Abstract
Cell fate is largely determined by interactions that occur at the interface between cells and their surrounding microenvironment. For this reason, especially in the field of tissue-engineering, there is a growing interest in developing techniques that allow evaluating cell–material interaction at the nanoscale, particularly focusing on cell adhesion processes. While for 2D culturing systems a consolidated series of tools already satisfy this need, in 3D environments, more closely recapitulating complex in vivo structures, there is still a lack of procedures furthering the comprehension of cell–material interactions. Here, the use of scanning electron microscopy coupled with a focused ion beam (SEM/FIB) for the characterization of cell interactions with 3D scaffolds obtained by different fabrication techniques is reported for the first time. The results clearly show the capability of the developed approach to preserve and finely resolve scaffold–cell interfaces highlighting details such as plasma membrane arrangement, extracellular matrix architecture and composition, and cellular structures playing a role in cell adhesion to the surface. It is anticipated that the developed approach will be relevant for the design of efficient cell-instructive platforms in the study of cellular guidance strategies for tissue-engineering applications as well as for in vitro 3D models.
Keywords: 3D materials, cell–material interface, focused ion beam, scaffold, scanning electron microscopy
A key role in the successful application of many biomedical platforms for prosthetics, artificial organs, and auxiliary devices is played by the interaction between cells and biomaterials.[1] In particular for implantable devices, optimal adhesion is crucial to avoid implant rejection and to ensure its complete integration into the host.[2] Such processes are regulated by the interaction of single cells with the biomaterial itself.[3,4] In the last decades, many efforts have been carried out to properly engineer the material surface and bulk properties, i.e., surface topography,[5] chemical functionalization,[6] and material stiffness[7,8] for delivering complex sets of cell-instructive signals capable to specifically affect cell fate.[9,10] The final goal in designing such platforms is, in fact, to create instructive adhesion areas capable of guiding several cell functions in a controlled way for a good implant integration. Those functions include migration, proliferation, differentiation, and the synthesis of endogenous extracellular matrix (ECM). In fact, particular attention has been dedicated to the development of 3D systems that better replicate the structural organization of living tissues and that, compared to 2D platforms, have been extensively demonstrated to affect cellular behavior in a more realistic way.[11,12] For instance, 3D matrices/scaffolds have been fabricated with different degrees of structural organization to correlate in vitro cells responses with diverse cell–material interactions.[13–16] In this context, in order to evaluate the effect of specific functional features upon cellular response, one can get fundamental informative cues from the investigation of the interface between cells and 3D biomaterials at the macro, micro, and nanoscales. Optical microscopy is effective in providing accurate characterization of cellular features, i.e., nucleus, actin filaments, focal adhesion, with a resolution down to sub-micrometers scale through direct imaging with fluorophores.[17,18] However, electron microscopy (EM) is the ultimate technique to achieve the highest resolution in the investigation of the cell–material interface. In fact, EM allows for the resolution of features in the nanometers range. In combination with fluorescence-based techniques, it could in fact substantially widen the overview on cell–material interactions adding important information related to cellular architecture in response to specific cell-instructive signals.[19] While traditional EM specimen preparation fits well with 2D cell–material systems,[20] it finds major limitations in the case of 3D matrices.[21] In resin embedding–based procedures coupled with mechanical sectioning, removal of the support material is required. This procedure can induce artifacts particularly at the contact area between cells and the material. While this is a common approach for cells on 2D materials, it is obviously incompatible with 3D materials since cells can grow in all directions and the removal of the material itself could cause the collapse of cellular components. In light of this, the ideal approach should 1) maintain the support material in place with the cells and 2) allow for direct sectioning. In fact, resin-embedded specimens together with the support material can be alternatively sectioned via focused ion beam (FIB).[22] However, also in this case, the presence of a large resin matrix does not allow the selection of a region of interest (ROI) while the whole specimen has to be sectioned.[23,24] Other methods have been established to characterize the interaction of 3D scaffold–like materials with cells involving hard drying procedures of specimens combined with scanning electron microscopy (SEM).[25,26] However, typical artifacts such as cracks and cell detachment can be visible and this is not representative of the actual cell–material interface. Indeed there is a lack of procedures which allow preservation of both the 3D material structure and the cell position, for subsequently performing selective sectioning for high-resolution investigation of the cell–3D material interface.
Recently, ultrathin plasticization (UTP) of cells has been successfully performed to overcome the aforementioned limitations in case of adherent cells on 2D and pseudo-3D materials (surface with protruding micro and nanostructures).[27–29] This technique grants the allocation of an ROI for a selective cross sectioning via FIB and a high resolution imaging (5–10 nm) via SEM, such that plasma membrane as well as intracellular compartments can be visualized.[30]
Here, we present the application of a UTP procedure for studying the interface between 3D polymeric architectures and cells at the nanoscale, by means of the EM technique. Complementarily to fluorescence-based microscopy (i.e., confocal), our approach can give insights on processes regulating cell–material 3D interactions at different length scales. In particular, we present two 3D scaffold types, which differ by spatial arrangement of their backbone (ordered vs nonordered) and their interaction with cells, showing the capability of the implemented technique in resolving structural features with nanometer resolution. In fact, this method allows for SEM of whole cells and in addition creating localized cross sections reveals, in detail, cellular components in the vicinity of the 3D material.
First, we fabricated polymeric scaffolds with two different geometries. By means of a 2-photon polymerization lithography system (Nanoscribe Photonic Professional GT, Nanoscribe GmbH), ordered scaffolds were obtained. A 4-layer example is shown in the SEM in Figure 1A.[31] Structures were fabricated processing a commercial negative photoresist using a constant power of 60 mW and a scan speed of 1000 μm s−1. The design consists of adjacent “cages” with ≈50 μm opening/height. 8 × 8 cages have been stitched in the x–y direction to form the individual layer (Figure 1B), that was in turn connected in the z-direction to form the entire structure. Adjacent cages were stitched together considering an overlap of ≈1 μm to ensure the necessary final structural stability. This serial fabrication approach allows us to obtain very complex and stable structures. Thanks to this strategy, it is also possible to change the overall dimensions of the structure without the need to revert to complex structural design iterations. Then, the scaffolds were sterilized and coated with fibronectin to encourage cell adhesion. Human glioblastoma astrocytoma (U87) cells were seeded on the ordered scaffolds (see Experimental Section).
Figure 1.
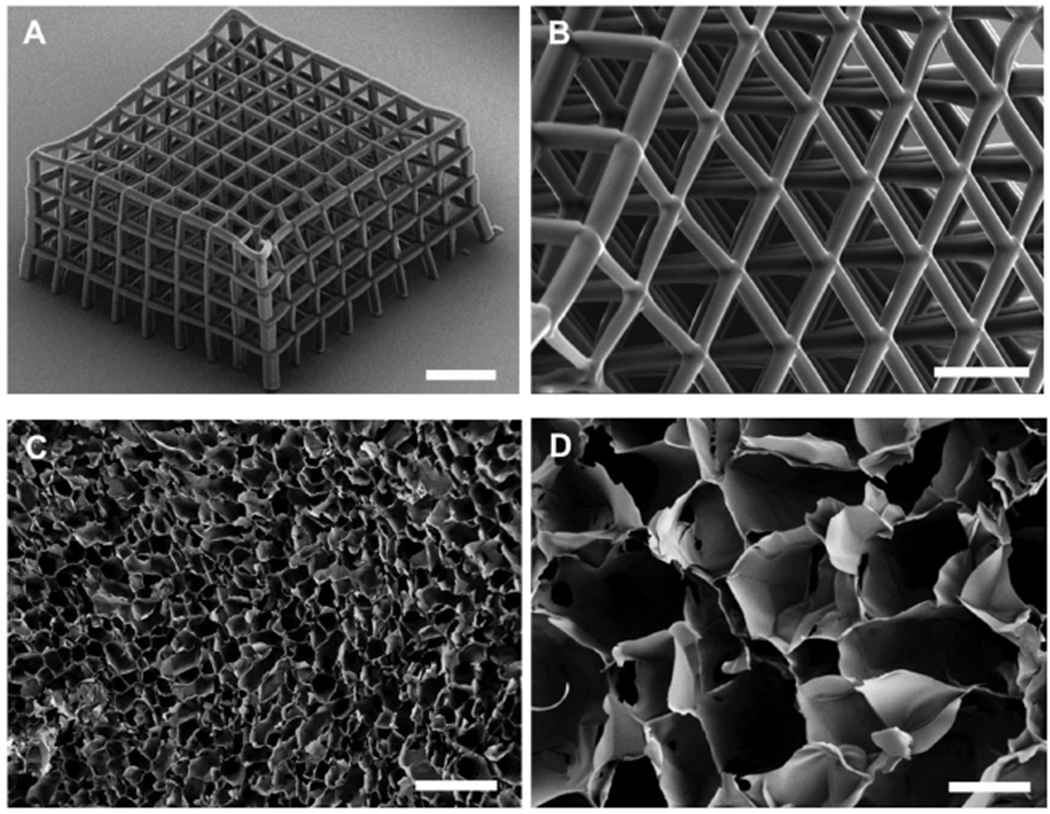
SEM characterization of fabricated scaffolds. A,B) Ormocomp ordered scaffolds fabricated by means of 2-photon patterning. C,D) PEDOT:PSS scaffolds prepared by the ice-templating technique. Scale bars: A) 150 μm, B) 50 μm, C) 400 μm, D) 100 μm, inset D) 200 μm.
Nonordered PEDOT:PSS scaffolds were fabricated, via the ice-templating technique.[25,32] Macroporous scaffolds were prepared following a slightly modified version of the procedure reported in previous studies.[25,32] An aqueous dispersion of PEDOT:PSS was prepared as previously reported[25] (described in Experimental Section), frozen at a specific rate and then the ice crystals were allowed to sublime under controlled conditions giving rise to a highly porous structure. Slices with 400 μm thickness were prepared using a Vibrating Blade Microtome (LEICA VT1200) and subsequently used for seeding human Adipose Derived Stem Cells (hADSC, Lonza). The ice-templating technique creates structures with interconnected pores displaying a broad range of diameters as shown in Figure 1C,D. The obtained cavities are irregular and, as it is possible to appreciate from the sample image in Figure 1D, pores have diameters in the range 100–250 μm, facilitating cell infiltration and media penetration. Cells were seeded taking care that the cell suspension was distributed homogeneously throughout the sample surface.
After cell culture, specimens were first observed via fluorescence microscopy (Figure 2B,E and Figure S1 in the Supporting Information) to preliminarily assert cells adhesion and spreading onto the investigated surfaces, by labeling cytoskeletal components and nuclei. Then, cells on scaffolds were investigated by SEM-FIB microscopy after the UTP procedure (Figure 2A). We tested two resins with different viscosities: EPON and SPURR (see Experimental Section). As a reference, viscosity measurements were run for both EPON and SPURR resins (Figure S2, Supporting Information) 0, 30, and 105 days after preparation. Such measurements were performed at room temperature with a rotational rheometer. Importantly, as reported from the viscosity measurements, these resins experience an increase in their viscosity with time, hindering the removal of the excess material in the final steps of the infiltration. It was thus crucial to proceed with samples preparation starting with freshly prepared resin mixtures. After the UTP embedding procedure, as shown in Figure 2C,D,F,G, no resin excess is present on the specimens such that both the material surface and individual cells are distinguishable.
Figure 2.
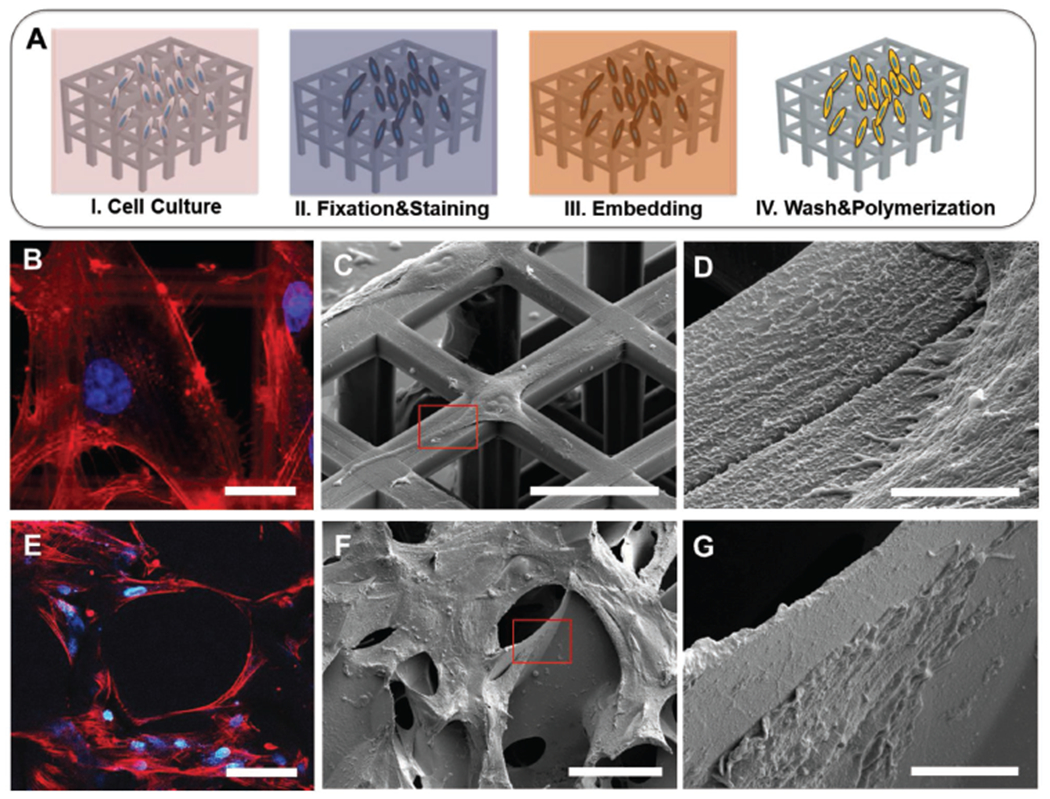
SEM characterization of plasticized cells on scaffolds. A) Schematic of UTP process on scaffolds. B) High magnification U87 glioma cells spreading on ordered scaffold with fluorescence staining of actin (red) and nuclei (blue). C,D) SEM images of plasticized U87 on ordered scaffold. E) hADCs spreading on nonordered scaffolds with fluorescence staining of actin (red) and nuclei (blue). F,G) Plasticized hADCs on nonordered scaffolds. Scale bars: B) 20 μm, C) 40 μm, D) 3 μm, E) 100 μm F) 100 μm, G) 10 μm.
Preliminarily the cell–scaffold interaction can be investigated with fluorescence-based imaging, where features like cytoskeleton architecture (i.e., actin filaments) and nuclei distribution could be visualized (Figure 2B,E) despite the material autofluorescence (Figure S3, Supporting Information). Subsequently, we performed the UTP procedure for EM characterization. Here, we observed in fact cell–material 3D interaction both at the macroscale, visualizing the cell layer conformation on a large ROI (Figure 2C,F) and, then, at higher magnification (micro and nanoscale), selectively imaging cell–material point of contacts (Figure 2D,G). From these images, we were able to easily evaluate how cells colonize and interact with the structural features of the scaffolds.
In particular, it is possible to directly observe cell alignment and stretching on the scaffold surfaces and how cells wrap their plasma membranes around the 3D structures. We were able to observe how cells seeded on macroporous scaffolds penetrate into pores and, in some cases, the presence of ECM as similarly shown in previous reports as this is of great importance for defining the cells fate and in directing cell differentiation.[33] The presence of a very thin layer of resin, as shown in the images, preserves cell integrity at different densities, leading to high quality observation of both sparse cells distributed on ordered scaffolds as well as denser, more complex cells architectures such as those on the nonordered scaffolds. Furthermore, for both scaffold types it is possible to resolve ultrastructures and cellular protrusions such as filopodia, fundamental features involved, for instance, in the formation of focal adhesion complexes (Figure 2D,G).
As mentioned earlier, when in contact with 3D scaffolds, cells show structural conformations which differ from those typically displayed by cells adhering onto 2D surfaces.[34] Moreover, these different types of interactions have been demonstrated to deeply influence cells tensional state and, as a consequence, their behavior.[12,34,35] Depending on the scaffold architecture, cells can partially adhere to the local surface area or even be suspended across the pore bridging two sites of the support material. These conditions reflect high tensional states of the membrane which could often result in cracks when specimens are prepared for EM with hard drying techniques.[16] This aspect limits the possibility to fully characterize cell–material interactions in complex cellular 3D systems and thus, the capability to properly evaluate the effect of the material geometry on cell adhesion processes.
The procedure developed allowed access and characterization of cells positioned in locations inaccessible by other approaches.[16] Here, considering a top view of an ordered scaffold with cells, we were indeed able to analyze three different relative positions of adherent cells in respect to the arms of the ordered scaffold: cells adherent to the wall surface (Figure 3A), cells suspended between two walls (Figure 3C), and cells partially attaching to one arm (Figure 3E). To further characterize the effect of structural features upon cell adhesion, we performed a FIB-based sectioning of both scaffold material and the cell body. Once ROIs were located, these were coated with a platinum layer first by electron beam–assisted deposition and then by ion beam–assisted deposition[28] to reach a final thickness of ≈1 μm (Figure S4, Supporting Information). Subsequently, the material close to the ROI was removed by digging trenches of depth of 10 μm in scaffold areas (nominal, as for silicon) as shown in Figure S5 in the Supporting Information. To further remove redeposited material (Figure S6, Supporting Information) created perpendicularly to the main direction of the square arm, a final polishing was performed by using a low ion beam current (≈0.7 nA–80 pA). The resulting polished sections are shown in Figure 3B,D,F. Remarkably, nuclei and plasma membrane have been resolved (Figure 3G). Moreover, mitochondria and 5–50 nm vesicular invaginating processes (i.e., caveolae) that are known to be regulated by cell-adhesion processes are visualized, resembling inward buddings as previously reported (Figure 3G).[28,30,35]
Figure 3.
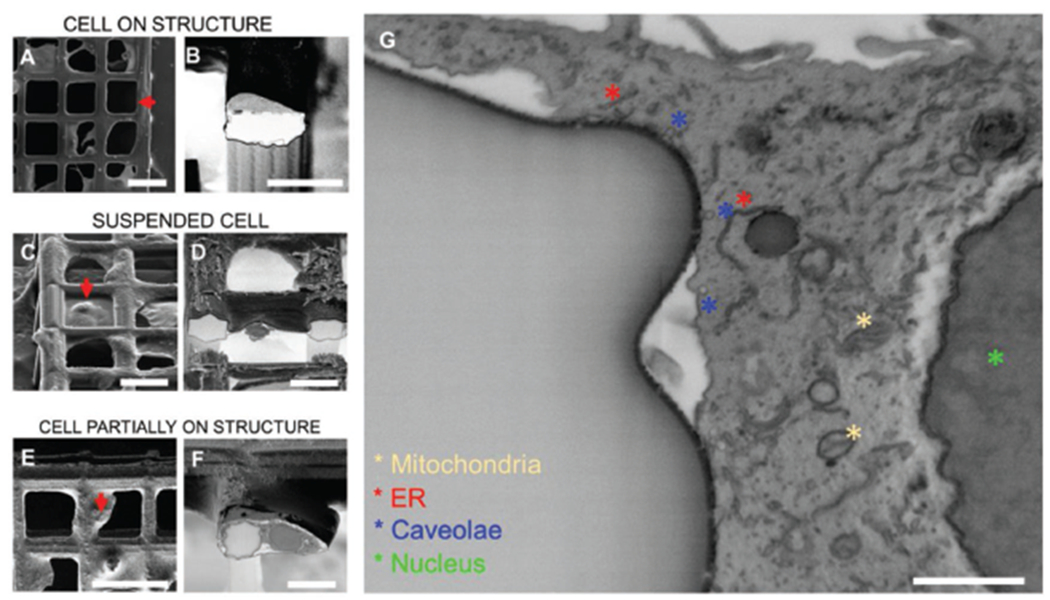
Cross sectioning of cells on ordered scaffolds. A) Top view of a cell growing on top of a square arm; B) 52° tilt view of cross section from position selected in (A); C) 52° tilt view of cell suspended and spreading between two arms; D) 52° tilt view of cross section from position selected in (C); E) top view of a cell partially attaching one arm and spreading over the perpendicular direction; F) 52° tilt view of cross section from position selected in (E); G) zoom-in of cross areas where cell located in (E),(F). (A), (C), (E) are acquired in secondary electrons mode. (B), (D), (F), (G) are acquired in backscattered mode and inverted. Scale bars: A) 50 μm, B) 20 μm, C) 30 μm, D) 20 μm, E) 50 μm, F) 20 μm, G) 1 μm.
Finally, the sectioning procedure has been performed on specimens with cells grown onto nonordered PEDOT:PSS scaffolds. In this case, cells freely colonize the scaffold surface and penetrate in the pores, depositing an abundant layer of ECM (Figure 4), necessary for cell adhesion, survival, cell-to-cell communication, and stability on the culture substrate properties.[33,36] Here, the UTP procedure allows visualization of both cells and ECM. In fact, only few nanometers of polymerized resin are left[27,28] in comparison to cellular and extracellular components which are tens of nanometers in thickness. Compared to ordered scaffolds, here even recent microscopy[37] approaches find major limitations in visualizing the scaffold core because of the material composition and complex morphology. While for ordered scaffolds it is possible to create cross sections following the geometry of the 3D structures and the relative position of cells on it, in the case of nonordered scaffolds the ROI is mainly located on the upmost layer of cells visible via SEM (Figure 4A,B). It is possible to perform nanometer sectioning and clearly observe the interface of multiple cells with the underlying scaffold for the evaluation of the adhesion processes. Interestingly, the approach developed allows for the resolution of details of the ECM deposited by cells (Figure 4C,D), giving also the possibility to easily distinguish different ECM elements like laminin, fibronectin, and collagen as also observed in transmission electron microscopy (TEM) characterizations.[37,38] Additionally, from such images cell spatial rearrangement and cell–cell interactions could also be resolved. In fact, two adjacent cells, as well as their contact with the surrounding ECM are visible (red arrows in Figure 4D). In particular, the brighter areas in Figure 4C,D reveal that residual resin is present in the inner areas, which is advantageous for preserving cell–cell position and the cellular structures as well as ECM components. Furthermore, focusing on the interface contact area, it is possible to completely resolve the plasma membrane approaching the surface area of the PEDOT:PSS cavity (Figure 4C,D).
Figure 4.
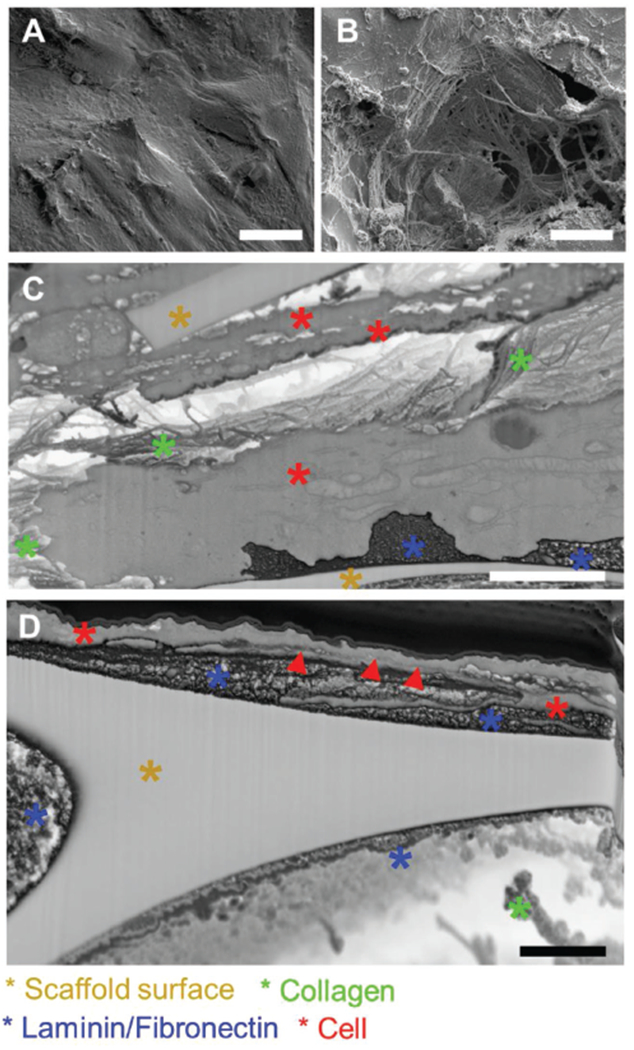
Cross sectioning of cells on nonordered scaffolds. A,B) High density plasticized adipose cells on PEDOT scaffold. C,D) Cross sections revealing cellular and extracellular component at the interface with the scaffold. Scale bars: A) 25 μm, B) 25 μm, C) 2 μm, D) 1 μm.
We have shown two types of scaffolds for tissue engineering fabricated by 2-photon lithography and ice-templating technique. The two fabrication methods lead to two distinct geometries, cage-like and random-distributed cavities, respectively. To investigate the influence of the different 3D scaffold architectures on cell–material interactions, we performed on the same samples, both confocal imaging and UTP, before imaging samples by SEM. Remarkably, the UTP procedure presented here, by limiting the volume of resin remaining in the samples after fixation and infiltration, allows the visualization of both scaffold surface and cells with nanoscale resolution. Moreover, the heavy metal staining allowed the resolution of intracellular components such as nuclei and plasma membrane, vesicles as well as ECM components. The unique possibility to fully characterize the interface both at the micro and nanoscale allows for an accurate evaluation of the effect of the properties of different materials (surface chemistry/geometry/stiffness), on the interaction of 3D scaffolds with the investigated cells. The high versatility of the approach developed allows for the investigation of a broad range of 3D scaffolds that could differ for both structural organization and material composition, performing observations from the tissue to the single cell level. This innovative approach opens the way for a deeper comprehension of the cell–material interaction in 3D environments that can be leveraged to rationally design more efficient new generation of tissue engineering materials and implants.
Supplementary Material
Acknowledgements
D. I. and F. A. P. have equally contributed to this work. F.S. acknowledges the Stanford Nanoshared Facility for the use of the dual beam machine Helios 600i, Michele Dipalo, and Francesco De Angelis for the use of dual beam machine Helios 650. D.I. acknowledges the support from Marie Skłodowska-Curie Individual Fellowship (IF-EF), (Grant Agreement No: 704175).
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Donata Iandolo, Department of Chemical Engineering and Biotechnology, University of Cambridge, UK.
Fabrizio A. Pennacchio, Center for Advanced Biomaterials for Healthcare, Istituto Italiano di Tecnologia, 80125, Italy
Valentina Mollo, Center for Advanced Biomaterials for Healthcare, Istituto Italiano di Tecnologia, 80125, Italy.
Domenico Rossi, Center for Advanced Biomaterials for Healthcare, Istituto Italiano di Tecnologia, 80125, Italy.
David Dannhauser, Center for Advanced Biomaterials for Healthcare, Istituto Italiano di Tecnologia, 80125, Italy.
Bianxiao Cui, Department of Chemistry, Stanford University, CA 94305, USA.
Roisin M. Owens, Department of Chemical Engineering and Biotechnology, University of Cambridge, UK
Francesca Santoro, Center for Advanced Biomaterials for Healthcare, Istituto Italiano di Tecnologia, 80125, Italy.
References
- [1].Langer R, Tirrell DA, Nature 2004, 428, 487. [DOI] [PubMed] [Google Scholar]
- [2].Polikov VS, Tresco PA, Reichert WM, J. Neurosci. Methods 2005, 148, 1. [DOI] [PubMed] [Google Scholar]
- [3].Anderson JM, Annu. Rev. Mater. Res 2001, 31, 81. [Google Scholar]
- [4].Rychly J, Nebe B, Cell Adhes. Migr 2009, 3, 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Khademhosseini A, Langer R, Borenstein J, Vacanti JP, Proc. Natl. Acad. Sci 2006, 103, 2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Albanese A, Tang PS, Chan WC, Annu. Rev. Biomed. Eng 2012, 14, 1. [DOI] [PubMed] [Google Scholar]
- [7].Trappmann B, Chen CS, Curr. Opin. Biotechnol 2013, 24, 948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Discher DE, Janmey P, Wang YL, Science 2005, 310, 1139. [DOI] [PubMed] [Google Scholar]
- [9].Lutolf MP, Hubbell JA, Nat. Biotechnol 2005, 23, 47. [DOI] [PubMed] [Google Scholar]
- [10].Ventre M, Netti PA, ACS Appl. Mater. Interfaces 2016, 8, 14896. [DOI] [PubMed] [Google Scholar]
- [11].Owens RM, Iandolo D, Wittmann KA, MRS Commun. 2017, 7, 287. [Google Scholar]
- [12].Pampaloni F, Reynaud EG, Stelzer EHK, Nat. Rev. Mol. Cell Biol 2007, 8, 839. [DOI] [PubMed] [Google Scholar]
- [13].Nisbet DR, Forsythe JS, Shen W, Finkelstein DI, Horne MK, J. Biomater. Appl 2009, 24, 7. [DOI] [PubMed] [Google Scholar]
- [14].Norman JJ, Desai TA, Ann. Biomed. Eng 2006, 34, 89. [DOI] [PubMed] [Google Scholar]
- [15].Bose S, Roy M, Bandyopadhyay A, Trends Biotechnol. 2012, 30, 546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Accardo A, Blatchè MC, Courson R, Loubinoux I, Thibault C, Malaquin L, Vieu C, Small 2017, 13, 1. [DOI] [PubMed] [Google Scholar]
- [17].Zhou M, Smith AM, Das AK, Hodson NW, Collins RF, Ulijn RV, Gough JE, Biomaterials 2009, 30, 2523. [DOI] [PubMed] [Google Scholar]
- [18].Entcheva E, Bien H, Yin L, Chung CY, Farrel M, Kostov Y, Biomaterials 2004, 25, 5753. [DOI] [PubMed] [Google Scholar]
- [19].Juignet L, Charbonnier B, Dumas V, Bouleftour W, Thomas M, Laurent C, Vico L, Douard N, Marchat D, Malaval L, Acta Biomater. 2017, 53, 536. [DOI] [PubMed] [Google Scholar]
- [20].Wrobel G, Holler M, Ingebrandt S, Dieulweit S, Sommerhage F, Bochem HP, Offenhausser A, J. R. Soc., Interface 2008, 5, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hanson L, Lin ZC, Xie C, Cui Y, Cui B, Nano Lett. 2012, 12, 5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Narayan K, Subramaniam S, Nat. Methods 2015, 12, 1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Denk W, Horstmann H, PLoS Biol. 2004, 2, e329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hoffpauir BK, Pope BA, Spirou GA, Nat. Protoc 2007, 2, 9. [DOI] [PubMed] [Google Scholar]
- [25].Inal S, Hama A, Ferro M, Pitsalidis C, Oziat J, Iandolo D, Pappa AM, Hadida M, Huerta M, Marchat D, Mailley P, Owens RM, Adv. Biosyst 2017, 1. [Google Scholar]
- [26].Xu CY, Inai R, Kotaki M, Ramakrishna S, Biomaterials 2004, 25, 877. [DOI] [PubMed] [Google Scholar]
- [27].Belu A, Schnitker J, Bertazzo S, Neumann E, Mayer D, Offenhausser A, Santoro F, J. Microsc 2016, 263, 78. [DOI] [PubMed] [Google Scholar]
- [28].Santoro F, Zhao W, Joubert LM, Duan L, Schnitker J, van de Burgt Y, Lou HY, Liu B, Salleo A, Cui L, Cui B, ACS Nano 2017, 11, 8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Santoro F, Van de Burgt Y, Keene ST, Cui B, Salleo A, ACS Appl. Mater. Interfaces 2017, 9, 39116. [DOI] [PubMed] [Google Scholar]
- [30].Zhao W, Hanson L, Lou HY, Akamatsu M, Chowdary PD, Santoro F, Marks JR, Grassart A, Drubin DG, Cui Y, Cui B, Nat. Nanotechnol 2017, 12, 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pennacchio FA, Fedele C, De Martino S, Cavalli S, Vecchione R, Netti PA, ACS applied materials & interfaces 2017, 10, 91. [DOI] [PubMed] [Google Scholar]
- [32].Wan AMD, Inal S, Williams T, Wang K, Leieux P, Estevez L, Giannelis EP, Fischbach C, Malliaras GG, Gourdon D, J. Mater. Chem. B 2015, 3, 5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Guneta V, Zhou Z, Tan NS, Sugii S, Wong MTC, Choong C, Biomater. Sci 2018, 6, 168. [DOI] [PubMed] [Google Scholar]
- [34].Eyckmans J, Chen CS, J. Cell Sci 2017, 130, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].von Erlach TC, Bertazzo S, Wozniak MA, Horejs CM, Maynard SA, Attwood S, Robinson BK, Autefage H, Kallepitis C, del RIo Hernandez A, Chen CS, Goldoni S, Stevens MM, Nat. Mater 2018, 17, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Di Maggio N, Piccinini E, Jaworski M, Trumpp A, Wendt DJ, Martin I, Biomaterials 2011, 32, 321. [DOI] [PubMed] [Google Scholar]
- [37].Lu H, Hoshiba T, Kawazoe N, Chen G, Biomaterials 2011, 32, 2489. [DOI] [PubMed] [Google Scholar]
- [38].Engel J, Furthmayr H, Methods Enzymol 1987, 145, 3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


