Abstract
Alphacoronavirus 1 (subgenus Tegacovirus, genus Alphacoronavirus, family Coronaviridae), which encompasses transmissible gastroenteritis virus (TGEV), feline coronavirus (FCoV) and canine coronavirus (CCoV), is an important pathogen that can cause severe gastroenteritis and is distributed worldwide. CCoV has two different genotypes: CCoV type I, which has a high identity with FCoV-I, and CCoV type II, which is divided into two subtypes, CCoV IIa (pantropic) and CCoV IIb, which is related to FCoV-II and has been involved in multiple recombination events. Between 2014 and 2018, 43 fecal samples from puppies and young dogs under 1 year of age with hemorrhagic enteritis and from 5 cats under 2 years of age with ascites or thoracic effusion were collected by a private veterinary practice in Bogotá, Colombia. A screening for Coronavirus via RT-PCR (nsp12) and PCR amplification of Canine protoparvovirus (VP1) revealed 27.1% (13/49) and 72.9% (35/49) positive samples, respectively. Positive samples for coronavirus were tested for M, N, S and the sequences grouped in the FCoV, CCoV-I and CCoV-IIb clusters that were distant from the pantropic type (IIa). The N gene formed two clusters, one exclusively with samples from this study in subtype II and another with strains in subtype I. For gene S (subtype I), the samples clustered with the Brazilian samples, while samples positive for S subtype IIb grouped into a cluster distinct from the other reference sequences. The prevalence of coronaviruses identified in this study is within the range reported by different countries worldwide.
Keywords: Genetics, Microbiology, Virology, Biocomputational method, Viral disease, Viral genetics, Phylogeny, Epidemiology, Coronavirus, Alphacoronavirus 1, Dogs, Cats, Colombia, Phylogenetic relationship
Genetics; Microbiology; Virology; Biocomputational method; Viral disease; Viral genetics; Phylogeny; Epidemiology; Coronavirus; Alphacoronavirus 1; Dogs; Cats; Colombia; Phylogenetic relationship.
1. Introduction
Alphacoronavirus 1 (Nidovirales: Coronaviridae: Coronavirinae: Alphacoronavirus: Tegacovirus) is a large enveloped virus species with a single-stranded positive-sense RNA genome that is 29 kb in size (Decaro et al., 2015), and this virus can infect dogs, cats and pigs. The 5′ 2/3 of the genome encodes multiple replicase proteins important for replication, with two overlapping open read frames, ORF 1a and 1ab, encoding 16 nonstructural proteins that generate the replicase complex (Decaro et al., 2015; Erles et al., 2007).
Located downstream of ORF1b are up to 11 ORFs that encode the 4 structural proteins spike glycoprotein (S), membrane (M), envelope (E) and nucleocapsid (N) and a variable set of accessory proteins (3a, 3b, 3c, 7a and 7b) (Decaro and Buonavoglia, 2011). Alphacoronavirus 1 encompasses the host-type species canine coronavirus (CCoV), feline coronavirus (FCoV), transmissible gastroenteritis virus (TGEV) and porcine respiratory virus (PRCoV) (Decaro and Buonavoglia, 2011; Ntafis et al., 2013).
Two genotypes of CCoV are known, CCoV-I and CCoV-II, with type II divided into the subtypes CCoV-IIa (classical strains) and CCoV-IIb (TGEV-like strains), with a putative recombination between CCoV-II and TGEV strains (Decaro et al., 2010, 2009). In addition, one unique subtype has emerged in the past decade and belongs to CCoV-IIa; this subtype is considered pantropic as it can cause enteric and systemic signs, reported in Belgium, Brazil, France, Greece, Hungary, Ireland, Italy, Romania and the Caribbean island of St. Kitts (De Barros et al., 2018; Decaro et al., 2010; Navarro et al., 2017; Ntafis et al., 2013; Pinto et al., 2014; Soma et al., 2011; Zicola et al., 2012).
Similarly, two genotypes of FCoV are also known: FCoV-I and FCoV-II. Type I CCoV and FCoV have been proposed to evolve from a common ancestral virus, while type II CCoV and FCoV strains arose from multiple recombination events (Regan et al., 2012). FCoV may be found as the mild enteric feline enteric coronavirus FCoV and as the highly virulent feline peritonitis virus (FIPV) (Jaimes and Whittaker, 2018).
This paper reports on the previously unknown molecular diversity of Alphacoronavirus 1 in cats and dogs in Colombia to provide phylogenetic information regarding this virus species in South America.
2. Material and methods
2.1. Samples
A total of 43 fecal samples from puppies and young dogs under 1 year of age with hemorrhagic enteritis and from 5 cats between 75 days and 2 years with ascites or thoracic effusion were collected from 2014 to 2018 in a private veterinary practice in Bogotá, Colombia.
2.2. RNA and DNA isolation
The fecal samples were prepared as a 20% (v/v) suspension in DEPC-treated water and clarified at 5,000×g for 15 min at 4 °C. The effusions were used undiluted. DNA was isolated from the fecal suspensions using guanidine-isothiocyanate (Chomczynski, 1993), and total RNA was extracted from the fecal suspensions and effusions with TRIzol Reagent™ (Life Technologies, Carlsbad, CA, USA).
2.3. Polymerase chain reaction
Complementary DNA (cDNA) was prepared for all samples with random primers and M-MLV Reverse Transcriptase™ (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions and used in PCR targeting pancoronavirus nsp12 (Escutenaire et al., 2007). All positive samples were submitted for the amplification of the partial M, N, S and 3b genes (De Barros et al., 2018).
All samples were tested for canine protoparvovirus CPV-VP1 (Demeter et al., 2009). GAPDH was used as an internal control in both the RNA and DNA samples (2019Leuteneger, 1999). Primers and amplicon sizes are shown in Table 1.
Table 1.
Primer sequences used for the screening and detection of the virus, the expected fragment sizes, and the references used.
| Primer | Target Gene | Sequence (5′-3′) | Fragment (bp) | Reference |
|---|---|---|---|---|
| PanCo11Fw | nsp12 | TGATGATGSNGTTGTNTGYTAYAA | 179 | (Escutenaire et al., 2007) |
| PanCo13Rv | GCATWGTRTGYTGNGARCARAATTC | |||
| CCV1| | M | TCCAGATATGTAATGTTCGG | 409 | (Pratelli et al., 1999) |
| CCV2 | TCTGTTGAGTAATCACCAGCT | |||
| CENP1 | N | CTCGTGGYCGGAAGAATAAT | 279 | (Erles and Brownlie, 2009) |
| CENP2 | GCAACCCAGAMRACTCCATC | |||
| EL1F | S I | CAAGTTGACCGTCTTATTATTACTGGTAG | 346 | (Pratelli et al., 2004) |
| EL1R | TCATATACGTACCATTATAGCTGAAGA | |||
| S5 | S IIb | TGCATTTGTGTCTCAGACTT | 694 | (Pratelli et al., 2004) |
| S6 | CCAAGGCCATTTTACATAAG | |||
| NSP3B-F | NSP3B | CTTGGTCTCTCTATTGTTGAAG | 200 | (De Barros et al., 2018) |
| NSP3B-R | GCGTTGCGTTTAGAATGG | |||
| CPV PPV F | VP1 | GACTTGTGCCTCCAGGTTAT | 420 | (Demeter et al., 2009) |
| CPV PPV R | GTTGAACTGCTCCATCACTC | |||
| GAPDH F | GAPDH | GCCGTGGAATTTGCCGT | 164 DNA | (Leutenegger et al., 1999) |
| GAPDH R | GCCATCAATGACCCCTTCAT | 82 RNA |
2.4. Phylogenetic analysis
Amplicons for S, M, and N were purified with Illustra ExoProStar 1-Step (GE Healthcare, Chalfont St. Giles, UK) and sequenced bidirectionally using BigDye Terminator v3.1 Cycle Sequencing Kit™ (Applied Biosystems) and ABI-3500 Genetic Analyzer™ (Applied Biosystems) following the manufacturer's instructions. Positions with Phred scores ≥20 were assembled with BioEdit 7.0.5.3 (Ibis Biosciences, Carlsbad, CA, USA), and the final sequences were aligned with homologous sequences from GenBank (accession numbers in Figures 1, 2, 3, and 4) using Clustal W in Bioedit 7.0.5.3. The nucleotide alignments were used to build a maximum likelihood tree (Tamura-Nei model) with 1,000 bootstrap replicates using MEGA 7 (Kumar et al., 2016), and CRCoV was used as a root for the trees because it is a Betacoronavirus. Recombination was assessed using RDP4 software (Muhire et al., 2015).
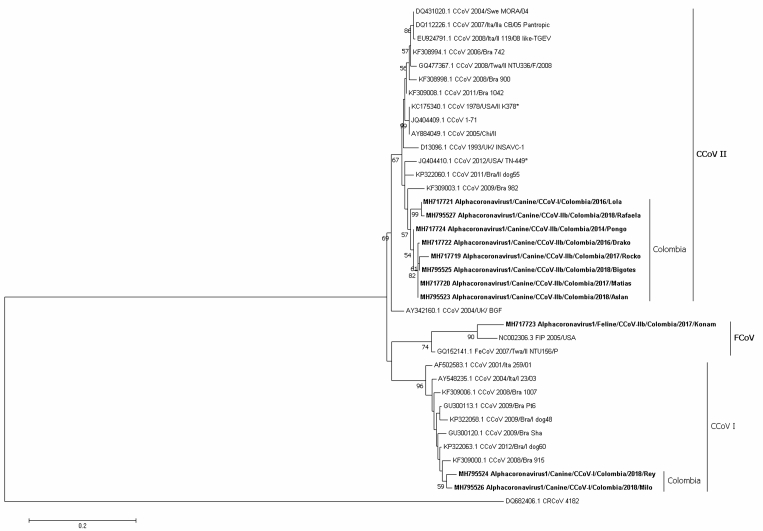
Figure 1.
Maximum likelihood of Alphacoronavirus 1 strains detected in Colombian dogs and a cat are shown in bold, based on the M gene (nt 409-818) of the JQ404409.1 sequence, while the root is represented by DQ682406.1 (CRCoV). Numbers represent the bootstrap values (1,000 replicates). The bar shows the number of substitutions per nucleotide per site.
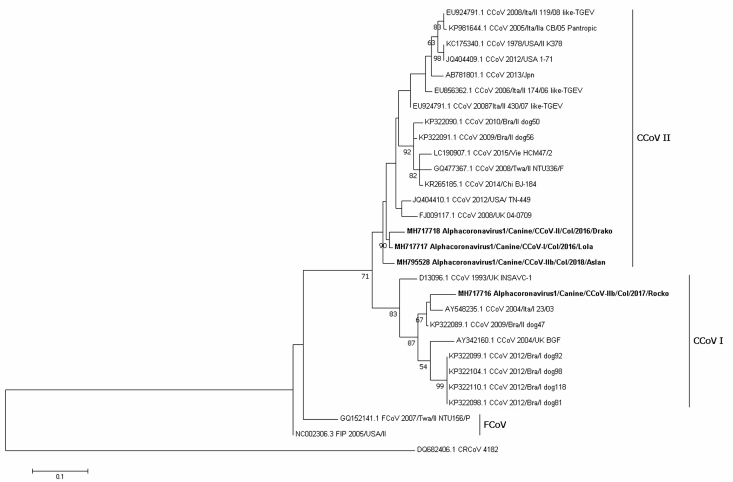
Figure 2.
Maximum likelihood tree of Alphacoronavirus 1 strains detected in Colombian dogs is shown in bold, based on the N gene (nt 68-347) of the JQ404409.1 sequence, while the root is represented by DQ482406.1 (CRCoV). Numbers represent the bootstrap values (1,000 replicates). The bar shows the number of substitutions per nucleotide per site.
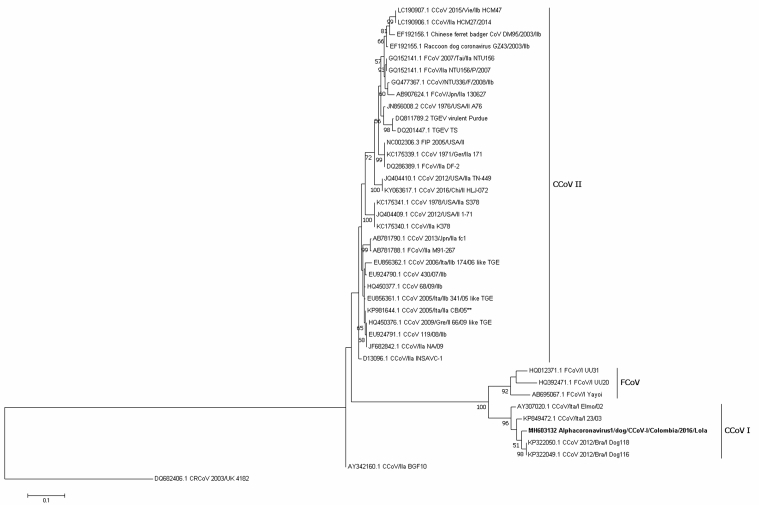
Figure 3.
Maximum likelihood tree of Alphacoronavirus 1 strains detected in Colombian dog is shown in bold, based on the S gene primers for CCoV-I (nt 3541-3886) of the KP849472.1 sequence, while the root is represented by DQ482406.1 (CRCoV). Numbers represent the bootstrap values (1,000 replicates). The bar shows the number of substitutions per nucleotide per site.
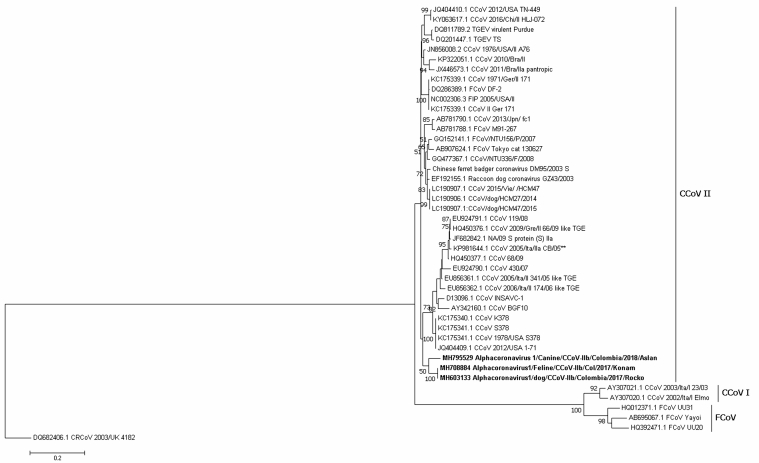
Figure 4.
Maximum likelihood tree of Alphacoronavirus 1 strains detected in Colombian dogs and cat are shown in bold, based on the S gene primers for CCoV-IIb (nt 3492-4185) of the JQ404409.1 sequence, while the root is represented by DQ482406.1 (CRCoV). Numbers represent the bootstrap values (1,000 replicates). The bar shows the number of substitutions per nucleotide per site.
3. Results
Twelve dog fecal samples and one cat effusion sample were positive for coronavirus after screening, while for carnivore protoparvovirus, 35 samples were positive, with 12 samples copositive (dogs) for coronavirus (A.1).
Regarding the M gene from the dog fecal samples, two subclusters were found for CCoV type II, one with the MH795525, MH717720 and MH795523 samples (bootstrap 83) and the other with the MH717721 and MH795527 samples (bootstrap 99), while the samples MH717724, MH717722 and MH717719 segregated in a more dispersed pattern within the Colombian cluster (identities ranging from 99.4 to 95.1%, Figure 1). The CCoV strains MH795526 and MH795524, however, were classified as CCoV I (identities ranging from 86.9 to 87.8%, Figure 1) in a subcluster containing only Colombian strains. Finally, the feline sample MH717723 from a cat with ascites was categorized into the FCoV type II cluster (identity = 92.1% with NC002306.3) (Figure 1).
For the N gene, one sample from a dog resulted in a sequence typed as the CCoV-I H717716 sample, which clustered closely to CCoV-I strains detected in Brazil in 2009 (KP322089.1) and in Italy in 2003 (AY548235.1), while three sequences were found to be related to CCoV-II MH717718 and MH717717 in one subcluster and MH795528 in a second subcluster (identities ranging from 90.1 to 91.7%, Figure 2), each of which was distinct from the pantropic CCoV type (DQ112226.1).
For the S gene, two primer sets (subtype I and IIb) were used, resulting in the identification of one sequence related to CCoV-I MH603132 (identity 0.962), which segregated into a cluster containing strains from Brazil from 2012 (Figure 3); three additional Colombian strains were classified as CCoV-II MH795529, MH708884, and MH603133 (identities ranging from 92 to 93.8%) in a cluster containing only Colombian strains that was distinct from pantropic CCoV-IIa (Figure 4).
All sequences generated in this study were submitted to GenBank under the accession numbers MH717719-24 and MH795523-27 (11 for the M gene), MH717716-18 and MH795528 (4 for the N gene) and MH603132-33, MH708884 and MH795529 (4 for the S gene).
Although no recombination events were found, notably, the vaccine strain D13096 INSAVC-1 was located in the CCoV-I type cluster when classified by the N gene; however, when based on the spike protein, this strain was located in the CCoV-II cluster (Figures 1, 3, and 4).
4. Discussion
Alphacoronavirus 1 is a major viral agent that causes gastrointestinal symptoms in dogs and cats. The detection frequency of CCoV in ill puppies in this study was low (27%) compared with that in other studies, with 65.1% of the samples positive for CCoV (Ntafis et al., 2013); some studies have shown low positive results of 4.8% (Navarro et al., 2017) for single infections, while other studies reported a prevalence of 20.3% for CCoV (Alves et al., 2018) and 18% for CPV in dogs (Wang et al., 2016). Infections with more than one virus are infrequent, and different studies have shown that the coinfection percentage varies between 20% (Soma et al., 2011) and 40% (Costa et al., 2014).
Both CCoV-I and CCoV-IIb have been found in dog fecal samples, and FCoV-II was found in an ascitic cat in Colombia in this study, showing that these types of Alphacoronavirus 1 are present and play a role in canine enteritis and feline infectious peritonitis in this country. As no previous reports on either the prevalence or sequences of these viruses in Colombia are available and due to limited sampling, a more conclusive evolutionary relationship cannot be drawn from the results, but as shown in the trees, Colombian CCoV strains are closely related to the Brazilian strains described by De Barros et al., 2018 based on the M, N, and SI genes evaluated in this study; however, these genes typically clustered into subclusters harboring the Colombian strains. Notably, subtypes IIa and IIb have been associated with fatal enteritis in puppies in the USA (Licitra et al., 2014).
Recombination in Alphacoronavirus 1 is a well-documented phenomenon, resulting mainly from copy choice related to the TRS motif (Decaro et al., 2010, 2009; Ntafis et al., 2013; Pérez et al., 2014; Terada et al., 2014); however, no recombination events were found among the Colombian Alphacoronavirus 1 strains in this study, possibly due to the limited number of sequences analyzed; thus, further studies are needed to clarify the pathogenicity of Alphacoronavirus in Colombia.
Nonetheless, the vaccine strain INSAVC-1 (D13096), a CCoV-IIa strain based on the spike protein (Decaro et al., 2013, 2007), was found herein in the CCoV-I cluster when classified by the N gene, suggesting intergenic homologous recombination (Terada et al., 2014).
Declarations
Author contribution statement
Santana-Clavijo, N. F: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Reyes Romero, D. P, de Souza Silva, S. O: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Arango Fajardo, D. F, Taniwaki, S. A: Performed the experiments; Analyzed and interpreted the data.
Velandia Muñoz, A: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Brandão, P. E: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by the CNPq (Brazilian National Board for Scientific and Technological Development) grant numbers 307291/2017-0 and 400604/2016-7 CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil) Finance Code 001.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We would like to thank the veterinarians at the Clinivet Veterinary Clinic and Magda Carrillo Veterinary Lab in Bogota, Colombia, for sample collection.
Appendix A. Supplementary data
Appendix Table 1.
Prevalence of Alphacoronavirus infection by age, sex and breed based on the clinical status of dogs and cats.
| Clinical status | Species | Status | No. of samples | CCoV Screening |
CCoV M |
CCoV N |
CCoV-I S |
CCoV-IIb S |
CPV VP1 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Rate (%) | Positive | Rate (%) | Positive | Rate (%) | Positive | Rate (%) | Positive | Rate (%) | Positive | Rate (%) | |||||
| Diarrhea | Dogs | Age | ≤3 months | 15 | 5 | 10,4 | 4 | 8,3 | 2 | 4,2 | 0 | 0 | 1 | 2,1 | 10 | 20,8 |
| 3–11 months | 27 | 6 | 14,6 | 6 | 12,5 | 2 | 4,2 | 1 | 2,1 | 1 | 2,1 | 24 | 50,0 | |||
| ≥1 year | 1 | 1 | 2,1 | 1 | 2,1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2,1 | |||
| Sex | Male | 14 | 8 | 18,8 | 8 | 16,7 | 3 | 6,3 | 0 | 0 | 2 | 4,2 | 22 | 45,8 | ||
| Female | 29 | 4 | 8,3 | 3 | 6,3 | 1 | 2,1 | 1 | 2,1 | 0 | 0 | 13 | 27,1 | |||
| Breed | Pure breed | 35 | 11 | 25,0 | 10 | 20,8 | 4 | 8,3 | 1 | 2,1 | 2 | 4,2 | 27 | 56,3 | ||
| Mix breed |
8 |
1 |
2,1 |
1 |
2,1 |
0 |
0 |
0 |
0 |
0 |
0 |
8 |
16,7 |
|||
| Ascites | Cats | Age | ≤3 month | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3–11 months | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| 1–2 years | 1 | 1 | 2,1 | 1 | 2,1 | 0 | 0 | 0 | 0 | 1 | 2,1 | 0 | 0 | |||
| Sex | Male | 3 | 1 | 2,1 | 1 | 2,1 | 0 | 0 | 0 | 0 | 1 | 2,1 | 0 | 0 | ||
| Female | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Breed | Pure breed | 1 | 1 | 2,1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Mix breed | 4 | 0 | 0 | 1 | 2,1 | 0 | 0 | 0 | 0 | 1 | 2,1 | 0 | 0 | |||
References
- Alves C.D.B.T., Granados O.F.O., da Budaszewski R.F., Streck A.F., Weber M.N., Cibulski S.P., Pinto L.D., Ikuta N., Canal C.W. Identification of enteric viruses circulating in a dog population with low vaccine coverage. Braz. J. Microbiol. 2018;1:4–8. doi: 10.1016/j.bjm.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P.A. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 1993;15(532–4):536–537. [PubMed] [Google Scholar]
- Costa M.E., Xavier de Castro T., de Oliveira Bottino F., de Nasser Cubel Garcia R.C. Molecular characterization of canine coronavirus strains circulating in Brazil. Vet. Microbiol. 2014;168:8–15. doi: 10.1016/j.vetmic.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Barros I.N., Pinto L.D., Oliveira S., Silva D.S. Canine coronavirus (CCoV), a neglected pathogen: molecular diversity of S, M, N and 3b genes. Host Viruses. 2018;5:1–6. [Google Scholar]
- Decaro N., Buonavoglia C. Canine coronavirus: not only an enteric pathogen. Vet. Clin. North Am. Small Anim. Pract. 2011;41:1121–1132. doi: 10.1016/j.cvsm.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Cordonnier N., Demeter Z., Egberink H., Elia G., Grellet A., Le Poder S., Mari V., Martella V., Ntafis V., Von Reitzenstein M., Rottier P.J., Rusvai M., Shields S., Xylouri E., Xu Z., Buonavoglia C. European surveillance for pantropic canine coronavirus. J. Clin. Microbiol. 2013;51:83–88. doi: 10.1128/JCM.02466-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., Campolo M., Lorusso A., Camero M., Elia G., Martella V., Cordioli P., Enjuanes L., Buonavoglia C. Recombinant canine coronaviruses related to transmissible gastroenteritis virus of swine are circulating in dogs. J. Virol. 2009;83:1532–1537. doi: 10.1128/JVI.01937-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., Elia G., Addie D.D., Camero M., Lucente M.S., Martella V., Buonavoglia C. Recombinant canine coronaviruses in dogs. Europe. Emerg. Inf. Disp. 2010;16:41–47. doi: 10.3201/eid1601.090726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., Elia G., Lanave G., Dowgier G., Colaianni M.L., Martella V., Buonavoglia C. Full-length genome analysis of canine coronavirus type I. Virus Res. 2015;210:100–105. doi: 10.1016/j.virusres.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Martella V., Elia G., Campolo M., Desario C., Cirone F., Tempesta M., Buonavoglia C. Molecular characterisation of the virulent canine coronavirus CB/05 strain. Virus Res. 2007;125:54–60. doi: 10.1016/j.virusres.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter Z., Gál J., Palade E.A., Rusvai M. Feline parvovirus infection in an Asian palm civet (Paradoxurus hermaphroditus) Vet. Rec. 2009;164:213–216. doi: 10.1136/vr.164.7.213. [DOI] [PubMed] [Google Scholar]
- Erles K., Brownlie J. Canine respiratory coronavirus: an emerging pathogen in the canine infectious respiratory disease complex. Vet. Clin. North Am. - Small Anim. Pract. 2008;38:815–825. doi: 10.1016/j.cvsm.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erles K., Shiu K.B., Brownlie J. Isolation and sequence analysis of canine respiratory coronavirus. Virus Res. 2007;124:78–87. doi: 10.1016/j.virusres.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escutenaire S., Mohamed N., Isaksson M., Thorén P., Klingeborn B., Belák S., Berg M., Blomberg J. SYBR Green real-time reverse transcription-polymerase chain reaction assay for the generic detection of coronaviruses. Arch. Virol. 2007;152:41–58. doi: 10.1007/s00705-006-0840-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., Whittaker G.R. Feline coronavirus: insights into viral pathogenesis based on the spike protein structure and function. Virology. 2018;517:108–121. doi: 10.1016/j.virol.2017.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary Genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutenegger C.M., Mislin C.N., Sigrist B., Ehrengruber M.U., Hoftmann-Lehmann R., Lutz H. Quantitive real-time PCR for the measurement of feline cytokine mRNA. Vet. Immunol. Immunopathol. 1999;71:291–305. doi: 10.1016/S0165-2427(99)00100-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licitra B.N., Duhamel G.E., Whittaker G.R. Canine enteric coronaviruses: emerging viral pathogens with distinct recombinant spike proteins. Viruses. 2014;6:3363–3376. doi: 10.3390/v6083363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhire B., Martin D.P., Murrell B., Khoosal A., Golden M. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:1–5. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro R., Nair R., Peda A., Aung M.S., Ashwinie G.S., Gallagher C.A., Malik Y.S., Kobayashi N., Ghosh S. Molecular characterization of canine parvovirus and canine enteric coronavirus in diarrheic dogs on the island of St. Kitts: first report from the Caribbean region. Virus Res. 2017;240:154–160. doi: 10.1016/j.virusres.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntafis V., Mari V., Decaro N., Papanastassopoulou M., Pardali D., Rallis T.S., Kanellos T., Buonavoglia C., Xylouri E. Canine coronavirus, Greece. Molecular analysis and genetic diversity characterization. Infect. Genet. Evol. 2013;16:129–136. doi: 10.1016/j.meegid.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez R., Calleros L., Marandino A., Sarute N., Iraola G., Grecco S., Blanc H., Vignuzzi M., Isakov O., Shomron N., Carrau L., Hernández M., Francia L., Sosa K., Tomás G., Panzera Y. Phylogenetic and genome-wide deep-sequencing analyses of canine parvovirus reveal co-infection with field variants and emergence of a recent recombinant strain. PloS One. 2014;9 doi: 10.1371/journal.pone.0111779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto L.D., Barros I.N., Budaszewski R.F., Weber M.N., Mata H., Antunes J.R., Boabaid F.M., Wouters A.T.B., Driemeier D., Brandão P.E., Canal C.W. Characterization of pantropic canine coronavirus from Brazil. Vet. J. 2014;202:659–662. doi: 10.1016/j.tvjl.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Decaro N., Tinelli A., Martella V., Elia G., Tempesta M., Cirone F., Buonavoglia C. Two genotypes of canine coronavirus simultaneously detected in the fecal samples of dogs with diarrhea. J. Clinic. Microbiol. 2004;42(4):1797–1799. doi: 10.1128/JCM.42.4.1797-1799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Tempesta M., Greco G., Martella V., Buonavoglia C. Development of a nested PCR assay for the detection of canine coronavirus. J. Virol. Method. 1999;80:11–15. doi: 10.1016/S0166-0934(99)00017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan A.D., Millet J.K., Tse L.P.V., Chillag Z., Rinaldi V.D., Licitra B.N., Dubovi E.J., Town C.D., Whittaker G.R. Characterization of a recombinant canine coronavirus with a distinct receptor-binding (S1) domain. Virology. 2012;430:90–99. doi: 10.1016/j.virol.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma T., Ohinata T., Ishii H., Takahashi T., Taharaguchi S., Hara M. Detection and genotyping of canine coronavirus RNA in diarrheic dogs in Japan. Res. Vet. Sci. 2011;90:205–207. doi: 10.1016/j.rvsc.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y., Matsui N., Noguchi K., Kuwata R., Shimoda H., Soma T., Mochizuki M., Maeda K. Emergence of pathogenic coronaviruses in cats by homologous recombination between feline and canine coronaviruses. PloS One. 2014;9 doi: 10.1371/journal.pone.0106534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li C., Guo D., Wei S., Geng Y., Wang E., Wang Z., Zhao X., Su M., Liu Q., Zhang S., Feng L., Sun D. Co-circulation of canine coronavirus I and IIa/b with high prevalence and genetic diversity in Heilongjiang Province, Northeast China. PloS One. 2016;11:1–11. doi: 10.1371/journal.pone.0146975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zicola A., Jolly S., Mathijs E., Ziant D., Decaro N., Mari V., Thiry E. Fatal outbreaks in dogs associated with pantropic canine coronavirus in France and Belgium. J. Small Anim. Pract. 2012;53:297–300. doi: 10.1111/j.1748-5827.2011.01178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]