Abstract
To evaluate whether the polygenic profile modifies the development of sporadic Alzheimer’s disease (sAD) and pathological biomarkers in cerebrospinal fluid (CSF), 462 sAD patients and 463 age-matched cognitively normal (CN) controls were genotyped for 35 single-nucleotide polymorphisms (SNPs) that are significantly associated with sAD. Then, the alleles found to be associated with sAD were used to build polygenic risk score (PRS) models to represent the genetic risk. Receiver operating characteristic (ROC) analyses and the Cox proportional hazards model were used to evaluate the predictive value of PRS for the sAD risk and age at onset. We measured the CSF levels of Aβ42, Aβ42/Aβ40, total tau (T-tau), and phosphorylated tau (P-tau) in a subgroup (60 sAD and 200 CN participants), and analyzed their relationships with the PRSs. We found that 14 SNPs, including SNPs in the APOE, BIN1, CD33, EPHA1, SORL1, and TOMM40 genes, were associated with sAD risk in our cohort. The PRS models built with these SNPs showed potential for discriminating sAD patients from CN controls, and were able to predict the incidence rate of sAD and age at onset. Furthermore, the PRSs were correlated with the CSF levels of Aβ42, Aβ42/Aβ40, T-tau, and P-tau. Our study suggests that PRS models hold promise for assessing the genetic risk and development of AD. As genetic risk profiles vary among populations, large-scale genome-wide sequencing studies are urgently needed to identify the genetic risk loci of sAD in Chinese populations to build accurate PRS models for clinical practice.
Electronic supplementary material
The online version of this article (10.1007/s12264-020-00469-8) contains supplementary material, which is available to authorized users.
Keywords: Alzheimer’s disease, Single nucleotide polymorphism, Polygenic risk score, Cerebrospinal fluid, Biomarker, Amyloid-beta, Tau
Introduction
Alzheimer’s disease (AD) is the most common neurodegenerative disorder and is closely related to the complex interaction among genes and environmental/lifestyle factors, among which heritability accounts for 58%–79% of the attribution for AD [1]. Apart from the apolipoprotein E (APOE) ε4 allele, which is the major susceptibility gene for sporadic AD (sAD) [2], a series of genome-wide association studies (GWASs) on AD dementia have identified a large number of single nucleotide polymorphisms (SNPs) with already known or hypothesized relationships to AD [3–11]. Most of these risk SNPs only exert minor effects on the susceptibility to AD. The polygenic risk score (PRS) determines the genetic risk for a disease by combining the effects of multiple genetic loci and has proved to be a promising strategy for identifying the genetic risk for sAD [12, 13].
Previous studies have demonstrated the value of PRS models for sAD risk prediction, and the age at onset has also been found to correlate with the PRS [14–16]. Because disease-risk genetic loci vary according to ethnicity, PRSs must be established for different ethnicities. Here, we hypothesized that if an individual’s PRS is associated with their disease liability, individuals having the highest PRSs may be the most likely to develop sAD, even at a young age. We built several PRS models to determine the contribution of the polygenic profile to the incidence risk and the age at onset of sAD in a Chinese cohort. Meanwhile, we analyzed the relationships between PRS and core cerebrospinal fluid (CSF) biomarkers of AD in an amyloid-beta (Aβ) deposition, pathologic tau, and neurodegeneration [AT(N)] scheme [17], with Aβ42, Aβ42/Aβ40, T-tau, and P-tau, to explore the impact of genetic risk on the pathology of AD.
Methods
Participants
A total of 462 sAD patients and 463 age-matched cognitively normal (CN) controls were recruited from Chongqing Daping Hospital from January 2015 to January 2019. Eligible participants were required (1) to have been diagnosed with sAD; (2) to be age-matched CN participants; and (3) to be willing to participate in the study. Participants were excluded for the following reasons: (1) a family history of dementia; (2) a concomitant neurologic disorder like head trauma or brain lesions that could potentially affect cognitive function, or other types of dementia; (3) severe cardiac, pulmonary, hepatic, or renal disease; and (4) mental illness (e.g., schizophrenia). The study was approved by the Institutional Review Board of Daping Hospital, and all participants and their caregivers provided informed consent.
Clinical Assessment and Diagnosis of sAD
Clinical assessment and diagnosis of sAD were performed following our previous protocol [18, 19]. All participants underwent clinical assessments that included medical history, physical examination, laboratory tests, APOE genotyping, and neuropsychological tests. Participants with abnormal cognition were further subjected to a brain CT/MRI investigation and blood tests for thyroxine, vitamin B12, folic acid, and HIV/syphilis to rule out metabolic and infectious reasons for cognitive decline.
The diagnosis of AD was made according to the criteria of the National Institute of Neurological and Communicative Diseases and Stroke and the Alzheimer’s Disease and Related Disorders Association (NINCDS–ADRDA) [20]; they included (1) insidious onset of symptoms, (2) a clear-cut history of worsening of cognition, and (3) prominent cognitive deficits in at least one of the following categories: amnestic presentation, language presentation, visuospatial presentation, or executive dysfunction. Patients were identified as having sAD if none of their first-degree relatives had dementia. CN participants had no memory complaints and performed within the normal range in the Mini-Mental State Examination (MMSE) [21] or the Montreal Cognitive Assessment [22].
SNP Selection and Genotyping
The SNPs reported in published GWASs and meta-analysis studies [5, 6, 9, 23, 24, 14] were initially included in the selection process. The alleles were excluded if (1) the minor allele frequency of the SNPs in the Chinese population was <0.05 (http://asia.ensembl.org/) or (2) the SNPs had been verified not to be associated with sAD risk in Chinese cohorts [25–29]. Detailed data for the selection and exclusion processes are provided in Table S1. Finally, a total of 35 SNPs in 18 candidate genes (including rs7412 and rs429358 in the APOE gene) were selected (Table S2).
Genotyping
Genotyping was conducted following a previously described method [30]. Briefly, genomic DNA was extracted from venous blood leukocytes using the Wizard genomic DNA purification kit (Promega, Madison, WI). Genotyping of the 35 SNPs was carried out with the multiplex polymerase chain reaction-ligase detection reaction method. For each SNP, the alleles were distinguished by the different fluorescent labels of allele-specific oligonucleotide probe pairs. Different SNPs were distinguished by different extended lengths at the 3’ end. All SNPs in the study had an overall call rate of >95%.
CSF Sampling and Analyses
A subgroup of 60 sAD and 200 CN participants underwent CSF sampling and analyses. In detail, in the sAD patients, CSF was sampled by the standard procedure [31]. In CN participants who had diseases of the urinary system, the CSF samples were collected during lumbar anesthesia before surgery for their diseases. Specifically, CSF samples free from any blood contamination were collected in polypropylene tubes by lumbar puncture, centrifuged at 1800 g at 4°C for 10 min within 1 h, and stored frozen at −80°C until analysis.
The levels of Aβ42, Aβ40, T-tau, and P-tau were determined using commercially available ELISA kits (Innotest, Fujirebio Europe, Ghent, Belgium), which have been widely used and validated in multiple studies and show good assay sensitivity and intra- and inter-assay precision. All measurements were made in one round of analysis with one batch of reagents by an experienced laboratory technician who was blinded to the clinical information. Our laboratory is a center of the Alzheimer’s Association quality control program [32] and is experienced in the examination of CSF biomarkers.
Statistical Analyses
Differences between groups were assessed by the two-sample independent t-test, the Mann-Whitney U test, the χ2 test, Fisher’s exact test, or analysis of variance according to the characteristics of the data. The data are expressed as the mean ± standard deviation (SD) for numerical variables or as the count (%) for categorical variables. All hypothesis-testing was two-sided, and statistical significance was defined as P < 0.05. All statistical computations were performed using SPSS version 19.0 (SPSS, Inc., Chicago, IL) or PLINK version 1.09 (http://www.cog-genomics.org/plink2), and all figures were created using a graphics package (GraphPad Software, Inc., San Diego, CA).
Single SNP Analyses
The allele and genotype distributions of the SNPs between the sAD patients and CN participants were analyzed using χ2 statistics [33]. The odds ratios (ORs; calculated relative to the common genotype) and 95% confidence intervals (CIs) were corrected for age (age at onset for sAD patients and age at inclusion for CN participants), sex, and APOE ε4 status (presence of one or two APOE ε4 alleles versus absence of the APOE ε4 allele) using logistic regression models (correction for APOE ε4 status was performed for all SNPs except those on the APOE gene).
Computation of PRSs
The SNPs associated with sAD (P < 0.05) in our cohort were selected to generate a PRS model (Model 1). For each participant, the PRS was calculated by summing the risk allele counts of the SNPs weighted by the natural logarithms of their respective ORs (calculated based on the present study). Given the strong effect of APOE genotypes on sAD and a recent systematic review, which suggested that including APOE in the PRS increased the AD prediction accuracy [13], APOE ε2/3/4 genotypes were incorporated into the PRS as special covariates with standard effects, namely, ε2/ε2 = 0.6, ε2/ε3 = 0.6, ε2/ε4 = 2.6, ε3/ε3 = 1.0, ε3/ε4 = 3.2, and ε4/ε4 = 14.9, as previously reported [34]. To build a more rigorous PRS model, only SNPs with a P-value threshold of 0.01 in the logistic regression analysis were included in Model 2. Because APOE is a critical gene for sAD, we also constructed Model 3 with APOE genotypes only.
The association of the PRS with sAD risk was tested by logistic regression, with age and sex as covariates. To evaluate the ability of PRS for case/control discrimination, receiver operating characteristic (ROC) analyses were performed by plotting the true positive rate against the false-positive rate. The area under the curve (AUC), sensitivity, and specificity with 95% CIs were calculated. Moreover, participants were partitioned into tertiles (two points at 33.33% and 66.67% divided the ordered distribution of PRSs into three parts, each containing a third of the population); the associations of the PRS with the age at onset and the cumulative incidence rate of sAD were reflected by a Cox proportional hazard model. Relationships between the PRSs and CSF biomarkers were assessed by Spearman correlation analyses. And the relationships were also evaluated with general linear models. Specifically, the PRS was used as the independent variable and the CSF biomarkers were used as dependent variables; the confounders age, sex, and APOE genotype were taken as covariates.
Results
Characteristics of the Study Participants and SNP Distributions
The characteristics of the participants are shown in Table 1. There were no significant differences in age (P = 0.17) between sAD patients and CN controls. sAD patients consisted of a higher proportion of females and APOE ε4 carriers and had lower MMSE scores. The CSF levels of Aβ42, Aβ40, and Aβ42/Aβ40 in the sAD group were lower than those in the control group (Aβ42: 632.60 ± 233.16 pg/mL vs 1265.39 ± 437.02 pg/mL, P < 0.001; Aβ40: 8519.27 ± 3846.22 pg/mL vs 11276.15 ± 4502.05 pg/mL, P < 0.001; Aβ42/Aβ40: 0.092 ± 0.092 vs 0.13 ± 0.088, P < 0.001). The CSF levels of T-tau and P-tau in the sAD group were higher than those in the control group (T-tau: 527.62 ± 443.62 pg/mL vs 219.74 ± 112.09 pg/mL, P < 0.001; P-tau: 70.03 ± 39.19 pg/mL vs 47.82 ± 16.72 pg/mL, P < 0.001).
Table 1.
Characteristics of the study participants.
| Characteristics | sAD (n = 462) | Control (n = 463) | P value |
|---|---|---|---|
| Age, mean (SD), years | 69.75 (9.84) | 68.85 (10.12) | 0.17 |
| Female, n (%) | 243 (52.6) | 193 (41.7) | 0.001 |
| MMSE score, mean (SD) | 14.41 (7.55) | 24.84 (3.97) | <0.001 |
| APOE ε4 carriers, n (%) | 191 (41.3) | 101 (21.8) | <0.001 |
| CSF Aβ42, pg/mL, mean (SD) | 632.60 (233.16) | 1265.39 (437.02) | <0.001 |
| CSF Aβ40, pg/mL, mean (SD) | 8519.27 (3846.22) | 11276.15 (4502.05) | <0.001 |
| CSF Aβ42/Aβ40, mean (SD) | 0.092 (0.092) | 0.13 (0.088) | <0.001 |
| CSF T-tau, pg/mL, mean (SD) | 527.62 (443.62) | 219.74 (112.09) | <0.001 |
| CSF P-tau, pg/mL, mean (SD) | 70.03 (39.19) | 47.82 (16.72) | <0.001 |
Differences between groups were assessed using Mann–Whitney U tests (for numerical variables) or χ2 tests (for categorical variables); sAD, sporadic Alzheimer’s disease; MMSE Mini-Mental State Examination; APOE, apolipoprotein E; CSF, cerebrospinal fluid.
Fourteen of the 35 SNPs were significantly associated with the risk of sAD (P < 0.05, Table 2): two (rs429358 and rs7412) on the APOE gene, two (rs6733839 and rs7561528) on the BIN1 gene, two (rs3865444 and rs3826656) on the CD33 gene, one (rs11771145) on the EPHA1 gene, four (rs561655, rs541458, rs10792832, and rs3851179) on the PICALM gene, two (rs11218343 and rs3781834) on the SORL1 gene, and one (rs2075650) on the TOMM40 gene. Only rs2075650 on the TOMM40 gene and rs429358 on the APOE gene remained significant after Bonferroni correction (P < 0.001). Information on the included SNPs (neighboring genes, chromosomes, minor alleles, minor allele frequencies, Hardy-Weinberg equilibrium values, and positions) and their allele and genotype frequencies are summarized in Tables S2–S4.
Table 2.
Allele distribution of the significant SNPs.
| SNP | Neighboring Gene | Risk allele | Risk allele frequency | OR (95% CI) | P value | |
|---|---|---|---|---|---|---|
| sAD | Control | |||||
| rs429358 | APOE | C | 0.25 | 0.12 | 2.55 (1.99–3.28) | <0.001 |
| rs7412 | APOE | T | 0.05 | 0.07 | 0.62 (0.42–0.92) | 0.02 |
| rs6733839 | BIN1 | T | 0.45 | 0.38 | 1.30 (1.08–1.57) | 0.007 |
| rs7561528 | BIN1 | A | 0.13 | 0.10 | 1.38 (1.03–1.85) | 0.032 |
| rs3865444 | CD33 | A | 0.17 | 0.22 | 0.72 (0.57–0.91) | 0.006 |
| rs3826656 | CD33 | A | 0.30 | 0.34 | 0.81 (0.66–0.98) | 0.03 |
| rs11771145 | EPHA1 | G | 0.48 | 0.43 | 1.25 (1.04–1.50) | 0.02 |
| rs561655 | PICALM | G | 0.43 | 0.49 | 0.78 (0.65–0.94) | 0.01 |
| rs541458 | PICALM | T | 0.47 | 0.53 | 0.79 (0.65–0.95) | 0.01 |
| rs10792832 | PICALM | A | 0.35 | 0.40 | 0.81 (0.67–0.98) | 0.03 |
| rs3851179 | PICALM | T | 0.35 | 0.39 | 0.81 (0.67–0.99) | 0.04 |
| rs11218343 | SORL1 | C | 0.26 | 0.31 | 0.78 (0.64–0.96) | 0.02 |
| rs3781834 | SORL1 | G | 0.19 | 0.23 | 0.79 (0.63–0.99) | 0.04 |
| rs2075650 | TOMM40 | G | 0.23 | 0.11 | 2.32 (1.79–3.00) | <0.001 |
The ORs and 95% CIs were adjusted for age, sex, and APOE ε4 status; SNP, single-nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Discriminative Performance of PRSs for sAD Patients and CN Controls
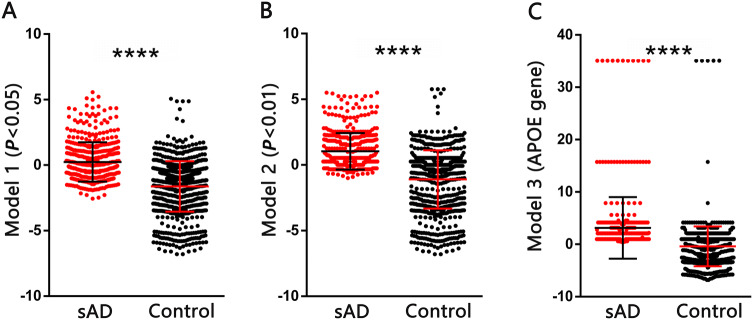
Three PRS models were developed (Table 3). As expected, the average PRSs in sAD patients were significantly higher than those in controls based on all three models (Mann-Whitney test, P < 0.0001, Fig. 1). Logistic regression analyses with adjustment for age and sex showed a positive relationship between sAD risk and PRS (OR > 1, Table 3). When we compared the discriminative ability of each PRS model by ROC curve analyses (Fig. 2), Model 1 had a sensitivity of 0.68 and a specificity of 0.57 (AUC = 0.66) and Model 2 had a sensitivity of 0.72 and a specificity of 0.49 (AUC = 0.65) (there was no significant difference between the two models). The sensitivity of Model 3 (0.61) based on the APOE gene alone was lower than that of the other models, but the specificity (0.78) was higher.
Table 3.
Logistic regressions for the associations of PRSs with sAD risk in the different PRS models.
| PRS models | P thresholds of SNPs | Logistic regressions | ROC curve analyses | |||
|---|---|---|---|---|---|---|
| OR (95% CI) | P | AUC (95% CI) | Sensitivity | Specificity | ||
| Model 1 | 0.05 | 1.58 (1.41–1.76) | <0.001 | 0.66 (0.63–0.70) | 0.68 | 0.57 |
| Model 2 | 0.01 | 1.56 (1.39–1.76) | <0.001 | 0.65 (0.61–0.68) | 0.72 | 0.49 |
| Model 3 | APOE genotype only | 1.20 (1.12–1.28) | <0.001 | 0.61 (0.58–0.65) | 0.42 | 0.78 |
The associations of the PRSs with sAD risk were tested by logistic regression adjusted by age and sex. PRS, polygenic risk score; SNP, single nucleotide polymorphism; sAD, sporadic Alzheimer’s disease.
Fig. 1.
PRSs in sAD patients and controls based on models 1 (A), 2 (B) and 3 (C). Differences between groups were assessed using Mann–Whitney U tests (error bars, SD; ****P < 0.0001; PRS, polygenic risk score; sAD, sporadic Alzheimer’s disease).
Fig. 2.
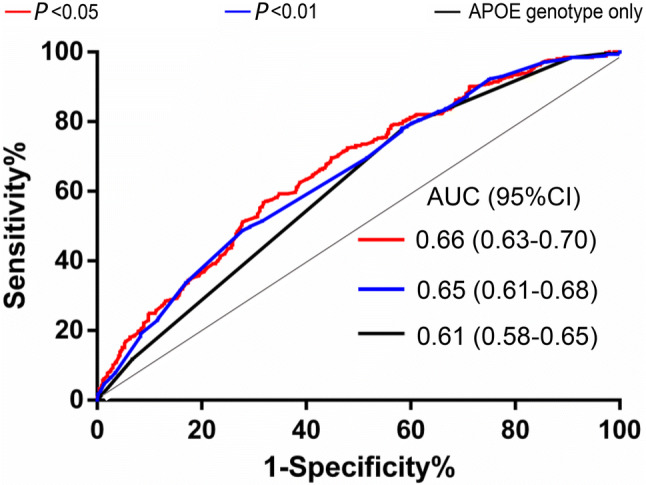
Discriminative ability (ROC curves) of different PRS models for sAD. PRSs were built with SNPs with P < 0.05 (red), P < 0.01 (blue), and APOE genotype only (black) (AUC, area under the curve; ROC, receiver operating characteristic curve; sAD, sporadic Alzheimer’s disease).
Predictive Ability of PRS Models for the Incidence Rate of sAD and Age at Onset
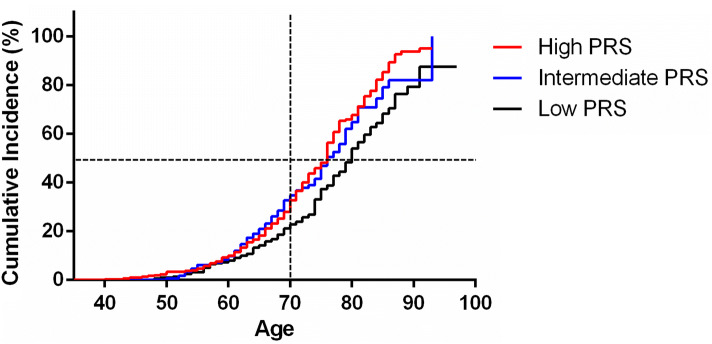
The modulation of PRS with the occurrence of sAD was evaluated using a Cox proportional hazards model (Fig. 3). The PRSs from Model 1 were chosen for the analysis. Participants were classified into three risk groups based on the PRS tertiles. The Log-Rank test revealed that a higher PRS was significantly associated with an earlier age at onset (high-PRS vs low-PRS, OR = 1.56, P = 0.0001, 95% CI: 1.26–1.97; intermediate-PRS vs low-PRS, OR = 1.39, P = 0.0076, 95% CI: 1.10–1.81). For example, in a cohort with a high-PRS, the expected age for 50% to develop sAD was ~75 years, earlier than in individuals with a low-PRS (the expected age for developing sAD in 50% was ~80 years). Moreover, the cumulative incidence rates in the high-PRS group were higher than those in the low-PRS group. For example, among two groups of 70-year-old individuals (with high-PRS or low-PRS), the percentage of sAD patients in the high-PRS group was higher than that in the low-PRS group (30% vs 20%).
Fig. 3.
Cumulative incidence rates of sAD in three genetic risk groups. Participants were partitioned into tertiles (low vs intermediate vs high PRS), with PRS cut-offs at 33.33% and 66.67% (PRS, polygenic risk score; sAD, sporadic Alzheimer’s disease).
Correlations Between PRS and AD Biomarkers in CSF
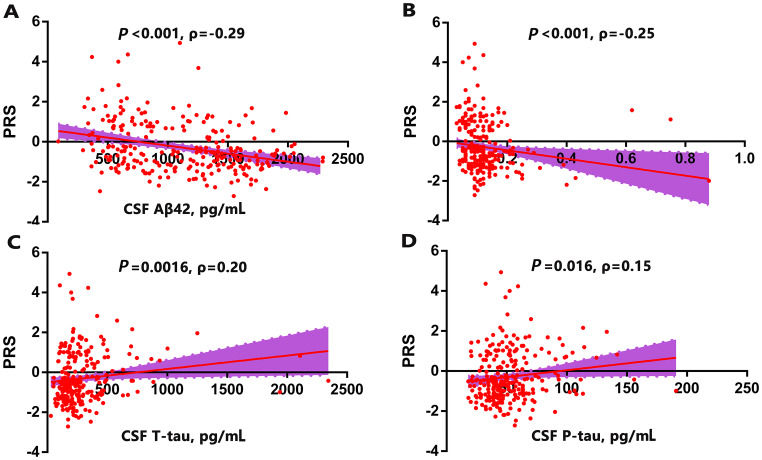
We analyzed the correlations between PRSs and the CSF levels of Aβ42, Aβ42/Aβ40, T-tau, and P-tau in the total cohort (Fig. 4), and used the PRSs from Model 1 for analysis. The CSF levels of Aβ42 and the Aβ42/Aβ40 ratio were inversely associated with the PRS (Aβ42: P < 0.001, Spearman ρ = −0.29; Aβ42/Aβ40 ratio: P < 0.001, Spearman ρ = −0.25), T-tau and P-tau were positively associated with the PRS (T-tau: P = 0.0016, Spearman ρ = 0.20; P-tau: P = 0.016, Spearman ρ = 0.15). The correlations remained similar for Aβ42 and T-tau (Aβ42, β = −0.31, P < 0.001; Aβ42/Aβ40 ratio, β = −0.13, P = 0.10; T-tau, β = 0.16, P = 0.032; P-tau, β = 0.13, P = 0.08) after adjusting for age, sex and APOE genotype with a general linear regression. The correlations in the CN control group were consistent with the total cohort (Table S5). Further, we partitioned the participants into three groups based on the tertiles of CSF Aβ42 level. The PRSs from Model 1 were used to differentiate individuals with the highest (third tertile) and lowest (first tertile) Aβ42 levels. The ability of PRS to determine the CSF level of Aβ42 was ~0.61 (AUC of the ROC curve) (Fig. 5), and increased to 0.69 when taking age and sex into account.
Fig. 4.
Correlations between PRSs and CSF biomarkers. A–D Scatterplots of PRS with (A) Aβ42, (B) Aβ42/Aβ40 ratio, (C) T-tau, and (D) P-tau (Spearman correlation coefficients (ρ) were used to assess the correlations; PRS, polygenic risk score; CSF, cerebrospinal fluid).
Fig. 5.
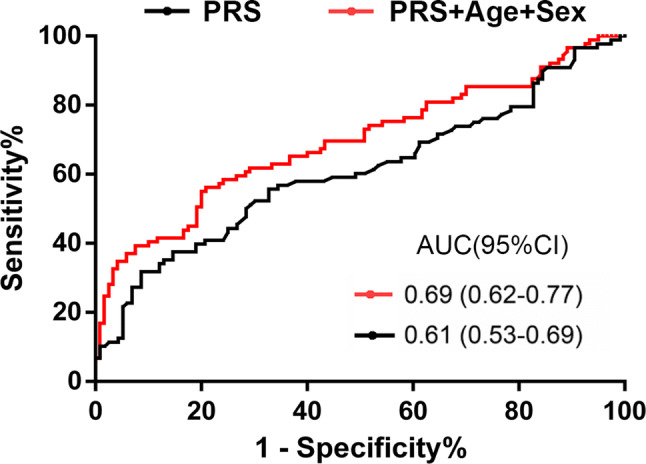
Discriminative ability of PRS model for CSF Aβ42 level. The AUC of the ROC curve was improved when taking age and sex into account (PRS, polygenic risk score; ROC, receiver operating characteristic curve; AUC, area under the curve; CI, confidence interval; CSF, cerebrospinal fluid).
Discussion
In this study, we explored the effects of genes on sAD development and the pathological process by screening and integrating AD-associated SNPs identified from large GWASs and building polygenic risk models. Only 14 of the 35 SNPs identified in other populations had significant correlations with sAD, and the PRSs based on the 14 SNPs were associated with the risk of sAD, age at onset, and CSF biomarkers in our Chinese cohort.
Three PRS models containing different numbers or categories of SNPs were built for case/control discrimination. We found no improvement in the discrimination when more SNPs were included in the PRS model. Thus, considering the expense of genotype sequencing, a PRS model based on fewer SNPs (SNPs with a P threshold of 0.01 in our study) would be more accessible. The PRS model built on the APOE genotype, which is associated with amyloid pathology in Chinese AD patients [35], had relatively higher specificity and lower sensitivity in case/control discrimination, confirming the hypothesis that sAD can be attributed to multiple genetic profiles rather than a single gene.
We used a Cox regression survival model for the age at onset analysis because it provides more power than a simple linear regression model. Consistent with previous findings [14–16], individuals with a high genetic risk (high-PRS) were more likely to develop sAD, and the time of onset was earlier than that in individuals with a low genetic risk (low-PRS), which suggests that the incidence risk of sAD and age at onset are modified by the polygenic profile. From a clinical perspective, although not ready for use in clinical practice, our PRS model has the potential to serve as a predictor for identifying seniors at risk for developing sAD at a given age and provides potential sAD patients with access to early diagnosis and treatment. Of course, additional studies with SNPs from Chinese GWASs are needed to strengthen the predictive power of PRS models.
The impact of genetic risk on the biomarkers of sAD can provide deep insight into the pathogenesis of the disease. We found a significant relationship between an increased PRS and decreased CSF levels of Aβ42 or the Aβ42/Aβ40 ratio as well as increased CSF levels of T-tau and P-tau, suggesting that the genetic profile modulates the pathogenesis of sAD. Because the pathological changes of AD begin 15–20 years before clinical presentation [36] and clinical trials of disease-modifying therapy at the preclinical stage are promising [37], the use of a polygenic model to identify individuals with abnormal level of CSF biomarkers seems valuable. In this study, we identified individuals with an extremely abnormal level of CSF Aβ42 using the PRS model we built. However, the model was not accurate enough to determine the sAD risk and its related pathology, which is reasonable because complex factors, including the environment and lifestyle, also contribute to the pathogenesis of AD [1]. Future studies elucidating these non-hereditary factors in individuals with genetic risk information may offer more valuable insight into the relationship between genes and AD pathology. Nevertheless, individuals who exhibit a ‘positive’ result from a genomic examination can apply for more accurate clinical, CSF, or imaging examinations, which will provide more accurate probabilistic assessments as to whether AD development is likely to occur.
The present study has three major limitations. First, the case/control approach assumes that controls do not develop sAD and considers the disease process to be a dichotomous variable; errors may exist because some controls may be at the preclinical stage of AD. Second, our AD participants were actually probable AD because the diagnosis was made based on the NINCDS–ADRDA criteria; results can be more accurate if AD is diagnosed with the assistance of biomarkers [18]. Third, the PRSs were based on SNPs identified in other ethnicities. Because the risk loci of sAD may differ among ethnicities, a PRS model built on SNPs identified in the Chinese population would provide more accurate prediction of the genetic risk of sAD in Chinese participants.
Conclusions
In the present study, several SNPs had significant correlations with sAD risk, age at onset, and its CSF biomarkers in our cohort, suggesting that PRS models hold promise for assessing the genetic risk of the development of AD. As genetic risk profiles vary among populations, large-scale genome-wide sequencing studies are urgently needed to identify the genetic risk loci of sAD in Chinese populations to build accurate PRS models for clinical practice.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank the patients and their families for their participation and dedication to research. This work was supported by the National Basic Research Development Program of Ministry of Science and Technology of China (2016YFC1306401) and the National Natural Science Foundation of China (91749206).
Competing interests
The authors declare that they have no competing interests.
Footnotes
Wei-Wei Li and Zhen Wang have contributed equally to this work.
Contributor Information
Li-Yong Chen, Email: mzkcly@aliyun.com.
Yan-Jiang Wang, Email: yanjiang_wang@tmmu.edu.cn.
References
- 1.Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 2.Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43:1467–1472. doi: 10.1212/WNL.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 3.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 8.Chouraki V, Seshadri S. Genetics of Alzheimer’s disease. Adv Genet. 2014;87:245–294. doi: 10.1016/B978-0-12-800149-3.00005-6. [DOI] [PubMed] [Google Scholar]
- 9.Miyashita A, Koike A, Jun G, Wang LS, Takahashi S, Matsubara E, et al. SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese. Koreans and Caucasians. PLoS One. 2013;8:e58618. doi: 10.1371/journal.pone.0058618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lacour A, Espinosa A, Louwersheimer E, Heilmann S, Hernandez I, Wolfsgruber S, et al. Genome-wide significant risk factors for Alzheimer’s disease: role in progression to dementia due to Alzheimer’s disease among subjects with mild cognitive impairment. Mol Psychiatry. 2017;22:153–160. doi: 10.1038/mp.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51:404–413. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun BL, Li WW, Zhu C, Jin WS, Zeng F, Liu YH, et al. Clinical research on Alzheimer’s disease: progress and perspectives. Neurosci Bull. 2018;34:1111–1118. doi: 10.1007/s12264-018-0249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stocker H, Mollers T, Perna L, Brenner H. The genetic risk of Alzheimer’s disease beyond APOE epsilon4: systematic review of Alzheimer’s genetic risk scores. Transl Psychiatry. 2018;8:166. doi: 10.1038/s41398-018-0221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karch CM, Goate AM. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2015;77:43–51. doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desikan RS, Fan CC, Wang Y, Schork AJ, Cabral HJ, Cupples LA, et al. Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score. PLoS Med. 2017;14:e1002258. doi: 10.1371/journal.pmed.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sleegers K, Bettens K, De Roeck A, Van Cauwenberghe C, Cuyvers E, Verheijen J, et al. A 22-single nucleotide polymorphism Alzheimer’s disease risk score correlates with family history, onset age, and cerebrospinal fluid Abeta42. Alzheimers Dement. 2015;11:1452–1460. doi: 10.1016/j.jalz.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Jack CR, Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14:535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li WW, Shen YY, Tian DY, Bu XL, Zeng F, Liu YH, et al. Brain amyloid-beta deposition and blood biomarkers in patients with clinically diagnosed Alzheimer’s disease. J Alzheimers Dis. 2019;69:169–178. doi: 10.3233/JAD-190056. [DOI] [PubMed] [Google Scholar]
- 19.Bu XL, Yao XQ, Jiao SS, Zeng F, Liu YH, Xiang Y, et al. A study on the association between infectious burden and Alzheimer’s disease. Eur J Neurol. 2015;22:1519–1525. doi: 10.1111/ene.12477. [DOI] [PubMed] [Google Scholar]
- 20.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–944. doi: 10.1212/WNL.34.7.939. [DOI] [PubMed] [Google Scholar]
- 21.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 22.Nasreddine ZS, Phillips NA, Bedirian V, Charbonneau S, Whitehead V, Collin I, et al. The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc. 2005;53:695–699. doi: 10.1111/j.1532-5415.2005.53221.x. [DOI] [PubMed] [Google Scholar]
- 23.Cuyvers E, Sleegers K. Genetic variations underlying Alzheimer’s disease: evidence from genome-wide association studies and beyond. Lancet Neurol. 2016;15:857–868. doi: 10.1016/S1474-4422(16)00127-7. [DOI] [PubMed] [Google Scholar]
- 24.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nature Genetics. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Li X, Ma G, Jiang Y, Liao M, Feng R, et al. CLU rs9331888 polymorphism contributes to Alzheimer’s disease susceptibility in Caucasian but not East Asian populations. Mol Neurobiol. 2016;53:1446–1451. doi: 10.1007/s12035-015-9098-1. [DOI] [PubMed] [Google Scholar]
- 26.Yu JT, Li L, Zhu QX, Zhang Q, Zhang W, Wu ZC, et al. Implication of CLU gene polymorphisms in Chinese patients with Alzheimer’s disease. Clin Chim Acta. 2010;411:1516–1519. doi: 10.1016/j.cca.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Xiao Q, Liu ZJ, Tao S, Sun YM, Jiang D, Li HL, et al. Risk prediction for sporadic Alzheimer’s disease using genetic risk score in the Han Chinese population. Oncotarget. 2015;6:36955–36964. doi: 10.18632/oncotarget.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Yu JT, Wang HF, Hao XK, Yang YF, Jiang T, et al. Association between NME8 locus polymorphism and cognitive decline, cerebrospinal fluid and neuroimaging biomarkers in Alzheimer’s disease. PLoS One. 2014;9:e114777. doi: 10.1371/journal.pone.0114777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu H, Zhu XC, Wang HF, Cao L, Tan MS, Tan CC, et al. Lack of Association between SLC24A4 polymorphism and late-onset Alzheimer’s disease in Han Chinese. Curr Neurovasc Res. 2016;13:239–243. doi: 10.2174/1567202613666160524144739. [DOI] [PubMed] [Google Scholar]
- 30.Zeng F, Shen C, Liu YH, Li J, Zhu J, Wang YR, et al. Genetic association between APP, ADAM10 gene polymorphism, and sporadic Alzheimer’s disease in the Chinese population. Neurotox Res. 2015;27:284–291. doi: 10.1007/s12640-015-9516-1. [DOI] [PubMed] [Google Scholar]
- 31.Blennow K, Hampel H, Weiner M, Zetterberg H. Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat Rev Neurol. 2010;6:131–144. doi: 10.1038/nrneurol.2010.4. [DOI] [PubMed] [Google Scholar]
- 32.Mattsson N, Andreasson U, Persson S, Arai H, Batish SD, Bernardini S, et al. The Alzheimer’s Association external quality control program for cerebrospinal fluid biomarkers. Alzheimers Dement. 2011;7(386–395):e386. doi: 10.1016/j.jalz.2011.05.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan L, Song Z, Deng X, Yang Z, Yang Y, Guo Y, et al. Genetic analysis of FBXO2, FBXO6, FBXO12, and FBXO41 variants in Han Chinese patients with sporadic Parkinson’s disease. Neurosci Bull. 2017;33:510–514. doi: 10.1007/s12264-017-0122-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. doi: 10.1001/jama.1997.03550160069041. [DOI] [PubMed] [Google Scholar]
- 35.Yang Q, Chen K, Zhang H, Zhang W, Gong C, Zhang Q, et al. Correlations between single nucleotide polymorphisms, cognitive dysfunction, and postmortem brain pathology in Alzheimer’s disease among Han Chinese. Neurosci Bull. 2019;35:193–204. doi: 10.1007/s12264-019-00343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mormino EC, Papp KV, Rentz DM, Schultz AP, LaPoint M, Amariglio R, et al. Heterogeneity in suspected non-Alzheimer disease pathophysiology among clinically normal older individuals. JAMA Neurol. 2016;73:1185–1191. doi: 10.1001/jamaneurol.2016.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheltens P, Blennow K, Breteler MM, de Strooper B, Frisoni GB, Salloway S, et al. Alzheimer’s disease. Lancet. 2016;388:505–517. doi: 10.1016/S0140-6736(15)01124-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.