Abstract
The conformational conversion of the cellular prion protein (PrPC) to the misfolded and aggregated isoform, termed scrapie prion protein (PrPSc), is key to the development of a group of neurodegenerative diseases known as transmissible spongiform encephalopathies (TSEs). Although the conversion mechanism is not fully understood, the role of gene polymorphisms in varying susceptibilities to prion diseases is well established. In ovine, specific gene polymorphisms in PrPC alter prion disease susceptibility: the Valine136-Glutamine171 variant (Susceptible structure) displays high susceptibility to classical scrapie while the Alanine136-Arginine171 variant (Resistant structure) displays reduced susceptibility. The opposite trend has been reported in atypical scrapie. Despite the differentiation between classical and atypical scrapie, a complete understanding of the effect of polymorphisms on the structural dynamics of PrPC is lacking. From our structural bioinformatics study, we propose that polymorphisms locally modulate the network of residue interactions in the globular C-terminus of the ovine recombinant prion protein while maintaining the overall fold. Although the two variants we examined exhibit a densely connected group of residues that includes both β-sheets, the β2-α2 loop and the N-terminus of α-helix 2, only in the Resistant structure do most residues of α-helix 2 belong to this group. We identify the structural role of Valine136Alanine and Glutamine171Arginine: modulation of residue interaction networks that affect the connectivity between α-helix 2 and α-helix 3. We propose blocking interactions of residue 171 as a potential target for the design of therapeutics to prevent efficient PrPC misfolding. We discuss our results in the context of initial PrPC conversion and extrapolate to recently proposed PrPSc structures.
Keywords: cellular prion protein, prion protein gene polymorphisms, molecular dynamics simulations, residue interaction network
Introduction
Prions are agents responsible for transmissible spongiform encephalopathies (TSE) (Prusiner, Scott, DeArmond, & Cohen, 1998), fatal neurodegenerative diseases in mammals. The most common form of prion disorders in humans is sporadic Creutzfeldt-Jakob disease (sCJD), which is reported to affect at least one person per million per year (Klug et al., 2013). The zoonotic nature of BSE in cattle (Bruce et al., 1997), and the zoonotic potential of CWD in cervids (Marsh, Kincaid, Bessen, & Bartz, 2005), and scrapie in ovine (Cassard et al., 2014), pose a public health risk. Prions, composed primarily of the misfolded and β-sheet rich aggregated prion protein, PrPSc, propagate information in the absence of specific nucleic acids (Burke et al., 2019). The conformational conversion of the host encoded version of the prion protein, PrPc, into the pathological isoform, PrPSc, is fundamental to prion formation (Deleault, Harris, Rees, & Supattapone, 2007). The mechanism of prion protein conformational conversion and propagation has been suggested to occur in other amyloidosis, such as Alzheimer’s and Parkinson’s diseases (Soto, 2012;Goedert, 2015). However, a complete molecular picture of the prion protein biological function, and of the mechanism of conversion, propagation and induced neurodegeneration remains elusive, despite the contributions of many (Sigurdson, Bartz, & Glatzel, 2019).
The cellular prion protein, PrPC, is an extracellular glycoprotein anchored via a glycosylphosphatidylinositol, GPI, molecule to the outer leaflet of the plasma membrane. The N-terminus of PrPC is highly unstructured and the C-terminus is globular with a well-defined secondary structure pattern: two short β-sheets and three α-helices. Previous works highlight the β–sheet 1 – α–helix 1 – β–sheet 2 fragment as key in nucleation and propagation of β–strand structures and in oligomer formation or polymerization process (Rezaei et al., 2005; Gao, Zhu, Zhang, Zhang, & Mei, 2018; Ji, Zhang, & Shen, 2005; DeMarco & Daggett, 2007; Blinov, Berjanskii, Wishart, & Stepanova, 2009). A second scenario proposes the unfolding of α–helix 2 and α–helix 3 as a key event in PrPC initial misfolding and oligomerization (Chakroun et al., 2013; Dima & Thirumalai, 2002; Tycko, Savtchenko, Ostapchenko, Makarava, & Baskakov, 2010; Adrover et al., 2010). A third scenario identifies the β2–α2 loop as key for structural transitions that may lead to misfolding (Gossert, Bonjour, Lysek, Fiorito, & Wuthrich, 2005; Sigurdson et al., 2009; Meli, Gasset, & Colombo, 2011; Gorfe & Caflisch, 2007).
The development of therapies to either cure, delay or prevent prion diseases is an active field of research. The lack of atomic-level structural information on PrPC conversion and PrPSc structure makes native PrPC an amenable target to identify druggable pathways (Rigoli, Spagnolli, Faccioli, Requena, & Biasini, 2019). The ovine prion protein PrPC is a model system to identify how polymorphisms modulate PrPC structural dynamics that leads to pathological conversion. The two reported forms of the disease, classical and atypical/Nor98 scrapie, show contrasting features (CFSPH, 2016): i) the number of detected strains causing classical scrapie is much greater than in atypical/Nor98, ii) classical is transmitted between hosts much more readily than atypical/Nor98, which is believed to be sporadic, and iii) the genotype of the host is distinctively linked to scrapie susceptibility. In classical scrapie, the Val136-Arg154-Glu171 (VRQ, termed “Susceptible” in this article) variant confers high scrapie susceptibility while the Ala136-Arg154-Arg171 (ARR, termed “Resistant” in this article) variant induces reduced susceptibility (Belt et al., 1995; Bossers, Schreuder, Muileman, Belt, & Smits, 1996; Ulvund, Bratberg, Osland, & Tranulis, 1999). In atypical/Nor98 scrapie, the susceptibility trend looks reversed: the ARR genotype is common while hosts with the VRQ genotype display reduced susceptibility. Although evidence suggests that tgHu mice intracerebrally inoculated with some classical scrapie isolates develop sCJD like phenotype (Cassard et al., 2014), no evidence has been reported of natural transmission of scrapie to humans (EFSA-BIOHAZ, 2015).
Studies suggest that the polymorphism at residue 136 (Ala in Resistant, Val in Susceptible) has a significant effect on disease susceptibility while the polymorphism at residue 171 (Arg in Resistant, Glu in Susceptible) modulates the incubation period (Goldmann, Hunter, Smith, Foster, & Hope, 1994). In vitro experiments differentiate the Susceptible and Resistant structure: First, the Susceptible structure shows higher thermal stability than the Resistant form. Second, although both variants form amyloids at acidic and physiological pH, the activation energy was greater in the Resistant form than in the Susceptible. Also, the β–sheet population of unfolding intermediates was found to be distinct in both structures(Rezaei et al., 2002). Third, the Susceptible form displays high conformational plasticity (Van der Rest, Rezaei, & Halgand, 2017). Taken together, the evidence lends support to the model of domains in PrPC, each with a distinct role in conversion and propagation (Goldmann et al., 1994), and the coexistence of multiple ovine PrPC to PrPSc conversion pathways.
Despite the evidence linking polymorphisms in PrPC to scrapie, an understanding of the effect of the polymorphisms on the structural dynamics of the C-terminus of PrPC is far from complete. The structural role of Val136Ala and Gln171Arg polymorphisms show competing effects: Valine (in the Susceptible structure) is more hydrophobic and has one more side chain dihedral angle that allows for a greater number of side chain orientations than alanine (in the Resistant structure). Glutamine (in the Susceptible structure) forms hydrogen bonds, while the electrostatics of arginine (in the Resistant structure) allows for hydrogen and ionic bonding. Additionally, the amphipathic arginine displays a longer hydrophobic side chain and one more dihedral angle than glutamine, which permits a greater number of side chain orientations.
X-ray crystallography studies (Eghiaian et al., 2004) indicate that the three-dimensional conformation of the Susceptible and Resistant structure display a similar overall fold. From this and molecular dynamics simulations studies (Bujdoso, Burke, & Thackray, 2005; Fitzmaurice et al., 2008), it has been proposed that the Susceptible structure is characterized with a greater number of hydrogen bonds on the protein surface, restricted conformations of the Asn162 and Arg139 side chains, decreased flexibility of the β2–α2 loop due to the Arg167-Gln171 hydrogen bond, increased length of β–sheet 1 and β–sheet 2, allele dependent local flexibility pattern in the α2-α3 loop, and reduced overall conformational flexibility with respect to the Resistant structure. However, the extent of the inter-dependent effects of Val136Ala and Gln171Arg on the structural dynamics of PrPC remains unknown, as well as how these effects could be interpreted to identify molecular targets for therapeutics.
Our study aims at providing an atomic level picture of the effect of Val136Ala and Gln171Arg on the network of protein residue interactions in ovine PrPC. Our analysis identifies the structural role of Val136Ala and Gln171Arg: modulation of the residue interaction network that affects the connectivity between α-helix 2 and α-helix 3. From this, we theorize on initial misfolding pathways of PrPC. We propose blocking interactions of residue 171 as a potential target for the design of therapeutics to prevent efficient PrPC misfolding.
Materials and methods
Each simulation system consisted of the cellular prion protein monomer (PrPC) solvated in aqueous solution. Two monomer ovine PrPC structures were simulated: i) The Val136-Arg154-Gln171 structure (“Susceptible”, pdb id: 1TQB) that confers high classical scrapie susceptibility and ii) the Ala136-Arg154-Arg171 structure (“Resistant”, pdb id: 1TQC) that induces resistance (Eghiaian et al., 2004). The structures deposited in the protein data bank correspond to variants of the ovine recombinant PrP (114 – 234) in complex with the VRQ14 Fab fragment. In our simulations, we used only the ovine recombinant PrP for which the Cartesian coordinates of the heavy atoms were deposited (residues 127 – 228). The glycan groups attached to residues Asn184 and Asn200 were not included in our model, as correlation between N-glycosylation and degree of susceptibility to scrapie diseases has not been found so far (Uslupehlivan, Deveci, & Ün, 2018). The AMBER ILDN (Lindorff-Larsen et al., 2010) force field was used for the protein, and the system was solvated using the TIP3P water model (Jorgensen, Chandrasekhar, Madura, Impey, & Klein, 1983). The protonation state of the residues was assigned to mimic a pH of 7. Counter ions were added to each system to represent a physiological ionic concentration (0.1 M). Eight independent 200-ns long simulations were monitored per structure and the last 100 ns of all trajectories were used for analysis. The aggregated simulation time per structure was 1.6 μs.
Each system was energy minimized, position restrained and equilibrated in the NVT ensemble in a stepwise manner at 2 fs, 3 fs, 4 fs, and 5 fs. Data acquisition simulations in the NPT ensemble were performed using a leapfrog integrator with a 5 fs time step. Group lists were generated using a 1.0 nm neighbor list cutoff. Electrostatic interactions were calculated using the smooth particle mesh Ewald method (Essmann et al., 1995) with a short-range cutoff of 1.0 nm, grid spacing of 0.12 nm, and fourth order interpolation. van der Waals interactions were modeled with a cutoff that was smoothly shifted to zero between 1.0 and 1.2 nm. Bonds were constrained using the LINCS algorithm (Hess, Bekker, Berendsen, & Fraaije, 1997). The temperature of the equilibration and data acquisition simulations was maintained at 310 K using the velocity rescale thermostat (Bussi, Donadio, & Parrinello, 2007) with a time constant of 0.1 ps. In the data acquisition simulations, the pressure was maintained at 1 bar using the Parinello-Rahman barostat (Parrinello & Rahman, 1980) with a time constant of 4 ps and a compressibility of 4.5 × 10–5 bar–1. All molecular dynamics simulations were performed and analyzed using Gromacs 4.x.(Hess, Kutzner, Spoel, & Lindahl, 2008). Secondary structure was assigned according to the DSSP algorithm (Kabsch & Sander, 1983).
Dynamic correlation network analysis was implemented using Bio3D (Skjaerven, Yao, Scarabelli, & Grant, 2014). Similar to published work (Sethi, Eargle, Black, & Luthey-Schulten, 2009; Leontiadou, Galdadas, Athanasiou, & Cournia, 2018; Li, Yao, & Grant, 2018), the method builds a weighted graph in which each node corresponds to a protein residue. Edges connecting a pair of nodes are defined to exist if the residues have a contact (distance between any heavy atom of the residue) within a cutoff of 0.45 nm for at least 75% of the aggregated trajectory. The weight of each edge corresponds to -log(|cij|), with cij the Pearson’s inner product cross-correlation value calculated from the aggregated trajectory of each structure. The Girvan-Newman method (Girvan & Newman, 2002) was used to partition the network into communities: residues belonging to the same community are highly interconnected while connections between communities are loose.
Linear mixed model regressions used the lme4 library (Bates, Mächler, Bolker, & Walker, 2015) as implemented in the R software.(R-Core-Team, 2013)
Results
Polymorphisms preserve the overall shape of the ovine PrPC protein
Figure 1 shows the x-ray crystallography structure of the globular C-terminus ovine PrPC protein corresponding to the Susceptible form. The Susceptible and Resistant structures show two short β–sheets, three α–helices and connecting unstructured loops (Eghiaian et al., 2004). The polymorphisms occur at two residue positions:
residue 136 (located in the β1–α1 loop) with valine in the Susceptible structure and the less hydrophobic alanine in the Resistant structure
residue 171 (located in the β2–α2 loop) with the polar glutamine in the Susceptible structure and the basic arginine in the resistant structure.
Figure 1.
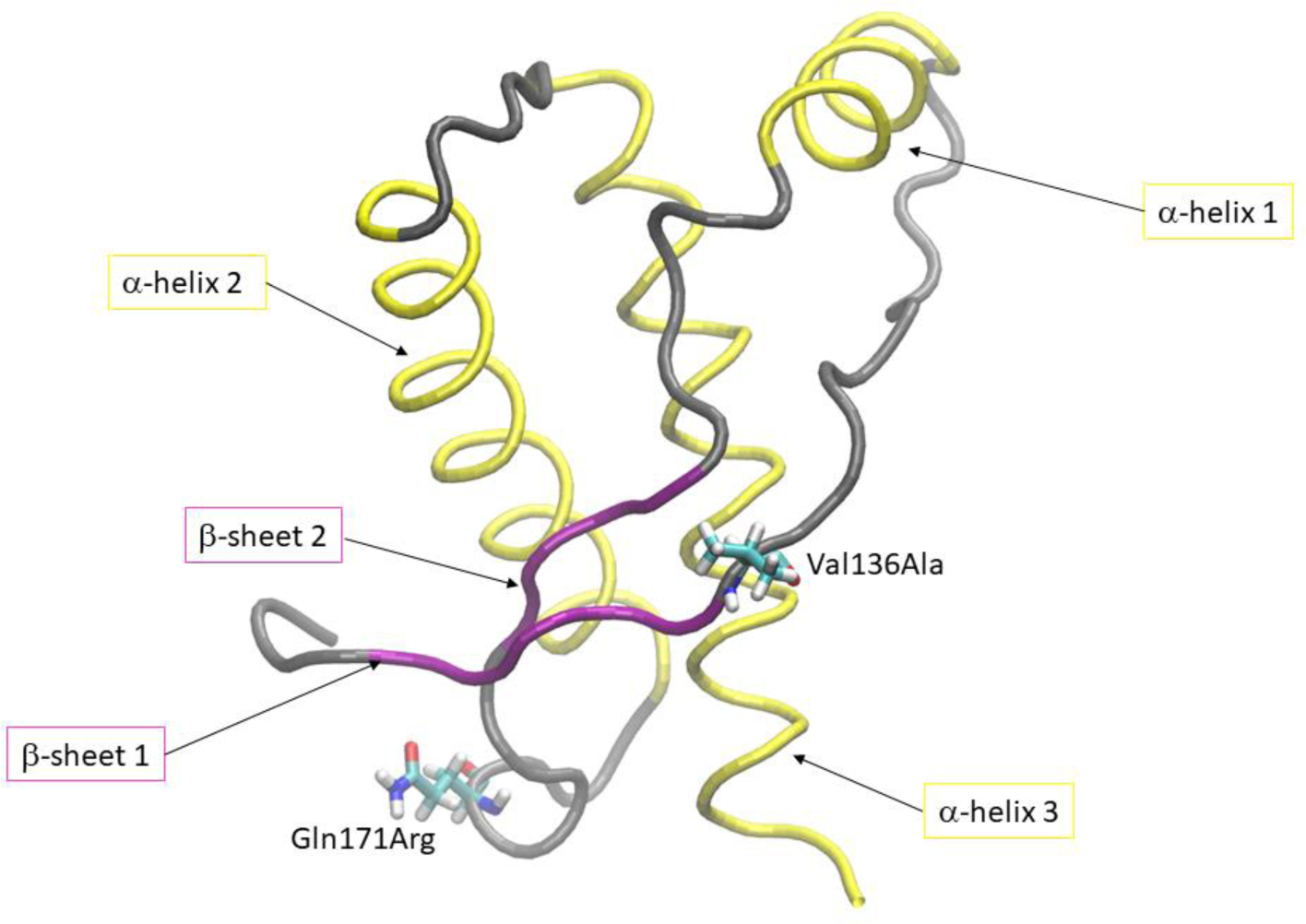
Three dimensional structure of ovine PrPC (pdb id: 1tqb). Secondary structure elements and polymorphisms at residue position 136 and 171 are highlighted.
The sixteen molecular dynamics trajectories monitored reached equilibrium as indicated by a Cα-position RMSD with respect to the initial structure of each trajectory less than 0.4 nm in the last 100 ns of each trajectory (see SI Figure 1). Standard structural descriptors indicate no strong effect on the overall shape of the folded ovine PrPC structure of Susceptible and Resistant due to polymorphisms. Figure 2A) shows that most conformations of the Susceptible (horizontal axis) and Resistant (vertical axis) structures are similar, within a Cα RMSD of 0.5 nm.
Figure 2.
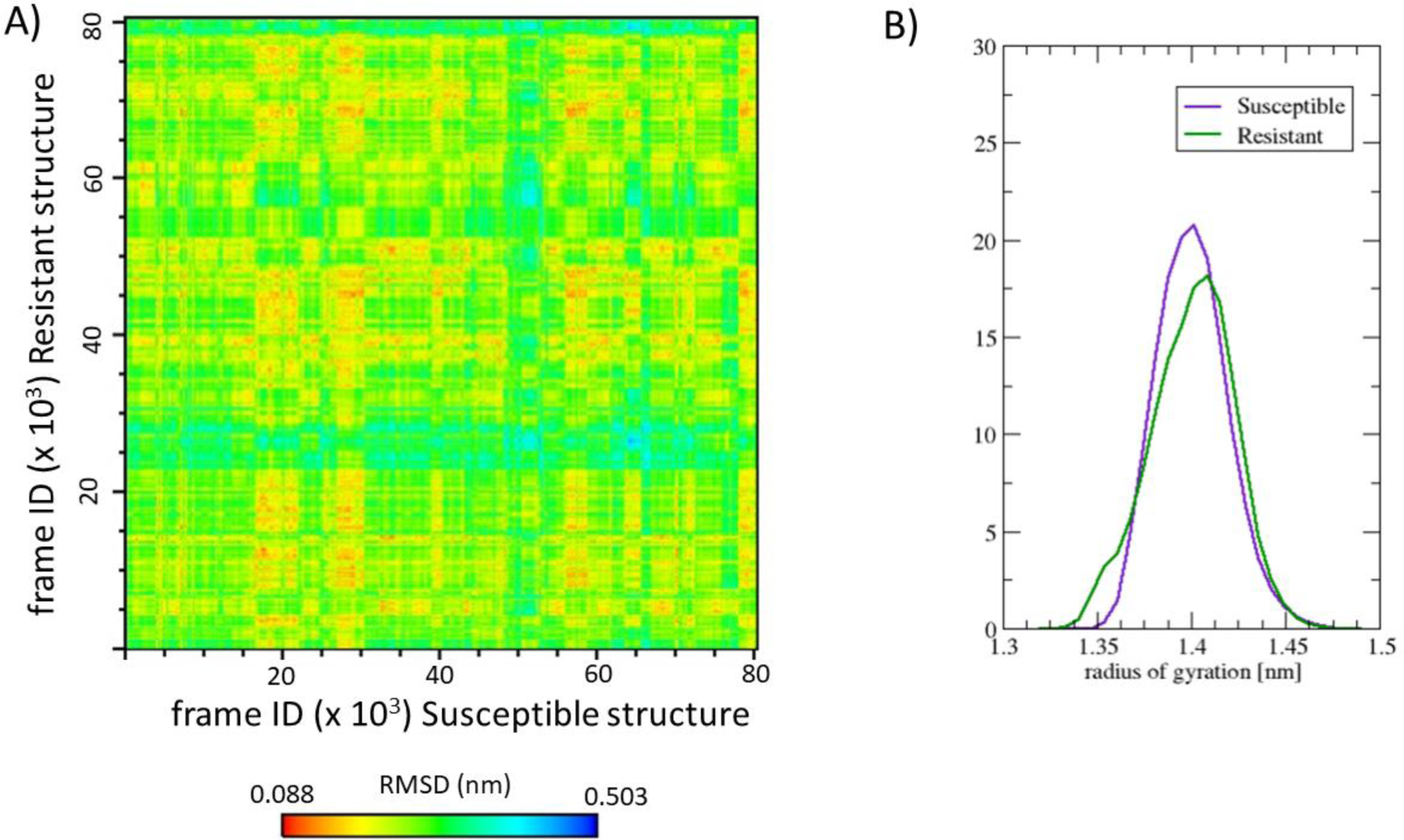
A) Matrix of Cα-position RMSD comparing all conformations of the Susceptible structure with the Resistant structure. B) Histogram of the radius of gyration distribution of the aggregated pool of conformations of the Susceptible and Resistant structure. The overall shape of the globular C-terminal ovine PrPC protein is preserved upon residue changes Val136Ala and Gln171Arg.
Figure 2B) shows that the distribution of Cα radius of gyration follows a similar trend in the Susceptible and Resistant structure. The Resistant and Susceptible structures sample a similar range of radius of gyration (see Figure 2B). Upon a mixed model regression analysis, we cannot conclude that the distributions of the radius of gyration are distinct, and our analysis therefore does not provide evidence of differential compactness. The trend of buried and exposed residues is the same in the Susceptible and Resistant structure (see SI Figure 2), except at the polymorphisms (Val136Ala and Gln171Arg), as expected. SI Figure 3 indicates that β-sheets and α-helices are preserved along the trajectories in the Susceptible and the Resistant structure.
However, the only difference in secondary structure is detected in the β2-α2 loop. The two-dimensional histogram of backbone dihedral angles of Gln171Arg shows one well-defined peak in the Susceptible structure (see Figure 3A)) while in the Resistant structure there are two peaks (see Figure 3B): one highly populated peak at the same location as the peak in Susceptible and one less populated peak. The profile of the distribution correlates with the secondary structure plot (see SI Figure 3): the β2-α2 loop in the Susceptible structure visits a 310-helix conformation with a greater relative population (89%) than in the Resistant structure (64%). Therefore, the peak in the backbone dihedral distribution corresponds to a population in β–turn shape. The distribution of backbone dihedral angles of Pro168, Gln171Arg, Tyr172, and Ser173 correlates with the greater percentage of 310-helix conformation in Susceptible than in Resistant (data not shown).
Figure 3.
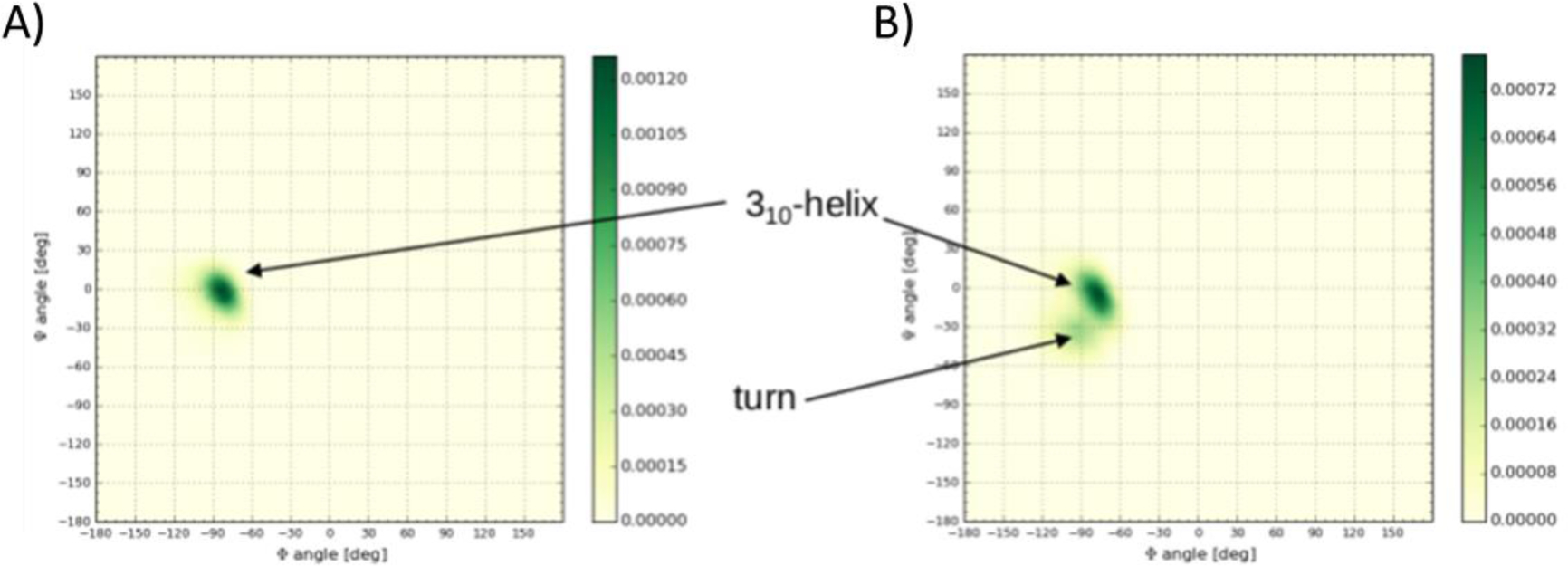
Backbone dihedral angle distribution of residue 171 in the A) Susceptible (Gln171) and B) Resistant (Arg171) structure. The β2-α2 loop samples to a greater extent a 310-helix conformation in the Susceptible structure than in the Resistant structure. In the Resistant structure, the β2-α2 loop populates a turn conformation in addition to the310-helix conformation.
Polymorphisms exhibit distinct local density of inter-residue connectivity
The dynamic cross-correlation matrix (Figure 4A) and B)) shows localized differences emerging in the Resistant structure, even though the overall profile is similar to the Susceptible. The matrix of the Resistant structure appears to be more modular in the sense of the existence of a greater number of “small spots” with strong correlation or anticorrelation.
Figure 4.
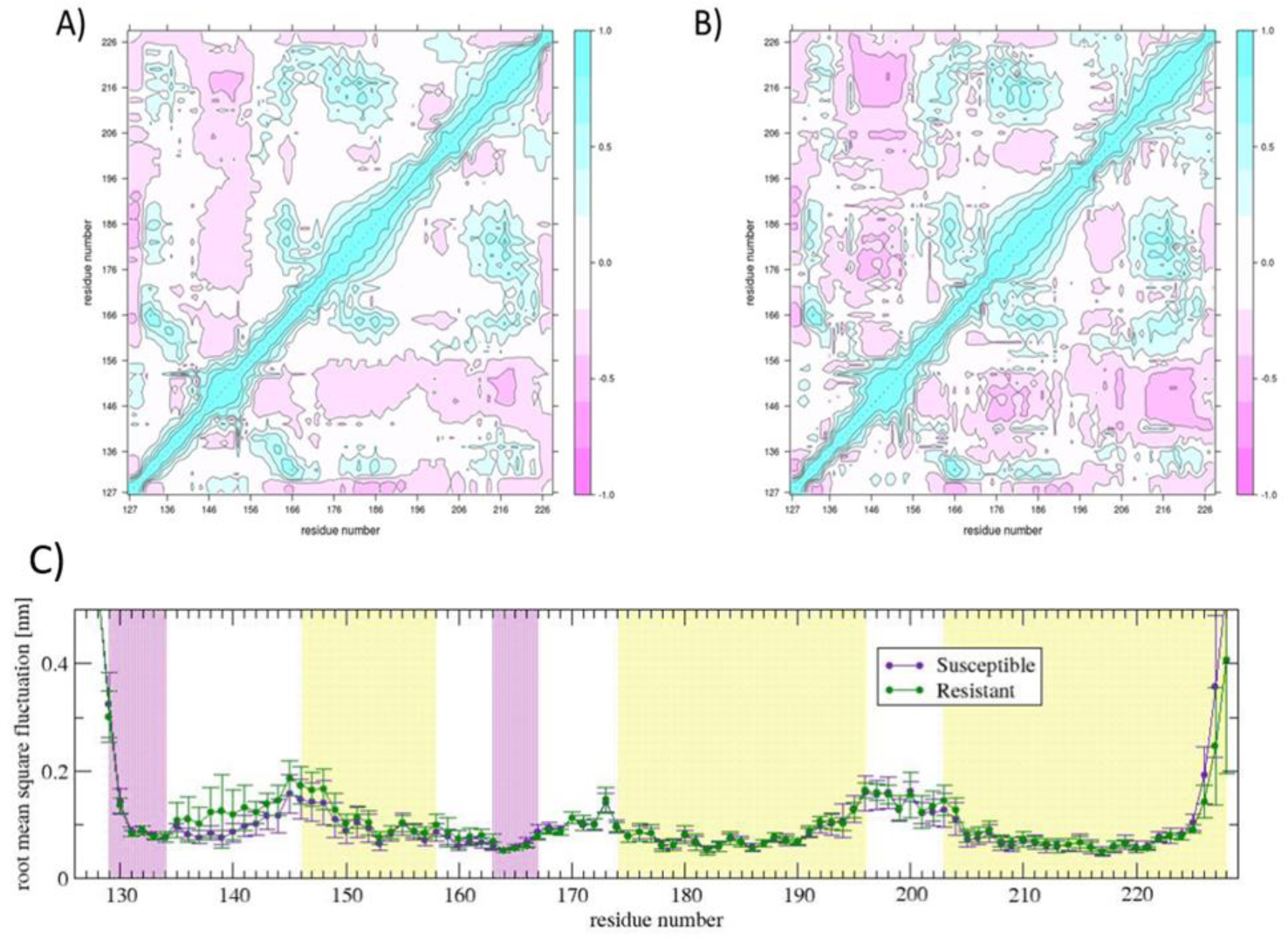
Dynamic cross correlation matrix of the A) Susceptible and B) Resistant structure; C) root mean square fluctuation plot of each residue of the Susceptible and Resistant structure. The local density of inter-residue connectivity, represented by the dynamic cross correlation matrix, indicates local differences when comparing the Susceptible with the Resistant structure. The most striking difference is the increased local conformational mobility in the β1-α1 loop in the Resistant structure.
Changes involving β1-α1 loop:
In the Resistant structure, the pattern of cross correlations changes the most, (|cijResistant – cijSusceptible| > 0.6), at β1-α1 loop (mainly residues Ser135,Val136Ala, Met137, Ser138, and Arg139): weaker correlations with the C-terminus of α1-β2 loop (Asn162) and beginning of β-sheet 2 (Gln163), and stronger anticorrelation with α-helix 2 (Val179), C-terminus of α2-α3 loop (Phe201, Thr202), and α-helix 3 (Val213, Gln214, Gln215, Met216, Ile218) as compared with the Susceptible structure. The response of the Resistant structure to the cross-correlation pattern may explain the gain in local conformational mobility seen in the β1-α1 loop, as shown in the root mean square fluctuation plot (Figure 4C)).
Changes involving β2–α2 loop:
The changes in the correlation profile of the β2–α2 loop (which includes polymorphism at position 171) are not as prominent as in the β1–α1 loop, with |cijResistant – cijSusceptible| > 0.4 but less than 0.55. The main difference shows in the change from anticorrelation (in the Susceptible structure) to correlation (in the Resistant structure) between the residues Pro168, Val169, and Asp170 in the β2–α2 loop and α–helix 2 (Ile185), and end of α–helix 3 (Gln226). The change from correlation (in the Susceptible structure) to anticorrelation (in the Resistant structure) shows between residue Ser173 and His143 (β1–α1 loop), and residues Asp170, Tyr172, and Ser 173 (in β2–α2 loop) and Ile218, Thr219 and Gln222 (in α–helix 3).
The Gln171Arg polymorphism shows two main changes (|cijResistant – cijSusceptible| > 0.3 but less than 0.4): from anticorrelation in the Susceptible structure to correlation in the Resistant structure with Arg139 (β1–α1 loop), and from correlation in the Susceptible structure to anticorrelation in the Resistant structure with Arg 211, Val213, and Gln222 (α–helix 3).
To picture the cross-correlation patterns, we built and clustered the residue network using the Newman-Girvan algorithm. A pattern of conserved and distinct inter-residue connectivity in Susceptible and Resistant emerged (see Figure 5A) and B)). The partitioning into communities indicates that the Susceptible and Resistant structure are distinct in their inter-residue connectivity. The largest preserved community in Susceptible and Resistant includes most residues in β1–α1 loop, α–helix1, N-terminus of α1–β2 loop, His 190 (located in α–helix 2), C-terminus of α2-α3 loop, and the first half of α–helix3 (backbone trace in yellow color in Figure 5A) and B)). Other preserved community corresponds to the C-terminus of α–helix 2 and N-terminus of α2-α3 loop (backbone trace in brown color in Figure 5A) and B)). Three other preserved communities have two or one residue members only (backbone trace in gray color in Figure 5A)) and B)).
Figure 5.
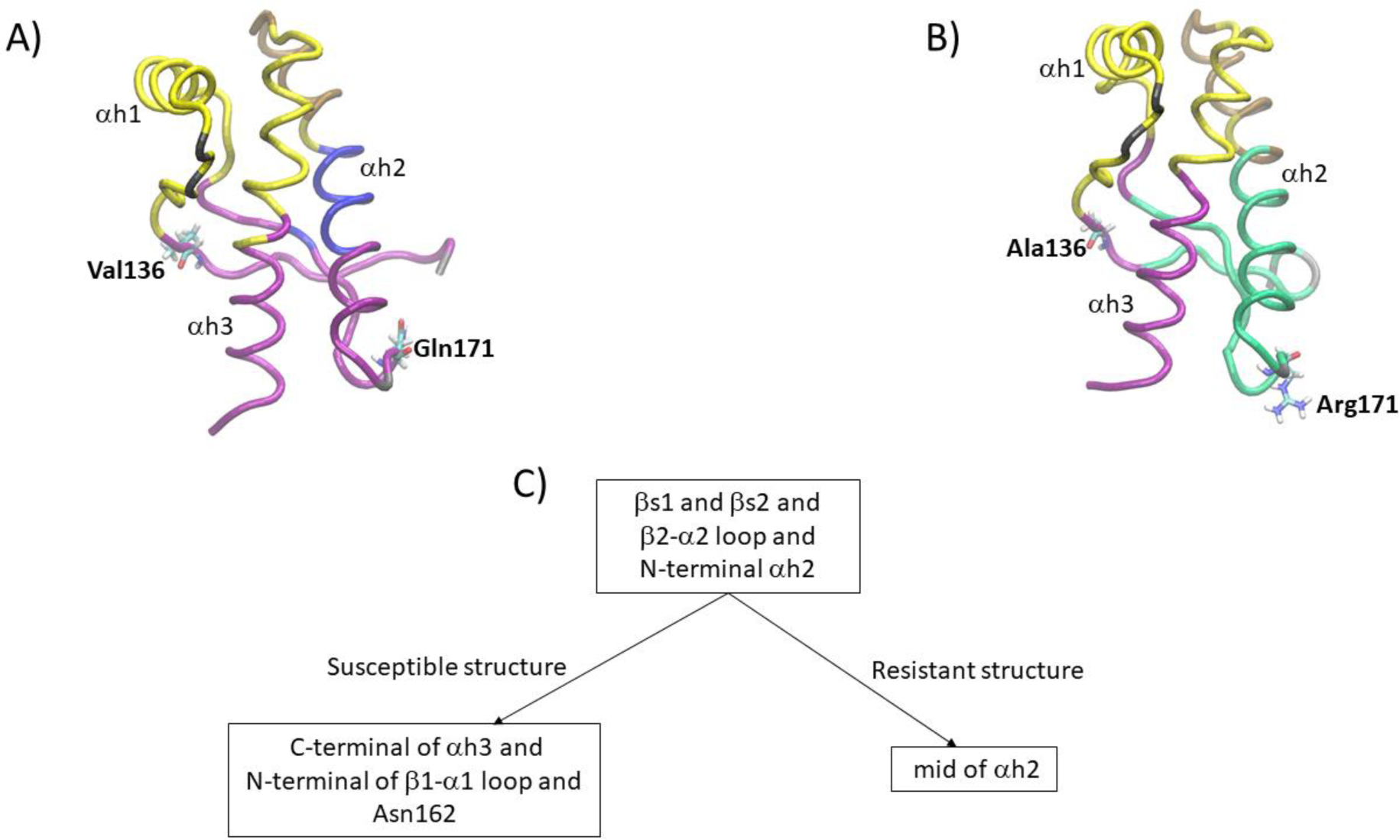
Representation of the partitioning into communities of the residue network of A) Susceptible and B) Resistant structure; C) illustration of the distinct partitioning of a key community of the Susceptible and Resistant structure. The β2–α2 loop of the Resistant structure shows stronger connectivity with the N-terminal of α–helix 2 than the Susceptible structure. The connectivity between the β–sheets and α–helix 3 present in the Susceptible structure is lost in the Resistant structure.
The communities that include polymorphisms are distinct in the Susceptible and Resistant structures. In the Susceptible structure, one large community includes β–sheet 1, N-terminus of β1–α1 loop, C-terminus of α1–β2 loop, β–sheet 2, β2–α2 loop, N-terminus α–helix 2, and the second half of α–helix 3 (backbone trace in purple color in Figure 5A)). Valine136 and Gln171 belong to this community. In contrast, in the Resistant structure Ala136 belongs to a community that includes the N-terminus of β1–α1 loop, C-terminus of α1–β2 loop, and second half of α–helix 3 (backbone trace in purple color in Figure 5A)); and Arg171 belongs to a community that includes β–sheet 1, β–sheet 2, β2–α2 loop, and the first 2/3 of α–helix 2 (backbone trace in green color in Figure 5B)). Another distinction shows in a small community in the Susceptible structure that includes Tyr165 (located in β–sheet 2) and residues 181 through 189 (located in α–helix 2; backbone trace in blue color in Figure 5A)); corresponding residues in the Resistant structure belong to a larger community (green color in Figure 5B)).
In summary, two communities in the Resistant structure, one including Ala136 and the other Arg171 (backbone trace in purple and green color in Figure 5B)) primarily correspond to one large community in the Susceptible structure that includes both Val136 and Gln171 (backbone trace purple color in Figure 5A)). The two main features of the partition are i) the connectivity between the β2–α2 loop, which includes the Gln171Arg polymorphism, and the loss of the C-terminus of α–helix 3 in the Resistant structure. Instead, the β2–α2 loop of the Resistant structure shows stronger connectivity with the N-terminus of α–helix 2, and ii) the connectivity between the β–sheets and α–helix 3 present in Susceptible is lost in the Resistant structure as well, likely due to the loss of connectivity between Ala136 and residues Asn162 and Gln163 in the Resistant structure.
Polymorphisms alter side chain packing of residues near or on β-sheet 2
To rationalize the effect of the polymorphisms on side chain connectivity, we examined how residues Val136Ala and Gln171Arg interact with other residues.
In the Susceptible structure, Val136 interacts with Asn162 and Gln163. Residues 136, 162, and 163 sample an equivalent range of backbone dihedral angles in the Susceptible and Resistant structure, indicating that the backbone conformation is not altered (data not shown) The distribution of side chain dihedral angles in Susceptible and Resistant shows differences in Asn162 (see Figure 6 A) and B)) but not in Gln163 (see SI Figure 4A)). The distinct population in dihedral angles within the same range of values suggests that the packing of the side chain of Asn162 in Resistant differentiates from that of the Susceptible structure. Figure 7 shows the distribution of distance between any pair of heavy atoms of two side chains. The distribution of Val136-Asn162 peaks at 0.44 nm in Susceptible, while the distribution of Ala136-Asn162 is broader, shows a shoulder at 0.38 nm and a peak at 0.46 nm (see Figure 7A)). The distribution of Met137 – Asn162 shows a non-negligible interaction, albeit with a small population, in Resistant but not in Susceptible (Figure 7B)). The distribution of Arg139 – Asn162 distance indicates that in the Resistant structure, the population of this interaction is reduced compared to the Susceptible structure. The loss of interaction corresponds to a hydrogen bond between the donor nitrogen atoms in Arg139 and the acceptor oxygen atom in Asn162, with 7% population in Resistant and 70% population in Susceptible; other interactions are hydrophobic in nature (see Figure 7C)). The distribution of Asn162-Met216 does not show changes in population for distances less than 0.5 nm (data not shown). Therefore, the changes in Asn162 side chain packing are due primarily to changes in the relative populations of hydrophobic interactions with residues Ala136 and Met137, and hydrogen bonding with Arg139.
Figure 6.
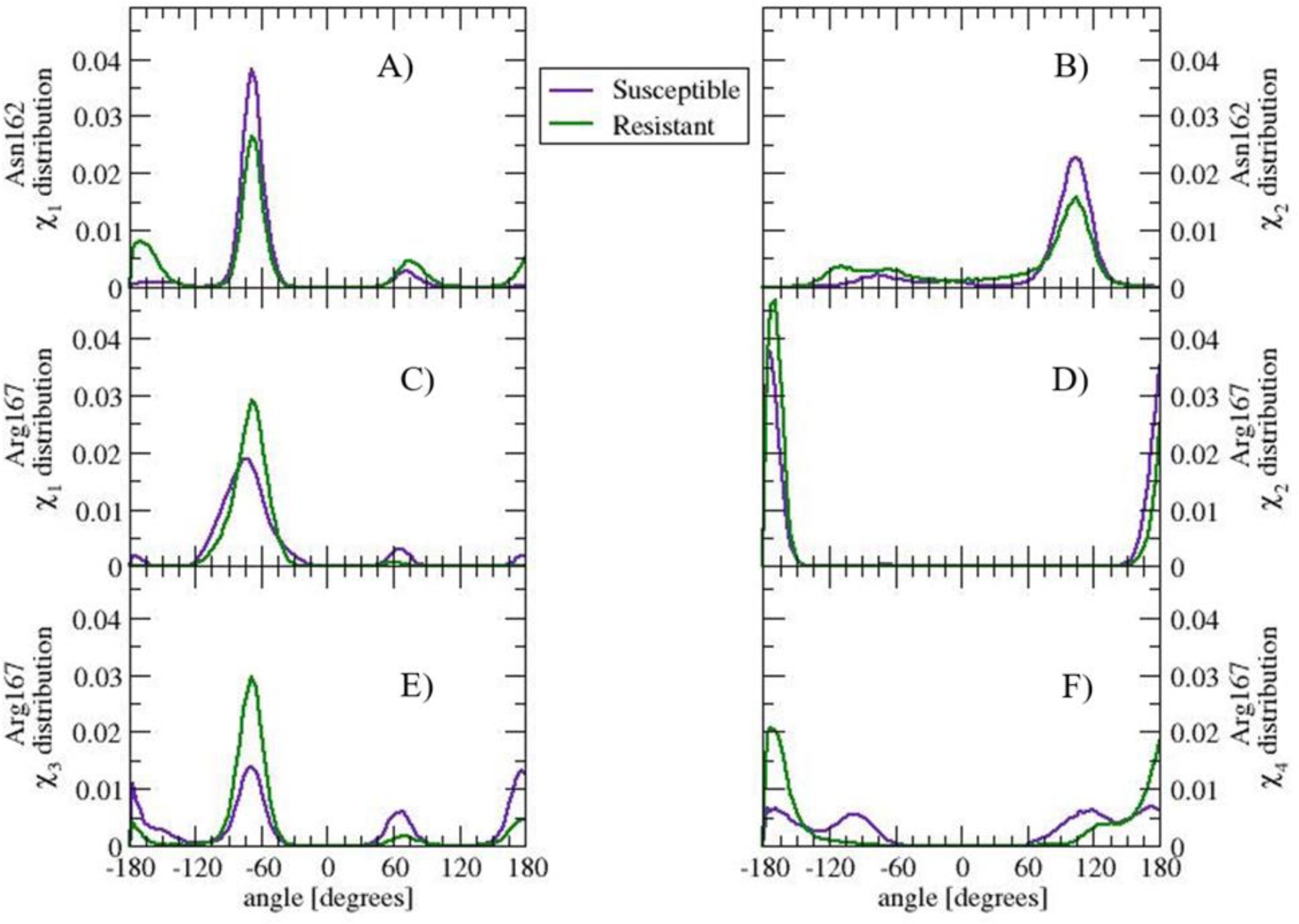
Side chain dihedral angle distribution of residue Asn162, A) χ1 and B) χ2), and Arg167, C) χ1, D) χ2, E)χ3, and F) χ4, in the Susceptible and Resistant structure. The side chain of Asn162 and Arg167 visit similar angle ranges albeit with different population in Susceptible and Resistant. The distinct angle distribution correlate with differences in the community partitioning in the Susceptible and Resistant structure.
Figure 7.
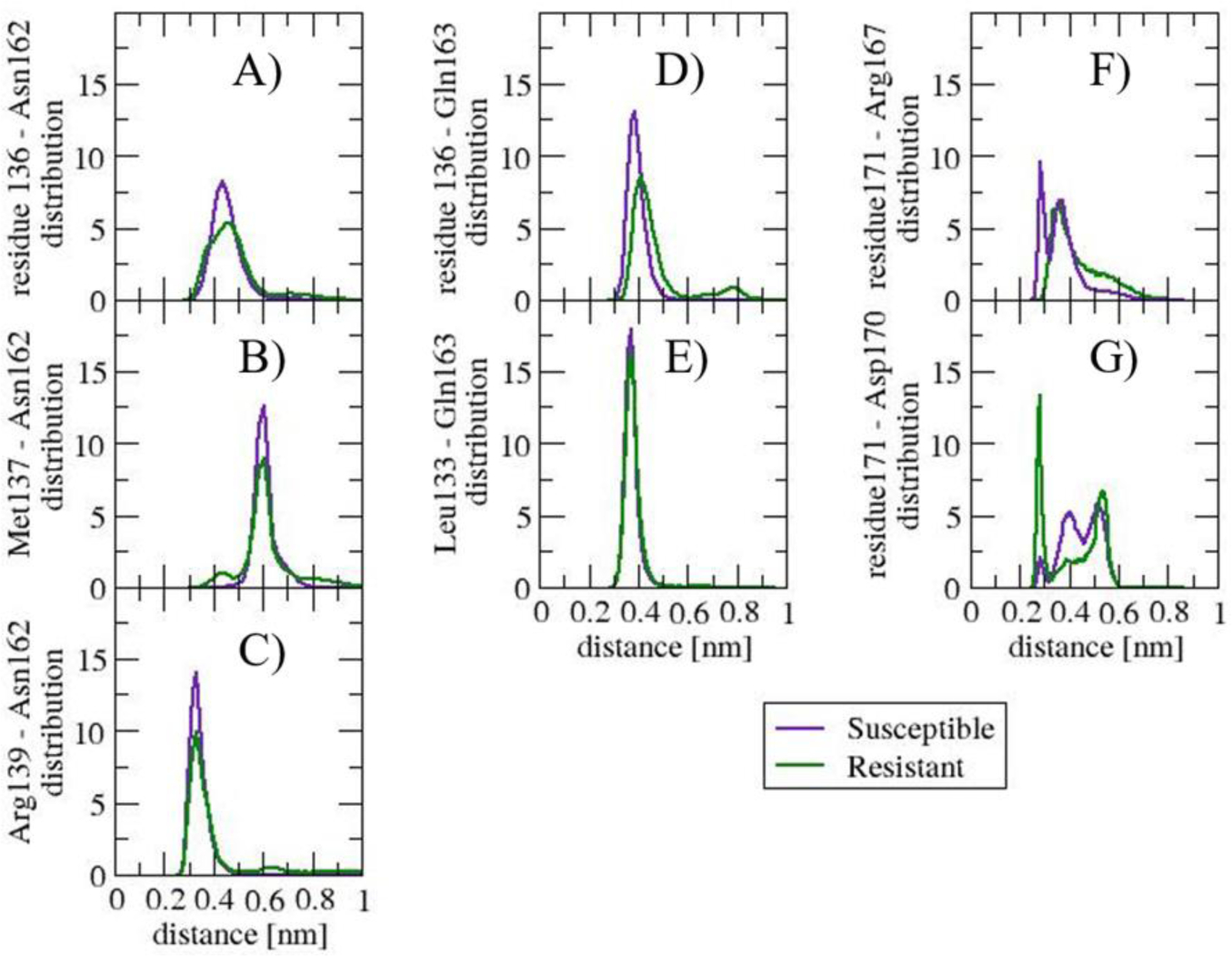
Distribution of distance between any pair of heavy atoms of two side chains: A) residue 136 – Asn162, B) Arg137 – Asn162, C) Arg139 - Asn162, D) residue 136 – Gln163, E) Leu133 – Gln163, F) residue 171 – Arg167, and G) residue 171-Asp170. The distribution of distance indicates what residue interactions are influenced by polymorphisms Val136Ala and Gln171Arg. The distinctions show in the β1-α1 loop interacting with residues Asn162 and Gln163, and in the side chain interactions in the β2-α2 loop.
The distribution of Val136-Gln163 peaks at 0.39 nm in Susceptible while the distribution Ala136-Gln163 peaks at 0.4 with a second peak at 0.79 nm (see Figure 7D)). The distribution of Leu133 – Gln163 distance is narrow and is similar between Susceptible and Resistant (see Figure 7E)). This interaction could explain the preserved distribution of side chain dihedral angles in Gln163.
In the Susceptible and Resistant structures, residue 171 interacts only with residues located nearby in primary sequence. Among the residues that interact with residue 171, the distribution of side chain dihedral angles is the most distinct in Arg167 (see Figure 6C) through F)). The change may be explained by the re-organization of side chain interaction pattern. In Susceptible, the profile of the distance distribution between Arg167 and Gln171 (see Figure 7F)) shows a well-defined peak at 0.28 nm, a minimum at 0.32 nm, and a second, less populated peak at 0.36 nm. The first peak is populated by pairs of atoms that form a hydrogen bond between Arg167 and Gln171. The second peak is populated by pairs of atoms interacting via van der Waals between Cδ and Cγ in Arg167 and side chain oxygen and nitrogen in Gln171. In the Resistant structure, however, the only peak in the Resistant structure distribution overlaps with the second peak in Susceptible; the peak is populated by Arg167-Arg171 van der Waals interactions. The electrostatic repulsion between the side chain nitrogen atoms in Arg167 and Arg171 in Resistant contributes to the shift in population.
Although the distribution of side chain dihedral angles of Asp170 is similar in the Susceptible and Resistant structures (see SI Figure 4B)), the profile of the distance distribution between Asp 170 and Gln171Arg is distinct. Figure 7G) shows a peak at 0.28 nm, which is much more populated in the Resistant than in the Susceptible structure. The peak corresponds to a salt bridge between Arg171 and Asp170. Additionally, within a heavy atom to heavy atom distance of 0.4 nm, there is a distinct network of side chain van der Waals interactions between the two residues. In the Resistant structure, Cβ–Cβ interactions are favored, while in the Susceptible structure, Cγ - Oxygen atoms interactions are favored. Residue Asp170 also interacts with Pro168, but the distribution of side chain distance does not change in the Susceptible structure when compared to the Resistant. The net effect of the salt bridge together with van der Waals interactions between Asp170 and residue171 may explain why the distribution of side chain dihedral angles in Asp170 is preserved in the Susceptible and Resistant structure.
Discussion
Our sixteen independent molecular dynamics simulations study maps the structural dynamics of two ovine PrPc polymorphisms that show distinct susceptibility to prion diseases: the Susceptible structure (VRQ) that displays high susceptibility to classical scrapie, and the Resistant structure (ARR) that shows low susceptibility. The opposite trend has been documented in atypical/Nor98 scrapie: The ARR structure is highly susceptible to atypical while the VRQ structure shows low susceptibility. Standard structural descriptors indicate that, within the time and length scale of our simulations, the conformations sampled by the Susceptible and Resistant structure exhibit similar trends in overall backbone geometry, compactness, and side chain solvent exposure. Our simulations do not provide evidence in regards to the differential compactness of the hydrophobic core (Rezaei et al., 2002) or side chain exposure to the solvent (Eghiaian et al., 2004), except at residues Val136Ala and Gln171Arg, when comparing the Susceptible and Resistant structures. Our simulations do not provide evidence on a specific structural role of residues Asn184 and Asn200, in line with the absent effect of N-glycosylation on differential prion protein conversion in scrapie (Uslupehlivan et al., 2018).
The β1–α1 loop shows higher local backbone mobility in the Resistant structure than in the Susceptible due to two factors: First, alanine136, in the Resistant structure, alters the side chain packing of Arg139 and Asn162, as identified by others (Bujdoso et al., 2005; Eghiaian et al., 2004) weakening the connections between the loop and residues in the C-terminus of the α1–β2 loop. And second, the β1–α1 loop becomes more anticorrelated in the Resistant structure with α–helix 3 and, to a lesser extent, with α–helix 2.
The β2–α2 loop shows a differentiated pattern of side chain interactions resulting from the residue 171 substitution: In the Resistant structure, the side chain of Arg171 salt bridges with Asp170 while Gln171 favors hydrogen bonding with Arg167 in the Susceptible structure, in agreement with others (Bujdoso et al., 2005; Eghiaian et al., 2004). To accommodate such interactions, the loop backbone rearranges and results in a shift in the 310-helix population, from dominant (in the Susceptible structure) to moderate (in the Resistant structure). Although in other mammalian PrPC the aromatic residue Tyr172 (residue 169 in mouse PrPC numbering) has been proposed to stabilize the 310-helix fold to hinder conversion (Damberger, Christen, Pérez, Hornemann, & Wüthrich, 2011; Kurt, Jiang, Bett, Eisenberg, & Sigurdson, 2014; Huang & Caflisch, 2015), the Arg167 – Tyr172, Tyr172 - Phe178 and Tyr172 – Asp181 interactions are similarly preserved in the Susceptible and Resistant structures. In addition, our simulations do not provide evidence of distinct backbone local mobility in the loop, in contrast to previous reports (Bujdoso et al., 2005). Therefore, our study indicates that interactions involving the residue at position 171 play a role in modulating 310-helix backbone preferences without a measurable effect on conformational flexibility.
From our analysis, we propose that residues 136 and 171 have a subtle but well-defined structural role: modulation of the residue connectivity of the β–sheets and β2–α2 loop with the rest of the protein structure. The change in the density of residue connections in ovine PrPC results in a distinct partitioning of the protein structure into groups of highly connected residues. Both structures, Susceptible and Resistant, show a high density of connections grouping β–sheet 1, β–sheet 2, β2–α2 loop and the N-terminus of α–helix 2. In the Susceptible structure, this group of residues connects with the N-terminus of β1–α1 loop, Asn162 and α–helix 3. In contrast, this group connects with most residues of α–helix 2 in the Resistant structure. Thus, our analysis does not lend support to the hypothesis that Arg171 destabilizes the junction between β–sheet 2 and α–helix 2 (Rezaei et al., 2002); instead, the interactions involving Arg171 result in a shift of the backbone population from 310-helix to β-turn in the Resistant structure. We interpret the re-arrangement of residue connectivity due to Arg171 as favoring interactions between the β2-α2 loop and α-helix 2 over interactions with α-helix 3.
Ovine polymorphisms display distinct structural dynamics and we speculate that PrPC conversion in Susceptible and Resistant occurs through distinct pathways. In a scenario in which either β–sheet 1 or β2–α2 loop is a spot that PrPSc recognizes to initiate misfolding, we argue that PrPC - PrPSc communication will be transmitted through different paths in each structure. In the Susceptible structure, the conformational perturbation templated by PrPSc will be transmitted more effectively along the large community that connects α–helix 3 with the N-terminus of β1–α1 loop, the β-sheets, the β2–α2 loop, and the N-terminus of α-helix 2 (see Figure 5A)). In the Resistant structure, on the other hand, because α-helix 2 and α-helix 3 are not highly interconnected (see Figure 5B)), PrPC conversion may follow a distinct route, perhaps less efficient, that is favored by the uncoupling of the helices. In an alternate scenario, if α-helix 1 is a hot spot of PrPC conversion, we would argue that initial misfolding would occur through similar routes in Susceptible and Resistant because most residues in β1-α1 loop, α-helix 1, and N-terminus of α1-β2 loop form a single community in both structures. In this case, PrPSc structure differentiation would show at a later step, perhaps in oligomer conformational re-arrangements as observed in experiments (Rezaei et al., 2002). Considering that the Resistant structure shows long incubation period in classical scrapie (Goldmann et al., 1994) and restricted PrPSc structural variability (Groschup M. H., Lacroux C., Buschmann A., Lühken G., Mathey J., Eiden M., Lugan S., Hoffmann C., Espinosa J.C., Baron T., Torres J. M., Erhardt G., 2007), we believe that Ala136 and Arg171 together constrain the conformational space that accommodates β-sheet rich aggregates.
In light of recently proposed PrPSc structures (Spagnolli et al., 2019), we speculate that Val136 would point toward the core of Rung-2 and is more efficient than Ala136 to pack with side chains in the adjacent rungs: Trp102 (Rung-1) and Asn174 (Rung-3). On the other hand, Gln171 would point toward the core of the fibril on Rung-3 and would pack with Leu133 (on Rung-2) and Glu203 (on Rung-4). In the case of the Resistant structure, Arg171 may add local side chain re-arrangements due to the electrostatics with nearby residues Arg167 (Rung-3) and Lys207 (Rung-4). A similar argument holds in a scenario of PIRIBS- PrPSc structures (Groveman et al., 2014), in which the positive charge of Arg171 demands re-organization of side chain interactions that may be less favorable than the stacking of polar Gln171. Stacking of monomers into in-register parallel β-sheets is, therefore, thermodynamically less favorable in the Resistant structure and could even result in yet other PrPSc architectures. Our interpretation is consistent with the current understanding of a mosaic of scrapie PrPSc structures (Groschup M. H., Lacroux C., Buschmann A., Lühken G., Mathey J., Eiden M., Lugan S., Hoffmann C., Espinosa J.C., Baron T., Torres J. M., Erhardt G., 2007).
Our study identifies a hot spot for future studies of potential targets in drug design. Most mammal PrPC sequences, regardless of the degree of prion disease susceptibility, show an alanine residue at residue position 136 (in human PrPC numbering: residue position 133), and mainly a glutamine or glutamic acid, and to a lesser frequency an arginine residue, at position 171 (in human PrPC numbering: residue position 168). We propose that blocking the Arg167-Gln171 hydrogen bond (or, equivalently the Arg167 – Glu171 salt bridge), or the Arg171 - Asp170 salt bridge re-arranges the density of side chain interactions of the β2-α2 loop with other parts of the protein in a manner such that PrPC conversion is withstood.
Supplementary Material
Acknowledgments
The authors acknowledge Noah Yoshida who performed preliminary analysis of the structures and the Creighton University Center for Undergraduate Research and Scholarship (CURAS) for partial funding. This work was made possible partly by grants from the National Institute for General Medical Science (NIGMS) (5P20GM103427), a component of the National Institutes of Health (NIH), and its contents are the sole responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
List of Abbreviations:
- PrPC
Cellular prion protein
- PrPSc
Scrapie prion protein
- TSEs
Transmissible spongiform encephalopathies
Footnotes
The authors declare no conflict of interests.
Supplemental Material
See additional file
References
- Adrover M, Pauwels K, Prigent S, de Chiara C, Xu Z, Chapuis C, … Rezaei H (2010). Prion fibrillization is mediated by a native structural element that comprises helices H2 and H3. The Journal of Biological Chemistry, 285(27), 21004–21012. 10.1074/jbc.M110.111815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, & Walker S (2015). Fitting Linear Mixed-Effects Models using lme4. Journal of Statistical Software, 67(1), 1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Belt PBGM, Muileman IH, Schreuder BEC, Bos-De Ruijter J, Gielkens ALJ, & Smits MA (1995). Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. Journal of General Virology, 76(3), 509–517. [DOI] [PubMed] [Google Scholar]
- Blinov N, Berjanskii M, Wishart DS, & Stepanova M (2009). Structural domains and main-chain flexibility in prion proteins. Biochemistry, 48(7), 1488–1497. 10.1021/bi802043h [DOI] [PubMed] [Google Scholar]
- Bossers A, Schreuder BEC, Muileman IH, Belt PBGM, & Smits MA (1996). PrP genotype contributes to determining survival times of sheep with natural scrapie. Journal of General Virology, 77(10), 2669–2673. 10.1099/0022-1317-77-10-2669 [DOI] [PubMed] [Google Scholar]
- Bruce ME, Will RG, Ironside JW, McConnell I, Drummond D, Suttie A, … Bostock CJ (1997). Transmissions to mice indicate that “new variant” CJD is caused by the BSE agent. Nature, 389(6650), 498–501. 10.1038/39057 [DOI] [PubMed] [Google Scholar]
- Bujdoso R, Burke DF, & Thackray AM (2005). Structural differences between allelic variants of the ovine prion protein revealed by molecular dynamics simulations. Proteins: Structure, Function and Genetics, 61(4), 840–849. 10.1002/prot.20755 [DOI] [PubMed] [Google Scholar]
- Burke C, Walsh D, Steele A, Agrimi U, Di Bari MA, Watts JC, & Supattapone S (2019). Full restoration of specific infectivity and strain properties from pure mammalian prion protein. PLoS Pathogens, 15(3), e1007662 10.1371/journal.ppat.1007662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G, Donadio D, & Parrinello M (2007). Canonical sampling through velocity rescaling. The Journal of Chemical Physics, 126(1), 014101 10.1063/1.2408420 [DOI] [PubMed] [Google Scholar]
- Cassard H, Torres J-M, Lacroux C, Douet J-Y, Benestad SL, Lantier F, … Andréoletti O (2014). Evidence for zoonotic potential of ovine scrapie prions. Nature Communications, 5(1), 5821 10.1038/ncomms6821 [DOI] [PubMed] [Google Scholar]
- CFSPH. (2016). Scrapie. Center for Food Security and Public Health Technical Factsheets - Iowa State University Center for Food Security and Public Health, 119 http://www.cfsph.iastate.edu/Factsheets/pdfs/scrapie.pdf [Google Scholar]
- Chakroun N, Fornili A, Prigent S, Kleinjung J, Dreiss CA, Rezaei H, & Fraternali F (2013). Decrypting prion protein conversion into a β-rich conformer by molecular dynamics. Journal of Chemical Theory and Computation, 9(5), 2455–2465. 10.1021/ct301118j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damberger FF, Christen B, Pérez DR, Hornemann S, & Wüthrich K (2011). Cellular prion protein conformation and function. Proceedings of the National Academy of Sciences of the United States of America, 108(42), 17308–17313. 10.1073/pnas.1106325108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault NR, Harris BT, Rees JR, & Supattapone S (2007). Formation of native prions from minimal components in vitro. Proceedings of the National Academy of Sciences of the United States of America, 104(23), 9741–9746. 10.1073/pnas.0702662104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarco M, & Daggett V (2007). Molecular Mechanism for Low pH Triggered Misfolding of the Human Prion Protein. Biochemistry, 46(11), 3045–3054. 10.1021/BI0619066 [DOI] [PubMed] [Google Scholar]
- Dima RI, & Thirumalai D (2002). Exploring the propensities of helices in PrPC to form β sheet using NMR structures and sequence alignments. Biophysical Journal, 83(3), 1268–1280. 10.1016/S0006-3495(02)73899-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA-BIOHAZ. (2015). Scientific Opinion on a request for a review of a scientific publication concerning the zoonotic potential of ovine scrapie prions. EFSA Journal, 13(8), 4197 10.2903/j.efsa.2015.4197 [DOI] [Google Scholar]
- Eghiaian F, Grosclaude J, Lesceu S, Debey P, Doublet B, Treguer E, … Knossow M (2004). Insight into the PrPC -> PrPSc conversion from the structures of antibody-bound ovine prion scrapie-susceptibility variants. Proceedings of the National Academy of Sciences, 101(28), 10254–10259. 10.1073/pnas.0400014101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, & Pedersen LG (1995). A smooth particle mesh Ewald method. The Journal of Chemical Physics, 103(19), 8577–8593. 10.1063/1.470117 [DOI] [Google Scholar]
- Fitzmaurice TJ, Burke DF, Hopkins L, Yang S, Yu S, Sy M-S, … Bujdoso R (2008). The stability and aggregation of ovine prion protein associated with classical and atypical scrapie correlates with the ease of unwinding of helix-2. The Biochemical Journal, 409(2), 367–375. 10.1042/BJ20071122 [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhu T, Zhang C, Zhang JZH, & Mei Y (2018). Comparison of the unfolding and oligomerization of human prion protein under acidic and neutral environments by molecular dynamics simulations. Chemical Physics Letters, 706, 594–600. 10.1016/j.cplett.2018.07.014 [DOI] [Google Scholar]
- Girvan M, & Newman MEJ (2002). Community structure in social and biological networks. Proceedings of the National Academy of Sciences of the United States of America, 99(12), 7821–7826. 10.1073/pnas.122653799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M (2015). Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled Aβ, tau, and α-synuclein. Science, 349(6248), 1255555 10.1126/science.1255555 [DOI] [PubMed] [Google Scholar]
- Goldmann W, Hunter N, Smith G, Foster J, & Hope J (1994). PrP Genotype and Agent Effects in Scrapie: Change In Allelic Interaction With Different Isolates of Agent in Sheep, a Natural Host of Scrapie. Journal of General Virology, 75(5), 989–995. 10.1099/0022-1317-75-5-989 [DOI] [PubMed] [Google Scholar]
- Gorfe AA, & Caflisch A (2007). Ser170 controls the conformational multiplicity of the loop 166–175 in prion proteins: implication for conversion and species barrier. The FASEB Journal, 21(12), 3279–3287. 10.1096/fj.07-8292com [DOI] [PubMed] [Google Scholar]
- Gossert AD, Bonjour S, Lysek DA, Fiorito F, & Wuthrich K (2005). Prion protein NMR structures of elk and of mouse/elk hybrids. Proceedings of the National Academy of Sciences, 102(3), 646–650. 10.1073/pnas.0409008102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groschup MH, Lacroux C, Buschmann A, Lühken G, Mathey J, Eiden M, Lugan S, Hoffmann C, Espinosa JC, Baron T, Torres JM, Erhardt G, A. O (2007). Classic Scrapie in Sheep with the ARR/ARR Prion Genotype in Germany and France. Emerging Infectious Diseases, 13(8), 1201–1207. 10.3201/eid1308.070077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groveman BR, Dolan MA, Taubner LM, Kraus A, Wickner RB, & Caughey B (2014). Parallel in-register intermolecular β-sheet architectures for prion-seeded prion protein (PrP) amyloids. Journal of Biological Chemistry, 289(35), 24129–24142. 10.1074/jbc.M114.578344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Bekker H, Berendsen HJC, & Fraaije JGEM (1997). LINCS: A Linear Constraint Solver for Molecular Simulations. J Comput Chem, 18(12), 1463–1472. [Google Scholar]
- Hess B, Kutzner C, Van Der Spoel D, & Lindahl E (2008). GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation, 4(3), 435–447. 10.1021/ct700301q [DOI] [PubMed] [Google Scholar]
- Huang D, & Caflisch A (2015). Evolutionary conserved Tyr169 stabilizes the β2-α2 loop of the prion protein. Journal of the American Chemical Society, 137(8), 2948–2957. 10.1021/ja511568m [DOI] [PubMed] [Google Scholar]
- Ji H-F, Zhang H-Y, & Shen L (2005). The Role of Electrostatic Interaction in Triggering the Unraveling of Stable Helix 1 in Normal Prion Protein. A Molecular Dynamics Simulation Investigation. Journal of Biomolecular Structure and Dynamics, 22(5), 563–570. 10.1080/07391102.2005.10507026 [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, & Klein ML (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics, 79(2), 926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Kabsch W, & Sander C (1983). Dictionary of protein secondary structure: Pattern recognition of hydrogen- bonded and geometrical features. Biopolymers, 22(12), 2577–2637. 10.1002/bip.360221211 [DOI] [PubMed] [Google Scholar]
- Klug GMJA, Wand H, Simpson M, Boyd A, Law M, Masters CL, … Collins SJ (2013). Intensity of human prion disease surveillance predicts observed disease incidence. Journal of Neurology, Neurosurgery and Psychiatry, 84(12), 1372–1377. 10.1136/jnnp-2012-304820 [DOI] [PubMed] [Google Scholar]
- Kurt TD, Jiang L, Bett C, Eisenberg D, & Sigurdson CJ (2014). A proposed mechanism for the promotion of prion conversion involving a strictly conserved tyrosine residue in the β2-α2 loop of PrPC. The Journal of Biological Chemistry, 289(15), 10660–10667. 10.1074/jbc.M114.549030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontiadou H, Galdadas I, Athanasiou C, & Cournia Z (2018). Insights into the mechanism of the PIK3CA E545K activating mutation using MD simulations. Scientific Reports, 8(1), 15544 10.1038/s41598-018-27044-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yao X-Q, & Grant BJ (2018). Comparative structural dynamic analysis of GTPases. PLOS Computational Biology, 14(11), e1006364 10.1371/journal.pcbi.1006364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, & Shaw DE (2010). Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Structure, Function, and Bioinformatics, 78(8), 1950–1958. 10.1002/prot.22711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh RF, Kincaid AE, Bessen RA, & Bartz JC (2005). Interspecies transmission of chronic wasting disease prions to squirrel monkeys (Saimiri sciureus). Journal of Virology, 79(21), 13794–13796. 10.1128/JVI.79.21.13794-13796.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meli M, Gasset M, & Colombo G (2011). Dynamic Diagnosis of Familial Prion Diseases Supports the β2-α2 Loop as a Universal Interference Target. PLoS ONE, 6(4), e19093 10.1371/journal.pone.0019093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello M, & Rahman A (1980). Crystal Structure and Pair Potentials: A Molecular-Dynamics Study. Physical Review Letters, 45(14), 1196–1199. 10.1103/PhysRevLett.45.1196 [DOI] [Google Scholar]
- Prusiner SB, Scott MR, DeArmond SJ, & Cohen FE (1998). Prion Protein Biology. Cell, 93(3), 337–348. 10.1016/S0092-8674(00)81163-0 [DOI] [PubMed] [Google Scholar]
- R-Core-Team. (2013). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.r-project.org/ [Google Scholar]
- Rezaei H, Choiset Y, Eghiaian F, Treguer E, Mentre P, Debey P, … Haertle T (2002). Amyloidogenic Unfolding Intermediates Differentiate Sheep Prion Protein Variants. Journal of Molecular Biology, 322(4), 799–814. 10.1016/S0022-2836(02)00856-2 [DOI] [PubMed] [Google Scholar]
- Rezaei H, Eghiaian F, Perez J, Doublet B, Choiset Y, Haertle T, & Grosclaude J (2005). Sequential Generation of Two Structurally Distinct Ovine Prion Protein Soluble Oligomers Displaying Different Biochemical Reactivities. Journal of Molecular Biology, 347(3), 665–679. 10.1016/J.JMB.2005.01.043 [DOI] [PubMed] [Google Scholar]
- Rigoli M, Spagnolli G, Faccioli P, Requena JR, & Biasini E (2019). Ok google, how could I design therapeutics against prion diseases? Current Opinion in Pharmacology, 44, 39–45. 10.1016/j.coph.2019.03.015 [DOI] [PubMed] [Google Scholar]
- Sethi A, Eargle J, Black AA, & Luthey-Schulten Z (2009). Dynamical networks in tRNA:protein complexes. Proceedings of the National Academy of Sciences of the United States of America, 106(16), 6620–6625. 10.1073/pnas.0810961106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdson CJ, Bartz JC, & Glatzel M (2019). Cellular and Molecular Mechanisms of Prion Disease. Annual Review of Pathology: Mechanisms of Disease, 14(1), 497–516. 10.1146/annurev-pathmechdis-012418-013109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdson CJ, Nilsson KPR, Hornemann S, Heikenwalder M, Manco G, Schwarz P, … Aguzzi A (2009). De novo generation of a transmissible spongiform encephalopathy by mouse transgenesis. Proceedings of the National Academy of Sciences, 106(1), 304–309. 10.1073/pnas.0810680105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skjaerven L, Yao XQ, Scarabelli G, & Grant BJ (2014). Integrating protein structural dynamics and evolutionary analysis with Bio3D. BMC Bioinformatics, 15, 399 10.1186/s12859-014-0399-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C (2012). Transmissible proteins: expanding the prion heresy. Cell, 149(5), 968–977. 10.1016/j.cell.2012.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnolli G, Rigoli M, Orioli S, Sevillano AM, Faccioli P, Wille H, … Requena JR (2019). Full atomistic model of prion structure and conversion. PLOS Pathogens, 15(7), e1007864 10.1371/journal.ppat.1007864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko R, Savtchenko R, Ostapchenko VG, Makarava N, & Baskakov IV (2010). The α-Helical C-Terminal Domain of Full-Length Recombinant PrP Converts to an In-Register Parallel β-Sheet Structure in PrP Fibrils: Evidence from Solid State Nuclear Magnetic Resonance. Biochemistry, 49(44), 9488–9497. 10.1021/bi1013134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulvund MJ, Bratberg B, Osland A, & Tranulis MA (1999). Prion protein gene polymorphisms in sheep with natural scrapie and healthy controls in Norway. Journal of General Virology, 80(4), 1073–1077. 10.1099/0022-1317-80-4-1073 [DOI] [PubMed] [Google Scholar]
- Uslupehlivan M, Deveci R, & Ün C (2018). In silico investigation of the prion protein glycosylation profiles in relation to scrapie disease resistance in domestic sheep (Ovis aries). Molecular and Cellular Probes, 42, 1–9. 10.1016/j.mcp.2018.09.004 [DOI] [PubMed] [Google Scholar]
- Van der Rest G, Rezaei H, & Halgand F (2017). Monitoring Conformational Landscape of Ovine Prion Protein Monomer Using Ion Mobility Coupled to Mass Spectrometry. Journal of The American Society for Mass Spectrometry, 28(2), 303–314. 10.1007/s13361-016-1522-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


