Abstract
In order to gain detailed insight into the biochemical behavior of proteins, researchers have developed chemical tools to incorporate new functionality into proteins beyond the canonical 20 amino acids. Important considerations regarding effective chemical modification of proteins include chemoselectivity, near stochiometric labeling, and reaction conditions that maintain protein stability. Taking these factors into account, we discuss an N-terminal labeling strategy that employs a simple two-step “one-pot” method using N-hydroxysuccinimide (NHS) esters. The first step converts a R-NHS ester into a more chemoselective R-thioester. The second step reacts the in situ generated R-thioester with a protein that harbors an N-terminal cysteine to generate a new amide bond. This labeling reaction is selective for the N-terminus with high stoichiometry. Herein, we provide a detailed description of this method and further highlight its utility with a large protein (>100 kDa) and labeling with a commonly used cyanine dye.
Keywords: N-hydroxysuccinimde ester, native chemical ligation, expressed protein ligation, protein labeling, cyanine dye
1. Introduction
There have been a vast number of protein semi-synthetic strategies developed to install new chemical functionality into proteins for their detailed analysis. At the heart of these chemical strategies are the goals to achieve site-specificity and near stochiometric modification. Nonselective protein labeling can result in either deleterious consequences to protein structure and function or heterogeneity that complicates mechanistic studies. A number of elegant strategies to chemically modify proteins have been published which have been the topic of numerous comprehensive reviews and books (Algar, Dawson, & Medintz, 2017; Hermanson, 2013; Koniev & Wagner, 2015; Sletten & Bertozzi, 2009). Examples of successful strategies that have been widely adopted by researchers include, but are not limited to using: (a) nonsense suppression technology for genetic incorporation of unnatural amino acids (Liu & Schultz, 2010; Noren, Anthony-Cahill, Griffith, & Schultz, 1989), (b) Staudinger ligation which reacts a phosphine with an azide to generate a new amide linkage (Lemieux, De Graffenried, & Bertozzi, 2003; Nilsson, Kiessling, & Raines, 2000, 2001; Saxon & Bertozzi, 2000), (c) click chemistry which can be copper-catalyzed cycloaddition of an azide and alkyne (numerous other iterations of this method has also been developed) (Baskin et al., 2007; Cravatt, Wright, & Kozarich, 2008; Kolb & Sharpless, 2003; Rostovtsev, Green, Fokin, & Sharpless, 2002; Speers & Cravatt, 2004; Tornoe, Christensen, & Meldal, 2002), (d) ketone or aldehyde condensation reactions such as hydrazone/hydrazide or oxime forming reactions (Bhat et al., 2018; Carrico, Carlson, & Bertozzi, 2007; Cornish, Hahn, & Schultz, 1996; Dirksen, Dirksen, Hackeng, & Dawson, 2006; O’Shannessy, Dobersen, & Quarles, 1984), (e) Michael acceptors such as maleimides (Friedmann, 1952; Friedmann, Marrian, & Simonreuss, 1949) or vinyl sulfones (Palmer, Rasnick, Klaus, & Bromme, 1995; Truce & Wellisch, 1952) that will selectively react with the sulfhydryl of cysteine, and (f) native chemical ligation which uses a transthioesterification followed by intramolecular rearrangement of a C-terminal thioester and N-terminal cysteine to generate a new peptide bond (Dawson & Kent, 2000; Dawson, Muir, Clark-Lewis, & Kent, 1994; Muir, Sondhi, & Cole, 1998).
Major modalities for protein labeling by biochemical investigators over the past century have involved selectively labeling amines or sulfhydryl groups in proteins. Primary amines in proteins consist of the α-amine of the N-terminus and the ε-amine of lysine, whereas cysteines contain a sulfhydryl group which serves as reactive handle that can be exploited for protein labeling. A number of amine reactive methods to label proteins have been reported including reductive amination with aldehydes (Antos & Francis, 2006; Chen, Disotuar, Xiong, Wang, & Chou, 2017; Glazer, 1970; Means & Feeney, 1990), nucleophilic substitution of activated esters like NHS esters to generate a new amide bond (Anderson, 1963; Brinkley, 1992; Hermanson, 2013), or the use of isothiocyanates to generate thiourea linkages (Edman, 1950; Riggs, Seiwald, Burckhalter, Downs, & Metcalf, 1958; Todrick & Walker, 1937). Due to a high abundance of lysines in proteins (~8%) (Brooks, Fresco, Lesk, & Singh, 2002), many of these strategies result in heterogeneous products. With this in mind, cysteine reactive probes have been more attractive for development because of their low abundance in proteins (< 1%) (Brooks et al., 2002), making it easier for selective incorporation of one’s functionality of choice. A few examples of cysteine reactive probes that are widely used include maleimides (Friedmann, 1952; Friedmann et al., 1949) or vinyl sulfones (Palmer et al., 1995; Truce & Wellisch, 1952) that undergo Michael addition, haloacetamides that generate thioether linkages (Dickens, 1933; Shevchenko, Wilm, Vorm, & Mann, 1996), thiol oxidation to generate disulfides (Chatterjee, McGinty, Fierz, & Muir, 2010; Chen, Ai, Wang, Haracska, & Zhuang, 2010; Gamblin et al., 2003; Laps, Sun, Kamnesky, & Brik, 2019), or β-elimination to form a reactive dehydroalanine followed by a coupling reaction (Chalker et al., 2011; Meledin, Mali, Singh, & Brik, 2016).
Another attractive site to selectively label is the N-terminus because of its singularity as compared to numerous lysines found in a single protein. However, such selective targeting is challenging because of the N-terminus’s cross-reactivity with many of the same chemical strategies used to label lysines. Nevertheless, chemical biologists have developed tools to overcome this obstacle by exploiting the lower pKa of the α-amine of the N-terminus as compared to the ε-amine of lysine and/or relying on the assistance of the side-chain of the N-terminal residue for chemoselectivity. A number of methods using these principles have been developed which have been extensively discussed in reviews by the Francis and Wagner groups (Koniev & Wagner, 2015; Rosen & Francis, 2017). Some examples include native chemical ligation (Dawson et al., 1994; Mali, Singh, Eid, & Brik, 2017; Muir et al., 1998), transamination of the N-terminal α-amine (Gilmore, Scheck, Esser-Kahn, Joshi, & Francis, 2006; Witus et al., 2010; Witus et al., 2013), thiazolidine formation from a cysteine and derivatized aldehyde (Zhang & Tam, 1996), or imidazolidinone formation with 2-pyridinecarboxyaldehhydes (MacDonald, Munch, Moore, & Francis, 2015). Selective N-terminal ligation can also be executed enzymatically. For example, sortase catalyzes a transpeptidation reaction with the specific sequence of LPXTG (Antos et al., 2009; Theile et al., 2013; Williamson, Fascione, Webb, & Turnbull, 2012) and subtiligase can facilitate the formation of a new peptide bond with either an oxyester or thioester peptide substrate (Abrahmsen et al., 1991; Chang, Jackson, Burnier, & Wells, 1994; Henager et al., 2016; Jackson et al., 1994). Alternatively, researchers have used an N-terminal Halo-tag or SNAP-tags for the addition of ligands of value in biochemical and cellular studies. While attractive, Halo-tag and SNAP-tag methods require the fusion of a large polypeptide (33 kDa or 20 kDa) to a protein of interest (Mohapatra, Lin, Feng, Basu, & Ha, 2019). The Halo-tag uses an engineered haloalkane dehalogenase to covalently attach a ligand with a haloalkane group by a nucleophilic displacement mechanism (Los et al., 2008). The SNAP-tag is derivatized from the DNA repair enzyme, O6-alkylguanine-DNA-alkyltransferase, that can react with O6-benzylguanine ligands to form a stable thioether linkage with the catalytic cysteine (Figure 1) (Cole, 2013).
Figure 1. Scheme for several N-terminal labeling methods.
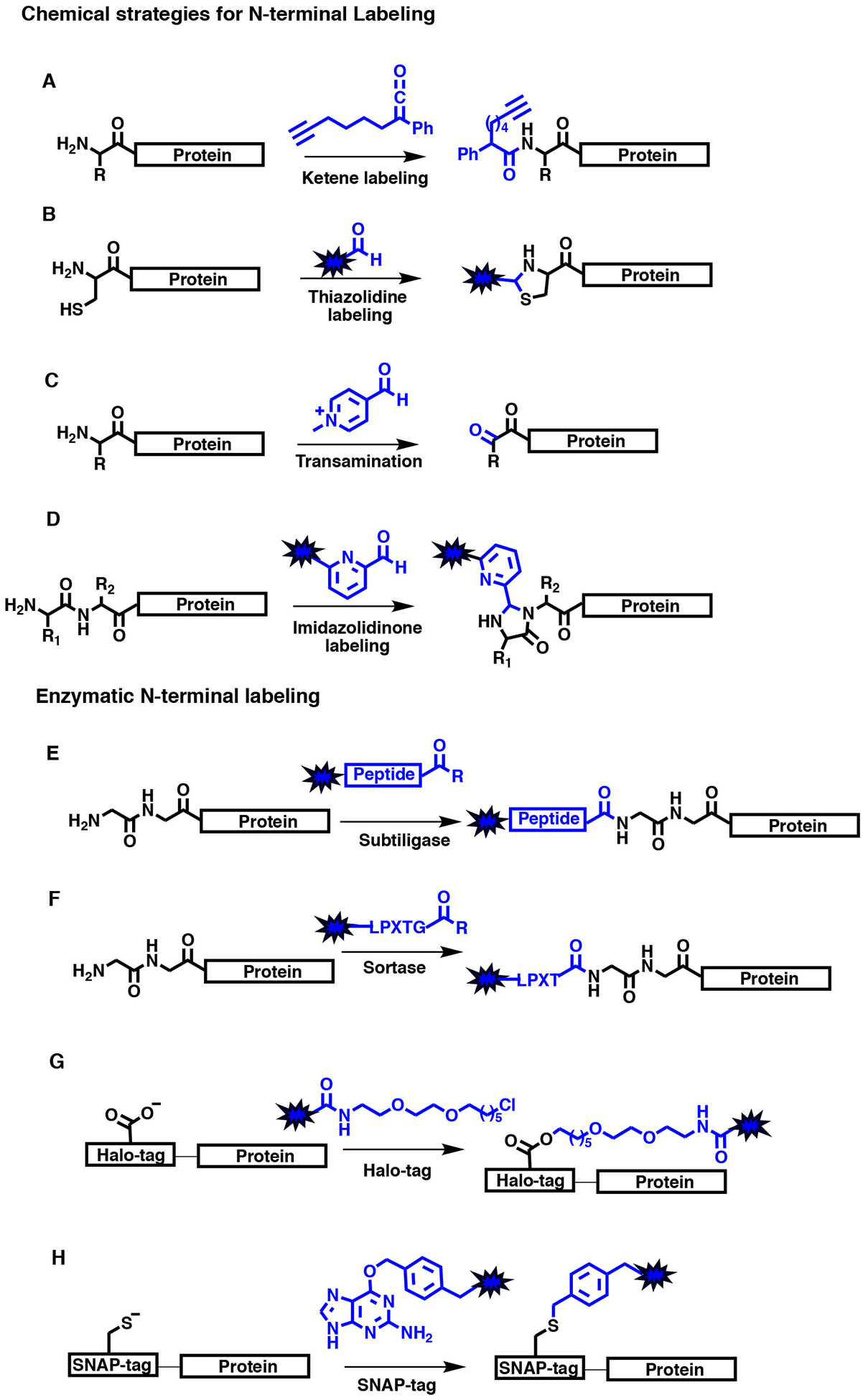
A. Ketene labeling. B. Condensation of aldehyde and N-terminal cysteine to generate thiazolidine. C. Transamination using pyridxal-5’-phosphate derivative (Rapoport’s salt) to generate a pyruvamide species. The second step reacts a hydroxylamine-probe to generate oxime. D. Imidazolidinone formation from 2-pyridinecarboxyladehyde-probes. E. Subtiligase-catalyzed N-terminal ligation. Peptide R group can either be oxyester or thioester. F. Sortase-catalyzed N-terminal ligation. G. Halo-tag. H. SNAP-tag. Blue represents new functionality installed into protein.
Herein, we discuss a recently reported simple strategy to selectively label protein N-termini, which relies on the reactivity of an N-terminal cysteine in a similar manner as NCL (Dempsey, Jiang, Kalin, Chen, & Cole, 2018b). In this approach, an R-NHS ester is converted to a R-thioester which can be further used to react with an N-terminal cysteine for chemoselective and near stochiometric labeling in a “one-pot” reaction. Advantages of this method are that it only provides a small chemical addition to the protein as compared to the much larger Halo-tag or SNAP-tags, it does not require any extensive synthetic ability or infrastructure, and can in principle utilize the vast number of commercially available NHS esters (> 1,000).
2. Native chemical ligation and expressed protein ligation
The N-terminal selective labeling strategy with NHS esters was inspired by the classical early methods of native chemical ligation (NCL) and expressed protein ligation (Dawson et al., 1994; Muir et al., 1998). The NCL reaction ligates two polypeptides, one containing a C-terminal thioester and the other an N-terminal cysteine, to form a new peptide bond. This reaction, pioneered by Wieland (Wieland, Bokelman, Bauer, Lang, & Lau, 1953), proceeds by a transthioesterification reaction, followed by an intramolecular rearrangement to generate the new amide bond (Figure 2a) with a cysteine at the ligation point. Originally, this work was developed to ligate synthetic peptides but later with the development of expressed protein ligation (EPL), this chemistry was applied to recombinant protein systems (Dawson et al., 1994; Muir et al., 1998). EPL uses a recombinantly expressed protein that is heterozygously fused to an intein, where the intein facilitates a reversible N-S acyl shift to generate a C-terminal thioester (Figure 2b). This thioester can be exploited with a small molecule thiol reagent, typically MESNa or MPAA, to undergo the first transthioesterification reaction to cleave the intein from a protein of interest to generate a new protein thioester. Following the intein cleavage, a synthetic peptide with an N-terminal cysteine and the modification of interest can be used to ligate to the newly formed protein thioester using NCL chemistry (Muir et al., 1998). These reactions are chemoselective and often proceed with high stoichiometry (>90% conversion). More recently, enzyme-catalyzed EPL was reported, which utilizes subtiligase to catalyze the ligation of a synthetic peptide and protein thioester (Henager et al., 2016). This method uses the same intein chemistry to generate the protein thioester but differs in the requirement of an N-terminal cysteine at the ligation point, which broadens its scope over classical EPL. Cysteines are one of the least abundant residues found in proteins (< 1%) (Brooks et al., 2002) and one may not be located in an advantageous position for chemical ligation of the synthetic peptide; therefore, a point mutation to introduce a Cys may be required for EPL (Chu et al., 2018). Usually a point mutation can be identified that faithfully mimics the wild-type protein function; however, in some instances this may not be feasible or its perturbation may not be revealed until later in your investigation as was observed in studies on the signaling protein PTEN (Henager et al., 2016; Henager, Henriquez, Dempsey & Cole, 2019a). PTEN is a lipid phosphatase that catalyzes the 3’-specific hydrolysis of phosphatidylinositol (3,4,5)-trisphosphate to generate PIP2, effectively counteracting the pro-growth PI3K/AKT signaling pathway (Maehama & Dixon, 1998; Maehama, Taylor, & Dixon, 2001; Worby & Dixon, 2014). PTEN is phosphorylated at four positions on its C-terminal tail, Ser380, Thr382, Thr383, and Ser385 which regulates its function by driving a conformational change from an open to closed state resulting in inhibited enzymatic function, reduced plasma membrane binding, and enhanced cellular stability (Bolduc et al., 2013; Chen, Dempsey, et al., 2016; Chen et al., 2016a, Dempsey & Cole, 2018a; Vazquez, Ramaswamy, Nakamura, & Sellers, 2000). Initially, a Y379C mutation was used for installation of the PTEN C-terminal tail peptide by EPL (Bolduc et al., 2013). However, it was later serendipitously discovered that Tyr379 contributed to stabilizing the closed autoinhibited state of tetraphosphorylated PTEN (Henager et al., 2016). Without the ability of restoring the wild-type sequence of PTEN by enzyme-catalyzed EPL it would have been challenging to uncover Tyr379’s contribution to conformational closure. Another advantage of enzyme-catalyzed EPL is that it can be more rapid than NCL which may be attractive for proteins with limited stability (Henager et al., 2016). These technological advances have helped advanced our understanding of how protein post-translational modifications influence their function. Furthermore, they have paved the way for installing unique chemical tools on proteins (e.g. crosslinking reagents, bisubstrate inhibitors, etc.) that can be applied in mechanistic and structural studies (Chen et al., 2016a; Shen & Cole, 2003; Shen, Hines, Schwarzer, Pickin, & Cole, 2005)
Figure 2. Scheme for NCL and EPL.
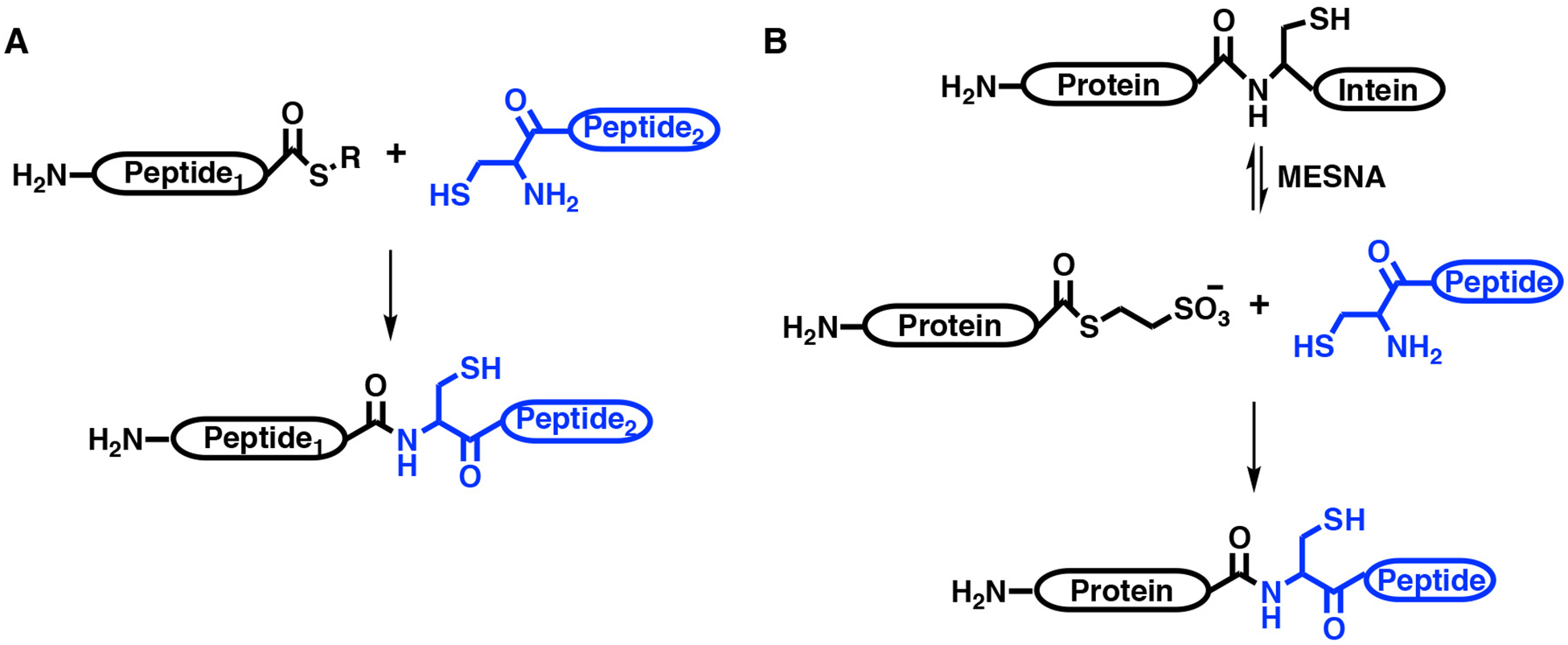
A. NCL (native chemical ligation) reaction. B. EPL (expressed protein ligation) reaction. Blue represents new ligated polypeptide installed into protein.
3. N-hydroxysuccinimide esters in protein labeling
One popular chemical tool that was developed for peptide and protein labeling is the use of N-hydroxysuccinimide esters which are reactive towards primary amines such as the α-amine of the N-terminus and the ε-amine of lysine that form stable amide linkages. Amino acid NHS ester building blocks were originally introduced because of their effectiveness in peptide synthesis as compared to other methods at that time (Anderson, 1963, 1964). Later NHS esters were adapted for peptide and protein labeling with useful compounds for pharmacological development or biochemical analysis such as biotin, fluorophores, PEGylation, fatty acids, etc. Although the labeling is generally selective for primary amines, lysines are one of the most abundant amino acids found in proteins. It is estimated that about 8% of a typical protein’s composition is made up of lysine residues, meaning an average sized protein of 450 amino acids (~50 kDa) would contain about 36 lysines (Brooks et al., 2002). Therefore, using NHS esters for protein modification can lead to heterogenous labeling and this may be deleterious to protein structure and function. However, using a more chemoselective reaction such as NCL for protein labeling can mitigate most risk associated with poorly controlled labeling (Dawson & Kent, 2000; Dawson et al., 1994; Muir et al., 1998). This has been employed by a number of groups who have generated synthetic peptides that incorporate the chemical modification of choice that also possess a C-terminal thioester. In recent studies, these modified thioester peptides contain a C-terminal hydrazide that can undergo nitrosation to generate an acyl azide which then can be exchanged with a thiol to make the final thioester (Fang, Wang, & Liu, 2012). Alternatively, such thioester peptides are synthesized on Dawson resin which can be converted to an imidazolidione (Nbz) using p-nitro chloroformate followed by a thiol exchange to make the final thioester (Blanco-Canosa & Dawson, 2008). Both strategies are very effective for the installation of an N-terminal peptide to an N-Cys-protein; however, these ligation methods require some experience in peptide synthesis and protein bioconjugation which have limited their widespread usage. In principle, small molecule thioesters that can react with N-terminal cysteines in a similar manner to NCL (Busch et al., 2008; Tolbert & Wong, 2002; Yi et al., 2011) would circumvent the challenges associated with standard protein semisynthesis. Unlike NHS esters, small molecule thioesters have not been generally commercially available. With this in mind, a simple strategy to selectively label the N-terminus of a protein with a two-step, “one-pot” reaction using NHS esters was recently reported (Dempsey et al., 2018b). The first step is a preincubation period where the NHS ester (R-NHS) is converted to a thioester through a transesterification reaction with a small thiol molecule, MESNa (R-MESNa). The second step is to add the in situ generated thioester to a protein with an exposed N-terminal cysteine facilitating specific labeling (Figure 3). A more detailed procedure for this is described in the subsequent section (Section 4). This strategy was initially demonstrated using three different proteins (GST, hUNG, and WWP2) in combination with three different NHS esters (fluorescein-NHS, rhodamine-NHS, and biotin-PEG4-NHS), which all showed similar selectivity (< 5% non-specific ligation) and high labeling yields (Dempsey et al., 2018b). Fluorescein-WWP2 was employed to analyze how PTEN phosphorylation influences the binding of WWP2 to PTEN and also its catalytic mechanism (Dempsey et al., 2018b). WWP2 is a HECT E3 ligase that catalyzes the ubiquitination of the number of substrates including itself and PTEN (Chen et al., 2017; Chen, Thomas, et al., 2016b; Jiang, Thomas, Chen, Chiang, & Cole, 2019; Maddika et al., 2011). This occurs through a sequential set of ubiquitin transfers that begin with the ATP-dependent loading of an E1 protein followed by transfer to an E2 enzyme, which is then transferred to the catalytic cysteine of WWP2, and then finally chemically installed onto a lysine of a protein substrate. The use of fluorescein labeled WWP2 in PTEN binding studies revealed how PTEN C-tail phosphorylation restricts its ubiquitination by WWP2 through weakening the protein-protein interaction, accounting for phospho-PTEN’s enhanced cellular stability (Dempsey et al., 2018b). In addition, the use of distinct labeling groups for generating wild-type and mutant forms of WWP2 were used to show that WWP2 catalyzes its autoubiquitination through an intramolecular ubiquitin transfer (Dempsey et al., 2018b). Thus, the recently developed NHS ester labeling strategy facilitated mechanistic studies into the regulation of two important proteins in cell biology.
Figure 3. Scheme for N-terminal labeling using R-NHS esters.
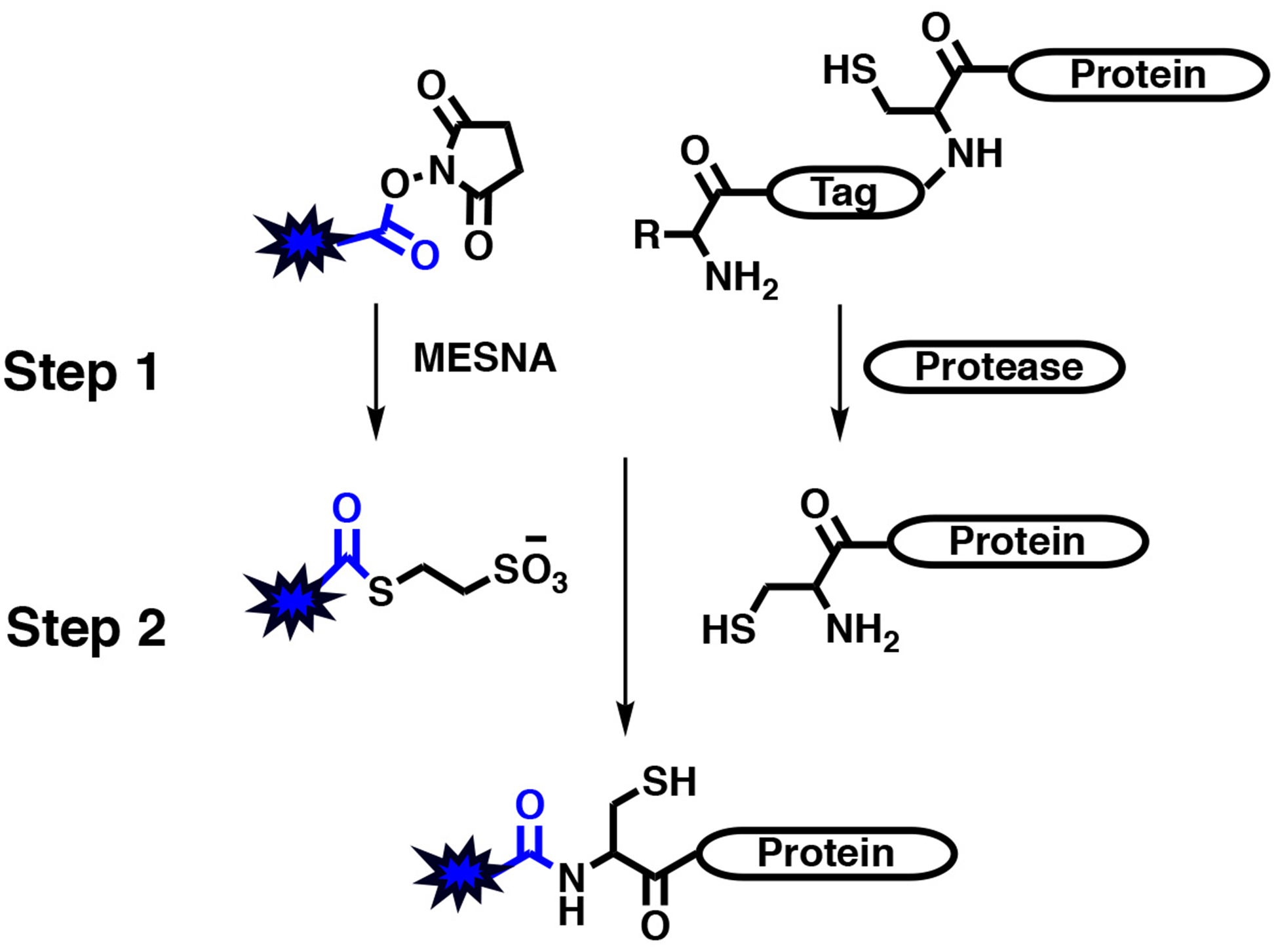
Step 1 is a transesterification reaction to convert the R-NHS ester into a R-MESNa thioester. Step two is the selective labeling of a protein with a N-terminal cysteine by the same mechanism as NCL/EPL. The proteolysis step to reveal an N-terminal cysteine can either be combined into the second step or done independently prior to labeling. Protease is either SUMO protease (ULP1) or TEV protease. New chemical functionality on protein shown in blue.
4. Protocol for N-terminal labeling with N-hydroxysuccinimide esters
Below we provide a detailed procedure to N-terminally label proteins with NHS esters along with some tips to maximize yield and minimize off-target labeling. This method is a simple “one-pot”, two-step reaction that does not require synthetic expertise to execute (Dempsey et al., 2018b). Moreover, this method can take advantage of the vast availability of commercial R-NHS esters (>1,000). One updated feature in this protocol is to include proteolysis of the tag in the same pot as the labeling reaction, which is validated in Section 7. With this in mind, we present two procedures that differ only at this step as described below:
- Express and purify protein that has an N-terminal tag that can be cleaved by SUMO protease or TEV protease that results in an N-terminal Cys (Kapust, Tozser, Copeland, & Waugh, 2002; Malakhov et al., 2004).
- For SUMO protease, install an N-terminal SUMO fusion with a Cys that follows the di-Gly motif.
- For TEV protease, install the following sequence ENLYFQC N-terminal to your protein of interest. There may be additional residues prior to this sequence such as a His-tag or GST-tag, but TEV protease will hydrolyze the peptide bond between the glutamine and cysteine.
- Proteolysis of the fusion-tag with TEV protease or SUMO protease to release the N-terminal cysteine. This step can be executed in two different ways:
- Procedure I – Discrete removal of the tag after purification to release N-terminal cysteine, further purify protein, and then dialyze/exchange protein into labeling reaction buffer.
- If cleaving the tag at this step, be conscious to minimize time between this step and labeling since an N-terminal Cys can be prone to adventitious oxidation. Consider using a sizing column as a buffer exchange to minimize time at this step.
- Procedure II – Directly dialyze protein into labeling reaction buffer without tag removal.
- Cleavage of the tag will occur in the same pot as the labeling reaction (see step 3).
- We recommend dialyzing the protein of interest into 100 mM HEPES pH 6.8 – 7.0 with 0.5 – 1 mM tris(2-carboxyethyl)phosphine (TCEP). Sodium phosphate is a fine alternative if HEPES needs to be avoided. TCEP should be used as a reducing reagent and not thiols such as dithiothreitol (DTT) or 2-mercaptoethanol (BME). DTT and BME may exchange with the newly formed MESNa thioester, which may hurt labeling yields. Note that the addition of salt if needed for protein stablity should not significantly impact the labeling reactions.
- Do not dialyze into a glycerol containing buffer. Lower reaction yields were observed for proteins in glycerol, likely due to impurities in the glycerol such as aldehydes and peroxides. After the labeling reaction is complete, glycerol can safely be added if needed.
- Initiate the first step of the “one-pot” reaction, transesterification of R-NHS ester to R-MESNa thioester:
- Make a solution of 100 mM HEPES pH 6.8–7.0, 0.5 – 1 mM TCEP, 500 mM MESNa, and 2.5 – 5 mM R-NHS ester and incubate them at room temperature for 3 – 6 hours, though three hours is typically sufficient for complete conversion to R-MESNa thioester. Please note that the concentration of the R-NHS ester is only important to attain your desired final concentration in the protein labeling reaction (~1 mM). Under these reaction conditions the initial transesterification reaction is under pseudo-first order condition, meaning the MESNa concentration is in large excess relative to the R-NHS ester; therefore, the reaction rate is dependent on the MESNa concentration.
- Make each reagent from fresh powder every time as NHS-esters are easily hydrolyzed and thiols will be oxidized over time. Do not keep solution stocks of these reagents. Also, avoid using DMSO to help solubilize R-NHS esters.
- Make sure pH is adjusted to 6.8 – 7.0. First make the 100 mM HEPES pH 6.8–7.0, 0.5 – 1 mM TCEP, 500 mM MESNA solution, check pH using a probe and then adjust accordingly. Following this, add the R-NHS ester and check the pH by spotting on pH paper.
- Vortex reaction vigorously making sure all R-NHS ester is in solution.
- Execute second step of the “one-pot” reaction, labeling N-Cys-protein with newly formed R-MESNa thioester:
- Add the newly formed R-MESNa thioester reaction mixture to a final concentration of 1 mM to your purified protein from step 2 and incubate on bench for 24 – 48 hours. If using “Procedure II” (step 2), also add the corresponding protease to cleave tag.
- The protein concentration should be sufficient to keep the protein stable unless it starts to approach or exceed the R-MESNa thioester concentration. In standard cases, it will fall well below the R-MESNa thioester concentration (1 mM) meaning this reaction is also pseudo-first order and the labeling rate is proportional to the R-MESNa thioester concentration.
- After the labeling reaction has reached the desired conversion (e.g., greater than 50%), immediately quench and remove the R-MESNa thioester.
- First quench the thioester with either 100 mM hydroxylamine, hydrazine, cysteine, or cysteamine at room temperature for at least an hour. Make sure to adjust the pH of the quenching solution to 7.
- After quenching, remove the ligation reagent by passing the mixture over a desalting column. If this is not feasible, then dialyze the sample overnight after making sure the R-MESNa is completely quenched. Be sure to keep the dialysis buffer at the same pH as that used in the labeling reaction.
- Following initial removal of the labeling reagent, pass the sample through a final sizing column to remove any remaining labeling reagent. At this step, the desired final buffer condition can be utilized.
5. Case study with USP7 labeling with biotin-PEG4-NHS.
The initial investigation of this NHS ester labeling strategy investigated three proteins which ranged in size from small to medium (GST – 26 kDa, UNG – 33 kDa, WWP2ΔC2 – 68 kDa) (Dempsey et al., 2018b). Here, we examine this method’s utility with a larger protein, USP7, which is 130 kDa. USP7 was engineered to have an N-terminal 8x-His-tag-SUMO fusion which unmasks an N-terminal cysteine after ULP1 (SUMO protease) cleavage (Figure 4a). USP7 is a deubiquitinase (DUB) enzyme of the ubiquitin specific Cys hydrolase family that catalyzes the hydrolysis of ubiquitin from a number of substrates including: PTEN (Morotti et al.; Song et al., 2008), E3 ligase MDM2 (Cummins & Vogelstein, 2004; Li, Brooks, Kon, & Gu, 2004), DNA methyltransferase DNMT1 (Du et al., 2010; Felle et al., 2011), and histone H2B (van der Knaap et al., 2005). We expressed USP7 in E. coli BL21(DE3) Rosetta cells and purified it using nickel affinity chromatography. We proceeded to selectively label USP7 with biotin-PEG4-NHS using Procedure I from Section 4 which first cleaves the 8x-His-SUMO fusion prior to the N-terminal labeling reaction. We evaluated the selectivity of the reaction by comparing cleaved and uncleaved USP7 by Western blot analysis with an Anti-biotin-HRP antibody. Our results demonstrate that this method maintains high selectivity for N-Cys labeling (Figure 4b). Despite USP7’s large size, the non-specific labeling for this reaction, assessed by comparing labeling of the uncleaved to the cleaved USP7, was less than 5%, in accord with the previous work on smaller proteins (Dempsey et al., 2018b). As USP7 contains 80 lysines and 19 cysteines that could theoretically be sites of nonspecific modification, labeling of less than 5% of uncleaved USP7 indicates that the average internal lysine or cysteine residue would be modified at lower than 0.1%, and this low level should be acceptable for the vast majority of biochemical studies. These data confirm that this method can be utilized for larger proteins and by extension, perhaps large protein complexes to interrogate their biochemical function.
Figure 4. Biotin-PEG4 labeling of USP7.
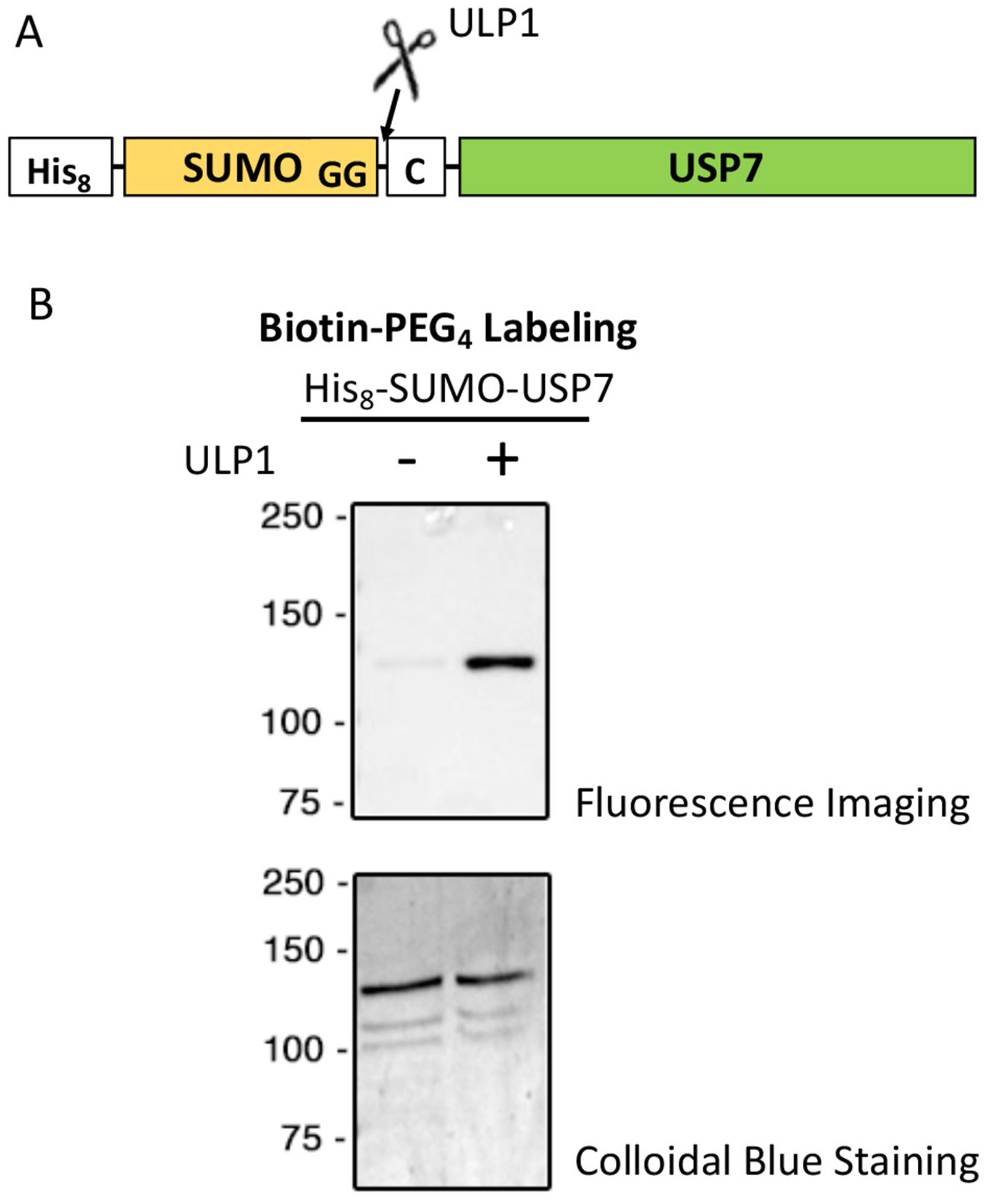
A. Diagram of expressed USP7 and how ULP1’s enzymatic function produces an N-terminal cysteine. USP7 has a N-terminal fused His8-SUMO with a cysteine following the di-glycine motif of SUMO. B. Preparation of Biotin-PEG4-USP7. Top panel is the Western image of ULP1 cleaved or uncleaved USP7 that was labeled by Biotin-PEG4 using an anti-biotin antibody. Labeling was executed using Procedure I for 24 hours. Labeling reaction quench with 100 mM hydroxylamine for 1 hour and then dialyzed overnight. Finally both USP7s were purified over a superdex200 column to remove any remaining labeling reagent. Bottom panel is the colloidal stained control showing equal loading on gel. Nonspecific labeling based on Western blot analysis is < 5%.
6. Sulfo-Cyanine5-NHS labeling of WWP2.
To further characterize the broad applicability of the NHS ester method, we thought it would be useful to evaluate the cyanine dye, sulfo-Cy5-NHS (Figure 5a, Lumiprobe). Cyanine dyes are frequently used in single molecule biochemical experiments (Mohapatra et al., 2019). Concerns have also been raised that they can undergo undesired covalent reaction with thiol reagents (Vaughan, Dempsey, Sun, & Zhuang, 2013). Cy5 harbors a conjugated system that separates its two indole rings and this may serve as an electrophilic acceptor of nucleophiles such as MESNa or TCEP. Products of such thiol or phosphine reactions could disrupt its spectral properties, although, it is generally believed that these linkages would be rapidly reversible because facile elimination reactions that restore extended conjugation can regenerate the fluorescent dyes. With these issues in mind, we tested the NHS ester labeling reaction using Procedure II from Section 4 with sulfo-Cy5-NHS and WWP2. Procedure II, introduced in this chapter, is a streamlined approach that concurrently cleaves the fusion-tag to generate an N-terminal cysteine and labels the protein with the newly formed R-MESNa thioester. In the protein labeling step of the reaction, we used 0.3 equivalent of TEV protease (4 μM) to 1 equivalent of GST-WWP2ΔC2 (12 μM), and removal of the GST-tag was complete in less than four hours (Figures 5b & 5c). This is an important parameter to consider with Procedure II because if this step is slow it will limit reaction yield or require extended reaction times. Additionally, the labeling reaction with 1 mM Cy5-MESNa tracked with similar kinetics and very low non-specific labeling (< 3%) as observed with other labeling reagents (Figure 5c) (Dempsey et al., 2018). Finally, the Cy5 labeled protein showed intense fluorescence suggesting that side reactions with the Cy5 group were inconsequential. Overall, these findings suggest that Cy5-labeling of proteins with this NHS ester strategy is robust.
Figure 5. Cy5 labeling of GST-WWP2ΔC2.
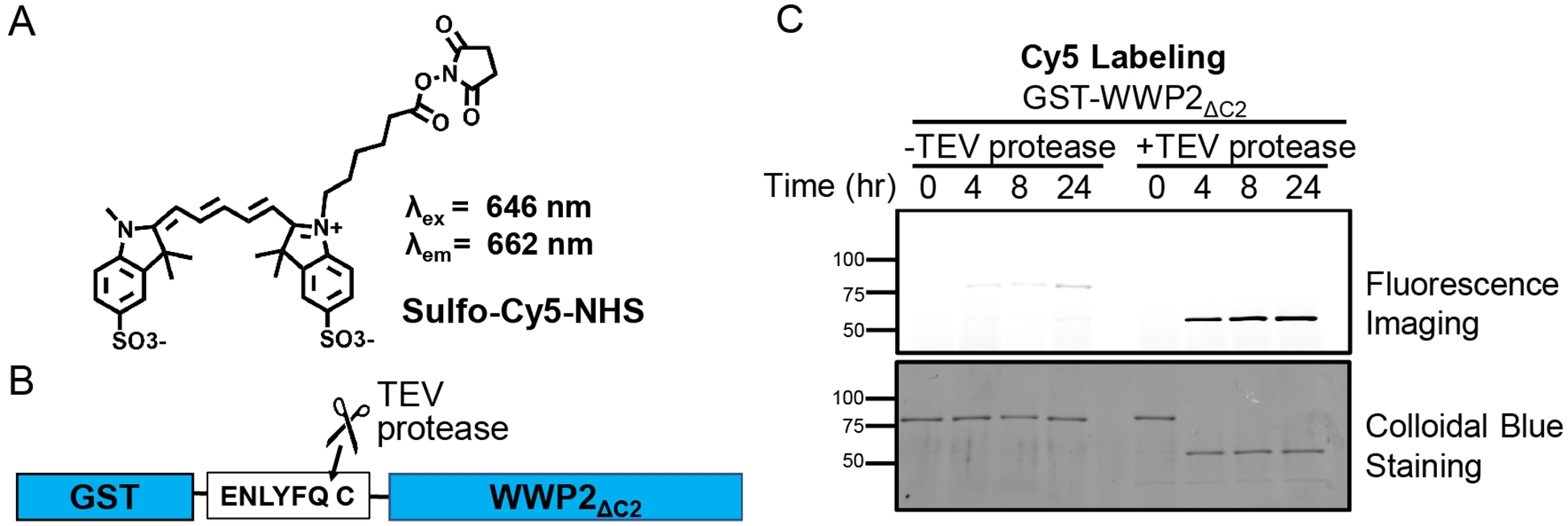
A. Sulfo-Cy5-NHS ester structure. B. Diagram showing the cleavage of GST-WWP2ΔC2 by TEV protease. C. Labeling of WWP2ΔC2 by Cy5 using Procedure II. Labeling reagent originated from sulfo-Cy5-NHS. GST-WWP2ΔC2 (12 μM) was mixed with TEV protease (4 μM) and sulfo-Cy5-NHS (1 mM) to start the labeling reaction. Different time points were taken at 0, 4, 8 and 24 hrs. Top gel corresponds to fluorescence imaging by a Typhoon FLA 9500 imager using its preset Cy5 fluorescent imaging program (excitation 635 nm, emission 670 nm). Each sample was quenched using 100 mM cysteamine for 10 min followed by immediate flash freezing and stored at −80°C until analysis. Bottom gel corresponds to the colloidal stained loading control. Nonspecific labeling based on fluorescence imaging is < 3%.
7. Summary and future directions
In conclusion, this chapter discusses how to use R-NHS esters for the specific labeling of proteins at their N-terminus. This method details a “one-pot” two-step reaction that converts an R-NHS ester into a thioester that can site-specifically label proteins that harbor an N-terminal cysteine. We have demonstrated the use of this method on proteins that are relatively large in size (USP7) and with potentially challenging R-NHS esters such as sulfo-Cy5-NHS, further establishing its versatility. Moreover, we have described a slightly modified protocol that streamlines the labeling method by including the protease cleave step into the “one-pot” reaction.
In the future, it will be worthwhile to investigate labeling with other R-NHS esters of interest to the biomedical community including: pegylated-NHS esters which may be useful for the pharmaceutical community, bivalent R-NHS esters that can drive protein dimerization, or immobilized R-NHS esters that can be used in microarray or surface plasmon resonance (SPR) experiments. Additionally, this method may be combined with other protein semi-synthesis strategies such as EPL to generate useful macromolecular reagents for biochemical or pharmacological investigation (Matico et al., 2019). Finally, it would be useful to see if this method could be expanded to in-vivo labeling.
Acknowledgments
We would like to thank NIH K99GM130961, NIH F32GM120855, T32GM095450 and NIH R01CA74305 for generous financial support.
References
- Abrahmsen L, Tom J, Burnier J, Butcher KA, Kossiakoff A, & Wells JA (1991). Engineering subtilisin and its substrates for efficient ligation of peptide bonds in aqueous solution. Biochemistry, 30, 4151–4159. [DOI] [PubMed] [Google Scholar]
- Algar WR Dawson PE; Medintz IL (2017). Chemoselective and bioorthogonal ligation reactions: concepts and application (1st ed.): Wiley. [Google Scholar]
- Anderson GW Z. JE; Callahan FM (1963). N-Hydroxysuccinimide ester in peptide synthesis. J. Am. Chem. Soc, 85, 3039–3039. [Google Scholar]
- Anderson GW Z. JE; Callahan FM (1964). The Use of Esters of N-Hydroxysuccinimide in Peptide Synthesis. J. Am. Chem. Soc, 86, 1839–1842. [Google Scholar]
- Antos JM, Chew GL, Guimaraes CP, Yoder NC, Grotenbreg GM, Popp MW, & Ploegh HL (2009). Site-specific N- and C-terminal labeling of a single polypeptide using sortases of different specificity. J. Am. Chem. Soc, 131, 10800–10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antos JM, & Francis MB (2006). Transition metal catalyzed methods for site-selective protein modification. Curr. Opin. Chem. Biol, 10, 253–262. [DOI] [PubMed] [Google Scholar]
- Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, et al. (2007). Copper-free click chemistry for dynamic in vivo imaging. Proc. Nat.l Acad. Sci. U. S. A, 104, 16793–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat S, Hwang Y, Gibson MD, Morgan MT, Taverna SD, Zhao Y, et al. (2018). Hydrazide mimics for protein lysine acylation to assess nucleosome dynamics and deubiquitinase action. J. Am. Chem. Soc, 140, 9478–9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Canosa JB, & Dawson PE (2008). An efficient Fmoc-SPPS approach for the generation of thioester peptide precursors for use in native chemical ligation. Angew Chem. Int. Ed. Engl, 47, 6851–6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc D, Rahdar M, Tu-Sekine B, Sivakumaren SC, Raben D, Amzel LM, et al. (2013). Phosphorylation-mediated PTEN conformational closure and deactivation revealed with protein semisynthesis. Elife, 2, e00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley M (1992). A brief survey of methods for preparing protein conjugates with dyes, haptens, and cross-linking reagents. Bioconjug. Chem, 3, 2–13. [DOI] [PubMed] [Google Scholar]
- Brooks DJ, Fresco JR, Lesk AM, & Singh M (2002). Evolution of amino acid frequencies in proteins over deep time: inferred order of introduction of amino acids into the genetic code. Mol. Biol. Evol, 19, 1645–1655. [DOI] [PubMed] [Google Scholar]
- Busch GK, Tate EW, Gaffney PR, Rosivatz E, Woscholski R, & Leatherbarrow RJ (2008). Specific N-terminal protein labelling: use of FMDV 3C pro protease and native chemical ligation. Chem. Commun, 3369–3371. [DOI] [PubMed] [Google Scholar]
- Carrico IS, Carlson BL, & Bertozzi CR (2007). Introducing genetically encoded aldehydes into proteins. Nat. Chem. Biol, 3, 321–322. [DOI] [PubMed] [Google Scholar]
- Chalker JM, Gunnoo SB, Boutureira O, Gerstberger SC, Fernandez-Gonzalez M, Bernardes GJL, et al. (2011). Methods for converting cysteine to dehydroalanine on peptides and proteins. Chem. Sci, 2, 1666–1676. [Google Scholar]
- Chang TK, Jackson DY, Burnier JP, & Wells JA (1994). Subtiligase: a tool for semisynthesis of proteins. Proc. Natl. Acad. Sci. U. S. A, 91, 12544–12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee C, McGinty RK, Fierz B, & Muir TW (2010). Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat. Chem. Biol, 6, 267–269. [DOI] [PubMed] [Google Scholar]
- Chen D, Disotuar MM, Xiong X, Wang Y, & Chou DH (2017). Selective N-terminal functionalization of native peptides and proteins. Chem. Sci, 8, 2717–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ai Y, Wang J, Haracska L, & Zhuang Z (2010). Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nat. Chem. Biol, 6, 270–272. [DOI] [PubMed] [Google Scholar]
- Chen Z, Dempsey DR, Thomas SN, Hayward D, Bolduc DM, & Cole PA (2016a). Molecular features of phosphatase and tensin homolog (PTEN) regulation by C-terminal phosphorylation. J. Biol. Chem, 291, 14160–14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Jiang H, Xu W, Li X, Dempsey DR, Zhang X, et al. (2017). A tunable brake for HECT ubiquitin ligases. Mol. Cell, 66, 345–357 e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Thomas SN, Bolduc DM, Jiang X, Zhang X, Wolberger C, & Cole PA (2016b). Enzymatic analysis of PTEN ubiquitylation by WWP2 and NEDD4–1 E3 ligases. Biochemistry, 55, 3658–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu N, Salguero AL, Liu AZ, Chen Z, Dempsey DR, Ficarro SB, et al. (2018). Akt kinase activation mechanisms revealed using protein semisynthesis. Cell, 174, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NB (2013). Site-specific protein labeling with SNAP-tags. Curr. Protoc. Protein Sci, 73, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish VW, Hahn KM, & Schultz PG (1996). Site-specific protein modification using a ketone handle. J. Am. Chem. Soc, 118, 8150–8151. [Google Scholar]
- Cravatt BF, Wright AT, & Kozarich JW (2008). Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem, 77, 383–414. [DOI] [PubMed] [Google Scholar]
- Cummins JM, & Vogelstein B (2004). HAUSP is required for p53 destabilization. Cell Cycle, 3, 689–692. [PubMed] [Google Scholar]
- Dawson PE, & Kent SB (2000). Synthesis of native proteins by chemical ligation. Annu. Rev. Biochem, 69, 923–960. [DOI] [PubMed] [Google Scholar]
- Dawson PE, Muir TW, Clark-Lewis I, & Kent SB (1994). Synthesis of proteins by native chemical ligation. Science, 266, 776–779. [DOI] [PubMed] [Google Scholar]
- Dempsey DR, & Cole PA (2018a). Protein chemical approaches to understanding PTEN lipid phosphatase regulation. Methods. Enzymol, 607, 405–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey DR, Jiang H, Kalin JH, Chen Z, & Cole PA (2018b). Site-specific protein ;abeling with N-Hydroxysuccinimide-esters and the analysis of ubiquitin ligase mechanisms. J. Am. Chem. Soc, 140, 9374–9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickens F (1933). Interaction of halogenacetates and SH compounds: The reaction of halogenacetic acids with glutathione and cysteine. The mechanism of iodoacetate poisoning of glyoxalase. Biochem. J, 27, 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen A, Dirksen S, Hackeng TM, & Dawson PE (2006). Nucleophilic catalysis of hydrazone formation and transimination: implications for dynamic covalent chemistry. J. Am. Chem. Soc, 128, 15602–15603. [DOI] [PubMed] [Google Scholar]
- Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, et al. (2010). DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci. Signal, 3, ra80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman P (1950). Method for determination of the amino acid sequence in peptides. Acta Chem. Scand, 4, 283–293. [Google Scholar]
- Fang GM, Wang JX, & Liu L (2012). Convergent chemical synthesis of proteins by ligation of peptide hydrazides. Angew Chem. Int. Ed. Engl, 51, 10347–10350. [DOI] [PubMed] [Google Scholar]
- Felle M, Joppien S, Nemeth A, Diermeier S, Thalhammer V, Dobner T, et al. (2011). The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res, 39, 8355–8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann E (1952). Spectrophotometric investigation of the interaction of glutathione with maleimide and N-ethylmaleimide. Biochim. Biophys. Acta, 9, 65–75. [DOI] [PubMed] [Google Scholar]
- Friedmann E, Marrian DH, & Simonreuss I (1949). Antimitotic action of maleimide and related substances. Br. J. Pharmacol. Chemother, 4, 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamblin DP, Garnier P, Ward SJ, Oldham NJ, Fairbanks AJ, & Davis BG (2003). Glycosyl phenylthiosulfonates (glyco-PTS): novel reagents for glycoprotein synthesis. Org. Biomol. Chem, 1, 3642–3644. [DOI] [PubMed] [Google Scholar]
- Gilmore JM, Scheck RA, Esser-Kahn AP, Joshi NS, & Francis MB (2006). N-terminal protein modification through a biomimetic transamination reaction. Angew Chem. Int. Ed. Engl, 45, 5307–5311. [DOI] [PubMed] [Google Scholar]
- Glazer AN (1970). Specific chemical modification of proteins. Annu. Rev. Biochem, 39, 101–130. [DOI] [PubMed] [Google Scholar]
- Henager SH, Chu N, Chen Z, Bolduc D, Dempsey DR, Hwang Y, et al. (2016). Enzyme-catalyzed expressed protein ligation. Nat. Methods, 13, 925–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henager SH, Henriquez S, Dempsey DR, & Cole PA (2019). Analysis of site-specific phosphorylation of PTEN by using enzyme-catalyzed expressed protein ligation. Chembiochem, doi: 10.1002/cbic.201900316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson G (2013). Bioconjugate Techniques (3rd ed.): Academic Press. [Google Scholar]
- Jackson DY, Burnier J, Quan C, Stanley M, Tom J, & Wells JA (1994). A designed peptide ligase for total synthesis of ribonuclease A with unnatural catalytic residues. Science, 266, 243–247. [DOI] [PubMed] [Google Scholar]
- Jiang H, Thomas SN, Chen Z, Chiang CY, & Cole PA (2019). Comparative analysis of the catalytic regulation of NEDD4–1 and WWP2 ubiquitin ligases. J. Biol. Chem, 294, 17421–17436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust RB, Tozser J, Copeland TD, & Waugh DS (2002). The P1’ specificity of tobacco etch virus protease. Biochem. Biophys. Res. Commun, 294, 949–955. [DOI] [PubMed] [Google Scholar]
- Kolb HC, & Sharpless KB (2003). The growing impact of click chemistry on drug discovery. Drug Discov. Today, 8, 1128–1137. [DOI] [PubMed] [Google Scholar]
- Koniev O, & Wagner A (2015). Developments and recent advancements in the field of endogenous amino acid selective bond forming reactions for bioconjugation. Chem. Soc. Rev, 44, 5495–5551. [DOI] [PubMed] [Google Scholar]
- Laps S, Sun H, Kamnesky G, & Brik A (2019). Palladium-mediated direct disulfide bond formation in proteins containing S-acetamidomethyl-cysteine under aqueous conditions. Angew Chem. Int. Ed. Engl, 58, 5729–5733. [DOI] [PubMed] [Google Scholar]
- Lemieux GA, De Graffenried CL, & Bertozzi CR (2003). A fluorogenic dye activated by the staudinger ligation. J. Am. Chem. Soc, 125, 4708–4709. [DOI] [PubMed] [Google Scholar]
- Li M, Brooks CL, Kon N, & Gu W (2004). A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell, 13, 879–886. [DOI] [PubMed] [Google Scholar]
- Liu CC, & Schultz PG (2010). Adding new chemistries to the genetic code. Annu. Rev. Biochem, 79, 413–444. [DOI] [PubMed] [Google Scholar]
- Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, et al. (2008). HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol, 3, 373–382. [DOI] [PubMed] [Google Scholar]
- MacDonald JI, Munch HK, Moore T, & Francis MB (2015). One-step site-specific modification of native proteins with 2-pyridinecarboxyaldehydes. Nat. Chem. Biol, 11, 326–331. [DOI] [PubMed] [Google Scholar]
- Maddika S, Kavela S, Rani N, Palicharla VR, Pokorny JL, Sarkaria JN, & Chen J (2011). WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol, 13, 728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T, & Dixon JE (1998). The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem, 273, 13375–13378. [DOI] [PubMed] [Google Scholar]
- Maehama T, Taylor GS, & Dixon JE (2001). PTEN and myotubularin: novel phosphoinositide phosphatases. Annu. Rev. Biochem, 70, 247–279. [DOI] [PubMed] [Google Scholar]
- Malakhov MP, Mattern MR, Malakhova OA, Drinker M, Weeks SD, & Butt TR (2004). SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genomics, 5, 75–86. [DOI] [PubMed] [Google Scholar]
- Mali SM, Singh SK, Eid E, & Brik A (2017). Ubiquitin signaling: chemistry comes to the rescue. J. Am. Chem. Soc, 139, 4971–4986. [DOI] [PubMed] [Google Scholar]
- Matico R, Szewczuk LM, Pietrak B, Chen S, Dul E, Bonnette WG, .et al. (2019). Modular protein ligation: a new paradigm as a reagent platform for preclinical drug discovery. Sci. Rep, 9, 13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means GE, & Feeney RE (1990). Chemical modifications of proteins: history and applications. Bioconjug. Chem, 1, 2–12. [DOI] [PubMed] [Google Scholar]
- Meledin R, Mali SM, Singh SK, & Brik A (2016). Protein ubiquitination via dehydroalanine: development and insights into the diastereoselective 1,4-addition step. Org. Biomol. Chem, 14, 4817–4823. [DOI] [PubMed] [Google Scholar]
- Mohapatra S, Lin CT, Feng XA, Basu A, & Ha T (2019). Single-molecule analysis and engineering of DNA motors. Chem. Rev [DOI] [PubMed] [Google Scholar]
- Morotti A, Panuzzo C, Crivellaro S, Carra G, Guerrasio A, & Saglio G (2015). HAUSP compartmentalization in chronic myeloid leukemia. Eur. J. Haematol, 94, 318–321. [DOI] [PubMed] [Google Scholar]
- Muir TW, Sondhi D, & Cole PA (1998). Expressed protein ligation: a general method for protein engineering. Proc. Natl. Acad. Sci. U. S. A, 95, 6705–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson BL, Kiessling LL, & Raines RT (2000). Staudinger ligation: a peptide from a thioester and azide. Org. Lett, 2, 1939–1941. [DOI] [PubMed] [Google Scholar]
- Nilsson BL, Kiessling LL, & Raines RT (2001). High-yielding Staudinger ligation of a phosphinothioester and azide to form a peptide. Org. Lett, 3, 9–12. [DOI] [PubMed] [Google Scholar]
- Noren CJ, Anthony-Cahill SJ, Griffith MC, & Schultz PG (1989). A general method for site-specific incorporation of unnatural amino acids into proteins. Science, 244, 182–188. [DOI] [PubMed] [Google Scholar]
- O’Shannessy DJ, Dobersen MJ, & Quarles RH (1984). A novel procedure for labeling immunoglobulins by conjugation to oligosaccharide moieties. Immunol. Lett, 8, 273–277. [DOI] [PubMed] [Google Scholar]
- Palmer JT, Rasnick D, Klaus JL, & Bromme D (1995). Vinyl sulfones as mechanism-based cysteine protease inhibitors. J. Med. Chem, 38, 3193–3196. [DOI] [PubMed] [Google Scholar]
- Riggs JL, Seiwald RJ, Burckhalter JH, Downs CM, & Metcalf TG (1958). Isothiocyanate compounds as fluorescent labeling agents for immune serum. Am. J. Pathol, 34, 1081–1097. [PMC free article] [PubMed] [Google Scholar]
- Rosen CB, & Francis MB (2017). Targeting the N-terminus for site-selective protein modification. Nat. Chem. Biol, 13, 697–705. [DOI] [PubMed] [Google Scholar]
- Rostovtsev VV, Green LG, Fokin VV, & Sharpless KB (2002). A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem. Int. Ed. Engl, 41, 2596–2599. [DOI] [PubMed] [Google Scholar]
- Saxon E, & Bertozzi CR (2000). Cell surface engineering by a modified Staudinger reaction. Science, 287, 2007–2010. [DOI] [PubMed] [Google Scholar]
- Shen K, Cole PA (2003). Conversion of a tyrosine kinase protein substrate to a high affinity ligand by ATP linkage. J. Am. Chem. Soc, 125, 16172–16173. [DOI] [PubMed] [Google Scholar]
- Shen K, Hines AC, Schwarzer D, Pickin KA, Cole PA (2005). Protein kinase structure and function analysis with chemical tools. Biochem. Biophys. Acta, 1754, 65–78. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, & Mann M (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem, 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Sletten EM, & Bertozzi CR (2009). Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem. Int. Ed. Engl, 48, 6974–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, & Pandolfi PP (2008). The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature, 455, 813–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speers AE, & Cravatt BF (2004). Profiling enzyme activities in vivo using click chemistry methods. Chem. Biol, 11, 535–546. [DOI] [PubMed] [Google Scholar]
- Theile CS, Witte MD, Blom AE, Kundrat L, Ploegh HL, & Guimaraes CP (2013). Site-specific N-terminal labeling of proteins using sortase-mediated reactions. Nat. Protoc, 8, 1800–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todrick A, & Walker E (1937). A note on the combination of cysteine with allyl isothiocyanate. Biochem. J, 31, 297–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert TJ, & Wong CH (2002). New methods for proteomic research: preparation of proteins with N-terminal cysteines for labeling and conjugation. Angew Chem. Int. Ed. Engl, 41, 2171–2174. [DOI] [PubMed] [Google Scholar]
- Tornoe CW, Christensen C, & Meldal M (2002). Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem, 67, 3057–3064. [DOI] [PubMed] [Google Scholar]
- Truce WE, & Wellisch E (1952). Michael type condensations with methyl vinyl sulfone. J. Am. Chem. Soc, 74, 2881–2884. [Google Scholar]
- van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, . et al. (2005). GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol. Cell, 17, 695–707. [DOI] [PubMed] [Google Scholar]
- Vaughan JC, Dempsey GT, Sun E, & Zhuang X (2013). Phosphine quenching of cyanine dyes as a versatile tool for fluorescence microscopy. J. Am. Chem. Soc, 135, 1197–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Ramaswamy S, Nakamura N, & Sellers WR (2000). Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell Biol, 20, 5010–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland T, Bokelmann E, Bauer L, Lang HU, Lau H (1953). Uber peptidsynthesen. 8. Mitteilung bildung von S-haltigen peptiden durch intramolekulare wanderung von aminoacylresten. Justus Liebigs Ann. Chem, 583, 129–149. [Google Scholar]
- Williamson DJ, Fascione MA, Webb ME, & Turnbull WB (2012). Efficient N-terminal labeling of proteins by use of sortase. Angew Chem. Int. Ed. Engl, 51, 9377–9380. [DOI] [PubMed] [Google Scholar]
- Witus LS, Moore T, Thuronyi BW, Esser-Kahn AP, Scheck RA, Iavarone AT, & Francis MB (2010). Identification of highly reactive sequences for PLP-mediated bioconjugation using a combinatorial peptide library. J. Am. Chem. Soc, 132, 16812–16817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witus LS, Netirojjanakul C, Palla KS, Muehl EM, Weng CH, Iavarone AT, & Francis MB (2013). Site-specific protein transamination using N-methylpyridinium-4-carboxaldehyde. J. Am. Chem. Soc, 135, 17223–17229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby CA, & Dixon JE (2014). Pten. Annu. Rev. Biochem, 83, 641–669. [DOI] [PubMed] [Google Scholar]
- Yi L, Sun H, Itzen A, Triola G, Waldmann H, Goody RS, & Wu YW (2011). One-pot dual-labeling of a protein by two chemoselective reactions. Angew Chem. Int. Ed. Engl, 50, 8287–8290. [DOI] [PubMed] [Google Scholar]
- Zhang L, & Tam JP (1996). Thiazolidine formation as a general and site-specific conjugation method for synthetic peptides and proteins. Anal. Biochem, 233, 87–93. [DOI] [PubMed] [Google Scholar]


