Abstract
Among mammals, placental invasion is correlated with vulnerability to malignancy. Animals with more invasive placentation (e.g. humans) are more vulnerable to malignancy. To explain this correlation, we propose the hypothesis of Evolved Levels of Invasibility: the evolution of invasibility of stromal tissue affects both, placental and cancer invasion. We provide evidence for this hypothesis using an in vitro model. We find that bovine endometrial and skin fibroblasts are more resistant to invasion than their human counterparts. Gene expression profiling identified genes with high expression in human but not in bovine fibroblasts. Knocking down a subset of them in human fibroblasts leads to stronger resistance to cancer cell invasion. Identifying the evolutionary determinants of stromal invasibility can provide significant insights to develop rational anti-metastatic therapeutics.
Placental invasion into the maternal endometrium of the uterus exhibits substantial similarities to early cancer dissemination into stroma1–4. These similarities have inspired the hypothesis of antagonistic pleiotropy5,6. According to this hypothesis, trophoblasts evolved the capacity to invade into the endometrium leading to invasive placentation. These mechanisms can become reactivated in cancer cells, leading to a predisposition to metastasis. This hypothesis implies that cancer malignancy should be limited to placental mammals, where invasive placentation first evolved. This prediction, however, is inconsistent with the fact that opossums, with ancestrally non-invasive placenta7,8, get invasive skin cancers 9. Here we explore an alternative scenario in which stromal cells of the uterus evolved to either resist or permit invasion, determining the outcome of placental invasiveness9.
The likelihood that the evolution of the stromal environment is driving the evolution of cancer malignancy is enhanced by the fact that the molecular mechanisms employed by cancer cells to metastasize are shared with other biological processes. For instance, the mechanisms regulating gastrulation, wound healing, extravasation by leukocytes etc., are shared with both trophoblast and cancer invasion6,10,11. This implies that invading cancer cells employ mechanisms that evolved much earlier than placental invasion, and therefore, the evolution of invasive placentation per se cannot be responsible for the origin of malignant cancer. It is important to note, however, that the invasiveness of the placenta continued to evolve after its origin. Placental invasion reverted to a non-invasive phenotype in several lineages of placental mammals, as well as evolved into an even higher degree of invasiveness in the great apes, which includes humans12–14. A complete loss of placental invasion evolved in the hoofed mammals, such as cows and horses and their relatives, and it is exactly in those animals that have lower malignancy rates for a variety of cancers9. In contrast, humans with very invasive placentas are highly vulnerable to melanoma malignancy.
Based on the arguments outlined above, we advance the hypothesis that evolutionary changes in the permissiveness, or resistance of uterine stromal cells to placental invasion are mechanistically linked to the vulnerability to cancer malignancy9. We term this hypothesis Evolved Levels of Invasibility (ELI) (Figure 1A), and experimentally test the ELI hypothesis using an in vitro model (Figure 1B). We demonstrate that human fibroblasts are more permissive to invasion by trophoblasts as well as cancer cells compared to their bovine counterparts. We then identify factors responsible for the resistance to invasion paving a way for therapeutic interventions. This study highlights how investigating evolutionarily processes may lead to the identification of therapeutic targets, pointing to the clinical potential of evolutionary analysis.
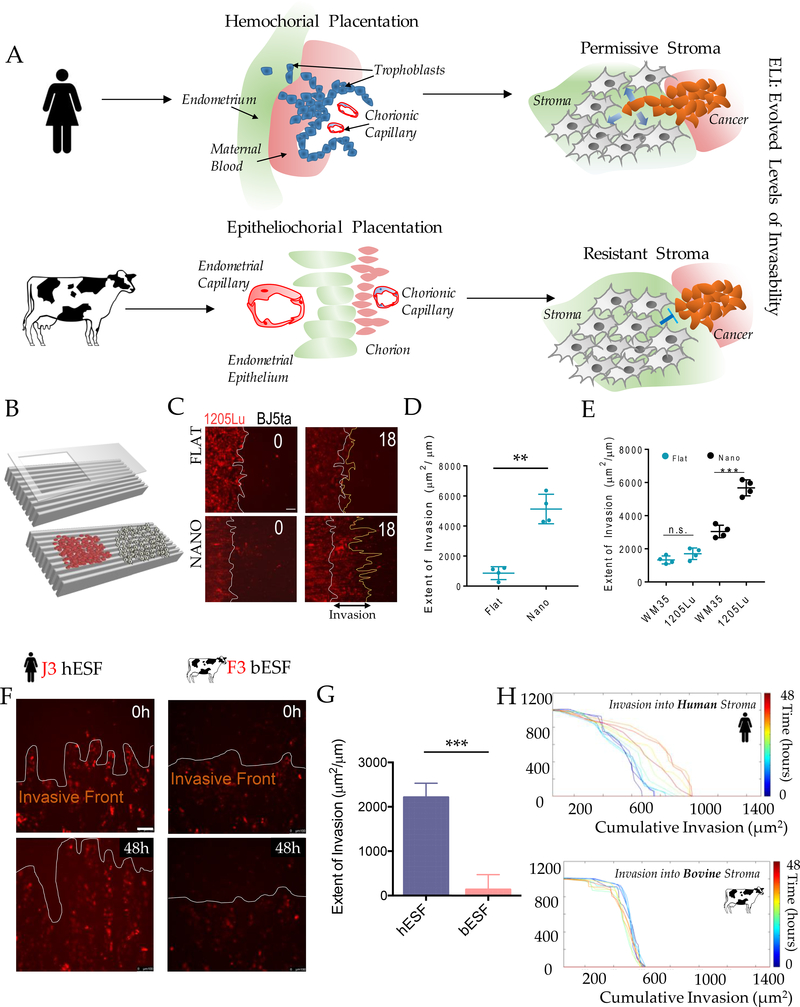
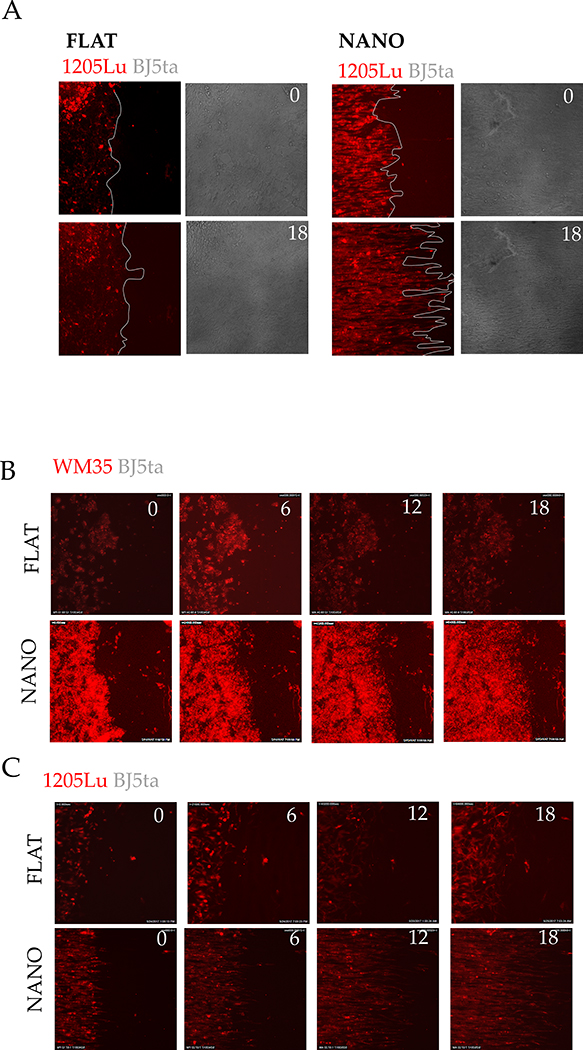
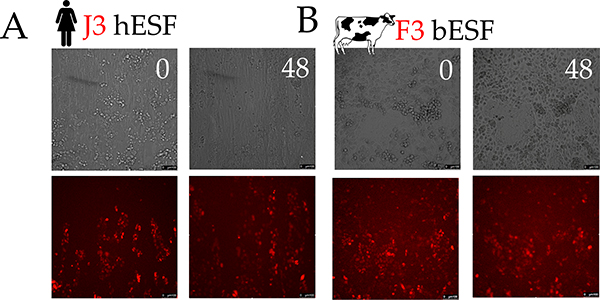
Figure 1. An experimental platform to test the hypothesis of Evolved Levels of Invasibility (ELI).
(A) Schematic describing the ELI hypothesis. Placentation in humans is hemochorial, wherein the placental trophoblasts invade into the maternal stroma reaching the blood supply. In contrast, in cows and other boroeutherians, placentation has recently evolved to be epitheliochorial, where the trophoblast epithelium attaches to the endometrial epithelium, but does not invade the maternal interstitium. The ELI paradigm states that bovine stroma has evolved to resist invasion compared to human stroma, and therefore secondarily limits cancer metastasis. (B) Schematic showing a cell patterning nanotextured platform to quantitatively and sensitively measure collective invasion into stroma; Stromal cells and invasive cells are patterned by a PDMS stencil into juxtaposed monolayers heterotypically interacting with each other, and imaged using live cell microscopy to observe collective cell invasion into the stroma. (C) Time course images showing invasion of 1205Lu malignant melanoma cells (red) into BJ5ta human skin fibroblasts (unlabeled) for 18 hours on a flat substrate vs a nanotextured substrate; Quantification of the extent of invasion per unit length of heterotypic intercellular interaction shown in (D). (E) Quantification of the extent of invasion of non-malignant WM35 and malignant 1205Lu melanoma into a monolayer of BJ5ta human skin fibroblasts on flat and nanotextured substrata. (F) Time course images showing invasion of human choriocarcinoma derived trophoblasts, J3 (red) into human endometrial stromal fibroblasts (unlabeled); and bovine trophoblasts, F3 (red) into bovine stromal fibroblasts (unlabeled) for 48 hours; See Figure S2 for phase-contrast and Supplementary Movies for dynamics of invasion; Quantification of the extent of invasion of trophoblasts into the respective stromal monolayer shown in (G). (H) Time course dynamic analysis showing cumulative invasion of J3 and F3 into respective species-specific endometrial stromal monolayers. In D, E, and G, n = 4 independent biological replicates; Statistical comparisons made using Student’s t-tests **: p < 0.01, ***: p < 0.001; Error bars denote standard error of the mean (s.e.m.).
Results
Collective cell invasion behavior can be modelled in an ECM-mimetic co-culture system
To test the ELI hypothesis, we developed a method for measuring collective cell invasion (CCI) into a lawn of stromal cells. The CCI into stroma is a complex process15,16,17. During infiltration and spread, invading cells interact with stroma, which modulates their invasive capacity, while experiencing contact guidance from the underlying Extracellular Matrix (ECM) fibers 1,18,19. Based on these considerations, we sought to mimic the stromal invasion process through a patterned co-culture of stromal and invasive cells on structured adhesion substrata. This approach was validated in previous cancer-stroma interaction studies20,21. The extent of invasion was measured as the average distance over which invasive cells have penetrated the stromal monolayer. The platform also enabled the sensitive measurement of cell invasion (Figure 1C).
We first compared the spread of malignant (1205Lu) and non-malignant (WM35) melanoma cells into a monolayer of human skin fibroblasts (BJ5ta) on substrata reproducing the nano-topographic features of aligned ECM fibers, or on flat surfaces. Aligned matrix fibers are a common feature in many tissues22. We found that mimicking aligned ECM matrix microenvironment allowed malignant 1205Lu melanoma cells to more extensively invade into skin fibroblast monolayer than non-malignant WM35 cells (Figure 1D–E, S1A). Furthermore, when 1205Lu and WM35 cells were compared on a flat surface, no statistically significant difference could be detected. In contrast, a significant difference between 1205Lu and WM35 was detectable in the ECM-mimetic platform (Figure 1E, S 1B, C), indicating that this assay has higher sensitivity in measuring stroma invasion compared to experiments on a flat surface.
Bovine endometrial stroma is resistant to trophoblast invasion
To explore the causes of species differences in placental invasion, we measured the invasion of a human choriocarcinoma (J3) cell line into human endometrial stromal fibroblasts (hESFs), and compared them to bovine trophoblasts (F3) invading into bovine endometrial stromal fibroblasts (bESFs). At 48 hours J3 cells invaded deeply into hESF layer, while the F3 invasion into bESFs was much more limited (Figure 1F–G, Supplementary Movies). Upon closer inspection we found that human J3 cells formed invasive forks, which propelled invasion into hESFs, while the bovine F3 showed no invasive forks (Figure 1F, G, H). These results recapitulate the in vivo species differences during embryo implantation 23–26.
In human placenta, extravillous trophoblasts (EVTs) constitute the most invasive cell type27,28. We therefore measured the invasion of human chorioblastoma cell line BeWo, the EVT cell line HTR8 and the bovine F3 cells invading into the layer of human or bovine stromal fibroblasts in all combinations (Figure 2A). BeWo cells, though being a carcinoma cell line that have been extensively used as model of human cyto-trophoblast cells29. The results showed that while hESF were invaded by all trophoblast cell lines, both human and bovine, bESF were more resistant to invasion (Figure 2B). The largest differences were found in the case of F3 invasion; with F3 invading rapidly into hESF monolayers, but are nearly completely halted by bESFs. This cross-species invasion experiments confirmed that the limited invasion in bESF is, to a large extent, a property of the stroma cells rather than the trophoblast cells (Figure 2B). In summary, the degree of trophoblast invasion is mostly controlled by the identity of the stromal cells rather than the invasive capacity of trophoblast cells, as assumed in the ELI hypothesis.
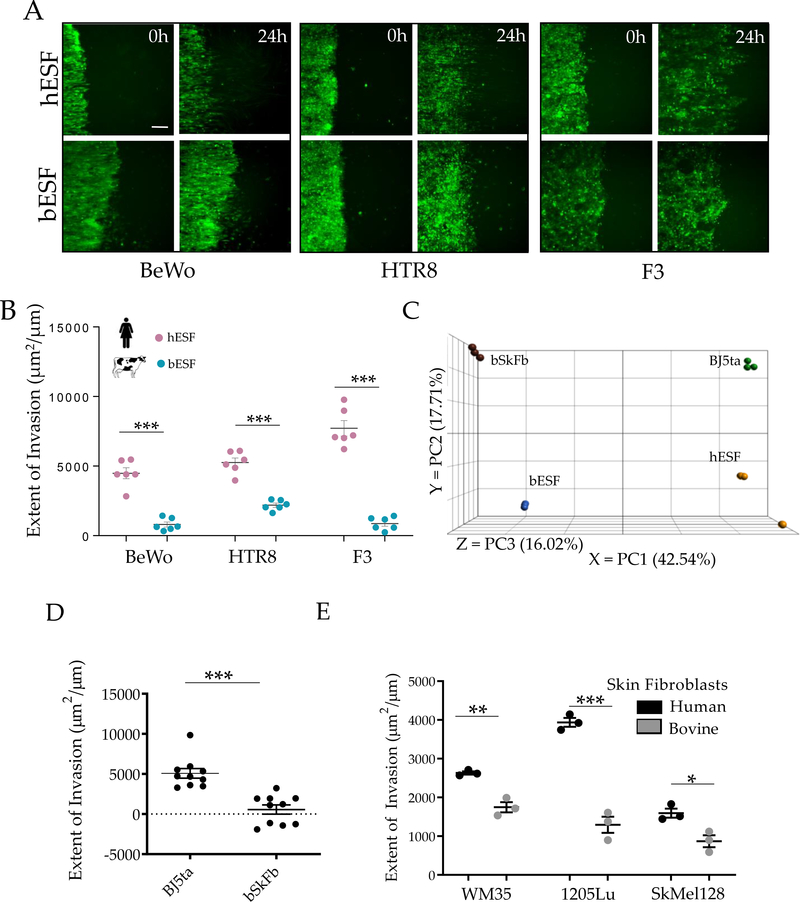
Figure 2. Bovine stroma resists trophoblast and melanoma invasion.
(A) Representative time course images of human trophoblasts BeWo, extravillous trophoblasts HTR8 (green) and bovine trophoblasts F3 (green) invading into endometrial stromal fibroblasts of the respective species for 18 hours; White traces show the boundary of the invasive fronts; Quantification of the extent of invasion for either trophoblasts shown in (B). (C) Principal Component Analysis (PCA) of the gene expression data from skin fibroblasts from human (BJ5ta) and bovine (bSkFb), and endometrial stromal fibroblasts from human (hESF) and bovine (bESF); 3 biological replicates were used for RNA Sequencing. (D) Extent of invasion of malignant A375 cells into BJ5ta and bSkFb monolayers measured over 24 hours. (E) Extent of invasion of other well characterized human melanoma cell lines into BJ5ta and bSkFb monolayers after 18 hours of observation. In B, C, E, n = 3, and in D, n = 8 independent biological replicates; Statistical comparisons made using Student’s t-tests *: p < 0.05, **: p < 0.01, ***: p < 0.001; Error bars denote standard error of the mean (s.e.m.).
Bovine skin fibroblasts resist melanoma invasion
The findings above leave open the possibility that the observed effects could be specific to the fetal-maternal interface rather than reflecting species differences in stromal invasibility in general. In humans, melanoma invasion into the surrounding stroma is a strong predictor of malignancy30. Would stromal compartments in other tissues than the uterus, e.g., the skin, show similar species-dependent properties as the uterine stromal cells? To address this question, we investigated whether gene expression profiles of human and bovine skin fibroblasts are more similar to those of their corresponding endometrial fibroblasts in the same species than they are to their homologous cell type in the other species.
The null hypothesis of independent transcriptome evolution of cell types predicts that corresponding (homologous) cell types should be more similar to each other than they are to a different cell type in the same species31. This is because, for instance, the human and bovine lineages diverged more recently than skin and endometrial fibroblasts have differentiated in evolution. This is necessarily true, because otherwise corresponding cell types would not be homologous32. On the other hand, if gene expression profiles in skin and endometrial fibroblasts co-evolved, the cells from the same species could be more similar than to the corresponding/homologous cell in the other species. The ELI hypothesis predicts that, in each species, endometrial fibroblasts and skin fibroblasts share the same level of invasion resistance because their gene expression profiles co-evolved, such that selection for higher or lower endometrial invasiveness has a parallel effect on the invasibility of skin fibroblasts. Therefore, we expect that the stromal cell types from the same species are more similar to each other than they are to corresponding cell types in the other species33.
To evaluate this expectation, we performed RNA sequencing on bovine and human skin fibroblasts, and compared them to human and bovine endometrial fibroblasts. Principal component analysis showed that the gene expression distance between skin fibroblasts and ESFs of the same species was less than the distance between corresponding cell types of different species (Figure 2C). The observed pattern is consistent with the hypothesis that gene expression in endometrial and skin fibroblasts indeed co-evolved due to pleiotropic effects of mutations on transcriptional regulation 31,34. In other words, the biology of skin and endometrial fibroblasts can be expected to be similar within species even though they differ between species.
We then measured invasion of malignant human melanoma cells (A375) into the skin fibroblasts from human (BJ5ta), and bovine (bSkFb), and found that bSkFb indeed resisted A375 invasion more strongly than human skin fibroblasts (Figure 2D). This greater invasion resistance of bSkFb was supported by a similarly high resistance of these cells to invasion by WM35, 1205Lu and SKMel28 melanoma cell lines (Figure 2E). A similar assay with bovine melanoma cells was precluded owing to lack of bovine melanoma cell lines35. Overall, these results indicate that bovine skin fibroblasts can resist cell invasion better than their human counterparts.
Human and bovine fibroblasts respond differently to trophoblast co-culture
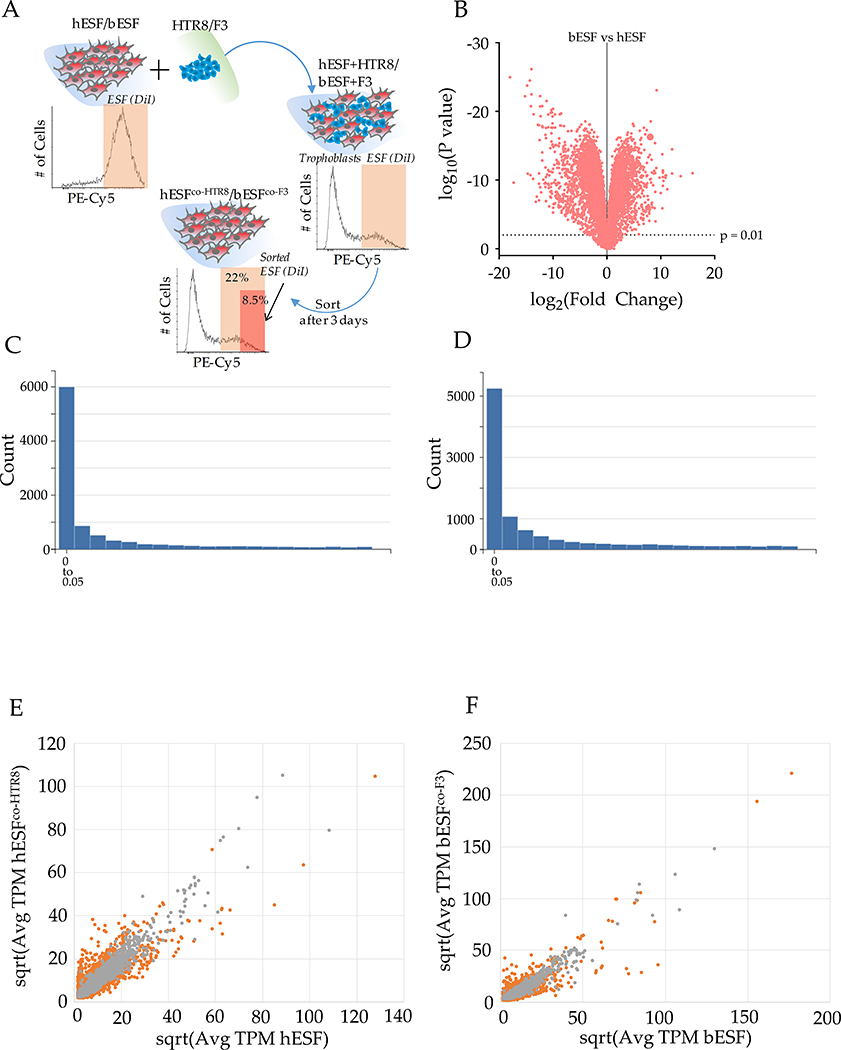
To understand the genetic underpinnings of ELI in bovine stromal cells, we collected RNA sequencing data from human and bovine endometrial fibroblasts, with and without co-culture with the respective trophoblasts. We focused on differentially expressed genes in stromal cells, both basally and upon interaction with species-specific trophoblasts. Endometrial fibroblasts of both species were labeled with the DiI dye, and co-cultured with equal number of unlabeled trophoblast cells, HTR8 and F3 for human and bovine cells respectively (Figure S3A). Co-cultures were maintained for 72 hours, and the cells were sorted using fluorescence activated cell sorting (FACS)20. Henceforth, the bovine cells will be referred to as bESFs and bESFco-F3 and human ESFs, hESFs and hESFco-HTR8.
A large number of genes were differentially expressed in bovine vs. human endometrial fibroblasts (Figure S3B), and ESFs responded strongly to co-culture with their cognate trophoblasts cells (Figure S3C–D). Furthermore, human and bovine ESFs responded differently to trophoblast co-culture (Figures 3A–B, Figures S3E–F). Human and bovine endometrial stromal cells differed substantially with respect to the number of genes affected by co-culture, with human cells displaying a larger number of differentially expressed genes (5,349 genes changed at p≤0.01, FDR=3.1×10−3) compared to bovine cells (3,101 genes at p≤0.01, FDR=7.01×10−3). Gene set analysis revealed genes belonging to chemotactic activity, cell motility, and metastasis ontologies at higher relative abundance in hESFco-H8 compared to bESFco-F3 (Figure 3B). These results suggest that species differences in invasibility may be caused by differential gene expression among the stromal cells in response to trophoblast cells.
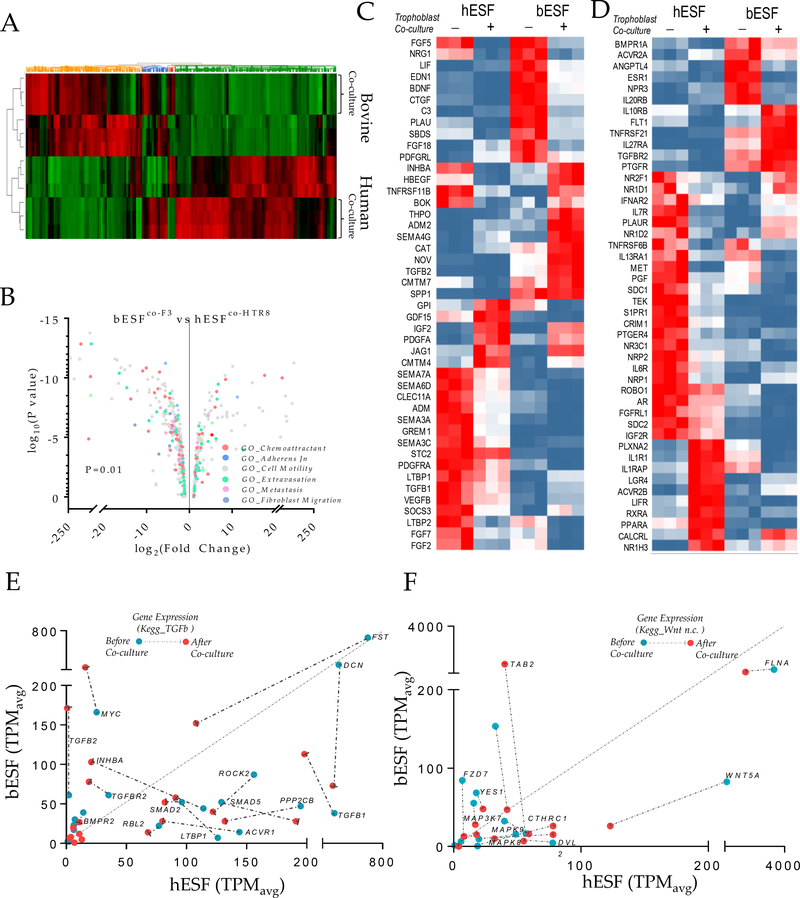
Figure 3. Transcriptomic analysis of bovine and human endometrial stromal fibroblasts reveals differential response to trophoblasts.
(A) Heat map of gene expression differences of human and bovine endometrial stromal fibroblasts with and without co-culture with HTR8 and F3 trophoblast cells respectively. (B) Volcano plot showing fold differences in the TPM values of bESFs vs hESFs co-cultured with the respective trophoblasts, color coded for relevant gene ontologies. (C-D) Heatmap showing expression of genes in hESFs and bESFs with or without co-culture with HTR8 and F3 respectively, belonging to the gene-sets chemokine (C) and chemokine receptors (D). TPM values of genes significantly different in hESFs and bESFs for KEGG pathways for TGFB (E), and non-cannonical WNT signaling (F); For each gene, TPM values before and after co-culture are shown in green, and red dots respectively; Arrows show the change in TPM values due to trophoblast co-culture; Also are shown coefficient of determination, R2 for the linear regression of TPM values between hESF and bESF, and the standard deviation of the residuals, Sy.x.
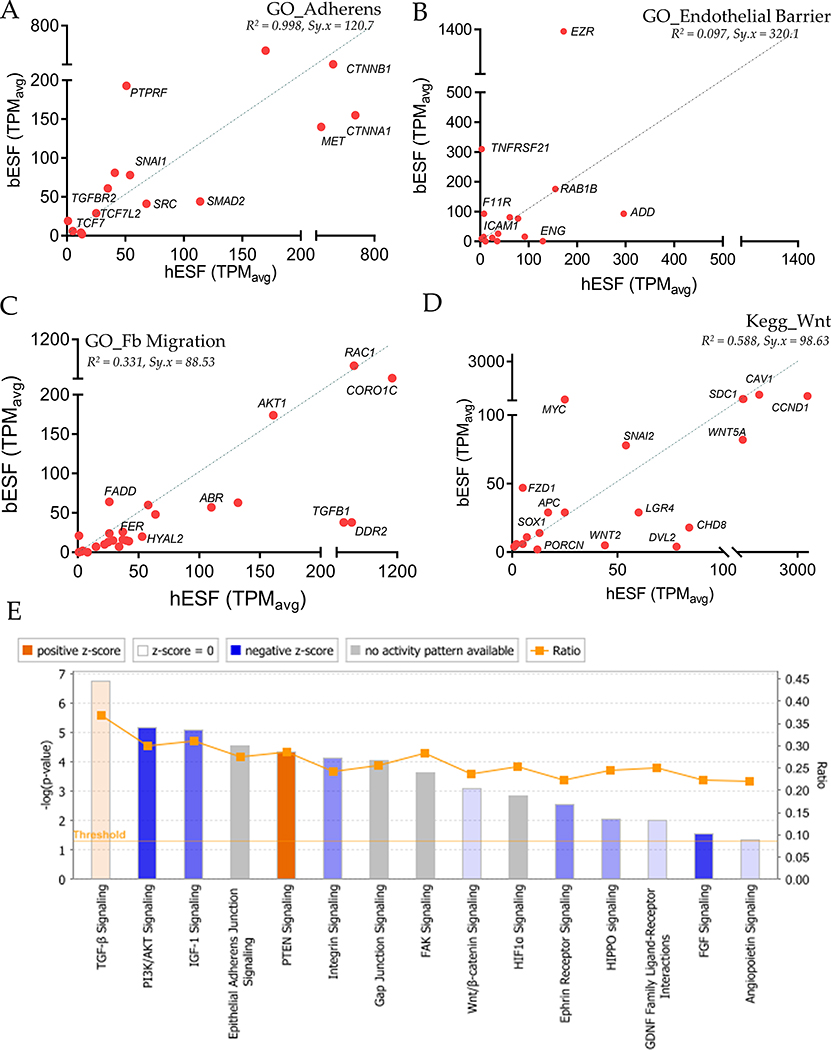
We found that human and bovine ESFs expressed markedly different sets of chemokine ligands (Figure 3C) and chemokine receptors (Figure 3D), although both were enriched in transcripts associated with angiogenesis. Genes showing elevated expression in hESFs included Fibroblast Growth Factors (FGFs), Vascular Endothelial Growth Factors (VEGFs), Semaphorins, members of the Transforming Growth Factor (TGF) family, as well as NRPs and ROBO1, whereas bESFs showed high expression of Endothelin (END1), Plasminogen activator (PLAU), Thrombopoeitin (THPO) and notably, Transforming Growth Factor beta 2 (TGFB2). In contrast, genes belonging to ontologies GO_Adherens Junction and Endothelial Barrier did not show systematic expression differences in favor of either hESF or bESF (Figure S4A–B). Finally, the genes related to GO_Fibroblast Migration tend to be expressed at lower levels in bESF vs. hESF (Figure S4C). These data point to the possibility that stromal response to trophoblast co-culture varies strongly between the species for genes that could play a role in the interaction between invading trophoblast cells and stromal cells, as opposed to genes which regulate basal stromal integrity. Differential expression of these genes may affect the invasibility phenotype in either species.
These analyses suggested that differences in the interactions between the stromal and invading cells were drivers of ELI in human and bovine. For instance, transforming Growth Factor (TGFB), and WNT signaling pathways are mediators of cancer stromal interaction36–41 and thus we explored the effect of trophoblast co-culture on the activation of these signaling pathways in order to see whether there are parallels between cancer-stroma and trophoblast-endometrial stroma interactions. Expression of genes in non-canonical WNT and TGFB pathways were indeed higher in hESFs compared with bESFs (Figure 3E, F). Ingenuity pathway analysis showed that both signaling pathways were differentially activated in bovine and human ESFs (Figure S4E). Although co-culture with trophoblasts resulted in reduction in expression of genes belonging to TGFΒ pathway for hESFs, they continued to remain many-fold higher when compared in bESFs. While TGF ligands TGFB1, and TGFB3, as well as receptors of the TGFΒ family, e.g. ACVR1 were highly expressed in hESFs, negative regulators like TGFβ2 and inhibin-A were expressed at higher levels in bESFs. Similarly, we observed that downstream transcription factors Smad 2, −3 and −5, as well as the co-SMAD, SMAD4, were expressed at higher levels in hESFs compared to bESFs (Figure 3E). Similarly, many members of the WNT signaling network displayed higher expression in hESF vs bESF (Figure 3F, S4D). These results supported a role for paracrine signaling between heterotypic cells during the invasion processes in both, the uterus and in tumor lesion, prompting us to further explore the role of the signaling networks in stromal invasion.
Selected gene knockdown in human fibroblasts increases resistance to melanoma invasion
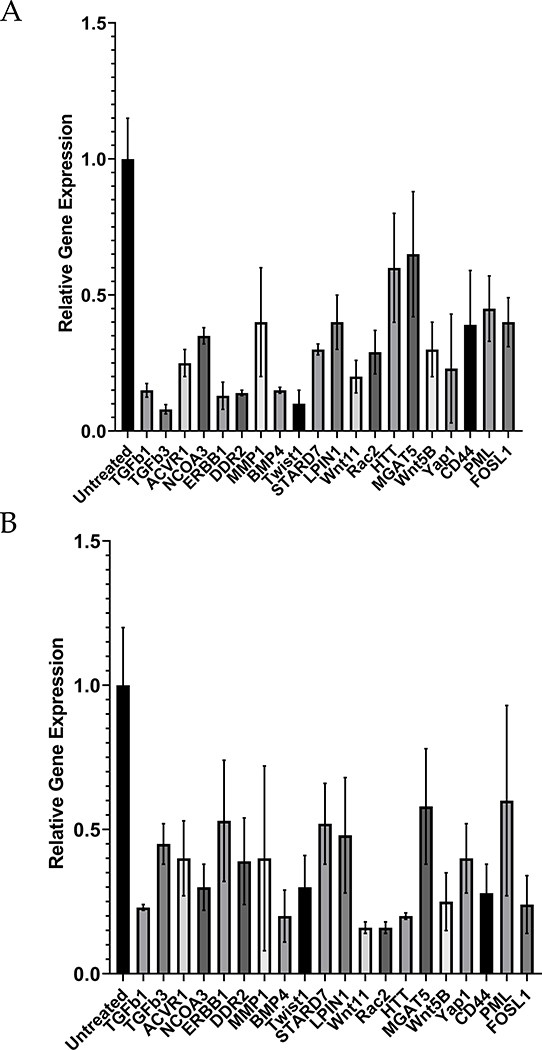
We hypothesized that modulating the differentially expressed genes in human stromal cells could induce them to become more resistant to invasion in human cells. We focused on genes related to WNT and TGFB signaling. Specifically, we selected 16 genes from the TGFB and WNT pathways that had higher expression in hESF or were upregulated in hESF in co-culture with trophoblast cells (Figure 4A). We then targeted these genes using a battery of siRNAs in hESFs and BJ5ta to test whether they could modulate stromal invasibility.
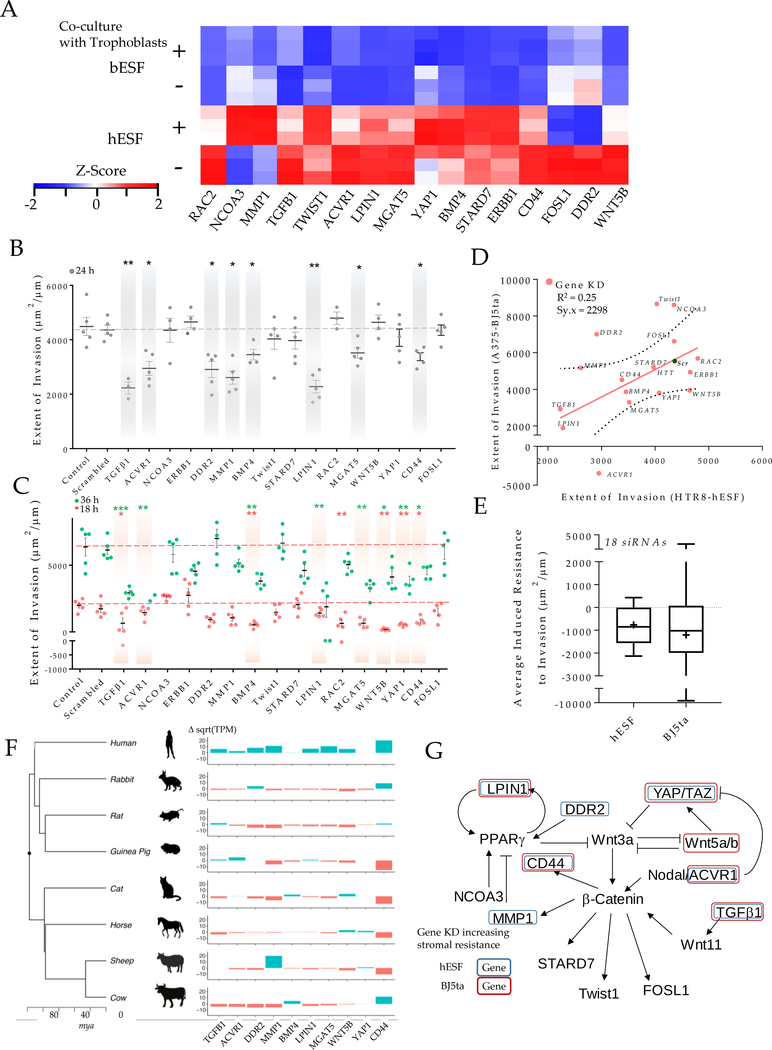
Figure 4. Induced resistance to invasion in human stroma by evolutionarily inspired gene silencing.
(A) Heatmap showing selected set of genes used for gene silencing in human stromal fibroblasts; Shown are z-score of TPM values for each transcript. (B-E) siRNA based gene knockdown in human stroma to induce expression similar to bESF increases resistance to invasion; (B) Extent of invasion of HTR8 cells into hESFs subjected to siRNA mediated gene silencing measured over 24 hours using nanotextured platform; (C) Extent of invasion measured for 18 hours (red), and 36 hours (green) using nanotextured platform for A375 invasion into BJ5ta subjected to siRNA induced silencing of individual genes; Each dot in B, C reflects an individual invasion observation, Error bars show s.e.m., Statistical comparisons made with scrambled siRNA using Student-t-test; *: P < 0.05, **: P < 0.01, ***: P < .0001. (D) Correlation shown between the extent of invasion of HTR8 into hESF, and A375 into BJ5ta for each siRNA based gene silencing; (E) Average extent of invasion in hESF and BJ5ta for the above 20 gene silencing normalized to the extent of invasion observed for control scrambled siRNA knockdown in either human stromal cells; Whiskers show min to max values; horizonal bar is the median; average values are indicated by +, box is 25 to 75 percentile. (F) Relative transcript levels of the selected genes in various boroeutherians species compared to their most common ancestor; red colored bars denote decrease, and green bars show difference in sqrt(TPM) values vs the common ancestor; mya refers to million years ago. (G) Network of WNT signaling and its interaction with TGFB pathway showing previously known interactions, mapped with the gene silencing induced resistance in hESF, and BJ5ta stromal cells.
Comparison of each siRNA-transfected stromal cell population was made with appropriate untransfected controls, as well as with cells transfected with control non-targeting siRNA (Figure S5A–B). HTR8 invasion into hESF was significantly reduced after 24 hours in eight out of 16 genes (Figure 4B). A375 invasion into BJ5ta skin fibroblast monolayer was observed for 18 hours and 36 hours after gene knockdown. Nine of these gene knockdowns significantly decreasing invasion into the skin fibroblast monolayer (Figure 4C), including members of WNT superfamily, TGFΒ ligands, as wells as less established targets and effectors of WNT signaling, e.g. STARD742, LPIN143–47, and YAP148. Furthermore, we detected a weak but significant correlation in the increase of stromal resistance to invasion following gene knockdowns in both hESF and BJ5ta (r2=0.25, p = 0.02), indicating that gene silencing enhances resistance to invasion in a similar manner in both human endometrial and skin fibroblasts (Figure 4D). Moreover, the average response for all gene knockdowns was an increase in resistance to invasion (Figure 4E). These results suggested that human skin fibroblasts could be induced to resist melanoma invasion and that knowledge of gene expression in cow fibroblasts can tell us how to achieve this.
Opposite evolutionary trends of vulnerability to malignancy in humans and bovines
In order to assess the evolutionary history of the expression of the genes identified above as important for ELI, we cultured the endometrial stromal fibroblasts from rabbit, rat, guinea pig, cat, horse and sheep to extend our analysis (Figure 4F). This taxon sample represents two clades of eutherian mammals, the Euarchontoglires, and the Laurasiatheria, together forming the clade of Boreoeutheria. We plotted the phylogenetic tree of these eight species49, along with the expression of these genes in skin fibroblasts compared to the inferred boreoeutherian ancestor (√TPMspecies - √TPMancestor).
We found, surprisingly, that humans evolved higher expression levels of invasibility enhancing genes (TGFB1, ACVR1, DDR2, LPIN1, CD44, MMP1), compared to the inferred boreotherian ancestor. The expression levels of these genes in human skin fibroblasts is higher than that of rabbit, rat and guinea pig. All of these species have hemochorial placentation and thus the higher expression level of these genes in humans likely evolved in the primate lineage. The limited data on a selected group of genes, therefore, supports the hypothesis of Evolved Levels of Invasibility (ELI) amongst mammals, and points to the possibility of increased stromal vulnerability in humans, and decreased stromal vulnerability in bovines.
Discussion
Mammalian species differ in their potential for tumorigenesis, as well as their vulnerability to cancer metastasis9,50. The comparative biology of cancer incidence across different animals has identified species specific tumor suppressor mechanisms explaining the variation in occurrence of tumors between species51,52,53,54,55,56. In a recent review, Constanzo and collaborators made a convincing case for a model where cancer progression in humans includes the re-activation of embryonic gene expression normally controlling placenta development and immune evasion57. Here, we focus on differences across species of the rates of cancer malignancy rather than differences in rates of tumorigenesis. For instance, melanoma does occurred in bovines and equines, but remains largely benign58–60; while it is highly malignant in humans. This correlates with the phenotype of the fetal-maternal interface, i.e. the degree of placental invasion during pregnancy.
siRNA-guided knockdown in human stromal fibroblasts revealed genes that impart stromal invasibility by melanoma and trophoblast cells (Figure 4G). These genes include TGFΒ ligands, consistent with results from colorectal cancer39. Other genes enhancing stromal invasibility are members of the non-canonical WNT pathway, as well as WNT signaling modulators, e.g. LPIN161, by directly regulating β-Catenin levels, e.g. YAP/TAZ48, or by being regulated by β-catenin induced transcription, e.g. CD4462. Of these, YAP1, a Hippo pathway target promoting tumor growth63, can also be regulated by non-canonical WNT signaling, and can in turn inhibit canonical WNT signaling48. Both TGFΒ64,65 and non-canonical WNT signaling66,67 are known to affect tumor progression and metastasis. TGFΒ1 and TGFΒ3, expressed at relatively higher levels in hESF, are also reported to regulate β-catenin activity, indicating that human stromal vulnerability to invasion may be influenced by paracrine signaling to invading cells through secreted ligands. Also of interest, many of the genes identified in our comparative gene expression screen have connections to metabolic regulation. These include genes encoding LIPIN1 which regulates triglyceride metabolism, 68 and MGAT5, known to regulate glucose uptake in tumor cells69.
The results presented here show that species differences in malignancy rates may, in part, be caused by species differences in invasibility of stromal cells. We found that bovine stroma evolved lower invasibility likely via decreased paracrine signaling through TGFΒ and WNT pathways. In particular, these results argue that TGFΒ secretion, and high non-canonical WNT signaling in stromal cells are causal factors explaining the high vulnerability of human stromal tissues to cancer invasion, at least in the case of melanoma. Comparative transcriptomic data across multiple additional species further suggests that the human lineage evolved higher expression of these genes enhancing tumor and trophoblast invasion and thus likely have evolved higher malignancy rates than in the common ancestor of eutherian mammals.
Our data support the ELI hypothesis, suggesting that differences in stromal gene expression between species are causal in determining both, the degree of embryo implantation as well as stromal resistance to early cancer dissemination. Epitheliochorial placentation has evolved multiple times from the pre-existing invasive placentation, suggesting evolutionary advantages of non-invasive placentation13,14,70 may drive corresponding differences in malignancy resistance. We further found that apes may also have evolved increased stromal receptivity to trophoblast invasion, and correlatively have evolved higher vulnerability to cancer malignancy. As extravillous trophoblasts have evolved recently in great apes, the endometrial stroma may also have increased invasibility to accommodate the more invasive trophoblast types71. Our ELI hypothesis, and its experimental validation, suggests that studying the genetic basis for evolved resistance to invasion can identify factors determining the stromal control of cancer progression and identify novel therapeutic targets.
Materials and Methods
Cell Sourcing
The study used a variety of cell sources for comparative analyses. Human ESFs were immortalized by the Charles Lockwood lab at Yale, and obtained from Gil Mor group72. F3 cells were previously established by Pfarrer group at the University of Veterinary Medicine, Hannover, Germany73,74. J3, HTR8/SVNeo, BeWo, and BJ5ta cells were obtained from ATCC.
bSkFb bovine (Bos Taurus) skin fibroblasts, as well as those from guinea pig (Cavia porcellus), sheep (Ovis aries), pig (Sus scrofa), and rabbit (Oryctolagus cuniculus) were isolated from the skin tissue of the face of a cow from Bristol Butchers. A small piece of skin was harvested, hair removed, and the sample was washed in PBS and cut into strips approximately 1.0 cm2. Dermis was separated from epidermis by enzymatic digestion (30 minutes in 0.25% trypsin at 37°C, followed by Dissociation Buffer (1 mg/ml collagenase, 1 mg/ml Dispase, 400 μg/ml DNaseI) for 45 minutes at 37°C). Epidermis was removed, and 2 mm pieces were cut from the dermis, and transferred to a 12-well plate and convered with media. Fibroblasts emerged from the explants, and grew to confluency in growth media. Extra tissue was removed, and cells subcultured for a few times using a cell scraper.
Bovine ESFs were isolated as follows: Uterine tissues were harvested from each species and primary ESF were isolated by enzymatic digestion. Uterus fragments, 2–3 mm in size, were created using scalpel, and digested with 0.25% trypsin-EDTA for 35 min at 37°C, followed by Dissociation Buffer (1 mg/ml collagenase, 1 mg/ml Dispase, 400 μg/ml DNaseI) for 45 minutes at 37°C. Cell clumps were homogenized by passage through a 22-gauge syringe followed by passage through a 40 μm nylon mesh filter to remove remaining clumps. A single cell suspension was obtained from the lysate, transferred to fresh growth medium and cultured in T25 flasks. To facilitate enrichment of fibroblasts versus epithelial cells, media was exchanged in each well after fifteen minutes in order to remove floating cells that had not yet attached while stromal fibroblasts had attached. Cells were grown to confluency and sub-passaged by scraping the cells off the surface to be split into two T25 flasks. At confluency cells were split through one more round of differential attachment. We used immunohistochemistry to test for abundance of vimentin (Santa Cruz, sc-6260) and cytokeratin (Abcam, ab9377) to validate fibroblast subtype in the isolated cells.
hESF, bSkFb and bESFs were tested for potential mycoplasma contamination, and were found to be free from it.
Cell culture
Human ESFs were grown in phenol-red free DMEM/F12 with high glucose (25 mM), supplemented with 10% charcoal stripped calf serum (Hyclone), and 1% antibiotic/antimycotic (Gibco). Decidualization was induced in ESFs with 0.5 mM 8-Br-cAMP (Sigma), and 0.5 mM of progesterone analog medroxy-progesterone acetate (MPA) for 96 hours in DMEM supplemented with 2% charcoal-stripped calf serum (Hyclone). BJ5ta (ATCC) cells were cultured in 80% DMEM and 20% MEM supplemented with 10% FBS, 1% antibiotic/antimycotic, and 0.01 mg/ml hygromycin. F3 cells were obtained from Dr. Pfarrer’s group, and were cultured in DMEM with high glucose, supplemented with 10% FBS, 1% antibiotic/antimycotic (Gibco). J3 cells were cultured in αMEM supplemented with 10% FBS, 1% antibiotic/antimycotic (Gibco), while HTR8/SVNeo (ATCC) were cultured in RPMI-1640 supplemented with 10% FBS, and 1% antibiotic/antimycotic (Gibco).
Fabrication of poly-urethane acrylate mold
Photoresist was spin coated on silicon wafers, and electron-beam lithography was used to nanopattern the wafers (JBX-9300FS, JEOL). After the photoresist was developed, exposed silicon was etched by deep-reactive ion etcher (STS ICP Etcher) resulting in formation of submicron parallel ridges. Residual photoresist was removed using ashing, and diced into silica masters for subsequent replica molding.
UV curable poly-urethane (PUA) was drop-wise dispensed onto the silicon master previously prepared, and contacted with polyethylene terephthalate (PET) film. Application of UV (λ= 200–400 nm, 100 mJ/cm2) for 1 minute was used to cure PUA, after which the mold was peeled off with a pair of tweezers and cured overnight under UV to terminate the residual acrylate groups. The process resulted in PUA mold of ~50 μm thickness.
Fabrication of nanotextured substrate
Previously prepared PET mold was used as a replica mold to transfer the topographic pattern on glass substrate using the technique of capillary force lithography. Briefly, glass substrate was cleaned using NaOH (0.1 M for 1 hour), washed with DIH2O, and dried under nitrogen stream. Primer (phosphoric acrylate and propylene glycol monomethyl ether acetate in a ratio of 1:10) was spin-coated on the coverslip as a thin layer, and baked for 20–30 minutes at 70oC. 150 μl of PEG-DA (molecular weight = 575) precursor was dispensed dropwise on the primed cover slip and the PET mold was placed reversibly. After the PUA precursor filled the submicron cavities by capillary action, the substrate was cured in UV (λ= 250–400 nm, 100 mJ/cm2) for 1 minute. The mold was peeled after polymerization using a pair of tweezers. The substrate resultant from the method consisted of nanotextured grooves with an expected elastic modulus of ~ 70 MPa. The substrate was cured again for an hour under UV to terminate residual active acrylate groups.
Cell patterning for stromal invasion assay
For cell patterning, we used a stereolithographic plastic mold to create a polydimethylsiloxane (PDMS) stencil. PDMS stencil was cast by mixing monomer and cross-linker in a ratio of 10:1, and cured at 80oC for 4 hours by placing on a pre-designed plastic mold. Stencils were washed using isopropyl alcohol and dried using N2 stream, placed on the nanotopographic substrate coated with laminin (25 μg/ml), and the device kept in vacuum to allow air under the stencil to be removed to avoid chance of leakage. DiI-labeled invasive cells were seeded on the stencil at a density of 106 cells/100μl, and allowed to attach and polarize for 8 hours. Unattached cells were washed off twice with PBS, and the stencil removed carefully using a pair of blunt-end tweezers. Unlabeled stromal cells were seeded at a density of 106 cells/100μl to attach to the area previously covered by PDMS stencil. After 6 hours of attachment, unattached cells were washed off, and the substrate mounted for live cell microscopy. To ensure selection of definitive fibroblasts, and to avoid selection of trophoblasts taking up dye from dead stromal cells and occasional heterotypic cell fusion events, we only collected DiIhigh cells. All experiments were conducted in similar culture conditions, by 1:1 mixture of the media in either comparable conditions.
Live Cell Fluorescence Microscopy
Phase-contrast and epifluorescence microscopy was performed using Leica DMi8 model, with an HC PL Fluotar 10x/0.30 Dry objective, and images were acquired using Leica LAS X Software, and processed using Fiji Image Analysis Software. Lumencor SpectraX was used as a source of light for excitation, and routed through a Rhodamine excitation filter cube consisting of Excitation 546 nm/ 10 nm (band-pass filter), Dichroic 560 nm (long-pass filter), and Emission 585 nm/ 40 nm (band-pass filter), and acquired using Andor EMCCD iXon Ultra 888 camera.
Invasion Analysis
Acquired sequential images for each condition were analyzed using Fiji Image Anlaysis Software after contrast enhancement. DiI labeled invasive cell fronts (trophoblasts or melanoma cells) were identified manually for each time point. Area occupied by DiI+ve cells was measured at each time point. Total invasion was calculated as follows:
Extent of invasion per unit measurement of interface was determined by dividing δArea by the length of initial intercellular interface
Fluorescence Assisted Flow Sorting
Cells were detached from the substrate using TrypLE solution (Gibco), quenched with excess medium, and washed thrice with phosphate-buffered saline (PBS). Isolated cells were suspended in 1% AlbuMAX (Gibco) dissolved in PBS and sorted using fluorescence assisted flow sorting (FACS). FACS was performed using BD FacsARIA II using PE.Cy5 channel, and analysis performed using FACSDiva 6.0. In order to increase the purity of ESFs collected after the co-culture experiments, and to account for possible uptake of dye from dead or dying stromal cells by the trophoblasts, as well as for occasional cell-fusion events, we only sorted DiIhi cells. Sorted cells were collected directly in RNASelect to limit RNA degradation. Even cells that were not in co-culture were subjected to the same sorting protocol.
siRNA Transfection and Characterization
For gene silencing experiments, stromal cells (BJ5ta, or hESFs) were cultured in 12 well plates transfected with at least two siRNAs (shown in table below) at a concentration of 50 nmoles per well. Lipofactamine RNAiMAX was used to transfect siRNAs, obtained from IDT.
| Gene Name | siRNA Duplex Name | Duplex Sequence 5’ | Duplex Sequence 3’ |
|---|---|---|---|
| LPIN1 | Hs.Ri.LPIN1.13.1 | GCGUCUACUUGGAUGACCUCACAGA | GCCGCAGAUGAACCUACUGGAGUGUCU |
| Hs.Ri.LPIN1.13.2 | AAGCAUUUUUUGUUCAAGAAACAGA | UCUUCGUAAAAAACAAGUUCUUUGUCU | |
| RAC2 | Hs.Ri.RAC2.13.1 | CCGUGUUUGACAACUAUUCAGCCAA | GUGGCACAAACUGUUGAUAAGUCGGUU |
| Hs.Ri.RAC2.13.2 | GGACUUUUGCUAUUGCAAAUAGAAA | GUCCUGAAAACGAUAACGUUUAUCUUU | |
| MGAT5 | Hs.Ri.MGAT5.13.1 | CUCUGACUUGUUGAGUAAUCAGUCA | GUGAGACUGAACAACUCAUUAGUCAGU |
| Hs.Ri.MGAT5.13.2 | CAUGACAGCUUAUGAUCUGAAGAAA | CAGUACUGUCGAAUACUAGACUUCUUU | |
| WNT11 | Hs.Ri.WNT11.13.1 | AGCCAAUAAACUGAUGCGUCUACAC | GUUCGGUUAUUUGACUACGCAGAUGUG |
| Hs.Ri.WNT11.13.2 | AAGGAAACCGACAAGAUUAAAAATA | GUUUCCUUUGGCUGUUCUAAUUUUUAU | |
| WNT5B | Hs.Ri.WNT5B.13.1 | CGGUUUCUCUCUGACAUUAAAUGCC | AUGCCAAAGAGAGACUGUAAUUUACGG |
| Hs.Ri.WNT5B.13.3 | GAGGAAGCUGUGCCAAUUGUACCAG | GUCUCCUUCGACACGGUUAACAUGGUC | |
| STARD7 | Hs.Ri.STARD7.13.2 | GGUAUCUCACUGAGCUAAUCUGGAA | AGCCAUAGAGUGACUCGAUUAGACCUU |
| Hs.Ri.STARD7.13.3 | GUCCCCACAAGUCAUUUGAUGAGAA | GGCAGGGGUGUUCAGUAAACUACUCUU | |
| YAP1 | Hs.Ri.YAP1.13.1 | GGUCAGAGAUACUUCUUAAAUCACA | GACCAGUCUCUAUGAAGAAUUUAGUGU |
| TGFB1 | Hs.Ri.TGFB1.13.1 | CAGCAACAAUUCCUGGCGAUACCTC | AUGUCGUUGUUAAGGACCGCUAUGGAG |
| Hs.Ri.TGFB1.13.2 | CUACUGUAGUUAGAUCUAUUUAUTG | GUGAUGACAUCAAUCUAGAUAAAUAAC | |
| TGFB3 | Hs.Ri.TGFB3.13.1 | CAACUUAGGUCUAGAAAUCAGCATT | AGGUUGAAUCCAGAUCUUUAGUCGUAA |
| Hs.Ri.TGFB3.13.2 | AAGUAUGAAUAUUACUCUCAAAATC | UGUUCAUACUUAUAAUGAGAGUUUUAG | |
| ACVR1 | hs.Ri.ACVR1.13.1 | GACUUUGACCAAAAUUGAUAAUUCC | UUCUGAAACUGGUUUUAACUAUUAAGG |
| hs.Ri.ACVR1.13.2 | CGCUACGGGGAAAAUGCAUUUUCTT | UUGCGAUGCCCCUUUUACGUAAAAGAA | |
| FOXO1 | hs.Ri.FOXO1.13.1 | GAUGUUUCUGACUAAUCUUAAAUCA | CACUACAAAGACUGAUUAGAAUUUAGU |
| hs.Ri.FOXO1.13.2 | GGAUAAUCUCAACCUUCUCUCAUCA | AACCUAUUAGAGUUGGAAGAGAGUAGU | |
| Twist1 | hs.Ri.TWIST1.13.1 | UUCUGAUAGAAGUCUGAACAGUUGT | GUAAGACUAUCUUCAGACUUGUCAACA |
| hs.Ri.TWIST1.13.2 | CUAUUUUAAAAUGGUAACAAUCAGA | GAGAUAAAAUUUUACCAUUGUUAGUCU | |
| NCOA3 | hs.Ri.NCOA3.13.1 | CUGCCAAUCUUAGUGAUAUUGACAA | UAGACGGUUAGAAUCACUAUAACUGUU |
| hs.Ri.NCOA3.13.3 | CCAGUGGAAUUGGUGAUUCUGAATT | CAGGUCACCUUAACCACUAAGACUUAA | |
| ERBB3 | hs.Ri.ERBB3.13.1 | GCCAUCUUCGUCAUGUUGAACUATA | AACGGUAGAAGCAGUACAACUUGAUAU |
| hs.Ri.ERBB3.13.2 | CACUGUACAAGCUCUACGAGAGGTG | CUGUGACAUGUUCGAGAUGCUCUCCAC | |
| DDR2 | hs.Ri.DDR2.13.1 | GUCAGUUACACCAAUCUGAAGUUTA | GACAGUCAAUGUGGUUAGACUUCAAAU |
| hs.Ri.DDR2.13.3 | CCACUCCAUCUGGACAUUUAAUGAA | ACGGUGAGGUAGACCUGUAAAUUACUU | |
| BMP4 | hs.Ri.BMP4.13.1 | CAGUCCUUGAGGAUAGACAGAUATA | CCGUCAGGAACUCCUAUCUGUCUAUAU |
| hs.Ri.BMP4.13.2 | CCUUGUUUUCUGUCAAGACACCATG | ACGGAACAAAAGACAGUUCUGUGGUAC | |
| MMP1 | hs.Ri.MMP1.13.1 | AGCAGACAUCAUGAUAUCUUUUGTC | GUUCGUCUGUAGUACUAUAGAAAACAG |
| hs.Ri.MMP1.13.3 | GUCAACCUUGUUUCUACUGUUUUAT | UUCAGUUGGAACAAAGAUGACAAAAUA | |
| CD44 | hs.Ri.CD44.13.1 | GCUCUGAGCAUCGGAUUUGAGACCT | UCGAGACUCGUAGCCUAAACUCUGGA |
RNA isolation and qRT-PCR
RNA was isolated using RNeasy micro kit (QIAGEN, Germany) and resuspended in 10 μL water. SuperScript IV VILO Master Mix (ThermoFisher, USA) was used to synthesize cDNA from 1μg mRNA and ezDNase was used to remove DNA after reverse transcription. Primers from Integrated DNA technologies (IDT, USA) were used for qRT-PCR reaction using SybrGreen (ThermoFisher, USA) to evaluate the efficiency of siRNA mediated knockdown. B2M and RPL0 were used as internal reference genes, and both were found to be consistent within samples after knockdown. qRT-PCR was performed in Quantstudio3 (ThermoFisher, USA) and relative quantitation was performed by comparative Ct.
RNA Sequencing
Agilent Bioanalyzer 2100 was used to determine RNA quality and RIN number of over 8 was observed in the samples. TruSeq RNA Library from illumina was used to prepare mRNA library. These libraries were sequenced on illumina HiSeq to generate 30–40million reads per sample (Single end 75 bp reads) and a high Q score was observed (Q>30) for the sequenced data. Alignment of reads and gene specific analysis was performed in Partek® Flow® software, version 5.0 Copyright ©; 2018 Partek Inc., St. Louis, MO, USA. Human cells data was aligned to reference genome (hg38) using STAR alignment (in Partek Flow) and bovine data was aligned using Bos Taurus assembly UMD_3.1.1/bosTau8. Quantification was performed using quantify to annotation model (Partek E/M) using Ensemble Transcripts release 85 (humans) or Ensemble-UMD3.1 gene annotation model. Normalization was performed using TPM (Transcripts Per Million75). Gene level comparison and statistical analysis was performed using ANOVA and significance of (adjusted) p=0.05 or less was considered for analysis. Gene lists comparison was performed by obtaining Ontology/pathway specific list from Gene Ontology76 or Kyoto Encyclopedia of Genes and Genomes(KEGG77)
Comparative transcriptome analysis
Raw sequence reads (single-ended 75bps) for endometrial stromal fibroblasts from rabbit (Oryctolagus cuniculus), rat (Rattus norvegicus), guinea pig (Cavia procellus), cat (Felis catus), horse (Equus caballus), and sheep (Ovis aries) were aligned to reference genome assemblies OryCun2.0, Rnor_6.0, Cavpor3.0, Felis_catus_8.0, Equcab2, and Oar_v3.1 respectively using Tophat2 (v2.1.1). Read counts for all genes were calculated using HTSeq (v0.6.1p1, https://htseq.readthedocs.io/en/release_0.11.1/index.html) with Python (v2.7) according to Ensembl gene annotation release 92 (horse) or release 89 (other species). Then transcripts per million (TPM) were calculated to estimate relative mRNA abundance75. To make gene expression levels comparable across species, we further normalized TPMs such that one-to-one orthologs have the same sum across species. The ortholog tables in this analysis were downloaded from Ensembl using BioMart tool. The ancestral state reconstruction for gene expression was performed on square root TPMs using function ‘ace’ from R package ‘APE’78,79.
Two genes that have been found to influence invasibility in our knockdown experiments (Figure 4B and 4C), YAP1 and MMP1, have one-to-many or no orthologs in some species in our analysis. YAP1 has two orthologs in cow, ENSBTAG00000039307 (TPM=183.4) and ENSBTAG00000047406 (TPM=96.3), and no ortholog in guinea pig. MMP1 has two orthologs in cow, ENSBTAG00000015818 (TPM=0.053) and ENSBTAG00000048029 (TPM=0.645), and three orthologs in rat, ENSRNOG00000055895 (TPM=0), ENSRNOG00000008881 (TPM= 0.157) and ENSRNOG00000032353 (TPM= 0.698). In these cases, the orthologs with the highest TPM were utilized to perform the ancestral state reconstruction.
Extended Data
Extended Data Figure 1. Nanotextured stromal invasion assay quantitatively and sensitively measures collective intrusion of cells into stroma.
(A) Time stamped images at 0 and 18 hours showing extent of invasion by DiI labeled 1205Lu cells (red) into BJ5ta stromal fibroblast monolayer (unlabeled) on flat and nanotextured substrata; Quantification shown in Fig. 1D. (B-C) Sensitive measurement of differences in invasion by nanotextured stromal invasion platform; Time stamped images at 0, 6, 12, and 18 hours showing extent of invasion by DiI labeled non-malignant WM35 (B), and malignant 1205Lu (C) cells into BJ5ta monolayer shown in both flat, and nanotextured cell patterned assay; Quantification shown in Fig. 1E.
Extended Data Figure 2. Human and bovine trophoblasts exhibit different invasive potential into endometrial stroma.
Time stamped images showing extent of invasion of human choriocarcinoma derived trophoblasts, J3 (red) (A), and bovine trophoblasts, F3 (red) (B) into their respective endometrial stromal fibroblast monolayers at 0, and 48 hours.
Extended Data Figure 3. Human and bovine endometrial stromal fibroblasts respond differently to co-culture with trophoblasts.
(A) Experimental plan to isolate endometrial cells after co-culture with respective trophoblast; (B) Volcano plot showing fold change in genes between bovine vs human endometrial stromal fibroblasts, along with their significance depicted in p value. (C) P-value distribution of t-tests comparing human ESFs with and without co-culture with HTR8 trophoblast cells. (D) P-value distribution of t-tests comparing bovine ESFs with and without co-culture with F3 trophoblast cells. In both C and D, note that thin right hand tail of the distribution, which indicates that a large number of genes are differentially expressed in response to the presence of the corresponding trophoblast cells. (E-F) Scatter plots showing relative TPM values of genes between (E) hESF and hESF co-cultured with HTR8, and (F) bESF and bESF co-cultured with F3; Red dots refer to individual gene transcripts abundance significantly different in between the compared conditions.
Extended Data Figure 4. Human and bovine endometrial stromal fibroblasts do not appear to differentially respond to their respective trophoblasts by regulating cell-cell adhesion.
Gene ontology analysis of individual genes belonging to GO_Adherens (A), GO_Endothelial Barrier (B), GO_Fibroblast Migration (C), and Kegg ontology for Wnt Signaling (D) for hESF and bESF shown with their relative TPM values; Also are shown the coefficient of determinant, R2 for the linear regression between TPM values of hESF and bESF in a given gene-sets, along with the standard deviation of the residuals, Sy.x. (E) Ingenuity Pathway Analysis of selected signaling pathways differentially activated in hESF and bESF; Shown are –log(p-values) of genes in the respective pathways differentially activated, while the color depicts the z-score.
Extended Data Figure 5. siRNA induced gene knockdown reduces transcript levels in hESFs and BJ5ta cells.
qRTPCR results showing the percentage knockdown of transcript levels in hESF (A) and BJ5ta (B), calculated using DDCt method, compared using GAPDH as housekeeping gene with scrambled siRNA as control. For both A, and B, n = 4 biological replicates.
Supplementary Material
Acknowledgements
Funding: this work was funded by National Cancer Institute Center Grant U54-CA209992.
Footnotes
Competing Interests Statement: the authors declare no competing interests.
Code Availability: the computational methods are described in the M&M. The computational methods are cited and no new code was used in this study.
Data and Materials Availability: data for the human-cow transcriptome comparison with and without co-culture with trophoblast cells is available under GSE136299 in the Gene Expression Omnibus (GEO) database of NCBI, https://www.ncbi.nlm.nih.gov/geo/. The comparative fibroblast gene expression data is available under PRJNA564062 under SUB6229748 and SUB6264591 on the Sequence Read Archive (SRA) of NCBI, https://www.ncbi.nlm.nih.gov/sra.
References
- 1.Chaturvedi P et al. Hypoxia-inducible factor-dependent breast cancer-mesenchymal stem cell bidirectional signaling promotes metastasis. The Journal of clinical investigation 123, 189–205, doi: 10.1172/JCI64993 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colegio OR et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559–563, doi: 10.1038/nature13490 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W & Huang P Cancer-stromal interactions: role in cell survival, metabolism and drug sensitivity. Cancer biology & therapy 11, 150–156 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luga V & Wrana JL Tumor-stroma interaction: Revealing fibroblast-secreted exosomes as potent regulators of Wnt-planar cell polarity signaling in cancer metastasis. Cancer research 73, 6843–6847, doi: 10.1158/0008-5472.CAN-13-1791 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Murray MJ & Lessey BA Embryo implantation and tumor metastasis: common pathways of invasion and angiogenesis. Seminars in reproductive endocrinology 17, 275–290, doi: 10.1055/s-2007-1016235 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Ferretti C, Bruni L, Dangles-Marie V, Pecking AP & Bellet D Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Human reproduction update 13, 121–141, doi: 10.1093/humupd/dml048 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Ratcliffe HL Incidence and Nature of Tumors in Captive Wild Mammals and Birds. Cancer research 17, 132–157 (1932). [Google Scholar]
- 8.Canfield PJ, Hartley WJ & Reddacliff GL Spontaneous proliferations in Australian marsupials--a survey and review. 2. Dasyurids and bandicoots. Journal of comparative pathology 103, 147–158 (1990). [DOI] [PubMed] [Google Scholar]
- 9.D’Souza AW & Wagner GP Malignant cancer and invasive placentation: A case for positive pleiotropy between endometrial and malignancy phenotypes. Evolution, medicine, and public health 2014, 136–145, doi: 10.1093/emph/eou022 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perry JK, Lins RJ, Lobie PE & Mitchell MD Regulation of invasive growth: similar epigenetic mechanisms underpin tumour progression and implantation in human pregnancy. Clin Sci (Lond) 118, 451–457, doi: 10.1042/CS20090503 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Bruning A, Makovitzky J, Gingelmaier A, Friese K & Mylonas I The metastasis-associated genes MTA1 and MTA3 are abundantly expressed in human placenta and chorionic carcinoma cells. Histochemistry and cell biology 132, 33–38, doi: 10.1007/s00418-009-0595-z (2009). [DOI] [PubMed] [Google Scholar]
- 12.Wildman DE et al. Evolution of the mammalian placenta revealed by phylogenetic analysis. Proc Natl Acad Sci U S A 103, 3203–3208, doi: 10.1073/pnas.0511344103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mess A & Carter AM Evolutionary transformation of fetal membrane characters in Eutheria with special reference to Afrotheria. J Exp Zool Part B (Mol Dev Evol) 306B, 140–163 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Elliot MG & Crespi BJ Phylogenetic evidence for early hemochorial placentation in eutheria. Placenta 30, 949–967, doi: 10.1016/j.placenta.2009.08.004 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Haeger A, Krause M, Wolf K & Friedl P Cell jamming: collective invasion of mesenchymal tumor cells imposed by tissue confinement. Biochim Biophys Acta 1840, 2386–2395, doi: 10.1016/j.bbagen.2014.03.020 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Shamir ER, Coutinho K, Georgess D, Auer M & Ewald AJ Twist1-positive epithelial cells retain adhesive and proliferative capacity throughout dissemination. Biol Open 5, 1216–1228, doi: 10.1242/bio.019703 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolf K et al. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol 9, 893–904, doi: 10.1038/ncb1616 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Lau TS et al. A loop of cancer-stroma-cancer interaction promotes peritoneal metastasis of ovarian cancer via TNFalpha-TGFalpha-EGFR. Oncogene 36, 3576–3587, doi: 10.1038/onc.2016.509 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bremnes RM et al. The role of tumor stroma in cancer progression and prognosis: emphasis on carcinoma-associated fibroblasts and non-small cell lung cancer. J Thorac Oncol 6, 209–217, doi: 10.1097/JTO.0b013e3181f8a1bd (2011). [DOI] [PubMed] [Google Scholar]
- 20.Howard JD et al. Notch signaling mediates melanoma-endothelial cell communication and melanoma cell migration. Pigment Cell Melanoma Res 26, 697–707, doi: 10.1111/pcmr.12131 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Kshitiz Afzal, J., Kim SY & Kim DH A nanotopography approach for studying the structure-function relationships of cells and tissues. Cell Adh Migr 9, 300–307, doi: 10.4161/cam.29359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim DH, Provenzano PP, Smith CL & Levchenko A Matrix nanotopography as a regulator of cell function. J Cell Biol 197, 351–360, doi: 10.1083/jcb.201108062 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo F Assessment of stromal invasion for correct histological diagnosis of early hepatocellular carcinoma. Int J Hepatol 2011, 241652, doi: 10.4061/2011/241652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khalil AA et al. Collective invasion in ductal and lobular breast cancer associates with distant metastasis. Clin Exp Metastasis 34, 421–429, doi: 10.1007/s10585-017-9858-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedl P & Alexander S Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 147, 992–1009, doi: 10.1016/j.cell.2011.11.016 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Cheung KJ et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc Natl Acad Sci U S A 113, E854–863, doi: 10.1073/pnas.1508541113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramsey EM The placenta : human and animal. (Praeger, 1982). [Google Scholar]
- 28.Wooding P & Burton G Comparative Placentation: Structures, Functions and Evolution. (Springer, 2008). [Google Scholar]
- 29.Orendi K et al. Placental and trophoblastic in vitro models to study preventive and therapeutic agents for preeclampsia. Placenta 32 Suppl, S49–54, doi: 10.1016/j.placenta.2010.11.023 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Bertero L et al. Eighth Edition of the UICC Classification of Malignant Tumours: an overview of the changes in the pathological TNM classification criteria-What has changed and why? Virchows Arch 472, 519–531, doi: 10.1007/s00428-017-2276-y (2018). [DOI] [PubMed] [Google Scholar]
- 31.Musser JM & Wagner GP Character trees from transcriptome data: Origin and individuation of morphological characters and the so-called “species signal”. J Exp Zool B Mol Dev Evol 324, 588–604, doi: 10.1002/jez.b.22636 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Wagner GP Homology, Genes and Evolutionary Innovation. (Princeton University Press, 2014). [Google Scholar]
- 33.Liang C, Musser JM, Cloutier A, Prum RO & Wagner GP Pervasive Correlated Evolution in Gene Expression Shapes Cell and Tissue Type Transcriptomes. Genome Biol Evol 10, 538–552, doi: 10.1093/gbe/evy016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang C, Musser JM, Cloutier A, Prum RO & Wagner GP Pervasive correlated evolution in gene expression shapes cell and tissue type transcriptomes. Genome Biol Evol, doi: 10.1093/gbe/evy016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garma-Avina A, Valli VE & Lumsden JH Cutaneous melanomas in domestic animals. J Cutan Pathol 8, 3–24 (1981). [DOI] [PubMed] [Google Scholar]
- 36.Meng XM, Nikolic-Paterson DJ & Lan HY TGF-beta: the master regulator of fibrosis. Nat Rev Nephrol 12, 325–338, doi: 10.1038/nrneph.2016.48 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Guido C et al. Metabolic reprogramming of cancer-associated fibroblasts by TGF-beta drives tumor growth: connecting TGF-beta signaling with “Warburg-like” cancer metabolism and L-lactate production. Cell Cycle 11, 3019–3035, doi: 10.4161/cc.21384 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vallee A, Lecarpentier Y, Guillevin R & Vallee JN Interactions between TGF-beta1, canonical WNT/beta-catenin pathway and PPAR gamma in radiation-induced fibrosis. Oncotarget 8, 90579–90604, doi: 10.18632/oncotarget.21234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calon A et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet 47, 320–329, doi: 10.1038/ng.3225 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Piersma B, Bank RA & Boersema M Signaling in Fibrosis: TGF-beta, WNT, and YAP/TAZ Converge. Front Med (Lausanne) 2, 59, doi: 10.3389/fmed.2015.00059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akhmetshina A et al. Activation of canonical Wnt signalling is required for TGF-beta-mediated fibrosis. Nat Commun 3, 735, doi: 10.1038/ncomms1734 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menke J et al. Circulating CSF-1 promotes monocyte and macrophage phenotypes that enhance lupus nephritis. J Am Soc Nephrol 20, 2581–2592, doi: 10.1681/ASN.2009050499 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takada I, Kouzmenko AP & Kato S Wnt and PPARgamma signaling in osteoblastogenesis and adipogenesis. Nat Rev Rheumatol 5, 442–447, doi: 10.1038/nrrheum.2009.137 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Ahn EH et al. Spatial control of adult stem cell fate using nanotopographic cues. Biomaterials 35, 2401–2410, doi: 10.1016/j.biomaterials.2013.11.037 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hubbi ME et al. A nontranscriptional role for HIF-1alpha as a direct inhibitor of DNA replication. Sci Signal 6, ra10, doi: 10.1126/scisignal.2003417 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suhail Y et al. Modeling intercellular transfer of biomolecules through tunneling nanotubes. Bull Math Biol 75, 1400–1416, doi: 10.1007/s11538-013-9819-4 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Zhang P, Takeuchi K, Csaki LS & Reue K Lipin-1 phosphatidic phosphatase activity modulates phosphatidate levels to promote peroxisome proliferator-activated receptor gamma (PPARgamma) gene expression during adipogenesis. J Biol Chem 287, 3485–3494, doi: 10.1074/jbc.M111.296681 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park HW et al. Alternative Wnt Signaling Activates YAP/TAZ. Cell 162, 780–794, doi: 10.1016/j.cell.2015.07.013 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meredith RW et al. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science 334, 521–524, doi: 10.1126/science.1211028 (2011). [DOI] [PubMed] [Google Scholar]
- 50.de Magalhaes JP & Costa J A database of vertebrate longevity records and their relation to other life-history traits. J Evol Biol 22, 1770–1774, doi: 10.1111/j.1420-9101.2009.01783.x (2009). [DOI] [PubMed] [Google Scholar]
- 51.Seluanov A et al. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc Natl Acad Sci U S A 106, 19352–19357, doi: 10.1073/pnas.0905252106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian X et al. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 499, 346–349, doi: 10.1038/nature12234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nunney L Lineage selection and the evolution of multistage carcinogenesis. Proc Biol Sci 266, 493–498, doi: 10.1098/rspb.1999.0664 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caulin AF, Graham TA, Wang LS & Maley CC Solutions to Peto’s paradox revealed by mathematical modelling and cross-species cancer gene analysis. Philos Trans R Soc Lond B Biol Sci 370, doi: 10.1098/rstb.2014.0222 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abegglen LM et al. Potential Mechanisms for Cancer Resistance in Elephants and Comparative Cellular Response to DNA Damage in Humans. JAMA 314, 1850–1860, doi: 10.1001/jama.2015.13134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sulak M et al. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. Elife 5, doi:ARTN e11994 10.7554/eLife.11994 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Costanzo V, Bardelli A, Siena S & Abrignani S Exploring the links between cancer and placenta development. Open Biol 8, doi: 10.1098/rsob.180081 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Curik I et al. Inbreeding and Melanoma in Lipizzan Horses. Agriculturae Conspectus Scientificus 65, 181–186 (2000). [Google Scholar]
- 59.Feldman WH A Study of the Tumor Incidence in the Lower Animals. Am J Pathol 2, 545–556 (1926). [PMC free article] [PubMed] [Google Scholar]
- 60.Priester WA & Mantel N Occurrence of tumors in domestic animals. Data from 12 United States and Canadian colleges of veterinary medicine. Journal of the National Cancer Institute 47, 1333–1344 (1971). [PubMed] [Google Scholar]
- 61.Bego T et al. Association of PPARG and LPIN1 gene polymorphisms with metabolic syndrome and type 2 diabetes. Med Glas (Zenica) 8, 76–83 (2011). [PubMed] [Google Scholar]
- 62.Wong NA & Pignatelli M Beta-catenin--a linchpin in colorectal carcinogenesis? Am J Pathol 160, 389–401 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao B, Li L, Lei Q & Guan KL The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev 24, 862–874, doi: 10.1101/gad.1909210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Padua D & Massague J Roles of TGFbeta in metastasis. Cell Res 19, 89–102, doi: 10.1038/cr.2008.316 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Wendt MK, Tian M & Schiemann WP Deconstructing the mechanisms and consequences of TGF-beta-induced EMT during cancer progression. Cell Tissue Res 347, 85–101, doi: 10.1007/s00441-011-1199-1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weeraratna AT et al. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell 1, 279–288 (2002). [DOI] [PubMed] [Google Scholar]
- 67.Kurayoshi M et al. Expression of Wnt-5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer research 66, 10439–10448, doi: 10.1158/0008-5472.CAN-06-2359 (2006). [DOI] [PubMed] [Google Scholar]
- 68.Kim JY, Kim G, Lim SC & Choi HS LPIN1 promotes epithelial cell transformation and mammary tumourigenesis via enhancing insulin receptor substrate 1 stability. Carcinogenesis 37, 1199–1209, doi: 10.1093/carcin/bgw104 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Mendelsohn R et al. Complex N-glycan and metabolic control in tumor cells. Cancer research 67, 9771–9780, doi: 10.1158/0008-5472.CAN-06-4580 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Wildman DE et al. Evolution of the mammalian placenta revealed by phylogenetic analysis. Proc. Natl. Acad. Sci USA 103, 3203–3208 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carter AM Evolution of placental function in mammals: the molecular basis of gas and nutrient transfer, hormone secretion, and immune responses. Physiol Rev 92, 1543–1576, doi: 10.1152/physrev.00040.2011 (2012). [DOI] [PubMed] [Google Scholar]
- 72.Kirkun G et al. A novel immortalized human endometrial stroma cell line with normal progestational response. Endocrinology 145, 2291–2296 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Haeger JD, Hambruch N, Dilly M, Froehlich R & Pfarrer C Formation of bovine placental trophoblast spheroids. Cells, tissues, organs 193, 274–284, doi: 10.1159/000320544 (2011). [DOI] [PubMed] [Google Scholar]
- 74.Hambruch N, Haeger JD, Dilly M & Pfarrer C EGF stimulates proliferation in the bovine placental trophoblast cell line F3 via Ras and MAPK. Placenta 31, 67–74, doi: 10.1016/j.placenta.2009.10.011 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Wagner GP, Kin K & Lynch VJ Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory in biosciences = Theorie in den Biowissenschaften 131, 281–285, doi: 10.1007/s12064-012-0162-3 (2012). [DOI] [PubMed] [Google Scholar]
- 76.The Gene Ontology C The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47, D330–D338, doi: 10.1093/nar/gky1055 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanehisa M & Goto S KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30, doi: 10.1093/nar/28.1.27 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Paradis E, Claude J & Strimmer K APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20, 289–290, doi: 10.1093/bioinformatics/btg412 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Paradis E Analysis of Rhylogenetics and Evolution with R. 2nd edn, (Springer, 2012). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.