Abstract
Histone acetyltransferase (HAT) p300 and its paralog CBP acetylate histone lysine sidechains and play critical roles in regulating gene transcription. The HAT domain of p300/CBP is a potential drug target for cancer. Through compound screening and medicinal chemistry, novel inhibitors of p300/CBP HAT with their IC50s as low as 620 nM were discovered. The most potent inhibitor is competitive against histone substrates and exhibits a high selectivity for p300/CBP. It inhibited cellular acetylation and had strong activity with EC50 of 1-3 μM against proliferation of several tumor cell lines. Gene expression profiling in estrogen receptor (ER)-positive breast cancer MCF-7 cells showed that inhibitor treatment recapitulated siRNA-mediated p300 knockdown, inhibited ER-mediated gene transcription, and suppressed expression of numerous cancer-related gene signatures. These results demonstrate that the inhibitor is not only a useful probe for biological studies of p300/CBP HAT, but also a pharmacological lead for further drug development targeting cancer.
Keywords: Histone acetyltransferase, p300/CBP, small-molecule inhibitor, Gene expression profiling, cancer therapy
Graphical Abstract
INTRODUCTION
Histone acetylation is one of the most important post-translational modifications. Acetylation of the sidechain amino group of a histone lysine residue neutralizes its positive charge (under physiological pH) and renders a more open DNA conformation to facilitate binding of transcription factors as well as other proteins for gene transcription, DNA replication and repair 1, 2. In addition, acetylated lysine can be recognized by a number of proteins (such as bromodomain-containing proteins) and serve as an anchor point in chromatin to form a transcription complex that regulates gene expression 3, 4.
Human p300 (E1A binding protein p300) and its paralog CBP [CREB (cAMP-response element binding protein) binding protein] are large proteins with ~2,400 amino acids 5-7, containing multiple structured domains including cysteine-histidine rich 1 (CH1), CREB-binding KIX, Bromodomain, histone acetyltransferase (HAT), CH3, and steroid receptor coactivator interaction (SID) domains. These structured domains, which share a high homology between p300 and CBP, are connected with less conserved intrinsically disordered regions (IDR). The CH1, KIX, CH3 and SID domains as well as IDR of p300/CBP are transactivation domains, which interact with a number of transcription factors (e.g., CREB, p53 and HIF-1) and transcription coactivators (e.g., steroid receptor coactivators) 8-10. Biologically, p300/CBP not only acetylates a lysine residue of histone (e.g., histone H3 lysine 27, or H3K27) and certain transcription factors (e.g., p53 and c-Myc) 7, 11, 12, it also serves as a hub protein to link and stabilize a transcription complex.
Previous studies have shown that HAT activity of p300/CBP is essential for many transcription factor-mediated gene transcription programs. For example, estrogen receptor (ER) or androgen receptor (AR) mediated gene transcription pathway is of importance in female or male development as well as in breast or prostate cancer, respectively. HAT activity of p300 has been found to be required for ER- or AR-mediated gene expression 8, 10, 13. In addition, HAT of p300/CBP is a potential drug target for cancer therapy. Although there have been debates on whether p300/CBP alone is a tumor suppressor or an oncogene, convincing evidence has shown that p300/CBP HAT is essential for breast and prostate cancer as well as other cancers with p300 overexpression or harboring a p300/CBP fusion oncogene 7, 14, 15.
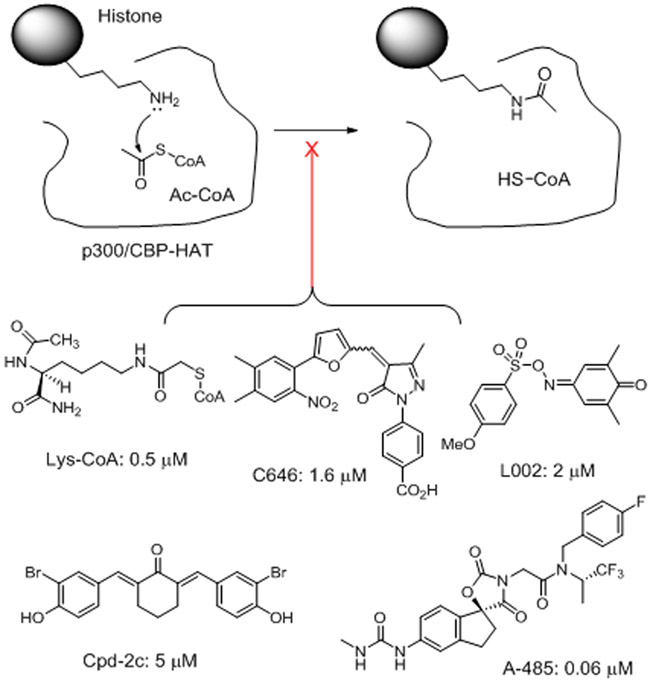
To date, several chemotypes of p300/CBP HAT inhibitors have been reported (Figure 1) 6, 7. However, except for recently disclosed compound A-485 16, other compounds are not drug-like probes for cellular or in vivo studies. Lys-CoA and its analogs are not cell-permeable17, 18. Compounds C646 19, L002 20 and Cpd-2c 21 contain a “PAINS” (pan-assay interfering compound) 22 or related structure, which are unfavorable for drug development or cell biology. A-485 is a potent inhibitor of p300/CBP HAT, which is competitive against the enzyme cofactor acetyl coenzyme A (Ac-CoA) 16. It showed strong activity against proliferation of several cancer cells. Nevertheless, given the low success rate in drug discovery, more chemotypes of p300 HAT inhibitors are needed. Here, we report discovery, structure activity relationships (SAR), biochemical and biological activities of a novel series of inhibitors of p300/CBP HAT that are competitive against the histone substrate.
Figure 1.
P300/CBP HAT catalyzed reaction and inhibitors with their IC50 values.
RESULTS
Inhibitor discovery.
A biochemical assay to determine the activity and inhibition of recombinant HAT domain of human p300 was developed, using the substrate histone H3 (1-21) peptide and the 3H-labeled cofactor Ac-CoA. We screened our proprietary compound library containing ~300 compounds, which were synthesized for SAR studies of lysine specific demethylase 1 (LSD1) 23 whose substrate is methylated histone H3 lysine 4 (H3K4). The rationale is that since the peptide sequence of H3K4 (ARTKQ) is similar to those of p300 HAT’s substrate H3K27 (AARKS), there is a higher likelihood to find an inhibitor from the LSD1 targeting compounds. The compound screen led to the discovery of thiophene-2-carbamide compound 1 (Table 1) that inhibited p300 HAT with an IC50 (concentration at which the enzyme activity is inhibited by 50%) value of 8.6 μM.
Table 1.
Structure and inhibitory activity of compounds 1-32.
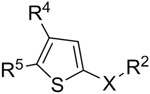 |
|||||
|---|---|---|---|---|---|
| Cpd | R4 | R5 | X | R2 | IC50 (μM) (% inhibition at 10 μM) |
| 1 | 4-t-Bu-Ph | = R4 | -CONH- | piperidin-4-ylmethyl | 8.6 ± 0.4 |
| 2 | 4-(furan-3-yl)-Ph | = R4 | -CONH- | same as above | 1.6 ± 0.2 |
| 3 | 4-methoxybiphenyl | = R4 | -CONH- | same as above | 2.8 ± 0.6 |
| 4 | 4-(furan-3-yl)-Ph | 4-t-Bu-Ph | -CONH- | same as above | 7.4 ± 0.8 |
| 5 | 4-aminomethyl-Ph | 4-t-Bu-Ph | -CONH- | same as above | 10.0 ± 1.4 |
| 6 | 4-(piperidin-1-ylmethyl)-Ph | 4-(furan-3-yl)-Ph | -CONH- | same as above | 1.6 ± 0.2 |
| 7 | 4-(piperazin-1-ylmethyl)-Ph | 4-t-Bu-Ph | -CONH- | same as above | >10 (12%) |
| 8 | furan-3-yl | 4-t-Bu-Ph | -CONH- | same as above | >10 (36%) |
| 9 | pyridin-3-yl | 4-t-Bu-Ph | -CONH- | same as above | >10 (31%) |
| 10 | 4-methoxybiphenyl | = R4 | -CH2NH- | same as above | 7.4 ± 1.0 |
| 11 | 4-(furan-3-yl)-Ph | = R4 | -CH2NH- | same as above | 1.7 ± 0.2 |
| 12 | 4-t-Bu-Ph | = R4 | -CH2NH- | same as above | 0.62 ± 0.2 |
| 13 | 4-t-Bu-Ph | = R4 | -CH2O- | same as above | >10 (0%) |
| 14 | 4-t-Bu-Ph | = R4 | -CH2NH- | piperidin-4-yl | 5.0 ± 1.6 |
| 15 | 4-t-Bu-Ph | = R4 | -CH2NH- | 2-(piperidin-4-yl)ethyl | 2.2 ± 0.4 |
| 16 | 4-t-Bu-Ph | = R4 | -CH2NH- | 3-aminopropyl | 3.0 ± 1.0 |
| 17 | 4-t-Bu-Ph | = R4 | -CH2NH- | 6-aminohexyl | 1.4 ± 0.3 |
| 18 | 4-t-Bu-Ph | = R4 | -CH2- | piperazin-1-yl | >10 (5%) |
| 19 | 4-t-Bu-Ph | = R4 | -CH2- | morpholin-4-yl | >10 (20%) |
| 20 | 4-t-Bu-Ph | = R4 | -CH2N(Me)- | piperidin-4-ylmethyl | 4.4 ± 1.6 |
| 21 | 4-t-Bu-Ph | = R4 | -CH2N(CHO)- | same as above | 7.0 ± 1.6 |
| 22 | 4-i-Pr-Ph | = R4 | -CH2NH- | same as above | 5.4 ± 1.4 |
| 23 | 4-Bu-Ph | = R4 | -CH2NH- | same as above | >10 (10%) |
| 24 | 4-(furan-3-yl)-Ph | 4-t-Bu-Ph | -CH2NH- | same as above | 5.8 ± 0.6 |
| 25 | 4-(furan-3-yl)-Ph | 4-(piperidin-1-ylmethyl)-Ph | -CH2NH- | same as above | 4.6 ± 1.4 |
| 26 | 4-t-Bu-Ph | 4-aminomethyl-Ph | -CH2NH- | same as above | 2.0 ± 0.5 |
| 27 | 4-aminomethyl-Ph | 4-t-Bu-Ph | -CH2NH- | same as above | >10 (35%) |
| 28 | 4-aminomethyl-Ph | = R4 | -CH2NH- | same as above | >10 (0%) |
| 29 | 4-hydroxymethyl-Ph | = R4 | -CH2NH- | same as above | >10 (22%) |
| 30 | furan-3-yl | = R4 | -CH2NH- | same as above | >10 (23%) |
| 31 | 3,4-dimethoxy-Ph | = R4 | -CH2NH- | same as above | >10 (0%) |
| 32 | 3-biphenyl | = R4 | -CH2NH- | same as above | >10 (18%) |
Synthesis.
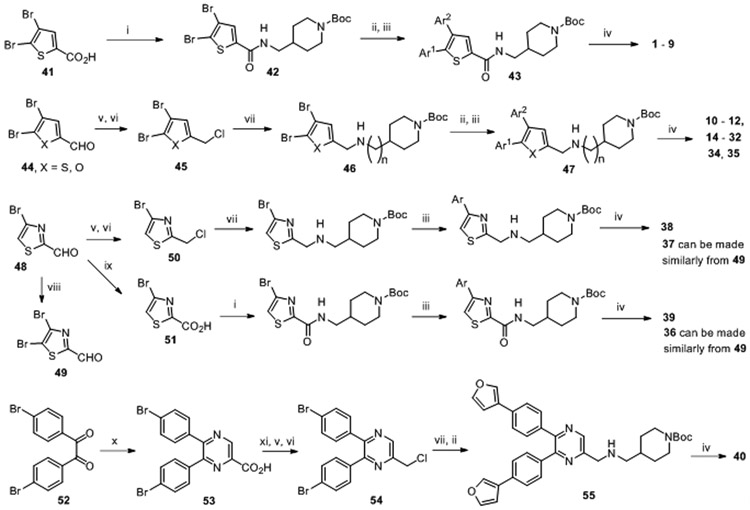
Given its seemingly drug-like structure, more derivatives of compound 1 were synthesized and tested for their activities against p300 HAT. General methods for synthesis of compounds 1-40 for SAR studies are shown in Scheme 1.
Scheme 1.
General synthetic methods for thiophene-containing compounds.a
aReagents and conditions: (i) 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, 1-hydroxybenzotriazole, 4-(aminomethyl)-1-BOC-piperidine, CH2Cl2, 96% yield; (ii) Ar1-B(OH)2, Pd(PPh3)4, Na2CO3, 80 °C, 80-90% yield; (iii) Ar2-B(OH)2, Pd(PPh3)4, Na2CO3, 100 °C, 80-90% yield; (iv) HCl in 1,4-dioxane, >90% yield; (v) NaBH4, MeOH; (vi) cyanuric chloride, DMF, 81% yield for the 2 steps; (vii) 4-(aminomethyl)-1-BOC-piperidine or other amine analogs, K2CO3, DMF, 60-85% yield; (viii) N-Bromosuccinimide, CH2Cl2; (ix) NaClO2, NaH2PO4, tert-butanol-H2O, 2-methyl-2-butene; (x) 2,3-diaminopropanoic acid, NaOH, MeOH, reflux, 55% yield; (xi) ClCO2Me, (iPr)2NEt, THF, 95% yield.
Synthesis of thiophene-2-carbamide compounds 1-9 was started from a reaction between 4,5-dibromo-thiophene-2-carboxylic acid (41) and 1-tert-butyloxycarbonyl (BOC) protected 4-(aminomethyl)piperidine, together with amide bond forming reagents 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and 1-hydroxybenzotriazole. The 5- and 4-Br groups of the thiophene-2-carbamide compounds (42) can undergo two successive Suzuki coupling reactions with different aryl boronic acids with a high selectivity 24: 5-Br was reacted first at a lower temperature of 80 °C, followed by 4-Br at 100 °C, to give compound 43. Removal of the BOC protecting group using HCl produced the target compounds 1 – 9. Synthesis of 2-aminomethylthiophene series of compounds 10-12 and 14-32 was started from 4,5-dibromo-thiophene-2-carbaldehyde (44, X = S), in which the 2-CHO group was reduced to an alcohol, followed by conversion to a -CH2Cl group by treatment with cyanuric chloride in DMF. The -Cl of the product 45 was next substituted with 4-(aminomethyl)-1-BOC-piperidine or other amine analogs. The resulting 2-substituted 4,5-dibromo-thiophene compound 46 was subjected to two selective Suzuki coupling reactions to produce, upon deprotection of BOC, the target 2-aminomethylthiophene series of compounds.
Furan-containing inhibitors 34 and 35 were prepared similarly from 4,5-dibromo-furan-2-carbaldehyde (44, X = O). All thiazole compounds were synthesized from 4-bromothiazole-2-carbaldehyde (48), which was treated with N-bromosuccinimide to produce di-Br compound 49. The aldehyde group of 48 and 49 was reduced and converted to a -CH2Cl group in compound 50, which was substituted with 1-BOC-4-(aminomethyl)piperidine followed by Suzuki coupling reactions to give, after deprotection, compounds 37 and 38. Oxidation of 48 (or 49) produced thiazole-2-carboxylic acid 51, which was reacted with 1-BOC-4-(aminomethyl)piperidine followed by Suzuki coupling reactions to afford thiazole compound 39 (or 36). For the synthesis of the pyrazine compound 40, di-4-bromobenzil (52) was refluxed with 2,3-diaminopropanoic acid in methanol to give 5,6-di-(4-bromophenyl)pyrazine-2-carboxylic acid (53), whose -CO2H group was acylated, reduced (with NaBH4) to -CH2OH, and converted to -CH2Cl (54). A substitution reaction with 4-(aminomethyl)-1-BOC-piperidine followed by Suzuki coupling reactions produced compound 55, which was treated with HCl to give compound 40.
Structure-activity relationship.
The structures and inhibitory activities of compounds 1-32 are shown in Table 1 and Supporting Information Chart S1-3 and Figure S1. Compound 1, having 4,5-di-substituted tert-butyl groups as well as a piperidine-containing amide group, had a moderate IC50 of 8.6 μM. Compound 2 with 4,5-di-substituted furan-3-yl groups was found to have significantly enhanced inhibitory activity with an IC50 of 1.6 μM, while compound 3 with two 4-methoxybiphenyl substituents showed slightly reduced inhibitory activity (IC50 = 2.8 μM). Compound 4 bearing a mixed tert-butyl and furanyl group also exhibited a compromised activity (IC50 = 7.4 μM). Compound 5 with a polar 4-aminomethylphenyl substituent rendered little activity change (IC50 = 10 μM). However, compound 6 with a piperidine-containing 4-substituent, which is less polar as compared to the polar primary amine in 5, showed an improved activity (IC50 = 1.6 μM). Compound 7 having a piperazine group was found to be almost inactive at 10 μM, showing an additional amino group (as compared to 6) is likely disfavored. Compounds 8 and 9 with no substituents in their 4-aromatic rings were found to have modest activity, inhibiting p300-HAT by ~30% at 10 μM, suggesting a substituent (e.g., furan-3-yl in 4) at this position is favorable.
Next, activity optimization of the 2-carboxamide sidechain was performed for compounds 10-21 (Table 1 and Chart S2). The first attempt was to replace the amide sidechain (in e.g., 1 and 2) with a piperidin-4-yl-methylaminomethyl group in compounds 10-12. While removal of the carbonyl group reduces the activity of compound 10 (IC50 = 7.4 μM) by ~50% (as compared to 3), compound 11 with an IC50 of 1.7 μM exhibited a comparable activity to that of 2. Of interest is that compound 12 showed a potent activity against p300-HAT with an IC50 of 620 nM, which is ~13× more active than its amide analog 1. Changing the -NH- linkage (in 10-12) to an -O- in compound 13 resulted in a complete loss of inhibitory activity, suggesting the -NH- (which is protonated at physiological pH) could be a key hydrogen bond donor. Compounds 14 and 15, whose 2-substituents are one -CH2- shorter and longer than that of 12, respectively, were found to possess ~8- and 3-fold activity reduction (IC50 = 5.0 and 2.2 μM). Compounds 16 and 17, which carry a linear, ω-amino-propyl and -hexyl side chain, respectively, are also strong inhibitors with IC50 values of 3.0 and 1.4 μM. Loss of activity was found for compounds 18 and 19 bearing a 2-substituent of piperazin-1-ylmethyl and morpholin-4-ylmethyl group, showing shorter cyclic 2-substituents are disfavored. Moreover, methylation and formylation of the middle -NH- group (in compound 12) produced compounds 20 and 21, respectively, which were found to have less inhibitory activities with IC50 values of 4.4 and 7.0 μM. These results suggest an -NH- or -CONH- linkage, which might be a hydrogen bond donor, is of importance to the inhibition.
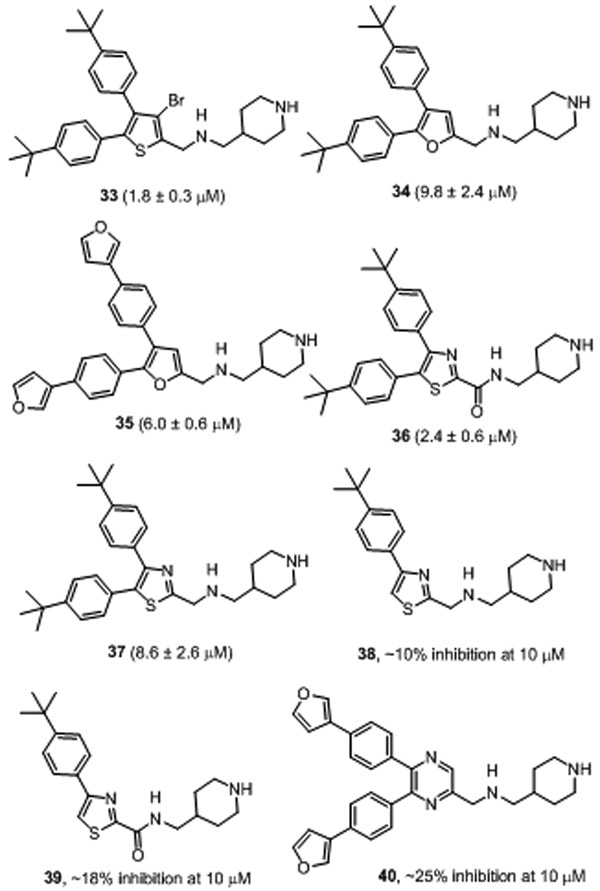
Compounds 22-32 (Table 1 and Chart S3) were synthesized to evaluate effects of varying 4- and/or 5-substituents of the most potent inhibitor 12. Replacing the tert-butyl with isopropyl groups in 22 (IC50 = 5.4 μM) resulted in ~8x activity reduction, suggesting a bulkier group is favorable for the 4- and 5-positions. Loss of activity for compound 23 with two n-butyl groups at these positions showed that long alkyl substituents are disfavored. Changing the 4-substituent to a furan-3-yl group in compound 24 (IC50 = 5.8 μM) as well as to a piperidin-1-ylmethyl group in 25 (IC50 = 4.6 μM) resulted in reduced activities, as compared to compounds 12 and 11. Compound 26 with a polar -CH2NH2 group at the 5-position (of the thiophene core) is still a strong inhibitor with an IC50 of 2.0 μM, although it exhibited ~3-fold activity reduction as compared to 12. However, moving the -CH2NH2 group to the 4-position in compound 27 suffered a major activity loss. Compound 28 with the amine group at both positions is inactive. Similarly, compound 29 with two -CH2OH groups is also a weak inhibitor. Activities for compounds 26-29 suggest polar groups are not favorable at 4- or 5-position. In addition, very low activity for compound 30 suggests a smaller aromatic ring at 4- and 5-positions is unfavored. Lack of inhibition for compounds 31 and 32 with meta-substitutions at the 4- and 5-positions indicated that para-substitution (e.g., those in compound 10) is more favorable for p300 HAT inhibition. Compound 33 (Chart 1) was synthesized with a -Br substituent at the 3-position of the thiophene core. It turned out to be still a strong inhibitor with an IC50 of 1.8 μM, while 3-Br reduces the inhibitory activity by ~3-fold (as compared to 12).
Chart 1.
Structures and inhibitory activity of compounds 33-40.
Finally, compounds 34-40 (Chart 1) were synthesized to study SARs for the central core. Furan analogs 34 and 35 with IC50 values of 4.9 and 3.0 μM, respectively, were found to be significantly less active than thiophene compounds 12 and 11, suggesting a furan core is less favored. While the thiazole compound 36 (IC50 = 2.4 μM) exhibited an improved activity as compare to its thiophene analog 1, the thiazole compound 37 (IC50 = 8.6 μM) is more than 13× less active than its thiophene analog 12. Loss of activity for compounds 38 and 39 shows that the 5-substituent is of importance to the inhibition of p300 HAT. Moreover, compound 40 with a 6-membered pyrazine core was found to be significantly less active than its thiophene analog 11. These results suggest thiophene is the most favorable aromatic core structure for this series of inhibitors.
Compound 12 is competitive against histone.
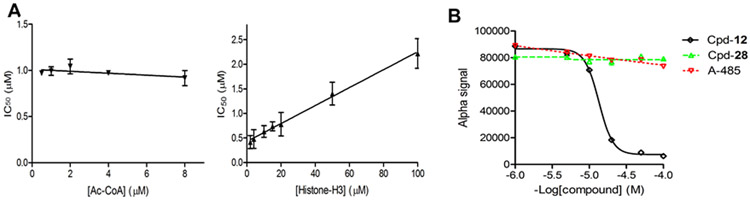
We next performed steady-state enzyme kinetics studies to investigate the mode of inhibition for the most potent inhibitor 12. Inhibitory activities of compound 12 were determined with varying concentrations of the cofactor Ac-CoA and the substrate histone. As shown in Figure 2A (left), IC50 values of 12 did not change with increasing concentrations of Ac-CoA (from 0.5 - 8 μM, or 0.07-1.2× Km 25), indicating compound 12 is likely a non-competitive inhibitor against the enzyme cofactor. Its inhibitory activities were found to be reduced in a linear manner (Figure 2A) when increasing concentrations of the substrate histone peptide (2 - 100 μM) were used in the assay, suggesting compound 12 is a competitive inhibitor against the substrate histone.
Figure 2.
Compound 12 is a competitive inhibitor of p300 HAT against the substrate histone. (A) Plots of IC50 values of 12 versus increasing concentrations of (left) Ac-CoA (0.5 - 8 μM, or 0.07-1.2× Km) and (right) histone H3 suggest the inhibitor is non-competitive against Ac-CoA and competitive against histone; (B) Alpha assay results show compound 12 can dose-dependently disrupt the binding between p300 HAT and histone H4. But such binding was not affected by inactive compound 28 as well as A-485 which is an inhibitor competitive against Ac-CoA.
Next, we used a reported Alpha (amplified luminescent proximity homogeneous assay) assay16 to confirm this finding. Upon excitation by a laser beam (680 nm), histone H4(1–21) peptide coated donor beads generate singlet oxygen radicals, which can travel for a very short distance (<200 nm) in the solution and cannot reach the free acceptor beads. The binding between the histone peptide and p300 HAT brings the donor and acceptor beads together, which allows the radicals to activate the p300 HAT coated acceptor beads and produce luminescence at 570 nm. Use of histone H4 is because the assay with histone H3 did not produce a luminescence signal, presumably due to a weaker binding between p300 HAT and histone H3. As shown in Figure 2B, compound 12 can disrupt the binding between the protein and histone H4, and reduce the Alpha-signal in a dose-dependent manner (IC50 = 13.5 μM). This is in contrast to Ac-CoA-competitive inhibitor A-48516, which did not affect the binding between p300 HAT and histone H4. In addition, such binding was not interfered by inactive compound 28 (Fig. 2B). Increasing concentrations of compound 12 did not significantly affect the Alpha-signals using a biotinylated His6 peptide that can directly link the donor and acceptor beads (Figure S2), showing 12 does not interfere with the singlet oxygen radicals or His6 tag. Taken together, these results indicate the inhibitor 12 is competitive against the substrate histone, which are of interest in that due to a different mode of inhibition, histone-competitive inhibitor 12 might have a different cellular activity or selectivity, as compared to Ac-CoA-competitive inhibitors A-485 and C646 16, 20.
Docking studies.
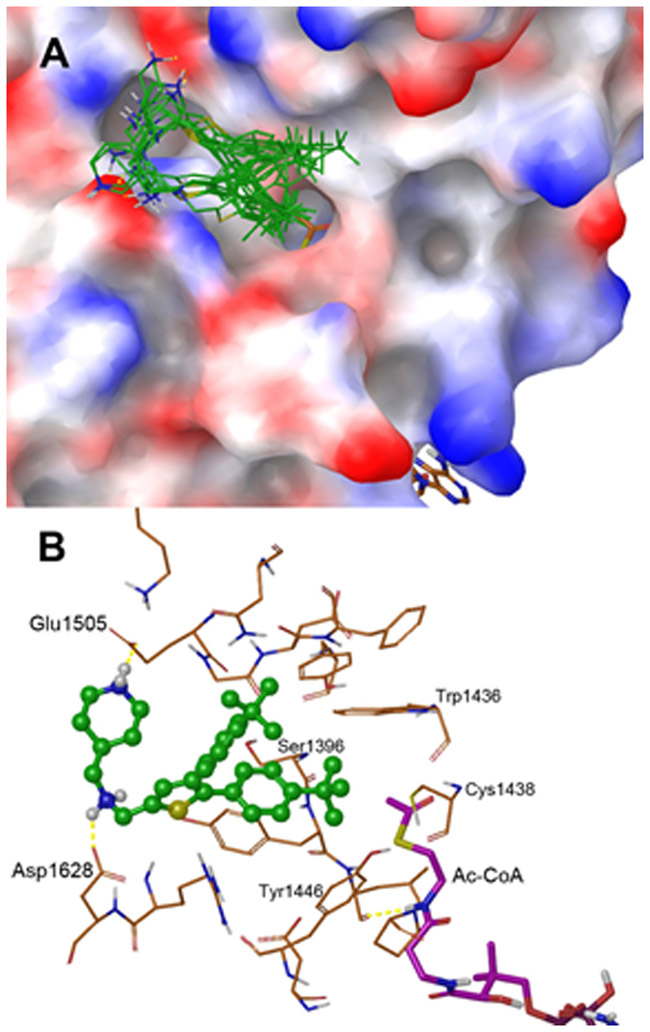
We next performed molecular modeling to find possible binding structures of the most potent inhibitor 12 in p300 HAT. The program Glide in Schrödinger small-molecule drug discovery software suite was used for the docking studies, with the crystal structure of human p300 HAT in complex with Ac-CoA (PDB code 4PZS) 26 as a template.
Since compound 12 is a competitive inhibitor against the substrate histone, Ac-CoA was treated as an integrate part of the protein. Upon protein structure preparation as well as docking grid generation, compound 12 was docked into the protein-Ac-CoA complex and the results are shown in Figure 3. The inhibitor can be favorably docked into the protein, as the 10 distinct docking conformations of compound 12 with the lowest energies exhibit similar binding features (Figure 3A). One of the tert-butyl group of these conformations is predicted to insert into and have favorable hydrophobic interactions with the so-called “lysine channel”, through which the histone lysine sidechain inserts into p300 HAT, attacks the carbonyl of Ac-CoA, and gets acetylated. Residues Trp1436, Cys1438, Tyr1446 and Ser1396 form the lysine channel of p300 HAT (Figure 3B). As shown in Figure 3B, the tert-butyl group is predicted to be ~3.5 Å from these residues with favorable van de Waals interactions. The central thiophene core and the two para-tert-butylphenyl groups of these structures have extensive hydrophobic interactions with either upper or lower surface of the protein (Figure 3A/B). In addition, the positively charged, two amino groups of many of these docking structures form hydrogen bonds and have favorable electrostatic interactions with Asp1628 and Glu1505, as exemplified in Figure 3B. These favorable binding features are consistent with the strong inhibitory activity of compound 12.
Figure 3.
Docking results for compound 12. (A) 10 docking conformations of 12 (with C atoms in green) with lowest energies in the crystal structure of p300-HAT (PDB: 4PZS, shown as an electrostatic surface) in complex with Ac-CoA (as a tube model with C atoms in brown); and (B) The lowest energy docking conformation of 12 (as a ball-and-stick model) with surrounding residues and Ac-CoA in p300-HAT (PDB: 4PZS). Hydrogen bonds are shown as yellow dotted lines.
Moreover, the docking results might be used to rationalize some of the observed SARs. For example, H-bond/electrostatic interactions between -NH- (positively charged at physiological pH) and Asp1628 are predicted to contribute significantly to the binding of compound 12. This could explain the loss of activity for compound 13 with an -O- linkage, which does not favorably interact with the (negatively charged) Asp1628 sidechain. Since an amide -CONH- mimics the positively charged -NH- in 12, many of the amide compounds (e.g., compounds 2 and 6 in Table 1) are potent inhibitors. Docking results also suggest favorable interactions of 12 with the lysine channel are important for tight binding. This could explain weak or no activities for compounds 30-32, which do not have a para-substituent or have a meta-substituent which might not favorably interact with the lysine channel residues of the protein.
Enzyme selectivity.
Humans have three classes of histone/protein lysine acetyltransferases with distinct conserved motifs and structures, including p300/CBP, Gcn5-related N-acetyltransferase (GNAT) and MYST (MOZ, Ybf2, Sas2 and Tip60) family of HATs. Compound 12 was tested for its activities against selected HATs from these three classes of HATs. CBP is a homolog of p300. The HAT domains of CBP and p300 exhibit 87% identity in sequence. PCAF (p300/CBP associating factor) is a member of the GNAT family, while Myst3 belongs to the MYST family of HATs. P300/CBP HATs have a distinct sequence and 3-dimensional structure from the GNAT and MYST HAT proteins. As shown in Table 2, compound 12 was found to be also a strong inhibitor of human CBP HAT with an IC50 of 1.2 μM, comparable to its IC50 against p300 HAT. However, compound 12 did not significantly inhibit the activity of PCAF and Myst3 even at 50 μM, showing a high selectivity for p300/CBP HAT. In addition, since the initial hit compound 1 was for SAR studies targeting LSD1, both compounds 1 and 12 did not significantly inhibit activity of LSD1 at 50 μM.
Table 2.
Inhibitory activities of compound 12 against HATs.
| IC50 (μM) | |
|---|---|
| P300-HAT | 0.62 ± 0.12 |
| CBP-HAT | 1.2 ± 0.13 |
| PCAF | >50 |
| Myst3 | >50 |
Inhibition of cellular histone acetylation.
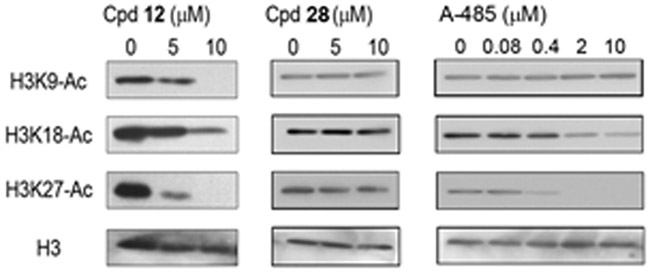
Ability of compound 12 to inhibit cellular p300/CBP HAT was evaluated. Also included in the experiment are an inactive compound 28 and a known inhibitor A-485. Kasumi-1 leukemia cells were treated with increasing concentrations of these compounds for 12h. Histone was extracted and subjected to electrophoresis separation and Western blot staining to determine levels of acetylation at various lysine residues. As shown in Figure 4, compound 12 was found to significantly inhibit cellular acetylation of histone H3K9, H3K18 and H3K27 at 5 and 10 μM, in a dose dependent manner. H3K27 seems to be the most sensitive cellular substrate of p300 HAT. Inactive compound 28 was found to have no significant activity against acetylation of these histone residues. As compared to A-485 (enzyme IC50 of 60 nM16), compound 12 is less active against acetylation of H3K27 and K18, which is consistent with the enzyme activities of these compounds. However, compound 12 seems to exhibit more potent cellular activity against acetylation of H3K9. These differences might be due to different mode of inhibition (competitive against histone vs. Ac-CoA) or cell permeability for the two compounds.
Figure 4.
Cellular activity of compounds 12, 28 and A-485 against acetylation of H3K9, H3K18 and H3K27.
Inhibition of tumor cell proliferation.
Activity of compound 12 was tested against proliferation of several tumor cell lines in which p300/CBP HAT is of importance. First, ER-mediated transcription is critical to ER+ breast cancer. It is an ordered, stepwise assembly of ER and coactivator proteins for gene expression 10, 27. Upon binding with an estrogen molecule in cytoplasm, ER undergoes conformational changes, forms a homodimer, and is translocated into the nucleus, where ER binds to estrogen response element, a short segment of DNA within a gene promoter. Next, a steroid receptor coactivator (SRC) protein binds to ER, followed by recruitment of p300 through its SRC-binding SID domain 9. P300 HAT activity is required for the ER-mediated gene expression, because a mutant p300 without the enzyme function failed to activate it 10. Given the critical role of p300-mediated histone acetylation in the ER signaling pathway, compound 12 was assessed for its activity against the ER+ breast cancer cell line MCF-7. In addition, ER antagonist tamoxifen is widely used in the clinic to treat ER+ breast cancer by suppressing ER mediated gene transcription. However, many patients eventually develop resistance to tamoxifen and die of the cancer. In our previous work 28, 29, we have developed MCF-7 derivative cells that were made resistant to tamoxifen. Activity of compound 12 was also tested against tamoxifen-resistant MCF-7 to find whether inhibition of HAT can affect growth of these cells. As summarized in Table 3, compound 12 exhibited strong antiproliferative activity against parent MCF-7 cells (EC50 = 2.8 μM) as well as tamoxifen-resistant cells (EC50 = 3.4 μM). These results show that inhibition of p300-mediated histone acetylation can inhibit growth of MCF-7 breast cancer cells and such activity is independent upon the status of tamoxifen resistance, suggesting the p300 HAT inhibitor could have potential clinical applications in breast cancer therapy.
Table 3.
Antiproliferative activity EC50 (μM) of compounds 12, 28, 31 and A-485.
| Cpd-12 | Cpd-28 | Cpd-31 | A-485 | |
|---|---|---|---|---|
| MCF-7 | 2.8 ± 0.1 | >30 | >30 | >30 |
| MCF-7 (Tam-R) | 3.4 ± 0.1 | >30 | >30 | >30 |
| PANC-1 | 1.0 ± 0.2 | >30 | >30 | >30 |
| MDA-PANC-28 | 2.8 ± 0.4 | >30 | >30 | >30 |
| Kasumi-1 | 2.6 ± 0.6 | >30 | 18.7 ± 0.1 | 0.33 ± 0.01 |
Moreover, p300 HAT activity has been reported to play important roles in pancreatic cancer 30 and acute myeloid leukemia caused by oncogene RUNX1-ETO 31. Activity of compound 12 was evaluated against two pancreatic cancer cells PANC-1 and MDA-PANC-28 and RUNX1-ETO leukemia cell line Kasumi-1. As shown in Table 3, compound 12 showed potent activities against proliferation of the two pancreatic cancer cells with EC50 values of 1.0 and 2.8 μM. It also inhibited growth of Kasumi-1 cells with an EC50 of 2.6 μM.
Inactive compounds 28 and 31 exhibited no inhibitory activities against proliferation of these cancer cells, supporting activity of compound 12 is related to inhibition of p300 HAT. Surprisingly, although A-485 exhibited more potent activity (EC50 = 0.33 μM) against Kasumi-1 leukemia cells, it had no significant activity against proliferation of breast and pancreatic cancer cells (Table 3). These differences might be due to different modes of inhibition (competitive against histone vs. Ac-CoA) or cell permeability for compound 12 and A-485.
Gene expression profiling.
RNA sequencing was used to investigate how treatment with p300 HAT inhibitor 12 affects gene expression in ER+ breast cancer MCF-7 cells. Upon starvation (i.e., culturing in charcoal treated FBS without hydrophobic hormones including estrogens) for 7 days, MCF-7 cells were supplemented with estradiol (10 nM), followed by treatment with compound 12 for 2 days. Total RNA from three groups of MCF-7 cells, including starved, control (with estradiol) and treated (with estradiol and compound 12) groups, was purified, prepared and sequenced. RNA sequencing data were mapped onto the human genome and gene expression profiles were determined and normalized. Genes with significantly changed expression levels were determined using a two-sided parametric t-test with the p value of <0.05. Gene set enrichment analysis (GSEA)32 was used to analyze significant gene expression changes between these three groups of MCF-7 cells.
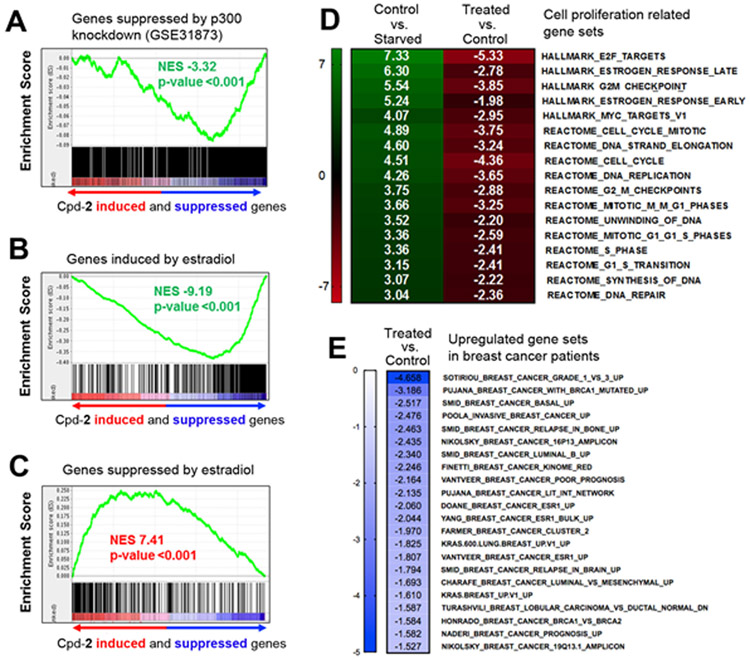
First, to determine whether the observed cellular activities of compound 12 are due to inhibition of p300 HAT, we used GSEA to compare the transcriptional profile of the 12-treated MCF-7 cells with a publicly available p300 transcriptional signature (GSE31873), which was derived from siRNA-mediated p300 knockdown in C4–2B prostate cancer cells33. As shown in Figure 5A, there is a strong concordance between the two transcriptional signatures: Genes suppressed by compound 12 were strongly enriched among those suppressed by p300 knockdown, with normalized enrichment score (NES) of −3.32 and p value of <0.001. This result demonstrates that treatment with the p300 HAT inhibitor mimics the transcriptional footprint of p300 depletion and supports that p300 HAT is the cellular target of compound 12.
Figure 5.
Gene set enrichment analysis (GSEA) results showing significant gene expression changes (p < 0.001) in ER+ MCF-7 cells by treatment with p300 HAT inhibitor 12, as compared to the starved (without estradiol) or control (with estradiol) group. (A) Inhibitor treatment caused significant downregulation of genes that were suppressed by siRNA-mediated p300 knockdown (GSE31873); (B-D) Results showing compound 12 counteracted estradiol in MCF-7 cells. Treatment with 12 significantly (B) suppressed expression of genes that were upregulated by estradiol, and (C) induced expression of genes that were downregulated by estradiol. (D, E) GSEA heatmaps of normalized enrichment scores (NES) for publicly available gene signatures (from the MSigDB database). (D) Inhibitor 12 caused significant downregulation of cancer-related gene sets that were induced by estradiol; (E) Inhibitor 12 suppressed expression of gene sets associated with breast cancer as well as the poor prognosis, progression, invasion and relapse of breast cancer.
Next, activity of compound 12 in the ER signaling pathway was analyzed. As compared to starved cells, supplementation with estradiol potently caused upregulation as well as downregulation of several publicly available ER-related gene signatures in the control group of MCF-7 cells (Figure S3). Importantly, treatment with compound 12 counteracted such estrogen-induced gene transcription: The gene set upregulated by estradiol was strongly downregulated upon treatment with 12 (Figure 5B, NES = −9.19 and p < 0.001), and the genes suppressed by estradiol were significantly upregulated by compound 12 (Figure 5C, NES = 7.41 and p < 0.001). These results are consistent with previous studies showing p300 HAT is essential for ER-mediated gene expression, and demonstrate that pharmacological inhibition of p300 HAT can offset the effects of estrogen in gene regulation.
Moreover, we investigated gene expression changes caused by compound 12 in the context of cancer biology and therapy. As shown in Figure 5D, global gene expression profiling analysis revealed that inhibition of p300 HAT by compound 12 counteracted estradiol and caused significant suppression of a number of estrogen-induced, cancer-related gene sets, including E2F and c-Myc gene signatures as well as those involved in cell proliferation, cell cycle, mitosis, DNA replication, DNA repair and self-renewal (stemness) (Figure S4). Specifically for breast cancer, treatment with compound 12 downregulated a number of gene signatures that have previously been shown to be upregulated in multiple breast cancer patient datasets (as compared to normal breast tissues), associated with poor prognosis, cancer progression, invasion and relapse (Figure 5E). Collectively, these GSEA results show the importance of p300 HAT in breast and other types of cancer, as well as the perspective for pharmacological inhibition of p300 HAT (by e.g., compound 12) in breast cancer therapy.
CONCLUSION
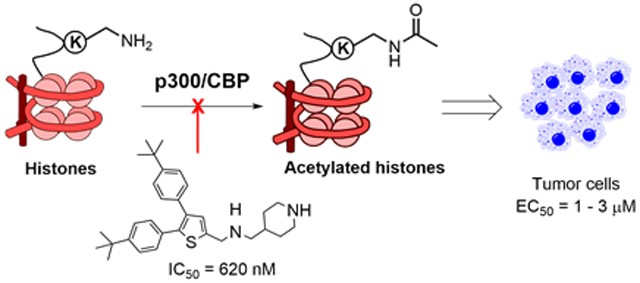
Histone acetylation by the HAT domain of p300/CBP has been found to play critical roles in many nuclear receptor-regulated signaling pathways, such as ER, AR and peroxisome proliferator-activated receptors (PPAR) 34. Dysfunction of these gene transcription programs has been found in a number of diseases such as cancer and obesity. Moreover, p300/CBP has been found to directly contribute to oncogenesis (e.g., in RUNX1-ETO leukemia) or be part of a fusion oncogene due to chromosome translocation 7, 14, 15. Therefore, drug-like inhibitors of p300/CBP HAT are needed, which could be useful chemical probes and potential therapeutics for these indications. In addition, p300 and CBP are large proteins (~2,400 amino acids) with multiple other domains playing important physiological roles. They also share a high degree of homology with many duplicate functions. Given these two points, cell-permeable, small-molecule inhibitors of the HAT domain of p300/CBP are particularly useful for studying biological functions of p300/CBP HAT, because observed activities by genetic knockdown/knockout of p300 or CBP might not be relevant.
Compound screening followed by medicinal chemistry studies have led to the discovery of tri-substituted thiophene compound 12 that is a novel inhibitor with an IC50 of 620 nM against p300 HAT and 1.2 μM against CBP HAT. It did not significantly inhibit two other major human HATs PCAF and Myst3 at 50 μM, showing a high selectivity. Biochemical studies showed that compound 12 is competitive against the substrate histone and non-competitive against the cofactor Ac-CoA, showing a distinct mode of inhibition from previous studied inhibitors C646 and A-485. Docking studies of 12 into the histone binding pocket of the p300 HAT structure provided possible binding models of the inhibitor in the protein. In addition, compound 12 was found to be cell-permeable and inhibit cellular acetylation at several histone lysine residues, with H3K27 being the most sensitive substrate. With EC50 of ~3 μM, compound 12 strongly inhibited proliferation of ER+ breast cancer cell line MCF-7, regardless of its sensitivity or resistance to the commonly used estrogen antagonist tamoxifen, suggesting the potential therapeutic application of this compound in the context of endocrine resistance. Compound 12 also exhibited strong activity against growth of several pancreatic cancer cells and RUNX1-ETO leukemia cell Kasumi-1, in which p300 HAT is known to be important 30, 31.
Global gene transcription profiling was performed to investigate the cellular target of compound 12 as well as the mechanism for its observed biological activities in MCF-7 cells. First, the overall transcriptional changes upon treatment with compound 12 recapitulated those caused by siRNA-mediated p300 knockdown. This result, together with its inhibitory activity in cellular histone lysine acetylation, support that p300 HAT is the cellular target of compound 12. Second, inhibition of p300 HAT activity by compound 12 significantly offset estrogen-induced gene expression, showing p300 HAT is essential for the ER activity. Moreover, treatment with compound 12 strongly downregulated a number of (general) cancer-related gene signatures, as well as gene sets that have been identified in the clinic for the progression, invasion, relapse and poor prognosis of breast cancer. These results, together with the compound’s potent antiproliferative activities against several types of cancer cells, suggest that pharmacological inhibition of p300 HAT is a useful therapeutic approach to cancer treatment.
In conclusion, our results demonstrate that compound 12 is not only a useful small-molecule probe for biological studies of p300/CBP HAT, but also a novel pharmacological lead for further drug development targeting breast and other types of cancer.
Experimental Section
All chemicals for synthesis were purchased from Alfa Aesar (Ward Hill, MA) or Aldrich (Milwaukee, WI). The identity of the synthesized compounds was characterized by 1H and 13C NMR on a Varian (Palo Alto, CA) 400-MR spectrometer and mass spectrometer (Shimadzu LCMS-2020). The identity of the potent inhibitors was confirmed with high resolution mass spectra (HRMS) using an Agilent 6550 iFunnel quadrupole-time-of-flight (Q-TOF) mass spectrometer with electrospray ionization (ESI). The purities of the final compounds were determined to be >95% with a Shimadzu Prominence HPLC using a Zorbax C18 (or C8) column (4.6 × 250 mm) monitored by UV at 254 nm.
General method A (reactions i-iv).
To a mixture of 4,5-Dibromothiophene-2-carboxylic acid 41 (2.86 g, 10 mmol), 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (1.71 g, 11 mmol), 1-hydroxybenzotriazole (1.48 g, 11 mmol) in dichloromethane (20 mL), 1-BOC-4-(aminomethyl)piperidine (2.57 g, 12 mmol) and triethylamine (2.8 mL, 20 mmol) in CH2Cl2 (10 mL) were added slowly at 0 °C. The reaction mixture was stirred at room temperature for 1 h and quenched by adding saturated NaHCO3 (50 mL). The product was extracted with dichloromethane (3 × 50 mL) and the combined organic layers were washed with water and brine, dried over Na2SO4. Upon removal of the solvent, the product was purified by column chromatography (silica gel, n-hexanes: ethyl acetate from 10:1 to 2:1) to give the amide 42 as a white solid (4.63 g, 96% yield).
A mixture of 42 (0.4 mmol), Ar1-boronic acid or its pinacol ester (0.42 mmol), tetrakis(tri-phenylphosphine)palladium (13.9 mg) and sodium carbonate (84.8 mg, 0.8 mmol) in 1,4-dioxane/H2O (6 mL, 5:1) were heated to 80 °C for 12 h. The reaction was then quenched with brine (10 mL). The product was extracted with diethyl ether (3 × 20 mL) and the combined organic layers were washed with water and brine, dried over Na2SO4. Upon removal of the solvent, the product was purified by column chromatography (silica gel, n-hexanes: ethyl acetate from 5:1 to 1:2) to give 5-Ar1 substituted thiophene product as a white solid (60-90% yield). The product thus obtained (0.2 mmol) was added into a solution of Ar2-boronic acid or its pinacol ester (0.21 mmol), tetrakis(triphenylphosphine)palladium (7 mg) and sodium carbonate (42.4 mg, 0.4 mmol) in 1,4-dioxane/H2O (5/1 mL). The mixture was heated to 100 °C for 12 h. Similar workup and purification gave 4,5-di-substituted thiophene compound 43 as a white solid (60-90% yield).
To a solution of 43 (0.1 mmol) in dichloromethane (2 mL) at 0 °C, HCl (0.1-0.2 mL, 4 N in 1,4-dioxane) was added slowly and then stirred at room temperature for 12 h. Upon removal of the solvent carefully, the residual oil was treated with anhydrous diethyl ether and vacuum dried to give the target compound (white powder) as a hydrochloric salt (90-100% yield).
General method B (reactions v-vii).
To a solution of 4,5-dibromothiophene-2-carboxaldehyde 44 (5.40 g, 20 mmol) in MeOH (40 mL), NaBH4 (0.79 g, 21 mmol) was added slowly 0 °C. The reaction mixture was stirred at room temperature for 1 h and quenched with water (50 mL). The product was extracted with diethyl ether (3 × 100 mL) and the combined organic layers were washed with water and brine, dried over Na2SO4. Upon removal of the solvent carefully, the residual oil was dried and used in the next step without purification. It was dissolved in DMF (20 mL) and the solution was cooled to 0 °C. Cyanuric chloride (3.69 g, 20 mmol) was added slowly at 0 °C and stirred at room temperature for 10 h. The reaction was quenched with saturated NaHCO3 (50 mL). The product was extracted with diethyl ether (3 × 100 mL) and the combined organic layers were washed with water and brine, dried over Na2SO4. Removal of the solvents in vacuo afforded compound 45 as a colorless oil, which is used without purification. To a solution of an amine (10 mmol) and potassium carbonate (1.38 g, 10 mmol) in DMF (10 mL), 45 (1.45 g, 5 mmol) in DMF (10 mL) was added slowly at 0 °C and the mixture was stirred at room temperature for 10 h. Upon quenching with saturated NaHCO3 (50 mL), the product was extracted with diethyl ether (3 × 50 mL) and the combined organic layers were washed with water and brine, dried over Na2SO4. A column chromatography (silica gel, hexanes: ethyl acetate from 40:1 to 2:1) for the residue oil gave compound 46 as a pale-yellow oil (60-85% yield for the three steps).
Suzuki coupling reactions of compound 46 as well as deprotection of the BOC group were performed as described above, to produce the target compounds.
General method C (reactions viii and ix).
4-Bromothiazole-2-carbaldehyde 48 (1.92 g, 10 mmol) and N-bromosuccinimide (NBS, 1.78 g, 10 mmol) were dissolved in dichloromethane (20 mL) and stirred at room temperature for 12 h. Upon removal of the solvent carefully, the residue was purified by column chromatography (silica gel, hexanes: ethyl acetate from 40:1 to 10:1) to give 4,5-dibromothiazole-2-carbaldehyde 49 as a white solid (433 mg, 16% yield). Conversion of aldehydes 48 and 49 to compounds 38 and 37 followed the reactions described above.
4,5-Dibromothiazole-2-carbaldehyde 49 (406 mg, 1.5 mmol) or 48, sodium chlorite (190 mg, 2.1 mmol) and 2-methylbut-2-ene (1.6 mL, 15 mmol) were dissolved by tBuOH/H2O (5/1 mL). Sodium dihydrogen phosphate (900 mg in 5 mL H2O) was added slowly into the reaction mixture. The mixture was stirred at rt for 12 h. The reaction was then quenched with brine (10 mL). The product was extracted with diethyl ether (3 × 10 mL) and the combined organic layers were washed with water and brine, dried over Na2SO4. The volatiles were removed in vacuo to afford acid 51 as a crude oil, which was used directly in the next step without further purification.
4,5-bis(4-(tert-Butyl)phenyl)-N-(piperidin-4-ylmethyl)thiophene-2-carboxamide hydrochloride (1) was prepared from 4,5-dibromothiophene-2-carboxylic acid, following the general method A, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 8.69 (br, 3H), 7.92 (s, 1H), 7.38-7.35 (m, 4H), 7.26-7.21 (m, 4H), 3.26-3.23 (m, 2H), 3.17 (br, 2H), 2.85-2.79 (m, 2H), 1.82-1.79 (m, 3H), 1.42-1.36 (m, 2H), and 1.27 (s, 18H); 13C NMR (100 MHz, DMSO-d6): 161.1, 150.8, 149.8, 141.5, 137.6 (2), 132.7, 131.4, 130.4, 128.4, 128.2, 125.6, 125.3, 43.8, 42.8, 34.4, 34.3, 33.7, 31.1, 31.0, and 26.22; MS (ESI) calcd for (C31H41N2OS)+ [M+H]+ 489.7, found 489.6.
4,5-bis(4-(Furan-3-yl)phenyl)-N-(piperidin-4-ylmethyl)thiophene-2-carboxamide hydrochloride (2) was prepared from 4,5-dibromothiophene-2-carboxylic acid, following the general method A, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 8.89 (br, 1H), 8.76 (br, 1H), 8.58 (br, 1H), 8.21 (s, 2H), 8.00 (s, 1H), 7.74 (s, 2H), 7.61-7.59 (m, 4H), 7.31-7.28 (m, 4H), 6.97 (s, 2H), 3.27-3.23 (m, 2H), 3.19 (br, 2H), 2.86-2.78 (m, 2H), 1.85-1.82 (m, 3H), and 1.42-1.36 (m, 2H); 13C NMR (100 MHz, DMSO-d6): 161.0, 147.1, 144.5, 141.6, 139.9, 139.6, 138.6, 138.0, 137.8, 133.9, 132.08, 131.96, 131.65, 131.54, 131.0, 130.8, 129.2, 129.0, 125.9, 125.7, 108.6, 43.8, 42.8, 33.8, and 26.2; MS (ESI) calcd for (C31H29N2O3S)+ [M+H]+ 509.6, found 509.5.
4,5-bis(4’-Methoxy-[1,1’-biphenyl]-4-yl)-N-(piperidin-4-ylmethyl)thiophene-2-carboxamide hydrochloride (3) was prepared from 4,5-dibromothiophene-2-carboxylic acid, following the general method A, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 8.91 (br, 1H), 8.79 (br, 1H), 8.62 (br, 1H), 8.02 (s, 1H), 7.70-7.56 (m, 8H), 7.38-7.35 (m, 4H), 7.02-6.99 (m, 4H), 3.78 (s, 6H), 3.27-3.23 (m, 2H), 3.19 (br, 2H), 2.87-2.79 (m, 2H), 1.85-1.82 (m, 3H), and 1.42-1.36 (m, 2H); 13C NMR (100 MHz, DMSO-d6): 161.1, 159.2, 159.0, 141.6, 139.6, 138.6, 138.1, 137.7, 133.8, 131.64, 131.57, 131.46, 131.38, 129.3, 129.2, 127.72, 127.66, 126.5, 126.2, 114.5, 114.4, 55.2, 43.8, 42.9, 33.8, 28.1, and 26.3; MS (ESI) calcd for (C37H37N2O3S)+ [M+H]+ 589.8, found 589.9.
5-(4-(tert-Butyl)phenyl)-4-(4-(furan-3-yl)phenyl)-N-(piperidin-4-ylmethyl)thiophene-2-carboxamide hydrochloride (4) was prepared from 4,5-dibromothiophene-2-carboxylic acid, following the general method A, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 8.90 (br, 1H), 8.76 (br, 1H), 8.61 (br, 1H), 8.21 (s, 1H), 7.97 (s, 1H), 7.92 (s, 1H), 7.74 (s, 1H), 7.59 (s, 1H), 7.36-7.23 (m, 6H), 6.97 (s, 2H), 3.27-3.23 (m, 2H), 3.18 (br, 2H), 2.85-2.79 (m, 2H), 1.83-1.80 (m, 3H), 1.40-1.36 (m, 2H), and 1.26 (s, 9H); 13C NMR (100 MHz, DMSO-d6): 161.1, 151.0, 144.4, 142.0, 139.6, 137.8, 137.5, 134.1, 131.0, 129.0, 128.44, 128.40, 128.3, 125.74, 125.70, 125.4, 108.6, 43.8, 42.9, 34.4, 33.8, 31.1, and 26.3; MS (ESI) calcd for (C31H35N2O2S)+ [M+H]+ 499.7, found 499.5.
4-(4-(Aminomethyl)phenyl)-5-(4-(tert-butyl)phenyl)-N-(piperidin-4-ylmethyl)thiophene-2-carboxamide hydrochloride (5) was prepared from 4,5-dibromothiophene-2-carboxylic acid, following the general method A, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, D2O): δ 7.76 (s, 1H), 7.25 (d, J = 7.6 Hz, 2H), 7.14 (d, J = 7.6 Hz, 2H), 6.97 (d, J = 7.3 Hz, 2H), 6.90 (d, J = 7.3 Hz, 2H), 4.03 (s, 2H), 3.45 (d, J = 11.7 Hz, 2H), 3.29-3.27 (m, 2H), 2.95 (t, J = 12.6 Hz, 2H), 1.98-1.94 (m, 3H), 1.52-1.42 (m, 2H), and 0.97 (s, 9H); 13C NMR (100 MHz, D2O): 163.4, 151.3, 143.8, 137.3, 136.1, 134.3, 131.6, 129.9, 129.4, 129.1, 128.5, 128.1, 125.4, 44.3, 43.6, 42.6, 33.9, 33.5, 30.5, and 26.1; MS (ESI) calcd for (C28H36N3OS)+ [M+H]+ 462.7, found 462.6.
5-(4-(Furan-3-yl)phenyl)-4-(4-(piperidin-1-ylmethyl)phenyl)-N-(piperidin-4-ylmethyl)thiophene-2-carboxamide hydrochloride (6) was prepared from 4,5-dibromothiophene-2-carboxylic acid, following the general method A, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 10.54 (br, 1H), 8.94 (br, 1H), 8.84 (br, 1H), 8.64 (br, 1H), 8.22 (s, 1H), 8.03 (s, 1H), 7.75 (s, 1H), 7.60 (d, J = 7.8 Hz, 4H), 7.36 (d, J = 7.8 Hz, 2H), 7.28 (d, J = 7.8 Hz, 2H), 6.97 (s, 1H), 4.24 (s, 4H), 3.25-3.24 (m, 4H), 3.19-3.16 (m, 2H), 2.86-2.78 (m, 4H), 1.83-1.78 (m, 6H), 1.70-1.66 (m, 1H), and 1.48-1.34 (m, 2H); 13C NMR (100 MHz, DMSO-d6): 161.0, 144.6, 142.2, 140.0, 138.2, 137.3, 136.6, 132.1, 131.8, 131.6, 131.5, 129.3, 129.0, 126.0, 125.2, 115.5, 108.6, 58.6, 51.7, 43.9, 42.9, 33.7, 26.3, 22.2, and 21.5; MS (ESI) calcd for (C33H38N3O2S)+ [M+H]+ 540.7, found 540.6.
5-(4-(tert-Butyl)phenyl)-4-(4-(piperazin-1-ylmethyl)phenyl)-N-(piperidin-4-ylmethyl)thiophene-2-carboxamide hydrochloride (7) was prepared from 4,5-dibromothiophene-2-carboxylic acid, following the general method A, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, CD3OD): δ 7.83 (s, 1H), 7.58 (d, J = 8.1 Hz, 2H), 7.42 (d, J = 8.1 Hz, 2H), 7.37 (d, J = 8.4 Hz, 2H), 7.23 (d, J = 8.4 Hz, 2H), 4.44 (s, 2H), 3.61 (br, 4H), 3.56 (br, 4H), 3.43 (d, J = 8.1 Hz, 2H), 3.35 (d, J = 6.0 Hz, 2H), 3.00 (t, J = 12.2 Hz, 2H), 2.03-1.98 (m, 3H), 1.55-1.45 (m, 2H), and 1.31 (s, 9H); 13C NMR (100 MHz, CD3OD): 164.3, 153.1, 145.8, 139.2, 138.5, 138.1, 134.2, 132.8, 132.3, 131.6, 130.9, 130.0, 126.9, 61.4, 58.7, 45.4, 45.0, 42.2, 35.6, 31.6, 28.5, and 27.8; MS (ESI) calcd for (C32H43N4OS)+ [M+H]+ 531.8, found 531.7.
5-(4-(tert-Butyl)phenyl)-4-(furan-3-yl)-N-(piperidin-4-ylmethyl)thiophene-2-carboxamide hydrochloride (8) was prepared from 4,5-dibromothiophene-2-carboxylic acid, following the general method A, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 8.90 (br, 1H), 8.73 (br, 1H), 8.59 (br, 1H), 7.97 (s, 1H), 7.65 (s, 1H), 7.44-7.34 (m, 5H), 6.29 (s, 1H), 3.27-3.24 (m, 2H), 3.17 (br, 2H), 2.84-2.80 (m, 2H), 1.83-1.78 (m, 3H), 1.40-1.37 (m, 2H), and 1.29 (s, 9H); 13C NMR (100 MHz, DMSO-d6): 161.4, 151.7, 144.1, 140.4, 138.2, 130.7, 130.5, 129.2, 129.1, 126.3, 126.0, 120.8, 110.6, 44.2, 43.2, 34.9, 34.1, 31.4, and 26.6; MS (ESI) calcd for (C25H31N2O2S)+ [M+H]+ 423.6, found 423.5.
5-(4-(tert-Butyl)phenyl)-N-(piperidin-4-ylmethyl)-4-(pyridin-3-yl)thiophene-2-carboxamide hydrochloride (9) was prepared from 4,5-dibromothiophene-2-carboxylic acid, following the general method A, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.00 (br, 1H), 8.92 (br, 1H), 8.74 (br, 1H), 8.70 (br, 1H), 8.17 (br, 2H), 7.85 (s, 1H), 7.61-7.55 (m, 2H), 7.40-7.26 (m, 4H), 3.27-3.18 (m, 4H), 2.83-2.80 (m, 2H), 1.84-1.79 (m, 3H), 1.42-1.39 (m, 2H), and 1.26 (s, 9H); 13C NMR (100 MHz, DMSO-d6): 161.2, 152.1, 145.2, 143.8, 143.3, 142.8, 139.2, 134.1, 132.2, 131.1, 129.5, 129.1, 126.6, 126.5, 44.3, 43.2, 34.9, 34.1, 31.4, and 26.6; MS (ESI) calcd for (C26H32N3OS)+ [M+H]+ 434.6, found 434.4.
1-(4,5-bis(4’-Methoxy-[1,1’-biphenyl]-4-yl)thiophen-2-yl)-N-(piperidin-4-ylmethyl)methanamine hydrochloride (10) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.46 (br, 1H), 9.39 (br, 1H), 8.90 (br, 1H), 8.72 (br, 1H), 7.69 (s, 1H), 7.63-7.54 (m, 8H), 7.34 (d, J = 8.2 Hz, 4H), 7.01 (d, J = 8.2 Hz, 4H), 4.41 (s, 2H), 3.78 (s, 6H), 3.29-3.26 (m, 2H), 2.94 (br, 2H), 2.87-2.79 (m, 2H), 2.08 (br, 1H), 1.98-1.92 (m, 2H), and 1.49-1.39 (m, 2H); 13C NMR (100 MHz, DMSO-d6): 159.2, 159.1, 139.4, 139.2, 138.6, 137.2, 134.2, 133.9, 131.7, 131.4, 129.4, 129.2, 127.76, 127.70, 126.9, 126.6, 126.4, 126.0, 114.54. 114.50, 55.3, 50.7, 44.7, 42.5, 30.60, 30.55, and 26.1; MS (ESI) calcd for (C37H39N2O2S)+ [M+H]+ 575.8, found 575.7.
1-(4,5-bis(4-(Furan-3-yl)phenyl)thiophen-2-yl)-N-(piperidin-4-ylmethyl)methanamine hydrochloride (11) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (yellow powder). 1H NMR (400 MHz, DMSO-d6): δ 9.54 (br, 2H), 9.03 (br, 1H), 8.88 (br, 1H), 7.46 (s, 1H), 7.16-7.12 (m, 8H), 4.35 (s, 2H), 3.26-3.23 (m, 2H), 2.92-2.88 (m, 2H), 2.84-2.79 (m, 2H), 2.54 (t, J =7.4 Hz, 4H), 2.09 (br, 1H), 1.97-1.93 (m, 2H), 1.52 (p, J = 7.4 Hz, 4H), 1.46-1.39 (m, 2H), 1.32-1.23 (m, 4H), and 0.87 (t, J = 7.4 Hz, 6H); 13C NMR (100 MHz, DMSO-d6): 144.5, 144.4, 139.8, 139.6, 139.1, 137.2, 134.04, 133.97, 131.79, 131.69, 131.62, 130.9, 129.2, 129.0, 126.0, 125.8, 125.3, 125.2, 108.58, 108.56, 50.6, 44.6, 42.4, 30.6, and 26.1; MS (ESI) calcd for (C31H31N2O2S)+ [M+H]+ 495.7, found 495.6.
1-(4,5-bis(4-(tert-Butyl)phenyl)thiophen-2-yl)-N-(piperidin-4-ylmethyl)methanamine hydrochloride (12) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.56 (br, 2H), 9.00 (br, 1H), 8.84 (br, 1H), 7.48 (s, 1H), 7.36 (d, J = 7.2 Hz, 4H), 7.20 (d, J = 7.2 Hz, 4H), 4.35 (s, 2H), 3.27-3.24 (m, 2H), 2.89 (br, 2H), 2.86-2.82 (m, 2H), 2.05 (br, 1H), 1.97-1.94 (m, 2H), 1.45-1.40 (m, 2H), and 1.27 (s, 18H); 13C NMR (100 MHz, DMSO-d6): 150.5, 149.6, 139.0, 137.0, 134.3, 132.8, 131.4, 130.6, 128.4, 128.2, 125.7, 125.4, 50.6, 44.6, 42.3, 34.42, 34.34, 31.13, 31.05, 30.6, and 26.1; MS (ESI) calcd for (C31H43N2S)+ [M+H]+ 475.8, found 475.7; HRMS (ESI+) calcd for C31H42N2S [M+H]+ 475.3147, found 475.3145.
4-(((4,5-bis(4-(tert-Butyl)phenyl)thiophen-2-yl)methoxy)methyl)piperidine hydrochloride (13).
Starting from 4,5-dibromothiophene-2-carboxaldehyde, reactions v and vi gave 2-chloromethyl-4,5-dibromothiophene, which reacted with sodium salt of 1-BOC-4-hydroxymethylpiperidine, to give 4,5-dibromothiophen-2-ylmethyl 1-BOC-piperidin-4-yl-methyl ether. Reactions ii and iii (General method B) produced compound 13 as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 8.72 (br, 1H), 8.38 (br, 1H), 7.36 (d, J = 8.1 Hz, 2H), 7.33 (d, J = 8.1 Hz, 2H), 7.48 (s, 1H), 7.21 (d, J = 8.1 Hz, 2H), 7.17 (d, J = 8.1 Hz, 2H), 5.05 (s, 2H), 3.27-3.23 (m, 4H), 2.86-2.77 (m, 2H), 1.78-1.75 (m, 2H), 1.62 (br, 1H), and 1.26 (s, 18H); 13C NMR (100 MHz, DMSO-d6): 150.4, 149.6, 138.8, 138.6, 136.7, 132.8, 131.7, 130.7, 128.4, 128.2, 125.6, 125.3, 65.0, 42.9, 40.8, 35.9, 34.4, 34.3, 31.10, 31.02, and 25.2; MS (ESI) calcd for (C31H42NOS)+ [M+H]+ 476.7, found 476.6.
N-((4,5-bis(4-(tert-Butyl)phenyl)thiophen-2-yl)methyl)piperidin-4-amine hydrochloride (14) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.85 (br, 2H), 9.24 (br, 1H), 8.99 (br, 1H), 7.50 (s, 1H), 7.36 (d, J = 7.2 Hz, 4H), 7.20 (d, J = 7.2 Hz, 4H), 4.40 (s, 2H), 2.95 (br, 3H), 2.30 (br, 2H), 1.95 (m, 2H), and 1.27 (s, 18H); 13C NMR (100 MHz, DMSO-d6): 150.6, 149.7, 139.1, 137.1, 134.3, 132.8, 131.2, 130.6, 128.4, 128.2, 125.7, 125.4, 51.4, 41.4, 34.42, 34.34, 31.14, 31.06, 28.9, and 24.9; MS (ESI) calcd for (C30H41N2S)+ [M+H]+ 461.7, found 461.6.
N-((4,5-bis(4-(tert-Butyl)phenyl)thiophen-2-yl)methyl)-2-(piperidin-4-yl)ethanamine hydrochloride (15) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.55 (br, 2H), 8.99 (br, 1H), 8.81 (br, 1H), 7.47 (s, 1H), 7.35 (br, 4H), 7.20 (br, 4H), 4.35 (s, 2H), 3.23 (br, 2H), 2.98 (br, 2H), 2.80 (br, 2H), 1.78 (br, 1H), 1.65 (br, 2H), 1.35 (br, 2H), and 1.26 (s, 18H); 13C NMR (100 MHz, DMSO-d6): 150.5, 149.6, 138.9, 137.0, 134.1, 132.7, 131.4, 130.5, 128.4, 128.2, 125.7, 125.4, 44.0, 43.8, 43.0, 34.39, 34.31, 31.3, 31.1, 31.0, 30.6, and 28.0; MS (ESI) calcd for (C32H45N2S)+ [M+H]+ 489.8, found 489.7.
N1-((4,5-bis(4-(tert-Butyl)phenyl)thiophen-2-yl)methyl)propane-1,3-diamine hydrochloride (16) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, CDCl3): δ 9.70 (br, 2H), 8.24 (br, 3H), 7.34 (s, 1H), 7.17-7.07 (m, 8H), 7.24 (d, J = 7.6 Hz, 4H), 4.22 (s, 2H), 3.29-3.20 (m, 2H), 2.47 (br, 2H), 1.34-1.32 (m, 2H), and 1.19 (s, 18H); 13C NMR (100 MHz, CDCl3): 150.6, 149.8, 140.8, 137.7, 135.0, 132.9, 130.8, 129.0, 128.8, 128.7, 125.5, 125.4, 45.5, 44.0, 37.8, 34.62, 34.57, 31.44, 31.36, and 29.8; MS (ESI) calcd for (C28H39N2S)+ [M+H]+ 435.7, found 435.6.
N1-((4,5-bis(4-(tert-Butyl)phenyl)thiophen-2-yl)methyl)hexane-1,6-diamine hydrochloride (17) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.34 (br, 2H), 7.87 (br, 3H), 7.44 (s, 1H), 7.38 (d, J = 8.2 Hz, 2H), 7.36 (d, J = 8.2 Hz, 2H), 7.21 (d, J = 8.2 Hz, 2H), 7.19 (d, J = 8.2 Hz, 2H), 4.35 (s, 2H), 2.94 (br, 2H), 2.76 (p, J = 7.4 Hz, 2H), 1.66 (p, J = 7.4 Hz, 2H), 1.54 (p, J = 7.4 Hz, 2H), 1.35-1.33 (m, 4H), and 1.27 (s, 18H); 13C NMR (100 MHz, DMSO-d6): 150.6, 149.7, 139.0, 137.0, 134.0, 132.7, 131.4, 130.5, 128.3, 128.2, 125.6, 125.4, 46.7, 46.1, 44.1, 34.4, 34.3, 31.1, 31.0, 26.7, 25.5, 25.3, and 25.2; MS (ESI) calcd for (C31H45N2S)+ [M+H]+ 477.8, found 477.6.
1-((4,5-bis(4-(tert-Butyl)phenyl)thiophen-2-yl)methyl)piperazine hydrochloride (18) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.61 (br, 1H), 9.50 (br, 2H), 7.44 (s, 1H), 7.38-7.34 (m, 4H), 7.22-7.19 (m, 4H), 4.48 (s, 2H), 3.33 (s, 4H), 2.50 (s, 4H), and 1.27 (s, 18H); 13C NMR (100 MHz, DMSO-d6): 150.6, 149.7, 140.0, 138.6, 137.1, 132.6, 131.7, 130.4, 128.4, 128.3, 125.7, 125.4, 52.6, 47.1, 40.8, 34.4, 34.3, 31.1, and 31.0; MS (ESI) calcd for (C29H39N2S)+ [M+H]+ 447.7, found 447.6.
4-((4,5-bis(4-(tert-Butyl)phenyl)thiophen-2-yl)methyl)morpholine hydrochloride (19) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, CDCl3): δ 13.34 (br, 1H), 7.45 (s, 1H), 7.30 (d, J = 7.6 Hz, 4H), 7.24 (d, J = 7.6 Hz, 4H), 4.46 (s, 2H), 4.30 (br, 2H), 4.01 (br, 2H), 3.46 (br, 2H), 3.08 (br, 2H), and 1.32 (s, 18H); 13C NMR (100 MHz, CDCl3): 151.3, 150.4, 142.2, 138.4, 136.6, 132.5, 130.3, 128.8, 128.6, 125.6, 125.5, 125.3, 63.8, 54.8, 50.6, 34.72, 34.64, 31.39, and 31.32; MS (ESI) calcd for (C29H38NOS)+ [M+H]+ 448.7, found 448.6.
1-(4,5-bis(4-(tert-Butyl)phenyl)thiophen-2-yl)-N-methyl-N-(piperidin-4-ylmethyl)methanamine hydrochloride (20) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.58 (br, 1H), 9.02 (br, 1H), 8.85 (br, 1H), 7.53 (s, 1H), 7.38-7.35 (m, 4H), 7.24-7.20 (m, 4H), 4.55 (s, 2H), 3.27-3.24 (m, 2H), 3.05-2.99 (m, 2H), 2.90-2.84 (m, 2H), 2.77 (s, 3H), 2.17-2.10 (m, 2H), 1.98-1.95 (m, 1H), 1.46-1.39 (m, 2H), and 1.27 (s, 18H); 13C NMR (100 MHz, DMSO-d6): 150.7, 149.7, 140.4, 137.2, 136.4, 132.6, 130.4, 128.4, 128.3, 128.2, 125.7, 125.4, 58.5, 52.6, 42.42, 42.37, 34.42, 34.33, 31.14, 31.05, 28.9, and 26.4; MS (ESI) calcd for (C32H45N2S)+ [M+H]+ 489.8, found 489.6.
N-((4,5-bis(4-(tert-Butyl)phenyl)thiophen-2-yl)methyl)-N-(piperidin-4-ylmethyl)formamide hydrochloride (21) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, CDCl3): δ 9.34 (br, 1H), 9.01 (br, 1H), 7.29-7.25 (m, 4H), 7.20-7.17 (m, 4H), 6.99 (s, 1H), 4.65 (s, 2H), 3.31-3.27 (m, 2H), 3.14-3.11 (m, 2H), 2.93-2.89 (m, 2H), 1.83-1.76 (m, 3H), 1.62-1.59 (m, 2H), and 1.29 (s, 18H); 13C NMR (100 MHz, CDCl3): 162.9, 154.8, 150.1, 138.7, 137.0, 136.5, 131.2, 130.8, 130.3, 128.7, 128.6, 125.5, 125.4, 52.7, 47.6, 47.5, 41.1, 34.6, 31.44, 31.36, 29.8, and 28.5; MS (ESI) calcd for (C32H43N2OS)+ [M+H]+ 503.8, found 503.6.
1-(4,5-bis(4-Isopropylphenyl)thiophen-2-yl)-N-(piperidin-4-ylmethyl)methanamine hydrochloride (22) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.67 (br, 2H), 9.13 (br, 1H), 9.03 (br, 1H), 7.50 (s, 1H), 7.20-7.15 (m, 8H), 4.34 (s, 2H), 3.26-3.23 (m, 2H), 2.90-2.83 (m, 4H), 2.75 (sep, J = 7.0 Hz, 2H), 2.10 (br, 1H), 1.99-1.94 (m, 2H), 1.47-1.40 (m, 2H), and 1.18 (d, J = 7.0 Hz, 12H); 13C NMR (100 MHz, DMSO-d6): 148.2, 147.3, 139.0, 137.1, 134.2, 133.1, 131.3, 130.9, 128.6, 128.5, 126.8, 126.5, 50.5, 44.5, 42.3, 33.1, 30.6, 28.1, 26.1, 23.8, and 23.7; MS (ESI) calcd for (C29H39N2S)+ [M+H]+ 447.7, found 447.6.
1-(4,5-bis(4-Butylphenyl)thiophen-2-yl)-N-(piperidin-4-ylmethyl)methanamine hydrochloride (23) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.54 (br, 2H), 9.03 (br, 1H), 8.88 (br, 1H), 7.46 (s, 1H), 7.16-7.12 (m, 8H), 4.35 (s, 2H), 3.26-3.23 (m, 2H), 2.90 (br, 2H), 2.84-2.79 (m, 2H), 2.54 (t, J = 7.4 Hz, 4H), 2.09 (br, 1H), 1.97-1.93 (m, 2H), 1.52 (p, J = 7.4 Hz, 4H), 1.46-1.39 (m, 2H), 1.32-1.23 (m, 4H), and 0.87 (t, J = 7.4 Hz, 6H); 13C NMR (100 MHz, DMSO-d6): 142.5, 141.5, 139.3, 137.3, 134.2, 133.0, 131.2, 130.8, 128.86, 128.77, 128.62, 128.56, 50.7, 44.7, 42.5, 34.6, 33.09, 33.00, 30.6, 26.1, 24.8, 21.9 (2), and 13.9 (2); MS (ESI) calcd for (C31H43N2S)+ [M+H]+ 475.8, found 475.7.
1-(5-(4-(tert-Butyl)phenyl)-4-(4-(furan-3-yl)phenyl)thiophen-2-yl)-N-(piperidin-4-ylmethyl)methanamine hydrochloride (24) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.51 (br, 2H), 8.95 (br, 1H), 8.79 (br, 1H), 8.08 (s, 1H), 7.89 (d, J = 8.1 Hz, 1H), 7.61-7.58 (m, 1H), 7.52 (s, 1H), 7.39-7.36 (m, 2H), 7.32 (d, J = 8.1 Hz, 1H), 7.24-7.19 (m, 4H), 6.97 (s, 1H), 4.38 (s, 2H), 3.28-3.25 (m, 2H), 2.93-2.88 (m, 2H), 2.86-2.82 (m, 2H), 2.08 (br, 1H), 1.98-1.95 (m, 2H), 1.47-1.42 (m, 2H), and 1.26 (s, 9H); 13C NMR (100 MHz, DMSO-d6): 146.5, 144.5, 139.6, 136.7, 134.1, 133.9, 131.6, 131.4, 129.0, 128.8, 128.4, 128.3, 127.4, 125.8, 125.7, 108.5, 50.6, 44.6, 42.4, 34.4, 31.0, 30.5, and 26.1; MS (ESI) calcd for (C31H37N2OS)+ [M+H]+ 485.7, found 485.6.
1-(4-(4-(Furan-3-yl)phenyl)-5-(4-(piperidin-1-ylmethyl)phenyl)thiophen-2-yl)-N-(piperidin-4-ylmethyl)methanamine hydrochloride (25) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 10.50 (br, 1H), 9.60 (br, 2H), 8.91 (br, 1H), 8.78 (br, 1H), 8.21 (s, 1H), 7.75 (s, 1H), 7.64-7.54 (m, 5H), 7.35 (d, J = 8.1 Hz, 2H), 7.24 (d, J = 8.1 Hz, 2H), 6.97 (s, 1H), 4.40 (s, 2H), 4.24 (s, 2H), 3.28-3.24 (m, 4H), 2.94-2.90 (m, 2H), 2.86-2.81 (m, 4H), 2.09 (br, 1H), 1.98-1.95 (m, 2H), 1.77 (br, 4H), 1.71-1.67 (m, 2H), and 1.48-1.39 (m, 2H); 13C NMR (100 MHz, DMSO-d6): 144.5, 139.6, 138.4, 137.6, 134.4, 134.1, 133.7, 132.1, 132.0, 131.5, 131.4, 129.0, 128.8, 128.7, 125.8, 108.6, 51.6, 50.6, 44.5, 42.3, 30.7, 30.5, 26.1, 22.1, and 21.4; MS (ESI) calcd for (C33H40N3OS)+ [M+H]+ 526.8, found 526.6.
1-(5-(4-(Aminomethyl)phenyl)-4-(4-(tert-butyl)phenyl)thiophen-2-yl)-N-(piperidin-4-ylmethyl)methanamine hydrochloride (26) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, D2O): δ 7.63 (s, 1H), 7.45 (d, J = 9.2 Hz, 2H), 7.42-7.39 (m, 4H), 7.28 (d, J = 9.2 Hz, 2H), 4.54 (s, 2H), 4.18 (s, 2H), 3.50-3.46 (m, 2H), 3.14 (br, 2H), 3.03 (t, J = 14.4 Hz, 2H), 2.15 (br, 1H), 2.07-2.04 (m, 2H), 1.57-1.48 (m, 2H), and 1.29 (s, 9H); 13C NMR (100 MHz, D2O): 144.2, 139.8, 138.3, 137.4, 134.0, 132.4, 130.2, 129.8, 129.1, 128.7, 125.6, 108.8, 50.7, 44.4, 43.1, 42.6, 30.7, 30.3, 25.8, and 22.1; MS (ESI) calcd for (C28H38N3S)+ [M+H]+ 448.7, found 448.8.
1-(4-(4-(Aminomethyl)phenyl)-5-(4-(tert-butyl)phenyl)thiophen-2-yl)-N-(piperidin-4-ylmethyl)-methanamine hydrochloride (27) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.73 (br, 1H), 9.12 (br, 1H), 9.00 (br, 1H), 8.50 (br, 2H), 8.28 (br, 2H), 7.55 (s, 1H), 7.47 (d, J = 7.9 Hz, 2H), 7.37 (d, J = 7.9 Hz, 2H), 7.28 (d, J = 7.9 Hz, 2H), 7.19 (d, J = 7.9 Hz, 2H), 4.35 (s, 2H), 4.00 (s, 2H), 3.24 (d, J = 12.7 Hz, 2H), 2.89-2.83 (m, 4H), 2.10 (br, 1H), 1.97 (d, J = 12.3 Hz, 2H), 1.50-1.42 (m, 2H), and 1.27 (s, 9H); 13C NMR (100 MHz, DMSO-d6): 150.7, 139.6, 136.6, 135.7, 134.2, 133.0, 130.5, 129.2, 128.7, 128.4, 125.7, 115.3, 50.5, 44.4, 42.3, 41.8, 34.4, 31.0, 30.5, and 26.1; MS (ESI) calcd for (C28H38N3S)+ [M+H]+ 448.7, found 448.8.
((5-(((Piperidin-4-ylmethyl)amino)methyl)thiophene-2,3-diyl)bis(4,1-phenylene))dimethanamine hydrochloride (28) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, D2O) δ 7.43 (s, 1H), 7.40 (br, 8H), 4.57 (s, 2H), 4.19 (s, 4H), 3.50 (d, J = 12.8 Hz, 2H), 3.15 (d, J = 6.8 Hz, 2H), 3.05 (t, J = 12.0 Hz, 2H), 2.18 (s, 1H), 2.08 (d, J = 14.0 Hz, 2H), and 1.59-1.50 (m, 2H); 13C NMR (100 MHz, D2O) 140.6, 137.9, 136.1, 133.9, 133.8, 132.6, 131.8, 130.5, 129.9, 129.6, 129.1, 129.0, 50.8, 45.4, 43.1, 42.64, 42.57, 30.7, and 25.8. MS (ESI) calcd for (C25H33N4S)+ [M+H]+ 421.6, found 421.5.
((5-(((Piperidin-4-ylmethyl)amino)methyl)thiophene-2,3-diyl)bis(4,1-phenylene))dimethanol hydrochloride (29) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.42 (br, 2H), 8.91 (br, 1H), 8.74 (br, 1H), 7.47 (s, 1H), 7.28 (d, J = 7.1 Hz, 2H), 7.26 (d, J = 7.1 Hz, 2H), 7.19 (d, J = 7.1 Hz, 2H), 7.17 (d, J = 7.1 Hz, 2H), 4.47 (s, 4H), 4.37 (s, 2H), 3.27-3.24 (m, 2H), 2.92 (br, 2H), 2.89-2.82 (m, 2H), 2.07 (br, 1H), 1.97-1.93 (m, 2H), and 1.48-1.39 (m, 2H); 13C NMR (100 MHz, DMSO-d6): 142.6, 141.7, 139.5, 137.4, 134.2, 134.0, 131.8, 131.4, 128.7, 128.4, 127.0, 126.8, 62.7, 62.6, 50.8, 44.8, 42.5, 30.6, and 26.1; MS (ESI) calcd for (C25H31N2O2S)+ [M+H]+ 423.6, found 423.5.
1-(4,5-di(Furan-3-yl)thiophen-2-yl)-N-(piperidin-4-ylmethyl)methanamine hydrochloride (30) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.61 (br, 2H), 9.09 (br, 1H), 8.96 (br, 1H), 7.88 (s, 1H), 7.78 (s, 2H), 7.71 (s, 1H), 7.48 (s, 1H), 6.52 (s, 1H), 6.48 (s, 1H), 4.32 (s, 2H), 3.27-3.24 (m, 2H), 2.87-2.82 (m, 4H), 2.08 (br, 1H), 1.97-1.94 (m, 2H), and 1.45-1.40 (m, 2H); 13C NMR (100 MHz, DMSO-d6): 144.2, 143.7, 140.9, 140.2, 133.1, 131.1, 129.8, 129.2, 120.1, 118.1, 111.1, 110.3, 50.4, 44.4, 42.3, 30.5, and 26.0; MS (ESI) calcd for (C19H23N2O2S)+ [M+H]+ 343.5, found 343.4.
1-(4,5-bis(3,4-Dimethoxyphenyl)thiophen-2-yl)-N-(piperidin-4-ylmethyl)methanamine hydrochloride (31) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.40 (br, 2H), 8.94 (br, 1H), 8.77 (br, 1H), 7.47 (s, 1H), 7.17-7.14 (m, 1H), 7.01-6.91 (m, 2H), 6.84 (d, J = 8.4 Hz, 1H), 6.78 (s, 1H), 6.74 (s, 1H), 4.34 (s, 2H), 3.75 (s, 3H), 3.73 (s, 3H), 3.56 (s, 3H), 3.54 (s, 3H), 3.28-3.25 (m, 2H), 2.92 (br, 2H), 2.88-2.82 (m, 2H), 2.07 (br, 1H), 1.98-1.91 (m, 2H), and 1.48-1.38 (m, 2H); 13C NMR (100 MHz, DMSO-d6): 148.8, 148.51, 148.48, 148.1, 136.9, 131.6, 131.5, 128.9, 128.8, 125.9, 121.4, 121.0, 112.54, 112.52, 112.0, 111.9, 55.69, 55.62, 55.59, 55.36, 50.7, 44.8, 42.4, 30.6, and 26.1; MS (ESI) calcd for (C27H35N2O4S)+ [M+H]+ 483.6, found 483.5.
1-(4,5-di([1,1’-Biphenyl]-3-yl)thiophen-2-yl)-N-(piperidin-4-ylmethyl)methanamine hydrochloride (32) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.50 (br, 2H), 8.88 (br, 1H), 8.68 (br, 1H), 7.65 (s, 2H), 7.52-7.31 (m, 17H), 4.42 (s, 2H), 3.29-3.26 (m, 2H), 2.95 (br, 2H), 2.90-2.82 (m, 2H), 2.08 (br, 1H), 1.98-1.95 (m, 2H), and 1.48-1.39 (m, 2H); 13C NMR (100 MHz, DMSO-d6): 140.7, 140.5, 139.8, 139.5, 139.4, 137.6, 136.0, 133.9, 129.7, 129.4, 128.95 (2), 128.90 (2), 127.9, 127.7, 127.6 (2), 127.24, 127.17, 126.6 (2), 126.5, 125.8, 50.5, 44.5, 42.3, 30.5, and 26.1; MS (ESI) calcd for (C35H35N2S)+ [M+H]+ 515.7, found 515.5.
1-(3-Bromo-4,5-bis(4-(tert-butyl)phenyl)thiophen-2-yl)-N-(piperidin-4-ylmethyl)methanamine hydrochloride (33) was prepared from 4,5-dibromothiophene-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.65 (br, 1H), 9.00 (br, 1H), 8.84 (br, 1H), 8.10 (br, 1H), 7.44 (d, J = 8.2 Hz, 2H), 7.31 (d, J = 8.2 Hz, 2H), 7.14 (d, J = 8.2 Hz, 2H), 7.11 (d, J = 8.2 Hz, 2H), 4.41 (s, 2H), 3.28-3.25 (m, 2H), 2.97 (br, 2H), 2.89-2.82 (m, 2H), 2.09 (br, 1H), 1.99-1.96 (m, 2H), 1.49-1.43 (m, 2H), 1.30 (s, 9H), and 1.22 (s, 9H); 13C NMR (100 MHz, DMSO-d6): 151.1, 150.4, 141.2, 137.0, 132.0, 129.95, 129.84, 128.0, 126.6, 125.6, 125.3, 118.0, 50.9, 44.7, 43.0, 42.3, 34.44, 34.40, 31.1, 30.9, and 26.0; MS (ESI) calcd for (C31H42BrN2S)+ [M+H]+ 554.7, found 554.6.
1-(4,5-bis(4-(tert-Butyl)phenyl)furan-2-yl)-N-(piperidin-4-ylmethyl)methanamine hydrochloride (34) was prepared from 4,5-dibromofuran-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.54 (br, 2H), 8.97 (br, 1H), 8.78 (br, 1H), 7.46-7.33 (m, 8H), 6.88 (s, 1H), 4.28 (s, 2H), 3.28-3.25 (m, 2H), 2.94 (br, 2H), 2.85-2.83 (m, 2H), 2.06 (br, 1H), 1.97-1.94 (br, 2H), 1.48-1.42 (m, 2H), 1.30 (s, 9H), and 1.27 (s, 9H); 13C NMR (100 MHz, DMSO-d6): 150.9, 150.0, 148.4, 144.8, 130.2, 127.8, 127.5, 125.8, 125.7, 125.5, 122.2, 116.2, 50.7, 42.8, 42.4, 34.49, 34.39, 31.15, 31.02, 30.5 and 26.0; MS (ESI) calcd for (C31H43N2O)+ [M+H]+ 459.7, found 459.6.
1-(4,5-bis(4-(Furan-3-yl)phenyl)furan-2-yl)-N-(piperidin-4-ylmethyl)methanamine hydrochloride (35) was prepared from 4,5-dibromofuran-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (yellow powder). 1H NMR (400 MHz, DMSO-d6): δ 9.57 (br, 2H), 8.96 (br, 1H), 8.78 (br, 1H), 8.24-8.21 (m, 1H), 7.78-7.74 (m, 2H), 7.70-7.58 (m, 5H), 7.55 (d, J = 8.0 Hz, 2H), 7.41 (d, J = 8.0 Hz, 2H), 6.99 (s, 1H), 6.96 (d, J = 8.0 Hz, 2H), 4.31 (s, 2H), 3.27-3.23 (m, 2H), 2.95 (br, 2H), 2.89-2.83 (m, 2H), 2.08 (br, 1H), 1.98-1.92 (m, 2H), and 1.49-1.38 (m, 2H); 13C NMR (100 MHz, DMSO-d6): 148.5, 145.2, 144.6, 139.9, 139.7, 131.6, 131.5, 128.9, 128.8, 128.7, 126.6, 126.1, 125.8, 125.4, 124.3, 122.6, 116.1, 114.7, 108.67, 108.61, 50.7, 42.9, 42.4, 30.6, and 26.1; MS (ESI) calcd for (C31H31N2O3)+ [M+H]+ 479.6, found 479.5.
4,5-bis(4-(tert-Butyl)phenyl)-N-(piperidin-4-ylmethyl)thiazole-2-carboxamide hydrochloride (36) was prepared from 4-bromothiozole-2-carboxaldehyde, following the general method A, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 8.96 (br, 1H), 8.85 (br, 1H), 8.53 (br, 1H), 7.48-7.42 (m, 4H), 7.38-7.33 (m, 4H), 3.26-3.20 (m, 4H), 2.82 (br, 2H), 1.87 (br, 1H), 1.82-1.76 (m, 2H), 1.42-1.36 (m, 2H), and 1.28 (s, 18H); 13C NMR (100 MHz, DMSO-d6): 161.0, 152.1, 151.3, 150.4, 137.8, 137.5, 131.7, 129.4, 128.9, 128.3, 126.4, 125.6, 44.2, 43.2, 34.9, 34.8, 34.1, 31.5, 31.4, and 26.6; MS (ESI) calcd for (C30H40N3OS)+ [M+H]+ 490.7, found 490.6.
1-(4,5-bis(4-(tert-Butyl)phenyl)thiazol-2-yl)-N-(piperidin-4-ylmethyl)methanamine hydrochloride (37) was prepared from 4-bromothiozole-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.83 (br, 2H), 9.13 (br, 1H), 8.98 (br, 1H), 7.44-7.31 (m, 8H), 4.57 (s, 2H), 3.27-3.24 (m, 2H), 3.03 (br, 2H), 2.85 (br, 2H), 2.10 (br, 1H), 1.98-1.95 (m, 2H), 1.55-1.50 (m, 2H), and 1.27 (s, 18H); 13C NMR (100 MHz, DMSO-d6): 151.4, 150.6, 148.5, 134.2, 131.7, 131.3, 129.0, 128.2, 128.0, 126.0, 125.2, 51.3, 46.8, 42.3, 34.52, 34.42, 31.07, 31.02, 30.6, and 28.1; MS (ESI) calcd for (C30H42N3S)+ [M+H]+ 476.7, found 476.5.
1-(4-(4-(tert-Butyl)phenyl)thiazol-2-yl)-N-(piperidin-4-ylmethyl)methanamine hydrochloride (38) was prepared from 4-bromothiozole-2-carboxaldehyde, following the general method B, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 9.77 (br, 2H), 9.01 (br, 2H), 8.01 (s, 1H), 7.86 (d, J = 8.4 Hz, 2H), 7.42 (d, J = 8.4 Hz, 2H), 4.55 (s, 2H), 3.27-3.24 (m, 2H), 3.03 (br, 2H), 2.85 (br, 2H), 2.10 (br, 1H), 1.98-1.95 (m, 2H), 1.47-1.39 (m, 2H), and 1.25 (s, 9H); 13C NMR (100 MHz, DMSO-d6): 160.0, 154.3, 150.9, 131.0, 125.9, 125.6, 116.0, 51.2, 46.8, 42.3, 34.4, 31.1, 30.6, and 26.0; MS (ESI) calcd for (C20H30N3S)+ [M+H]+ 344.5, found 344.6.
4-(4-(tert-Butyl)phenyl)-N-(piperidin-4-ylmethyl)thiazole-2-carboxamide hydrochloride (39) was prepared from 4-bromothiozole-2-carboxaldehyde, following the general method A, as a hydrochloric acid salt (white powder). 1H NMR (400 MHz, DMSO-d6): δ 8.99 (br, 1H), 8.90 (br, 1H), 8.68 (br, 1H), 8.34 (s, 1H), 7.98 (d, J = 8.2 Hz, 2H), 7.49 (d, J = 8.2 Hz, 2H), 3.27-3.19 (m, 4H), 2.88-2.79 (m, 2H), 1.91 (br, 1H), 1.83-1.79 (m, 2H), 1.67-1.63 (m, 2H), and 1.31 (s, 9H); 13C NMR (100 MHz, DMSO-d6): 163.5, 159.4, 153.9, 151.2, 130.8, 126.1, 125.5, 118.8, 43.8, 42.9, 34.4, 31.1, 28.1, and 26.3; MS (ESI) calcd for (C20H28N3OS)+ [M+H]+ 358.5, found 358.4.
N-((5,6-bis(4-(Furan-3-yl)phenyl)pyrazin-2-yl)methyl)piperidin-4-amine hydrochloride (40).
A mixture of 2,3-diaminopropanoic acid hydrochloride (600.4 mg, 4.26 mmol), 1,2-bis(4-bromophenyl)ethane-1,2-dione 52 (1.745 g, 4.26 mmol) and NaOH (677.2 mg, 16.93 mmol) in methanol (25 mL) were refluxed for 2 h. The mixture was cooled to 25℃ and air was bubbled through the solution for 2 days, after which pH was adjusted to 2 by HCl. Upon removal of the solvents, the product was extracted with diethyl ether (3 × 50 mL) and the combined organic phases were dried, filtered and evaporated under reduced pressure. The crude product 53 was dissolved in THF (22 mL) followed by addition of methyl chloroformate (0.22 mL, 2.8 mmol) and diisopropylethylamine (0.6 mL, 3.5 mmol). The reaction mixture was stirred at room temperature for 5h. Methanol (1.8 mL) was then added followed by NaBH4 (537.2 mg, 14.2 mmol) in small portions at 0℃. Stirring was continued at 0℃ for 1h. The product was extracted with ether and purified with flash chromatography (silica gel, hexane/ethyl acetate = 5/1) to give the corresponding alcohol (509 mg, 52% for two steps), which was converted to compound 40 as a pale yellow powder using the reactions vi, vii, ii and iv described in general methods A and B. 1H NMR (400 MHz, DMSO-d6): δ 8.94 (br, 2H), 8.72 (br, 1H), 8.69 (s, 1H), 8.24 (br, 2H), 7.92 (d, J = 7.1 Hz, 2H), 7.75 (s, 2H), 7.62 (d, J = 7.7 Hz, 4H), 7.45 (d, J = 4.5 Hz, 2H), 7.43 (d, J = 4.5 Hz, 2H), 6.99 (s, 2H), 5.26 (s, 2H), 3.21 (d, J = 10.2 Hz, 2H), 2.94 (br, 2H), 2.79-2.75 (m, 2H), 1.76 (d, J = 14.2 Hz, 2H), 1.69 (br, 1H), and 1.33-1.26 (m, 2H); 13C NMR (100 MHz, DMSO-d6): 156.0, 150.48, 150.44, 149.5, 144.5, 140.4, 140.02, 139.99, 136.63, 136.57, 132.3, 130.1, 130.0, 129.7, 128.0, 126.4, 125.3, 125.2, 108.58, 108.56, 53.4, 45.3, 42.7, 33.9, and 26.0; MS (ESI) calcd for (C31H31N4O2)+ [M+H]+ 491.6, found 491.5.
Inhibition of p300-HAT and other HATs.
The HAT domain (1195-1673) of human p300 was cloned, inserted into pGEX-KG vector and the DNA sequence was verified by sequencing. The p300-HAT expression plasmid was transformed into E. coli BL21-CodonPlus strain (Agilent) and cultured at 37 °C in LB medium containing ampicillin (50 μg/mL) and chloramphenicol (34 μg/mL). After the optical density of the bacterial culture reached ~0.9 at 600 nm, p300 HAT expression was induced by adding 300 μM isopropylthiogalactoside (IPTG) at 18 °C for 48 hours. Cells were collected, lysed, centrifuged for 20 min at 20,000 rpm. The supernatant was subjected to column chromatography with glutathione sepharose resins. The recombinant GST-p300-HAT fusion protein was obtained in ~90% purity (SDS-PAGE) by elution with 10 mM of glutathione solution, which was further purified using a Superdex 200 gel filtration column chromatography. CBP, PCAF and Myst3 HATs were obtained using similar methods.
To determine inhibitory activity, a compound with concentrations ranging from 100 nM to 10 μM was incubated with p300-HAT (10 nM) in 20 μL of 50 mM phosphate buffer (pH = 7.0) containing 0.01% Brij-35 for 10 min at 25 °C. Histone H3 peptide (ARTKQTARKSTGGKAPRKQLA) (20 μM) and Acetyl-CoA (1 μM 3H-Ac-CoA and 19 μM Ac-CoA) were added to initiate the reaction. After 30 min at 25 °C, the reaction was stopped by adding 6 N formic acid (5 μL). 20 μL of reaction mixture was then transferred to a small piece of P81 filter paper (Whatman) that binds histone H3 peptide. The filter paper was washed 3x with 50 mM NaHCO3, dried, and transferred into a scintillation vial containing 2 mL of scintillation cocktail. Radioactivity on the filter paper was measured with a Beckman LS-6500 scintillation counter. IC50 values were obtained by using a standard sigmoidal dose response curve fitting in Prism (version 5.0, GraphPad Software, Inc., La Jolla, CA). IC50 values were the mean values from at least three experiments.
Alpha assay.
We followed a published method16 to investigate whether compound 2 can disrupt the binding of p300 HAT and histone H4 peptide. In brief, the assay was performed in a 384-well plate using His6-tagged P300 HAT (125 nM) coated nickel chelate acceptor beads (5 μL, Perkin Elmer), biotinylated H4 peptide [SGRGKGGKGLGKGGAKRHRKVLRGG-K(Biotin)-NH2] (30 nM) coated streptavidin donor beads (5 μL, Perkin Elmer), and varying concentrations of 12 in a PBS buffer with 0.5 % BSA (final volume of 25 μL). Upon incubation for 1h, Alpha signal was determined (laser excitations at 680 nm and reading at 570 nm) using a Tecan Spark microplate reader. The IC50 values were determined using the sigmoidal dose-response fitting in the program of Prism 5.0 (GraphPad). The biotinylated His6 peptide (Perkin Elmer # 6760302) was used as a reference to eliminate the possible interference of compounds on the signal generated by singlet oxygen transfer.
Molecular modeling.
Docking studies were performed with Schrödinger small-molecule drug discovery software suite (Schrödinger, LLC, New York, NY, 2017), using our previous published methods 23, 35. The crystal structure of p300-HAT in complex with Ac-CoA (PDB: 4PZS) was prepared using the module “protein preparation wizard” in Maestro with the default protein parameters. Hydrogen atoms were added and water molecules were extracted. Ac-CoA was included in the protein structure for docking. Hydrogen bonds were optimized, the partial charges were assigned, and the protein structure was energy-minimized using OPLS-2005 force field. A receptor grid was generated using the program Glide without any constraints. Compound 12 was constructed, energy-minimized using OPLS-2005 force field in Maestro and then docked into the prepared protein structure using Glide (docking parameters: standard-precision and dock flexibly).
Western blot.
106 Kasumi-1 cells/well were incubated with compound 12 at 0, 5 and 10 μM for 12 hours. Histone proteins were extracted using EpiQuik total histone extraction kit (Epigentek), according to the manufacturer’s protocol. Equal amounts of histones (2 μg) were separated on SDS-PAGE and transferred to PVDF membranes. The blots were probed with primary antibodies against H3K9Ac, H3K18Ac, H3K27Ac and H3 (Cell Signaling), followed by anti-rabbit IgG (Thermo Scientific) secondary antibodies.
Cell growth inhibition.
The anti-proliferation assays for non-breast cancer cells were performed using our previous method 36-38. In brief, for Kasumi-1 cells, 106 cells per well were added into 96-well plates and cultured with increasing concentrations of a compound in RPMI-1640 medium supplemented with 20% fetal bovine serum and penicillin (100 U/mL) and streptomycin (100 μg/mL) at 37 °C in a 5% CO2 atmosphere with 100% humidity. For solid tumor cells, 105 cells per well were added into 96-well plates and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum and penicillin (100 U/mL) and streptomycin (100 μg/mL) overnight. Upon addition of increasing concentrations of a compound, cells were incubated for 5 days. Cell viability was assessed by using an XTT assay kit (Roche) for leukemia cells or an MTT assay (Sigma) for attachment cells. For the breast cancer MCF-7 cells, culture and inhibition assays were performed according to our previous methods 28, 29. Compound EC50 values were calculated from dose response curves using Prism 5.0.
RNA extraction.
Upon starvation (culturing in phenol red-free DMEM medium with 10% charcoal treated FBS to remove hydrophobic hormones such as estrogens) for 7 days, MCF-7 cells were treated with the p300 inhibitor (6 μM) or DMSO control, in the presence or absence of (10 nM) estradiol (E2) for 48 hours. Total RNA was extracted using RNeasy Plus Mini Kit (Qiagen), according to the manufacturer’s instruction.
Library Preparation.
The RNA integrity for each sample was assessed with a RNA 6000 Nano chip on a 2100 Bioanalyzer (Agilent; Santa Clara, CA). The average RNA integrity score for the sample set was 6.48. Sequencing libraries were prepared using the TruSeq Stranded Total RNA Library Prep Kit (Illumina, Inc; San Diego, CA). Concisely, ribosomal RNA (rRNA) was depleted from total RNA where the remaining (ribo-depleted RNA) was purified, fragmented for 7.5 minutes, and primed for cDNA synthesis. Blunt-ended cDNA was generated after first and second strand synthesis. Adenylation of the 3’ blunt-ends was followed by adapter ligation prior to the enrichment of the cDNA fragments. Final library quality control was carried out by measuring the fragment size on a DNA1000 chip on a 2100 BioAnalyzer (Agilent; Santa Clara, CA). The average library yielded an insert bp size of 258. The concentration of each library was determined by quantitative PCR (qPCR) by the KAPA Library Quantification Kit for Next Generation Sequencing (KAPA Biosystems; Woburn, MA).
Sequencing.
Libraries were normalized to 50 nmol/L in 10 mM Tris-Cl, pH8.5 with 0.1% Tween 20 then pooled evenly. The 50 nmol/L library pool was diluted to 2 nmol/L with 10 mM Tris-Cl, pH8.5 with 0.1% Tween 20. The pooled libraries were denatured with 0.05N NaOH and diluted to 20 pmol/L. Cluster generation of the denatured libraries was performed according to the manufacture’s specifications (Illumina, Inc; San Diego, CA) utilizing the HiSeq Rapid PE Cluster Kit v2 chemistry and flow cell. Libraries were clustered appropriately with a 1% PhiX spike-in. Sequencing-by-synthesis (SBS) was performed on a HiSeq 2500 utilizing v2 chemistry with paired-end 101 bp reads and a 6 bp index read culminating in an average output of 20 million paired-end reads (or 40 million total reads) per sample. Sequence read data were processed and converted to FASTQ format by Illumina BaseSpace analysis software (v2.0.13).
Bioinformatics Analysis.
RNAseq data files were trimmed using Trim Galore! 39, 40. Then the data was mapped using Hisat2 onto the human reference genome build UCSC hg38/GRCh38and sorted using samtools 41, 42. Gene expression was assessed and fragments per kilobase of transcript per million (FPKM) values were determined using Stringtie 41. Gene expression profiles were normalized using the quantile normalization method. Significantly changed genes were determined using a two-sided parametric t-test, with significance assessed at p<0.05, only genes for which at least one sample exceeded 1 FPKM in expression were considered. Enriched pathways were inferred using the Gene Set Enrichment (GSEA) method, and the Molecular Signature Database (MSigDB) pathway collection.
Supplementary Material
Acknowledgment.
This work was supported by the Cancer Prevention and Research Institute of Texas (CPRIT) grants RP150129 and RP180177 to Y.S. It was also partly supported in part by the Breast Cancer Research Foundation grants 16-142; 17-143 ,18-145 (to R.S.), Susan G. Komen for the Cure Foundation Promise Grants (PG12221410 to R.S.), Department of Defense (W81XWH-14-1-0326 to X.F.), and the NCI SPORE P50CA58183 and P50CA186784 grants (to R.S. and N.M.). The gene expression profiling was supported by the CPRIT grant RP170005, the Prostate Cancer Foundation Grant 18YOUN09 to (S.K.) and NCI SPORE P50 CA186784 (N.M.). The authors also acknowledge the assistance of the Dan L Duncan Cancer Center Core resources (supported by the NCI Cancer Center Support Grant P30CA125123).
ABBREVATIONS:
- Ac-CoA
acetyl coenzyme A
- AR
androgen receptor
- CBP
CREB (cAMP-response element binding protein) binding protein
- CH1
cysteine-histidine rich 1
- CoA
coenzyme A
- ER
estrogen receptor
- GNAT
Gcn5-related N-acetyltransferase
- GSEA
Gene set enrichment analysis
- H3K4
Histone H3 lysine 4
- H3K9
Histone H3 lysine 9
- H3K27
Histone H3 lysine 27
- LSD1
lysine specific demethylase 1
- MYST
MOZ, Ybf2, Sas2 and Tip60p300
- NES
normalized enrichment score
- E1A
binding protein p300
- PCAF
p300/CBP associating factor
- SAR
structure activity relationship
- SID
steroid receptor coactivator interaction
- SRC
steroid receptor coactivator
Footnotes
Supporting Information Available. Chart S1-S3, Figure S1-S4, representative HPLC traces and 1H and 13C NMR spectra for compound 12 and PDB file for docking models. This material and Molecular Formula Strings for all compounds are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jones PA; Baylin SB The epigenomics of cancer. Cell 2007, 128, 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kouzarides T Chromatin modifications and their function. Cell 2007, 128, 693–705. [DOI] [PubMed] [Google Scholar]
- 3.Belkina AC; Denis GV BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer 2012, 12, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Z; Yik JH; Chen R; He N; Jang MK; Ozato K; Zhou Q Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 2005, 19, 535–545. [DOI] [PubMed] [Google Scholar]
- 5.Schiltz RL; Mizzen CA; Vassilev A; Cook RG; Allis CD; Nakatani Y Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J Biol Chem 1999, 274, 1189–1192. [DOI] [PubMed] [Google Scholar]
- 6.Dancy BM; Cole PA Protein lysine acetylation by p300/CBP. Chem Rev 2015, 115, 2419–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang F; Marshall CB; Ikura M Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition. Cell Mol Life Sci 2013, 70, 3989–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson AB; O’Malley BW Steroid receptor coactivators 1, 2, and 3: critical regulators of nuclear receptor activity and steroid receptor modulator (SRM)-based cancer therapy. Mol Cell Endocrinol 2012, 348, 430–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dilworth FJ; Chambon P Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene 2001, 20, 3047–3054. [DOI] [PubMed] [Google Scholar]
- 10.Kraus WL; Manning ET; Kadonaga JT Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol Cell Biol 1999, 19, 8123–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu W; Shi XL; Roeder RG Synergistic activation of transcription by CBP and p53. Nature 1997, 387, 819–823. [DOI] [PubMed] [Google Scholar]
- 12.Vervoorts J; Luscher-Firzlaff JM; Rottmann S; Lilischkis R; Walsemann G; Dohmann K; Austen M; Luscher B Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep 2003, 4, 484–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma H; Hong H; Huang SM; Irvine RA; Webb P; Kushner PJ; Coetzee GA; Stallcup MR Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol Cell Biol 1999, 19, 6164–6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavau C; Du C; Thirman M; Zeleznik-Le N Chromatin-related properties of CBP fused to MLL generate a myelodysplastic-like syndrome that evolves into myeloid leukemia. EMBO J 2000, 19, 4655–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gervais C; Murati A; Helias C; Struski S; Eischen A; Lippert E; Tigaud I; Penther D; Bastard C; Mugneret F; Poppe B; Speleman F; Talmant P; VanDen Akker J; Baranger L; Barin C; Luquet I; Nadal N; Nguyen-Khac F; Maarek O; Herens C; Sainty D; Flandrin G; Birnbaum D; Mozziconacci MJ; Lessard M Acute myeloid leukaemia with 8p11 (MYST3) rearrangement: an integrated cytologic, cytogenetic and molecular study by the groupe francophone de cytogenetique hematologique. Leukemia 2008, 22, 1567–1575. [DOI] [PubMed] [Google Scholar]
- 16.Lasko LM; Jakob CG; Edalji RP; Qiu W; Montgomery D; Digiammarino EL; Hansen TM; Risi RM; Frey R; Manaves V; Shaw B; Algire M; Hessler P; Lam LT; Uziel T; Faivre E; Ferguson D; Buchanan FG; Martin RL; Torrent M; Chiang GG; Karukurichi K; Langston JW; Weinert BT; Choudhary C; de Vries P; Van Drie JH; McElligott D; Kesicki E; Marmorstein R; Sun C; Cole PA; Rosenberg SH; Michaelides MR; Lai A; Bromberg KD Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 2017, 550, 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau OD; Kundu TK; Soccio RE; Ait-Si-Ali S; Khalil EM; Vassilev A; Wolffe AP; Nakatani Y; Roeder RG; Cole PA HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol Cell 2000, 5, 589–595. [DOI] [PubMed] [Google Scholar]
- 18.Kwie FH; Briet M; Soupaya D; Hoffmann P; Maturano M; Rodriguez F; Blonski C; Lherbet C; Baudoin-Dehoux C New potent bisubstrate inhibitors of histone acetyltransferase p300: design, synthesis and biological evaluation. Chem Biol Drug Des 2011, 77, 86–92. [DOI] [PubMed] [Google Scholar]
- 19.Bowers EM; Yan G; Mukherjee C; Orry A; Wang L; Holbert MA; Crump NT; Hazzalin CA; Liszczak G; Yuan H; Larocca C; Saldanha SA; Abagyan R; Sun Y; Meyers DJ; Marmorstein R; Mahadevan LC; Alani RM; Cole PA Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem Biol 2010, 17, 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H; Pinello CE; Luo J; Li D; Wang Y; Zhao LY; Jahn SC; Saldanha SA; Chase P; Planck J; Geary KR; Ma H; Law BK; Roush WR; Hodder P; Liao D Small-molecule inhibitors of acetyltransferase p300 identified by high-throughput screening are potent anticancer agents. Mol Cancer Ther 2013, 12, 610–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costi R; Di Santo R; Artico M; Miele G; Valentini P; Novellino E; Cereseto A Cinnamoyl compounds as simple molecules that inhibit p300 histone acetyltransferase. J Med Chem 2007, 50, 1973–1977. [DOI] [PubMed] [Google Scholar]
- 22.Baell J; Walters MA Chemistry: Chemical con artists foil drug discovery. Nature 2014, 513, 481–483. [DOI] [PubMed] [Google Scholar]
- 23.Wu F; Zhou C; Yao Y; Wei L; Feng Z; Deng L; Song Y 3-(Piperidin-4-ylmethoxy)pyridine containing cmpounds are potent inhibitors of lysine specific demethylase 1. J Med Chem 2016, 59, 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pereira R; Furst A; Iglesias B; Germain P; Gronemeyer H; de Lera AR Insights into the mechanism of the site-selective sequential palladium-catalyzed cross-coupling reactions of dibromothiophenes/dibromothiazoles and arylboronic acids. Synthesis of PPARbeta/delta agonists. Org Biomol Chem 2006, 4, 4514–4525. [DOI] [PubMed] [Google Scholar]
- 25.Liu X; Wang L; Zhao K; Thompson PR; Hwang Y; Marmorstein R; Cole PA The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature 2008, 451, 846–850. [DOI] [PubMed] [Google Scholar]
- 26.Maksimoska J; Segura-Pena D; Cole PA; Marmorstein R Structure of the p300 histone acetyltransferase bound to acetyl-coenzyme A and its analogues. Biochemistry 2014, 53, 3415–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demarest SJ; Martinez-Yamout M; Chung J; Chen H; Xu W; Dyson HJ; Evans RM; Wright PE Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature 2002, 415, 549–553. [DOI] [PubMed] [Google Scholar]
- 28.Fu X; Jeselsohn R; Pereira R; Hollingsworth EF; Creighton CJ; Li F; Shea M; Nardone A; De Angelis C; Heiser LM; Anur P; Wang N; Grasso CS; Spellman PT; Griffith OL; Tsimelzon A; Gutierrez C; Huang S; Edwards DP; Trivedi MV; Rimawi MF; Lopez-Terrada D; Hilsenbeck SG; Gray JW; Brown M; Osborne CK; Schiff R FOXA1 overexpression mediates endocrine resistance by altering the ER transcriptome and IL-8 expression in ER-positive breast cancer. Proc Natl Acad Sci U S A 2016, 113, E6600–E6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison G; Fu X; Shea M; Nanda S; Giuliano M; Wang T; Klinowska T; Osborne CK; Rimawi MF; Schiff R Therapeutic potential of the dual EGFR/HER2 inhibitor AZD8931 in circumventing endocrine resistance. Breast Cancer Res Treat 2014, 144, 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ono H; Basson MD; Ito H P300 inhibition enhances gemcitabine-induced apoptosis of pancreatic cancer. Oncotarget 2016, 7, 51301–51310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L; Gural A; Sun XJ; Zhao X; Perna F; Huang G; Hatlen MA; Vu L; Liu F; Xu H; Asai T; Deblasio T; Menendez S; Voza F; Jiang Y; Cole PA; Zhang J; Melnick A; Roeder RG; Nimer SD The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science 2011, 333, 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A; Tamayo P; Mootha VK; Mukherjee S; Ebert BL; Gillette MA; Paulovich A; Pomeroy SL; Golub TR; Lander ES; Mesirov JP Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences 2005, 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ianculescu I; Wu DY; Siegmund KD; Stallcup MR Selective roles for cAMP response element-binding protein binding protein and p300 protein as coregulators for androgen-regulated gene expression in advanced prostate cancer cells. J Biol Chem 2012, 287, 4000–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin Q; Yu LR; Wang L; Zhang Z; Kasper LH; Lee JE; Wang C; Brindle PK; Dent SY; Ge K Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J 2011, 30, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng B; Yao Y; Liu Z; Deng L; Anglin JL; Jiang H; Prasad BV; Song Y Crystallographic investigation and selective inhibition of mutant isocitrate dehydrogenase. ACS Med Chem Lett 2013, 4, 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu F; Jiang H; Zheng B; Kogiso M; Yao Y; Zhou C; Li XN; Song Y Inhibition of cancer-associated mutant isocitrate dehydrogenases by 2-thiohydantoin compounds. J Med Chem 2015, 58, 6899–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Z; Yao Y; Zhou C; Chen F; Wu F; Wei L; Liu W; Dong S; Redell M; Mo Q; Song Y Pharmacological inhibition of LSD1 for the treatment of MLL-rearranged leukemia. J Hematol Oncol 2016, 9:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L; Deng L; Chen F; Yao Y; Wu B; Wei L; Mo Q; Song Y Inhibition of histone H3K79 methylation selectively inhibits proliferation, self-renewal and metastatic potential of breast cancer. Oncotarget 2014, 5, 10665–10677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011, 17, 3. [Google Scholar]
- 40.Andrews S FastQC A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. (accessed March 27, 2018). [Google Scholar]
- 41.Pertea M; Kim D; Pertea GM; Leek JT; Salzberg SL Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 2016, 11, 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H; Handsaker B; Wysoker A; Fennell T; Ruan J; Homer N; Marth G; Abecasis G; Durbin R The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.