Abstract
Exosomes are a type of extracellular vesicles (EVs) secreted by almost all cells, with a diameter range of 30–150 nm and a lipid bilayer membrane. Exosomes are now considered as vital mediators of intercellular communication and participate in multiple cellular processes, such as signal transduction and antigen presentation. Recently, circular RNAs (circRNAs), a novel class of noncoding RNAs (ncRNAs), have been found to be abundant and stable in exosomes. Increasing evidence indicates that exosome-derived circRNAs act as signaling molecules to regulate cancer growth, angiogenesis, invasion, metastasis, and sensitivity to chemotherapy. Moreover, circulating exosomal circRNAs can reflect the progression and malignant characteristics of cancer, implying their great potential as promising, non-invasive biomarkers for cancer diagnosis and prognosis. In this review, we summarize the recent progress on the functional roles of exosomal circRNAs in cancer progression, discussing their potential as promising biomarkers and therapeutic targets in cancer. Comprehensive elucidation of molecular mechanisms relevant to the implications of exosomal circRNAs in cancer progression will be conducive to the development of innovative diagnostic and therapeutic approaches in cancer.
Keywords: exosomes, circular RNAs, cancer pathogenesis, non-invasive biomarkers, therapeutic targets
Graphical Abstract
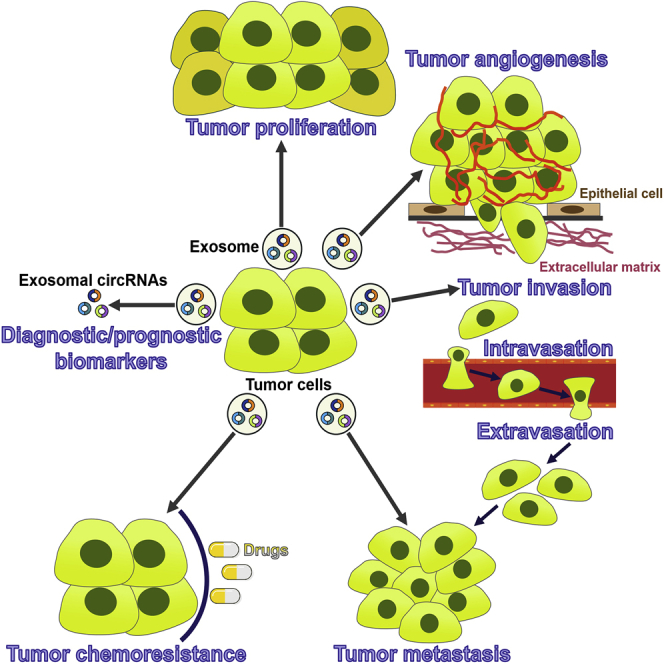
Exosomal circular RNAs (circRNAs) are involved in tumor cell growth, angiogenesis, invasion, metastasis, and chemoresistance. However, many gaps in our understanding of the relationship between exosomal circRNAs and cancer biology exist. This review gathers current knowledge regarding the origination, expression, functions, and clinical implications of exosomal circRNAs in cancer.
Main Text
Exosome biology has received great attention in recent years. Exosomes are lipid bilayer-enclosed, nano-sized extracellular vesicles (EVs) that are released by various types of cells.1,2 These nano-vesicles are intraluminal vesicles (ILVs) derived from the endolysosomal system and released into the extracellular space by fusion of multivesicular bodies (MVBs) with the cellular membrane.3,4 More importantly, exosomes can be readily accessible in most of bodily fluids, including blood, urine, saliva, and breast milk.5,6 Exosomes carry a broad repertoire of constituents derived from the original cells, including lipids, proteins, RNAs, DNAs and noncoding RNAs (ncRNAs).7, 8, 9 These exosomal cargos can be transferred from donor cells to recipient cells.10,11 Thus, exosomes mediate the intercellular exchange of critical information and contents in both physiological and pathological processes. Recently, the role of exosomes in carcinogenesis and cancer progression has been intensively studied. Cancer-derived exosomes can transfer cancer-specific molecules to other cells and induce malignant transformation of target cells.12 In addition, they mediate the communication between cancer cells and their microenvironment, contributing to establishing a fertile soil that supports cancer development.13
Circular RNAs (circRNAs) have been confirmed to be enriched and stable in exosomes and can exert their functions after exosomes reach neighboring or distant cells.14 circRNAs are a novel type of endogenous ncRNAs that exist in all eukaryotic cells and are generated through a specific form of alternative splicing, known as back-splicing.15 Since circRNAs do not have 5′ caps and 3′ polyadenylated tails, they are more stable than linear RNAs.16 However, circRNAs were initially regarded as non-functional byproducts of aberrant RNA splicing.17 With the development of high-throughput RNA sequencing (RNA-seq) technology, circRNAs have been proven to be widely present in eukaryotic cells.15 The discovery of numerous circRNAs transforms them into a hotspot in the ncRNA field. The biological functions of circRNAs have been gradually disclosed. Although some reports have shown that certain circRNAs act as molecular sponges for microRNAs (miRNAs),18, 19, 20 several studies have indicated that some circRNAs do not serve as miRNA sponges.21,22 It seems that miRNA inhibition is not a general feature of circRNAs. Moreover, circRNAs are crucial regulators of alternative splicing, transcriptional events, and post-transcriptional events. circRNAs can work as protein sponges or decoys to indirectly modulate their functions. Notably, circRNAs have been verified to be associated with the occurrence and development of cancer. Guarnerio et al.23 found that the fusion-circRNAs (f-circPR and f-circM9) derived from cancer-associated chromosomal translocations promoted the proliferation and transformation of mouse embryonic fibroblasts (MEFs). Both in vitro and in vivo evidence demonstrated that f-circM9 was oncogenic and favored leukemia progression. In addition, f-circM9 conferred resistance to arsenic trioxide in leukemic cells. circRNAs may be promising biomarkers and therapeutic targets in cancer owing to their high abundance, stability, and conservation.24 Cells can deliver circRNAs by encapsulating them into exosomes. Increasing evidence has indicated that exosomal circRNAs possess a multitude of functions resulting in cancer cell proliferation, invasion, metastasis, and chemoresistance.25, 26, 27 In addition, exosomal circRNAs can be detected in bodily fluids.28 Circulating exosomal circRNAs can reflect the malignant features of cancer. Thus, exosomal circRNAs are likely to be exploited as novel non-invasive biomarkers and prospective targetable factors in cancer. In this review, we summarize the research progression of exosomal circRNAs in cancer pathogenesis, as well as their potential as promising biomarkers and therapeutic targets in cancer. Increasing knowledge of the effects of exosomal circRNAs on cancer biology will be helpful for both revealing molecular mechanisms underlying cancer pathogenesis and further developing diagnostic and therapeutic approaches in cancer. Finally, we also discuss further directions for research into the relationship between exosomal circRNAs and cancer, which require to be addressed to favor the translation of exosomal circRNA-related research into clinical practice.
Classification and Characteristics of EVs
EVs are a heterogeneous family of membrane-bound vesicles shed from almost all cells.29 Cells can release distinct types of EVs that are highly heterogeneous in size, properties, molecular content, biogenetic origin, and biological activity.30 Initially, EVs were considered as cellular debris and a disposal mechanism to discard unwanted materials from cells.31 However, EVs are now understood to act as important vehicles of intercellular communication by shuttling biological information to neighboring or distant cells.32, 33, 34 EVs can be internalized into recipient cells via diverse endocytic mechanisms, including caveolin-mediated, clathrin-dependent, and clathrin-independent endocytosis, as well as by membrane fusion, phagocytosis, micropinocytosis, and lipid raft-mediated internalization.35, 36, 37 Lipids, proteins, and proteoglycans that present on the surface of EVs and recipient cells, as well as changes in environmental stressors, may determine the manner of endocytic uptake of EVs.38, 39, 40 At present, there is no consensus on the classification of EVs due to heterogeneity. Based on their origin and cargo, EVs can now be divided into four broad categories: exosomes, microvesicles (MVs), apoptotic bodies, and oncosomes.41,42 All of these EV subpopulations are involved in intercellular communication and have important roles in immune regulation.43, 44, 45 Note, however, that the classification of EVs into four categories may be oversimplified. New developments on the identification and characterization of different EV subpopulations may be conducive to improving the criteria for classification.
Exosomes are a type of 30- to 150-nm extracellular vehicles secreted by most cells, including immune cells, stem cells, and cancer cells.46 Exosomes are generated by exocytosis of MVBs.47 Exosomes are enriched for endosomal proteins, including tetraspanins (CD9, CD63, and CD81), apoptosis-linked gene-2 interacting protein X (ALIX), and tumor susceptibility gene 101 (TSG101), which are used as exosomal markers.48 Exosomes play an important role in waste disposal and intercellular communication.49 In contrast to exosomes, MVs are large vesicles with a size ranging from 100 to 1,000 nm in diameter.50 They are generated by the outward budding and fission from the plasma membrane.51 MVs carry transmembrane proteins common for the plasma membrane such as integrins and selectins.30 The process of MV biogenesis is not well characterized. The MV biogenesis can be triggered by calcium influx into the parent cells as well as release of intracellular calcium.52,53 This eventually leads to alternations in transbilayer lipid distribution and membrane budding. Reorganization of the actin cytoskeleton through the Ras homolog gene family membrane A (RhoA)-dependent signaling pathway also triggers MV generation.54 Cytoskeleton components (actin and microtubules), molecular motors (kinesins and myosins), and fusion machinery (soluble N-ethylmaleimide-sensitive factor attachment protein receptors [SNAREs] and tethering factors) may be involved in the process of MV formation.55 Similarly to exosomes, the secretion of MVs is also partially dependent on the endosomal sorting complex required for transport (ESCRT) family and requires the formation of lipid-rich microdomains at the plasma membrane.56,57 ADP-ribosylation factor 6 (ARF6) is identified to trigger MV release by remodeling the cytoskeleton.58 Adenosine 5′-triphosphate (ATP)-mediated activation of ionotropic purinergic (P2X) receptors causes the rearrangement of the plasma membrane and is likely to participate in MV shedding.59 The uptake of MVs seems to be an energy-dependent process and can be inhibited at lower temperatures.38,39,60 The well-studied function of MVs is procoagulation owing to their ability to deliver tissue factor, a transembrane molecule that initiates the extrinsic coagulation cascade and thrombus formation.61 Apoptotic bodies are heterogeneous vesicles that are released from cells undergoing programmed cell death.62 Plasma membrane blebbing and nuclear fragmentation during apoptosis lead to the formation of apoptotic bodies.63 Apoptotic bodies are by-products of cell disassembling, with a broad range of diameters (50–5,000 nm), which separates them from other EVs that are mostly produced by normal viable cells.64 Unlike exosomes and MVs, apoptotic bodies contain nuclear fractions and intact organelles.65 Apoptotic bodies can be recognized and engulfed by phagocytes.66 The well-characterized protein markers of apoptotic bodies include thrombospondin, complement component 3b (C3b), annexin V, and histone.67 Rho-associated kinase I (ROCK I) regulates the formation of membrane blebs and re-localization of fragmented DNA into blebs and apoptotic bodies.68 The release of apoptotic bodies can promote the clearance of apoptotic cells.69
Oncosomes are a relatively novel type of EVs and have been defined as oncogenic EVs or exosomes that mediate transport of pro-tumorigenic factors.70 The term oncosome was initially used to describe EVs with a diameter ranging from 100 to 400 nm shed from brain tumor cells.12 These vesicles promoted malignant transformation through the delivery of oncogenic cargos to other cancer cells. Atypically large EVs (1–10 μm in diameter) were reported to emanate from prostate cancer cells in response to activation of oncogenic signals, and their formation correlated with cell migration.42 These gigantic EVs are referred to as large oncosomes due to their unusual size. The formation of large oncosomes is generally observed in aggressive and migratory cancer cells with an amoeboid phenotype.71, 72, 73 Large oncosomes contain abundant bioactive molecules, including enzymes involved in cell metabolism, cytokeratin 18 (CK18), miRNAs, long noncoding RNAs (lncRNAs), metalloproteinases, and caveolin-1.71,73 Similar to MVs, oncosomes and large oncosomes are created by outward budding of the plasma membrane.42,73 The shedding of large oncosomes can be induced by the small guanosine triphosphatase (GTPase) ARF6 or depletion of the cytoskeletal regulator diaphanous-related formin 3 (DIAPH3).71 The release of large oncosomes is also correlated with the abnormal expression of several oncoproteins, including caveolin-1, heparin-binding epidermal growth factor-like growth factor (HB-EGF), and myristoylated protein kinase B (MyrAkt1).42 The formation and secretion of large oncosomes facilitate the migration and invasion of cancer cells.72
EVs encapsulate a variety of molecular constitutes such as proteins, RNAs, and lipids.74 The mechanisms responsible for cargo sorting into EVs are yet to be systematically deciphered. Post-translational modifications may confer specific characteristics to proteins and thus control their sorting into EVs. For instance, ubiquitination or SUMOylation of specific proteins may affect their packaging and secretion.75,76 In contrast, ISGylation regulates exosome release by promoting lysosomal degradation of MVB proteins.77 Alternatively, protein cargos are selectively transported into EVs via specific mechanisms that involve the ESCRT machinery, tetraspanins, and lipids.78, 79, 80 Specific sequences in RNA and certain proteins can dictate RNA sorting into EVs. It has been reported that SUMOylated hnRNPA1 recognizes the GAGAG motif in certain miRNAs and thus selectively targets these miRNAs into EVs.81 The exact mechanism by which lipids are sorted into EVs is still obscure. It is likely that lipid sorting is associated with the yield and size of EVs.82 Collectively, the incorporation of various constituents into EVs is a highly regulated process. Further studies are required to elucidate the complex mechanisms that control cargo sorting into EVs.
Exosome Biogenesis and Uptake
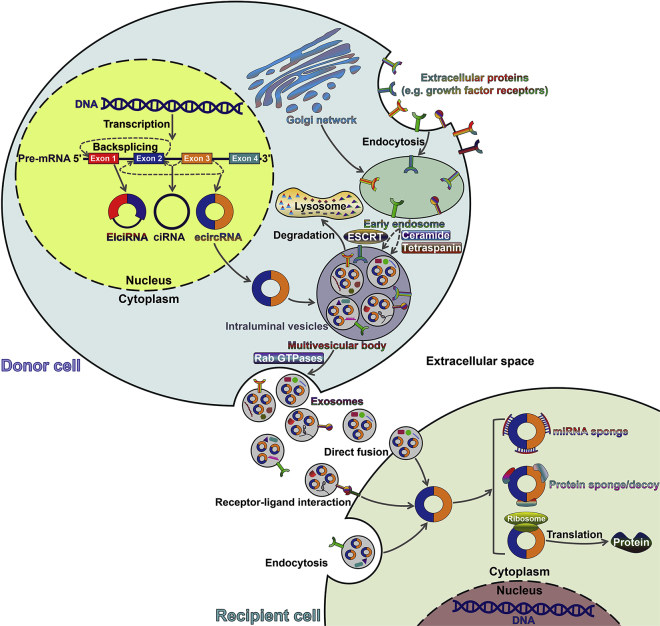
Exosomes are composed of a lipid bilayer membrane enclosing various constituents of the parent cells. They are present in nearly all bodily fluids, such as urine, blood, saliva, and cerebrospinal fluid.83, 84, 85, 86 Exosomes play an important role in intercellular communication. Exosomes are generated by the endocytic pathway. The biogenesis of exosomes initiates when the cellular membrane invaginates to form an early endosome (Figure 1).31 During early endosome maturation, ILVs are formed via inward budding of the endosomal membrane, which results in the formation of MVBs or late endosomes.87,88 During this process, portions of the cytosolic contents are engulfed within the ILVs, while specific proteins are incorporated into the invaginated membranes.65,89 MVBs can be trafficked to lysosomes, where their contents undergo degradation.90 Alternatively, MVBs can fuse with the cellular membrane, releasing ILVs into the extracellular space.3,4 The released ILVs are referred to as exosomes.
Figure 1.
Formation and Delivery of Exosomal circRNAs
circRNAs are formed through the back-splicing of pre-mRNAs. circRNAs are commonly divided into three types according to their component. EIciRNAs are circularized with both exons and introns. ciRNAs are derived from introns. ecircRNAs are composed of exons only. ecircRNAs generally locate in the cytoplasm, while EIciRNA and ciRNAs mainly accumulate in the nucleus to promote transcription of their parent genes. Exosomes are nano-vesicles of endocytic origin. Exosome biogenesis begins with the formation of an early endosome from the cellular plasma. With the assistance of the Golgi network, early endosomes mature into late endosomes. Intraluminal vesicles (ILVs) accumulate within late endosomes by inward budding of the endosomal membrane, leading to the formation of a multivesicular body (MVB). During this process, a variety of proteins (e.g., receptors and tetraspanins), lipids (e.g., ceramide and cholesterol), and ncRNAs (e.g., miRNAs, lncRNAs, and circRNAs) are packed into ILVs. An MVB can fuse with the cellular membrane to release its ILVs as exosomes into the extracellular milieu. Exosome biogenesis involves both ESCRT-dependent and ESCRT-independent pathways. MVB trafficking and fusion with the cellular membrane can be coordinated by Rab GTPases. Alternatively, an MVB can fuse with the lysosome for degradation. The released exosomes can be delivered into recipient cells through three mechanisms, that is, direct membrane fusion, receptor-ligand interaction, and endocytosis. Exosomes shuttle bioactive contents including circRNAs from donor cells to recipient cells. In recipient cells, exosomal circRNAs execute crucial biological functions by acting as sponges or decoys to sequester miRNAs/proteins. Exosomal circRNAs may also have translation potential. EIciRNA, exon-intron circRNA; ciRNA, circular intronic RNA; ecircRNA, exonic circRNA; ESCRT, endosomal sorting complex required for transport; GTPase, guanosine triphosphatase.
The ESCRT pathway plays a vital role in exosome biogenesis. The ESCRT machinery consists of four multi-protein complexes, namely ESCRT-0, -I, -II, and -III. These ESCRTs work cooperatively to favor MVB formation, vesicle budding, and cargo assortment. ESCRT-0 is responsible for recognizing and sequestering ubiquitinated proteins in the late endosome membrane. ESCRT-0 then recruits ESCRT-1 to the endosomal membrane, which in turn triggers assembly of the ESCRT-II/-III complex.91,92 Both ESCRT-I and ESCRT-II initiate the budding of MVBs and propel the enzymatic de-ubiquitination of cargo proteins before ILV formation. After that, ESCRT-III and vacuolar protein sorting protein 4 (VPS4) drive the scission of the membrane buds to form ILVs. VPS4 is also involved in the dissociation and recycling of the ESCRT machinery. The implication of syndecan and its cytoplasmic adaptor syntenin in ESCRT-dependent exosome generation has been identified. The association of syndecans with syntenin triggers its interaction with CD63 and ALIX, leading to intraluminal budding of the endosomal membrane.93 Interestingly, exosome formation can also occur in the absence of the ESCRT machinery, suggesting the existence of ESCRT-independent mechanisms.94 It has been reported that exosome biogenesis can be induced by the production of ceramide, rather than the ESCRT pathway.95 Neutral sphingomyelinase 2 (nSMase2) that prompts ceramide synthesis is capable of regulating exosome biogenesis and release.96 Exosomal tetraspanin proteins also play an important role in selective sorting of biomolecules. For instance, CD63 is highly present in exosomes and mediates the sorting of premelanosome protein (PMEL) into ILVs.97 CD81 is involved in cargo sorting of tetraspanin ligands, such as Rac GTPase.98 As stated above, numerous molecules are implicated in exosome biogenesis. The detailed mechanisms await more intensive investigation. The effects of internal and external stimuli on exosome biogenesis and release need to be further explored.
The contributory factors that determine MVB fate remain to be identified. Type I interferon (IFN-I) can block exosome release by inducing protein ISGylation of the MVB protein TSG101.77,99 Accordingly, MVB protein ISGylation contributes to the fusion of MVBs with the lysosome. The transport of MVBs to the cellular membrane requires molecular and cytoskeletal motors. Moreover, the release of exosomes involves various proteins, including coat protein complex (COP) I and II, SNAREs, and GTPases.55 Moreover, the Rab GTPase family is implicated in membrane trafficking regulation, dominating vesicle budding, vesicle transport, and membrane fusion.100 For example, Rab35 mediates MVB docking to the cellular membrane in neuralgia cells.101 Rab27a and Rab27b participate in the recruitment of MVBs to the cellular membrane.102 In addition, the tumor suppressor p53 and its downstream effector, tumor suppressor-activated pathway 6 (TSAP6), are able to increase exosome release.103 Upon release from parent cells, exosomes remain aggregated and can be associated with the cellular membrane by tetherin.104 Exosome can be taken up by recipient cells via three mechanisms, that is, direct membrane fusion, receptor-ligand interaction, and endocytosis. Specific exosomal contents derived from donor cells may determine their affinity with certain types of cells. Substantial investigations are demanded to figure out how exosomes are directed to target cells.
Functional Roles of Exosomes
Exosomes released from parent cells were originally proposed as a mechanism through which cells expel unwanted or unnecessary cellular components. Nevertheless, during the past decade, exosomes have been proven to play an important role in the exchange of materials between cells. Exosomes can reprogram the recipient cells they encounter though their bioactive constituents. Exosomes can exert their functions as vehicles of both physiological and pathological messengers. The biological functions of exosomes from different cell types vary and mainly depend on their contents.
Exosomes have been reported to promote cell proliferation and tissue regeneration. Stem cells are characterized by secretion of exosomes with such function.105 Mesenchymal stem cell (MSC)-derived exosomes could induce fibroblast proliferation.106 MSC-derived exosomes induced cardiac tissue growth and regeneration.107 The regenerative property of stem cell-derived exosomes will provide new therapeutic options for tissue repair and regeneration. Normal cell-derived exosomes also induce tissue regeneration. For instance, exosomes released by hepatocytes could deliver sphingosine kinase 2 (SK2) to form sphingosine-1-phosphate (S1P) within target hepatocytes, thus contributing to hepatocyte proliferation and liver regeneration.108 Currently, the biological function of exosomes in tissue regeneration has not been adequately disclosed. It is necessary to elucidate which components in exosomes can promote tissue repair and regeneration.
The role of exosomes as immune regulators has been defined. Exosomes serve an important role in orchestrating innate immune responses through different pathways. Exosomes possess pro-inflammatory activities in the innate immune system. Exosomes secreted by mycobacteria-infected macrophages could transfer bacterial components to uninfected macrophages, thereby initiating a pro-inflammatory response.109 Tumor cells exposed to stress released exosomes expressing heat shock protein 70 (HSP70).110,111 These HSP70-positive exosomes could drive natural killer (NK) cell activation and pro-inflammatory cytokine production by macrophages. Dendritic cell (DC)-derived exosomes harbored various tumor necrosis factor (TNF) superfamily members (Fas ligand [FasL], TNF, and TNF-related apoptosis-inducing ligand [TRAIL]) on their surface, which directly activated NK cells to reinforce their cytotoxic activity.112 Interestingly, exosomes secreted by immunocytes can directly exert effector functions. NK cell-secreted exosomes were reported to cause the lysis and elimination of tumor cells.113
Alternatively, immunocyte exosomes play a regulatory role in adaptive immune responses. Exosomes can present antigenic peptides in conjunction with major histocompatibility complex (MHC) molecules to T cells.114 Accordingly, exosomes serve as immunostimulatory factors in T cell responses. Exosomes secreted by almost all cells harbor MHC class I molecules that are capable of activating CD8+ T cells.115 DC-derived exosomes could transport the peptide-MHC class I complex and co-stimulatory molecule CD80 to non-specific CD4+ T cells, which in turn activated naive/antigen-specific CD8+ cytotoxic T lymphocytes (CTLs).116 Exosomes secreted by activated human peripheral CD3+ T cells promoted cytokine production and proliferation of CD8+ T cells.117 Human B cell-derived exosomes directly provided MHC class II complexes required for activation of CD4+ T cells.118 On the contrary, exosomes exert suppressive roles in T cell activation. CD4+CD25+FOXP3+ T cells secreted exosomes bearing the anti-inflammatory mediator CD73, thus suppressing the proliferation of recipient CD4+ T cells.119 Exosomes derived from activated CD4+ T cells inhibited CD4+ T cell proliferation and CD8+ CTL responses.120 CD4+ T cells and human B cell-derived lymphoblastoid cell lines (LCLs) secreted FasL-positive exosomes that induced apoptosis in recipient T cells.121, 122, 123 In addition, CD8+ T cell-mediated antitumor responses were suppressed by exosomes derived from CD8+CD25+ regulatory T cells (Tregs).124 It can be concluded that exosomes play dual roles in T cell responses. The function of exosomes in immune regulation is worthy of further exploration. It is of great importance in illuminating composition differences between immunostimulatory and immunosuppressive exosomes.
Apart from their essential roles in physiological conditions, exosomes also play a critical role in disease progression, especially in cancer pathogenesis.125 The Rab GTPase family members are constitutively active or even overexpressed in cancer cells.126,127 Consequently, cancer cells release more exosomes into the extracellular milieu than do normal cells.128 Owing to the identification of unique contents within cancer-derived exosomes, the roles of exosomes in cancer progression have come into the spotlight. Cancer-derived exosomes function as significant mediators in the interaction and communication between cancer cells and other cells in the tumor microenvironment.129 Particularly, cancer-released exosomes can enter the circulation to function at distant sites, hence propelling cancer development. Studies have demonstrated that exosomes play a pivotal role in cancer growth, invasion, angiogenesis, and metastasis by delivering oncogenic molecules.130,131 Cancer-derived exosomes help cancer cells to evade immune attack by impairing the function of effector T cells or inducing the apoptosis of activated T cells.132,133 Cancer-secreted exosomes induce vascular leakiness to create a pre-metastatic niche.134 They remodel the extracellular matrix (ECM) to support tumor growth.135,136 In addition, cancer-derived exosomes function to discharge chemotherapeutic agents, thereby conferring chemoresistance in cancer cells.137 Exosomal cargos also compete with anticancer drugs to interact with the therapeutic targets.138 Intriguingly, cancer cells shed exosomes to transmit their chemoresistant phenotype to recipient cancer cells.139 Cancer cells can secrete their exosomes into the circulation, resulting in increased concentrations of exosomes in cancer patients relative to healthy individuals.140 The amount and composition of exosomal cargos vary between cancer patients and healthy controls. In a previous study, eight specific miRNAs (miR-21, miR-141, miR-200a, miR-200b, miR-200c, miR-203, miR-205, and miR-214) were identified in circulating exosomes from patients with ovarian cancer (OC), but they could not be detected in circulating exosomes from normal controls.140 Another study showed that the mean concentration of exosomal miRNAs was significantly higher in patients with lung adenocarcinoma (LUAD) than in healthy controls.141 Importantly, the miRNA signatures of circulating exosomes paralleled those of tumor-derived exosomes. The miRNA profiling of exosomes might be used as a screening tool for cancer diagnosis. Moreover, significant differences were also observed between the protein concentration of circulating exosomes for the LUAD group and the control group.141 Likewise, the fraction of circulating exosomes in patients with colorectal cancer (CRC) was significantly higher than in healthy controls.142 Notably, high levels of circulating exosomes correlated with poor prognosis parameters and short overall survival in CRC patients. Thus, exosomes may represent promising prognostic biomarkers in cancer. Furthermore, it has been proposed that exosomes can act as an efficient delivery platform for targeted transfer of anticancer drugs to cancer cells. Saari et al.143 revealed that prostate cancer cell-derived exosomes were capable of delivering paclitaxel into their parental cells via an endocytic pathway, leading to release of the drug into the cell cytosol. Remarkably, exosome-mediated delivery could enhance the cytotoxic effects of paclitaxel. Recently, tumor exosome-sheathed doxorubicin (DOX)-loaded nanoparticles (DOX@E-PSiNPs) were developed as a drug carrier for targeted cancer chemotherapy.144 DOX@E-PSiNPs exhibited enhanced tumor accumulation, tumor penetration, and cross-reactive cellular uptake by cancer cells and cancer stem cells (CSCs). These features endowed DOX@E-PSiNPs with augmented in vivo DOX enrichment in total tumor cells and side population cells with characteristics of CSCs, thus resulting in strong anticancer activity and CSC reduction in tumor-bearing mice models. Given the great potential of exosomes as non-invasive biomarkers and therapeutic nano-sized carriers, regulation of exosome biogenesis, modification of exosomal composition, and improvement of cell-targeting specificity may represent hopeful means for clinical cancer treatment.
Exosome-Enclosed Substances
The molecular constituents in exosomes consist of lipids, proteins, and nucleic acids. The components of exosomes can be different from parent cells owing to the selective incorporation of cargos into exosomes. Exosomes released by the same cell type can contain different constituents. The composition of exosomes may vary depending on whether the donor cells are exposed to distinct stimuli or stressors.145, 146, 147 Harmati et al.148 compared the miRNA content of exosomes secreted by nasopharyngeal carcinoma (NPC) cells cultured under normal conditions and in the presence of DOX. They found that the diversity of exosomal miRNAs was increased in DOX-treated NPC cells compared with the control. de Jong et al.149 reported that exposure of endothelial cells to hypoxia caused the change of both mRNA and protein compositions in their exosomes. The lipid compositions of exosomes are derived from those of the plasma membrane, such as sphingomyelin, cholesterol, phosphatidylserine, and hexosylceramide.7 The exosomes contain cytosolic, endosomal, plasma, and nuclear proteins. Proteins enriched in exosomes are composed of diverse HSPs, cytoskeletal proteins, endosome-related proteins (annexin, flotillin, and SNARE), as well as the components of the ESCRT machinery (ESCRT complexes, ALIX, and TSG101). Exosomes also incorporate various cell surface molecules that render them capable of attaching to different cell receptors.150 Molecules associated with antigen presentation, such as CD1, MHC class I, and MHC class II, are found to be highly abundant in exosomes.151 Exosomes also harbor adhesion molecules, tetraspanin proteins (CD9, CD63, CD81, and CD82), and co-stimulatory molecules. These compositions endow exosomes with the immunoregulatory capability. Multiple exosomal protein components appear to mirror the exosome biogenesis pathway, while other protein compositions may be abundant in exosomes as a consequence of their increased levels in parent cells.
Emerging studies have verified that exosomes are rich in multiple RNA species, such as mRNAs, miRNAs, and lncRNAs.10,152 Recently, circRNAs have been proven to be highly present in exosomes.14 These findings open up a new avenue of research on the biological functions of exosomes. The RNA profiles of exosomes differ from those of donor cells. Exosomes of immune cells and cancer cells had unique miRNA signatures that were not merely a reflection of the miRNA contents of the parental cells.8,153 Exosomal RNAs are biologically functional and can influence the transcriptome of target cells. Valadi et al.10 compared the mRNA and miRNA expression profiles between mouse mast cells and their exosomes. A total of 270 mRNA transcripts could only be detected in mast cell-derived exosomes, but they were not detectable in the parental cells. Approximately 121 miRNAs were identified in mast cell-derived exosomes. Moreover, there were significant differences in the expression of mRNAs and miRNAs from exosomes versus their parental cells. Intriguingly, the mRNA transcripts from mouse mast cell-derived exosomes were transferable to human mast cells. The transferred exosomal mRNAs could be translated after entering the target cells and thus affected their biological function. Farahani et al.154 characterized miRNA transcripts from chronic lymphocytic leukemia (CLL) and their exosomes. The results indicated that a variety of miRNAs, including miR-1290, miR-202-3p, and miR-628-3p, were differentially expressed in CLL-derived exosomes versus donor cells. CLL-derived exosomes could be taken up by stromal cells, resulting in altered expression of multiple genes in recipient cells. In addition, stromal cells incubated with CLL-derived exosomes displayed enhanced proliferation compared with control cells. Thus, CLL-derived exosomes could affect the transcriptome and behavior of recipient stromal cells. Preferential sorting of particular RNA species occurs within exosomes, implying that cells possess sorting mechanisms for exosomal RNAs.155 More studies should be conducted to reveal the sorting mechanisms for exosomal RNA species.
Origin of Exosomal circRNAs
Li et al.14 first revealed the presence and enrichment of circRNAs within cancer-derived exosomes by RNA-seq analysis. Remarkably, cancer-derived exosomal circRNAs were able to enter the circulation. The circulating exosomal circRNAs could differentiate patients with colon cancer from healthy individuals. CDR1as circRNA is known to serve as a miR-7 sponge.19 The sorting of CDR1as to exosomes seemed to be affected, at least in part, by changes of miR-7 levels in parental cells.14 Nevertheless, it is unclear whether the sorting of other exosomal circRNAs is controlled by altered levels of their associated miRNAs. Further studies are needed to confirm and expand these emerging findings. Exosomal circRNAs can be transported to recipient cells to exert their biological functions. For instance, CDR1as-overexpressing exosomes could abrogate the suppressive effect of miR-7 on recipient cell proliferation.14 circNFIX was transferred by incorporation into exosomes, and exosomal circNFIX from temozolomide-resistant glioma cells conferred drug resistance to recipient-sensitive cells through the enhancement of cell migration and invasion and the suppression of cell apoptosis.27 Another study also showed that colon cancer cell-derived circRNAs could be packaged into exosomes.156 Moreover, circRNAs might be enriched in colon cancer cell-secreted exosomes. It was reported that 1,147 and 1,385 exosomal circRNAs were deregulated in patients with metastatic and localized breast cancer (BCa) in comparison with healthy controls (Table 1).157 Meanwhile, 480 exosomal circRNAs were found to be differentially expressed in metastatic BCa patients compared with patients with localized disease. The expression profile of exosomal circRNAs in patients with endometrial cancer was also explored.158 As a result, 209 upregulated and 66 downregulated circRNAs were found in serum exosomes from patients with endometrial cancer compared with those from normal controls. These deregulated circRNAs might be involved in signaling pathways that were associated with cancer migration and invasion. A total of 453 differentially expressed circRNAs, consisting of 274 upregulated and 179 downregulated circRNAs, were discovered in exosomes from the plasma of patients with pancreatic ductal adenocarcinoma (PDAC) relative to healthy volunteers.159 It was proposed that these exosomal circRNAs might regulate PDAC pathogenesis by targeting miRNAs in recipient cells. Recently, 182 exosomal circRNAs were reported to be differentially expressed in the plasma from LUAD patients compared with healthy controls.160 These circulating exosomal circRNAs held great promise as non-invasive biomarkers in lung cancer. The levels of 22 circRNAs were altered in serum exosomes from patients with papillary thyroid carcinoma relative to healthy controls by high-throughput sequencing analysis.161 These circRNAs might be associated with various signaling pathways, such as the thyroid hormone signaling cascade and the phosphoinositide 3-kinase (PI3K)/Akt pathway. Further study on the expression profile of exosomal circRNAs in cancer would contribute to elucidating the detailed mechanisms underlying carcinogenesis and cancer progression.
Table 1.
Deregulated Expression of Exosomal circRNAs in Cancer Patients
| Cancer Type | Source of Exosomes | Scope | No. of Deregulated circRNAs |
References | |
|---|---|---|---|---|---|
| Upregulated | Downregulated | ||||
| Breast cancer | serum | metastatic cancer versus normal | 1,061 | 86 | 157 |
| localized cancer versus normal | 1,084 | 301 | |||
| metastatic versus localized | 369 | 111 | |||
| Endometrial cancer | serum | cancer versus normal | 209 | 66 | 158 |
| Pancreatic ductal adenocarcinoma | plasma | cancer versus normal | 274 | 179 | 159 |
| Lung adenocarcinoma | plasma | cancer versus normal | 105 | 77 | 160 |
| Papillary thyroid carcinoma | serum | cancer versus normal | 3 | 19 | 161 |
In addition to cancer cells, exosomal circRNAs also originate from activated human platelets.162 circRNAs could be selectively packaged and released within platelet-derived exosomes. The association between exosomal circRNAs and the nervous system was previously verified. circRNAs were shown to be differentially expressed in exosomes from mice brain extracellular space following traumatic brain injury.163 Intriguingly, these circRNAs might be involved in the development of the nervous system and the transmission of nerve signals. Exosomal circRNAs were present in cerebrospinal fluid from patients with immune-mediated demyelinating disease.164 The deregulation of circRNAs might be linked with the onset and progression of this disease. Additionally, adipocytes are able to secret exosomal circRNAs. A single-exon circRNA, hsa_circ_0075932, was highly expressed in human normal adipose tissue.165 hsa_circ_0075932 exhibited a markedly promoting effect on inflammation and apoptosis in dermal keratinocytes. circ-DB released by adipocytes was found to prompt cell growth and reduce DNA damage in hepatocellular carcinoma (HCC) by targeting miR-34a and activating the ubiquitin-specific protease 7 (USP7)/cyclin A2 signaling pathway.25 Many gaps in our current understanding of the relationship between circRNAs and exosome remain. The molecular mechanisms that control the sorting process of exosomal circRNAs remain elusive. In-depth investigations on circRNAs and their associated miRNAs are required to verify the impact of miRNA abundance on circRNA sorting into exosomes. Intriguingly, numerous RNA-binding proteins (RBPs), including ribosomal proteins and elongation factors, were identified to be present in exosomes.166 Several RBPs are responsible for sorting RNAs with specific binding motifs into exosomes.155,167 It is proposed that RBPs may direct RNA sorting by interacting with MVB and/or membrane microdomains where the exosomes form.168,169 Given that circRNAs can interact with RBPs,170,171 it can be hypothesized that RBPs may regulate the process of circRNA sorting. However, more efforts should be made to identify RBPs that are involved in exosomal assortment of circRNAs. The intricate mechanisms by which circRNAs are selectively sorted into exosomes warrant further elucidation. Furthermore, it is essential to reveal the biological functions of exosomal circRNAs under both physiological and pathological conditions. Additional work is required to figure out whether the selective package of exosomal circRNAs is a consequence of preservation of cellular homeostasis or disease progression.
Emerging Roles of Exosomal circRNAs in Cancer
The aberrant expression of exosomal circRNAs has been identified in various types of cancer, including gastric cancer (GC), CRC, pancreatic cancer (PC), HCC, cholangiocarcinoma (CCA), small-cell lung cancer (SCLC), and urogenital system tumor. Emerging studies through gain- and loss-of-function strategies suggest that exosomal circRNAs are involved in the progression of these cancers (Table 2). Exosomal circRNAs may also represent promising biomarkers and therapeutic targets in cancer. Nonetheless, the study of the role of exosomal circRNAs in cancer is still on the way. The mechanisms by which exosomal circRNAs function in cancer warrant further investigation.
Table 2.
Expression and Function of Exosomal circRNAs in Different Cancers
| Exosomal circRNAs | Cancer Type | Expression | Functions | References |
|---|---|---|---|---|
| ciRS-133 | gastric cancer | upregulated | promotes cancer-associated cachexia | 172 |
| circNRIP1 | gastric cancer | upregulated | prompts EMT and metastasis | 173 |
| circ-RanGAP1 | gastric cancer | upregulated | potential prognostic biomarker | 174 |
| hsa_circ_0000419 | gastric cancer | downregulated | diagnostic/prognostic biomarker | 175 |
| hsa_circ_0065149 | gastric cancer | downregulated | diagnostic biomarker | 176 |
| circ-KIAA1244 | gastric cancer | downregulated | potential prognostic biomarker | 177 |
| ciRS-122 | colorectal cancer | upregulated | confers chemoresistance | 178 |
| hsa_circ_0000338 | colorectal cancer | upregulated | potential indicator of chemotherapy response | 179 |
| hsa_circ_0004771 | colorectal cancer | upregulated | diagnostic biomarker | 180 |
| circ-IARS | pancreatic cancer | upregulated | facilitates cell invasion and metastasis | 181 |
| circ-PDE8A | pancreatic ductal adenocarcinoma | upregulated | potential diagnostic/prognostic biomarker | 182 |
| circRNA_100284 | hepatocellular carcinoma | upregulated | accelerates cell cycle and facilitates cell proliferation | 183 |
| hsa_circ_0051443 | hepatocellular carcinoma | downregulated | induces cell cycle arrest and promotes cell apoptosis | 184 |
| circPTGR1 | hepatocellular carcinoma | upregulated | fosters cell metastasis | 185 |
| circRNA-100338 | hepatocellular carcinoma | upregulated | promotes cell angiogenesis and metastasis | 26 |
| circ-0000284 | cholangiocarcinoma | upregulated | boosts cell proliferation and migration | 187 |
| FECR1 | small-cell lung cancer | upregulated | potential prognostic biomarker | 188 |
| circWHSC1 | ovarian cancer | upregulated | enhances cell metastasis | 189 |
| circ_0044516 | prostate cancer | upregulated | potential diagnostic/prognostic biomarker | 190 |
| circPRMT5 | urothelial carcinoma of the bladder | upregulated | potential prognostic biomarker | 191 |
GC
circRNAs that play a regulatory role in GC progression have been found to be present in GC-derived exosomes. circRNA sponge for miR-133 (ciRS-133) was upregulated in exosomes isolated from the plasma of GC patients.172 GC cells could deliver ciRS-133 into preadipocytes via exosomes. Exosomal ciRS-133 facilitated the differentiation of preadipocytes into brown-like cells by inhibiting miR-133 and activating PR domain containing protein 16 (PRDM16). Depletion of ciRS-133 decreased cancer cachexia in tumor-implanted mice, reducing oxygen consumption and heat production. Exosomal ciRS-133 might be involved in cancer-associated cachexia. circNRIP1 was abundantly expressed in GC cells, and it prompted the proliferation, migration, and invasion of GC cells via activating the Akt1/mammalian target of rapamycin (mTOR) axis by sponging miR-149-5p.173 circNRIP1 could be transmitted between GC cells via exosomes, and exosomal circNRIP1 fostered epithelial-mesenchymal transition (EMT) and tumor metastasis in vivo. The impact of exosomal circNRIP1 on the Akt1/mTOR signaling cascade in recipient cells should be explored. The molecular mechanism associated with the role of exosomal circNRIP1 in GC remains to be further elucidated. Similarly, circ-RanGAP1 was remarkably upregulated in GC tissues and plasma exosomes of GC patients.174 The level of exosomal circ-RanGAP1 was higher in preoperative GC patients than in postoperative patients or healthy controls. Exosomal circ-RanGAP1 might be used as a prognostic biomarker for GC (Figure 2). circ-RanGAP1 promoted GC cell invasion and metastasis by sponging miR-877-3p to upregulate vascular endothelial growth factor A (VEGFA). The plasma exosomes from GC patients could promote the migration and invasion of GC cells. Nevertheless, it is uncertain whether circ-RanGAP1 within GC-derived exosomes contributed to their effect on the biological behaviors of recipient GC cells. Further experimental studies should be carried out to validate the functional role of exosomal circ-RanGAP1 in GC progression.
Figure 2.
The Clinical Value of Exosomal circRNAs in Gastric Cancer
Several circRNAs (circ-RanGAP1, hsa_circ_0000419, hsa_circ_0065149, and circ-KIAA1244) have been found to be deregulated in plasma exosomes of GC patients. These exosomal circRNAs may be used as non-invasive biomarkers for GC diagnosis and prognosis. GC, gastric cancer.
The diagnostic and prognostic values of exosomal circRNAs in GC were explored in previous reports. For instance, the expression of hsa_circ_0000419 was dramatically decreased in GC cells, cancer tissues, and plasma from GC patients compared with normal controls.175 Plasma hsa_circ_0000419 was closely associated with tumor stage, invasion, and metastasis. Plasma hsa_circ_0000419 also showed a good diagnostic performance for GC. There was no significant difference between the expression level of hsa_circ_0000419 in plasma and corresponding plasma exosomes. To summarize, hsa_circ_0000419 might act as a new biomarker for GC diagnosis and prognosis. The clinical values of exosomal hsa_circ_0000419 are worthy of further validation. The expression of hsa_circ_0065149 was significantly downregulated in plasma exosomes of early GC patients compared with healthy controls.176 Exosomal hsa_circ_0065149 showed higher sensitivity and specificity in early GC screening than did traditional clinical biomarkers. Therefore, hsa_circ_0065149 in plasma exosomes might serve as a useful indicator for early diagnosis of GC. The level of circ-KIAA1244 was decreased in GC tissues, plasma, and cells relative to controls.177 The downregulation of circ-KIAA1244 in plasma was negatively associated with tumor-node-metastasis (TNM) stage, lymphatic metastasis, and overall survival of GC patients. Plasma circ-KIAA1244 might release in the form of exosomes and thus stably existed in plasma protected by exosomes. GC-secreted circ-KIAA1244 might represent an exploitable circulating marker for GC screening. Although these reports have suggested that exosomal circRNAs have great potential as diagnostic and prognostic biomarkers for cancer, more clinical studies are required to confirm these experimental results. Compared with other ncRNAs that exhibit the potential to function as cancer biomarkers, circRNAs have more superior characteristics, including high stability and conservation. Therefore, exosomal circRNAs may be more suitable for use as prospective cancer biomarkers. Furthermore, the diagnostic efficacy of the combination of exosomal circRNAs and traditional biomarkers should be investigated.
CRC
Exosomal circRNAs may serve as intercellular signaling molecules to transmit chemoresistance from drug-resistant CRC cells to sensitive ones. Recently, Wang et al.178 revealed that exosomes from oxaliplatin-resistant CRC cells delivered ciRS-122 to sensitive cells. In vitro and in vivo studies showed that the transferred ciRS-122 accelerated glycolysis and enhanced resistance to oxaliplatin in sensitive CRC cells by sponging miR-122 to upregulate pyruvate kinase M2 (PKM2). ciRS-122 might be a potential target for the treatment of drug-resistant CRC. Exosomal circRNAs may represent potential biomarkers for predicting chemotherapy resistance in CRC patients. hsa_circ_0000338 was upregulated in exosomes derived from oxaliplatin/fluorouracil/leucovorin (FOLFOX)-resistant CRC cells as compared to sensitive cell-derived exosomes.179 Likewise, the expression level of hsa_circ_0000338 was higher in serum exosomes of FOLFOX-resistant patients than in FOLFOX-sensitive patients. Additional work is essential to validate the potential of exosomal hsa_circ_0000338 as a biomarker for early diagnosis of chemoresistance among CRC patients. The expression level of hsa_circ_0000338 was increased in sensitive CRC cells co-cultured with resistant cells compared with control sensitive cells. However, there is still a lack of experimental evidence for the occurrence of exosome-mediated hsa_circ_0000338 shuttle between drug-resistant CRC cells and sensitive cells. Further studies are warranted to support this assumption. The loss-of-function study showed that knockdown of hsa_circ_0000338 increased the viability of FOLFOX-resistant CRC cells under 5-fluorouracil (5-FU) exposure. However, the role of exosomal hsa_circ_0000338 in CRC is still unknown. Thus, it is necessary to explore the possibility of the involvement of exosomal hsa_circ_0000338 in CRC chemoresistance. Exosomal hsa_circ_0004771 was significantly upregulated in the serum of CRC patients compared to healthy individuals and patients with benign intestinal diseases.180 Moreover, circulating exosomal hsa_circ_0004771 could efficiently distinguish CRC patients from healthy controls. It was closely correlated with TNM stage and cancer metastasis in CRC patients. Thus, circulating exosomal hsa_circ_0004771 might be a new biomarker for early diagnosis of CRC. More clinical studies in large cohorts are needed to confirm its diagnostic significance.
PC
circ-IARS was highly expressed in cancer tissues and plasma exosomes of patients with metastatic PC.181 Overexpression of circ-IARS in human umbilical vein endothelial cells (HUVECs) could increase the permeability of endothelial monolayer cells by sponging miR-122 to elevate RhoA activity. Remarkably, PC cell-secreted circ-IARS could be delivered into HUVECs by exosome transportation, which led to the increase of vascular endothelial permeability and enhancement of PC cell metastasis. Further progress in elucidating the biological functions and underlying mechanisms of exosomal circRNAs in PC development will provide evidence and potential utility for innovative therapeutic strategies for PC. The potential of exosomal circRNAs as PC biomarkers has also been investigated. circ-PDE8A could favor the invasive growth of PDAC cells by regulating the miR-338/metastasis-associated in colon cancer 1 (MACC1)/mesenchymal-epithelial transition factor (MET) pathway.182 circ-PDE8A could be transmitted between PDAC cells via exosomes. Exosomal circ-PDE8A derived from PDAC cells even entered into blood circulation. Plasma exosomal circ-PDE8A was correlated with PDAC progression and prognosis. Collectively, exosomal circ-PDE8A might serve as a prospective biomarker of PDAC diagnosis or progression.
HCC
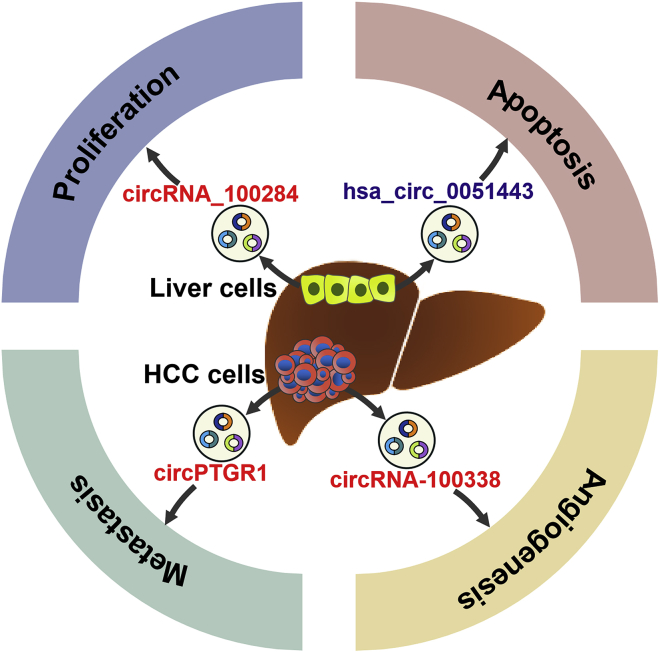
Several exosomal circRNAs have been reported to participate in the occurrence and development of HCC (Figure 3). circRNA_100284 was upregulated in arsenite-transformed normal liver cells.183 circRNA_100284 could be shuttled from transformed liver cells to normal cells via exosomes. In recipient cells, exosomal circRNA_100284 accelerated the cell cycle and promoted cell proliferation by sponging miR-217 to upregulate enhancer of zeste homolog 2 (EZH2) and cyclin D1. These findings suggested that exosomal circRNA_100284 served as intercellular signaling molecules during arsenite-induced hepatocarcinogenesis. hsa_circ_0051443 was expressed at low levels in the plasma exosomes and tissues from HCC patients compared to healthy controls.184 Exosomal hsa_circ_0051443 served as a useful marker for differentiating HCC patients from healthy controls. hsa_circ_0051443 was transported from normal cells to HCC cells via exosomes and inhibited the malignant characteristics of HCC cells by regulating the cell cycle and apoptosis. Exosomal hsa_circ_0051443 might function as a mediator of intercellular communication during HCC carcinogenesis. In HCC cell models, hsa_circ_0051443 was shown to increase the expression of pro-apoptotic Bcl-2 antagonist killer 1 (BAK1) by sponging miR-331-3p. Exosomal hsa_circ_0051443 stimulated BAK1 expression and suppressed the growth of HCC xenograft tumors in nude mice. However, an important question remains as to whether exosomal hsa_circ_0051443 functions in recipient cells by acting as a miR-331-3p sponge. Additional work is needed to elucidate the molecular mechanism by which exosomal hsa_circ_0051443 regulates HCC cell apoptosis. circPTGR1 could promote the migration and metastasis of HCC cells through the miR-449a/MET axis.185 circPTGR1 was highly expressed in serum exosomes from HCC patients and correlated with the clinical stage and prognosis in HCC patients. High metastatic HCC cells conferred this potential on non-metastatic and low-metastatic cells via circPTGR1-enriched exosomes. Exosomal circPTGR1 could promote HCC progression in vivo. It is not clear whether the transferred circPTGR1 exerts its effect on recipient HCC cells via the same mechanism as cellular circPTGR1. Further research should be carried out to investigate the molecular mechanism underlying the contribution of exosomal circPTGR1 to HCC progression. A recent report indicated that exosomal circRNA-100338 was more abundant in highly metastatic HCC cells than that in lowly metastatic cells.26 Exosomal circRNA-100338 significantly increased the invasive ability of HCC cells. circRNA-100338 could be transferred from HCC cells to HUVECs via exosomes, thus affecting the proliferation, angiogenesis, and permeability of recipient HUVECs. In HUVECs transfected with biotin-labeled circRNA-100338, the internalized circRNA was found to interact with NOVA2, an RBP regulating vascular development.186 In vivo evidence indicated that exosomal circRNA-100338 promoted HCC metastasis by regulating angiogenesis. Exosomal circRNA-100338 might be involved in the crosstalk between HCC cells and HUVECs. Additionally, high expression of exosomal circRNA-100338 in serum might be correlated with cancer progression and poor prognosis in HCC patients. These findings demonstrated that exosomal circRNAs might offer considerable promise for therapeutic intervention in HCC. However, there are a very limited number of studies exploring the roles of exosomal circRNAs in HCC progression. The exact functions of exosomal circRNAs in HCC are worthy of deep exploration.
Figure 3.
The Role of Exosomal circRNAs in Hepatocellular Carcinoma
Liver cell-secreted exosomal circRNAs (circRNA_100284 and hsa_circ_0051443) are capable of regulating the proliferation and apoptosis of HCC cells. HCC cell-released exosomal circRNAs (circPTGR1 and circRNA-100338) facilitate HCC angiogenesis and metastasis. HCC, hepatocellular carcinoma.
CCA
A previous study has suggested the potential role of exosomal circ-0000284 in CCA development.187 circ-0000284 was upregulated in cancer tissues and plasma exosomes from CCA patients compared with healthy controls. circ-0000284 could increase the proliferation, migration, and invasion of CCA cells through interaction with miR-637 that targeted lymphocyte antigen 6 complex locus E (LY6E). Moreover, CCA cells transported circ-0000284 to surrounding cells by secreting exosomes. CCA-derived exosomal circ-0000284 facilitated the proliferation and migration of recipient cells and inhibited their apoptosis. Although the expression and potential function of exosomal circ-0000284 were preliminarily revealed in recipient cells, no further information was provided with regard to the underlying mechanism by which it regulated the biological functions of recipient normal cells. The mechanism involving miR-637 sponging needs to be more clearly elucidated. At present, most of the studies reported in CCA focus on the deregulation and clinical significance of circRNAs rather than performing an in-depth analysis of their functions. Further mechanistic investigation is warranted to increase our knowledge about the biological implications of exosomal circRNAs in the initiation and development of CCA.
SCLC
The high expression of FLI1 exonic circRNAs (FECRs) in SCLC tissues was positively associated with the metastatic features of SCLC.188 Exosomal FECR1 was expressed at high levels in the serum of SCLC patients as compared with normal controls. Importantly, the level of serum exosomal FECR1 was closely correlated with poor survival and clinical response to chemotherapy in SCLC patients. Serum exosomal FECR1 might be clinically useful as a biomarker to track the progression of SCLC. Although the oncogenic role of FECRs has been verified in SCLC cells, further efforts are required to clarify whether exosomal FECRs are implicated in intercellular communication during SCLC progression.
Urogenital System Tumor
circWHSC1 was highly expressed in OC tissues compared to normal tissues.189 circWHSC1 favored the proliferation and invasion of OC cells by acting as a molecular sponge of miR-145 and miR-1182. Peritoneal mesothelial cells acted as recipient cells and took up circWHSC1-enriched exosomes released by OC cells. Exosomal circWHSC1 increased the expression of mucin 1 (MUC1) in recipient cells and facilitated peritoneal dissemination, which might promote OC progression. Targeting circWHSC1 may represent an innovative therapeutic option for OC treatment. circ_0044516 was significantly upregulated in exosomes from prostate cancer cells and patients compared with the controls.190 The diagnostic or prognostic value of exosomal circ_0044516 needs to be validated. In prostate cancer cells, circ_0044516 acted as a miR-29a-3p sponge. circ_0044516 suppressed the proliferation and metastasis of prostate cancer cells. The biological function of exosomal circ_0044516 in prostate cancer should be verified in future studies. The expression of circPRMT5 was higher in urothelial carcinoma of the bladder (UCB) tissues than in matched nontumor tissues.191 Its upregulation was positively correlated with advanced clinical stage and worse survival in UCB patients. Moreover, circPRMT5 fostered the EMT process in UCB cells via the miR-30c/SNAIL1/E-cadherin pathway. circPRMT5 was also highly expressed in serum and urine exosomes from UCB patients as compared with normal controls. The level of circPRMT5 in serum and urine exosomes positively correlated with cancer metastasis and progression in UCB patients. Exosomal circPRMT5 might serve as a prognostic biomarker for UCB patients. Our knowledge of the characteristics and function of exosomal circRNAs in urogenital system tumor is very limited. Hence, more efforts should be made to gain a better understanding of their potential as valuable biomarkers and therapeutic targets in this disease.
Conclusions and Future Perspectives
Although there are many studies on exosomal circRNAs, their formation and sorting mechanisms remain largely unknown. At present, two assumptions of exosomal circRNAs have been proposed. One is that exosomes can deliver circRNAs to target cells and protect them from clearance. Oppositely, the other is that exosomes help to reduce the accumulation of circRNAs within cells. Further studies are essential to fully discover key regulators of circRNA fate decisions. Additionally, the mechanisms relevant to the enrichment and degradation of exosomal circRNAs need comprehensive elucidation. It is possible that circRNAs are enriched in the cytoplasm and passively incorporated into exosomes. Alternatively, cells may adopt certain mechanisms to actively deliver intracellular circRNAs into exosomes. Despite their resistance to exonucleolytic degradation, circRNAs may harbor specific endonuclease sites and could be degraded in a coordinated manner. The balance between circRNA biogenesis and degradation may be influenced by dynamic cellular states or external stimuli. The biogenesis/degradation pathways of exosomal circRNAs under different conditions should be illuminated in further studies. In addition, improved knowledge on the nature of exosomes will facilitate the disclosure of detailed mechanisms involved in the sorting, trafficking, and loading of circRNAs in exosomes.
Notably, the selective sorting of circRNAs into exosomes may form a significant mechanism underlying the regulation of cancer progression. Cancer cells exploit exosomes into transferring their circRNAs as a way to control the behaviors of target cells. Accordingly, the sorting process of exosomal circRNAs in cancer cells is another unanswered question that needs to be addressed. Compared with exosomal miRNAs and lncRNAs, many gaps in our current understanding of the relationship between exosomal circRNAs and cancer science exist. A wide variety of exosomal circRNAs have been identified to be aberrantly expressed in different cancers, including gastric, colorectal, pancreatic, liver, bile duct, lung, and urogenital system cancer. Exosomal circRNAs are involved in the proliferation, migration, invasion, metastasis, and chemoresistance of cancer cells. However, several challenges regarding circRNAs in exosomes need to be addressed. First, it is controversial whether the function of exosomes is only associated with their circRNA cargos. Isolated exosomes are commonly contaminated with other molecules, including lipoproteins and ribonucleoprotein complexes, which may result in inaccurate analysis of exosomal circRNAs.192 Current separation methods include centrifugation, filtration, polymeric precipitation, and immunoaffinity isolation.193 Distinct isolation techniques lead to discrepancies in the results of downstream analysis. The separation method can be chosen based on the question being addressed. The use of an appropriate method may improve the accuracy of results to a certain extent. Nevertheless, more advanced techniques that produce high yields of pure exosomes should be developed. Second, efficient methods for examining circRNA transmission between cells are in urgent need. The transportation of exosomal circRNAs between cells has been indirectly validated by measuring the expression levels of circRNAs in both donor and recipient cells. Fluorescence signal amplification by a confocal imaging system may be an efficient approach for directly studying exosome-mediated circRNA delivery. Third, due to their circular structure and sequence overlap with linear RNA counterparts, it is difficult to precisely define the expression and function of exosomal circRNAs. Advances in our understanding of circRNA biology will facilitate the identification of the biological function of exosomal circRNAs. Finally, the lack of appropriate models has been a major obstacle to studying the role of exosomal circRNAs in cancer. Overexpression or knockdown of a specific circRNA in parent cells can lead to corresponding expression changes of this circRNA in their exosomes.26 Alterations in recipient cell behavior may be partially attributed to artificial regulation of exosomal circRNA expression. Gain- and loss-of-function experiments have been conducted to determine the role of exosomal circRNAs in cancer biology. However, further studies are needed to explore whether upregulation or downregulation of a specific circRNA in cells can have an influence on the composition, content, and characteristics of their exosomes. Additionally, in vitro cell model systems cannot adequately simulate exosomal circRNA-mediated intercellular communication in vivo. A large quantity of exosomes incorporated into cultured cells in vitro may amplify the genuine impact of their circRNA cargos in recipient cells. It remains to verify whether exosomal circRNAs actually function in vivo. Therefore, convenient in vivo model systems should be developed to accurately uncover the function of exosomal circRNAs in intercellular communication.
The mechanisms underlying the regulatory roles of exosomal circRNAs in cancer remain to be further elucidated. It has been reported that exosomal ncRNAs function to reprogram cells within the tumor microenvironment.194,195 The regulatory function of exosomal circRNAs in remodeling the tumor microenvironment deserves thorough research. Also, the impact of exosomal circRNAs on tumor immune evasion is a crucial area that needs exhaustive exploration. A single circRNA may interact with different miRNAs and thus regulate the expression of multiple target genes that take part in cancer-associated signaling cascades. Accordingly, the genuine function of exosomal circRNAs in cancer must be ascertained. Alternatively, different circRNAs can interact with the same miRNA. It is essential to investigate whether diverse circRNAs in exosomes competitively bind to specific miRNAs. The accurate mechanisms that coordinate the interaction between circRNAs and their target miRNAs in exosomes should be expounded. It is known that cellular circRNAs are involved in multiple key processes during cancer pathogenesis. Given that there is no evidence to indicate the functional discrepancy between intracellular and exosomal circRNAs, it is essential to figure out whether these circRNAs act synergistically in the process of cancer progression. In addition, cancer-derived exosomes also contain other types of ncRNAs, such as miRNAs and lncRNAs.196 These types of ncRNAs also play a role in cancer development.197,198 Both circRNAs and lncRNAs can act as molecular sponges for miRNAs. Exosomal circRNAs may interfere with the action of exosomal miRNAs in cancer development. It is possible that circRNAs and lncRNAs compete with each other for interacting with miRNAs. Thus, in-depth investigations should be conducted to delineate the complicated interplays among these exosomal ncRNAs during cancer progression.
Exosomal circRNAs can be released by various cancer cells and can be found in bodily fluids. Importantly, the expression profiles of exosomal circRNAs differ between cancer patients and healthy controls. Aberrant expression of cancer-derived exosomal circRNAs has been linked with the development of various cancers. Therefore, exosomal circRNAs may be used as molecular diagnostic and prognostic biomarkers in cancer for their stability, conservation, and specificity. However, developing exosomal circRNAs as cancer biomarkers would encounter considerable difficulties and obstacles that impede their clinical application. It is intriguing whether the expression signatures of exosomal circRNAs can reflect those of original cancer cells. Much work is needed to compare circRNA expression profiles between cancer cells and their exosomes. Current procedures for exosome extraction and purification are time-consuming and expensive, which are not suitable for high-throughput detection in the clinical setting. Moreover, it is not clear whether current approaches for exosome extraction and purification have an impact on the expression profile of exosomal circRNAs. More efforts are required to develop simple and efficient methods for isolation and purification of exosomes. The low abundance of circRNAs in exosomes has made their detection challenging. Thus, superior quantitative approaches should be created to accurately detect the expression level of circRNAs in exosomes. Furthermore, both cancer cells and normal cells can shed exosomes. The commonly utilized biomarkers including TGS101, CD9, CD63, and CD81 are not cancer-specific. It is essential to identify additional biomarkers for cancer-derived exosomes. This will contribute to differentiating circRNAs derived from tumor cells from those originated from the tumor microenvironment. The clinical significance of exosomal circRNAs in cancer diagnosis, therapy, and prognosis needs to be adequately examined. Further studies in big cohorts and in distinct kinds of cancer are indispensable to confirm the specificity and accuracy of exosomal circRNA analysis. Cancer type-specific or stage-specific exosomal circRNAs must be identified to ensure the precision of detection and the effectiveness of therapy. Since some circRNAs can act as molecular sponges for miRNAs and proteins, exosomal circRNAs are likely to be applied as promising carriers for targeted drug delivery. Loading exosomes with therapeutic circRNAs may represent a feasible approach for treating cancer. Despite their promising prospects, there is still a long way to go to reach the goal of developing exosomal circRNA-based cancer diagnostic and therapeutic strategies.
Author Contributions
M.W. and K.W. conceived this article. F.Y. collected the related papers. M.W. drew the figures and wrote the manuscript. P.L. and K.W. revised the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (no. 81701991) and the Applied Basic Research Program of Qingdao, China (no. 17-1-1-59-jch).
Contributor Information
Man Wang, Email: wangman@qdu.edu.cn.
Kun Wang, Email: wangk696@163.com.
References
- 1.Melo S.A., Sugimoto H., O’Connell J.T., Kato N., Villanueva A., Vidal A., Qiu L., Vitkin E., Perelman L.T., Melo C.A. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vestad B., Llorente A., Neurauter A., Phuyal S., Kierulf B., Kierulf P., Skotland T., Sandvig K., Haug K.B.F., Øvstebø R. Size and concentration analyses of extracellular vesicles by nanoparticle tracking analysis: a variation study. J. Extracell. Vesicles. 2017;6:1344087. doi: 10.1080/20013078.2017.1344087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harding C., Heuser J., Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983;97:329–339. doi: 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan B.T., Teng K., Wu C., Adam M., Johnstone R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985;101:942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzales P.A., Pisitkun T., Hoffert J.D., Tchapyjnikov D., Star R.A., Kleta R., Wang N.S., Knepper M.A. Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol. 2009;20:363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lässer C., Alikhani V.S., Ekström K., Eldh M., Paredes P.T., Bossios A., Sjöstrand M., Gabrielsson S., Lötvall J., Valadi H. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J. Transl. Med. 2011;9:9. doi: 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subra C., Laulagnier K., Perret B., Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie. 2007;89:205–212. doi: 10.1016/j.biochi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Xiao D., Ohlendorf J., Chen Y., Taylor D.D., Rai S.N., Waigel S., Zacharias W., Hao H., McMasters K.M. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS ONE. 2012;7:e46874. doi: 10.1371/journal.pone.0046874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sansone P., Savini C., Kurelac I., Chang Q., Amato L.B., Strillacci A., Stepanova A., Iommarini L., Mastroleo C., Daly L. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci. USA. 2017;114:E9066–E9075. doi: 10.1073/pnas.1704862114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 11.Villarroya-Beltri C., Gutiérrez-Vázquez C., Sánchez-Madrid F., Mittelbrunn M. Analysis of microRNA and protein transfer by exosomes during an immune synapse. Methods Mol. Biol. 2013;1024:41–51. doi: 10.1007/978-1-62703-453-1_4. [DOI] [PubMed] [Google Scholar]
- 12.Al-Nedawi K., Meehan B., Micallef J., Lhotak V., May L., Guha A., Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 13.Kumar B., Garcia M., Weng L., Jung X., Murakami J.L., Hu X., McDonald T., Lin A., Kumar A.R., DiGiusto D.L. Acute myeloid leukemia transforms the bone marrow niche into a leukemia-permissive microenvironment through exosome secretion. Leukemia. 2018;32:575–587. doi: 10.1038/leu.2017.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei X., Fang Z., Guo L. Predicting circRNA-disease associations based on improved collaboration filtering recommendation system with multiple data. Front. Genet. 2019;10:897. doi: 10.3389/fgene.2019.00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 18.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 20.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 21.You X., Vlatkovic I., Babic A., Will T., Epstein I., Tushev G., Akbalik G., Wang M., Glock C., Quedenau C. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guarnerio J., Bezzi M., Jeong J.C., Paffenholz S.V., Berry K., Naldini M.M., Lo-Coco F., Tay Y., Beck A.H., Pandolfi P.P. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 24.Maass P.G., Glažar P., Memczak S., Dittmar G., Hollfinger I., Schreyer L., Sauer A.V., Toka O., Aiuti A., Luft F.C., Rajewsky N. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. (Berl.) 2017;95:1179–1189. doi: 10.1007/s00109-017-1582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H., Deng T., Ge S., Liu Y., Bai M., Zhu K., Fan Q., Li J., Ning T., Tian F. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38:2844–2859. doi: 10.1038/s41388-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Huang X.Y., Huang Z.L., Huang J., Xu B., Huang X.Y., Xu Y.H., Zhou J., Tang Z.Y. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J. Exp. Clin. Cancer Res. 2020;39:20. doi: 10.1186/s13046-020-1529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding C., Yi X., Wu X., Bu X., Wang D., Wu Z., Zhang G., Gu J., Kang D. Exosome-mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett. 2020;479:1–12. doi: 10.1016/j.canlet.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Bai H., Lei K., Huang F., Jiang Z., Zhou X. Exo-circRNAs: a new paradigm for anticancer therapy. Mol. Cancer. 2019;18:56. doi: 10.1186/s12943-019-0986-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mastoridis S., Bertolino G.M., Whitehouse G., Dazzi F., Sanchez-Fueyo A., Martinez-Llordella M. Multiparametric analysis of circulating exosomes and other small extracellular vesicles by advanced imaging flow cytometry. Front. Immunol. 2018;9:1583. doi: 10.3389/fimmu.2018.01583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kowal J., Arras G., Colombo M., Jouve M., Morath J.P., Primdal-Bengtson B., Dingli F., Loew D., Tkach M., Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnstone R.M., Adam M., Hammond J.R., Orr L., Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J. Biol. Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 32.Henderson M.C., Azorsa D.O. The genomic and proteomic content of cancer cell-derived exosomes. Front. Oncol. 2012;2:38. doi: 10.3389/fonc.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratajczak J., Miekus K., Kucia M., Zhang J., Reca R., Dvorak P., Ratajczak M.Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 34.Skog J., Würdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M., Curry W.T., Jr., Carter B.S., Krichevsky A.M., Breakefield X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hemler M.E. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 2003;19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- 36.Feng D., Zhao W.L., Ye Y.Y., Bai X.C., Liu R.Q., Chang L.F., Zhou Q., Sui S.F. Cellular internalization of exosomes occurs through phagocytosis. Traffic. 2010;11:675–687. doi: 10.1111/j.1600-0854.2010.01041.x. [DOI] [PubMed] [Google Scholar]
- 37.Tian T., Wang Y., Wang H., Zhu Z., Xiao Z. Visualizing of the cellular uptake and intracellular trafficking of exosomes by live-cell microscopy. J. Cell. Biochem. 2010;111:488–496. doi: 10.1002/jcb.22733. [DOI] [PubMed] [Google Scholar]
- 38.Christianson H.C., Svensson K.J., van Kuppevelt T.H., Li J.P., Belting M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. USA. 2013;110:17380–17385. doi: 10.1073/pnas.1304266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Escrevente C., Keller S., Altevogt P., Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 2011;11:108. doi: 10.1186/1471-2407-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parolini I., Federici C., Raggi C., Lugini L., Palleschi S., De Milito A., Coscia C., Iessi E., Logozzi M., Molinari A. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009;284:34211–34222. doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merchant M.L., Rood I.M., Deegens J.K.J., Klein J.B. Isolation and characterization of urinary extracellular vesicles: implications for biomarker discovery. Nat. Rev. Nephrol. 2017;13:731–749. doi: 10.1038/nrneph.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Vizio D., Kim J., Hager M.H., Morello M., Yang W., Lafargue C.J., True L.D., Rubin M.A., Adam R.M., Beroukhim R. Oncosome formation in prostate cancer: association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res. 2009;69:5601–5609. doi: 10.1158/0008-5472.CAN-08-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller L., Mitsuhashi M., Simms P., Gooding W.E., Whiteside T.L. Tumor-derived exosomes regulate expression of immune function-related genes in human T cell subsets. Sci. Rep. 2016;6:20254. doi: 10.1038/srep20254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacKenzie A., Wilson H.L., Kiss-Toth E., Dower S.K., North R.A., Surprenant A. Rapid secretion of interleukin-1β by microvesicle shedding. Immunity. 2001;15:825–835. doi: 10.1016/s1074-7613(01)00229-1. [DOI] [PubMed] [Google Scholar]
- 45.Winau F., Weber S., Sad S., de Diego J., Hoops S.L., Breiden B., Sandhoff K., Brinkmann V., Kaufmann S.H., Schaible U.E. Apoptotic vesicles crossprime CD8 T cells and protect against tuberculosis. Immunity. 2006;24:105–117. doi: 10.1016/j.immuni.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 46.Wortzel I., Dror S., Kenific C.M., Lyden D. Exosome-mediated metastasis: communication from a distance. Dev. Cell. 2019;49:347–360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 47.Bebelman M.P., Bun P., Huveneers S., van Niel G., Pegtel D.M., Verweij F.J. Real-time imaging of multivesicular body-plasma membrane fusion to quantify exosome release from single cells. Nat. Protoc. 2020;15:102–121. doi: 10.1038/s41596-019-0245-4. [DOI] [PubMed] [Google Scholar]
- 48.Mathivanan S., Fahner C.J., Reid G.E., Simpson R.J. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–D1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Devhare P.B., Sasaki R., Shrivastava S., Di Bisceglie A.M., Ray R., Ray R.B. Exosome-mediated intercellular communication between hepatitis C virus-infected hepatocytes and hepatic stellate cells. J. Virol. 2017;91 doi: 10.1128/JVI.02225-16. e02225-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jayachandran M., Miller V.M., Heit J.A., Owen W.G. Methodology for isolation, identification and characterization of microvesicles in peripheral blood. J. Immunol. Methods. 2012;375:207–214. doi: 10.1016/j.jim.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akuthota P., Carmo L.A., Bonjour K., Murphy R.O., Silva T.P., Gamalier J.P., Capron K.L., Tigges J., Toxavidis V., Camacho V. Extracellular microvesicle production by human eosinophils activated by “inflammatory” stimuli. Front. Cell Dev. Biol. 2016;4:117. doi: 10.3389/fcell.2016.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bucki R., Bachelot-Loza C., Zachowski A., Giraud F., Sulpice J.C. Calcium induces phospholipid redistribution and microvesicle release in human erythrocyte membranes by independent pathways. Biochemistry. 1998;37:15383–15391. doi: 10.1021/bi9805238. [DOI] [PubMed] [Google Scholar]
- 53.Pasquet J.M., Dachary-Prigent J., Nurden A.T. Calcium influx is a determining factor of calpain activation and microparticle formation in platelets. Eur. J. Biochem. 1996;239:647–654. doi: 10.1111/j.1432-1033.1996.0647u.x. [DOI] [PubMed] [Google Scholar]
- 54.Li B., Antonyak M.A., Zhang J., Cerione R.A. RhoA triggers a specific signaling pathway that generates transforming microvesicles in cancer cells. Oncogene. 2012;31:4740–4749. doi: 10.1038/onc.2011.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai H., Reinisch K., Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 56.Nabhan J.F., Hu R., Oh R.S., Cohen S.N., Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc. Natl. Acad. Sci. USA. 2012;109:4146–4151. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Del Conde I., Shrimpton C.N., Thiagarajan P., López J.A. Tissue-factor-bearing microvesicles arise from lipid rafts and fuse with activated platelets to initiate coagulation. Blood. 2005;106:1604–1611. doi: 10.1182/blood-2004-03-1095. [DOI] [PubMed] [Google Scholar]
- 58.Muralidharan-Chari V., Clancy J., Plou C., Romao M., Chavrier P., Raposo G., D’Souza-Schorey C. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr. Biol. 2009;19:1875–1885. doi: 10.1016/j.cub.2009.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas L.M., Salter R.D. Activation of macrophages by P2X7-induced microvesicles from myeloid cells is mediated by phospholipids and is partially dependent on TLR4. J. Immunol. 2010;185:3740–3749. doi: 10.4049/jimmunol.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morelli A.E., Larregina A.T., Shufesky W.J., Sullivan M.L., Stolz D.B., Papworth G.D., Zahorchak A.F., Logar A.J., Wang Z., Watkins S.C. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104:3257–3266. doi: 10.1182/blood-2004-03-0824. [DOI] [PubMed] [Google Scholar]
- 61.Bastida E., Ordinas A., Escolar G., Jamieson G.A. Tissue factor in microvesicles shed from U87MG human glioblastoma cells induces coagulation, platelet aggregation, and thrombogenesis. Blood. 1984;64:177–184. [PubMed] [Google Scholar]
- 62.Jiang L., Paone S., Caruso S., Atkin-Smith G.K., Phan T.K., Hulett M.D., Poon I.K.H. Determining the contents and cell origins of apoptotic bodies by flow cytometry. Sci. Rep. 2017;7:14444. doi: 10.1038/s41598-017-14305-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Atkin-Smith G.K., Tixeira R., Paone S., Mathivanan S., Collins C., Liem M., Goodall K.J., Ravichandran K.S., Hulett M.D., Poon I.K. A novel mechanism of generating extracellular vesicles during apoptosis via a beads-on-a-string membrane structure. Nat. Commun. 2015;6:7439. doi: 10.1038/ncomms8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jeppesen D.K., Hvam M.L., Primdahl-Bengtson B., Boysen A.T., Whitehead B., Dyrskjøt L., Orntoft T.F., Howard K.A., Ostenfeld M.S. Comparative analysis of discrete exosome fractions obtained by differential centrifugation. J. Extracell. Vesicles. 2014;3:25011. doi: 10.3402/jev.v3.25011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Théry C., Boussac M., Véron P., Ricciardi-Castagnoli P., Raposo G., Garin J., Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 66.Canbay A., Feldstein A.E., Higuchi H., Werneburg N., Grambihler A., Bronk S.F., Gores G.J. Kupffer cell engulfment of apoptotic bodies stimulates death ligand and cytokine expression. Hepatology. 2003;38:1188–1198. doi: 10.1053/jhep.2003.50472. [DOI] [PubMed] [Google Scholar]
- 67.Hristov M., Erl W., Linder S., Weber P.C. Apoptotic bodies from endothelial cells enhance the number and initiate the differentiation of human endothelial progenitor cells in vitro. Blood. 2004;104:2761–2766. doi: 10.1182/blood-2003-10-3614. [DOI] [PubMed] [Google Scholar]
- 68.Coleman M.L., Sahai E.A., Yeo M., Bosch M., Dewar A., Olson M.F. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat. Cell Biol. 2001;3:339–345. doi: 10.1038/35070009. [DOI] [PubMed] [Google Scholar]
- 69.Torr E.E., Gardner D.H., Thomas L., Goodall D.M., Bielemeier A., Willetts R., Griffiths H.R., Marshall L.J., Devitt A. Apoptotic cell-derived ICAM-3 promotes both macrophage chemoattraction to and tethering of apoptotic cells. Cell Death Differ. 2012;19:671–679. doi: 10.1038/cdd.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meehan B., Rak J., Di Vizio D. Oncosomes—large and small: what are they, where they came from? J. Extracell. Vesicles. 2016;5:33109. doi: 10.3402/jev.v5.33109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Di Vizio D., Morello M., Dudley A.C., Schow P.W., Adam R.M., Morley S., Mulholland D., Rotinen M., Hager M.H., Insabato L. Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. Am. J. Pathol. 2012;181:1573–1584. doi: 10.1016/j.ajpath.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morello M., Minciacchi V.R., de Candia P., Yang J., Posadas E., Kim H., Griffiths D., Bhowmick N., Chung L.W., Gandellini P. Large oncosomes mediate intercellular transfer of functional microRNA. Cell Cycle. 2013;12:3526–3536. doi: 10.4161/cc.26539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Minciacchi V.R., You S., Spinelli C., Morley S., Zandian M., Aspuria P.J., Cavallini L., Ciardiello C., Reis Sobreiro M., Morello M. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget. 2015;6:11327–11341. doi: 10.18632/oncotarget.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abraham A., Krasnodembskaya A. Mesenchymal stem cell-derived extracellular vesicles for the treatment of acute respiratory distress syndrome. Stem Cells Transl. Med. 2020;9:28–38. doi: 10.1002/sctm.19-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blot V., Perugi F., Gay B., Prévost M.C., Briant L., Tangy F., Abriel H., Staub O., Dokhélar M.C., Pique C. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J. Cell Sci. 2004;117:2357–2367. doi: 10.1242/jcs.01095. [DOI] [PubMed] [Google Scholar]
- 76.Kunadt M., Eckermann K., Stuendl A., Gong J., Russo B., Strauss K., Rai S., Kügler S., Falomir Lockhart L., Schwalbe M. Extracellular vesicle sorting of α-Synuclein is regulated by sumoylation. Acta Neuropathol. 2015;129:695–713. doi: 10.1007/s00401-015-1408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Villarroya-Beltri C., Baixauli F., Mittelbrunn M., Fernández-Delgado I., Torralba D., Moreno-Gonzalo O., Baldanta S., Enrich C., Guerra S., Sánchez-Madrid F. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat. Commun. 2016;7:13588. doi: 10.1038/ncomms13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Raiborg C., Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 79.Mazurov D., Barbashova L., Filatov A. Tetraspanin protein CD9 interacts with metalloprotease CD10 and enhances its release via exosomes. FEBS J. 2013;280:1200–1213. doi: 10.1111/febs.12110. [DOI] [PubMed] [Google Scholar]
- 80.Yuyama K., Sun H., Mitsutake S., Igarashi Y. Sphingolipid-modulated exosome secretion promotes clearance of amyloid-β by microglia. J. Biol. Chem. 2012;287:10977–10989. doi: 10.1074/jbc.M111.324616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Villarroya-Beltri C., Gutiérrez-Vázquez C., Sánchez-Cabo F., Pérez-Hernández D., Vázquez J., Martin-Cofreces N., Martinez-Herrera D.J., Pascual-Montano A., Mittelbrunn M., Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Haraszti R.A., Didiot M.C., Sapp E., Leszyk J., Shaffer S.A., Rockwell H.E., Gao F., Narain N.R., DiFiglia M., Kiebish M.A. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles. 2016;5:32570. doi: 10.3402/jev.v5.32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pisitkun T., Shen R.F., Knepper M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Caby M.P., Lankar D., Vincendeau-Scherrer C., Raposo G., Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 85.Ogawa Y., Miura Y., Harazono A., Kanai-Azuma M., Akimoto Y., Kawakami H., Yamaguchi T., Toda T., Endo T., Tsubuki M., Yanoshita R. Proteomic analysis of two types of exosomes in human whole saliva. Biol. Pharm. Bull. 2011;34:13–23. doi: 10.1248/bpb.34.13. [DOI] [PubMed] [Google Scholar]
- 86.Emelyanov A., Shtam T., Kamyshinsky R., Garaeva L., Verlov N., Miliukhina I., Kudrevatykh A., Gavrilov G., Zabrodskaya Y., Pchelina S., Konevega A. Cryo-electron microscopy of extracellular vesicles from cerebrospinal fluid. PLoS ONE. 2020;15:e0227949. doi: 10.1371/journal.pone.0227949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stoorvogel W., Strous G.J., Geuze H.J., Oorschot V., Schwartz A.L. Late endosomes derive from early endosomes by maturation. Cell. 1991;65:417–427. doi: 10.1016/0092-8674(91)90459-c. [DOI] [PubMed] [Google Scholar]
- 88.Klumperman J., Raposo G. The complex ultrastructure of the endolysosomal system. Cold Spring Harb. Perspect. Biol. 2014;6:a016857. doi: 10.1101/cshperspect.a016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wubbolts R., Leckie R.S., Veenhuizen P.T., Schwarzmann G., Möbius W., Hoernschemeyer J., Slot J.W., Geuze H.J., Stoorvogel W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem. 2003;278:10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 90.Futter C.E., Pearse A., Hewlett L.J., Hopkins C.R. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J. Cell Biol. 1996;132:1011–1023. doi: 10.1083/jcb.132.6.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Katzmann D.J., Babst M., Emr S.D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 92.Babst M., Katzmann D.J., Snyder W.B., Wendland B., Emr S.D. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 2002;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 93.Baietti M.F., Zhang Z., Mortier E., Melchior A., Degeest G., Geeraerts A., Ivarsson Y., Depoortere F., Coomans C., Vermeiren E. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 94.Stuffers S., Sem Wegner C., Stenmark H., Brech A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic. 2009;10:925–937. doi: 10.1111/j.1600-0854.2009.00920.x. [DOI] [PubMed] [Google Scholar]
- 95.Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 96.Kosaka N., Iguchi H., Hagiwara K., Yoshioka Y., Takeshita F., Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013;288:10849–10859. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.van Niel G., Charrin S., Simoes S., Romao M., Rochin L., Saftig P., Marks M.S., Rubinstein E., Raposo G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell. 2011;21:708–721. doi: 10.1016/j.devcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Perez-Hernandez D., Gutiérrez-Vázquez C., Jorge I., López-Martín S., Ursa A., Sánchez-Madrid F., Vázquez J., Yáñez-Mó M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 2013;288:11649–11661. doi: 10.1074/jbc.M112.445304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hoshino D., Kirkbride K.C., Costello K., Clark E.S., Sinha S., Grega-Larson N., Tyska M.J., Weaver A.M. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 2013;5:1159–1168. doi: 10.1016/j.celrep.2013.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 101.Hsu C., Morohashi Y., Yoshimura S., Manrique-Hoyos N., Jung S., Lauterbach M.A., Bakhti M., Grønborg M., Möbius W., Rhee J. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J. Cell Biol. 2010;189:223–232. doi: 10.1083/jcb.200911018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ostrowski M., Carmo N.B., Krumeich S., Fanget I., Raposo G., Savina A., Moita C.F., Schauer K., Hume A.N., Freitas R.P. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 103.Yu X., Harris S.L., Levine A.J. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 2006;66:4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
- 104.Edgar J.R., Manna P.T., Nishimura S., Banting G., Robinson M.S. Tetherin is an exosomal tether. eLife. 2016;5:e17180. doi: 10.7554/eLife.17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yáñez-Mó M., Siljander P.R., Andreu Z., Zavec A.B., Borràs F.E., Buzas E.I., Buzas K., Casal E., Cappello F., Carvalho J. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shabbir A., Cox A., Rodriguez-Menocal L., Salgado M., Van Badiavas E. Mesenchymal stem cell exosomes induce proliferation and migration of normal and chronic wound fibroblasts, and enhance angiogenesis in vitro. Stem Cells Dev. 2015;24:1635–1647. doi: 10.1089/scd.2014.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lai R.C., Arslan F., Lee M.M., Sze N.S., Choo A., Chen T.S., Salto-Tellez M., Timmers L., Lee C.N., El Oakley R.M. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. (Amst.) 2010;4:214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 108.Nojima H., Freeman C.M., Schuster R.M., Japtok L., Kleuser B., Edwards M.J., Gulbins E., Lentsch A.B. Hepatocyte exosomes mediate liver repair and regeneration via sphingosine-1-phosphate. J. Hepatol. 2016;64:60–68. doi: 10.1016/j.jhep.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bhatnagar S., Schorey J.S. Exosomes released from infected macrophages contain Mycobacterium avium glycopeptidolipids and are proinflammatory. J. Biol. Chem. 2007;282:25779–25789. doi: 10.1074/jbc.M702277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gastpar R., Gehrmann M., Bausero M.A., Asea A., Gross C., Schroeder J.A., Multhoff G. Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res. 2005;65:5238–5247. doi: 10.1158/0008-5472.CAN-04-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vega V.L., Rodríguez-Silva M., Frey T., Gehrmann M., Diaz J.C., Steinem C., Multhoff G., Arispe N., De Maio A. Hsp70 translocates into the plasma membrane after stress and is released into the extracellular environment in a membrane-associated form that activates macrophages. J. Immunol. 2008;180:4299–4307. doi: 10.4049/jimmunol.180.6.4299. [DOI] [PubMed] [Google Scholar]
- 112.Munich S., Sobo-Vujanovic A., Buchser W.J., Beer-Stolz D., Vujanovic N.L. Dendritic cell exosomes directly kill tumor cells and activate natural killer cells via TNF superfamily ligands. OncoImmunology. 2012;1:1074–1083. doi: 10.4161/onci.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhu L., Kalimuthu S., Gangadaran P., Oh J.M., Lee H.W., Baek S.H., Jeong S.Y., Lee S.W., Lee J., Ahn B.C. Exosomes derived from natural killer cells exert therapeutic effect in melanoma. Theranostics. 2017;7:2732–2745. doi: 10.7150/thno.18752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Raposo G., Nijman H.W., Stoorvogel W., Liejendekker R., Harding C.V., Melief C.J., Geuze H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lynch S., Santos S.G., Campbell E.C., Nimmo A.M., Botting C., Prescott A., Antoniou A.N., Powis S.J. Novel MHC class I structures on exosomes. J. Immunol. 2009;183:1884–1891. doi: 10.4049/jimmunol.0900798. [DOI] [PubMed] [Google Scholar]
- 116.Hao S., Bai O., Li F., Yuan J., Laferte S., Xiang J. Mature dendritic cells pulsed with exosomes stimulate efficient cytotoxic T-lymphocyte responses and antitumour immunity. Immunology. 2007;120:90–102. doi: 10.1111/j.1365-2567.2006.02483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wahlgren J., Karlson Tde.L., Glader P., Telemo E., Valadi H. Activated human T cells secrete exosomes that participate in IL-2 mediated immune response signaling. PLoS ONE. 2012;7:e49723. doi: 10.1371/journal.pone.0049723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Muntasell A., Berger A.C., Roche P.A. T cell-induced secretion of MHC class II-peptide complexes on B cell exosomes. EMBO J. 2007;26:4263–4272. doi: 10.1038/sj.emboj.7601842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Smyth L.A., Ratnasothy K., Tsang J.Y., Boardman D., Warley A., Lechler R., Lombardi G. CD73 expression on extracellular vesicles derived from CD4+ CD25+ Foxp3+ T cells contributes to their regulatory function. Eur. J. Immunol. 2013;43:2430–2440. doi: 10.1002/eji.201242909. [DOI] [PubMed] [Google Scholar]
- 120.Zhang H., Xie Y., Li W., Chibbar R., Xiong S., Xiang J. CD4+ T cell-released exosomes inhibit CD8+ cytotoxic T-lymphocyte responses and antitumor immunity. Cell. Mol. Immunol. 2011;8:23–30. doi: 10.1038/cmi.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Alonso R., Mazzeo C., Mérida I., Izquierdo M. A new role of diacylglycerol kinase α on the secretion of lethal exosomes bearing Fas ligand during activation-induced cell death of T lymphocytes. Biochimie. 2007;89:213–221. doi: 10.1016/j.biochi.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 122.Cai Z., Yang F., Yu L., Yu Z., Jiang L., Wang Q., Yang Y., Wang L., Cao X., Wang J. Activated T cell exosomes promote tumor invasion via Fas signaling pathway. J. Immunol. 2012;188:5954–5961. doi: 10.4049/jimmunol.1103466. [DOI] [PubMed] [Google Scholar]
- 123.Klinker M.W., Lizzio V., Reed T.J., Fox D.A., Lundy S.K. Human B cell-derived lymphoblastoid cell lines constitutively produce Fas ligand and secrete MHCII+FasL+ killer exosomes. Front. Immunol. 2014;5:144. doi: 10.3389/fimmu.2014.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xie Y., Zhang X., Zhao T., Li W., Xiang J. Natural CD8+25+ regulatory T cell-secreted exosomes capable of suppressing cytotoxic T lymphocyte-mediated immunity against B16 melanoma. Biochem. Biophys. Res. Commun. 2013;438:152–155. doi: 10.1016/j.bbrc.2013.07.044. [DOI] [PubMed] [Google Scholar]
- 125.Stefanius K., Servage K., de Souza Santos M., Gray H.F., Toombs J.E., Chimalapati S., Kim M.S., Malladi V.S., Brekken R., Orth K. Human pancreatic cancer cell exosomes, but not human normal cell exosomes, act as an initiator in cell transformation. eLife. 2019;8:e40226. doi: 10.7554/eLife.40226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cheng K.W., Lahad J.P., Kuo W.L., Lapuk A., Yamada K., Auersperg N., Liu J., Smith-McCune K., Lu K.H., Fishman D. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat. Med. 2004;10:1251–1256. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- 127.Wang T., Gilkes D.M., Takano N., Xiang L., Luo W., Bishop C.J., Chaturvedi P., Green J.J., Semenza G.L. Hypoxia-inducible factors and RAB22A mediate formation of microvesicles that stimulate breast cancer invasion and metastasis. Proc. Natl. Acad. Sci. USA. 2014;111:E3234–E3242. doi: 10.1073/pnas.1410041111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Melo S.A., Luecke L.B., Kahlert C., Fernandez A.F., Gammon S.T., Kaye J., LeBleu V.S., Mittendorf E.A., Weitz J., Rahbari N. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ham S., Lima L.G., Chai E.P.Z., Muller A., Lobb R.J., Krumeich S., Wen S.W., Wiegmans A.P., Möller A. Breast cancer-derived exosomes alter macrophage polarization via gp130/STAT3 signaling. Front. Immunol. 2018;9:871. doi: 10.3389/fimmu.2018.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.He M., Qin H., Poon T.C., Sze S.C., Ding X., Co N.N., Ngai S.M., Chan T.F., Wong N. Hepatocellular carcinoma-derived exosomes promote motility of immortalized hepatocyte through transfer of oncogenic proteins and RNAs. Carcinogenesis. 2015;36:1008–1018. doi: 10.1093/carcin/bgv081. [DOI] [PubMed] [Google Scholar]
- 131.Beckham C.J., Olsen J., Yin P.N., Wu C.H., Ting H.J., Hagen F.K., Scosyrev E., Messing E.M., Lee Y.F. Bladder cancer exosomes contain EDIL-3/Del1 and facilitate cancer progression. J. Urol. 2014;192:583–592. doi: 10.1016/j.juro.2014.02.035. [DOI] [PubMed] [Google Scholar]
- 132.Andreola G., Rivoltini L., Castelli C., Huber V., Perego P., Deho P., Squarcina P., Accornero P., Lozupone F., Lugini L. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J. Exp. Med. 2002;195:1303–1316. doi: 10.1084/jem.20011624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huber V., Fais S., Iero M., Lugini L., Canese P., Squarcina P., Zaccheddu A., Colone M., Arancia G., Gentile M. Human colorectal cancer cells induce T-cell death through release of proapoptotic microvesicles: role in immune escape. Gastroenterology. 2005;128:1796–1804. doi: 10.1053/j.gastro.2005.03.045. [DOI] [PubMed] [Google Scholar]
- 134.Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K., Becker A., Hoshino A., Mark M.T., Molina H. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mu W., Rana S., Zöller M. Host matrix modulation by tumor exosomes promotes motility and invasiveness. Neoplasia. 2013;15:875–887. doi: 10.1593/neo.13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bandari S.K., Purushothaman A., Ramani V.C., Brinkley G.J., Chandrashekar D.S., Varambally S., Mobley J.A., Zhang Y., Brown E.E., Vlodavsky I., Sanderson R.D. Chemotherapy induces secretion of exosomes loaded with heparanase that degrades extracellular matrix and impacts tumor and host cell behavior. Matrix Biol. 2018;65:104–118. doi: 10.1016/j.matbio.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Muralidharan-Chari V., Kohan H.G., Asimakopoulos A.G., Sudha T., Sell S., Kannan K., Boroujerdi M., Davis P.J., Mousa S.A. Microvesicle removal of anticancer drugs contributes to drug resistance in human pancreatic cancer cells. Oncotarget. 2016;7:50365–50379. doi: 10.18632/oncotarget.10395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Safaei R., Larson B.J., Cheng T.C., Gibson M.A., Otani S., Naerdemann W., Howell S.B. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol. Cancer Ther. 2005;4:1595–1604. doi: 10.1158/1535-7163.MCT-05-0102. [DOI] [PubMed] [Google Scholar]
- 139.Zhang J., Zhang H.D., Yao Y.F., Zhong S.L., Zhao J.H., Tang J.H. β-Elemene reverses chemoresistance of breast cancer cells by reducing resistance transmission via exosomes. Cell. Physiol. Biochem. 2015;36:2274–2286. doi: 10.1159/000430191. [DOI] [PubMed] [Google Scholar]
- 140.Taylor D.D., Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 141.Rabinowits G., Gerçel-Taylor C., Day J.M., Taylor D.D., Kloecker G.H. Exosomal microRNA: a diagnostic marker for lung cancer. Clin. Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 142.Silva J., Garcia V., Rodriguez M., Compte M., Cisneros E., Veguillas P., Garcia J.M., Dominguez G., Campos-Martin Y., Cuevas J. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer. 2012;51:409–418. doi: 10.1002/gcc.21926. [DOI] [PubMed] [Google Scholar]
- 143.Saari H., Lázaro-Ibáñez E., Viitala T., Vuorimaa-Laukkanen E., Siljander P., Yliperttula M. Microvesicle- and exosome-mediated drug delivery enhances the cytotoxicity of Paclitaxel in autologous prostate cancer cells. J. Control. Release. 2015;220(Pt B):727–737. doi: 10.1016/j.jconrel.2015.09.031. [DOI] [PubMed] [Google Scholar]
- 144.Yong T., Zhang X., Bie N., Zhang H., Zhang X., Li F., Hakeem A., Hu J., Gan L., Santos H.A., Yang X. Tumor exosome-based nanoparticles are efficient drug carriers for chemotherapy. Nat. Commun. 2019;10:3838. doi: 10.1038/s41467-019-11718-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Van Niel G., Mallegol J., Bevilacqua C., Candalh C., Brugière S., Tomaskovic-Crook E., Heath J.K., Cerf-Bensussan N., Heyman M. Intestinal epithelial exosomes carry MHC class II/peptides able to inform the immune system in mice. Gut. 2003;52:1690–1697. doi: 10.1136/gut.52.12.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Crenshaw B.J., Kumar S., Bell C.R., Jones L.B., Williams S.D., Saldanha S.N., Joshi S., Sahu R., Sims B., Matthews Q.L. Alcohol modulates the biogenesis and composition of microglia-derived exosomes. Biology (Basel) 2019;8:25. doi: 10.3390/biology8020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Clayton A., Turkes A., Navabi H., Mason M.D., Tabi Z. Induction of heat shock proteins in B-cell exosomes. J. Cell Sci. 2005;118:3631–3638. doi: 10.1242/jcs.02494. [DOI] [PubMed] [Google Scholar]
- 148.Harmati M., Tarnai Z., Decsi G., Kormondi S., Szegletes Z., Janovak L., Dekany I., Saydam O., Gyukity-Sebestyen E., Dobra G. Stressors alter intercellular communication and exosome profile of nasopharyngeal carcinoma cells. J. Oral Pathol. Med. 2017;46:259–266. doi: 10.1111/jop.12486. [DOI] [PubMed] [Google Scholar]
- 149.de Jong O.G., Verhaar M.C., Chen Y., Vader P., Gremmels H., Posthuma G., Schiffelers R.M., Gucek M., van Balkom B.W. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J. Extracell. Vesicles. 2012;1:18396. doi: 10.3402/jev.v1i0.18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Beach A., Zhang H.G., Ratajczak M.Z., Kakar S.S. Exosomes: an overview of biogenesis, composition and role in ovarian cancer. J. Ovarian Res. 2014;7:14. doi: 10.1186/1757-2215-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lamparski H.G., Metha-Damani A., Yao J.Y., Patel S., Hsu D.H., Ruegg C., Le Pecq J.B. Production and characterization of clinical grade exosomes derived from dendritic cells. J. Immunol. Methods. 2002;270:211–226. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- 152.Ogawa Y., Tsujimoto M., Yanoshita R. Next-generation sequencing of protein-coding and long non-protein-coding RNAs in two types of exosomes derived from human whole saliva. Biol. Pharm. Bull. 2016;39:1496–1507. doi: 10.1248/bpb.b16-00297. [DOI] [PubMed] [Google Scholar]
- 153.Mittelbrunn M., Gutiérrez-Vázquez C., Villarroya-Beltri C., González S., Sánchez-Cabo F., González M.A., Bernad A., Sánchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Farahani M., Rubbi C., Liu L., Slupsky J.R., Kalakonda N. CLL exosomes modulate the transcriptome and behaviour of recipient stromal cells and are selectively enriched in miR-202-3p. PLoS ONE. 2015;10:e0141429. doi: 10.1371/journal.pone.0141429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shurtleff M.J., Temoche-Diaz M.M., Karfilis K.V., Ri S., Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife. 2016;5:e19276. doi: 10.7554/eLife.19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dou Y., Cha D.J., Franklin J.L., Higginbotham J.N., Jeppesen D.K., Weaver A.M., Prasad N., Levy S., Coffey R.J., Patton J.G., Zhang B. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci. Rep. 2016;6:37982. doi: 10.1038/srep37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang J., Zhang Q., Zhou S., Xu H., Wang D., Feng J., Zhao J., Zhong S. Circular RNA expression in exosomes derived from breast cancer cells and patients. Epigenomics. 2019;11:411–421. doi: 10.2217/epi-2018-0111. [DOI] [PubMed] [Google Scholar]
- 158.Xu H., Gong Z., Shen Y., Fang Y., Zhong S. Circular RNA expression in extracellular vesicles isolated from serum of patients with endometrial cancer. Epigenomics. 2018;10:187–197. doi: 10.2217/epi-2017-0109. [DOI] [PubMed] [Google Scholar]
- 159.Li Q., Geng S., Yuan H., Li Y., Zhang S., Pu L., Ge J., Niu X., Li Y., Jiang H. Circular RNA expression profiles in extracellular vesicles from the plasma of patients with pancreatic ductal adenocarcinoma. FEBS Open Bio. 2019;9:2052–2062. doi: 10.1002/2211-5463.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chen F., Huang C., Wu Q., Jiang L., Chen S., Chen L. Circular RNAs expression profiles in plasma exosomes from early-stage lung adenocarcinoma and the potential biomarkers. J. Cell. Biochem. 2020;121:2525–2533. doi: 10.1002/jcb.29475. [DOI] [PubMed] [Google Scholar]
- 161.Yang C., Wei Y., Yu L., Xiao Y. Identification of altered circular RNA expression in serum exosomes from patients with papillary thyroid carcinoma by high-throughput sequencing. Med. Sci. Monit. 2019;25:2785–2791. doi: 10.12659/MSM.915658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Preußer C., Hung L.H., Schneider T., Schreiner S., Hardt M., Moebus A., Santoso S., Bindereif A. Selective release of circRNAs in platelet-derived extracellular vesicles. J. Extracell. Vesicles. 2018;7:1424473. doi: 10.1080/20013078.2018.1424473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhao R.T., Zhou J., Dong X.L., Bi C.W., Jiang R.C., Dong J.F., Tian Y., Yuan H.J., Zhang J.N. Circular ribonucleic acid expression alteration in exosomes from the brain extracellular space after traumatic brain injury in mice. J. Neurotrauma. 2018;35:2056–2066. doi: 10.1089/neu.2017.5502. [DOI] [PubMed] [Google Scholar]
- 164.He J., Ren M., Li H., Yang L., Wang X., Yang Q. Exosomal circular RNA as a biomarker platform for the early diagnosis of immune-mediated demyelinating disease. Front. Genet. 2019;10:860. doi: 10.3389/fgene.2019.00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang X., Chen L., Xiao B., Liu H., Su Y. Circ_0075932 in adipocyte-derived exosomes induces inflammation and apoptosis in human dermal keratinocytes by directly binding with PUM2 and promoting PUM2-mediated activation of AuroraA/NF-κB pathway. Biochem. Biophys. Res. Commun. 2019;511:551–558. doi: 10.1016/j.bbrc.2019.02.082. [DOI] [PubMed] [Google Scholar]
- 166.Epple L.M., Griffiths S.G., Dechkovskaia A.M., Dusto N.L., White J., Ouellette R.J., Anchordoquy T.J., Bemis L.T., Graner M.W. Medulloblastoma exosome proteomics yield functional roles for extracellular vesicles. PLoS ONE. 2012;7:e42064. doi: 10.1371/journal.pone.0042064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Santangelo L., Giurato G., Cicchini C., Montaldo C., Mancone C., Tarallo R., Battistelli C., Alonzi T., Weisz A., Tripodi M. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016;17:799–808. doi: 10.1016/j.celrep.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 168.Statello L., Maugeri M., Garre E., Nawaz M., Wahlgren J., Papadimitriou A., Lundqvist C., Lindfors L., Collén A., Sunnerhagen P. Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PLoS ONE. 2018;13:e0195969. doi: 10.1371/journal.pone.0195969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gibbings D.J., Ciaudo C., Erhardt M., Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 170.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 171.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 172.Zhang H., Zhu L., Bai M., Liu Y., Zhan Y., Deng T., Yang H., Sun W., Wang X., Zhu K. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int. J. Cancer. 2019;144:2501–2515. doi: 10.1002/ijc.31977. [DOI] [PubMed] [Google Scholar]
- 173.Zhang X., Wang S., Wang H., Cao J., Huang X., Chen Z., Xu P., Sun G., Xu J., Lv J., Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer. 2019;18:20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lu J., Wang Y.H., Yoon C., Huang X.Y., Xu Y., Xie J.W., Wang J.B., Lin J.X., Chen Q.Y., Cao L.L. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38–48. doi: 10.1016/j.canlet.2019.11.038. [DOI] [PubMed] [Google Scholar]
- 175.Tao X., Shao Y., Lu R., Ye Q., Xiao B., Ye G., Guo J. Clinical significance of hsa_circ_0000419 in gastric cancer screening and prognosis estimation. Pathol. Res. Pract. 2020;216:152763. doi: 10.1016/j.prp.2019.152763. [DOI] [PubMed] [Google Scholar]
- 176.Shao Y., Tao X., Lu R., Zhang H., Ge J., Xiao B., Ye G., Guo J. Hsa_circ_0065149 is an indicator for early gastric cancer screening and prognosis prediction. Pathol. Oncol. Res. 2020;26:1475–1482. doi: 10.1007/s12253-019-00716-y. [DOI] [PubMed] [Google Scholar]
- 177.Tang W., Fu K., Sun H., Rong D., Wang H., Cao H. circRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol. Cancer. 2018;17:137. doi: 10.1186/s12943-018-0888-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang X., Zhang H., Yang H., Bai M., Ning T., Deng T., Liu R., Fan Q., Zhu K., Li J. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 2020;14:539–555. doi: 10.1002/1878-0261.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hon K.W., Ab-Mutalib N.S., Abdullah N.M.A., Jamal R., Abu N. Extracellular vesicle-derived circular RNAs confers chemoresistance in colorectal cancer. Sci. Rep. 2019;9:16497. doi: 10.1038/s41598-019-53063-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Pan B., Qin J., Liu X., He B., Wang X., Pan Y., Sun H., Xu T., Xu M., Chen X. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front. Genet. 2019;10:1096. doi: 10.3389/fgene.2019.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li J., Li Z., Jiang P., Peng M., Zhang X., Chen K., Liu H., Bi H., Liu X., Li X. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J. Exp. Clin. Cancer Res. 2018;37:177. doi: 10.1186/s13046-018-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Li Z., Yanfang W., Li J., Jiang P., Peng T., Chen K., Zhao X., Zhang Y., Zhen P., Zhu J., Li X. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237–250. doi: 10.1016/j.canlet.2018.04.035. [DOI] [PubMed] [Google Scholar]
- 183.Dai X., Chen C., Yang Q., Xue J., Chen X., Sun B., Luo F., Liu X., Xiao T., Xu H. Exosomal circRNA_100284 from arsenite-transformed cells, via microRNA-217 regulation of EZH2, is involved in the malignant transformation of human hepatic cells by accelerating the cell cycle and promoting cell proliferation. Cell Death Dis. 2018;9:454. doi: 10.1038/s41419-018-0485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chen W., Quan Y., Fan S., Wang H., Liang J., Huang L., Chen L., Liu Q., He P., Ye Y. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119–128. doi: 10.1016/j.canlet.2020.01.022. [DOI] [PubMed] [Google Scholar]
- 185.Wang G., Liu W., Zou Y., Wang G., Deng Y., Luo J., Zhang Y., Li H., Zhang Q., Yang Y., Chen G. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine. 2019;40:432–445. doi: 10.1016/j.ebiom.2018.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Giampietro C., Deflorian G., Gallo S., Di Matteo A., Pradella D., Bonomi S., Belloni E., Nyqvist D., Quaranta V., Confalonieri S. The alternative splicing factor Nova2 regulates vascular development and lumen formation. Nat. Commun. 2015;6:8479. doi: 10.1038/ncomms9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wang S., Hu Y., Lv X., Li B., Gu D., Li Y., Sun Y., Su Y. Circ-0000284 arouses malignant phenotype of cholangiocarcinoma cells and regulates the biological functions of peripheral cells through cellular communication. Clin. Sci. (Lond.) 2019;133:1935–1953. doi: 10.1042/CS20190589. [DOI] [PubMed] [Google Scholar]
- 188.Li L., Li W., Chen N., Zhao H., Xu G., Zhao Y., Pan X., Zhang X., Zhou L., Yu D. FLI1 exonic circular RNAs as a novel oncogenic driver to promote tumor metastasis in small cell lung cancer. Clin. Cancer Res. 2019;25:1302–1317. doi: 10.1158/1078-0432.CCR-18-1447. [DOI] [PubMed] [Google Scholar]
- 189.Zong Z.H., Du Y.P., Guan X., Chen S., Zhao Y. circWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J. Exp. Clin. Cancer Res. 2019;38:437. doi: 10.1186/s13046-019-1437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Li T., Sun X., Chen L. Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J. Cell. Biochem. 2020;121:2118–2126. doi: 10.1002/jcb.28239. [DOI] [PubMed] [Google Scholar]
- 191.Chen X., Chen R.X., Wei W.S., Li Y.H., Feng Z.H., Tan L., Chen J.W., Yuan G.J., Chen S.L., Guo S.J. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin. Cancer Res. 2018;24:6319–6330. doi: 10.1158/1078-0432.CCR-18-1270. [DOI] [PubMed] [Google Scholar]
- 192.Gheinani A.H., Vögeli M., Baumgartner U., Vassella E., Draeger A., Burkhard F.C., Monastyrskaya K. Improved isolation strategies to increase the yield and purity of human urinary exosomes for biomarker discovery. Sci. Rep. 2018;8:3945. doi: 10.1038/s41598-018-22142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Coumans F.A.W., Brisson A.R., Buzas E.I., Dignat-George F., Drees E.E.E., El-Andaloussi S., Emanueli C., Gasecka A., Hendrix A., Hill A.F. Methodological guidelines to study extracellular vesicles. Circ. Res. 2017;120:1632–1648. doi: 10.1161/CIRCRESAHA.117.309417. [DOI] [PubMed] [Google Scholar]
- 194.Baroni S., Romero-Cordoba S., Plantamura I., Dugo M., D’Ippolito E., Cataldo A., Cosentino G., Angeloni V., Rossini A., Daidone M.G., Iorio M.V. Exosome-mediated delivery of miR-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis. 2016;7:e2312. doi: 10.1038/cddis.2016.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Liu J., Ding D., Jiang Z., Du T., Liu J., Kong Z. Long non-coding RNA CCAT1/miR-148a/PKCζ prevents cell migration of prostate cancer by altering macrophage polarization. Prostate. 2019;79:105–112. doi: 10.1002/pros.23716. [DOI] [PubMed] [Google Scholar]
- 196.Jenjaroenpun P., Kremenska Y., Nair V.M., Kremenskoy M., Joseph B., Kurochkin I.V. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ. 2013;1:e201. doi: 10.7717/peerj.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang Z., Xing T., Chen Y., Xiao J. Exosome-mediated miR-200b promotes colorectal cancer proliferation upon TGF-β1 exposure. Biomed. Pharmacother. 2018;106:1135–1143. doi: 10.1016/j.biopha.2018.07.042. [DOI] [PubMed] [Google Scholar]
- 198.Li Z., Jiang P., Li J., Peng M., Zhao X., Zhang X., Chen K., Zhang Y., Liu H., Gan L. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene. 2018;37:3822–3838. doi: 10.1038/s41388-018-0237-9. [DOI] [PubMed] [Google Scholar]