Dear Editor,
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disease characterized by movement disorder, progressive dementia, and psychiatric and behavioral changes. It is caused by unstable expanded CAG trinucleotide repeats in exon 1 of the huntingtin (HTT) gene, located on chromosome 4p16.3 [1]. The number of CAG repeats is inversely correlated with the onset age of HD, with more expanded CAG repeats correlated with an earlier onset [2]. In previous studies, we have confirmed the inverse correlation between the onset age and the CAG repeat size in Chinese HD patients [3, 4].
It has been reported that the expanded CAG trinucleotides are unstable during germline transmission, tend to expand in paternal transmission, but are more stable in maternal transmission [5]. Some studies have suggested that an intra- or flanking sequence of the HTT gene influences the CAG instability, thus causing the different prevalence rates of the disease [6–8]. A CCG polymorphism is adjacent to the CAG triplet repeat of the HTT gene, and is associated with the presence or absence of GAG repeats at codon 2642 (∆2642) [9]. There is a significant difference in the allele frequencies of CCG polymorphisms among populations of different ancestries [9, 10].
In Caucasian populations, it is thought that a CCG7 polymorphism is associated with expanded CAG alleles [2]. However, in Japanese populations, CCG10 is considered to be associated with the expanded CAG alleles [10]. The different distributions of CCG polymorphisms suggest that they may be a risk factor for CAG instability and cause the varied prevalence. However, only two studies have reported the distribution frequencies of CCG polymorphisms in limited numbers of HD patients in mainland China (32 and 53 cases) [11, 12]. Furthermore, the CCG repeat sizes influencing the CAG instability during germline transmission have never been reported either in Chinese or Caucasian populations. Therefore, we aimed to use parent-child pairs to investigate the correlation between CAG and CCG repeats.
A total of 253 individuals (204 patients manifesting HD and 49 pre-symptomatic mutation carriers) with expanded CAG repeats within the HTT gene from 201 HD families were enrolled between February 2008 and May 2019 from three centers located in Southeastern China (the Second Affiliated Hospital of Zhejiang University School of Medicine, Huashan Hospital of Fudan University, and the First Affiliated Hospital of Fujian Medical University). Among them, 193 cases had already participated in our previous study for a different research purpose [4]. Clinical data were analyzed retrospectively and 52 parent-child pairs were collected: 49 HD parents and 52 offspring. In addition, 248 unrelated control individuals were included. This study was approved by the local Ethics Committees of the above three hospitals. Written consent was given by participants or their legally authorized caregivers. Genomic DNA was extracted from peripheral blood and genetic testing for HTT was carried out using the previously reported method [13]. Statistical analysis was conducted using SPSS 20.0 (SPSS Inc., Chicago, IL). A P-value < 0.05 was considered statistically significant. To compare two continuous variables, T tests, Wilcoxon tests, or Mann-Whitney tests were used. For categorical variables, Pearson or Fisher’s exact χ2 test was used.
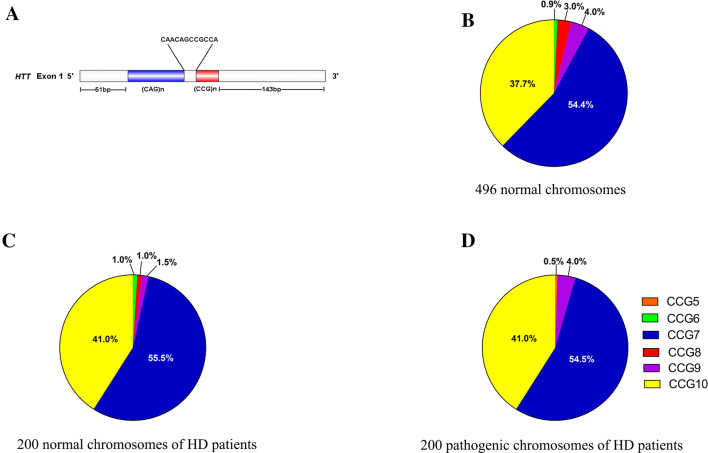
The frequency distribution of CCG repeats was determined in 200 unrelated patients manifesting HD (members of one family were all pre-symptomatic mutation carriers) and 248 unrelated normal controls. A total of six different CCG polymorphisms (CCG5, 6, 7, 8, 9, and 10) were detected (Fig. 1). Among the 496 normal chromosomes from 248 normal controls, CCG7 repeat alleles were most frequent (n= 270, 54.4%) followed by CCG10 (n= 187, 37.7%), and the remaining 39 cases (7.9%) were CCG6, CCG8, and CCG9 (Fig. 1B). Among the normal alleles of 200 unrelated HD patients (Fig. 1C), 111 (55.5%) were CCG7, 82 (41.0%) were CCG10, and the remaining 7 (3.5%) were CCG6, CCG8, and CCG9. Among the 200 pathogenic alleles, 109 (54.5%) were CCG7, 82 (41.0%) were CCG10, and the remaining 9 (4.5%) were CCG5 and CCG9 (Fig. 1D). We found that the distribution characteristics of CCG polymorphisms in the normal chromosomes were almost same as those of the pathogenic chromosomes.
Fig. 1.
Frequency distributions of CCG repeat sizes in general and HD populations. A Locations of the CAG and CCG polymorphism regions. B–D Frequency distribution of CCG repeats on the normal chromosomes from the general population (B, n = 496), on the normal chromosomes from the HD population (C, n = 200), and on the pathogenic chromosomes from the HD population (D, n = 200).
To explore whether the CCG repeat sizes change during germline transmission, we analyzed the inter-generational changes of CCG repeat sizes in 52 parent–child pairs, including 24 father-child and 28 mother–child pairs. During father–child transmission, inter-generational changes of CCG repeats occurred in 29.2% (7/24) of meiosis, including 20.8% (5/24) expansions and 8.4% (2/24) contractions. The remaining 70.8% (17/24) were not changed. However, in mother-child transmission, only 7.1% (2/28) showed contraction and none shown expansion. The percentage of CCG changes did not differ significantly between paternal and maternal transmission (P = 0.06).
As CCG7 and CCG10 were the predominant polymorphisms in both the general and HD populations, we only compared the CAG sizes of the CCG7 and CCG10 groups between the two populations. Among normal controls, the number of CAG repeats was 17.5 ± 2.0 (range 15–31) in the CCG7 group (n = 270) and 17.4 ± 1.7 (range 9–31) in the CCG10 group (n = 187) (P = 0.54). Among the normal alleles of HD patients, the number of CAG repeats was 17.7 ± 1.7 (range 15–26) in the CCG7 group (n = 111) and 17.4 ± 1.6 (range 9–24) in the CCG10 group (n = 82). Although the former was slightly larger than the latter, no significant difference was found (P = 0.19). Among the pathogenic alleles of HD patients, the number of CAG repeats was 45.0 ± 4.4 (range 40–66) in the CCG7 group (n = 109) and 45.0 ± 5.8 (range 39–74) in the CCG10 group (n = 82). No difference was found in the mean CAG repeat size between CCG7 and CCG10 (P = 0.91).
As CAG repeat sizes in the CCG7 and CCG10 groups did not differ at the population level, we tested the relationship at the inter-generational level. Among the 52 parent-child pairs, CCG7 accounted for 48.1% (25/52), CCG10 for 32.7% (17/52), and CCG6, CCG8 and CCG9 for the remaining 19.2% (10/52) (Table 1). Since the parent-child pairs of CCG6, CCG8 and CCG9 were too small, we only compared the CCG7 and CCG10 polymorphisms. In the total of 42 parent-child pairs, no statistical difference of △CAG change was found between the CCG7 (0.8 ± 2.5) and CCG10 (3.5 ± 8.7) groups (P = 0.14). Of note, parents with larger CAGs were more likely to have larger △CAG repeats [5], so we compared the CAG repeats of parents between the CCG7 and CCG10 groups but found no significant difference (P = 0.46). In addition, a previous study reported that the gender of the affected parent is a risk factor for CAG instability [4], so we subdivided the parent-child pairs according to the gender of the parents to compare the number of △CAG repeats between the CCG7 and CCG10 polymorphisms. In paternal transmission, although the △CAG of the CCG7 group (1.6 ± 3.2) was lower than that of CCG10 (6.6 ± 13.2), it did not reach statistical significance (P = 0.29). In maternal transmission, the △CAG of the CCG7 group (0.3 ± 1.9) was similar to that of the CCG10 group (1.4 ± 2.4) (P = 0.21).
Table 1.
Comparisons of inter-generational changes of CAG repeat length between CCG7 and CCG10 groups.
| CCG7 | CCG10 | P value | |
|---|---|---|---|
| Pairs (P/M) | 25 (9/16) | 17 (7/10) | |
| Parent | 44.6 ± 2.5 | 43.8 ± 3.8 | 0.46 |
| Offspring | 45.4 ± 3.2 | 47.4 ± 12.0 | 0.42 |
| △CAG | 0.8 ± 2.5 | 3.5 ± 8.7 | 0.14 |
| Paternal | 43.4 ± 1.4 | 44.6 ± 5.7 | 0.57 |
| Offspring | 45.0 ± 3.5 | 51.1 ± 18.6 | 0.34 |
| △CAG | 1.6 ± 3.2 | 6.6 ± 13.2 | 0.29 |
| Maternal | 45.2 ± 2.8 | 43.3 ± 1.9 | 0.07 |
| Offspring | 45.5 ± 3.1 | 44.7 ± 3.1 | 0.53 |
| △CAG | 0.3 ± 1.9 | 1.4 ± 2.4 | 0.21 |
| P value | 0.23# | 0.34# |
M: maternal inheritance; P: paternal inheritance; △CAG: the inter-generational changes of CAG repeats.
#: Comparison between the △CAG of father-child transmission and mother-child transmission in each CCG group.
Different populations had different CCG polymorphisms. In Caucasian HD patients, CCG7 polymorphism accounted for 94.4% of the expanded alleles and for 5.6% of the normal alleles (Table S1) [2]. And in the Japanese population, 84.5% of pathogenic alleles were associated with CCG10 but only 15.5% with CCG7 [14]. However, in our current study in mainland China, the CCG7 (54.5%) and CCG10 (41.0%) repeats were similar and the CCG10 repeats had a significantly higher proportion than that reported in the Caucasian populations [2]. The discrepancy between Chinese and Caucasians may be due to the different genetic backgrounds [10]. Many SNPs have been reported to have different distributions between Chinese and Caucasian populations, like the delta 2642 polymorphisms. This may be caused by different founders in the Chinese and Caucasian populations, as supported by haplotype analysis [6–8]. On the other hand, the CCG distributions differ between populations in Japan and mainland of China [2]. A plausible explanation might be the smaller sample size of Japanese.
Here, we first studied the correlation between the inter-generational CCG change and CAG repeats, which have rarely been explored either in Caucasian or Chinese populations. The △CAG of the CCG10 group was higher than that of the CCG7 group, but did not reach a significant level, perhaps because of the small sample size. Therefore, a larger cohort is needed to further support the results.
In summary, to our knowledge, this study is the first to analyze the correlation between CCG polymorphisms and CAG repeats in the HTT region at the inter-generational level. We concluded that the distribution of CCG polymorphisms in China differs from that in Caucasians, but CCG polymorphisms did not influence the CAG instability although CCG10 showed a tendency towards CAG expansion during germline transmission. A larger cohort is needed to explore this relationship.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We sincerely thank the participants for their help and willingness to take part in this study. We especially thank Wan-Qing Xu for help with grammar and expression. This work was supported by the Key Research and Development Project of Zhejiang Province, China (2019C03039), and the Research Foundation for Distinguished Scholars of Zhejiang University, China (188020-193810101/089).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Hong-Rong Cheng and Xiao-Yan Li have contributed equally to this work.
Contributor Information
Hong-Lei Li, Email: lihonglei@zju.edu.cn.
Zhi-Ying Wu, Email: zhiyingwu@zju.edu.cn.
References
- 1.MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, Srinidhi L, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- 2.Squitieri F, Andrew SE, Goldberg YP, Kremer B, Spence N, Zeisler J, et al. DNA haplotype analysis of Huntington disease reveals clues to the origins and mechanisms of CAG expansion and reasons for geographic variations of prevalence. Hum Mol Genet. 1994;3:2103–2114. doi: 10.1093/hmg/3.12.2103. [DOI] [PubMed] [Google Scholar]
- 3.Li XY, Zhang YB, Xu M, Cheng HR, Dong Y, Ni W, et al. Effect of apolipoprotein E genotypes on Huntington’s disease phenotypes in a Han Chinese population. Neurosci Bull. 2019;35:756–762. doi: 10.1007/s12264-019-00360-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li HL, Li XY, Zhang YB, Dong Y, Cheng HR, Gan SR, et al. Clinical and genetic profiles in Chinese patients with Huntington’s disease: A ten-year multicenter study in China. Aging Dis. 2019;10:1003–1011. doi: 10.14336/AD.2018.0911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kremer B, Almqvist E, Theilmann J, Spence N, Telenius H, Goldberg YP, et al. Sex-dependent mechanisms for expansions and contractions of the CAG repeat on affected Huntington disease chromosomes. Am J Hum Genet. 1995;57:343–350. [PMC free article] [PubMed] [Google Scholar]
- 6.Li XY, Li HL, Dong Y, Gao B, Cheng HR, Ni W, et al. Haplotype analysis encompassing HTT gene in Chinese patients with Huntington’s disease. Eur J Neurol. 2020;27:273–279. doi: 10.1111/ene.14072. [DOI] [PubMed] [Google Scholar]
- 7.Kay C, Collins J, Skotte JH, Southwell AL, Warby SC, Caron NS, et al. Huntingtin Haplotypes provide prioritized target panels for allele-spectic silencing in Huntington disease of European ancestry. Mol Ther. 2015;11:1759–1771. doi: 10.1038/mt.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warby SC, Montpetit A, Hayden AR, Carroll JB, Butland SL, Visscher H, et al. CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am J Hum Genet. 2009;84:351–366. doi: 10.1016/j.ajhg.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hecimovic S, Klepac N, Vlasic J, Vojta A, Janko D, Skarpa-Prpic I, et al. Genetic background of Huntington disease in Croatia: Molecular analysis of CAG, CCG, and Delta2642 (E2642del) polymorphisms. Hum Mutat. 2002;20:233. doi: 10.1002/humu.9055. [DOI] [PubMed] [Google Scholar]
- 10.Morovvati S, Nakagawa M, Osame M, Karami A. Analysis of CCG repeats in Huntingtin gene among HD patients and normal populations in Japan. Arch Med Res. 2008;39:131–133. doi: 10.1016/j.arcmed.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 11.Ma M, Yang Y, Shang H, Su D, Zhang H, Ma Y, et al. Evidence for a predisposing background for CAG expansion leading to HTT mutation in a Chinese population. J Neurol Sci. 2010;298:57–60. doi: 10.1016/j.jns.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 12.Zhang BR, Tian J, Yan YP, Yin XZ, Zhao GH, Wu ZY, et al. CCG polymorphisms in the huntingtin gene have no effect on the pathogenesis of patients with Huntington’s disease in mainland Chinese families. J Neurol Sci. 2012;312:92–96. doi: 10.1016/j.jns.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 13.Dong Y, Sun YM, Liu ZJ, Ni W, Shi SS, Wu ZY. Chinese patients with Huntington’s disease initially presenting with spinocerebellar ataxia. Clin Genet. 2013;83:380–383. doi: 10.1111/j.1399-0004.2012.01927.x. [DOI] [PubMed] [Google Scholar]
- 14.Masuda N, Goto J, Murayama N, Watanabe M, Kondo I, Kanazawa I, et al. Analysis of triplet repeats in the huntingtin gene in Japanese families affected with Huntington’s disease. J Med Genet. 1995;32:701–705. doi: 10.1136/jmg.32.9.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.