Highlights
-
•
The prevalence of infected patients with SARS-CoV-2 or other viruses is unknown.
-
•
We analyzed SARS-CoV-2 and 17 viruses in 191 patients with cold-like symptom in Japan.
-
•
21 % of patient (n = 40) was infected at least one virus, including SARS-CoV-2 (n = 8).
-
•
No influenza virus was observed in this study.
-
•
The data implies different infectivity between influenza and SARS-CoV2.
Keywords: SARS-CoV-2, Influenza, Covid-19, PCR, Infection, FilmArray
Abstract
Background
Severe acute respiratory coronavirus 2 (SARS-CoV-2) has spread and caused death worldwide. Preventive measures and infection control are underway, and some areas show signs of convergence. Other viruses in addition to SARS-CoV-2 cause cold-like symptoms and spread in the winter. However, the extent to which SARS-CoV-2, influenza viruses and other causative viruses have prevailed since implementing preventive measures is unclear.
Objectives
We aim to investigate the incidence of causative viruses and pathogens in patients.
Study design
We collected 191 nasopharyngeal swabs from patients with cold-like symptoms in Japan. All samples were subjected to multiplex PCR with the FilmArray Respiratory Panel and reverse transcription PCR (RT-PCR) to detect SARS-CoV-2.
Results
FilmArray Respiratory Panel analysis detected at least one virus in 32 of 191 patients with cold-like symptoms (21 %). Of these, we frequently identified human rhinoviruses/enteroviruses (5.8 %, n=11), human metapneumovirus (3.7 %, n=7), coronavirus 229E (2.1 %, n=4) and coronavirus OC43 (1.6 %, n=3); while no influenza viruses were detected. RT-PCR analysis detected SARS-CoV-2 (4.2 %, n=8) in patients who were not infected with the aforementioned respiratory viruses.
Conclusions
Co-infection with SARS-CoV-2 and other viruses was not observed. Causative viruses remain prevalent after implementing preventive measures. SARS-CoV-2 differs from influenza viruses in its infectivity.
1. Introduction
The new emergent coronavirus, severe acute respiratory coronavirus 2 (SARS-CoV-2), broke out in Wuhan, China [1]. The virus immediately spread worldwide, causing many Covid-19 cases and deaths. SARS-CoV-2 resides in the upper respiratory tract and spreads via droplets from coughing or sneezing. The World Health Organization recommends washing hands with soap, maintaining social distancing, and avoiding crowded places to prevent the spread of viral infections [2]. In addition to these preventive measures, most countries recommend travel restrictions and curfews.
Influenza viruses, rhinoviruses, adenoviruses, coronaviruses (229E, OC43, NL63 and HKU1) and the human metapneumovirus cause common cold-like symptoms [3]. Of these viruses, influenza viruses are an annual epidemic in the winter. Owing to the development of treatments and vaccines, the mortality rate from influenza viruses is low.
In December 2019, the SARS-CoV-2 outbreak coincided with the influenza viral epidemic. Seasonal influenza activity in Japan was lower in 2020 than in previous years [4]. However, whether SARS-CoV-2 is more infectious than other viruses remains unknown. Here, we performed genetic analyses to determine which viruses were spreading in the Japanese population. This epidemiologic study shows the infectability of each virus after implementing social preventive measures against SARS-CoV-2.
2. Materials and methods
2.1. Patients and samples
We studied 191 patients exhibiting cold-like symptoms from 10 March 2020 to 7 May 2020 in our district 100–150 km west of Tokyo, Japan. Nasopharyngeal samples were collected from all patients using cotton swabs and universal transport media (Copan, Murrieta, CA, USA). The Institutional Review Board at Yamanashi Central Hospital approved this study and the use of an opt-out consent method (G-2019-30). The requirement for written informed consent was waived. Participation in the study by patients was optional.
2.2. FilmArray Respiratory Panel
To analyze the viral species related to patients’ respiratory diseases, we performed multiplex PCR targeting 17 viruses and three bacterial species using the FilmArray Respiratory Panel (bioMérieux, Marcy-l'Etoile, France) per the manufacturer’s instructions. Briefly, buffer and 300 μl from the nasopharyngeal swab were injected into the FilmArray pouch. The reaction proceeded automatically on the FilmArray Torch system. The limit of detection was listed as previously reported (Table 1 ) [5].
Table 1.
The list of limit of detection of FilmArray Respiratory Panel.
| Viruses and bacteria | Standard strains | Limit of detection (TCID50/mL) |
|---|---|---|
| Adenovirus | AdVC1 | 100 |
| AdVC2 | 100 | |
| AdVE4 | 100 | |
| AdVC6 | 100 | |
| Coronavirus 229E | ATCC VR-740 | 4 |
| Coronavirus HKU1 | Clinical Specimen | 1.9 × 106 RNA copies/mL |
| Coronavirus NL63 | NR-470 | 5 |
| Coronavirus OC43 | ATCC VR-759 | 600 |
| Human rhinovirus/ Enterovirus | HRV Type 1A | 1 |
| Echovirus 6 | 30,000 | |
| Human metapneumovirus | hMPV-16, IA10-2003 (Type A1) | 2 |
| Human rhinovirus | Type 1A | 1 |
| Influenza A H1 (Influenza A H1N1) | A/Brisbane/59/07 | 200 |
| A/New Caledonia/20/99 | 2000 | |
| Influenza A H1-2009 (Influenza A H1N1-2009) | A/SwineNY/03/2009 | 100 |
| Influenza A H3 (Influenza A H3N2) | A/Wisconsin/67/2005 | 5 |
| A/Port Chalmers/1/73 | 50 | |
| Influenza B | B/FL/04/06 | 60 |
| B/Taiwan/2/62 | 60 | |
| Parainfluenza virus 1 Type 1 | Type 1 | 500 |
| Parainfluenza virus 2 Type 2 | Type 2 | 10 |
| Parainfluenza virus 3 Type 3 | Type 3 | 10 |
| Parainfluenza virus 4 Type 4 | Type 4 (subtype A) | 5000 |
| Respiratory syncytial virus | Type A | 2 |
| Bordetella pertussis | A639 | 4000 CFU/mL |
| Chlamydophila pneumoniae | TW183 | 3000 DNA copies/mL |
| Mycoplasma pneumoniae | M129 (Type 1) | 30 |
TCID50, Tissue Culture Infectious Dose 50; CFU, colony forming unit
2.3. Reverse transcription PCR (RT-PCR) analysis
The total nucleic acid was automatically isolated from the nasopharyngeal swabs using the MagMax Viral/Pathogen Nucleic Acid Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA) on KingFisher Duo Prime (Thermo Fisher Scientific) as previously described [6,7]. To detect SARS-CoV-2, we performed RT-PCR as previously described [7,8]. The RT-PCR assays were conducted on a StepOnePlus Real-Time PCR System with the following cycling conditions: 50 °C for 5 min for reverse transcription, 95 °C for 20 s, and 45 cycles of 95 °C for 3 s and 60 °C for 30 s. The threshold cycle (Ct) value was assigned to each PCR reaction, and the amplification curve was visually assessed.
3. Results
Of 191 patients, 32 (17 %) were infected at least one causative virus, including human rhinovirus/enterovirus (n = 11), human metapneumovirus (n = 7), coronavirus 229E (n = 4), coronavirus OC43 (n = 3), adenovirus (n = 2), respiratory syncytial virus (n = 2) and coronavirus NL63 (n = 1) (Table 2 ). Two patients were simultaneously infected with two viruses each, one with adenovirus and human rhinovirus/enterovirus and one with adenovirus and human metapneumovirus (Table 2). However, no influenza A, A/H1, A/H3, A/H1-2009 or B viruses were detected. Thus, influenza viruses either have a very low prevalence in our district or were prevented by the intensive measures taken against coronaviruses.
Table 2.
Numbers of patients infected with SARS-CoV-2 and other respiratory infection viruses (n = 40).
| Viruses | n |
|---|---|
| Human rhinovirus/enterovirus | 11 |
| SARS-CoV-2* | 8 |
| Human metapneumovirus | 7 |
| Coronavirus 229E | 4 |
| Coronavirus OC43 | 3 |
| Adenovirus | 2 |
| Respiratory syncytial virus | 2 |
| Coronavirus NL63 | 1 |
| Adenovirus and human rhinovirus/enterovirus | 1 |
| Adenovirus and human metapneumovirus | 1 |
| Coronavirus HKU1 | 0 |
| Influenza A | 0 |
| Influenza A/H1 | 0 |
| Influenza A/H1-2009 | 0 |
| Influenza A/H3 | 0 |
| Influenza B | 0 |
| Parainfluenza 1 | 0 |
| Parainfluenza 2 | 0 |
| Parainfluenza 3 | 0 |
| Parainfluenza 4 | 0 |
| Bacteria | |
| Bordetella pertussis | 0 |
| Chlamydophila pneumoniae | 0 |
| Mycoplasma pneumoniae | 0 |
| Total | 40 |
SARS-CoV-2, severe acute respiratory coronavirus 2.
SARS-CoV-2 was detected by RT-PCR.
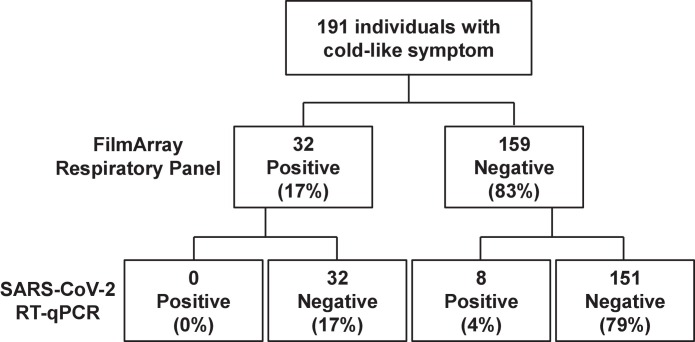
RT-PCR performed on samples from the same cohort of 191 patients detected that 8 were infected with SARS-CoV-2. The FilmArray Respiratory Panel showed that these 8 patients were uninfected with any other causative virus (Fig. 1 ). Thus, the SARS-CoV-2 infection prevalence was 4 %, while the influenza viral prevalence was 0 % in this cohort (Fig. 1). This comparative analysis suggests that SARS-CoV-2 was much more infectious than the influenza viruses were under the same conditions in the same region.
Fig. 1.
Viral infection status among 191 patients.
Flowchart showing the number of patients infected with causative viruses. A FilmArray Respiratory Panel detected 32 positive and 159 negative patients. RT-PCR analysis detected SARS-CoV-2 in 8 patients.
The respiratory panel detected that 17 % of the cohort (32/191 patients) were infected with causative viruses. Seven viral species were detected, none of which were influenza viruses. At the start of the coronavirus epidemic, the infectivity of SARS-CoV-2 was unknown compared to that of influenza viruses. Our data implies that influenza viral spread would be suppressed under the preventive measure.
4. Discussion
This study evaluated the differences in infectivity between SARS-CoV-2 and influenza viruses. Infection prevention measures, such as handwashing and mask wearing, led to fewer influenza infections [9]. Mask wearing is a common practice among Japanese in the winter. After Japanese government announced on the citizen to take preventive measures, “behavioural changes” (i.e. avoid crowded places, wearing a mask, handwashing with soap and hand sanitizers) were as clearly observed at the end of March [10]. Sakamoto et.al. analyzed epidemiological data from 2014 to 2020 and showed that the number of seasonal influenza cases was lower in 2020 than in previous years in Japan [4]. These observations along with the current results suggest different infectious activity levels between seasonal influenza viruses and SARS-CoV2.
We revealed the infection rate of respiratory viruses as well as SARS-CoV-2 (Table 2). Recent study showed surgical mask is important to prevent viral shedding [9]. In particular, detection rate of common coronavirus and influenza virus decreased in droplet and aerosol after waring mask, but less effective for rhinovirus [9]. Concordant with these results, our data showed highest infection rate is caused by the rhinovirus / enterovirus. Common coronavirus such as 229E, OC43 and NL63 were detected in a few patients compared to SARS-CoV-2. Although it still unknown to what extent face mask has the same effects in SARS-CoV-2 to common coronavirus, it could be important for preventing airborne transmission [11].
Although this was a prospective study, there were limitations. First, this study was conducted by a single-center in our district. National registry indicated a marked reduction in influenza viral infections at the end rather than at the beginning of winter [4], which coincided with our study. Second, it needs a comparator group. A retrospective and/or prospective study with data before and after preventive measure against would answer the difference of infectivity among viruses. Third, the influenza vaccination status was not investigated. The less infection rate of influenza virus would be explained by vaccination. Instead, we examined the statistical surveys of vaccination rates for all Japanese citizens published by the government. Overall trend shows the vaccination rate from 2007 to 2017 is almost 50 % (range: 48.2 %–55.9 %). Although the latest data are not yet available, it is likely that the vaccination rate would not change significantly in 2020. In this context, further studies should verify whether preventive social measures reduced the infection prevalence of influenza viral transmission.
This study showed that taking stringent measures may prevent influenza viruses, which have more strongly affected human life for a longer time.
Funding
This study was supported by a Grant-in-Aid for the Genome Research Project from Yamanashi Prefecture (to M.O. and Y.H.), the Japan Society for the Promotion of Science (JSPS) KAKENHI Early-Career Scientists JP18K16292 (to Y.H.), Grant-in-Aid for Scientific Research (B) 20H03668 (to Y.H.), a Research Grant for Young Scholars (to Y.H.), the YASUDA Medical Foundation (to Y.H.), the Uehara Memorial Foundation (to Y.H.), and Medical Research Grants from the Takeda Science Foundation (to Y.H.).
Availability of data and material
The datasets in the current study are available from the corresponding author upon reasonable request.
Declaration of Competing Interest
None.
Acknowledgments
We thank the medical and ancillary hospital staff and the patients for consenting to participate. We thank Traci Raley, MS, ELS, from Edanz Group (https://en-author-services.edanzgroup.com/) for editing a draft of this manuscript.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019;2020(382):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . 2020. Coronavirus Disease (COVID-19) Advice for the Public.https://wwwwhoint/emergencies/diseases/novel-coronavirus-2019/advice-for-public [Google Scholar]
- 3.Gaunt E.R., Hardie A., Claas E.C., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakamoto H., Ishikane M., Ueda P. Seasonal influenza activity during the SARS-CoV-2 outbreak in Japan. JAMA. 2020;323(19):1969–1971. doi: 10.1001/jama.2020.6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poritz M.A., Blaschke A.J., Byington C.L., Meyers L., Nilsson K., Jones D.E., Thatcher S.A., Robbins T., Lingenfelter B., Amiott E. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirotsu Y., Maejima M., Nakajima M., Mochizuki H., Omata M. Environmental cleaning is effective for the eradication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in contaminated hospital rooms: a patient from the diamond princess cruise ship. Infect. Control Hosp. Epidemiol. 2020:1–8. doi: 10.1017/ice.2020.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirotsu Y., Maejima M., Shibusawa M., Nagakubo Y., Hosaka K., Amemiya K., Sueki H., Hayakawa M., Mochizuki H., Omata M. medRxiv; 2020. Pooling RT-PCR Test of SARS-CoV-2 for Large Cohort of ‘healthy’ and Infection-Suspected Patients: a Prospective and Consecutive Study on 1,000 Individuals. [Google Scholar]
- 8.Hirotsu Y., Mochizuki H., Omata M. Double-Quencher Probes Improved the Detection Sensitivity of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by One-Step RT-PCR. J. Virol. Methods. 2020 doi: 10.1016/j.jviromet.2020.113926. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leung N.H.L., Chu D.K.W., Shiu E.Y.C., Chan K.H., McDevitt J.J., Hau B.J.P., Yen H.L., Li Y., Ip D.K.M., Peiris J.S.M. Respiratory virus shedding in exhaled breath and efficacy of face masks. Nat. Med. 2020;26:676–680. doi: 10.1038/s41591-020-0843-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muto K., Yamamoto I., Nagasu M., Tanaka M., Wada K. Japanese citizens’ behavioral changes and preparedness against COVID-19: an online survey during the early phase of the pandemic. PLoS One. 2020;15 doi: 10.1371/journal.pone.0234292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang R., Li Y., Zhang A.L., Wang Y., Molina M.J. Identifying airborne transmission as the dominant route for the spread of COVID-19. Proc. Natl. Acad. Sci. U. S. A. 2020;117:14857–14863. doi: 10.1073/pnas.2009637117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets in the current study are available from the corresponding author upon reasonable request.