Abstract
The pen or gladius of the squid is an internalized shell and serves as a site of attachment for important muscle groups and as a protective barrier for the visceral organs. The pen’s durability and flexibility is derived from its unique composition of chitin and protein. We report the characterization of the structure, development, and composition of pens from Doryteuthis pealeii. The nanofibrils of the polysaccharide, β-chitin, are arranged in an aligned configuration in only specific regions of the pen. Chitin is secreted early in development enabling us to characterize the changes in pen morphology prior to hatching. The chitin and proteins are assembled in the shell sac surrounded by fluid which has a significantly different ionic composition than squid plasma. Two groups of proteins are associated with the pen, those on its surface and those embedded within the pen. Only twenty proteins are identified as embedded within the pen. Embedded proteins are classified into 6 groups including chitin associated, protease, protease inhibitors, intracellular, extracellular matrix and those that are unknown. The pen proteins share many conserved domains with proteins from other chitinous structures. We conclude that the pen is one of the least complex, load-bearing, chitin-rich structures currently known, and is amenable to further studies to elucidate natural construction mechanisms using chitin and protein.
Keywords: squid pen, Doryteuthis pealeii, chitin, shell sac, chitin binding domain
Introduction
The pen of the squid is considered an internalized shell, homologous with the heavily mineralized external shells of other molluscs (Naef, 1928;Young et al., 1998). Pens are most commonly transparent, tough, flexible, non-mineralized skeletal elements comprised of chitin and proteins. Along its dorsal surface it supports the mantle muscles, whose expansion and contraction produces the jet propulsion of squid locomotion (Williams, 1909;Gosline and DeMont, 1985;Thompson et al., 2014), powerful enough to launch squid into flight (O’Dor, 2013). On its ventral surface, muscles insert that control the siphon and give direction to the jet, control the rotational motion of the head and its retraction into the mantle cavity, and support the musculature of digestive organs (Williams, 1909;Young et al., 1998;Thompson et al., 2016). However, some squid, like the bobtail squid, that live a benthic existence, form no pen (Cole and Hall, 2009;Lee et al., 2009), and others, such as the bigfin reef squid, have a pen with some degree of mineralization (Subhapradha et al., 2013).
The pen is valuable as a source of information for the life history of the squid, as well as a marine resource of commerce. The pen enlarges over the life cycle of the squid, incorporating stable environmental isotopes, providing a record of an individual’s age and also its feeding and migratory behavior (Bizikov, 1995;Ruiz-Cooley et al., 2010;Lorrain et al., 2011;Merten et al., 2017). The pen is also a rich source of β-chitin that has many commercial applications (Vournakis et al., 2003;Di Martino et al., 2005;Ma et al., 2011;Fernandez and Ingber, 2014;Das et al., 2015) and many efforts report characterization of the purified chitin (Chaussard and Domard, 2004;Lavall et al., 2007;Cortizo et al., 2008;Ianiro et al., 2014;Cuong et al., 2016). However, pen construction and its protein composition have largely been ignored.
In other organisms where external shells are constructed with chitin-protein composites, shells can be heavily mineralized, as in decapods, gastropods, and bivalves (Raabe et al., 2005;Marin et al., 2008;Fernandez and Ingber, 2013;Suzuki and Nagasawa, 2013). Recent studies attempt to characterize the proteins associated with chitin by using transcriptomic and proteomic approaches (Marin et al., 2013). Many of the recovered proteins are unknown and for many others it is difficult to decide whether a particular protein is involved in the assembly, organization and nucleation of the inorganic matrix or involved with the structure and biosynthesis of the chitin-protein matrix or both (Marin et al., 2008). Furthermore, many other chitin-protein composites are sclerotized, hardened through dehydration and covalent cross-linking of structural macromolecules including insect cuticles and wings, and cephalopod beaks (Andersen, 2010;Fernandez and Ingber, 2013;Tan et al., 2015). Extraction and identification of the cross-linked proteins is very challenging (Fernandez and Ingber, 2013;Tan et al., 2015). In order to learn how chitin and proteins are assembled into 3-dimensional structures, it would be ideal to study a chitin-protein composite without mineralization or sclerotization, in a model organism that can be studied at the cellular and molecular level from its embryonic origin to adulthood. The pen of Doryteuthis pealeii, is one such example.
The squid pen is constructed by, and contained within, the closed shell sac, an epithelium which completely surrounds the pen (Williams, 1909;Hopkins and Boletzky, 1994). In the squid, Loligo vulgaris, it was suggested that the ventral shell sac epithelium is primarily responsible for secretion of the materials that make up the pen, and that the dorsal epithelium plays a more significant role in anchoring the pen to the mantle musculature (Hopkins and Boletzky, 1994). These conclusions are based on differences in cellular morphology and organelle quantity between the ventral and dorsal regions of the shell sac rather than direct evidence of the secretion of the pen constituents (Hopkins and Boletzky, 1994). In fact, much of the evidence regarding the secretion and organization of chitin and proteins in the pen, is based on various imaging methods and analogy to known structures rather than on the identification of individual proteins and how they interact with other proteins and/or chitin (Hunt and Nixon, 1981;Hunt and El Sherief, 1990;Wu et al., 2003;Yang et al., 2014;Cuong et al., 2016).
Pens from various species of squid contain by weight, 25-49% chitin and 43-75% protein with very low ash (inorganic composition) after thermo-gravimetric analysis (Hunt and Nixon, 1981;Wu et al., 2003;Chaussard and Domard, 2004;Lavall et al., 2007;Cortizo et al., 2008;Cuong et al., 2016). In the small samples of the pen that were studied, chitin nanofibrils appear wrapped in a protein layer (Raabe et al., 2005;Yang et al., 2014;Merzendorfer et al., 2016) and α-helical protein coils and β-chitin nanofibrils, are aligned parallel to the long axis of the pen (Yang et al., 2014). While a unidentified collagen has been inferred to surround pens of Illex argentinus (Wu et al., 2003), no other proteins associated with the pen have been identified. Amino acid analysis has been performed and indicates that pen proteins from three different species are enriched with hydrophobic amino acids (Hackman, 1960;Hunt and Nixon, 1981;Wu et al., 2003;Cuong et al., 2016).
The identity of the pen proteins and the mechanisms by which they assemble with the chitin nanofibrils to construct and repair this complex 3-D structure remain unknown. Here, we use microscopy to describe the chitin-protein organization in whole adult pens and the developmental morphology of embryonic pens. We determine the inorganic composition of the extracellular fluid surrounding the pen to begin to understand how the pen remains without mineralization. We also report and discuss the proteins that are associated with adult pens and compare them to the proteins associated with other chitinous structures.
Materials and Methods
Collection and preparation of biological materials
Pens were removed from the shell sacs of freshly sacrificed adult squid (Doryteuthis pealeii). An incision in the mantle was made along the midline and the pen was removed by peeling back the mantle and gently pulling on the pen. Two methods were used for removing adhering tissue. Method 1 (M1) involved removing all visible soft tissue by rubbing pens with gloved hands under deionized (DI) water. Method 2 (M2) involved rinsing pens with isotonic saline while scrubbing the pen surface with a small, plastic bristled brush. Dehydration of whole pens was performed in a short period of time using a vacuum evaporator. Pulverized pens were chopped into a fine powder with a blade grinder (Capresso, Closter, N.J.).
Embryonic squid and hatchlings were collected from squid egg capsules that were deposited in the squid holding tanks of the Marine Resource Center (MBL, Woods Hole, MA) between June and September, and held at ambient temperature. Embryos were staged according to Arnold (1965).
Shrimp shells appear similar to squid pen with regard to low covalent cross-linking and low mineralization. Therefore, they were used to compare protein extraction and general composition with the squid pen despite the fact that shrimp shells contain α-chitin rather than β-chitin in the squid pen. The shells were obtained from chilled white shrimp (Litopenaeus setiferus). We used M2 to remove cellular material from the shrimp shells after the shells were peeled away from the body. Shells were dried and pulverized as described above.
Microscopy
We examined pen construction using polarized light, fluorescence, and electron microscopy. Images of dried pens (M1) were obtained with transmitted light that was unpolarized or polarized to aid in identification of chitin-protein alignment in whole pens. Four individual images were assembled to make each composite image. The squid pen was placed between crossed polarizer and analyzer filters during illumination with polarized light. Filters were mounted on a Zeiss Discovery microscope equipped with a color digital camera. Polarizers were rotated to achieve the maximum extinction (dark in the absence of anisotropic material). Isotropic materials remain dark when rotated between polarizers. Anisotropic materials become brighter at set intervals when rotated between polarizers. In order to obtain the maximal (brightest) birefringence signal, the long axis of the pen is oriented at 45° to the transmission axis of the polarizer. The anisotropic structures in the pen produce Newton interference colors. The hue corresponds to the magnitude of birefringence retardation and can be estimated by the Michel Levy Color Chart (Oldenbourg and Shribak, 2010).
Fluorescence microscopy of pen development was performed by staining pen chitin with Calcofluor white (Sigma Aldrich, St. Louis, M.O.) at a final concentration of 0.01%. Specimens were fixed in phosphate buffered saline (PBS) with 2% paraformaldehyde and rinsed with 0.1% Triton X-100 in PBS to remove membranes. Samples were rinsed in PBS before staining with Calcofluor white for 1 hour and destained for 1 hour with multiple exchanges of PBS. Images were collected using a Zeiss two-photon LSM platform configured on an inverted Observer Z1 microscope. Embryos and hatchlings were positioned dorsal side down in PBS and z-stacks of images were collected of the curved pen. Calcofluor was excited with a Coherent Chameleon II laser at 705 nm and the emitted light was collected with an internal detector, 420-492 nm. Rhodamine phalloidin (ThermoFisher, Waltham, MA) was used to stain mantle actin using 540 nm/565 nm, excitation/emission in order to determine the association between the pen and the mantle. Stacks of images were projected onto a single plane using the Z-projection function in ImageJ software (Rasband, 1997-2018).
To identify finer detail on the surface of dehydrated pens we used scanning electron microscopy (S.E.M.) with a Zeiss SUPRA 40vp Gemini Scanning Electron Microscope. Pens were attached to aluminum specimen mount (Ted Pella Inc. cat.# 16111-9, Redding, CA) using double sticky carbon tabs (Ted Pella, Inc. cat.# 16084-20, Redding, CA) and sputter coated with a 6 nm layer of platinum using a Leica EM MED020 high vacuum coating system. Images were captured with the Zeiss SMARTSEM software. Dorsal and ventral surfaces of the squid pens were investigated.
Shell sac ion composition
The ionic composition of the extracellular fluid surrounding the pen was determined to understand the chemical environment in which the pen is constructed. Extracellular fluid was diluted 100 fold and the free ionic concentration was determined, taking into account the dilution. Water content from freshly isolated pens was determined by weighing the hydrated pens in 15mL conical tubes before and after vacuum evaporation of the water in each pen. Deionized water (DI), equal to 100 times the original water content was added back to each tube to rehydrate each pen and dilute the ionic composition before analyzing the solute levels. Ion chromatography and colorimetric assays were performed at the Water Resources Research Center at the University of New Hampshire (Durham, NH). We report the mean ± the standard deviation of the free ion concentration for 5 whole pens.
Shell sac transcriptome
A shell sac transcriptome of Doryteuthis pealeii was prepared by excising both medial and lateral sections (25 mm2) of the ventral shell sac from freshly isolated and dissected adult squid. Total RNA was extracted from the tissue using the RNAqueous kit (Thermofisher Scientific) and converted into libraries using polyA selection with the Illumina TruSeq Stranded mRNA Library preparation Kit LT. Libraries were pooled prior to sequencing. Samples were sequenced using an Illumina HiSeq 2000 flow cell using TrueSeq SBS Version 3 reagents. Illumina sequencing was perfomed using 2 x 150 bp paired-end sequencing cycles. Base calling was done by Illumina Real Time Analysis (RTA) v1.17.21.3 and output of RTA was demultiplexed and converted to FastQ with Illumina Bcl2fastq v1.8.4. Sequencing data from all samples were pooled for assembly. Data went through two stages of read trimming: quality based trimming using Trimmomatic v0.33 followed by K-mer spectral analysis to remove low abundance k-mers using the Khmer 2.0 package. FastQC v0.11.3 was used to check data quality before and after trimming. After filtering, high-quality fragments were used for de novo transcriptome assembly using Trinity v6.0.2. SeqClean was used to trim polyA tails and remove low complexity sequences. For functional annotation, assembled transcripts were blasted against the SWISS-PROT database and best hits with p-values less than 10−3 were selected. All sequencing reads have been deposited into the NCBI small reads archive under the following accession numbers: SRX817965 for the marginal (lateral) epithelial layer and SRX817964 for the ventral (medial) epithelial layer.
RACE
In order to obtain more complete sequence information for the protein with greatest relative abundance we used rapid amplification of cDNA ends (RACE). Total RNA was extracted from the tissue of the lateral, ventral shell sac using RNeasy (Qiagen). cDNA with 5’ and 3’ adapter regions was made using the FirstChoice RLM-RACE kit (Thermofisher Scientific). RACE was achieved using 3’ and 5’ RACE primers provided with the kit and internal primers for the sequence listed below.
| Forward | Reverse |
|---|---|
| GTGCCCCTGGTACTCTCTATT | ATGTGGGCAACAACAGGTTTAT |
| CCTGAAGAACCCGCCAAAT | CGGCTGATACACAAACCTTTT |
| GCCCCAGGAACCGCTTATT |
PCR was performed with LongAmp Taq 2X Master Mix (NEB, Ipswich, MA). Nested priming was performed to reduce nonspecific amplification prior to sequencing. Gel extraction and DNA purification with Monarch DNA gel extraction (NEB) and Monarch PCR and DNA cleanup (NEB) was used, when necessary, to clean up PCR products prior to sequencing by GenScript.
Pen protein extraction and identification
We determined the relative protein content of the pens by comparing the dry weight of whole and pulverized pens before and after treatment with 1 N NaOH to hydrolyze the proteins and minimize deacetylation of chitin (Chaussard and Domard, 2004;Ianiro et al., 2014). Pulverized pens (0.25 g) were dissolved in 40 mL of 1N NaOH and incubated overnight at either room temperature or 95° C. The next day the material was rinsed with NaOH to remove the amino acids before rinsing with DI H2O to remove the NaOH. The wet material was vacuum dried and reweighed. We report the mean percent weight ± the standard deviation.
Whole proteins were removed from pulverized pens with treatments of either 8 M guanidine HCl, or 8 M urea with 5% acetic acid. Pulverized pens (0.25 -0.5g) were incubated in 2 exchanges of 10 mL of extraction buffer for 30 minutes each with gentle agitation. We pelleted the chitin with centrifugation and precipitated proteins from the supernatant using 90% ethanol (guanidine) or 80% acetone (urea). Proteins were resuspended in loading buffer for separation using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE). The lane was cut into fragments to contain all proteins and prepared for shipment according to the instructions provided by the Mass Spectrometry and Proteomics Resource Laboratory (MSPRL) (Harvard University, Cambridge MA). Protein fragments, were made using in-gel digestion with trypsin, separated using HPLC, and identified with the help of MS/MS. The spectral data of the peptides was correlated with sequences from the squid transcriptome to identify pen transcripts using the Proteome Discoverer search engine. All data were filtered to 1% FDR. BLAST was used in Uniprot and NCBI to identify homologous proteins and identify gene ontology (GO) terms, conserved domains and protein similarity. The procedure above would not extract proteins that are covalently bound to chitin and present in other chitinous structures (Andersen, 2010;Tan et al., 2015).
We attempted to remove covalently bound pen proteins using chitin oxidation (Vold and Christensen, 2005) and also selective protein hydrolysis (Tan et al., 2015). Prior to extraction of covalently bound proteins, noncovalently bound proteins were removed by rinsing eight times with 8 M guanidine followed by eight rinses with acetate buffer. This method removed noncovalently bound proteins as assessed by the loss of a protein pellet after the precipitation step. We attempted to remove covalent proteins from chitin using chitin oxidation with 10 mM periodate (Sigma-Aldrich, St. Louis, MO) in 100 mM acetate buffer (pH 4.5) for 30 min at room temperature with gentle agitation. A fraction of the pen chitin is deacetylated and periodate exposure hydrolyzes the deacetylated chitin (Vold and Christensen, 2005). This periodate treatment is gentle on proteins (Woodward et al., 1985). Periodate liberated proteins were precipitated in acetone, separated with SDS PAGE, excised and identified. We also attempted to remove covalently bound proteins using hydroxylamine treatment as it was previously successful in releasing some proteins from the squid beak (Tan et al., 2015). Noncovalently bound proteins were removed from pulverized pen as described above and remaining material was treated with hydroxylamine using the procedure described by Tan et al., (2015). Proteins were precipitated and identified.
Results
Pen structure and development
A whole pen from an adult D. pealeii is illuminated with transmitted unpolarized light (Fig. 1A) and transmitted polarized light (Fig. 1B). The illustration of the adult squid in Fig. 1 shows the location and orientation of the pen (black). The pen is bilaterally symmetric with a thicker, narrow anterior and a broad thinner posterior region known as the vane. The rachis or spine, runs the length of the pen but is thicker toward the anterior. Illumination with polarized light results in bright colors (Fig. 1B). This indicates that the microstructure of the pen is highly birefringent, having two refractive indices, depending on the orientation of the polarization plane. When the polarizer and analyzer are removed, the pen is transparent (Fig. 1A), indicating that there is no pigmentation, no restriction of light by minerals, and the color is created by birefringence only. The greatest birefringence appears in the rachis, anterior region of the pen and lateral edges of the vane. The lack of color in the body of the vane indicates that there is much less birefringence. The rachis and narrow anterior region of the pen have the greatest alignment of macromolecules, however, the body of the vane is less uniform in chitin-protein alignment.
Figure 1.
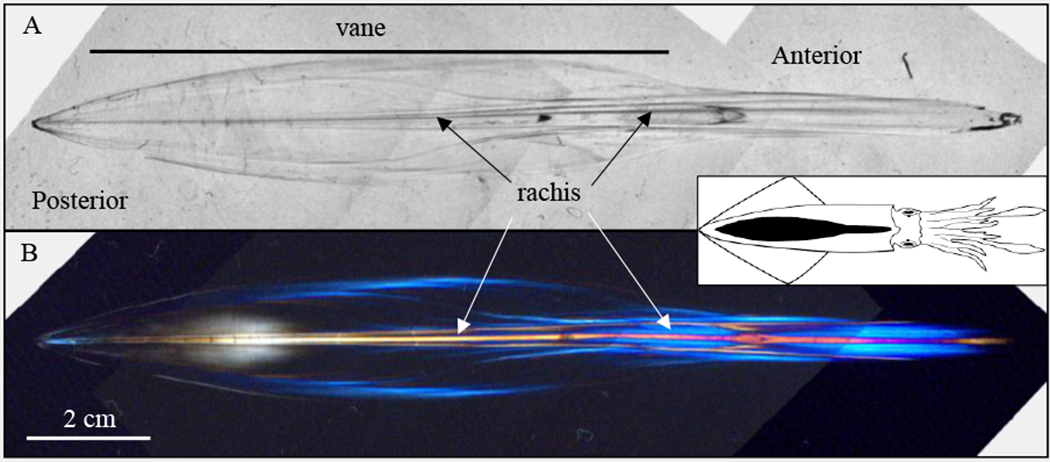
Transmitted light images of an adult pen. (A) Unpolarized, transmitted light illumination of a dried pen reveals a transparent combination of chitin and protein. (B) Polarized light illumination of the pen reveals a great degree of birefringence (colored regions) along the long axis of the pen. The inset shows an illustration of an adult D. pealeii from the dorsal view with the same orientation and with its pen colored black.
The pen is secreted in layers which is visualized with S.E.M. Removal of a small superficial layer of the pen exposes multiple layers beneath, and in close proximity, that are identified by white arrows (Fig. 2A). The shell sac epithelium, indicated by the white triangles, adheres tightly to the dorsal surface of the pen even after attempts to remove it from the pen with M2 (Fig. 2B).
Figure 2.
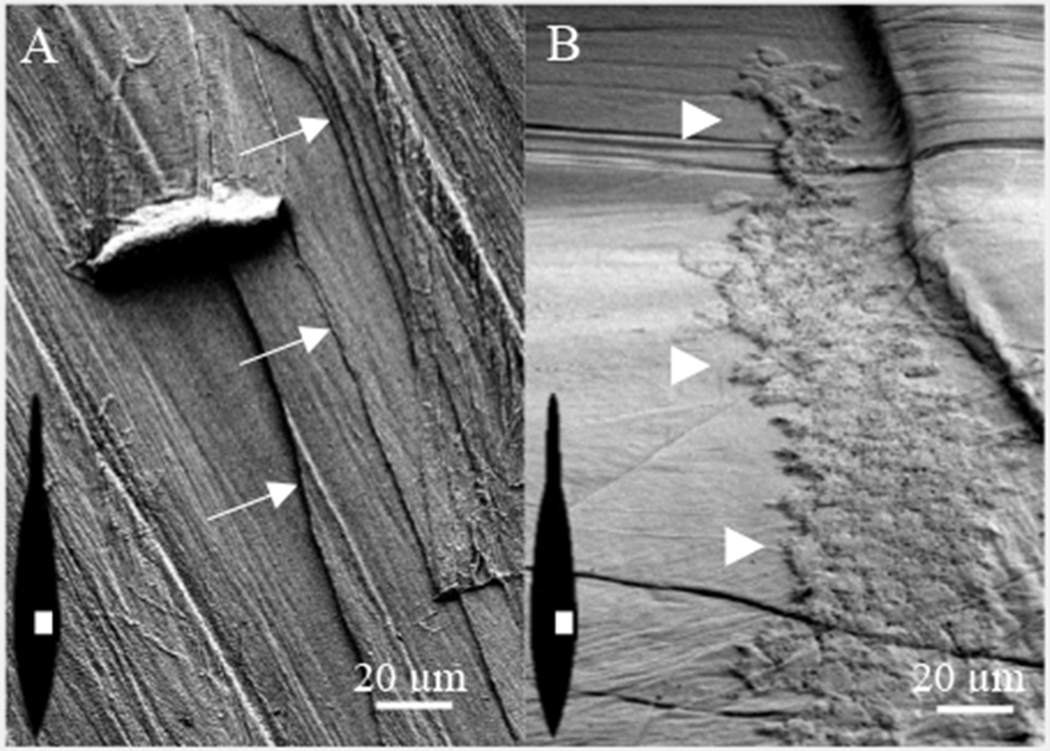
S.E.M of pen surface. (A) The pen is composed of multiple layers identified by white arrows. (B) A region of the shell sac epithelium remains attached to the pen. White triangles identify the left perimeter. The illustrations within the images show the pen orientation (black) and region (white) used to collect the micrographs in A and B.
The birefringence of the adult pen encouraged us to follow development of the embryonic pen in vivo with polarized light. However, the birefringence of the overlaying mantle skeletal muscle overwhelmed the birefringence of the chitin and protein in the pen (not shown). Instead, we characterized pen development using the fluorescent chitin stain, Calcofluor white. Calcofluor selectively stains squid pen with weak staining of surrounding tissues (Fig. 3A–E). While emybronic development of the mantle begins at stage 19 (Arnold, 1965, 1990), we observed the first sign of chitin secretion beneath the mantle at stage 23 (Fig. 3A). Development of the narrow, anterior pen appears first (Fig. 3A). However, further elongation occurs simultaneously with elaboration of the lateral edges of the vane, forming first an inverted ‘V’ at stage 25-26 (Fig. 3B) along the anterior-posterior axis and later an inverted ‘Y’ at stage 27 (Fig. 3C) as the anterior region elongates more rapidly than the posterior. Continued elaboration at stage 28 (Fig. 3D) is associated with elongation of both the narrow, anterior pen and the lateral edges of the wider, posterior vane. The central, most posterior segment of the vane invaginates (Fig. 3D, red arrow). A small, superficial cluster of cells just posterior to this invagination (gray arrow), in a position where the Hoyle organ has been identified (Arnold, 1990;Cyran et al., 2018), stains more brightly with the Calcofluor dye (Fig. 3D) than other surrounding cells. The posterior invagination is no longer present prior to hatching (Fig. 3E). The thinner layers of chitin in the lateral vane stain more faintly than the thicker, central rachis. At hatching, the entire pen structure appears to be a miniature form of the pen in the adult shown above (Fig. 1A). In the hatchling pen, the skeletal muscle of the mantle (red, inset to Fig. 3F) attaches along the lateral edges of the pen (green, inset to Fig. 3F). The pen expands laterally, beneath the edges of the skeletal muscle of the mantle. In embryos as in adults (Ruiz-Cooley et al., 2010;Lorrain et al., 2011) the anterior region of the pen elongates more rapidly than the vane.
Figure 3.
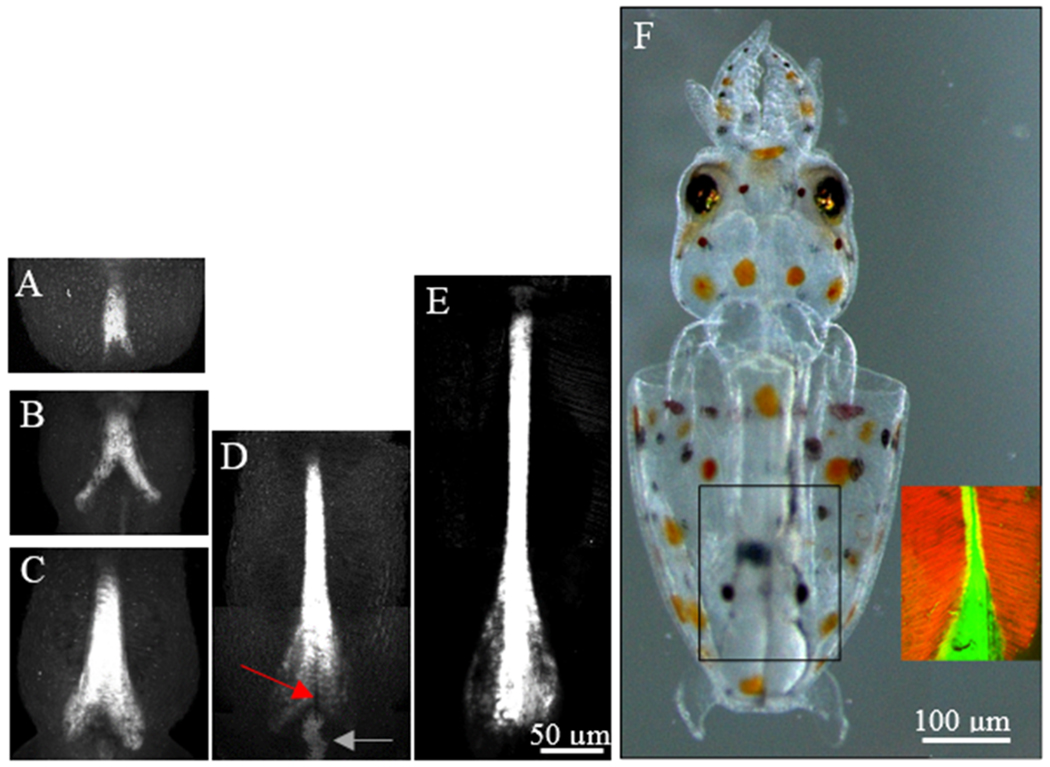
Elongation of the developing pen. Fluorescence imaging of Calcofluor white reveals pen development in fixed embryos. (A-C) Early stages show elaboration of chitin toward the anterior and the posterior vane. (D-E) In later stages, the anterior extends significantly longer while the vane expands and rounds off along the posterior edge. (F) In the hatchling, the actin filaments of the mantle (red) attach to the edges of the rachis and expanded vane of the pen (green) and do not completely encircle the pen.
Shell sac ion composition
The images presented above show that pens from D. pealeii lack significant mineralization. Does the shell sac insulate the pen from mineralization, or do the pen macromolecules prevent mineralization? To begin to address this question we determined the inorganic composition of the extracellular fluid surrounding the pen. We find that nearly half the weight of freshly extracted pens is water, 48.5 ± 2.6% (n=10 whole pens). The shell sac enclosed fluid contains twice the concentration of free Ca2+, the same concentration of K+ and a little more than half as much Na+, Mg2+, and Cl- compared to squid plasma that is similar to seawater (Table 1). Based on the concentration of these inorganic ions, the shell sac fluid is close to 60% of the osmolality of the squid plasma. The shell sac maintains an ionic environment that is chemically distinct from the plasma and extracellular fluid in the squid. Despite the lack of mineralization, Ca2+ is enriched in the shell sac, leading us to hypothesize that protein modifications help to prevent mineralization of the pen.
Table 1.
Ionic composition of select fluids.
| Solute | Seawatera | Squid Plasmaa | Shell Sac | Extrapallialb Fluid |
|---|---|---|---|---|
| Na+ | 460 | 440 | 235.6 ± 21.7 | 442.3 ± 16.2 |
| K+ | 10 | 20 | 21.1 ± 3.2 | 9.5 ± 1.1 |
| Ca2+ | 10 | 10 | 19.5 ± 1.0 | 11.1 ± 1.4 |
| Mg2+ | 53 | 54 | 36.4 ± 3.2 | 58.3 ± 6.6 |
| Cl− | 540 | 560 | 311.5 ± 27.3 | 476.3 ± 14.4 |
| SO42− | --- | --- | 2.5 ± 0.6 | 23.6 ± 3.0 |
| PO4− | --- | --- | 4.3 ± 1.4 | --- |
(Hodgkin, 1958)
Pen protein extraction and characterization
As the β-chitin of the pen has emerged as a renewable commercial resource, its associated proteins are stripped away to enable applications for raw chitin (Ianiro et al., 2014;Cuong et al., 2016). The proteins have largely been ignored. In order to measure the total contribution of protein to the pen, alkaline hydrolysis of pulverized pens (M1 and M2) was used to remove proteins. When the treatment was performed at room temperature we found that D. pealeii pens are composed of 33.9 ± 0.01% chitin and 66.1 ± 0.01% nonchitinous material (n=6). The nonchitinous material is dominated by proteins (Hunt and Nixon, 1981). We also performed alkaline hydrolysis at 95°C yielding a slightly elevated estimate of nonchitinous material 71.2 ± 0.5% (n=4), which may reflect either a greater hydrolysis of proteins than performed at room temperature or may reflect deacetylation of chitin into chitosan.
In order to identify microscopic differences between whole pens with and without proteins (extracted using guanidine) we imaged the lateral edges of the vane of treated pens with with S.E.M. The fiber alignment running top to bottom (Fig. 4) is parallel to the anterior-posterior axis. Under lower magnification (200x) the pen surface with proteins (Fig. 4A) is difficult to discriminate from the pen surface without proteins (Fig. 4B). However, under higher magnification (1500x), the pen surface with proteins (Fig. 4C) is roughened compared to the smooth, glassy appearance of the surface of the pen obtained in the absence of proteins (Fig. 4D).
Figure 4.
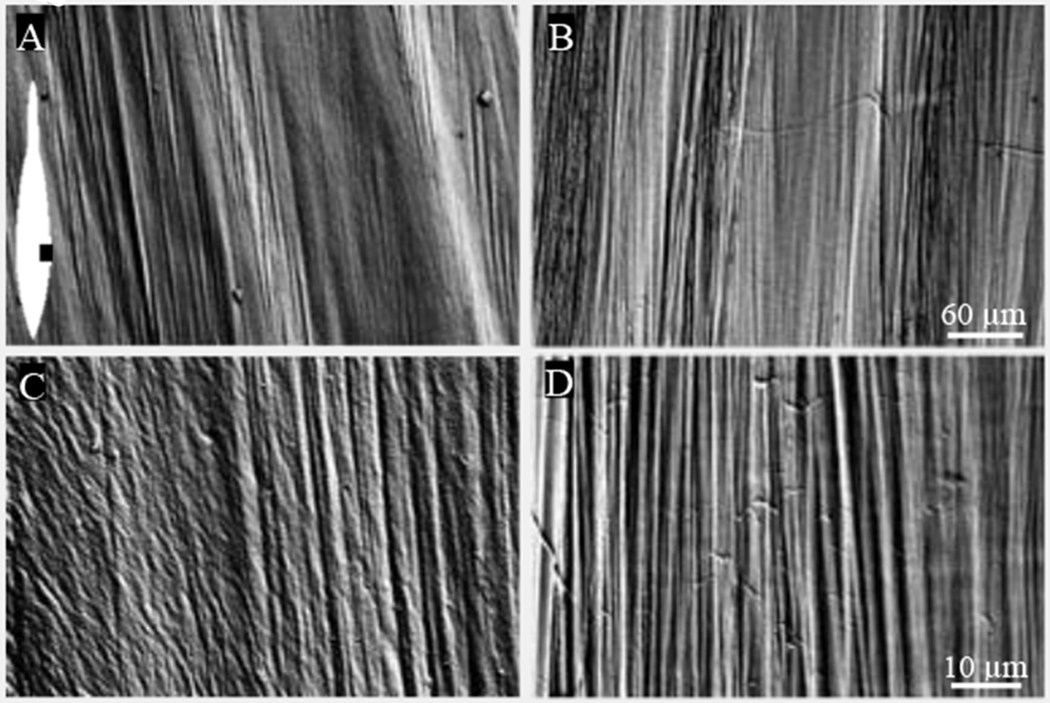
Surface structure of the lateral pen in the presence and absence of proteins. (A,B) Low magnification S.E.M shows little difference in surface structure between an untreated (A) and a protein-extracted pen (B). The inset in A shows an illustration of the pen (white) with similar orientation and the region of the pen (black) used to collect the micrographs. (C,D) Higher magnification shows a change from a roughened, fibrous coating in the untreated pen (C) to a smooth, glassy appearance in the protein-extracted pen (D).
We further characterized the organic composition of pens by extracting proteins for identification. Protein extraction with either guanidine or acidic urea led to similar banding patterns when compared with SDS-PAGE, however, guanidine treatment showed greater quantities of extracted protein. Therefore, guanidine treatment was used for further extractions.
Using the partial sequences from the shell sac transcriptome and multiple iterations of protein extraction methods we explored the identity of pen proteins using MS/MS of proteins excised from the polyacrylamide gels. Removal of proteins from pens (M1) led to identification of 309 shell sac sequences from the protein fragments. We also observe that guanidine extraction removes only 36.3 ± 0.02% (n =3), of the mass of the nonchitinous material compared to alkaline hydrolysis, leading us to hypothesize that a majority of the pen proteins are either covalently cross-linked to the chitin fibers or immobilized within the chitin nanofibril meshwork. Some of the most abundant proteins of this extraction include actin, myosin and tropomyosin, along with a long list of other intracellular proteins indicating substantial cellular material from tissues outside of the pen including the mantle muscle.
In order to reduce proteins from tissues outside of the pen we took a more rigorous approach to removing them by scrubbing pens with a plastic bristled brush (M2). After this treatment, the number of intracellular proteins was significantly reduced and is shown in Fig. 5. The identified pen proteins are listed in Table S1, available online, according to the area of the protein fragments in the ion chromatograms. Of the 144 unique transcripts identified by protein fragments, over 60% are intracellular proteins, leading us to conclude that the brushed pens still contained a substantial amount of attached cellular material. This statement is supported by S.E.M. in Fig. 2B.
Figure 5.
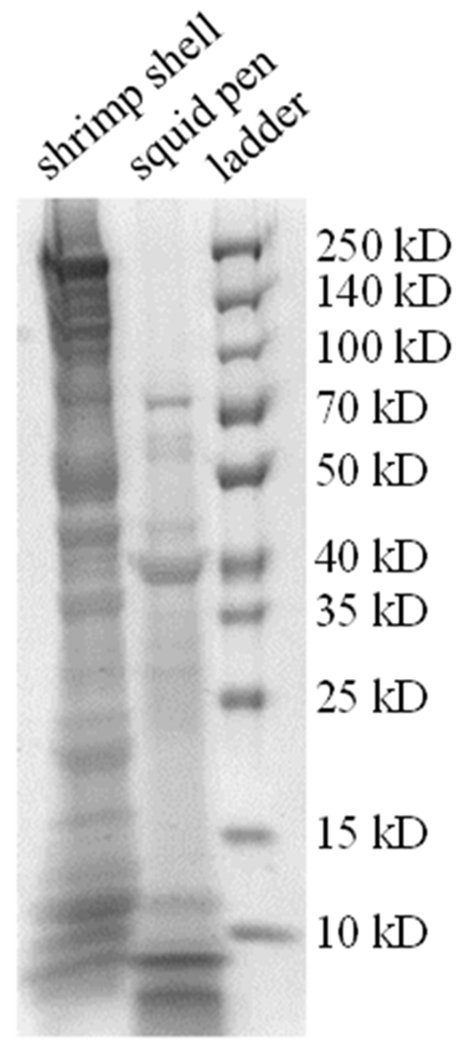
SDS PAGE of guanidine extracted proteins from squid pens and shrimp shells. The extracted proteins from the pens are shown for comparison against extracted proteins from white shrimp shell, a mineralized chitinous structure.
A further attempt to remove intracellular proteins was made by removing the superficial proteins with repeated rinses of guanidine followed by rinses with acetate buffer (pH 4.5). The remaining material was submerged in 10 mM periodate in acetate buffer, which oxidizes carbon-carbon bonds of the chitin glucose backbone (Vold and Christensen, 2005) and should release proteins trapped between the chitin nanofibrils. With these combined treatments, only twenty proteins were extracted from the interior of the pulverized pen fragments. All of these proteins were identified in the two earlier protein extractions, and are in relatively high abundance in the whole pen protein extraction listed in Table S1, available online. Table S2, available online, contains the protein fragments used in combination with the transcriptome to identify the proteins.
In order to further explore the possibility that other pen proteins may be covalently linked to the chitin nanofibrils we used hydroxylamine, a treatment which cleaves peptide bonds between asparagine and glycine. This method was successful in identifying proteins covalently cross-linked in the beak of the Humboldt squid (Tan et al., 2015). After removing noncovalently bound proteins with guanidine, pulverized pens (M2) were exposed to hydroxylamine and the protein fragments were isolated and identified. Covalently bound proteins that were not liberated by the guanidine treatments alone but released due to hydroxylamine treatment, would yield unique proteins. However, no unique proteins were identified in this manner. The results of the periodate and hydroxylamine-based protein extractions, lead us to conclude that pen proteins are not covalently bound to the chitin but rather remain trapped within the tight fibrous mesh of the chitin nanofibrils and are not fully released by guanidine treatments.
We classified the twenty predicted proteins (PPs) embedded within the pen into 6 different groups; chitin associated, unknown, protease inhibitor, intracellular, protease, and extracellular matrix. We designate the proteins as PPs due to the fact that the transcriptome nucleotide sequences used to identify them are incomplete. In Table 2, PPs are listed first by group and then by relative abundance within that group according to the mass spectrometry results. Except for PP 1 and the unknown PPs 9-13, the e-scores for amino acid alignment were less than 10−10 when compared with sequences on Uniprot.
Table 2.
Proteins embedded in the pen.
| PP | Transcript ID | Category | GO Terms | Conserved Domains | Protein Similarity | Relative Abundance | Area |
|---|---|---|---|---|---|---|---|
| 1 | 52508.tr74186 | chitin associated | chitin binding, chitin metabolic process | CBD2 (4) | unknown | 1 | 2.4*109 |
| 2 | 38507.tr70062 | chitin associated | hydrolase activity (acting on carbon-nitrogen but not peptides) | NodB homology | chitin deacetylase | 5 | 1.0*107 |
| 3 | 49689.tr48900 | chitin associated | chitin binding, chitin metabolic process, | CBD2 (1), Glyco-18 |
chitinase | 6 | 9.2*106 |
| 4 | 54994.tr85459 | chitin associated | chitin binding, chitin metabolic process | CBD2 (6) | chondroitin proteoglycan 2 | 7 | 9.0*106 |
| 5 | 59952.tr135549 | chitin associated | chitin binding, chitin metabolic process | CBD2 (2), vWFA, C-type lectin | perlucin, protein PIF | 9 | 3.3*106 |
| 6 | 54996.tr85459 | chitin associated | chitin binding | CBD2 (1) | chitin binding | 10 | 3.2*106 |
| 7 | 54997.tr85459 | chitin associated | chitin binding, chitin metabolic process, | CBD2 (5) | uncharacterized | 13 | 1.8*106 |
| 8 | 94003.tr63364 | chitin associated | chitin binding, chitin metabolic process | CBD2 (1) | CBP-1 gene | 20 | 5.3*105 |
| 9 | 31568.tr142139 | unknown | none | none | none | 2 | 7.1*108 |
| 10 | 27116.tr45562 | unknown | none | none | none | 3 | 2.9*108 |
| 11 | 55008.tr156806 | unknown | none | none | none | 8 | 6.1*106 |
| 12 | 66474.tr139103 | unknown | none | none | none | 15 | 1.1*106 |
| 13 | 32788.tr132141 | unknown | none | none | none | 19 | 6.2*105 |
| 14 | 29086.tr108669 | protease inhibitor | serine-type endopeptidase inhibitor activity, negative regulation of endopeptidase activity | BPTI/Kunitz inhibitor (2) | uncharacterized | 11 | 3.0*106 |
| 15 | 32979.tr123994 | protease inhibitor | none | Thyroglobulin type I (1) | testican-3 | 14 | 1.4*106 |
| 16 | 26810.tr140091 | protease inhibitor | serine-type endopeptidase inhibitor activity, negative regulation of endopeptidase activity | BPTI/Kunitz inhibitor (2) | uncharacterized | 17 | 8.8*105 |
| 17 | 65783.tr161718 | intracellular | none | Fact-Spt16_Nlob, SPT16, Rtt106, | FACT complex | 16 | 1.0*106 |
| 18 | 47072.tr157054 | intracellular | structural molecule activity, intermediate filament | none | omega-crystallin | 18 | 6.4*105 |
| 19 | 47505.tr84302 | protease | metalloendopeptidase activity, metallopeptidase activity, proteolysis | peptidase M12B | ADAMTS | 4 | 3.7*107 |
| 20 | 59603.tr98346 | extracellular matrix | extracellular matrix structural constituent, collagen trimer | fibrillar collagen NC1, vWFC | collagen | 12 | 2.3*106 |
CBD2 - chitin binding type 2 domain, vWF(A,C) - von Willebrand factor domain, BPTI - bovine pancreatic trypsin inhibitor, Glyco-18 - Glycoside hydrolase family 18,
The largest group of PPs, 1-8, is associated with chitin binding or chitin modification including chitin cross-linking proteins, a chitinase and a chitin deacetylase. PPs 1 and 4-8 are homologous to proteins with multiple chitin binding domains (CBDs) but have weaker homology to chitinases or chitin deacetylases that may identify them as chitin cross-linking proteins. PPs 1, 4 and 7 are homologous to proteins with 10 - 25 CBDs. The number of CBDs contained within each of the partial transcripts, is listed in parentheses under the Conserved Domains column, for the chitin binding proteins in Table 2.
The translated ORF for PP 1 contains four chitin binding domains (CBDs) and has highest homology with chitin binding proteins from the sea squirt, Ciona. Regions of PP 1 do not show sufficient homology with the conserved CBD2 domain to be flagged during a conserved domain search. In part, this may be due to the fact that the CBDs of PP 1 contain 4 cysteines rather than 6. The cysteines form disulfide bridges that are reported to promote binding to the chitin (Shen and Jacobs-Lorena, 1998;Tjoelker et al., 2000). The amino acid alignment shows a repeating, conserved M*CAPG region just before the 4th cysteine of the Ciona sequences. (* represents amino acids with aromatic side groups)
PP 5 and 8 are chitin binding proteins with sequence homology to proteins involved with nacre formation and nucleating the inorganic component of mollusc shells (Blank et al., 2003) and a chitin binding protein found in the sclerotized beak of the giant squid (Tan et al., 2015), respectively. While PP 6 has greatest homology with proteins containing domains with hydrolase activity and activity for hydrolyzing O-glycosyl compounds, common to the chitin modifying proteins, the partial sequence of the transcript contains only a CBD2 so we list it conservatively, as a chitin binding protein.
Two other chitin associated proteins, have homology with enzymes leading to chitin hydrolysis or deacetylation. PP 2, the protein with the 5th greatest relative abundance, is homologous to a deacetylase. Chitin deacetylases remove the 2 carbon, acetyl group and expose an amine which is positively charged at neutral pH. This may help prevent mineralization of the pen by repelling Ca2+ but also might promote ionic interactions with surrounding negatively charged proteins. In the absence of a deacetylase, insect cuticle is less compact and animals die at molting (Yu et al., 2016). PP 3, has a conserved glycoside hydrolase domain (GH18) common to chitinases, and also a chitin binding domain. While this protein has homology with chitotriosidase, a chitinase involved with the innate immune system in vertebrates, we think that its relative abundance and location within the pen indicates that it may be playing a role in remodeling of the pen chitin. Whether it still acts as a chitinase deep within the pen or has some important secondary structural role, remains unanswered.
The second largest group contains five PPs, 9-13, and is composed of proteins with low homology to any known proteins based on low e-scores or are homologous to other uncharacterized proteins with no conserved domains. Large numbers of shell proteins remain unknown for similar reasons (Mann and Jackson, 2014;Jackson et al., 2015). Proteins with the second and third greatest relative abundance belong to this category.
Three proteins, PP 14-16, have homology with protease inhibitors. PPs 14 and 16 contain multiple, bovine pancreatic trypsin inhibitor (BPTI)/Kunitz domains. These domains are involved with inhibition of serine proteases, act as anti-coagulant factors, and are involved in defense against microbial pathogens (Ranasinghe and McManus, 2013). A protein with similar domains is required for collagen biosynthesis in the chitinous cuticle of C. elegans (Stepek et al., 2010). PP 15, however, has homology to testican-3, a secreted proteoglycan which is involved in regulation of extracellular protease cascades (Nakada et al., 2001).
Two intracellular proteins, 17 and 18, were found in relatively low abundance after the periodate treatment. PP 17, a FACT complex, is a nuclear histone chaperone involved with replication, transcription and DNA repair (Winkler and Luger, 2011). In the pen, the FACT complex may be a result of remaining shell sac epithelium. PP 18, an intermediate filament protein, has homology to omega crystallin, a protein found in the lens and cornea of the eye (Vasiliou et al., 2013).
PP 19 appears to be a lone protease embedded in the pen from the ADAMTS family (a disintegrin and metalloproteinase with thrombospondin motif) and is present in 4th greatest relative abundance. These proteases are secreted proteins known for modulating extracellular proteins including processing of procollagen to collagen, and cleavage of extracellular proteoglycans and von Willebrand factor (Porter et al., 2005). PP 19 has highest homology with ADAMTS8 which has been shown to cleave extracellular proteoglycans and possess anti-angiogenic properties (Vazquez et al., 1999;Collins-Racie et al., 2004). ADAMTS proteins are involved with ecdysis, shedding of the outer cuticle, and metamorphosis in the silkworm (Ote et al., 2005;Kawasaki et al., 2018). Remodeling of the squid pen may be similar to remodeling of the insect cuticle.
PP 20 is homologous to collagen, an extracellular matrix protein found in load-bearing structures such as tendons or ligaments. While collagen was inferred as a surface protein on the pen of I. argentinus (Wu et al., 2003) our results indicate that collagen is also embedded within the pen. Its relative abundance increased significantly between total pen protein, Table S1, available online, and proteins extracted after periodate treatment, Table 2. This indicates that collagen is found in relatively higher abundance within the pen than on the surface of the pen. PP 20 may interact with PP 5, which has both chitin and collagen binding domains, to crosslink chitin and collagen within the pen.
A number of other interesting proteins surrounding the pen were identified prior to the periodate extraction of proteins. These proteins are listed in Table 3, including 7 more chitin associated proteins, 2 more proteins with BPTI/Kunitz domains, many forms of collagen, and proteins with homology to shell matrix proteins including galaxin and nidogen.
Table 3.
Superficial pen proteins.
| Transcript ID | Category | GO Terms | Conserved Domains | Protein Similarity | *Relative Abundance | Area |
|---|---|---|---|---|---|---|
| 45265.tr155449 | chitin associated | hydrolase activity (acting on carbon-nitrogen but not peptide bonds) | NodB homology | chitin deacetylase | 18 | 5.2*108 |
| 65756.tr73867 | chitin associated | chitin binding, chitin metabolic process, extracellular region, integral membrane component | SEA, CBD2 | 32 | 5.0*107 | |
| 39347.tr275096 | chitin associated | catalytic activity, hydrolase activity (acting on carbon-nitrogen but not peptides) | NodB homology chitinase, chitin binding, chitin catabolic process, extracellular region | chitin deacetylase | 37 | 4.2*107 |
| 55680.tr97066 | chitin associated | integral part of membrane, chitin binding, chitin metabolic process, extracellular region, | CBD2 | 62 | 1.3*107 | |
| 43085.tr17217 | chitin associated | hydrolase activity, chitin binding | Glyco_18 | acetylchitobiase | 124 | n.a. |
| 9423.tr17450 | chitin associated | chitin binding, chitin metabolic process, extracellular region, integral membrane component, calcium ion binding | CBD2, SEA, EGF-like | 138 | n.a. | |
| 51377.tr107208 | chitin associated | serine-type endopeptidase inhibitor activity, chitin binding, peptidase activity, peptidase inhibitor activity, | WAP, BPTI/Kunitz inhibitor, Ig-like, PLAC, CBD2, Fibronectin type-III, | papilin | 143 | n.a. |
| 56294.tr390985 | protease inhibitor | serine-type endopeptidase inhibitor activity, negative regulation of endopeptidase activity | BPTI/Kunitz inhibitor | uncharacterized | 29 | 6.2*107 |
| 58162.tr107664 | plasma membrane | membrane, integral component of membrane | EGF-like | nidogen | 69 | 1.1*107 |
| 61874.tr9545 | plasma membrane | calcium ion binding, cell-matrix adhesion, membrane | NIDO,Nidogen G2 Beta-barrel, EGF-like, LDL-receptor class B | nidogen | 121 | 5.6*105 |
| 63972.tr52534 | extracellular | none | none | galaxin | 33 | 4.9*107 |
| 41827.tr29969 | extracellular | none | none | galaxin | 35 | 4.6*107 |
| 29662.tr119876 | extracellular matrix | calcium ion binding | vWFA, EGF-like | collagen | 79 | 8.0*106 |
| 62922.tr246504 | extracellular matrix | none | Lam G Domain | collagen α1 | 80 | 7.6*106 |
| 59233.tr145596 | extracellular matrix | extracellular matrix constituent, collagen trimer, | Collagen IV NC1, 4 disulfide bonds | collagen | 90 | 5.2*106 |
| 43429.tr227065 | extracellular matrix | extracellular matrix structurual constituent, collagen trimer | Collagen IV NC1 | collagen α2 (IV) | 99 | 4.0*106 |
| 3733.tr66993 | extracellular matrix | none | vWFA | collagen | 127 | n.a. |
Relative abundance with relation to all proteins extracted with guanidine after M2.
SEA, Sea urchin sperm protein, Enterokinase, Agrin
vWF(A) von Willebrand Factor A,
WAP-type ‘four-disulfide core’ domain
Discussion
Microscopic examination of adult and embryonic pens of D. pealeii supports the absence of sclerotization or mineralization as found in pens from other species of squid (Subhapradha et al., 2013). The birefringence of the D. pealeii pen (Fig. 1B) and the scanning electron micrographs (Figs. 2 and 4) support an anterior-posterior alignment of the chitin-protein composite along the rachis, the narrow anterior portion of the pen, and edges of the vane similar to the alignment in other species (Yang et al., 2014). The pen does not show the ‘twisted plywood’ arrangement of crustacean exoskeletons (Raabe et al., 2005) and is not porous or pitted as found in insects (Schroder-Turk et al., 2011;Kaya and Baran, 2015). In a single layer, the chitin nanofibrils appear in close juxtaposition even after removal of surface proteins (Fig. 4D).
The lamellar structure of the pen (Fig. 2A) is very similar to that found in the near-tip region of the squid beak (Miserez et al., 2007) and the shells of the sea snail (Bezares et al., 2012), with very little space between layers. The tight arrangement of nanofibrils in a single layer, remains even after alkaline hydrolysis of proteins, however the layers of chitin now appear disconnected (Wu et al., 2003;Ianiro et al., 2014). Based on these observations we hypothesize that chitin nanofibril interactions between layers have greater dependence on proteins than nanofibril interactions within a layer.
We have shown that the embryonic shell sac, mantle and pen are very thin, enabling 3D imaging of pen morphology using Calcofluor white (Fig. 3) in combination with multi-photon microscopy. In vivo gene knockdown and Calcofluor imaging will provide insight into the function of some of the proteins involved with pen construction. Moreover, transgene expression of fluorescently-tagged proteins should enable stage-dependent localization of important proteins.
The ionic environment in which chitin nanofibrils and proteins assemble is distinct from squid plasma and extracellular fluid (Table 1). The inorganic solutes in the plasma with the greatest osmotic activity, Na+ and Cl−, are reduced in the pen. While this may indicate that the shell sac fluid is hypotonic to plasma, it may also indicate that these inorganic solutes have been replaced with small organic solutes (Wright, 1995). Further analysis could confirm this hypothesis.
An unexpected finding is that the Ca2+ concentration surrounding the non-mineralized pen is twice as great as the Ca2+ concentration in the extrapallial fluid between the shell and the mantle epithelium of three marine bivalve molluscs that produce mineralized shells, Table 1 (Crenshaw, 1972). Even with the higher surrounding Ca2+ concentration, the pen remains uncalcified. This may be due to the relatively larger fraction of hydrophobic amino acids and reduction of the acidic amino acids as reported in pens from other species (Hackman, 1960;Hunt and Nixon, 1981;Wu et al., 2003;Cuong et al., 2016). For comparison, mineralized mollusc shells and bone contain proteins enriched with acidic amino acids, and amino acids containing negatively charged sulfates and phosphates which are used to concentrate Ca2+ and are necessary for nucleation on the structured carboxylate domains (Weiner and Hood, 1975;Addadi et al., 1987;George and Veis, 2008). Consistent with this hypothesis is that the free sulfate concentration in the pen is 10 fold lower than the average concentration in the extrapallial fluid of the bivalve molluscs listed in Table 1 (Crenshaw, 1972). Further experiments to obtain full length pen proteins for D. pealeii will be necessary to confirm this hypothesis.
Chitin nanofibrils of pens have been characterized as being surrounded by proteins with helical and coiled-coil structures (Yang et al., 2014). We identified seven proteins embedded in the pen with these properties including collagen, which is a supercoiled, triple helix (Beck and Brodsky, 1998). We also found other proteins which have α-helical secondary structures including ADAMTS (Gerhardt et al., 2007), chitinase (Hahn et al., 2000), chitin deacetylase (Liu et al., 2017), proteins with Kunitz domains (Dai et al., 2012) and omega crystallin (Vasiliou et al., 2013). Additional helical or coiled secondary structures may be found in the five proteins that have not been identified.
The largest groups of proteins associated with the pen are the chitin associated proteins. Seven of the twenty embedded proteins (Table 2) and an additional four proteins associated with the surface of the pen (Table 3) have chitin binding domains. PP 1, the protein embedded in the pen with the greatest relative abundance, appears to possess multiple chitin binding domains but does not appear to have catalytic domains for remodeling the secreted chitin, similar to five other proteins embedded in the pen. Additionally, no unique proteins were extracted from the pulverized pen during periodate or hydroxylamine treatment, leading us to conclude that proteins in the pen are not covalently crosslinked to chitin. Considering that the strength of the pen is derived from its careful construction of chitin and protein in the absence of sclerotization, a number of different chitin binding proteins might have different uses. For example, shorter chitin binding proteins with few chitin binding domains might crosslink individual chitin nanofibrils, while longer proteins with many more chitin binding domains might bundle multiple chitin nanofibrils as postulated for the chitinous peritrophic matrix of the invertebrate intestinal lining (Merzendorfer et al., 2016).
We hypothesize that a significant amount of chitin and protein modification is occurring after the macromolecules are released from the ventral shell sac. This is based on the chitin modifying proteins that were present embedded in the pen and on the surface of the pen and also, the presence of a protease, known for modulating extracellular matrix proteins, and protease inhibitors. Understanding these mechanisms will help to clarify the complex biomolecular self-assembly used to construct such a highly polarized structure by so many different cells in the epithelium.
In the pens of L. vulgaris, proline was reported to be in its hydrophobic form and was not hydroxylated (Hackman, 1960;Hunt and Nixon, 1981) as commonly found in collagen. This may be due to the fact that proline is enriched in the more abundant proteins in the pen and overwhelms the hydroxyproline from the collagen which appears at relatively lower levels in the pen, Table 2. Alternatively, the proline on the collagen may not be hydroxylated. Microbial collagen and recombinant collagen still form triple helices in the absence of hydroxylated proline (Perret et al., 2001;Mohs et al., 2007). The increased hydrophobicity of the collagen in this state, may increase interactions with the hydrophobic acetyl groups of chitin and also reduce hydration of collagen although it does not prevent folding. Like proline, glycine is also enriched (Hackman, 1960;Hunt and Nixon, 1981;Wu et al., 2003;Cuong et al., 2016). Proline and glycine are often found at turns or in protein loop structures consistent with the observation that pen proteins surrounding the chitin have helical and coiled-coil structures (Yang et al., 2014).
When compared to other chitinous structures, the pen appears to be one of the simplest arrangements of chitin and protein which is flexible and still maintains its strength. The protein composition of the squid pen shares a great deal of similarity with proteins from other chitinous structures. However, in the absence of sclerotization and mineralization, the proteins are easier to collect, identify and study. Further studies will help elucidate the complex mechanisms of chitin-protein structure and function, and the self-assembly that occurs to make the highly organized pen. Comparison of uncalcified squid pens with calcified squid pens (Subhapradha et al., 2013) or cuttlefish bone (Le Pabic et al., 2017) would also be useful in understanding protein modifications used to promote biomineralization. A completed D. pealeii genome will be most beneficial in helping to characterize and identify the unknown proteins described above.
Supplementary Material
Figure 6.
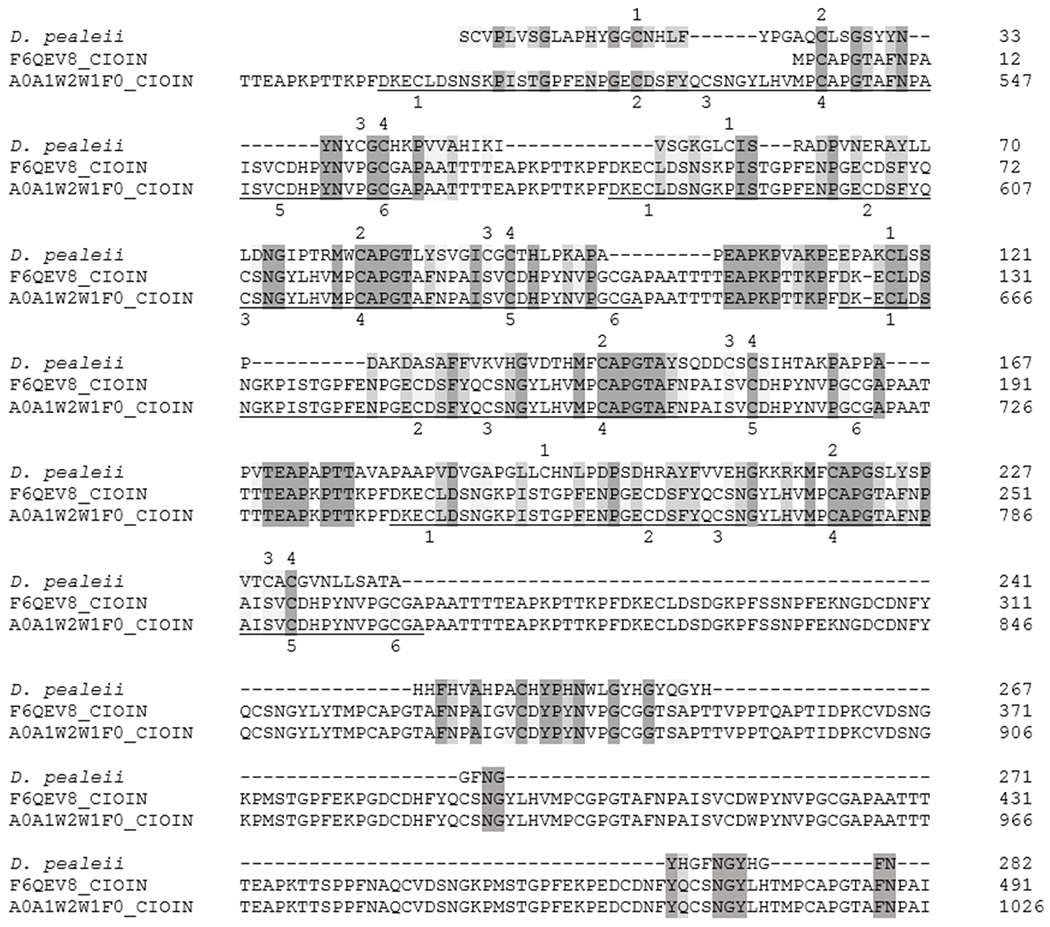
Sequence alignment between the protein with greatest relative abundance in the D. pealeii pen (PP 1) and homologous C. intestinalis sequences (F6QEV8 length 570, A0A1W2W1F0, length 1105). Numbers at the end of each line indicate the amino acid position in the sequence. Dark shading represents conserved residues while lighter shading represents conservation between groups with strongly similar properties. The underlined regions are repeating, CBD2 domains. Numbers above the D. pealeii sequence mark the 4 conserved cysteine residues while the numbers below the C. intestinalis sequences mark the 6 conserved cysteine residues in the CBD2 domains.
Acknowledgements
We thank John Dowling for financial support. We thank Kasia Hammar and Louie Kerr of the MBL Central Microscopy Facility for help obtaining scanning electron micrographs. We thank Bogdan Budnik and Renee Robinson from the MSPRL for their help and advice with protein identification. We thank Shin-Yi Marzano and Chenchen Feng of SDSU for help with RACE. Funding for this work was provided by the Eugene and Millicent Bell Fellowship Fund in Tissue Engineering (MM), an ABS Undergraduate Research Award (KB), NIH grant R01 GM101701 (MS), NSF IOS1557748 (JR) and BSF 2013094 (JR).
Literature Cited
- Addadi L, Moradian J, Shay E, Maroudas NG, and Weiner S. 1987. A chemical model for the cooperation of sulfates and carboxylates in calcite crystal nucleation: Relevance to biomineralization. Proc. Natl. Acad. Sci. U. S. A 84:2732–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SO 2010. Insect cuticular sclerotization: A review. Insect Biochem. Mol. Biol 40:166–178. [DOI] [PubMed] [Google Scholar]
- Arnold JM 1965. Normal embryonic stages of the squid Loligo pealeii (Lesueur). Biol. Bull 128:24–32. [Google Scholar]
- Arnold JM 1990. Embryonic development of the squid. Pp. 77–90 in Squid as Experimental Animals, Gilbert DL, Adelman WJ and Arnold JM, eds. Springer, Boston, MA. [Google Scholar]
- Beck K, and Brodsky B. 1998. Supercoiled protein motifs: The collagen triple-helix and the α-helical coiled coil. J. Struct. Biol 122:17–29. [DOI] [PubMed] [Google Scholar]
- Bezares J, Asaro RJ, and Lubarda VA. 2012. Core structure of aligned chitin fibers within the interlamellar framework extracted from Haliotis rufescens nacre. Part I: implications for growth and mechanical response. Theor. Appl 39:343–363. [Google Scholar]
- Bizikov VA 1995. Growth of Sthenoteuthis oualaniensis, using a new method based on gladius microstructure. ICES Mar. Sci. Symp 199:445–458. [Google Scholar]
- Blank S, Arnoldi M, Khoshnavaz S, Treccani L, Kuntz M, Mann K, Grathwohl G, and Fritz M. 2003. The nacre protein perlucin nucleates growth of calcium carbonate crystals. J. Microsc 212:280–291. [DOI] [PubMed] [Google Scholar]
- Chaussard G, and Domard A. 2004. New aspects of the extraction of chitin from squid pens. Biomacromolecules 5:559–564. [DOI] [PubMed] [Google Scholar]
- Cole AG, and Hall BK. 2009. Cartilage differentiation in cephalopod molluscs. Zoology 112:2–15. [DOI] [PubMed] [Google Scholar]
- Collins-Racie LA, Flannery CR, Zeng W, Corcoran C, Annis-Freeman B, Agostino MJ, Arai M, DiBlasio-Smith E, Dorner AJ, Georgiadis KE, Jin M, Tan X-Y, Morris EA, and LaVallie ER. 2004. ADAMTS-8 exhibits aggrecanase activity and is expressed in human articular cartilage. Matrix Biol 23:219–230. [DOI] [PubMed] [Google Scholar]
- Cortizo MS, Berghoff CF, and Alessandrini JL. 2008. Characterization of chitin from Illex argentinus squid pen. Carbohydr. Polym 74:10–15. [Google Scholar]
- Crenshaw MA 1972. The inorganic composition of molluscan extrapallial fluid. Biol. Bull 143:506–512. [DOI] [PubMed] [Google Scholar]
- Cuong HN, Minh NC, Hoa NV, and Trung TS. 2016. Preparation and characterization of high purity β-chitin from squid pens (Loligo chenisis). Int. J. Biol. Macromol 93:442–447. [DOI] [PubMed] [Google Scholar]
- Cyran N, Palumbo A, Klepal W, Vidal EAG, Staedler Y, Schönenberger J, and von Byern J. 2018. The short life of the Hoyle organ of Sepia officinalis: formation, differentiation and degradation by programmed cell death. Hydrobiologia 808:35–55. [Google Scholar]
- Dai S. x., Zhang A, and Huang J. 2012. Evolution, expansion and expression of the Kunitz/BPTI gene family associated with long-term blood feeding in Ixodes scapularis. BMC Evol. Biol 12:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SN, Madhuprakash J, Sarma PVSRN, Purushotham P, Suma K, Manjeet K, Rambabu S, El Gueddari NE, Moerschbacher BM, and Podile AR. 2015. Biotechnological approaches for field applications of chitooligosaccharides (COS) to induce innate immunity in plants. Crit. Rev. Biotechnol 35:29–43. [DOI] [PubMed] [Google Scholar]
- Di Martino A, Sittinger M, and Risbud MV. 2005. Chitosan: A versatile biopolymer for orthopaedic tissue-engineering. Biomaterials 26:5983–5990. [DOI] [PubMed] [Google Scholar]
- Fernandez JG, and Ingber DE. 2013. Bioinspired chitinous material solutions for environmental sustainability and medicine. Adv. Funct. Mater 23:4454–4466. [Google Scholar]
- Fernandez JG, and Ingber DE. 2014. Manufacturing of large-scale functional objects using biodegradable chitosan bioplastic. Macromol. Mater. Eng 299:932–938. [Google Scholar]
- George A, and Veis A. 2008. Phosphorylated proteins and control over apatite nucleation, crystal growth, and inhibition. Chem. Rev 108:4670–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt S, Hassall G, Hawtin P, McCall E, Flavell L, Minshull C, Hargreaves D, Ting A, Pauptit RA, Parker AE, and Abbott WM. 2007. Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains. J. Mol. Biol 373:891–902. [DOI] [PubMed] [Google Scholar]
- Gosline JM, and DeMont ME. 1985. Jet-propelled swimming in squids. Sci. Am 252:96–103. [Google Scholar]
- Hackman RH 1960. Studies on chitin IV. The occurrence of complexes in which chitin and protein are covalently linked. Aust. J. Biol. Sci 13:568–577. [Google Scholar]
- Hahn M, Hennig M, Schlesier B, and Hohne W. 2000. Structure of the jack bean chitinase. Acta Crystallogr. Sect. D. Biol. Crystallogr D56:1096–1099. [DOI] [PubMed] [Google Scholar]
- Hopkins B, and Boletzky S. 1994. The fine morphology of the shell sac in the squid genus Loligo (Mollusca: Cephalopoda) features of a modified Conchiferan program. The Veliger 37:344–357. [Google Scholar]
- Hunt S, and El Sherief A. 1990. A periodic structure in the ‘pen’ chitin of the squid Loligo vulgaris. Tissue Cell 22:191–197. [DOI] [PubMed] [Google Scholar]
- Hunt S, and Nixon M. 1981. A comparative study of protein composition in the chitin-protein complexes of the beak, pen, sucker disc, radula and oesophageal cuticle of cephalopods. Comp. Biochem. Physiol 68B:535–546. [Google Scholar]
- Ianiro A, Giosia MD, Fermani S, Samori C, Barbalinardo M, Valle F, Pellegrini G, Biscarini F, Zerbetto F, Calvaresi M, and Falini G. 2014. Customizing properties of β-chitin in squid pen (gladius) by chemical treatments. Mar. Drugs 12:5979–5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DJ, Mann K, Haussermann V, Schilhabel MB, Luter C, Griesshaber E, Schmahl W, and Worheide G. 2015. The Magellania venosa biomineralizing proteome: A window into brachiopod shell evolution. Genome Biol. Evol 7:1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki H, Manickam A, Shahin R, Ote M, and Iwanaga M. 2018. Expression of matrix metalloproteinase genes during basement membrane degradation in the metamorphosis of Bombyx mori. Gene 638:26–35. [DOI] [PubMed] [Google Scholar]
- Kaya M, and Baran T. 2015. Description of a new surface morphology for chitin extracted from wings of cockroach (Periplaneta americana). Int. J. Biol. Macromol 75:7–12. [DOI] [PubMed] [Google Scholar]
- Lavall RL, Assis OBG, and Campana-Filho SP. 2007. β-chitin from the pens of Loligo sp.: Extraction and characterization. Bioresour. Technol 98:2465–2472. [DOI] [PubMed] [Google Scholar]
- Le Pabic C, Marie A, Marie B, Percot A, Bonnaud-Ponticelli L, Lopez PJ, and Luquet G. 2017. First proteomic analyses of the dorsal and ventral parts of the Sepia officinalis cuttlebone. J. Proteomics 150:63–73. [DOI] [PubMed] [Google Scholar]
- Lee PN, Callaerts P, and de Couet HG. 2009. The embryonic development of the Hawaiian bobtail squid. Cold Spring Harbor Protocols 11:pdb.ip77. [DOI] [PubMed] [Google Scholar]
- Liu Z, Gay LM, Tuveng TR, Agger JW, Westereng B, Mathiesen G, Horn SJ, Vaaje-Kolstad G, van Aalten DMF, and Eijsink VGH. 2017. Structure and function of a broad-specificity chitin deacetylase from Aspergillus nidulans FGSC A4. Sci. Rep 7:1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain A, Arguelles J, Alegre A, Bertrand A, Munaron J-M, Richard P, and Cherel Y. 2011. Sequential isotopic signature along gladius highlights contrasted individual foraging strategies of jumbo squid (Dosidicus gigas). PLOS 1 6:e22194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Burger C, Hsiao BS, and Chu B. 2011. Ultrafine polysaccharide nanofibrous membranes for water purification. Biomacromolecules 12:970–976. [DOI] [PubMed] [Google Scholar]
- Mann K, and Jackson DJ. 2014. Characterization of the pigmented shell-forming proteome of the common grove snail Cepaea nemoralis. BMC Genomics 15:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin F, Luquet G, Marie B, and Medakovic D. 2008. Molluscan shell proteins: primary structure, origin and evolution. Curr. Top. Dev. Biol 80:209–276. [DOI] [PubMed] [Google Scholar]
- Marin F, Marie B, Hamada SB, Ramos-Silva P, Le Roy N, Guichard N, Wolf SE, Montagnani C, Joubert C, Piquemal D, Saulnier D, and Gueguen Y. 2013. ‘Shellome’: Proteins involved in mollusc shell biomineralization-diversity, functions Pp. 149–166 in Recent Advances in Pearl Research, Watabe S, Maeyama K and Nagasawa H, eds. Terrapub. [Google Scholar]
- Merten V, Christiansen B, Javidpour J, Piatkowski U, Puebla O, Gasca R, and Hoving H-JT. 2017. Diet and stable isotope analyses reveal the feeding ecology of the orangeback squid Sthenoteuthis pteropus (Steenstrup 1855) (Mollusca, Ommastrephidae) in the eastern tropical Atlantic. PLoS One 12: e0189691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzendorfer H, Kelkenberg M, and Muthukrishnan S. 2016. Peritrophic Matrices Pp. 255–324 in Extracellular Composite Matrices in Arthropods, Cohen E and Moussian B, eds. Springer, Cham. [Google Scholar]
- Miserez A, Li Y, Waite JH, and Zok F. 2007. Jumbo squid beaks: Inspiration for design of robust organic composites. Acta Biomater 3:139–149. [DOI] [PubMed] [Google Scholar]
- Mohs A, Silva T, Yoshida T, Amin R, Lukomski S, Inouye M, and Brodsky B. 2007. Mechanism of stabilization of a bacterial collagen triple helix in the absence of hydroxyproline. J. Biol. Chem 282:29757–29765. [DOI] [PubMed] [Google Scholar]
- Naef A 1928. Die Cephalopoden (Embryologie). Fauna Flora Golf Neapel 35:1–357. [Google Scholar]
- Nakada M, Yamada A, Takino T, Miyamori H, Takahashi T, Yamashita J, and Sato H. 2001. Suppresion of membrane-type 1 matrix metalloproteinase (MMP)-mediated MMP-2 activation and tumor invasion by testican 3 and its splicing variant gene product, N-Tes. Cancer Res 61:8896–8902. [PubMed] [Google Scholar]
- O’Dor RK 2013. How squid swim and fly. Can. J. Zool 91:413–419. [Google Scholar]
- Oldenbourg R, and Shribak M. 2010. Microscopes Pp. 1–62 in Handbook of Optics, Third Edition, Volume I: Geometrical and Physical Optics, Polarized Light, Components and Instruments, Bass M, ed. McGraw-Hill. [Google Scholar]
- Ote M, Mita K, Kawasaki H, Kobayashi M, and Shimada T. 2005. Characteristics of two genes encoding proteins with an ADAM-type metalloprotease domain, which are induced during the molting periods in Bombyx mori. Arch. Insect Biochem. Physiol 59:91–98. [DOI] [PubMed] [Google Scholar]
- Perret S, Merle C, Bernocco S, Berland P, Garrone R, Hulmes DJS, Theisen M, and Ruggiero F. 2001. Unhydroxylated triple helical collagen I produced in transgenic plants provides new clues on the role of hydroxyproline in collagen folding and fibril formation. J. Biol. Chem 276:43693–43698. [DOI] [PubMed] [Google Scholar]
- Porter S, Clark IM, Kevorkian L, and Edwards DR. 2005. The ADAMTS metalloproteinases. Biochem. J 386:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raabe D, Sachs C, and Romano P. 2005. The crustacean exoskeleton as an example of a structurally and mechanically graded biological nanocomposite material. Acta Mater 53:4281–4292. [Google Scholar]
- Ranasinghe S, and McManus DP. 2013. Structure and function of invertebrate Kunitz serine protease inhibitors. Immunology 39:219–227. [DOI] [PubMed] [Google Scholar]
- Rasband WS 1997-2018. ImageJ. [Online]. U.S. National Institutes of Health, Bethesda, MD, Available: https://imagej.nih.gov/ij/ [2018, Sept. 1]. P.^Pp. [Google Scholar]
- Ruiz-Cooley RI, Villa EC, and Gould WR. 2010. Ontogenetic variation of d13C and d15N recorded in the gladius of the jumbo squid Dosidicus gigas: geographic differences. Mar. Ecol. Prog. Ser 399:187–198. [Google Scholar]
- Schroder-Turk GE, Wickham S, Averdunk H, Brink F, Fitz Gerald JD, Poladian L, Large MCJ, and Hyde ST. 2011. The chiral structure of porous chitin within the wing-scales of Callophrys rubi. J. Struct. Biol 174:290–295. [DOI] [PubMed] [Google Scholar]
- Shen ZC, and Jacobs-Lorena M. 1998. A type I peritrophic matrix protein from the malaria vector Anopheles gambiae binds to chitin - Cloning, expression, and characterization. J. Biol. Chem 273:17665–17670. [DOI] [PubMed] [Google Scholar]
- Stepek G, McCormack G, and Page AP. 2010. The kunitz domain protein BLI-5 plays a functionally conserved role in cuticle formation in a diverse range of nematodes. Mol. Biochem. Parasitol 169:1–11. [DOI] [PubMed] [Google Scholar]
- Subhapradha N, Ramasamy P, Shanmugam V, Madeswaran P, Srinivasan A, and Shanmugam A. 2013. Physiochemical characterization of β-chitosan from Sepioteuthis lessoniana gladius. Food Chem 141:907–913. [DOI] [PubMed] [Google Scholar]
- Suzuki M, and Nagasawa H. 2013. Mollusk shell structures and their formation mechanism. Can. J. Zool 91:349–366. [Google Scholar]
- Tan YP, Hoon S, Guerette PA, Wei W, Ghadban A, Hao C, Miserez A, and Waite JH. 2015. Infiltration of chitin by protein coacervates defines the squid beak mechanical gradient. Nat. Chem. Biol 11:488–495. [DOI] [PubMed] [Google Scholar]
- Thompson JT, Lavalva SM, and Loiacono M. 2016. A multifuntion muscle in squid. Biol. Bull 231:225–235. [DOI] [PubMed] [Google Scholar]
- Thompson JT, Shelton RM, and Kier WM. 2014. The length-force behavior and operating length range of squid muscle vary as a function of position in the mantle wall. J. Exp. Biol 217:2181–2192. [DOI] [PubMed] [Google Scholar]
- Tjoelker LW, Gosting L, Frey S, Hunter CL, Trong HL, Steiner B, Brammer H, and Gray PW. 2000. Structural and functional definition of the human chitinase chitin-binding domain. J. Biol. Chem 275:514–520. [DOI] [PubMed] [Google Scholar]
- Vasiliou V, Thompson DC, Smith C, Fujita M, and Chen Y. 2013. Aldehyde dehydrogenases: From eye crystallins to metabolic disease and cancer stem cells. Chem.-Biol. Interact 202:2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Hastings G, Ortega M-A, Lane TF, Oikemus S, Lombardo M, and Iruela-Arispe ML. 1999. METH-1, a human ortholog of ADAMTS-1, and METH-2 are members of a new family of proteins with angio-inhibitory activity. J. Biol. Chem 274:23349–23357. [DOI] [PubMed] [Google Scholar]
- Vold IMN, and Christensen BE. 2005. Periodate oxidation of chitosans with different chemical compositions. Carbohydr. Res 340:679–684. [DOI] [PubMed] [Google Scholar]
- Vournakis JN, Demcheva M, Whitson AB, Finkielsztein S, and Connolly RJ. 2003. The RDH bandage: hemostasis and survival in a lethal aortotomy hemorrhage model. J. Surg. Res 113:1–5. [DOI] [PubMed] [Google Scholar]
- Weiner S, and Hood L. 1975. Soluble protein of the organic matrix of mollusk shells: A potential template for shell formation. Science 190:987–989. [DOI] [PubMed] [Google Scholar]
- Williams LW 1909. The anatomy of the common squid Loligo pealii, Lesuer; Brill, Leiden. [Google Scholar]
- Winkler DD, and Luger K. 2011. The histone chaperone FACT: Structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem 286:18369–18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward MP, Young WW Jr., and Bloodgood RA. 1985. Detection of monoclonal antibodies specific for carbohydrate epitopes using periodate oxidation. J. Immunol. Methods 78:143–153. [DOI] [PubMed] [Google Scholar]
- Wright PA 1995. Nitrogen excretion: Three end products, many physiological roles. J. Exp. Biol 198:273–281. [DOI] [PubMed] [Google Scholar]
- Wu S-D, Wu C-S, and Chen H-C. 2003. Cuticle structure of squid Illex argentinus pen. Fish. Sci 69:849–855. [Google Scholar]
- Yang F-C, Peters RD, Dies H, and Rheinstadter MC. 2014. Hierarchical, self-similar structure in native squid pen. Soft Matter 10:5541–5549. [DOI] [PubMed] [Google Scholar]
- Young RE, Vecchione M, and Donovan DT. 1998. The evolution of coleoid cephalopods and their present biodiversity and ecology. S. Afr. J. Mar. Sci./S.-Afr. Tydskr. Seewet 20:393–420. [Google Scholar]
- Yu R, Liu W, Li D, Zhao X, Ding G, Zhang M, Ma E, Zhu KY, Li S, Moussian B, and Zhang J. 2016. Helicoidal organization of chitin in the cuticle of the migratory locust requires the function of the chitin deacetylase2 enzyme (LmCDA2). J. Biol. Chem 291:24352–24363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


