Abstract
Lipid-nanodiscs have been shown to be an exciting innovation as a membrane-mimicking system for studies on membrane proteins by a variety of biophysical techniques, including NMR spectroscopy. Although NMR spectroscopy is unique in enabling the atomic-resolution investigation of dynamic structures of membrane-associated molecules, it, unfortunately, suffers from intrinsically low sensitivity. The long data acquisition often used to enhance the sensitivity is not desirable for sensitive membrane proteins. Instead, paramagnetic relaxation enhancement (PRE) has been used to reduce NMR data acquisition time or to reduce the amount of sample required to acquire an NMR spectra. However, the PRE approach involves the introduction of external paramagnetic probes in the system, which can induce undesired changes in the sample and on the observed NMR spectra. For example, the addition of a paramagnetic ions, as frequently used, can denature the protein via direct interaction and also through sample heating. In this study, we show how the introduction of paramagnetic tags on the outer belt of polymer-nanodiscs can be used to speed-up data acquisition by significantly reducing the spin-lattice relaxation (Tl) times with minimum-tono alteration of the spectral quality. Our results also demonstrate the feasibility of using different types of paramagnetic ions (Eu3+, Gd3+, Dy3+, Er3+, Yb3+) for NMR studies on lipid-nanodiscs. Experimental results characterizing the formation of lipid-nanodiscs by the metal-chelated polymer, and their increased tolerance toward metal ions are also reported.
Keywords: Polymer-nanodiscs, SMA, PRE, paramagnetic NMR, lanthanides
Graphical Abstract
Introduction
Membrane proteins are ubiquitous in every cell and perform a plethora of fundamental functions for the living organisms[l], Despite their importance, there is still a large gap in the structural studies of membrane proteins when compared to their soluble protein counterparts. This is mainly because of the difficulties in mimicking native lipid membranes and using the membrane mimetics to stabilize native-like folding and function of membrane proteins[2]. While detergents are still in use in the purification and structural studies of membrane proteins, it is known that detergents are capable of denaturing membrane proteins. Other well-utilized model membranes are bicelles and liposomes, which also have limitations[3]. Lipid nanodiscs, in recent years, have shown great potential in the structural biology of membrane proteins.[1,4–10], Nanodiscs are disc-shaped lipid bilayer patches surrounded by an amphiphilic macromolecule that can be made up of membrane scaffold proteins (MSPs)[1,11–13], peptides[14], or synthetic-polymers [5,9,15–22], Although different types of nanodiscs are used, most MSP and peptide based nanodiscs show several limitations that hinder the characterization of reconstituted membrane proteins by several biophysical techniques [18]. On the other hand, the synthetic polymer-based nanodiscs have been shown to provide a number of flexibilities and choices, enabling a variety of applications. Many different types of polymer belts have been reported in the literature, and they have been used to demonstrate that polymer based nanodisc technology is suitable for the study of biomacromolecules by most biophysical techniques including both solution and solid-state NMR spectroscopy[15,23–28], While NMR is unique in rendering the measurement of atomic-resolution dynamics at different time scales, it, however, has the significant disadvantage of being a relatively lowsensitivity technique[29]. This disadvantage is amplified by the instability or scarce amount of most biologically interesting membrane proteins and also to characterize short-lived intermediates such as amyloid oligomers. This low sensitivity is most commonly overcome by either expensive isotopic labeling and encumberingly long experimental times which is not desirable for heat-sensitive proteins[29–35]. Another solution is to increase the concentration of the sample, but it is often untenable for many protein systems due to the potential instability and aggregation issues of the proteins such as amyloid proteins. The sensitivity of NMR spectroscopy can also be enhanced by dynamic nuclear polarization (DNP) technique. This methodology is increasingly used and has already offered significant contributions in the fields of structural biology and materials [36–42], However, one of the most important ways to increase the sensitivity of NMR is by the addition of paramagnetic dopants to utilize the paramagnetic relaxation enhancement (PRE)[43–50].
PRE effect can be used to enhance NMR sensitivity by shortening the spin-lattice (T1) relaxation properties of the nuclei[51,52], Briefly, relaxation is a process in which the thermal equilibrium of the nuclear spin states is regained after the perturbation of applied radio-frequency (RF) pulses. The relaxation of multiple-spin systems can be an extremely complex process to describe, which can be further complicated by chemical exchange and local motions; but it can be phenomenologically described by two of the major relaxation mechanisms: longitudinal (or spin-lattice or T1) and transverse (or spin-spin or T2) relaxations as defined by the Bloch equations[53], PRE-NMR involves the introduction of a paramagnetic salt into the sample or external paramagnetic-tags in the investigated system. Based on the chosen probe, the PRE effect can alter the relaxation in different ways[52,54–56]. Unfortunately, it is not possible to separately influence T1 and T2 values; so, while affecting one of the relaxation parameters, it is inevitable to change the other. This aspect can cause undesired effects on the spectra, such as line-broadening (related to T2-shortening) and peak-shifts (due to hyperfine shift )[43,50,57–60] that can negatively affect the overall quality of the resultant spectra. Additionally, sample preparation is complicated by a further step of "tagging," which need not always proceed to completion[57].
Since the PRE effect can be used to speed-up the Tl process, it can be utilized to accelerate data acquisition by reducing the recycle delay between successive scans used for signal averaging in an NMR experiments. In addition, the paramagnetic effect induced shift in resonance frequency (known as hyperfine shift and composed of two contributions: the contact and the pseudo-contact shifts or PCS) can be used to obtain structural information such as topological analysis and distance measurements since paramagnetic effects are typically proportional to l/rn (n = 6 for PRE and 3 for PCS), where r is the distance between the paramagnetic tag and the investigated nuclei[61–63].
The versatility and flexibility of paramagnetic methods are benefited from a large pool of paramagnetic tags to choose from and the ways of incorporation to access the sites of interest[64–73]. Indeed, the use of diamagnetic and paramagnetic ions bound to specific positions or solvent-exposed surfaces of macromolecules or supramolecular aggregates both in solution and solid-state can reveal structural and topological information.
We recently reported a nanodisc forming synthetic polymer, SMA-EA-DOTA[74], In addition to the already reported advantages of forming polymer-based lipid-nanodiscs, this copolymer enables the introduction of paramagnetic metal ions to the DOTA units in the polymer located on the outer rim of polymer-nanodiscs. Thus, this DOTA-unit containing polymer avoids the presence of free paramagnetic ions in solution and the use of paramagnetically-tagged lipids[52,75]. As a result, an SMA-EA-DOTA polymer-based nanodisc offers a planar lipid bilayer to reconstitute membrane protein(s) without the interference from the added paramagnetic metal ions. The aim of this study is to reduce the spin-lattice relaxation time (T1) of the sample with a minimum line broadening due to a reduced spin-spin relaxation time (T2) by using the paramagnetic metals chelated to the DOTA units of the polymers in nanodiscs. Due to the size flexibility of polymer nanodiscs[76], a wide variety of NMR conditions can be utilized for solution NMR, partial alignment for RDC measurements by solution NMR, and a higher degree of alignment for solid-state NMR experiments[77]. Because of this flexibility, a library of a variety of metal ions for differing conditions of PRE is needed. In addition, the introduction of paramagnetic metals can enable the applications of EPR experiments[78,79]. In this study, we report a systematic investigation of the PRE effects from five different paramagnetic metal ions (Eus+, Gd5+, Dy3+, Er3+, Yb3+) for NMR applications[50,57,60,80–85].
Materials and Methods
1. Reagents and Materials
Poly(Styrene-co-Maleic Anhydride)-Cumene terminated, SMAnh, Mw ~ 1.6 kDa, anhydrous 1Methyl2-pyrrolidone (NMP), 2-Aminoethanol (EA), Triethylamine, Trifluoroacetic Acid (TFA), Diethyl Ether (Et20), Hydrochloric Acid (HCI), Sodium Hydroxide (NaOH), 4-(2-Hydroxyethyl)Piperazine-l-ethanesulfonic acid (HEPES), Sodium Chloride (NaCI), Europium(lll) Chloride Hexahydrate (EuCI3-6H20), Gadolinium(lll) Chloride Hexahydrate (GdCI3-6H20), Dysprosium(lll) Chloride Hexahydrate (DyCI3-6H20), Erbium(lll) Chloride Hexahydrate (ErCI3-6H20), Ytterbium(lll) Chloride Hexahydrate (YbCI3-6H20) were purchased from SigmaAldrich® (St. Louis, Missouri). l,4,7,10-Tetraazacyclododecane-l,4,7-tris(t-butyl-acetate)-10(aminoethylacetamide) was purchased from Macrocyclics® Inc. (Plano, Texas). l,2-dimyristoyl-sn-glycero-3phosphocholine (DMPC) was purchased from Avanti® Polar Lipids, Inc. (Alabaster, Alabama).
2. Polymer synthesis and characterization
2.1.
SMA-EA copolymer was synthesized, purified, and characterized according to the procedure described in the literature[76].
2.1.
SMA-EA-DOTA copolymer was synthesized, as described previously[74]. In short, 1 g of SMAnh, 0.435 g of 1,4,7,10-Tetraazacyclododecane-l,4,7-tris(t-butyl-acetate)-10-(aminoethylacetamide) (amino-DOTA), and 1 mL of Triethylamine were dissolved in 60 ml of anhydrous NMP and heated to 70 °C under continuous stirring (Step-1). The addition of an excess of ethanolamine in the presence of an extra 1 mL of triethylamine for 2 more hours at the same temperature in the same round bottom flask completed the ǹucleophilic ring-opening reaction (Step-2). The final product was precipitated using 1 M HCI. To deprotect the chelator units, the polymer was dissolved in TFA and reacted for 2 hours at room temperature under gentle stirring (Step-3). Finally, the product was precipitated using diethyl ether and washed multiple times with deionized water prior to lyophilization. The 3-step reaction yielded about 600 mg. The reaction scheme is presented in Figure S1.
2.3.
The newly synthesized polymer was characterized by FT-IR. The shift of the carbonyl stretching frequency from 1770 cm−1 to 1702 cm−1 indicates the conversion of the anhydride to the amide, confirming the success of the reaction. FT-IR spectra are reported in Figure S2.
3. Polymer nanodiscs preparation
Stock solutions of each copolymer (SMA-EA and SMA-EA-DOTA) were obtained by dissolving the desired amount of powder in a 0.1 M NaOH solution. The pH was then adjusted to 7.4 using 1 M HCI. Polymer-based lipid nanodiscs were prepared by mixing the desired quantity of DMPC lipids and polymers in the ratio 1:1 by weight from 20 mg/mL stock solutions. Each sample was incubated overnight at room temperature prior to its use.
4. Polymer nanodiscs characterization
4.1. Size Exclusion Chromatography (SEC)
Polymer-based phospholipid nanodiscs were purified by SEC using a self-packed Sephadex 200 16/600 column operated on a GE Healthcare® AKTA purifier. Samples were eluted at room temperature and at a flow rate of 1 mL/min. The elution was monitored using a UVdetector at λ = 254 nm. Chromatograms are shown in Figure S3.
4.2. Dynamic Light Scattering (DLS)
All the DLS experiments were performed on a Wyatt Technology® DynaPro NanoStar® using a 1 μL quartz MicroCuvette. The size distribution profiles for both DMPC: SMA-EA 1:1 w/w and DMPC: SMA-EA-DOTA 1:1 w/w polymer-lipid nanodiscs used in this study are reported in Figure 1d.
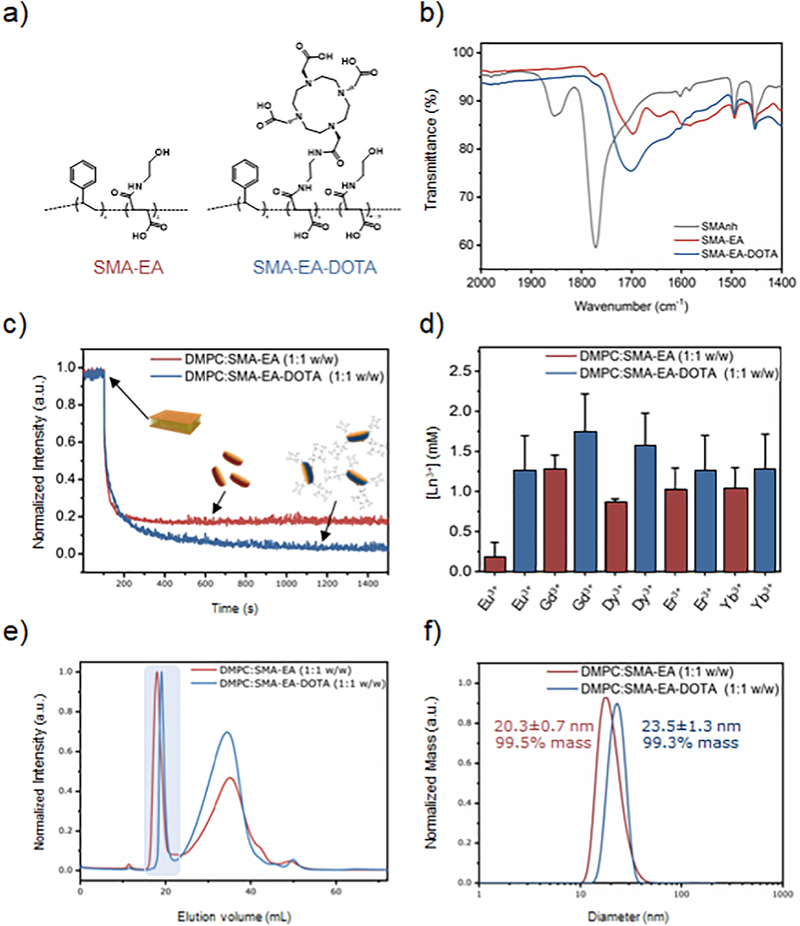
Figure 1. Characterization of polymers and polymer nanodiscs.
a) Molecular structures of SMA-EA and SMA-EADOTA. b) FT-IR spectra of the starting material (SMAnh) and synthetic polymers. FT-IR results confirm the functionalization of the starting material (SMAnh in dark gray) and similarities among SMA-EA (in red) and SMA-EADOTA (in blue). Full spectra are included in the Supporting Information (Figure S2). c) Dissolution of multilamellar vesicles (MLVs) by SMA-EA (red) and by SMA-EA-DOTA (blue) for a 1:1 lipid:polymer weight ratio, d) Tolerance of DMPC:SMA-EA 1:1 w/w and DMPC: SMA-EA-DOTA 1:1 w/w macro-nanodiscs against different metal ions. Size exclusion chromatography (e) and DLS (f) profiles for 1:1 w/w ratio of DMPC:SMA-EA and DMPC:SMA-EA-DOTA samples. The DLS profiles were obtained after SEC purification for the nanodiscs fraction highlighted in (e).
4.3. Static Light Scattering (SLS)
All the SLS experiments were performed using a 4 mL cuvette (1 cm optical path) under continuous stirring at 25°C on a FluoroMax-4® Spectrofluorometer from Horiba Scientific®. The excitation wavelength was set at 400 nm while the emission wavelength was set at 404 nm, and the slit was set to 2 nm.
a). Solubilization Experiments
The time-dependent solubilization of the DMPC suspension in 10 mM HEPES buffer 50 mM NaCI was monitored by the intensity of scattered light at a 90° angle. The solubilization power of two different polymers SMA-EA and SMA-EA-DOTA was tested on a 1 mg/mL DMPC MLVs solution. The amount of polymer added was equivalent (1:1 w/w ratio) for all of them. Data are shown in Figure 1c.
b). Metal ion titrations
Polymer-based lipid nanodiscs tolerance toward the investigated cations was tested by titrating a 1 mg/mL solution of DMPC: copolymer 1:1 w/w nanodiscs in 10 mM HEPES buffer at pH = 7.40 with a 4 M solution of each metal. The results are shown in Figure 1b.
4.4. NMR Spectroscopy
Solution NMR experiments were performed at 11.75 T on a 500 MHz Bruker Avance III HD spectrometer. NMR samples were prepared according to the procedure described in the “Nanodisc Formation” section and adding the desired concentration of metal ions (0, 5 × 10−4, 2.5 × 10−2, 5 × 10−2, 0.125, 0.250, 0.500, 1.250, and 2.500 mM), then lyophilized for 24 hours prior to resuspension in 600μL of D2O and then transferred to 5 mm Norrell® Sample Vault Series™ glass tubes and placed in a commercial 5 mm quadruple resonance 2H/1H/15N/13C Bruker round-coil TXI™ 500 SB probe. The experiments were performed in D2O at neutral pH at three different temperatures, 15, 25, and 35°C. Each sample was made using 4 mg of lipids, and an equal amount (by weight) of SMA-EA-DOTA polymer, titrated at pH 7.4 to obtain a DMPC: SMA-EA-DOTA (1:1 w/w) system. Each concentration of each paramagnetic metal investigated was prepared individually and tested on three sequential different experiments.
a). 1H-NMR
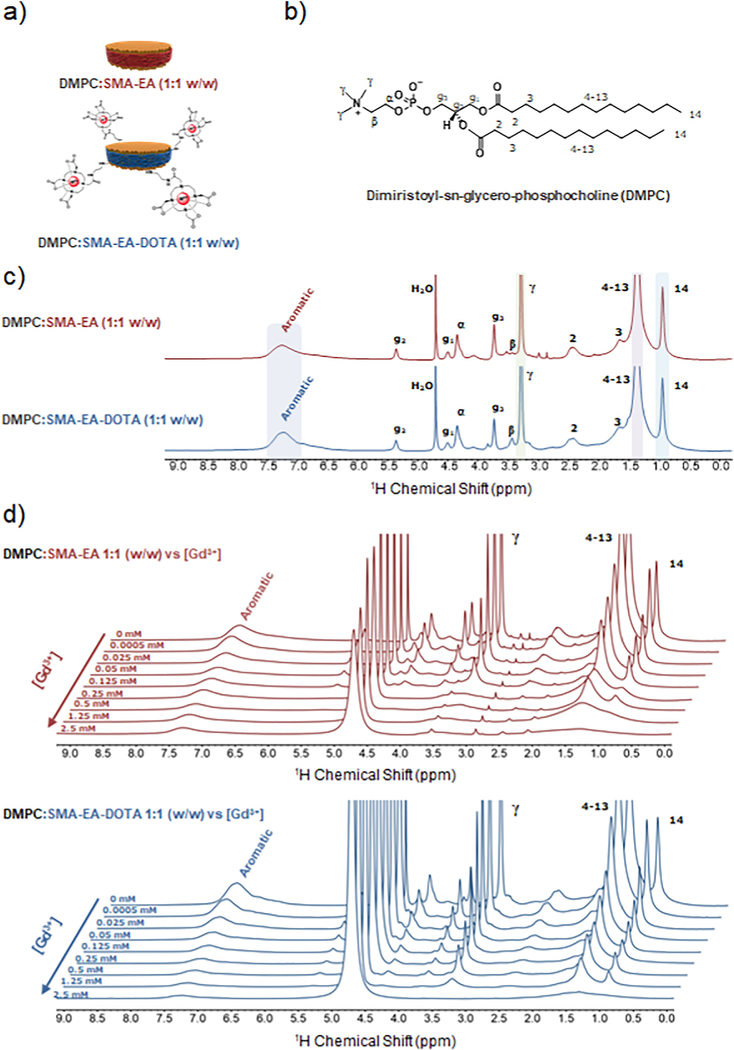
1H spectra were recorded by collecting 32 scans with a spectral width of 25 ppm. The transmitter frequency offset was set at 4.7 ppm. Acquisition time and relaxation delay were respectively set at 0.8 s and 1.0 s. 1H-NMR spectra for DMPC:SMA-EA 1:1 (w/w) and DMPC:SMA-EADOTA 1:1 (w/w) are shown in Figure 3c.
Figure 3. NMR spectra of nanodiscs in the presence of Gd3+ ions.
a) Schematic representation of a macro-nanodisc, b) Molecular structure of l,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC). c) 1H NMR spectra of 1:1 w/w DMPC:SMA-EA (red) and DMPC:SMA-EA-DOTA (blue) macro-nanodiscs, d) 1H NMR spectra of 1:1 w/w DMPC: SMA-EA (red) and SMA-EA-DOTA (blue) nanodiscs titrated with the indicated amount of Gd3+ ions. All NMR were obtained using 4 mg of lipids in 10 mM phosphate buffer (pH = 7.4) in 100% D2O at 35°C.
b). Inversion recovery experiment
To measure T1, an inversion recovery experiment was performed. Fifteen data points from 0.001 s to 10.0 s were collected by acquiring 8 scans with a spectral width of 10 ppm. The transmitter frequency offset was set at 4.7 ppm. Acquisition time and relaxation delay were respectively set at 0.001 s and 10 s. Data are shown in Figures S6–S11.
c). Data Processing
Data have been processed using both Bruker Topspin™ 3.2 and Mestrelab Research S.L. MestReNova™ software was used to integrate the peaks of interest.
Results and Discussion
Formation of SMA-EA-DOTA lipid-nanodiscs
SMA-EA-DOTA copolymer was synthesized according to the procedure as briefly described above[74]. Figure 1a shows the chemical structures of SMA-EA and SMA-EA-DOTA co-polymers (la). FT-IR characterization (Figure 1b) confirms the functionalization of SMA-EA and DOTA, as reported previously.[74] The ability to solubilize l,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) lipid vesicles (or aggregates) was tested at room temperature (~25 °C). For this study, about 20–25 nm size polymer-nanodiscs formed by the addition of a 1:1 lipids-to-polymer weight ratio (1:1 w/w) was used (Figure 1(e and f)). Data shown in Figure 1c confirms similar solubilization capabilities of SMA-EA and SMA-EA-DOTA copolymers. Since only a small number of DOTA units per polymer chain were introduced, both SMA-EA and SMA-EA-DOTA have comparable nanodiscs forming capabilities as shown by the experimental results in Figure 1. Additionally, dynamic light scattering (DLS) profiles (Figure 1f) after size-exclusion chromatography (SEC) purification (Figure 1e) confirm the formation of nanodiscs and their isolation. These large-size nanodiscs (>20 nm) are also known as "macro-nanodiscs" which can be aligned in the presence of an external magnetic field as demonstrated in the previous studies and are useful for solid-state NMR studies.[76]
SMA-EA-DOTA nanodiscs are more stable than SMA-EA nanodiscs in the presence of metals
The tolerance of the polymer nanodiscs toward metal ions was examined using static light scattering (SLS) experiments as described above. The SLS experimental results shown in Figure 1d demonstrate the enhanced-tolerance of DOTA containing polymer nanodiscs in the presence of metal ions. In addition to the above-mentioned FT-IR data (Figure 1), the SLS results confirm the successful functionalization of SMA-EA with DOTA. Because of the presence of DOTA groups in SMA-EA-DOTA, the high binding affinity for various lanthanides renders a near complete chelation and thus the high tolerance observed in the presence of various metal ions as shown in Figure 1d[86,87].
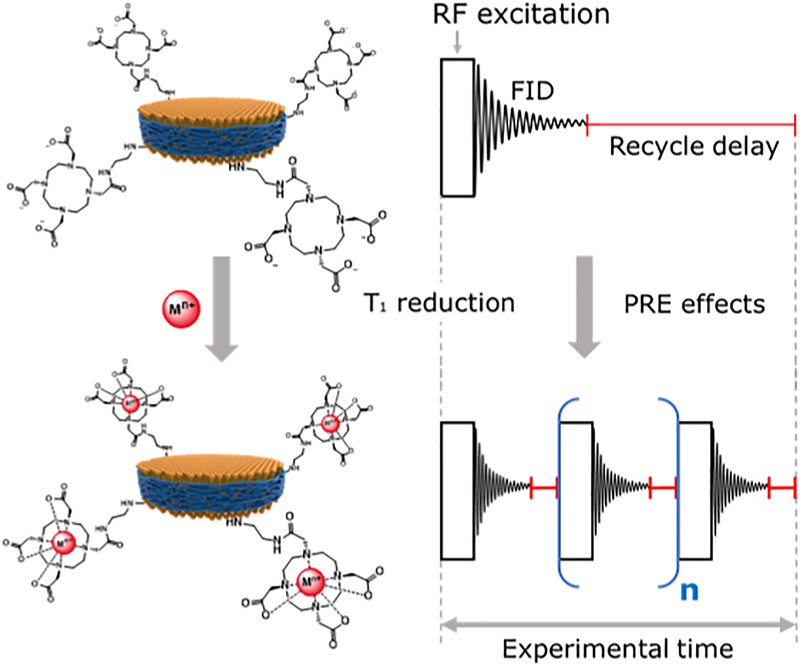
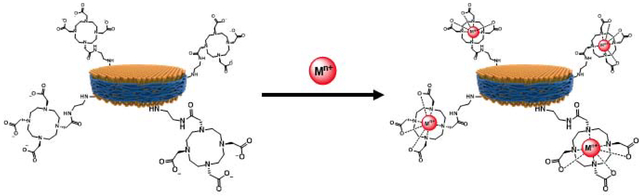
Figure 2 shows a schematic representation of the chelation of paramagnetic cations to SMA-EADOTA co-polymer nanodiscs and its use for rapid NMR data acquisition. A first successful application of Cu2+ based PRE-NMR with this polymer system was recently reported from our research group which also probed the interaction between polymer nanodiscs and G-quadruplex[74], In this study, the use of paramagnetic properties of five additional trivalent cations from elements of the f-block of the periodic table (Eu3+, Gd3+, Dy3+, Er3+, Yb3+) is investigated by measuring the spin-lattice (Tl) relaxation times of protons for various concentrations of the metal ions.
Figure 2. Use of paramagnetic metal-chelated polymers to speed-up NMR data acquisition.
A schematic representation of how SMA-EA-DOTA copolymer-based nanodiscs chelated with paramagnetic metals can be used to reduce T1 relaxation times and to shorten the recycle delay between the successive scans in NMR data acquisition.
Based on the assignment of NMR peaks as reported previously[74,88], it is possible to assign the 1H peaks observed from lipid and polymer components of nanodiscs: 1 ppm for protons from the lipid acyl (-CH3), 1.4 ppm for protons from the lipid acyl (-CH2-), 3.3 ppm for protons from the lipid head quaternary ammonium (-CH3, γ), and 7.3 ppm for protons from the aromatic styrene group of the polymer. The structure of DMPC and peak assignment of 1H NMR spectra for both DMPC:SMA-EA and DMPC:SMA-EADOTA polymer-based nanodiscs are shown in Figure 3c. The various paramagnetic metals used in the present study (Eu3+, Gd3+, Dy3+, Er3+, Yb3+) were chosen among the 14 f-block elements to be representative of the magnetic differences reported in the literature[50,82,84,88,89].
Figure 3d shows 1H NMR spectra of SMA-EA and SMA-EA-DOTA nanodiscs containing DMPC lipid; and the indicated concentration of Gd3+ ions. Here, using two different types of nanodisc systems (with anc without the DOTA) of comparable sizes (Figure 1e and 1f), one noticeable difference we observed was that significant line-broadening occurred at a much lower Gd3+ ions concentration (0.25 mM) for the SMA-E/5 polymer nanodiscs as compared to that observed for SMA-EA-DOTA. The severe line-broadening observec for the lipid headgroup’s methyl protons of SMA-EA polymer nanodiscs is most likely due to the presence ol a higher population of free Gd3+ ions that directly bind to the zwitterionic lipid headgroup. On the othei hand, the SMA-EA-DOTA polymer containing nanodiscs attract the Gd3+ ions from the sample to be chelated to the DOTA unit on the polymer belt of the nanodisc and avoid the line-broadening effects. The inversion-recovery NMR experiments were performed to measure the T1 values of protons in order tc examine the effect of the added paramagnetic metal ions in the sample (see Materials and Methods). The T1 values were measured by integrating the observed 1H NMR peaks of interest and then fitting the experimentally measured data to the following equation:
| (1) |
Where Mz(τ) is the z-component of the magnetization dependent on time τ, Mz,eq is the z-magnetization al thermal equilibrium, τ is the time between the 180° and 90° pulses, and Tl is the spin-lattice relaxatior time.
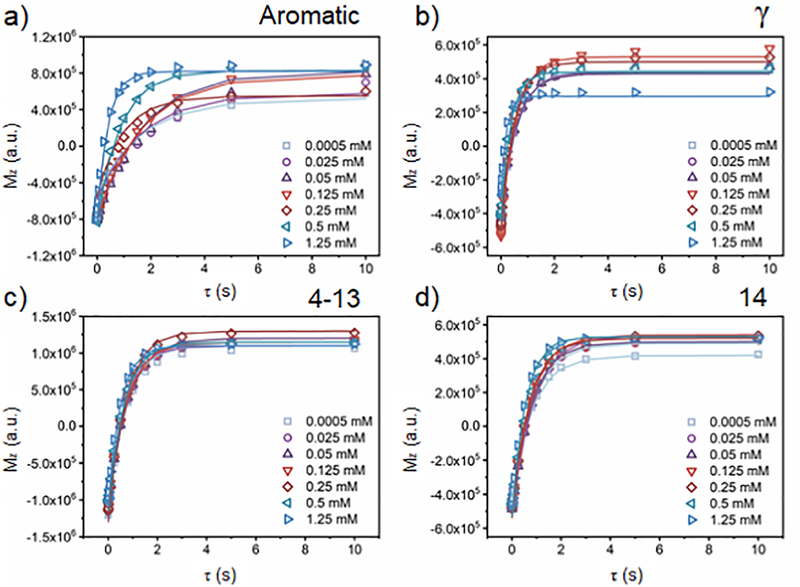
Figure 4 shows the fitting curves obtained for the 1:1 w/w DMPC:SMA-EA-DOTA nanodiscs sample foi various concentrations of Gd3+ ions. These experiments were also carried out for the other paramagnetic metal ions, and the results are included in the Supporting Information (Figures S6–S11).
Figure 4. Measurement of T1 for protons.
Spin-inversion NMR experimental data obtained from 1:1 w/w DMPC:SMA-EA-DOTA nanodiscs to determine Tl values of protons for varying concentrations of Gd3+ as indicated. Equation 1 was used to obtain the best-fitting values given in Table S2.
The resulting T1 of the four-aforementioned peaks (aromatic, γ, methylene and methyl groups) are plotted against [Gd3+] for both DMPC:SMA-EA (1:1 w/w) and DMPC:SMA-EA-DOTA (1:1 w/w) polymer-nanodiscs samples in Figures S6–S7. Figure 5 shows the T1 values for the four 1H peaks of interest obtained for the different metal ions. By comparing the different lanthanide trivalent cations ([Ln3+]) at the same molar concentration, different effects are noted. When [Ln3+] = 0.5 mM, Gd3+, followed by Dy3+, showed the largest Trreduction for all of the peaks as compared to a reference diamagnetic system made up of DMPC:SMA-EA-DOTA 1:1 (w/w). The aromatic peak from the styrene fraction of the polymer is the most affected by the paramagnetic ions due to the proximity of the styrene group to the DOTA uniťs chelated metals on the nanodiscs belt. Gd3+ and Dy3+ ions show T1 times reduced respectively by 1.9- and 1.3-times. Even though Gd3+ ions provided the greatest T1 relaxation effect at the lowest concentration among all the investigated metals, other metal ions, like Dy34, had little to no significant line-broadening effect up to 1.25 mM (Figure S8 and Table S3). Metal ions such as Er3+, Eu3+, and Yb3+ showed significant line-broadening at the highest concentration investigated (2.5 mM) but no effects on T1-reduction were observed (Figures S7Sll and Tables S2–S6). Data for [Er3+] = 2.5 mM are not shown because the sample showed instability.
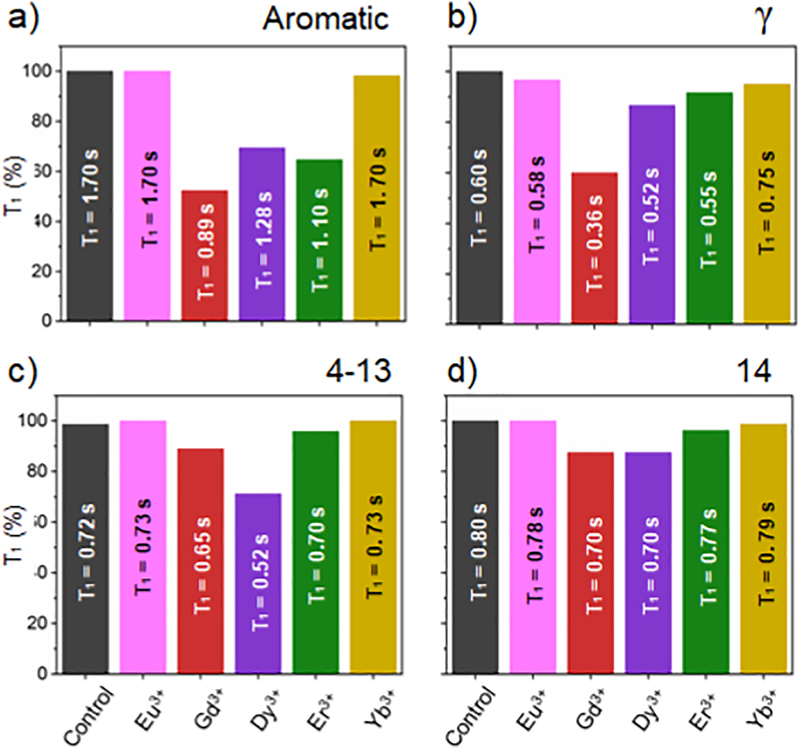
Figure 5. Efficiency of paramagnetic metal ions in shortening T1.
T1 comparison among the different investigated metal ions at [Ln3+] = 0.5 mM. The black bar represents the data obtained from a control sample, 1:1 (w/w) DMPC: SMA-EA-DOTA without paramagnetic metal ions. Data shown for 1H peaks of: a) styrene/aromatic group, b) y methyl groups in the quaternary ammonium of DMPC, c) the methylene groups from the acyl chains (C4-C13) of DMPC, and d) the terminal acyl-chain methyl groups (C14) of DMPC. Each of the NMR samples used in these measurements consisted of 4 mg of DMPC in 10 mM Phosphate buffer pH = 7.4 in D2O.
NMR peaks from the DMPC lipid are also T1 enhanced by the presence of the paramagnetic tags on the belt but to a lesser extent, especially for Eu3+, Er3+, and Yb3+ ions (Figure 5). Particularly Gd3+ ions shorten T1 values of protons from γ, methylene (C4-C13), and methyl-groups (C14) by respectively ~48%, ~40%, ~10%, and ~12% respectively. While line-broadening was observed for Gd3+ ions, the effects observed for other metals at 0.5 mM are on the following order: Eu3+ < Yb3+ < Er3+ < Dy3+. Eu3+ and Yb3+ do not show any T1-reduction. Er3+ ions showed an important paramagnetic relaxation enhancement on the aromatic peak from styrene groups (T1-reduction of ~35% at a [Er3+] = 0.5 mM) but weak-to-negligible effects for y, methylene (4–13), and methyl (14) peaks. The fact that DMPC’s acyl chains are less affected by the paramagnetic tags may be attributed to the large size of the nanodisc investigated) ~20 nm. We expect the PRE effect to be higher for very large macro-nanodiscs under high viscous conditions as they donť tumble fast and therefore the unaveraged 1H-1H dipolar couplings should aid the PRE effect from the metals present on the belt. Such results have been observed for large bicelles that align magnetically[90]. Overall, the effect of paramagnetic metals on shortening Ti can be ranked as Gd3+> Dy3+> Er3+> Eu3+> Yb3+. Figures S7–S11 show the stacking of ID 1H NMR spectra and the T1 fitting curves. Tables S1–S6 report the experimentally measured T1 values.
Because of the large size of the nanodiscs, within the spectral resolution, the observed proton NMR spectra do not show any shift in the observed resonance frequency when the nanodiscs are loaded with any of the lanthanides even for those that are considered as "shifting-agents" such as Eu3+, Dy3+, Er3+[50], Spectra and full titrations with the fitting of the experimental data are included in the Supporting Information (Figure S12).
Conclusions
SMA-based nanodiscs have shown to be a great innovation in biochemistry and biophysics, and are widely used as a membrane-mimicking system to investigate membrane proteins through several techniques. NMR spectroscopy, as a valuable non-disruptive technique, offers structural and dynamic information. Unfortunately, limitations such as its intrinsically low sensitivity result in long acquisition times to enhance the signal-to-noise ratio of NMR spectra. PRE can be used to reduce the acquisition times effectively but involves the introduction of external probes in the system and may cause undesired line-broadening in the spectra, when compared to the diamagnetic counterpart. To overcome these limitations, we have demonstrated the efficiency of SMA-EA-DOTA copolymer in T1-reduction. This modification of SMA-EA copolymer allows the use of PRE effects in nanodiscs samples. Particularly, the introduction of chelating units that strongly bind paramagnetic metals on the outer belt of the nanodiscs avoids the addition of paramagnetic dopants directly in buffer solutions, in the membrane protein of interest, and in the bilayer components. SMA-EA-DOTA-nanodiscs represent a much less invasive approach toward the preservation of the integrity of the sample, offering PRE effects in a native-like environment.
As demonstrated in this study, this approach can be used to speed up NMR data acquisition (up to ~50%) with minimum-to-no alteration of the spectral quality due to spin-spin relaxation enhancement. A comparison of the effects of different paramagnetic metals shows that Gd3+ and Dy3+ can be successfully used to shorten T1, and so, the recycle delay of NMR experiments. We believe that these results can broaden the applications of polymer-nanodiscs in the investigation of membrane proteins in a native-like environment, using both solution and solid-state NMR spectroscopy. Additionally, paramagnetically-labeled nanodiscs can be used for both dynamic nuclear polarization (DNP) NMR and electron paramagnetic resonance (EPR) studies. Both techniques require the presence of paramagnetic tags and could benefit from this improved version of polymer-nanodiscs.
Supplementary Material
Highlights.
SMA-EA-DOTA shows effective chelation of a variety of metal ions.
Polymer chelated paramagnetic lanthanide ions enable T1-reduction.
Eu3+, Gd3+, Dy3+, Er3+, and Yb3+ were investigated for PRE effects.
Acknowledgments
This research was supported by the National Institute of Health (NIH, GM084018 to A.R.). We thank Dr. Vojc Kocman for the help with NMR experiments and Dr. Thirupathi Ravula for providing the SMA-EA copolymer.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Denisov IG, Sligar SG, Nanodiscs in Membrane Biochemistry and Biophysics, Chem. Rev. 117 (2017)4669–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rawson S, Davies S, Lippiat JD, Muench SP, The changing landscape of membrane protein structural biology through developments in electron microscopy, Mol. Membr. Biol. 33 (2016) 12–22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Durr UHN, Gildenberg M, Ramamoorthy A, The magic of bicelles lights up membrane protein structure, Chem. Rev. 112 (2012) 6054–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bayburt TH, V Grinkova Y, Sligar SG, Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins, Nano Lett. 2 (2002) 853–856. [Google Scholar]
- [5].Knowles TJ, Finka R, Smith C, Lin YP, Dafforn T, Overduin M, Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer, J. Am. Chem. Soc. 131 (2009)7484–7485. [DOI] [PubMed] [Google Scholar]
- [6].Gulati S, Jamshad M, Knowles TJ, Morrison KA, Downing R, Cant N, Collins R, Koenderink JB, Ford RC, Overduin M, Kerr ID, Dafforn TR, Rothnie AJ, Detergent-free purification of ABC (ATPbinding-cassette) transporters, Biochem. J. 461 (2014) 269–278. [DOI] [PubMed] [Google Scholar]
- [7].Swainsbury DJK, Scheidelaar S, Foster N, van Grondelle R, Killian JA, Jones MR, The effectiveness of styrene-maleic acid (SMA) copolymers for solubilisation of integral membrane proteins from SMA-accessible and SMA-resistant membranes, Biochim. Biophys. Acta - Biomembr. 1859 (2017)2133–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Swainsbury DJK, Proctor MS, Hitchcock A, Cartron ML, Qian P, Martin EC, Jackson PJ, Madsen J, Armes SP, Hunter CN, Probing the local lipid environment of the Rhodobacter sphaeroides cytochrome be 1 and Synechocystis sp. PCC 6803 cytochrome b 6 f complexes with styrene maleic acid, Biochim. Biophys. Acta - Bioenerg. 1859 (2018) 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dorr JM, Koorengevel MC, Schafer M, V Prokofyev A, Scheidelaar S, van der Cruijsen EAW, Dafforn TR, Baldus M, Killian JA, Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs., Proc. Natl. Acad. Sci. U. S. A.Ill (2014)18607–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dörr JM, Scheidelaar S, Koorengevel MC, Dominguez JJ, Schäfer M, van Walree CA, Killian JA, The styrene-maleic acid copolymer: a versatile tool in membrane research, Eur. Biophys. J. 45 (2016) 3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bayburt TH, Carlson JW, Sligar SG, Reconstitution and imaging of a membrane protein in a nanometer-size phospholipid bilayer, J. Struct. Biol 123 (1998) 37–44. [DOI] [PubMed] [Google Scholar]
- [12].Denisov IG, Sligar SG, Nanodiscs for structural and functional studies of membrane proteins, Nat. Struct. Mol. Biol. 23 (2016) 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hagn F, Nasr ML, Wagner G, Assembly of phospholipid nanodiscs of controlled size for structural studies of membrane proteins by NMR, Nat. Publ. Gr 13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Toraya S, Nishimura K, Naito A, Dynamic structure of vesicle-bound melittin in a variety of lipid chain lengths by solid-state NMR, Biophys. J. 87 (2004) 3323–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Stroud Z, Hall SCL, Dafforn TR, Purification of membrane proteins free from conventional detergents: SMA, new polymers, new opportunities and new insights, Methods. 147 (2018) 106–117.. [DOI] [PubMed] [Google Scholar]
- [16].Dominguez Pardo JJ, Dörr JM, Iyer A, Cox RC, Scheidelaar S, Koorengevel MC, Subramaniam V, Killian JA, Solubilization of lipids and lipid phases by the styrene-maleic acid copolymer, Eur. Biophys. J. 46 (2017) 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Oluwole AO, Danielczak B, Meister A, Babalola JO, Vargas C, Keller S, Solubilization of Membrane Proteins into Functional Lipid-Bilayer Nanodiscs Using a Diisobutylene/Maleic Acid Copolymer, Angew. Chemie - Int. Ed 56 (2017) 1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Danielczak B, Meister A, Keller S, Influence of Mg2+ and Ca2+ on nanodisc formation by diisobutylene/maleic acid (DIBMA) copolymer, Chem. Phys. Lipids. 221 (2019) 30–38. [DOI] [PubMed] [Google Scholar]
- [19].Lee SC, Knowles TJ, Postis VLG, Jamshad M, Parslow RA, Lin YP, Goldman A, Sridhar P, Overduin M, Muench SP, Dafforn TR, A method for detergent-free isolation of membrane proteins in their local lipid environment, Nat. Protoc. 11 (2016) 1149–1162. [DOI] [PubMed] [Google Scholar]
- [20].Rajesh S, Knowles T, Overduin M, Production of membrane proteins without cells or detergents., N. Biotechnol. 28 (2011) 250–4. [DOI] [PubMed] [Google Scholar]
- [21].Overduin M, Klumperman B, Advancing membrane biology with poly(styrene-co-maleic acid)-based native nanodiscs, Eur. Polym. J. 110 (2019) 63–68. [Google Scholar]
- [22].Burridge KM, Harding BD, Sahu ID, Kearns MM, Stowe RB, Dolan MT, Edelmann RE, Dabney-Smith C, Page RC, Konkolewicz D, Lorigan GA, Simple Derivatization of RAFT-Synthesized Styrene-Maleic Anhydride Copolymers for Lipid Disk Formulations, Biomacromolecules. 21 (2020) 1274–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yasuhara K, Arakida J, Ravula T, Ramadugu SK, Sahoo B, Kikuchi J.l., Ramamoorthy A, Spontaneous Lipid Nanodisc Fomation by Amphiphilic Polymethacrylate Copolymers, J. Am. Chem. Soc.139(2017)18657–18663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hardin NZ, Ravula T, Di Mauro G, Ramamoorthy A, Hydrophobic Functionalization of Polyacrylic Acid as a Versatile Platform for the Development of Polymer Lipid Nanodisks, Small. 15 (2019) 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Barnaba C, Sahoo BR, Ravula T, Medina-Meza IG, Im S-C, Anantharamaiah GM, Waskell L, Ramamoorthy A, Cytochrome-P450-lnduced Ordering of Microsomal Membranes Modulates Affinity for Drugs, Angew. Chemie. 130 (2018) 3449–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ravula T, Hardin NZ, Ramadugu SK, Ramamoorthy A, PH Tunable and Divalent Metal Ion Tolerant Polymer Lipid Nanodiscs, Langmuir. 33 (2017) 10655–10662. [DOI] [PubMed] [Google Scholar]
- [27].Ravula T, Hardin NZ, Bai J, Im SC, Waskell L, Ramamoorthy A, Effect of polymer charge on functional reconstitution of membrane proteins in polymer nanodiscs, Chem. Commun. 54 (2018) 9615–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lindhoud S, Carvalho V, Pronk JW, Aubin-Tam ME, SMA-SH: Modified Styrene-Maleic Acid Copolymer for Functionalization of Lipid Nanodiscs, Biomacromolecules. 17 (2016) 1516–1522. [DOI] [PubMed] [Google Scholar]
- [29].Fujiwara T, Ramamoorthy A, How Far Can the Sensitivity of NMR Be Increased?, Annu. Reports NMR Spectrosc. 58 (2006) 155–175. [Google Scholar]
- [30].Verardi R, Traaseth NJ, Masterson LR, V Vostrikov V, Veglia G, Isotope Labeling for Solution and Solid-State NMR Spectroscopy of Membrane Proteins, in: Atreya HS, Isot. Labeling Biomol. NMR, Springer Netherlands, Dordrecht, 2012: pp. 35–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tugarinov V, Kanelis V, Kay LE, Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy, Nat. Protoc. 1 (2006) 749–754. [DOI] [PubMed] [Google Scholar]
- [32].Lacabanne D, Meier BH, Bockmann A, Selective labeling and unlabeling strategies in protein solid-state NMR spectroscopy, J. Biomol. NMR. 71 (2018) 141–150. [DOI] [PubMed] [Google Scholar]
- [33].Opella SJ, Marassi FM, Structure determination of membrane proteins by NMR spectroscopy, Chem. Rev. 104 (2004) 3587–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lee JH, Okuno Y, Cavagnero S, Sensitivity enhancement in solution NMR: Emerging ideas and new frontiers, J. Magn. Reson. 241 (2014) 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Grage SL, Xu X, Schmitt M, Wadhwani P, Ulrich AS, 19F-labeling of peptides revealing long-range NMR distances in fluid membranes, J. Phys. Chem. Lett. 5 (2014) 4256–4259. [DOI] [PubMed] [Google Scholar]
- [36].Niedbalski P, Parish C, Wang Q, Kiswandhi A, Hayati Z, Song L, Lumata L, 13C Dynamic Nuclear Polarization Using a Trimeric Gd3+ Complex as an Additive, J. Phys. Chem. A. 121 (2017) 5127–5135. [DOI] [PubMed] [Google Scholar]
- [37].Niedbalski P, Parish C, Wang Q, Hayati Z, Song L, Martins AF, Sherry AD, Lumata L, Transition Metal Doping Reveals Link between Electron Ti Reduction and 13C Dynamic Nuclear Polarization Efficiency, J. Phys. Chem. A. 121 (2017) 9221–9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Su Y, Andreas L, Griffin RG, Magic angle spinning NMR of proteins: high-frequency dynamic nuclear polarization and 1H detection, Annu. Rev. Biochem. 84 (2015) 465–497. [DOI] [PubMed] [Google Scholar]
- [39].Thurber KR, Tycko R, Perturbation of nuclear spin polarizations in solid state NMR of nitroxidedoped samples by magic-angle spinning without microwaves, J. Chem. Phys. 140 (2014) 184201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bruno F, Francischello R, Bellomo G, Gigli L, Flori A, Menichetti L, Tenori L, Luchinat C, Ravera E, Multivariate Curve Resolution for 2D solid-state NMR spectra., Anal. Chem. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yamamoto K, Caporini MA, Im SC, Waskell L, Ramamoorthy A, Transmembrane Interactions of Full-length Mammalian Bitopic Cytochrome-P450-Cytochrome-b5 Complex in Lipid Bilayers Revealed by Sensitivity-Enhanced Dynamic Nuclear Polarization Solid-state NMR Spectroscopy, Sci. Rep. 7 (2017)1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Matsuki Y, Idehara T, Fukazawa J, Fujiwara T, Advanced instrumentation for DNP-enhanced MAS NMR for higher magnetic fields and lower temperatures, J. Magn. Reson. (2016) 107–115. [DOI] [PubMed] [Google Scholar]
- [43].Pell AJ, Pintacuda G, Grey CP, Paramagnetic NMR in solution and the solid state, Prog. Nucl. Magn. Reson. Spectrosc. Ill (2019) 1–271. [DOI] [PubMed] [Google Scholar]
- [44].Kocman V, Di Mauro GM, Veglia G, Ramamoorthy A, Use of paramagnetic systems to speed-up NMR data acquisition and for structural and dynamic studies, Solid State Nucl. Magn. Reson. 102 (2019)36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Parthasarathy S, Yoo B, McElheny D, Tay W, Ishii Y, Capturing a reactive state of amyloid aggregates: NMR-based characterization of copper-bound alzheimer disease amyloid (β-fibrils in a redox cycle, J. Biol. Chem. 289 (2014) 9998–10010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ashbrook SE, Jaroniec CP, NMR spectroscopy of paramagnetic solids, Solid State Nucl. Magn. Reson. 104(2019)101625. [DOI] [PubMed] [Google Scholar]
- [47].Mukhopadhyay D, Nadaud PS, Shannon MD, Jaroniec CP, Rapid Quantitative Measurements of Paramagnetic Relaxation Enhancements in Cu(ll)-Tagged Proteins by Proton-Detected Solid-State NMR Spectroscopy, J. Phys. Chem. Lett. 8 (2017) 5871–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Perez A, Gaalswyk K, Jaroniec CP, MacCallum JL, High Accuracy Protein Structures from Minimal Sparse Paramagnetic Solid-State NMR Restraints, Angew. Chemie - Int. Ed. 58 (2019) 6564–6568. [DOI] [PubMed] [Google Scholar]
- [49].Theint T, Xia Y, Nadaud PS, Mukhopadhyay D, Schwieters CD, Surewicz K, Surewicz WK, Jaroniec CP, Structural Studies of Amyloid Fibrils by Paramagnetic Solid-State Nuclear Magnetic Resonance Spectroscopy, J. Am. Chem. Soc. 140 (2018) 13161–13166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bertini I, Luchinat C, Parigi G, Ravera E, NMR of Paramagnetic Molecules: applications to metallobiomolecules and models, Second Edition, Elsevier, Amsterdam, 2017. [Google Scholar]
- [51].Cai S, Seu C, Kovacs Z, Sherry AD, Chen Y, Sensitivity enhancement of multidimensional NMR experiments by paramagnetic relaxation effects, J. Am. Chem. Soc. 128 (2006) 13474–13478. [DOI] [PubMed] [Google Scholar]
- [52].Yamamoto K, Vivekanandan S, Ramamoorthy A, Fast NMR data acquisition from bicelles containing a membrane-associated peptide at natural-abundance, J. Phys. Chem. B. 115 (2011) 12448–12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bloch F, Nuclear induction, Phys. Rev. 70 (1946) 460–474. [Google Scholar]
- [54].Tesch DM, Nevzorov AA, Sensitivity enhancement and contrasting information provided by free radicals in oriented-sample NMR of bicelle-reconstituted membrane proteins, J. Magn. Reson. 239 (2014)9–15. [DOI] [PubMed] [Google Scholar]
- [55].Li W, Celinski VR, Weber J, Kunkel N, Kohlmann H, Homogeneity of doping with paramagnetic ions by NMR, Phys. Chem. Chem. Phys. 18 (2016) 9752. [DOI] [PubMed] [Google Scholar]
- [56].Lo AYH, Sudarsan V, Sivakumar S, Van Veggel F, Schurko RW, Multinuclear solid-state NMR spectroscopy of doped lanthanum fluoride nanoparticles, J. Am. Chem. Soc. 129 (2007) 4687–4700. [DOI] [PubMed] [Google Scholar]
- [57].Nitsche C, Otting G, Chapter 2: Intrinsic and Extrinsic Paramagnetic Probes, in: New Dev. NMR, 2018: pp. 42–84. [Google Scholar]
- [58].Yagi H, Banerjee D, Graham B, Huber T, Goldfarb D, Otting G, Gadolinium Tagging for HighPrecision Measurements of 6 nm Distances in Protein Assemblies by EPR, J. Am. Chem. Soc. 133 (2011) 10418–10421. [DOI] [PubMed] [Google Scholar]
- [59].Potapov A, Yagi H, Huber T, Jergic S, Dixon NE, Otting G, Goldfarb D, Nanometer-scale distance measurements in proteins using Gd3+ spin labeling, J. Am. Chem. Soc. 132 (2010) 9040–9048. [DOI] [PubMed] [Google Scholar]
- [60].Otting G, Prospects for lanthanides in structural biology by NMR, J. Biomol. NMR. 42 (2008) 1–9. [DOI] [PubMed] [Google Scholar]
- [61].Iwahara J, Tang C, Marius Clore G, Practical aspects of 1H transverse paramagnetic relaxation enhancement measurements on macromolecules, J. Magn. Reson. 184 (2007) 185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Solomon I, Relaxation Processes in a System of Two Spins, Phys. Rev. 99 (1955) 559–565. [Google Scholar]
- [63].Bloembergen N, Proton relaxation times in paramagnetic solutions, J. Chem. Phys. 27 (1957) 572–573.. [Google Scholar]
- [64].Strickland M, Catazaro J, Rajasekaran R, Strub MP, O’Hern C, Bermejo GA, Summers MF, Marchant J, Tjandra N, Long-Range RNA Structural Information via a Paramagnetically Tagged Reporter Protein, J. Am. Chem. Soc. 141 (2019) 1430–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kooshapur H, Schwieters CD, Tjandra N, Conformational Ensemble of Disordered Proteins Probed by Solvent Paramagnetic Relaxation Enhancement (sPRE), Angew. Chemie - Int. Ed. 57 (2018) 13519–13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Miao Q, Liu WM, Kock T, Blok A, Timmer M, Overhand M, Ubbink M, A Double-Armed, Hydrophilic Transition Metal Complex as a Paramagnetic NMR Probe, Angew. Chemie - Int. Ed. 58 (2019)13093–13100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Camacho-Zarco AR, Munari F, Wegstroth M, Liu WM, Ubbink M, Becker S, Zweckstetter M, Multiple paramagnetic effects through a tagged reporter protein, Angew. Chemie - Int. Ed. 54 (2015) 336–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Liu W-M, Skinner SP, Timmer M, Blok A, Hass MAS, Filippov DV, Overhand M, Ubbink M, A Two-Armed Lanthanoid-Chelating Paramagnetic NMR Probe Linked to Proteins via Thioether Linkages, Chem. - A Eur. J. 20 (2014) 6256–6258. [DOI] [PubMed] [Google Scholar]
- [69].Nitsche C, Otting G, Pseudocontact shifts in biomolecular NMR using paramagnetic metal tags, Prog. Nucl. Magn. Reson. Spectrosc. 98–99 (2017) 20–49. [DOI] [PubMed] [Google Scholar]
- [70].Orton HW, Otting G, Accurate Electron-Nucleus Distances from Paramagnetic Relaxation Enhancements, J. Am. Chem. Soc. 140 (2018) 7688–7697. [DOI] [PubMed] [Google Scholar]
- [71].Pilla KB, Otting G, Huber T, 3D Computational Modeling of Proteins Using Sparse Paramagnetic NMR Data, in: Keith JM, Bioinforma. Vol. II Struct. Funct. Appl, Springer New York, New York, NY, 2017: pp. 3–21. [DOI] [PubMed] [Google Scholar]
- [72].Esposito G, Lesk AM, Molinari H, Motta A, Niccolai N, Pastore A, Probing protein structure by solvent perturbation of nuclear magnetic resonance spectra, J. Mol. Biol. 224 (1992) 659–670. [DOI] [PubMed] [Google Scholar]
- [73].Madl T, Mulder FAA, Chapter 10: Small Paramagnetic Co-solute Molecules, New Dev. NMR. 2018Janua (2018)283–309. [Google Scholar]
- [74].Hardin NZ, Kocman V, Di Mauro GM, Ravula T, Ramamoorthy A, Metal-Chelated Polymer Nanodiscs for NMR Studies, Angew. Chemie - Int. Ed. 58 (2019) 17246–17250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Viola-Villegas N, Doyle RP, The coordination chemistry of 1,4,7,10-tetraazacyclododecaneN,N,N,N-tetraacetic acid (H4 DOTA): Structural overview and analyses on structure-stability relationships, Coord. Chem. Rev. 253 (2009) 1906–1925. [Google Scholar]
- [76].Ravula T, Ramadugu SK, Di Mauro GM, Ramamoorthy A, Bioinspired, Size-Tunable Self-Assembly of Polymer-Lipid Bilayer Nanodiscs, Angew. Chemie Int. Ed. 56 (2017) 11466–11470. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [77].Ravula T, Ramamoorthy A, Magnetic Alignment of Polymer Macro-Nanodiscs Enables ResidualDipolar-Coupling-Based High-Resolution Structural Studies by NMR Spectroscopy, Angew. Chemie Int. Ed. 58 (2019) 14925–14928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Mahawaththa MC, Lee MD, Giannoulis A, Adams LA, Feintuch A, Swarbrick JD, Graham B, Nitsche C, Goldfarb D, Otting G, Small neutral Gd(iii) tags for distance measurements in proteins by double electron-electron resonance experiments, Phys. Chem. Chem. Phys. 20 (2018) 23535–23545. [DOI] [PubMed] [Google Scholar]
- [79].Gamble Jarvi A, Cunningham TF, Saxena S, Efficient localization of a native metal ion within a protein by Cu2+-based EPR distance measurements, Phys. Chem. Chem. Phys. 21 (2019) 10238–10243.. [DOI] [PubMed] [Google Scholar]
- [80].Corsi DM, Platas-lglesias C, Van Bekkum H, Peters JA, Determination of paramagnetic lanthanide(lll) concentrations from bulk magnetic susceptibility shifts in NMR spectra, Magn. Reson. Chem. 39 (2001) 723–726. [Google Scholar]
- [81].Bertmer M, Paramagnetic solid-state NMR of materials, Solid State Nucl. Magn. Reson. 81 (2017) 1–7.. [DOI] [PubMed] [Google Scholar]
- [82].Bertini I, Luchinat C, Parigi G, Magnetic susceptibility in paramagnetic NMR, Prog. Nucl. Magn. Reson. Spectrosc. 40 (2002) 249–273. [Google Scholar]
- [83].Pintacuda G, Vaara J, Bertini I, Park M, Banci L, Bertini I, Bertini I, Bertini I, Luchinat C, NMR of Paramagnetic Molecules in Biological Systems, (2001) 1–2. [Google Scholar]
- [84].Suturina EA, Mason K, Geraldes CFGC, Chilton NF, Parker D, Kuprov I, Lanthanide-induced relaxation anisotropy, Phys. Chem. Chem. Phys. 20 (2018) 17676–17686. [DOI] [PubMed] [Google Scholar]
- [85].Bertini I, Janik MBL, Lee YM, Luchinat C, Rosato A, Magnetic susceptibility tensor anisotropies for a lanthanide ion series in a fixed protein matrix, J. Am. Chem. Soc. 123 (2001) 4181–4188. [DOI] [PubMed] [Google Scholar]
- [86].Kumar K, Chang CA, Francesconi LC, Dischino DD, Malley MF, Gougoutas JZ, Tweedle MF, Synthesis, Stability, and Structure of Gadolinium(lll) and Yttrium(lll) Macrocyclic Poly(amino carboxylates), Inorg. Chem. 33 (1994) 3567–3575. [Google Scholar]
- [87].Karraker DG, Coordination of trivalent lanthanide ions, J. Chem. Eduction. 47 (1970) 424–430. [Google Scholar]
- [88].V Dvinskikh S, Zimmermann H, Maliniak A, Sandström D, Measurements of motionally averaged heteronuclear dipolar couplings in MAS NMR using R-type recoupling, J. Magn. Reson. 168 (2004) 194–201. [DOI] [PubMed] [Google Scholar]
- [89].Kurzen H, Bovigny L, Bulloni C, Daul C, Electronic structure and magnetic properties of lanthanide 3 + cations, Chem. Phys. Lett. 574 (2013) 129–132. [Google Scholar]
- [90].Yamamoto K, Xu J, Kawulka KE, Vederas JC, Ramamoorthy A, Use of a Copper-Chelated Lipid Speeds Up NMR Measurements from Membrane Proteins, J. Am. Chem. Soc. 132 (2010) 6929–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.