foster City, CA).
Method Details
Modeling
A model of the elongating SARS-CoV-2 polymerase complex was generated based on a homology model which used the cryo-EM structure of apo SARS-CoV as a template (PDB: 6NUR; Kirchdoerfer and Ward, 2019) as described previously (Gordon et al., 2020b). RNA primer and template, a substrate ATP and two catalytic Mg2+ ions were oriented in the structure based on alignment to a ternary X-ray structure of HCV NS5B (PDB: 4WTD; Appleby et al., 2015). The structure was then optimized with a series of constrained minimizations. To this structure we aligned the new cryo-EM structure of the SARS-CoV-2 replication complex. As the SARS-CoV-2 structure does not significantly differ from the SARS-CoV structure, rather than a complete replacement of the model, we incorporated only those residues that had not been resolved in the previous structure (residues 31-116). Additional optimization, particularly of the RNA in the exit channel, was done following the previously outlined procedures. After optimization of the ATP structure, RDV-TP was modeled into the active site and minimized. Models of the V557L and F480L mutants and the other coronavirus models reported here were generated based on this final model. All work was carried out using Prime and Macromodel (Schrödinger, LLC, New York, NY, 2020). 3D coordinates of the SARS-CoV-2/RDV-TP model are provided in the Supplementary Material.
Sequence alignments
Coronavirus nsp12 sequence alignment was generated using CLC Workbench (QIAGEN) from sequences downloaded from the NCBI website (Accession numbers MN985325.1 and MT123290.1) and from GISAID’s EpiFlu Database (Elbe and Buckland-Merrett, 2017; Shu and McCauley, 2017): Accession ID EPI_ISL_402124; virus name hCoV-19/Wuhan/WIV04/2019; Location: Asia / China / Hubei / Wuhan; Collection date 2019-12-30 Originating lab Wuhan Jinyintan Hospital; Submitting lab Wuhan Institute of Virology, Chinese Academy of Sciences; Authors Peng Zhou, Xing-Lou Yang, Ding-Yu Zhang, Lei Zhang, Yan Zhu, Hao-Rui Si, Zhengli Shi. Accession ID EPI_ISL_412028; virus name hCoV-19/Hong Kong/VM20001061/2020; Location Asia / Hong Kong; Collection date 2020-01-22; Originating lab Hong Kong Department of Health; Submitting lab: School of Public Health, The University of Hong Kong; Authors Dominic N.C. Tsang, Daniel K.W. Chu, Leo L.M. Poon, Malik Peiris.
Replication in different cell types
Vero E6, Vero, Calu3 2B4, and Huh7 cells were seeded in 24 well plates and allowed to adhere for 24 h. Cells were adsorbed with 100 μl SARS-CoV-2 in gel saline for 30 minutes (min) at 37°C with manual rocking every 10 min. Virus inoculum was removed, cells were washed in PBS, and 0.5 mL medium was added to each well. Supernatant was collected at 0, 24, 48, and 72 h post-infection, and infectious viral titer in supernatants was determined by plaque assay.
Antiviral activity assays
Vero E6 cells were seeded at 1 × 105 cells per well, and Calu3 2B4 cells were seeded at 2 × 105 cells per well in 24-well plates (Corning). Cells were allowed to adhere for 16-24 h. Drugs were dissolved in DSMO and serially diluted in DMSO to achieve 1000x final concentration. Equal volumes of each 1000x concentration were further diluted 1000-fold in medium up to 2 h before start of the infection. Cells were adsorbed at MOI = 0.01 PFU/cell with SARS-CoV-2 in gel saline for 30 min at 37°C. Plates were rocked manually to redistribute the inoculum every 10 minutes. Viral inoculum was removed, and cells were washed with pre-warmed PBS+/+ (Corning) for 5 minutes. PBS+/+ was removed, and medium containing dilutions of RDV, GS-441524, or vehicle (DMSO) was added. Cells were incubated at 37°C. At 48 (Vero E6) or 72 (Calu3 2B4) hpi, supernatants were harvested and processed for plaque assay and RT-qPCR.
Huh7 cells were plated at a density of 8 × 104 cells per well. Twenty-four hours later, fresh medium was added. Triplicate wells of cells were infected for 1 h at 37°C with SARS1/SARS2-RdRp-nLUC or SARS-CoV-nLUC diluted 1:100 in culture medium. Virus was removed, cultures were rinsed once with medium, and fresh medium containing dilutions of RDV or vehicle (DMSO) was added. DMSO (0.05%) was constant in all conditions. At 48 hpi, virus replication was measured by nLUC assay (Promega) using a SpectraMax plate reader (Molecular Devices).
Before infection, HAE cultures (approximately 1 × 106 cells per well) were washed with phosphate-buffered saline (PBS) and moved into air-liquid interface medium containing various doses of RDV ranging from 0.00260 to10 mM (final DMSO, < 0.05%). Cultures were infected with SARS-CoV-2 clinical isolate (2019-nCoV/USA-WA1/2020) at MOI = 0.5 PFU/cell for 2 h at 37°C, after which virus was removed and cultures were washed three times with PBS, followed by incubation at 37°C for 48 h. The apical surface of each culture was washed with PBS and collected for virus titration, measured as plaque-forming units (PFU) as previously described for SARS-CoV (Scobey et al., 2013; Sims et al., 2005).
Cytotoxicity Assays
Cells were seeded at a density of 15,000 cells/well (Vero E6) or 30,000 cells/well (Calu3 2B4) in a white-wall clear-bottom 96-well plate (Corning) and incubated at 37°C overnight. Medium was removed, and serial dilutions of drug in medium were added to each well (see “Antiviral activity assays”). Cytotoxicity was determined using CellTiterGlo Cell Viability Assay (Promega) according to manufacturer’s instructions at 48 h (Vero E6) or 72 h (Calu3). HAE cultures were treated with the same concentration range of drug in Transwell plates (Corning). Cytotoxicity in HAE was previously determined by RT-qPCR of TRIzol-extracted RNA (Sheahan et al., 2017)
Quantification of infectious viral titer
Approximately 1 × 106 Vero E6 cells/well were seeded in 6-well tissue culture plates (Corning) and allowed to grow to confluence for 48 h. Medium was removed, and 200 μL of 10-fold serial dilutions of virus-containing supernatants in gel saline were adsorbed in duplicate for 30 min at 37°C. Plates were rocked manually to redistribute inoculum every 10 minutes. Cells were overlaid with a 1:1 mixture of 2x DMEM and 2% agar in ddH2O and incubated at 37°C. Plaques were enumerated in unstained monolayers at 48-72 hpi using a light box.
Quantification of viral RNA copy number
Cell supernatants were harvested in TRIzol LS reagent (Invitrogen), and RNA was purified following phase separation by chloroform as recommended by the manufacturer. RNA in the aqueous phase was collected and further purified using PureLink RNA Mini Kits (Invitrogen) according to manufacturer’s protocol. Viral RNA was quantified by reverse-transcription quantitative PCR (RT-qPCR) on a StepOnePlus Real-Time PCR System (Applied Biosystems) using TaqMan Fast Virus 1-Step Master Mix chemistry (Applied Biosystems). SARS-CoV-2 N gene RNA was amplified using forward (5′-GACCCCAAAATCAGCGAAAT) and reverse (5′-TCTGGTTACTGCCAGTTGAATCTG) primers and probe (5′- FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1) designed by the United States Centers for Disease Control and Prevention (oligonucleotides produced by IDT, cat# 10006606). RNA copy numbers were interpolated from a standard curve produced with serial 10-fold dilutions of N gene RNA. Briefly, SARS-CoV-2 N gene positive control plasmid (IDT, cat# 10006625) served as template to PCR-amplify a 1280 bp product using forward (5′-TAATACGACTCACTATAGGGATGTCTGATAATGGACCCCA) and reverse (5′- TTAGGCCTGAGTTGAGTCAG) primers that appended a T7 RNA polymerase promoter to the 5′ end of the complete N ORF. PCR product was column purified (Promega) for subsequent in vitro transcription of N RNA using mMESSAGE mMACHINE T7 Transcription Kit (ThermoFisher Scientific) according to manufacturer’s protocol. N RNA was purified using RNeasy mini kit (QIAGEN) according to manufacturer’s protocol, and copy number was calculated using SciencePrimer.com cop number calculator.
In vitro metabolism of RDV and GS-441524
Calu3 2B4 or Vero E6 cells were seeded in a 6-well plate at 8.0 × 105 or 3.5 X 105 cells/well, respectively. Twenty-four hours later, cell culture media was replaced with media containing 1 μM RDV (GS-5734) or GS-441524 and incubated at 37°C. Differentiated HAE cultures from two healthy donors (MatTek Corporation; Ashland, MA) were maintained with media replacement every other day for 1 week. The HAE donors were 56- and 62-year-old females of the same race. At the time of treatment, media was replaced on the basal side of the transwell HAE culture, while the apical surface media was replaced with 200 μL media containing 1 μM RDV. At 8, 24 and 48h post drug addition to all cultures, cells were washed 3 times with ice-cold tris-buffered saline, scraped into 0.5 mL ice-cold 70% methanol and stored at −80°C. Extracts were centrifuged at 15,000 x g for 15 minutes and supernatants were transferred to clean tubes for evaporation in a MiVac Duo concentrator (Genevac). Dried samples were reconstituted in mobile phase A containing 3 mM ammonium formate (pH 5) with 10 mM dimethylhexylamine (DMH) in water for analysis by LC-MS/MS, using a multi-stage linear gradient from 10% to 50% acetonitrile in mobile phase A at a flow rate of 300 μL/min. Analytes were separated using a 50 × 2 mm, 2.5 μm Luna C18(2) HST column (Phenomenex) connected to an LC-20ADXR (Shimadzu) ternary pump system and HTS PAL autosampler (LEAP Technologies). Detection was performed on a Qtrap 6500+ (AB Sciex) mass spectrometer operating in positive ion and multiple reaction monitoring modes. Analytes were quantified using a 7-point standard curve ranging in concentration from 0.156 to 40 pmol prepared in extracts from untreated cells. For normalization by cell number, multiple untreated Calu3 or Vero E6 culture wells were counted at each time point. HAE cells were counted at the 24-h time point and the counts for other time points were determined by normalized to endogenous ATP levels for accuracy.
Formulations for in vivo studies
RDV was solubilized at 2.5 mg/mL in vehicle containing 12% sulfobutylether-β-cyclodextrin sodium salt in water (with HCl/NaOH) at pH 5.0.
In vivo efficacy studies
All animal experiments were performed in accordance with the University of North Carolina at Chapel Hill Institutional Animal Care and Use Committee policies and guidelines. To achieve a pharmacokinetic profile similar to that observed in humans, we performed therapeutic efficacy studies in Ces1c −/− mice (stock 014096, The Jackson Laboratory), which lack a serum esterase not present in humans that dramatically reduces RDV half-life (Sheahan et al., 2017). 17 week-old female Ces1c −/− mice were anaesthetized with a mixture of ketamine/xylazine and intranasally infected with 103 PFU SARS1/SARS2-RdRp in 50 μL. One dpi, vehicle (n = 7) and RDV (n = 7) dosing was initiated (25 mg/kg subcutaneously) and continued every 12 h until the end of the study at five dpi. To monitor morbidity, mice were weighed daily. Pulmonary function testing was performed daily by whole body plethysmography (WBP) (Data Sciences International) (Sheahan et al., 2017). At five dpi, animals were sacrificed by isoflurane overdose, lungs were scored for lung hemorrhage, and the inferior right lobe was frozen at −80°C for viral titration via plaque assay on Vero E6 cells. Lung hemorrhage is a gross pathological phenotype readily observed by the naked eye and driven by the degree of virus replication, where lung coloration changes from pink to dark red (Sheahan et al., 2017, 2020a). For the plaque assay, 5 × 105 Vero E6 cells/well were seeded in 6-well plates. The following day, medium was removed, and monolayers were adsorbed at 37°C for one h with serial dilutions of sample ranging from 10−1 to 10−6. Cells were overlayed with 1X DMEM, 5% Fetal Clone 2 serum, 1 × antibiotic-antimycotic, 0.8% agarose. Viral plaques were enumerated three days later.
Quantification and Statistical Analysis
Mathematical and statistical analyses
The EC50 value was defined in GraphPad Prism 8 as the concentration at which there was a 50% decrease in viral replication relative to vehicle alone (0% inhibition). Curves were fitted based on four parameter non-linear regression analysis. All statistical tests were executed using GraphPad Prism 8. Statistical details of experiments are described in the figure legends.
Acknowledgments
This project was funded in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services awards 1U19AI142759 (Antiviral Drug Discovery and Development Center) (to M.R.D. and R.S.B.), R01AI132178 and R01AI132178-03S1 (to T.P.S. and R.S.B.), and R01AI108197 (to M.R.D. and R.S.B.). D.R.M. was funded by T32AI007151 and a Burroughs Wellcome Fund Postdoctoral Enrichment Program award. The Marsico Lung Institute Tissue Procurement and Cell Culture Core is supported by NIH grant DK065988 and Cystic Fibrosis Foundation grant BOUCHE15RO. We also are grateful for support from the Dolly Parton COVID-19 Research Fund, the VUMC Office of Research, and the Elizabeth B. Lamb Center for Pediatric Research at Vanderbilt University. The graphical abstract was created with BioRender. We thank Dr. Natalie Thornburg at the Centers for Disease Control and Prevention in Atlanta, Georgia for providing the stock of SARS-CoV-2 used in this study. We thank VUMC and UNC Environmental Health and Safety personnel for ensuring that our work is performed safely and securely. We also thank Facilities Management personnel for tireless commitment to excellent facility performance and our grant management teams for administrative support of our research operations.
Author Contributions
A.J.P. and T.P.S. conceived, designed, and performed experiments and management and coordinated responsibility for research activity planning and execution. A.J.P., J.K.P., J.P.B., and T.P.S. wrote the manuscript. A.S.G., A.S., S.R.L., K.H.D., B.L.Y., M.L.A., L.J.S., J.D.C., X.L., T.M.H., K.G., D.R.M., A.J.B., R.L.G., J.K.P., V.D.P., J.P., B.M., D.B., and E.M. performed experiments. T.P.S., D.R.M., R.S.B., A.J.P., J.D.C., and M.R.D. secured funding. D.P.P. and T.C. provided reagents. J.D.C., J.Y.F., J.P.B., D.P.P., T.C., R.S.B., and M.R.D. edited the manuscript and provided expertise and feedback.
Declaration of Interests
The authors affiliated with Gilead Sciences, Inc. are employees of the company and may own company stock.
Published: July 7, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107940.
Supplemental Information
References
- Adachi T., Chong J.-M., Nakajima N., Sano M., Yamazaki J., Miyamoto I., Nishioka H., Akita H., Sato Y., Kataoka M. Clinicopathologic and Immunohistochemical Findings from Autopsy of Patient with COVID-19, Japan. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2609.201353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. MBio. 2018;9:e00221. doi: 10.1128/mBio.00221-18. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M.L., Pruijssers A.J., Chappell J.D., Gribble J., Lu X., Andres E.L., Bluemling G.R., Lockwood M.A., Sheahan T.P., Sims A.C. Small-Molecule Antiviral β-d-N4-Hydroxycytidine Inhibits a Proofreading-Intact Coronavirus with a High Genetic Barrier to Resistance. J. Virol. 2019;93:e01348-19. doi: 10.1128/JVI.01348-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleby T.C., Perry J.K., Murakami E., Barauskas O., Feng J., Cho A., Fox D., 3rd, Wetmore D.R., McGrath M.E., Ray A.S. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015;347:771–775. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- Arabi Y.M., Balkhy H.H., Hayden F.G., Bouchama A., Luke T., Baillie J.K., Al-Omari A., Hajeer A.H., Senga M., Denison M.R. Middle East Respiratory Syndrome. N. Engl. J. Med. 2017;376:584–594. doi: 10.1056/NEJMsr1408795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S. Remdesivir for the Treatment of Covid-19 – Preliminary Report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2007764. Published online May 22, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., McGreig J.E., McLaughlin K.-M., Masterson S.G., Widera M., Krähling V., Ciesek S., Wass M.N., Michaelis M., Cinatl J. SARS-CoV-2 and SARS-CoV differ in their cell tropism and drug sensitivity profiles. bioRxiv. 2020 doi: 10.1101/2020.04.03.024257. [DOI] [Google Scholar]
- Brown A.J., Won J.J., Graham R.L., Dinnon K.H., 3rd, Sims A.C., Feng J.Y., Cihlar T., Denison M.R., Baric R.S., Sheahan T.P. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res. 2019;169:104541. doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-Yeung M., Xu R.-H. SARS: epidemiology. Respirology. 2003;8(Suppl):S9–S14. doi: 10.1046/j.1440-1843.2003.00518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. Pathogenicity and transmissibility of 2019-nCoV-A quick overview and comparison with other emerging viruses. Microbes Infect. 2020;22:69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho A., Saunders O.L., Butler T., Zhang L., Xu J., Vela J.E., Feng J.Y., Ray A.S., Kim C.U. Synthesis and antiviral activity of a series of 1′-substituted 4-aza-7,9-dideazaadenosine C-nucleosides. Bioorg. Med. Chem. Lett. 2012;22:2705–2707. doi: 10.1016/j.bmcl.2012.02.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy K.-T., Wong A.Y., Kaewpreedee P., Sia S.-F., Chen D., Hui K.P.Y., Chu D.K.W., Chan M.C.W., Cheung P.P., Huang X. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. USA. 2020;117:6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S. Is the expression of deoxynucleoside kinases and 5′-nucleotidases in animal tissues related to the biological effects of nucleoside analogs? Curr. Med. Chem. 2013;20:4241–4248. doi: 10.2174/0929867311320340004. [DOI] [PubMed] [Google Scholar]
- Fulcher M.L., Gabriel S., Burns K.A., Yankaskas J.R., Randell S.H. Well-differentiated human airway epithelial cell cultures. Methods Mol. Med. 2005;107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P., Peersen O.B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA. 2010;107:22505–22510. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.-X. Compassionate Use of Remdesivir for Patients with Severe Covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with 2019 Novel Coronavirus Disease, United States. Emerg. Infect. Dis. 2020;26(6) doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Washington State 2019-nCoV Case Investigation Team First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., Kato T., Lee R.E., Yount B.L., Mascenik T.M. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020 doi: 10.1016/j.cell.2020.05.042. Published online May 27, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung I.F.N., Cheng V.C.C., Wu A.K.L., Tang B.S.F., Chan K.H., Chu C.M., Wong M.M.L., Hui W.T., Poon L.L.M., Tse D.M.W. Viral loads in clinical specimens and SARS manifestations. Emerg. Infect. Dis. 2004;10:1550–1557. doi: 10.3201/eid1009.040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon S., Ko M., Lee J., Choi I., Byun S.Y., Park S., Shum D., Kim S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020;64:e00819-20. doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koczor C.A., Torres R.A., Lewis W. The role of transporters in the toxicity of nucleoside and nucleotide analogs. Expert Opin. Drug Metab. Toxicol. 2012;8:665–676. doi: 10.1517/17425255.2012.680885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M.K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A.L., Flint M., McMullan L.K., Siegel D., Clarke M.O. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017;7:43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehellou Y., Rattan H.S., Balzarini J. The ProTide Prodrug Technology: From the Concept to the Clinic. J. Med. Chem. 2018;61:2211–2226. doi: 10.1021/acs.jmedchem.7b00734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Gralinski L.E., Baric R.S., Ferris M.T. New Metrics for Evaluating Viral Respiratory Pathogenesis. PLoS ONE. 2015;10:e0131451. doi: 10.1371/journal.pone.0131451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.-Y., Donaldson E.F. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. USA. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulangu S., Dodd L.E., Davey R.T., Jr., Tshiani Mbaya O., Proschan M., Mukadi D., Lusakibanza Manzo M., Nzolo D., Tshomba Oloma A., Ibanda A., PALM Writing Group. PALM Consortium Study Team A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. N. Engl. J. Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumtaz N., Jimmerson L.C., Bushman L.R., Kiser J.J., Aron G., Reusken C.B.E.M., Koopmans M.P.G., van Kampen J.J.A. Cell-line dependent antiviral activity of sofosbuvir against Zika virus. Antiviral Res. 2017;146:161–163. doi: 10.1016/j.antiviral.2017.09.004. [DOI] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10(12, Suppl):S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runfeng L., Yunlong H., Jicheng H., Weiqi P., Qinhai M., Yongxia S., Chufang L., Jin Z., Zhenhua J., Haiming J. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol. Res. 2020;156:104761. doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): A Review. JAMA. 2020 doi: 10.1001/jama.2020.6019. Published online April 13, 2020. [DOI] [PubMed] [Google Scholar]
- Scobey T., Yount B.L., Sims A.C., Donaldson E.F., Agnihothram S.S., Menachery V.D., Graham R.L., Swanstrom J., Bove P.F., Kim J.D. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2013;110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A., Le N.T.-T., Selisko B., Eydoux C., Alvarez K., Guillemot J.-C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Res. 2020;178:104793. doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Leist S.R., Schäfer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., 3rd, Stevens L.J. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12:eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims A.C., Baric R.S., Yount B., Burkett S.E., Collins P.L., Pickles R.J. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 2005;79:15511–15524. doi: 10.1128/JVI.79.24.15511-15524.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobart C.C., Sexton N.R., Munjal H., Lu X., Molland K.L., Tomar S., Mesecar A.D., Denison M.R. Chimeric exchange of coronavirus nsp5 proteases (3CLpro) identifies common and divergent regulatory determinants of protease activity. J. Virol. 2013;87:12611–12618. doi: 10.1128/JVI.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020;94:e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson B.N., Feldmann F., Schwarz B., Meade-White K., Porter D.P., Schulz J., van Doremalen N., Leighton I., Yinda C.K., Pérez-Pérez L. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.15.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T., Hill T.E., Yoshikawa N., Popov V.L., Galindo C.L., Garner H.R., Peters C.J., Tseng C.-T.K. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS ONE. 2010;5:e8729. doi: 10.1371/journal.pone.0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Fritz E.A., Hensley L.E., Jahrling P.B., Prentice E., Denison M.R., Geisbert T.W., Baric R.S. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2003;100:12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The pdb file containing SARS-CoV-2 model showing RDV-TP in the active site has been deposited to Mendeley Data: https://doi.org/10.17632/x3sw5z2bg9.1.