Abstract
Pathogen persistence in host communities is influenced by processes operating at the individual host to landscape-level scale, but isolating the relative contributions of these processes is challenging. We developed theory to partition the influence of host species, habitat patches, and landscape connectivity on pathogen persistence within metacommunities of hosts and pathogens. We used this framework to quantify the contributions of host species composition and habitat patch identity on the persistence of an amphibian pathogen across the landscape. By sampling over 11,000 hosts of six amphibian species, we found that a single host species could maintain the pathogen in 91% of observed metacommunities. Moreover, this dominant maintenance species contributed, on average, twice as much to landscape-level pathogen persistence compared to the most influential source patch in a metacommunity. Our analysis demonstrates substantial inequality in how species and patches contribute to pathogen persistence, with important implications for targeted disease management.
Keywords: maintenance species, reservoir species, source-sink dynamics, metacommunity, metapopulaton, Batrachochytrium dendrobatidis, endemic, Pseudacris regilla, hotspots, chytrid fungus
Introduction
Many pathogens of conservation and health concern infect multiple host species and occur on landscapes composed of interacting host communities (i.e. ‘disease metacommunities’; Paull et al. 2012; Miller et al. 2013; Johnson et al. 2015). Heterogeneity among species within a habitat patch and among patches on a landscape can interact in non-additive ways to influence pathogen invasion and persistence (Lloyd-Smith et al. 2005; Johnson et al. 2015; North & Godfray 2017). At the patch-scale, host species-level differences in population densities, contact rates, and shedding rates dictate contributions to infection dynamics within the community (Mihaljevic et al. 2014; Webster et al. 2017; Wilber et al. 2019). At the landscape-scale, characteristics of habitat patches and their degree of connectivity can influence pathogen invasion success and persistence (McCallum 2008; Schreiber & Lloyd-Smith 2009; Arino 2009). This creates a challenging yet foundational question: from a control standpoint, what are the relative contributions of heterogeneities across scales in determining pathogen persistence (McCallum & Dobson 2002; Paull et al. 2012)? For instance, whether disease management should prioritize strategies such as quarantine, culling, habitat modification, or targeted vaccination will depend critically on the relative influence of specific patches (hotspots) versus specific species (maintenance or amplification hosts).
While both variation among host species and across habitat patches influences pathogen invasion and persistence, how these factors interact remains unknown. In a multi-species, single-patch system, ‘maintenance’ host species are those that can independently maintain a pathogen and contribute to its spillover into other host species (De Castro & Bolker 2005; McCallum 2012; Webster et al. 2017). However, in a multi-species, multi-patch system, labeling species as maintenance or spillover hosts is made challenging by the added influence of variability among habitat patches –a species potential to maintain a pathogen (i.e. ‘maintenance potential’) may vary among habitats due to changes in community structure or the physical environment (Haydon et al. 2002; Rudge et al. 2013; Roberts & Heesterbeek 2020). This context-dependent variability in species maintenance potentials further leads to variability in the potential of individual patches to support a pathogen (i.e. ‘source potential’), including whether they are ‘source’ patches capable of independently maintaining a pathogen in isolation from all other patches (McCallum 2008; Schreiber & Lloyd-Smith 2009). As a result, how a pathogen spreads across the landscape will depend, in part, on the degree to which the maintenance potential of each host species varies with –or is moderate by –patch location.
While the potential of habitat patches to contribute infections across the landscape is ultimately a function of the maintenance potentials of its constituent species, patches can further influence pathogen persistence through their connectivity to other patches in the metacommunity (Park et al. 2001; Schreiber & Lloyd-Smith 2009; North & Godfray 2017). For example, depending on the locations of source and sink patches on the landscape, high enough pathogen or host dispersal rates can actually hinder the ability of a pathogen to invade a metapopulation that would otherwise be invasible at lower dispersal rates (Schreiber & Lloyd-Smith 2009; Jousimo et al. 2014). Thus, correctly accounting for patch connectivity and pathogen dispersal can fundamentally alter predictions about pathogen invasion across the landscape. Despite this theoretical understanding, few studies have empirically quantified the links among species maintenance potential, patch source potential, and patch connectivity to understand pathogen persistence within a metacommunity (Penczykowski et al. 2015). This is a critical next step for understanding the drivers of pathogen dynamics in multi-species, multi-patch disease systems, which is arguably a common feature of many emerging infections of importance for conservation or society.
The maintenance potential and source potential of a species and patch, respectively, can be defined in terms of the fundamental recruitment number R0. For a single host species in a single patch, R0 defines the number of secondary infections produced over the lifetime of an average infected individual in a susceptible population (Diekmann et al. 1990). When R0 > 1, a pathogen can invade a susceptible host population (Keelling & Rohani 2008). In a multi-species, multi-patch system, there is a hierarchy of R0 values: species-level R0, patch-level R0, and landscape-level R0,L (Figure 1A). Maintenance species and source patches are defined by scale-specific values of R0 > 1, while landscape-level R0,L combines the species- and patch-level R0 values to determine when a pathogen can invade the host metacommunity (i.e. R0,L > 1, Fig. 1A; Arino et al. 2005, but see Cross et al. (2005); North & Godfray (2017)). Theoretically, the species-, patch-, and landscape-level R0 values, coupled with information on species connectivity and patch connectivity, provide all the information necessary to understand pathogen invasion and persistence within host-pathogen metacommunities. Empirically, however, the parameters required to calculate species- and patch-level R0 values can be difficult to estimate, particularly for multiple species across multiple patches. Fortunately, recent work indicates that many of these difficult-to-estimate parameters, such as the absolute values of transmission coefficients, can be replaced by more commonly estimated parameters such as prevalence and parameter ratios (Rudge et al. 2013; Fenton et al. 2015). While these approaches have been applied to understand the maintenance potential of hosts in multi-species systems (Rudge et al. 2013; Fenton et al. 2015), they have yet to be extended to multi-patch, multi-species systems.
Figure 1: A.
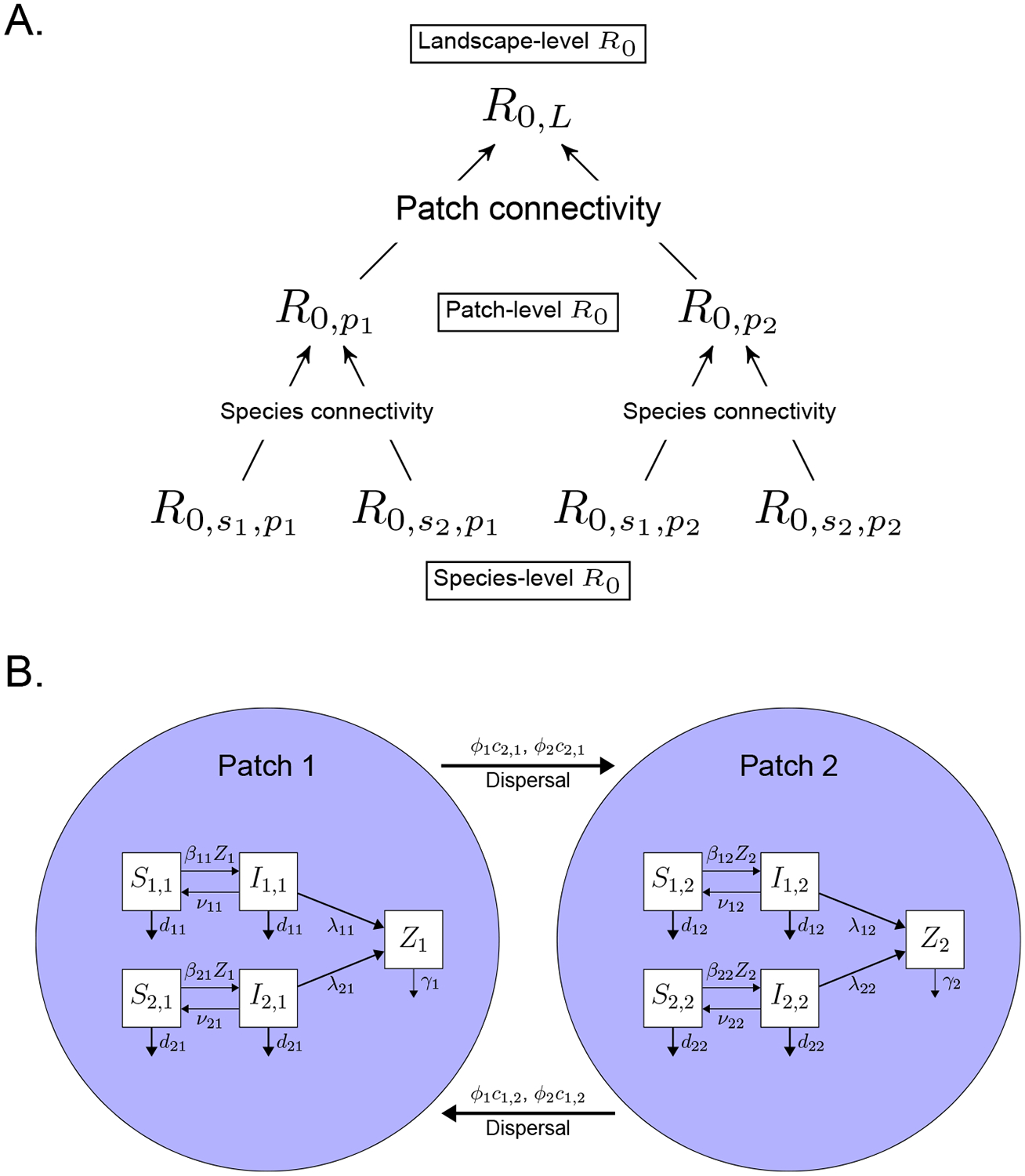
The partitioning of a multi-species, multi-patch system into species-level , patch-level , landscape-level R0,L, species connectivity (e.g. the off-diagonals of a Who-Acquired-Infection-From-Whom matrix, Dobson 2004), and patch connectivity. In this example, there are two species and two patches on the landscape. B. The multi-species, multi-patch pathogen model used to partition the importance of maintenance species and source patches on pathogen persistence in a metacommunity (equation 1). The diagram uses two species and two patches as an example.
Here, we first develop multi-patch, multi-species models of host-pathogen metacommunities and then confront these models with commonly-collected pathogen data to address three questions: (1) Are the relative maintenance potentials of host species consistent across habitat patches? (2) How do patch connectivity and pathogen dispersal affect species maintenance and patch source potential in a metacommunity? and (3) What are the relative contributions of maintenance species compared to source patches for determining pathogen persistence on the landscape? To quantify the contributions of species and patches on pathogen persistence, we focused on interactions between amphibian host species and the fungal pathogen Batrachochytrium dendrobatidis (Bd), which is the causative agent of the disease chytridiomycosis and declines in hundreds of amphibian species worldwide (Kilpatrick et al. 2010; Scheele et al. 2019). Between 2013 and 2018, we compiled infection information on over 11,000 hosts comprising six species of amphibians persisting endemically with Bd across 139 habitat patches to parameterize a multi-species, multi-patch model and answer our three questions. The theory we develop provides a quantitative means to assess the contribution of species, patches, and connectivity to pathogen persistence across scales in empirical host-pathogen metacommunities, which is essential information for identifying and implementing effective management strategies.
Materials and Methods
Study system and data collection
Between 2013 and 2018, we examined the within-season Bd maintenance potential of amphibian species from 77 metacommunities in the East Bay Region of California, USA (Fig. S1). We considered six species of wetland-breeding amphibians: Pseudacris regilla (Baird and Girard, 1852) (Pacific tree frog), Anaxyrus boreas (Baird and Girard, 1852) (western toad), Rana catesbeiana (Shaw, 1802) (American bullfrog), Rana draytonii (Baird and Girard, 1852) (California red-legged frog), Taricha torosa (Rathke, 1833) (California newt), and T. granulosa (Skilton, 1849) (rough-skinned newt). Adult amphibians of all species typically breed in ponds from January to late spring and co-occur as larvae between May and August (Fig. S1, Stebbins & McGinnis 2012). The length of the larval period varies among species (Fig. S1, Johnson et al. 2012). Larvae of the native species (P. regilla, A. boreas, R. draytonii, T. granulosa, and T. torosa) typically mature and leave the pond within the same year, while the non-native R. catesbeiana usually requires two or more years to reach metamorphosis (Stebbins & McGinnis 2012). Amphibian communities persist endemically with Bd across multiple interconnected ponds and wetlands with little evidence of Bd-associated pathology. Available information on the biology of these amphibian species coupled with the feasibility of standardized sampling protocols for Bd infection and host density (e.g. Johnson et al. 2012; Joseph et al. 2016; Stutz et al. 2018), make this amphibian-Bd system ideal to link with multi-species, multi-patch pathogen models.
We defined a metacommunity as a potentially interconnected network of ponds and wetlands among which amphibians could move. Each pond represented a patch in the metacommunity. We defined metacommunities such that they closely corresponded to administratively delineated parks and properties (Johnson et al. 2016). Spatially, this meant that all ponds were no more than two kilometers away from at least one other pond within a metacommunity. Our rationale was that properties provided a connected stretch of habitat within which amphibians could potentially disperse. In addition, we considered the same spatial metacommunity sampled over multiple years as different spatiotemporal metacommunities. We assumed that between season amphibian migrations and pond dynamics (e.g. ponds drying) largely uncoupled pathogen dynamics between years. The 77 metacommunities were comprised of between one to 26 ponds with an average of six ponds per metacommunity. There were 139 unique ponds sampled across six years and, on average, each pond was sampled in four years, for a total of 496 unique pond-by-year combinations.
From May through August in 2013–2018, crews sampled amphibian communities over the course of two visits per pond. During the first visit (early summer), standardized dipnet surveys were used to estimate the density of larval amphibians for each species (for details see Johnson et al. 2013; Joseph et al. 2016). During the second visit (mid summer), we used standardized methods to swab the skin of metamorphosing anurans or late-stage larvae (caudates) to assess Bd infection status and Bd load using quantitative polymerase chain reaction (qPCR) with a standardized TaqMan assay (Boyle et al. 2004; Hyatt et al. 2007). qPCR for Bd was run on each sample in triplicate to quantify measurement error (DiRenzo et al. 2018). As the Bd and density sampling was all within a season, we focused our analysis on within-season Bd dynamics and not between season dynamics. While adult amphibians were present and occasionally captured during surveys, the sampling protocols were not designed to sample adult amphibians. We discuss the implications of excluding adults in Appendix S1.
A multi-species, multi-patch model of pathogen dynamics
We developed a dynamic model to address our three questions regarding the contributions of host species and habitat patches to Bd persistence on the landscape within a season. We considered a multi-species, multi-patch (S)usceptible-(I)nfected-(S)usceptible model with infection from an environmental zoospore pool Z for host species s = 1, … , H and patches p = 1, … , P (Fig. 1B). Bd is transmitted between hosts via a motile, aquatic zoospore stage (Longcore et al. 1999). Consistent with previous Bd models, we assumed that amphibians acquired infection directly from an aquatic zoospore pool into which infected amphibians shed Bd zoospores (Mitchell et al. 2008; Briggs et al. 2010). We did not directly model Bd load as there was little evidence for load-dependent mortality in this system. We did, however, use Bd load as a proxy for shedding rates of infectious zoospores, as described below.
The multi-species, multi-patch model we consider is (Fig. 1B)
| (1) |
where Ssp and Isp are the densities of susceptible and infected hosts, respectively, of species s in patch p. Zp is the density of zoospores in the zoospore pool in patch p. The term βspZp is the force of infection for species s in patch p. λip is the shedding rate of Bd zoospores into the environment of species i in patch p. γp is the patch-specific decay rate of the zoospores in the environment. We assumed all hosts in a patch share the same pathogen pool and that the pathogen pool is well-mixed. The parameter νsp is the species and patch-specific recovery rate of an infected host. Host birth rate is given by the generic function f(Nsp) where Nsp = Ssp + Isp and is species and patch specific. Host death rate is given by dsp and is species and patch specific.
The parameter ϕs is the within-season dispersal rate for host species s (i.e. the rate at which individuals of species s leave a patch) and cjp is the probability that a host moves from patch p to patch j. The P × P matrix C contains cjp movement probabilities and is irreducible –all patches were accessible to all other patches in a finite time (Arino 2009). We assumed that both infected and susceptible individuals can disperse, that infection does not affect dispersal, and that infection status does not change during dispersal. Finally, Ap is the area of patch p. The area ratio converts the total number of individuals leaving patch j in a time step to the appropriate density units in patch p.
Species-level
Given equation 1, species-level of species s in patch p is given by , where is the density of susceptible hosts of species s in patch p before infection arrives and bsp is the loss rate from the infected class such that bsp = dsp + νsp (Fig. 1B). Note that if we included Bd-induced mortality at some constant rate αsp, this would be additively included into bsp.
At equilibrium, algebraic manipulations of equation 1 show that
| (2) |
The variable is the equilibrium Bd prevalence and is the equilibrium density of species s in patch p. If it holds, the equilibrium assumption is useful because one can calculate without needing hard-to-estimate parameters such as species-specific absolute transmission rates (Rudge et al. 2013; Fenton et al. 2015). One can instead use more commonly collected parameters such as host density, Bd prevalence and Bd infection load. In Appendix S2, we discuss why an approximate equilibrium assumption is appropriate for the within-season dynamics of this amphibian-Bd system.
Finally, a useful property of equation 2 is that the ratio between two species-level values from the same patch p depends only on the parameters relating to the two species being compared (Appendix S1). As a result, if one does not have the necessary data on other community members that are potentially important for the persistence of Bd, one can still analyze the contribution of each species to pathogen persistence, relative to the other species that have been sampled.
Linking empirical data to maintenance species and source patches
We fitted statistical models that accounted for false absences to estimate Bd load, Bd prevalence and host density for species s in patch p (models described in Appendix S3; Miller et al. 2012; Joseph et al. 2016; DiRenzo et al. 2018). We assumed that host zoospore shedding rate was proportional to Bd load (DiRenzo et al. 2014) and estimated the shedding rate ratios for species s and i in patch p as the ratio between estimated mean Bd loads for species s and i in patch p. We calculated using equation 2, propagating the uncertainty in the parameter estimates. The results we present use the median species-level estimates. Finally, because our model assumed that amphibians were sharing the same pool of zoospores, patch-level could be directly calculated as (Rudge et al. 2013).
Question 1: Are the maintenance potentials of host species consistent across patch locations?
We began our analysis with the assumption that patches were unconnected within a metacommunity (i.e. ϕs = 0). We then calculated relative and absolute values for all species across across patch-by-year combinations. Under the assumption of no connectivity, the only parameters needed to calculate species-specific were Bd prevalence, relative density, and relative shedding rates for the different amphibian species within a patch (equation 2). If one species consistently had higher relative values than another, this was evidence that relative species maintenance potential for that species pair was independent of patch location. We used a two-sided binomial test to test if relative maintenance potentials of species were consistent across patches by comparing whether one species consistently had a larger species-level than another species.
Question 2: How does patch connectivity affect species maintenance and patch source potential?
Equation 2 shows that accounting for the connectivity of patches can change our conclusions about the consistency of species maintenance potential and patch source potential within a metacommunity. Here we give a summary of how we included connectivity into our model. The complete methods are described in Appendix S4.
The key unknown connectivity parameter in the model was the ratio between species-specific dispersal rate and the loss rate from the infected class, rsp = ϕs/bsp. This parameter describes the expected number of patches to which an infected individual of species s that disperses from patch p moves to over its time infected. As this parameter could not be uniquely inferred from snapshot data, there were many values for rsp that were equally “plausible” given observed patterns of prevalence, Bd loads, and host density. By “plausible” we mean that for all species and patches in the metacommunity. In other words, if for a value of rsp, then the observed levels of prevalence, Bd loads, and host density were not consistent with an endemic equilibrium state of the multi-species, multi-patch model. For example, high levels of host dispersal would tend to homogenize species-level density and pathogen prevalence among patches, such that observing highly variable equilibrium host densities and prevalences among patches at endemic equilibrium would be inconsistent with high levels of host dispersal.
We explored how species maintenance potential and patch source potential changed across plausible values of rsp, compared to an assumption of no dispersal (i.e. rsp = 0). For each metacommunity with H species and P patches, we drew 10,000 parameter sets of plausible H × P rsp parameters and computed the species-level for all species and patches using equation 2 (Appendix S4). For each species s in patch p, we ensured that we drew a plausible rsp value (i.e. one such that ) by setting equation 2 to zero and solving for rsp = ϕs/bsp (Appendix S4). For the results presented below, we used both the median values from the 10,000 plausible parameter sets and the maximally connected plausible parameter set. We defined the maximally connected plausible parameter set for a metacommunity as the set where each rsp was at its maximum plausible value (Appendix S4).
Question 3: What are the contributions of maintenance species compared to source patches for pathogen persistence?
For our final question, we sought to quantify the relative contributions of maintenance species and source patches to landscape-level R0,L. For each metacommunity, we calculated how much landscape-level R0,L changed when we removed a particular species in the metacommunity compared to when we removed the most influential source patch (see Appendix S5 for details). We performed this simulated removal experiment over the plausible sets of rsp values for a metacommunity. We defined the most influential source patch in a metacommunity as the patch with the largest patch-level , given a set of plausible rsp parameters. We performed this in silico removal experiment on 61 metacommunities that had more than one habitat patch and more than one amphibian species.
Results
Patterns of host density, Bd prevalence, and Bd load across patches
Overall, P. regilla was observed in 82% of patch-by-year combinations (405 / 496), T. torosa in 67% (334 / 496), T. granulosa in 28% (137 / 496), A. boreas in 28% (138 / 496), R. draytonii in 13% (65 / 496), and R. catesbeiana in 11% (56 / 496). Median species richness per patch was between 2 and 3, depending on the year. P. regilla and T. torosa were present in 74 and 70 of 77 metacommunities, respectively, while R. draytonii, T. granulosa, A. boreas, and R. catesbeiana were all found in less than 50% of the 77 metacommunities (37, 36, 32, and 27 metacommunities, respectively). Pseudacris regilla and T. torosa also had higher estimated larval densities relative to R. draytonii, A. boreas, T. granulosa, and R. catesbeiana, although density estimates varied substantially across years (Fig. 2A).
Figure 2: A.
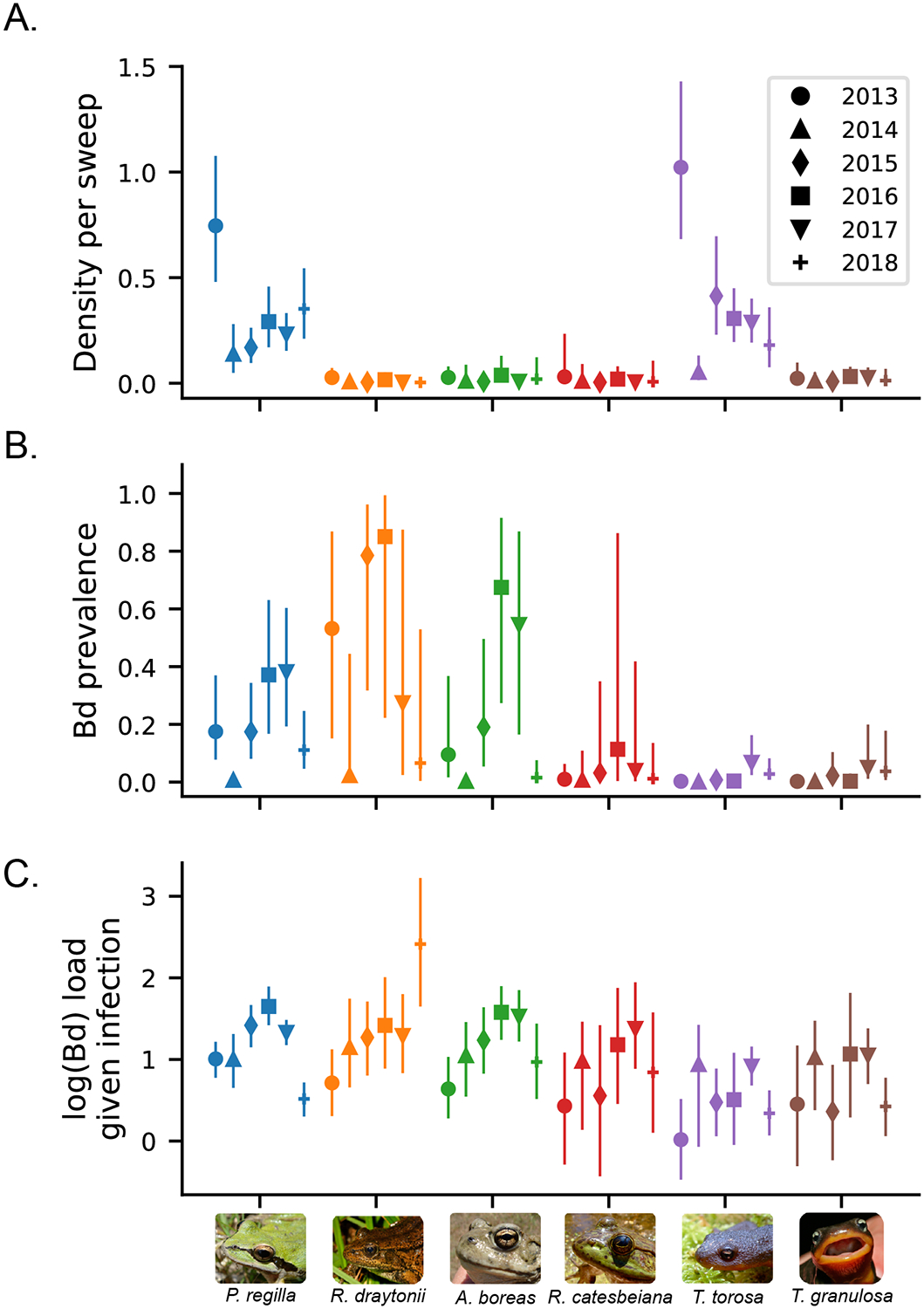
Median estimated amphibian density per dip net sweep after accounting for false absences across six years, 139 patches and six amphibian species. B. Median estimated prevalence after accounting for false absences. C. Median estimated mean log(Bd) load conditional on infection after accounting for measurement error. For all figures, the error bars are 95% credible intervals about the estimated median. Different shapes represent different years. The species on the x-axis are Pacific tree frog (P. regilla), western toad (A. boreas), American bullfrog (R. catesbeiana), California red-legged frog (R. draytonii), California newt (T. torosa), and rough-skinned newt (T. granulosa).
Bd was detected in 73% of patches that were sampled from 2013–2018. Rana draytonii generally had the highest Bd prevalence, followed by P. regilla and A. boreas (Fig. 2B). Observed prevalence was the lowest for T. torosa and T. granulosa (Fig. 2B). Prevalence estimates varied across years, with 2014 showing a substantially lower prevalence for most species (Fig. 2B). Within a year, mean Bd load given infection was generally significantly higher in A. boreas, P. regilla, and R. draytonii compared to T. granulosa and T. torosa (95% credible intervals of log load differences between these species were significantly different than 0, but not in year 2014; Fig. 2C). Loads detected in R. catesbeiana were generally not significantly different from those in other species.
Question 1: Are the maintenance potentials of host species consistent across patch locations?
Within a patch, P. regilla was consistently the most important amphibian host species for the persistence of Bd:
The relative of P. regilla was larger than other amphibian species in 82% of the patches in which the amphibian species co-occurred (489 / 600 instances of P. regilla co-occurring with other amphibian species within a patch; Fig. 3). P. regilla was statistically more likely to have a higher species-level than all five species with which it co-occurred (Fig. 3). Of the 111 instances where P. regilla had a lower relative to another species in the community, 38% were with A. boreas, 29% were with T. torosa, 19% were with R. draytonii, and 8% were with R. catesbeiana.
Figure 3:
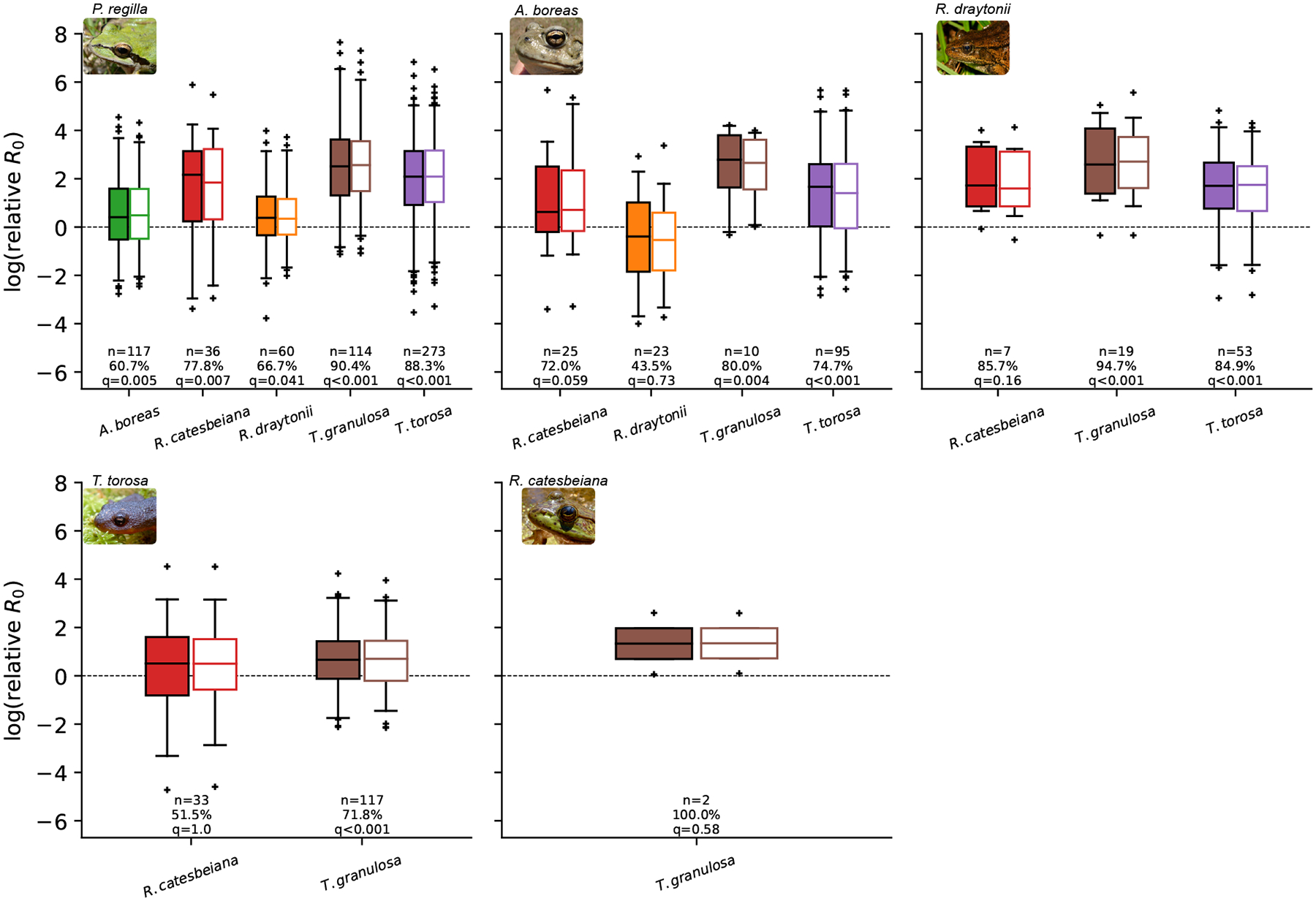
Relative species-level values within a patch calculated using equation 2 with no connectivity (filled boxplots) and using the median from the plausible set of dispersal rate to loss of infected rate ratios rsp (unfilled boxplots). As an example of the labeling, the “A. boreas” x-label of the plot titled “P. regilla” shows the distribution of the ratios of P. regilla values to A. boreas values for patches where P. regilla and A. boreas were both present. A value larger than zero indicates that the relative maintenance potential of P. regilla is greater than A. boreas for that comparison. ‘n’ gives the number of patches where both species were found, ‘%’ gives the percent of comparisons where relative log values were greater than zero, and ‘q’ gives the p-value corrected for false discovery rate (Benjamini & Hochberg 1995) from a binomial test with the null hypothesis that species are equally likely to have a higher species-level within a patch. The bars give the median relative values, the boxes given the upper and lower quartiles, the whiskers give the 2.5 and 97.5 percentiles, and “+”s show points outside of these percentiles.
The majority of host communities had at least one maintenance host species and P. regilla was almost always a maintenance host when it was present:
Of the 496 patch-by-year combinations sampled, 126 had estimates where all species-specific values were less than one, but patch-level was greater than one (i.e. an obligate host community, Fig. 4). In 66 of these 126 obligate host communities, Bd was not empirically observed, but low levels of Bd load and prevalence were inferred given a non-zero probability of Bd detection error from the Bd load model (see Appendix S3, Fig. 4). Of the 370 communities that were not obligate host communities, 9% had multiple host species with (i.e. facultative communities). Eighty-eight percent of facultative communities were comprised of P. regilla and either A. boreas or R. draytonii. The other 91% of non-obligate communities (338 communities) had only one species with (i.e. spillover communities) and all other species (if any were present) had . In the non-obligate communities where P. regilla was present, P. regilla was a maintenance host 89% of the time (267 / 300; Fig. 4).
Figure 4:
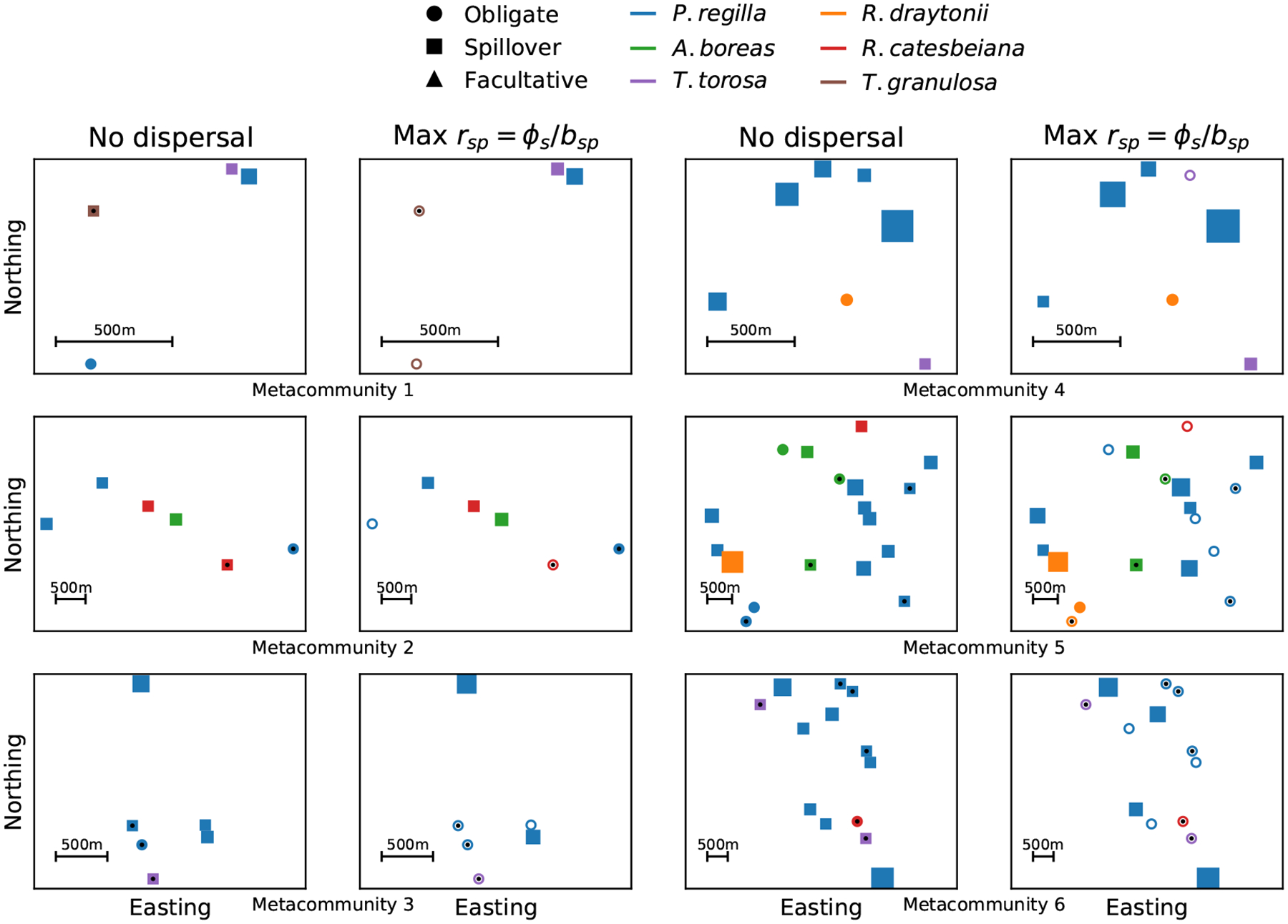
Six representative metacommunities given no dispersal and maximum plausible connectivity for each species in a metacommunity (Max rsp = ϕs/bs). Each point represents the spatial location of a patch within the metacommunity using a UTM projection. The color of the point indicates which amphibian has the highest species-level in the patch. If the point is filled, the patch-level is greater than 1 and the patch is a source patch. If the point is not filled, the patch-level is less than 1 and the patch is a sink. The shape of the point indicates what type of community is found in the patch. Circle = an obligate community where for all species, Square = A spillover community where for only one species, and Triangle = a facultative community where for more than one species. The size of the point represents a scaled measure of patch-level when patch-level . Finally, points with small black dots indicate patches where Bd was not observed for any species. Our statistical model for Bd load accounted for detection error, such that there was some probability that Bd was present, but at low prevalence in these patches. We used the model-predicted prevalence given detection error when making inference for these patches (Appendix S3).
Question 2: How does patch connectivity affect species maintenance and patch source potential?
Under plausible levels of connectivity, multiple source patches contributed to Bd persistence and P. regilla was the dominant maintenance host:
Under the assumption of no connectivity, all patches within a metacommunity had to be, by definition, source patches –if Bd was present and endemic and the patch was not connected to any other patch then it must be a source patch. However, connectivity can alter the relative maintenance and source potential of a species and patch, respectively. We found that across the plausible parameter space of connectivity, the importance of P. regilla as a maintenance host within patches was largely unchanged (Fig. 3). Under plausible levels of connectivity, the relative of P. regilla was larger than other amphibian species in 81% (483 / 600 combinations) of the patches in which the amphibian species co-occurred (Fig. 3).
While species maintenance potential did not change across plausible connectivity parameters, patch source potential did (e.g. Fig. 4). However, even under the maximally connected plausible parameter scenario 51 of the 62 metacommunities with more than one patch had two or more source patches contributing to Bd persistence (e.g. Fig. 4).
Question 3: What are the contributions of maintenance species compared to source patches for pathogen persistence?
Removing P. regilla from metacommunities led to larger decreases in landscape-level R0,L than removing the most influential source patch:
Over the plausible range of connectivity, removing P. regilla led to, on average, a 53% larger reduction in landscape-level R0,L compared to removing the largest source patch (95% confidence interval from single sample t-test: [33%, 85%], Fig. 5). In contrast, removing any of the other five amphibian species was significantly less effective, on average, at reducing landscape-level R0,L than removing the most influential source patch (Fig. 5). In six of the 61 metacommunities with more than one patch and one species, removing the most influential source patch reduced landscape-level R0,L more than removing any particular species (Fig. 5).
Figure 5:
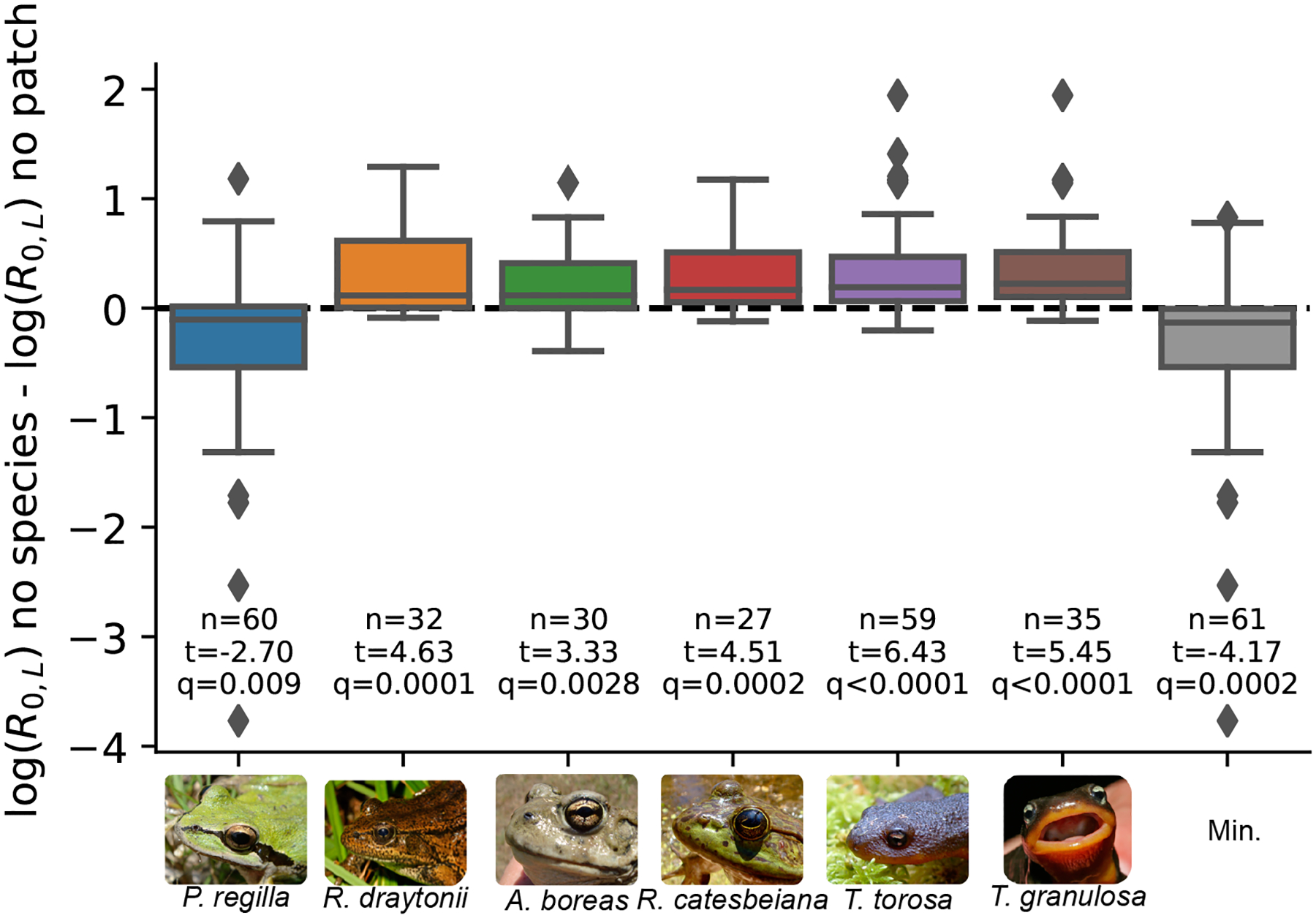
The effect of removing a species on landscape-level R0,L compared to removing the most influential source patch for 61 metacommunities with at least two patches and two species. Negative values indicate a larger reduction in landscape-level R0,L when a species is removed compared to when the most influential source patch is removed from the metacommunity. The sample sizes give the number of metacommunities out of 61 where a species was present. The t-statistics are from single sample t-tests testing the null hypothesis that the ratio is significantly different than zero. The q value is the significance value of the single sample t-test, after adjusting for multiple comparisons using the false discovery rate correction (Benjamini & Hochberg 1995). The gray boxplot “Min.” shows the minimum ratio across all species within a metacommunity. The dashed line indicates where removing a species and removing the most influential source patch had the same effect on landscape-level R0,L.
Discussion
Understanding how different scales of heterogeneity in host-parasite systems interact can improve the efficiency of managing human and wildlife diseases (Lloyd-Smith et al. 2005; Paull et al. 2012; Webster et al. 2017). By identifying the relative effects of super-spreading individuals (Lloyd-Smith et al. 2005), highly competent host species (Kilpatrick et al. 2006), or source patches on the landscape (Paull et al. 2012), the effort required to mitigate pathogen impacts can be greatly reduced. In this study, we used a widely-applicable theoretical framework to isolate different scales of heterogeneity contributing to pathogen persistence in multi-species, multi-patch host-pathogen systems. We linked this framework to empirical pathogen data from over 11,000 hosts comprising six species across 77 metacommunities to identify to roles of species, patches and connectivity on pathogen persistence at the landscape-level. We found that pathogen persistence in multi-species, multi-patch metacommunities was primarily driven by a single maintenance host species, rather than particular source patches and among patch connectivity. Our study contributes to broader theory on host-pathogen dynamics by illustrating that even in host-pathogen systems with multiple sources of heterogeneity (e.g. species-level and patch-level heterogeneity), a single source of heterogeneity can disproportionately contribute to pathogen persistence.
A key challenge that multi-species communities pose for disease management is that the identity of maintenance species can vary across habitat patches, making pathogen management strategies habitat-dependent (Rudge et al. 2013; Webster et al. 2017). Quantifying the degree to which maintenance potential depends on the species and not the habitat can help identify when specific-specific management can reduce disease on the landscape. In our study, we found that the Pacific tree frog (P. regilla) was consistently the dominant maintenance host across patches. This was reflected in higher relative values of species-level when P. regilla co-occurred with other amphibian species. However, we did find that in some communities other amphibian species were predicted to be maintenance hosts. Of particular interest in amphibian-Bd systems is the effect that the invasive American bullfrog R. catesbeiana has on Bd persistence (Garner et al. 2006; Adams et al. 2017). We found that, while infected with Bd in this system, bullfrogs were not consistently more important relative maintenance hosts than P. regilla, A. boreas, or R. draytonii when these species co-occurred. Moreover, while bullfrogs were predicted to be maintenance hosts in 17 patches under a no connectivity scenario, bullfrogs did not remain a maintenance host in six of these source patches when we included patch connectivity. This was in contrast to the patches where A. boreas and R. draytonii were maintenance hosts and remained maintenance hosts with or without connectivity. Taken together, our results suggest that bullfrogs are not disproportionately more influential on within-season Bd dynamics in an average patch than other amphibian species found in this system. However, given the multi-year tadpole stages of bullfrogs we cannot rule out their importance in between-season Bd dynamics.
Empirical studies often identify host maintenance potential using independent comparisons of host characteristics such as prevalence, pathogen load, disease-induced mortality, and host density (e.g. Reeder et al. 2012; Stockwell et al. 2016; Brannelly et al. 2018; Hudson et al. 2019). While a useful approach, the challenge with independently using these characteristics to identify maintenance hosts is that it becomes hard to compare maintenance potential among multiple species within a community. For example, in our study, while P. regilla had a higher density than R. draytonii in all patches where they co-occurred, it tended to have a lower Bd prevalence and load relative to R. draytonii; as a result, P. regilla was the more important maintenance host in only 65% of these patches. Previous work in multi-species systems has shown how these commonly-collected characteristics can be linked to an established quantitative measure of maintenance potential, R0 (Rudge et al. 2013; Fenton et al. 2015), and we generalized this approach to multi-species, multi-patch host-pathogen systems. Note that computing R0 within and across habitat patches does require assumptions that need to be checked (Keelling & Rohani 2008; Fenton et al. 2015). However, when done systematically it provides an unambiguous way to relate characteristics that are suggestive of a maintenance host to a quantitative measure of maintenance potential across species and patches.
Identifying host maintenance potential in multi-host communities can have important conservation implications for managing disease impacts. While Bd was not a cause of conservation concern in our system, it is in many other multi-species amphibian communities (Scheele et al. 2019). In Central and South America, for example, amphibians have experienced drastic Bd-induced declines and particular species have been implicated as disproportionately contributing to infection risk (Schloegel et al. 2010; DiRenzo et al. 2014). However, we are not aware of any studies in amphibian-Bd systems that have quantified maintenance hosts by synthesizing the multiple dimensions of host and pathogen characteristics into a single, theoretically-supported metric of maintenance potential: species-level R0 (see Canessa et al. 2019, for an example with the pathogen Batrachochytrium salamandrivorans). The approach developed in Fenton et al. (2015) and extended here provides a feasible way to use data often collected in amphibian-Bd systems with multiple host species to promote theoretically informed amphibian management where Bd is a conservation concern.
When habitat patches are unconnected, identifying species maintenance potential is key for understanding pathogen dynamics within a patch (Fenton et al. 2015). However, when patches are connected, heterogeneity in species maintenance potential across habitat patches can lead to heterogeneity in patch source potential, which can interact with patch connectivity through source-sink dynamics to affect pathogen persistence within a metacommunity (Schreiber & Lloyd-Smith 2009; North & Godfray 2017). Equation 2 and Fig. 4 illustrate this point –depending on the level of connectivity and variability in host prevalence and density across patches, species-level values can increase or decrease as connectivity increases and can do so at different rates across species and patches. Thus, for example, hosts that were predicted to be maintenance species at low levels of connectivity may no longer be maintenance species at high levels of connectivity. Testing the effects of connectivity on our predictions of species maintenance potential, we found minimal changes in relative species maintenance potential compared to no connectivity. While we did observe significant changes in patch source potential as connectivity increased, even at maximum plausible levels of connectivity we found that 82% (51 / 62) of the amphibian-Bd metacommunities observed in this system were most consistent with a weakly connected network of more than one source patch.
Pathogen dynamics within a metacommunity are driven by processes operating across scales (Paull et al. 2012). However, most empirical applications of epidemiological theory have considered the role of processes operating at a single scale on emergent disease dynamics (e.g. how does individual heterogeneity in contact rates affect pathogen invasion? Bansal et al. 2007; Schreiber & Lloyd-Smith 2009; Rudge et al. 2013). The theoretical developments in this study, combined with a large spatial dataset, provided us the unique opportunity to test the relative contributions of species- and patch-level processes to disease persistence in a metacommunity. When we systematically removed either the dominant maintenance species or source patch in a metacommunity in silico, we found that removing the dominant maintenance species on average reduced landscape-level R0,L twice as much as removing the primary source patch in the metacommunity. As P. regilla was the dominant maintenance species in most metacommunities, removing P. regilla was the most effective strategy for reducing landscape-level R0,L for Bd. Note that our in silico removal of a species does not necessarily mean killing the species. Any mechanism that removed the potential for a species to contribute to Bd transmission, such as treatment, could similarly affect landscape-level R0,L.
There were two reasons for the larger effects of P. regilla removal than habitat patch removal on landscape-level R0,L. First, because P. regilla maintenance potential was consistent across patches, removing P. regilla consistently removed the most important species for Bd persistence within a patch. Second, because P. regilla was widely dispersed on the landscape, the source potentials of many patches within a metacommunity were affected by P. regilla removal. In contrast, just removing a widely-dispersed species (e.g. T. torosa) or a species with high maintenance potential (e.g. R. draytonii) was significantly less effective at reducing R0,L than removing the most influential source patch. These results highlight the importance of identifying how consistent species maintenance potentials are across habitat patches, as this can help determine whether landscape-level management of pathogens might be most effective at the species or patch scale.
The model we develop is applicable to other multi-species, multi-patch systems and is amenable to asking additional theoretical questions to further unravel how multiple sources of heterogeneity drive host-pathogen dynamics. Just as with the multi-species models that our approach extends (Rudge et al. 2013; Fenton et al. 2015), our model is particularly useful at the interface between theory and data because hard-to-estimate rates such as transmission, pathogen decay, and dispersal are not needed. Therefore, multi-patch, multi-species models can be more easily linked with commonly-collected empirical data to identify how different scales of heterogeneity affect pathogen persistence.
Supplementary Material
Acknowledgments
For host sampling and fieldwork, we thank T. McDevitt-Galles, W. Moss, D. Calhoun, T. Riepe, K. Leslie, and the many other individuals who helped with data collection. Access to properties and wetlands was kindly provided by East Bay Regional Parks District, East Bay Municipal Utility District, Santa Clara County Parks, and multiple private landowners. M. Hamilton and Z. Harlow of the Blue Oak Ranch Reserve (University of California) provided logistical support, laboratory space, and housing for field sampling. Thanks to R. Chen, F. Pfab, S. Sambado, I. Russell and three anonymous reviewers for useful feedback on the manuscript. We also thank R. Chen for processing qPCR samples. Funding was provided by the David and Lucile Packard Foundation, the National Science Foundation (DEB 1149308 and DEB 1754171), the US Fish and Wildlife Services Endangered Species Conservation and Recovery Grant Program, and the NIH/NSF Ecology and Evolution of Infectious Diseases program (R01GM109499 and R01GM135935).
Footnotes
Data accessibility statement: Data are available from the Dryad Digital Repository: https://doi.org/10.25349/D9W59R
Contributor Information
Mark Q. Wilber, Email: mark.wilber@lifesci.ucsb.edu.
Pieter T. J. Johnson, Email: pieter.johnson@colorado.edu.
Cheryl J. Briggs, Email: briggs@lifesci.ucsb.edu.
References
- Adams AJ, Kupferberg SJ, Wilber MQ, Pessier AP, Grefsrud M, Bobzien S, Vredenburg VT & Briggs CJ (2017). Extreme drought, host density, sex, and bullfrogs influence fungal pathogen infection in a declining lotic amphibian. Ecosphere, 8, e01740. [Google Scholar]
- Arino J (2009). Diseases in metapopulations. In: Modeling the Dynamics of Infectious Disease, Series in Contemporary Applied Mathematics (eds. Ma Z, Zhou Y & Wu J). World Scientific, Singapore, pp. 64–122. [Google Scholar]
- Arino J, Davis JR, Hartley D, Jordan R, Miller JM & van den Driessche P (2005). A multi-species epidemic model with spatial dynamics. Math Med Biol, 22, 129–142. [DOI] [PubMed] [Google Scholar]
- Bansal S, Grenfell BT & Meyers LA (2007). When individual behaviour matters: Homogeneous and network models in epidemiology. J R Soc Interface, 4, 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y & Hochberg Y (1995). Controlling the False Discovery Rate: A practical and powerful approach to multiple testing. J Roy Stat Soc B Met, 57, 289–300. [Google Scholar]
- Boyle DG, Boyle DB, Olsen V, Morgan JAT & Hyatt AD (2004). Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis Aquat Organ, 60, 141–8. [DOI] [PubMed] [Google Scholar]
- Brannelly LA, Webb RJ, Hunter DA, Clemann N, Howard K, Skerratt LF, Berger L & Scheele BC (2018). Non-declining amphibians can be important reservoir hosts for amphibian chytrid fungus. Anim Conserv, 21, 91–101. [Google Scholar]
- Briggs CJ, Knapp RA & Vredenburg VT (2010). Enzootic and epizootic dynamics of the chytrid fungal pathogen of amphibians. P Natl Acad Sci USA, 107, 9695–9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canessa S, Bozzuto C, Pasmans F & Martel A (2019). Quantifying the burden of managing wildlife diseases in multiple host species. Conserv Biol, 33, 1131–1140. [DOI] [PubMed] [Google Scholar]
- Cross PC, Lloyd-Smith JO, Johnson PLF & Getz WM (2005). Duelling timescales of host movement and disease recovery determine invasion of disease in structured populations. Ecol Lett, 8, 587–595. [Google Scholar]
- De Castro F & Bolker B (2005). Mechanisms of disease-induced extinction. Ecol Lett, 8, 117–126. [Google Scholar]
- Diekmann O, Heesterbeek JAP & Metz JAJ (1990). On the definition and the computation of the basic reproduction ratio R0 in models for infectious diseases in heterogeneous populations. J Math Biol, 28, 365–382. [DOI] [PubMed] [Google Scholar]
- DiRenzo GV, Campbell Grant EH, Longo AV, Che-Castaldo C, Zamudio KR & Lips KR (2018). Imperfect pathogen detection from non-invasive skin swabs biases disease inference. Methods Ecol Evol, 9, 380–389. [Google Scholar]
- DiRenzo GV, Langhammer PF, Zamudio KR & Lips KR (2014). Fungal infection intensity and zoospore output of Atelopus zeteki, a potential acute chytrid supershedder. PLoS ONE, 9, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A (2004). Population dynamics of pathogens with multiple host species. Am Nat, 164, 64–78. [DOI] [PubMed] [Google Scholar]
- Fenton A, Streicker DG, Petchey OL & Pedersen AB (2015). Are all hosts created equal? Partitioning host species contributions to parasite persistence in multihost communities. Am Nat, 186, 610–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner TWJ, Perkins MW, Govindarajulu P, Seglie D, Walker S, Cunningham AA & Fisher MC (2006). The emerging amphibian pathogen Batrachochytrium dendrobatidis globally infects introduced populations of the North American bullfrog, Rana catesbeiana. Dis Aquat Organ, 2, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon DT, Cleaveland S, Taylor LH & Laurenson MK (2002). Identifying reservoirs of infection: A conceptual and practical challenge. Emerg Infect Dis, 8, 1468–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson MA, Griffiths RA, Martin L, Fenton C, Adams S-L, Blackman A, Sulton M, Perkins MW, Lopez J, Garcia G, Tapley B, Young RP & Cunningham AA (2019). Reservoir frogs: Seasonality of Batrachochytrium dendrobatidis infection in robber frogs in Dominica and Montserrat. PeerJ, 7, e7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt AD, Boyle DG, Olsen V, Boyle DB, Berger L, Obendorf D, Dalton A, Kriger K, Hero M, Hines H, Phillott R, Campbell R, Marantelli G, Gleason F & Colling A (2007). Diagnostic assays and sampling protocols for the detection of Batrachochytrium dendrobatidis. Dis Aquat Organ, 73, 175–192. [DOI] [PubMed] [Google Scholar]
- Johnson PT, De Roode JC & Fenton A (2015). Why infectious disease research needs community ecology. Science, 349, 1259504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PTJ, Preston DL, Hoverman JT & Richgels KLD (2013). Biodiversity decreases disease through predictable changes in host community competence. Nature, 494, 230–233. [DOI] [PubMed] [Google Scholar]
- Johnson PTJ, Rohr JR, Hoverman JT, Kellermanns E, Bowerman J & Lunde KB (2012). Living fast and dying of infection: Host life history drives interspecific variation in infection and disease risk. Ecol Lett, 15, 235–242. [DOI] [PubMed] [Google Scholar]
- Johnson PTJ, Wood CL, Joseph MB, Preston DL, Haas SE & Springer YP (2016). Habitat heterogeneity drives the host-diversity-begets-parasite-diversity relationship: Evidence from experimental and field studies. Ecol Lett, 19, 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph MB, Stutz WE & Johnson PT (2016). Multilevel models for the distribution of hosts and symbionts. PLoS ONE, 11, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousimo J, Tack AJM, Ovaskainen O, Mononen T, Susi H, Tollenaere C & Laine A-L (2014). Ecological and evolutionary effects of fragmentation on infectious disease dynamics. Science, 344, 1289–1294. [DOI] [PubMed] [Google Scholar]
- Keelling M & Rohani P (2008). Modeling Infectious Diseases in Humans and Animals. Princeton University Press, Princeton, New Jersey. [Google Scholar]
- Kilpatrick AM, Briggs CJ & Daszak P (2010). The ecology and impact of chytridiomycosis: An emerging disease of amphibians. Trends Ecol Evol, 25, 109–118. [DOI] [PubMed] [Google Scholar]
- Kilpatrick AM, Daszak P, Jones MJ, Marra PP & Kramer LD (2006). Host heterogeneity dominates West Nile virus transmission. P Roy Soc B-Biol Sci, 273, 2327–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Smith JO, Schreiber SJ, Kopp PE & Getz WM (2005). Superspreading and the effect of individual variation on disease emergence. Nature, 438, 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longcore JE, Pessier AP & Nichols DK (1999). Batrachochytrium dendrobatidis gen. et sp. nov., a chytrid pathogenic to amphibians. Mycologia, 91, 219–227. [Google Scholar]
- McCallum H (2012). Disease and the dynamics of extinction. Philos T Roy Soc B, 367, 2828–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum H & Dobson A (2002). Disease, habitat fragmentation and conservation. P Roy Soc B-Biol Sci, 269, 2041–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum HI (2008). Landscape structure, distrubance, and disease dynamics In: Infectious Disease Ecology: Effects of Ecosystems on Disease and of Disease on Ecosystems (eds. Ostfeld RS, Keesing F & Eviner VT). Princeton University Press, Princeton, New Jersey. [Google Scholar]
- Mihaljevic JR, Joseph MB, Orlofske SA & Paull SH (2014). The scaling of host density with richness affects the direction, shape, and detectability of diversity-disease relationships. PLoS ONE, 9, e97812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DAW, Talley BL, Lips KR & Campbell Grant EH (2012). Estimating patterns and drivers of infection prevalence and intensity when detection is imperfect and sampling error occurs. Methods Ecol Evol, 3, 850–859. [Google Scholar]
- Miller RS, Farnsworth ML & Malmberg JL (2013). Diseases at the livestock-wildlife interface: Status, challenges, and opportunities in the United States. Prev Vet Med, 110, 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KM, Churcher TS, Garner TWJ & Fisher MC (2008). Persistence of the emerging pathogen Batrachochytrium dendrobatidis outside the amphibian host greatly increases the probability of host extinction. P Roy Soc B-Biol Sci, 275, 329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North AR & Godfray HCJ (2017). The dynamics of disease in a metapopulation: The role of dispersal range. J Theor Biol, 418, 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park AW, Gubbins S & Gilligan CA (2001). Invasion and persistence of plant parasites in a spatially structured host population. Oikos, 94, 162–174. [Google Scholar]
- Paull SH, Song S, McClure KM, Sackett LC, Kilpatrick a. M. & Johnson PTJ (2012). From superspreaders to disease hotspots: linking transmission across hosts and space. Front Ecol Environ, 10, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penczykowski RM, Laine A-L & Koskella B (2015). Understanding the ecology and evolution of host parasite interactions across scales. Evol Appl, 9, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder NMM, Pessier AP & Vredenburg VT (2012). A reservoir species for the emerging amphibian pathogen Batrachochytrium dendrobatidis thrives in a landscape decimated by disease. PLoS ONE, 7, e33567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MG & Heesterbeek JA (2020). Characterizing reservoirs of infection and the maintenance of pathogens in ecosystems. J R Soc Interface, 17, 20190540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge JW, Webster JP, Lu D-B, Wang T-P, Fang G-R & Basanez M-G (2013). Identifying host species driving transmission of schistosomiasis japonica, a multihost parasite system, in China. P Natl Acad Sci USA, 110, 11457–11462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheele BC, Pasmans F, Skerratt LF, Berger L, Martel A, Beukema W, Acevedo AA, Burrowes PA, Carvalho T, Catenazzi A, la Riva I, Fisher MC, Flechas SV, Foster CN, Frías-Álvarez P, Garner TWJ, Gratwicke B, Guayasamin JM, Hirschfeld M, Kolby JE, Kosch TA, La Marca E, Lindenmayer DB, Lips KR, Longo AV, Maneyro R, McDonald CA, Mendelson J, Palacios-Rodriguez P, Parra-Olea G, Richards-Zawacki CL, Rödel M-O, Rovito SM, Soto-Azat C, Toledo LF, Voyles J, Weldon C, Whitfield SM, Wilkinson M, Zamudio KR & Canessa S (2019). Amphibian fungal panzootic causes catastrophic and ongoing loss of biodiversity. Science, 363, 1459–1463. [DOI] [PubMed] [Google Scholar]
- Schloegel LM, Ferreira CM, James TY, Hipolito M, Longcore JE, Hyatt AD, Yabsley M, Martins AM, Mazzoni R, Davies AJ & Daszak P (2010). The North American bullfrog as a reservoir for the spread of Batrachochytrium dendrobatidis in Brazil. Anim Conserv, 13, 53–61. [Google Scholar]
- Schreiber SJ & Lloyd-Smith JO (2009). Invasion dynamics in spatially heterogeneous environments. Am Nat, 174, 490–505. [DOI] [PubMed] [Google Scholar]
- Stebbins RC & McGinnis SM (2012). Field Guide to Amphibians and Reptiles of California: Revised Edition. University of California Press, Berkeley, CA. [Google Scholar]
- Stockwell MP, Bower DS, Clulow J & Mahony MJ (2016). The role of non-declining amphibian species as alternative hosts for Batrachochytrium dendrobatidis in an amphibian community. Wildlife Res, 43, 341–347. [Google Scholar]
- Stutz WE, Blaustein AR, Briggs CJ, Hoverman JT, Rohr JR & Johnson PT (2018). Using multi-response models to investigate pathogen coinfections across scales: Insights from emerging diseases of amphibians. Methods Ecol Evol, 9, 1109–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster JP, Borlase A & Rudge JW (2017). Who acquires infection from whom and how? Disentangling multi-host and multimode transmission dynamics in the elimination’ era. Philos T Roy Soc B, 372, 20160091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilber MQ, Pepin KM, Campa III H, Hygnstrom SE, Lavelle MJ, Xifara T, VerCauteren KC & Webb CT (2019). Modeling multi-species and multi-mode contact networks: implications for persistence of bovine tuberculosis at the wildlife-livestock interface. J Appl Ecol, 56, 1471–1481. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


