Abstract
Reticuloendotheliosis virus (REV) is an avian retrovirus that causes an oncogenic, immunosuppressive, and runting-stunting syndrome in avian hosts. The co-infection of REV and Marek’s disease virus (MDV), an oncogenic herpesvirus in chickens, further increases disease severity and reduces MDV vaccine efficacy. The clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system has successfully been used against pathogens in mammalian cells. However, the large size of the CRISPR-Cas9 coding sequences makes its in vivo delivery challenging. Here, following the design of a panel of single-guided RNAs targeting REV, we demonstrate that CRISPR/Cas9 can efficiently mediate the editing of the long terminal repeats of REV, resulting in the inhibition of viral protein expression. The CRISPR-Cas9 system disrupts the integrated proviral genome and provides defense against new viral infection and replication in chicken cells. Moreover, by constructing recombinant MDV carrying CRISPR-Cas9 components using an attenuated MDV vaccine strain as the vector, we efficiently delivered the CRISPR-Cas9 system into chickens, and the MDV-delivered CRISPR-Cas9 drastically reduced REV viral load and significantly diminished REV-associated symptoms. To our knowledge, this is the first study establishing avian retrovirus resistance in chickens utilizing herpesvirus-delivered CRISPR-Cas9, which provides a novel and effective strategy against viral infections.
Keywords: avian retrovirus, Marek’s disease virus, viral infection, CRISPR-Cas9, vaccine delivery
Graphical Abstract
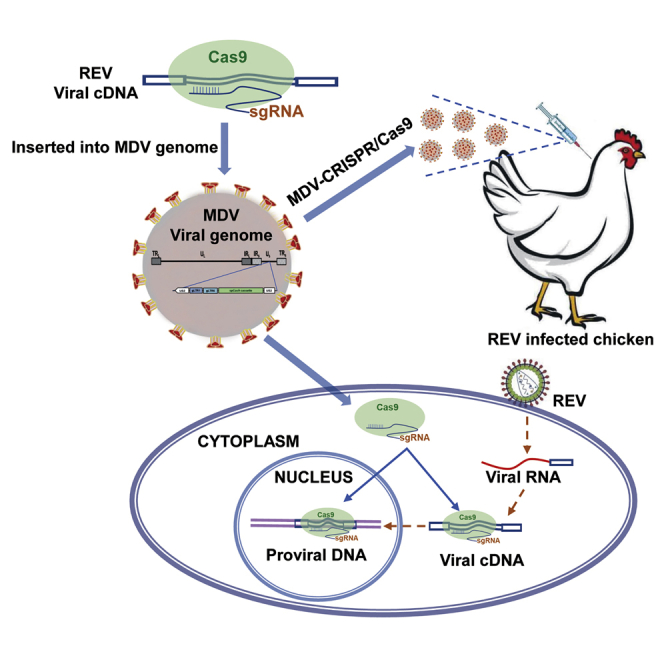
The CRISPR-Cas9 system has successfully been used against pathogens in mammalian cells. However, the large size of the CRISPR-Cas9 coding sequences makes its in vivo delivery challenging. This study established avian retrovirus resistance in chickens utilizing herpesvirus-delivered CRISPR-Cas9, which provides a novel and effective strategy against viral infections.
Introduction
Reticuloendotheliosis virus (REV) is an avian retrovirus that infects multiple avian hosts, resulting in reticuloendotheliosis (RE), an avian disease mainly characterized by immunosuppression, runting-stunting syndrome, and chronic lymphomas.1,2 Along with Marek’s disease virus (MDV) and avian leukosis virus, REV represents a third distinct group of avian viral neoplasms.3 REV early infection typically causes atrophy of thymus and bursa of Fabricius, resulting in poor immune responses to other avian vaccines and an increased susceptibility to additional infections.4,5 REV is transmitted both horizontally and vertically,6 and can be present as a contaminant in a variety of poultry biologics and vaccines.7,8 Previous studies have reported REV antibodies in 3.3%–25% of chicken flocks,9 and REV seropositive rate has reached 30%–40% in some parts of China.10, 11, 12 However, no commercial REV vaccine is currently available.4
Recently, an engineered clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system has been developed for genome editing in mammalian cells.13, 14, 15 It uses a single-guided RNA (sgRNA) that directs the nuclease Cas9 to the complementary target sequence present immediately upstream of a protospacer adjacent motif (PAM) sequence of NGG. Upon the guidance of sgRNA, Cas9 protein cleaves the targeted DNA at a position three nucleotides from the PAM, resulting in double-stranded breaks (DSBs). The DSBs are repaired by the error-prone non-homologous end joining machinery, generating sequence changes and disturbing the function of target DNA. The CRISPR-Cas9 system has been successfully applied against pathogens including hepatitis B virus,16,17 human papillomavirus,18,19 Epstein-Barr virus,20,21 and HIV-1.22, 23, 24, 25, 26, 27 While this system is promising, the large size of the CRISPR-Cas9 coding sequences makes its in vivo delivery difficult.28
Marek’s disease virus is a highly pathogenic and oncogenic herpesvirus, which causes Marek’s disease (MD), a highly contagious malignant T cell lymphomatosis in chickens.29 Vaccines based on the attenuated MDV serotype 1 have been used for protection against MD.30 In addition to the losses caused by REV or MDV infection alone, co-infection of REV and MDV has more severe implications in chickens.31, 32, 33, 34 Epidemiological studies showed that the REV-positive rate in MDV-positive clinical samples ranged from 11.7% to 16.4% annually between 2010 and 2016.4 It has been reported that immune responses to the MD vaccine were drastically reduced by REV contamination,35 and that REV and MDV co-infection significantly increased disease severity and reduced MD vaccine efficacy.4 Additionally, the long terminal repeat (LTR) region of REV could be integrated into MDV genome during REV and MDV co-infection, which increases the potential for MDV transmission.36, 37, 38
The aim of this study was to assess sgRNAs targeting the REV genome and identify targets that prevent REV infection. MDV has a large genome with several regions that are suitable for the insertion of foreign genes, making it attractive for the development of live viral vectored vaccines for poultry diseases.39, 40, 41 Moreover, the co-infection and integration of REV LTR into MDV genome indicates that REV and MDV could infect the same host cells in vivo. We therefore investigated the potential of MDV as a CRISPR-Cas9 delivery system in chickens. The approach presented here may provide a new strategy against avian retroviral infectious diseases.
Results
Efficient Targeting of the REV Genome with Single and Dual gRNAs
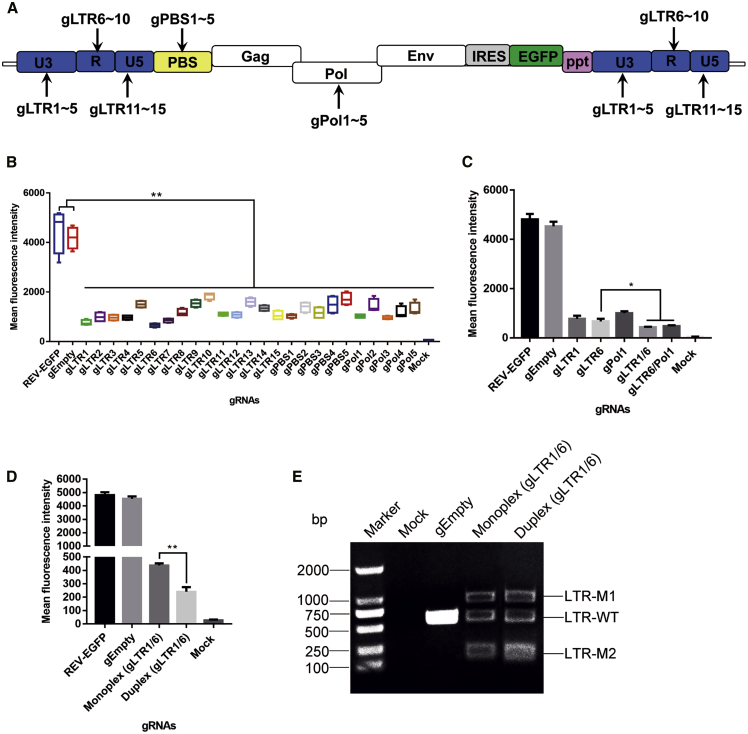
The REV proviral sequence contains two identical LTRs, each with a typical retroviral U3-R-U5 organization, which is critical for retrovirus expressional regulations (Figure 1A). The 5′ LTR of REV is followed by a primer binding site (PBS) leader region where the RNA polymerase attaches for initiation of transcription of the whole genome RNA. By efficiently predicting and excluding off-target effects on the chicken genome, we selected 25 top-ranked sgRNAs among the candidates targeting LTR, PBS, and the highly conserved Pol gene (Figure 1A; Table S1). To test their antiviral activity, we transfected DF-1 cells, a chicken fibroblast cell line, with plasmids expressing REV-EGFP, Cas9, and one of the REV-targeting sgRNAs. 5 days after transfection, the mean fluorescence intensity (MFI) of GFP expression was analyzed by flow cytometry. In DF-1 cells, a significant reduction in GFP expression was observed by the REV-specific gRNAs and Cas9 due to gRNA-Cas9-induced cleavage of the REV-EGFP plasmid (Figure 1B). However, this reduction did not occur when we used the empty gRNA vector with Cas9 (Figure 1B). We also noted that gRNAs targeting the R region (especially gLTR6) were more effective than others, indicating the importance of the LTR-R region in REV expression.
Figure 1.
Screening of Effective gRNAs-Cas9 Targeting REV Proviral DNA
(A) Proviral DNA of reticuloendotheliosis virus (REV) reporter virus with the position of gRNAs tested in this study. (B) Flow cytometry analyses of the mean fluorescence intensity of EGFP in DF-1 cells transfected with a REV-EGFP reporter together with plasmids expressing Cas9 and a variety of gRNAs against the REV genome. (C) Comparison of antiviral disruption with single or double gRNAs performed in DF-1 cells transfected with REV-EGFP. (D) Duplex gRNAs-Cas9 in an all-in-one vector exhibited stronger inhibition of EGFP expression from the REV genome. (E) PCR genotyping of gLTR1/6 using primers to amplify the DNA fragment covering the REV LTR U3/R/U5 regions. ∗p < 0.05; ∗∗p < 0.01.
To investigate the effect of dual gRNAs targeting the REV genome, we paired gLTR6 targeting the R region with a second sgRNA targeting either the U3 region (gLTR1) or the Pol gene (gPol1). As shown in Figure 1C, combinations of different gRNAs further reduced GFP expression from plasmid REV-EGFP compared to the single gRNA. In addition, to ensure maximum efficiency of REV excision and suppression, we constructed an all-in-one plasmid expressing a pair of sgRNA (gLTR1 and gLTR6) together with Cas9. The duplex gRNAs/Cas9 induced a more complete reduction of GFP levels compared to the co-transfection of two separate single gRNA-Cas9, which is likely due to the co-expression of these three components in the same cell (Figure 1D). Cleavage of the REV-EGFP proviral genome was confirmed by PCR genotyping, which showed a clear reduction in the wild-type band generated by PCR primers across the LTR (LTR-WT; Figure 1E). Moreover, the combined expression of gLTR1 and gLTR6 in the cells yields an 887 bp fragment via deletion of the entire 5′ to 3′-LTR-spanning viral genome caused by gLTR1/6-guided cleavage at both LTRs (LTR-M1), and a residual 221 bp fragment via excision of a 322 bp LTR sequence between the gLTR1- and gLTR6-targeting sites (LTR-M2; Figure 1E). Taken together, these results indicate that the selected gRNAs can efficiently target REV genome and inhibit viral gene expression.
Disruption of Integrated REV Provirus in AVOL-2 Cells by CRISPR-Cas9
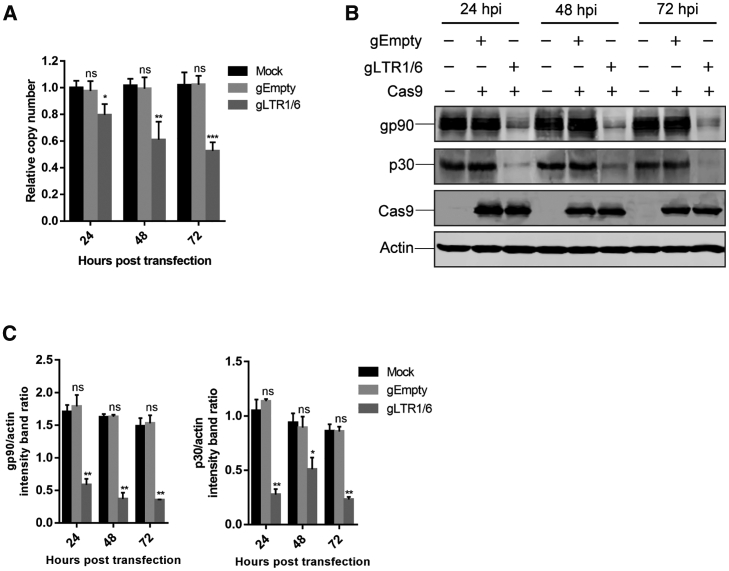
To examine whether the REV proviral genome can be disrupted from the cellular genome by CRISPR-Cas9, we used AVOL-2 cells that harbored integrated REV proviral DNA, as a model. We transfected AVOL-2 cells with a duplex gRNAs and Cas9-EGFP expression plasmid, and the positively transfected cells were collected by flow cytometry for further analyses. The results of quantitative real-time PCR showed that the relative amount of REV proviral DNA was greatly decreased by the anti-LTR gRNAs, but not gRNA-empty CRISPR-Cas9 treatment (Figure 2A). Correspondingly, a clear reduction in viral gene expression was observed in AVOL-2 cells transfected with the duplex gRNAs/Cas9; in comparison, the empty gRNA-Cas9 exerted no effect on viral expression (Figures 2B and 2C). These results indicate that the CRISPR-Cas9 system can disrupt the integrated proviral DNA and inhibit viral protein expression from the host cell chromosome.
Figure 2.
Disruption of Integrated REV Provirus with CRISPR-Cas9
AVOL-2 cells that harbored integrated REV proviral DNA were transfected with plasmids expressing duplex gRNAs (gLTR1/6) and Cas9-EGFP. The positively transfected cells were collected by flow cytometry at 24 h intervals post-transfection for further analyses. (A) REV proviral copy number quantitation by quantitative real-time PCR. (B) Western blot analyses of REV gp90 and p30 expression at different days after transfection of gRNAs-Cas9 into AVOL-2 cells. β-actin was used as a protein loading control. (C) The band intensity ratios of gp90/actin and p30/actin were normalized to the control. ns, no significant difference; ∗p < 0.05; ∗∗p < 0.01.
Inhibition of REV Replication by CRISPR-Cas9
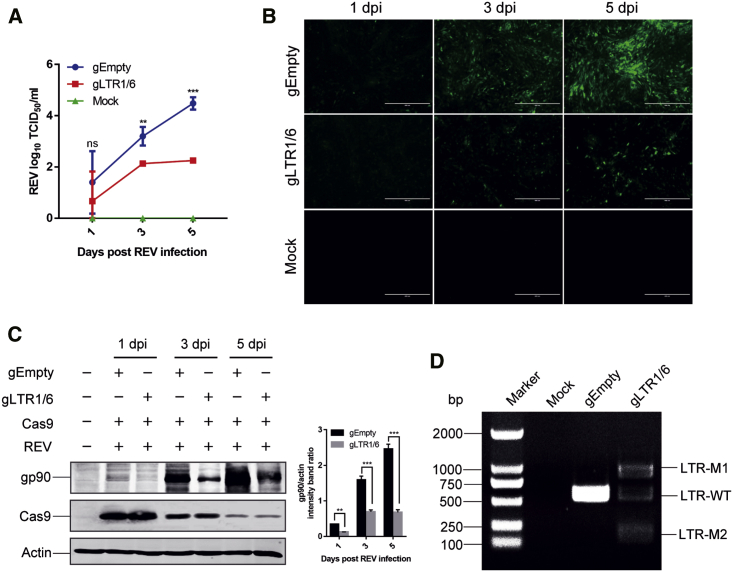
We next tested whether the duplex gRNAs-Cas9 system can protect cells against REV infection. As shown in Figure 3A, although REV infection increased over time in the control gRNA group, the cells with the duplex gRNAs targeting the U3 and R regions of the REV LTR maintained a low virus titer during infection, indicating that they were immunized effectively against new REV infection. The results further showed that the number of REV-infected cells and the viral gene expression was markedly reduced by gLTR1/6 together with Cas9, compared to the controls (Figures 3B and 3C). In addition, PCR genotyping exhibited efficient excision of the entire 5′ to 3′-LTR-spanning viral genome (LTR-M1) or the 322 bp gLTR1/6 site-spanning LTR fragment (LTR-M2; Figure 3D). The results suggest that the duplex gRNAs-Cas9 system is highly effective in suppressing REV replication in chicken cells.
Figure 3.
Inhibition of REV Replication in CEFs by CRISPR-Cas9
Chicken embryo fibroblasts (CEFs) were transfected with Cas9 together with gLTR1/6 or empty gRNA plasmids for 24 h and then infected with REV. (A) The kinetics of REV replication was tested in CEFs. (B) The REV-infected CEFs and mock control cells were stained with anti-gp90 antibody and gp90 expression was detected by an immunofluorescence assay. (C) Western blot analysis of gp90 expression from CEFs treated with CRISPR-Cas9. (D) PCR amplification of REV proviral DNA with primers flanking the LTR region. ns, no significant difference; ∗∗p < 0.01; ∗∗∗p < 0.001.
Delivery of the CRISPR-Cas9 System by Marek’s Disease Virus
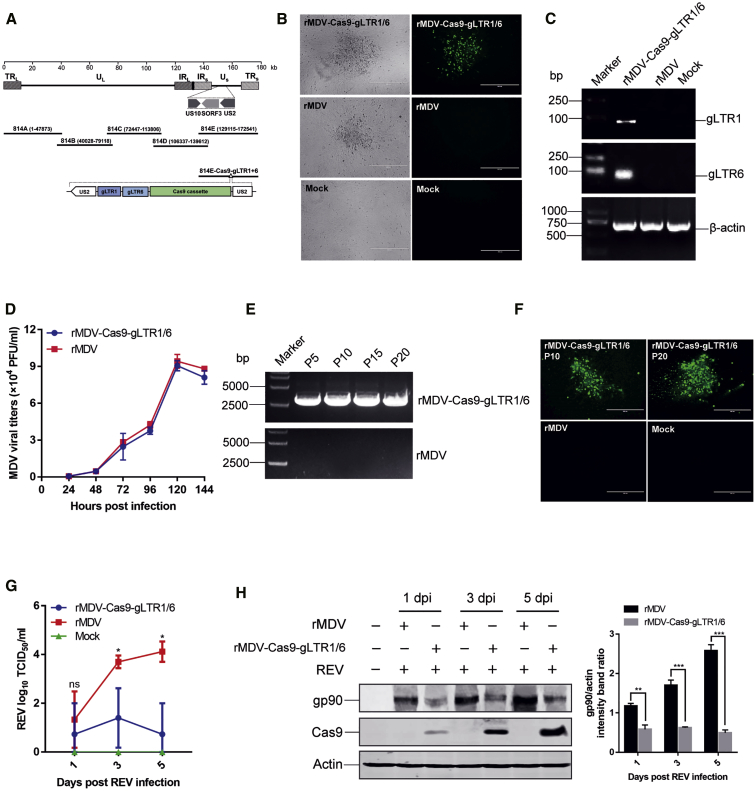
Considering the co-infection of REV and MDV in the same cells in vitro and in vivo, and the superiority of the herpesvirus vector in bearing the insertion of a large size of foreign genes, we sought to deliver the CRISPR-Cas9 components using an attenuated MDV vaccine strain (814 strain) that has been widely used in the field for MD prevention for decades with proven safety and efficacy.39 With a previously constructed fosmid-based rescue system, the duplex gRNAs and Cas9 expression cassettes were successfully inserted into the US2 site of the MDV genome (Figure 4A). Apparent cytopathic effects were observed at 3 days post-transfection (Figure 4B). Following infection of chicken embryo fibroblasts (CEFs) with the recombinant virus, Cas9 and gRNAs expression were confirmed by immunofluorescence and RT-PCR, respectively (Figures 4B and 4C). Further analysis demonstrated that the growth kinetics and magnitude of the recombinant MDV containing CRISPR-Cas9 were very similar to those of the parental virus (Figure 4D), indicating that the insertion of CRISPR-Cas9 in MDV genome did not affect the replication of the parental MDV vaccine strain. PCR amplification and sequencing of CRISPR-Cas9 inserted in the recombinant MDV passaged 20 times confirmed that the recombinant MDV had good genetic stability (Figure 4E). Cas9 expression from the serially passaged viruses was also confirmed by immunofluorescence with anti-Cas9 antibodies (Figure 4F).
Figure 4.
Construction and Evaluation of Recombinant MDV Expressing REV-Targeting CRISPR-Cas9
(A) Diagram of the recombinant Marek’s disease virus (MDV) vector carrying CRISPR-Cas9 components. (B) A typical cytopathic effect caused by the recombinant MDV in CEFs (left), and Cas9 expression from rMDV-Cas9-gLTR1/6 detected through immunofluorescence using an anti-Cas9 antibody (right). Scale bar, 400 μm. (C) RT-PCR-based detection of gLTR1, gLTR6, and β-actin RNA in cells inoculated with recombinant MDV expressing gRNAs and Cas9. β-actin is used as the RNA loading control. (D) Comparison of the replication kinetics of the recombinant MDV and the parental virus in cell cultures. (E) PCR amplification of the Cas9 gene from the recombinant MDV passaged 5, 10, 15, and 20 times in CEFs. (F) Detection of Cas9 expression from the recombinant virus passaged 10 or 20 times in CEFs with indirect immunofluorescence assay. (G and H) CEFs were first inoculated with rMDV-Cas9-gLTR1/6 or the parental MDV for 24 h, and then infected with REV. The kinetics of REV replication were tested in CEFs (G). REV gp90 expression in CEFs treated with CRISPR-Cas9 was detected by western blotting (H). β-actin was used as a protein loading control. The band intensity ratio of gp90/actin was normalized to the control. ns, no significant difference; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
To test whether the duplex gRNAs-Cas9 expressed by the recombinant MDV can confer REV resistance, we first inoculated CEFs with rMDV-Cas9-gLTR1/6 and then infected them with WT REV. The results showed that the REV titers increased gradually in the control cells inoculated with the parental MDV (rMDV), while the REV replication in CEFs inoculated with rMDV-Cas9-gLTR1/6 was effectively reduced compared to the controls (Figure 4G). Correspondingly, the viral gene gp90 expression in cells with rMDV-Cas9-gLTR1/6 was markedly decreased compared to cells inoculated with the parental MDV (Figure 4H). These results indicated that the recombinant MDV expressing REV-targeting gRNAs-Cas9 could sufficiently inhibit REV replication in cells.
Inhibition of REV Infection in Chickens by MDV-Delivered CRISPR-Cas9
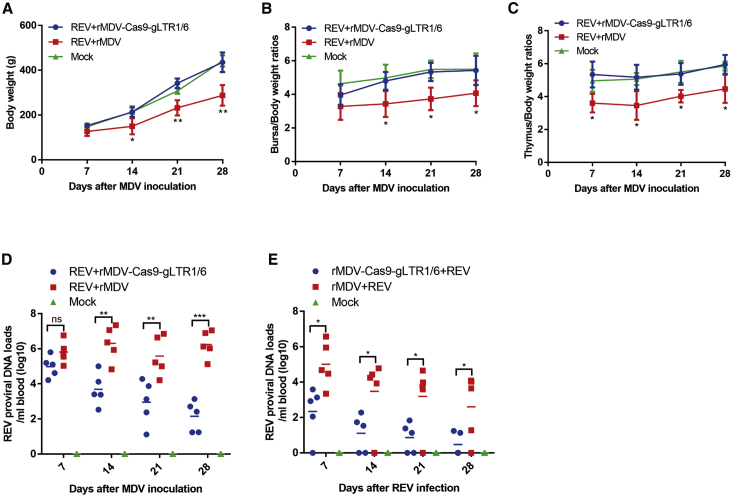
We next examined whether the duplex gRNAs-Cas9 delivered by the MDV vector could prevent against REV infection in chickens. In animal experiment 1, 1-day-old specific-pathogen-free (SPF) chicks were first challenged with REV and then inoculated with rMDV-Cas9-gLTR1/6 or the WT parental MDV 7 days later. As shown in Figure 5A, the chickens infected with REV at 1 day of age and inoculated with the parental MDV later had significantly slower weight gain than the mock controls; however, inoculation of rMDV-Cas9-gLTR1/6 relieved the typical runting-stunting syndrome caused by REV infection, resulting in a similar weight gain as the mock control birds. Compared to the mock controls, the chickens challenged with REV and inoculated with the parental MDV showed atrophied bursae and thymuses, while these signs were not observed in the rMDV-Cas9-gLTR1/6 group (Figures 5B and 5C). Moreover, inoculation with rMDV-Cas9-gLTR1/6 clearly reduced the viremia rate and viral shedding after REV infection, compared to the challenge controls (Table 1). In addition, quantitative real-time PCR analyses demonstrated that the REV proviral DNA loads were decreased in chickens inoculated with rMDV-Cas9-gLTR1/6 (Figure 5D). These results suggested that rMDV-Cas9-gLTR1/6 could inhibit REV replication in the REV pre-infected chickens and prevent the symptoms caused by REV early infection.
Figure 5.
Prevention of REV Infection by MDV-Delivered CRISPR-Cas9 in Chickens
(A–D) 1-day-old chicks were first challenged with REV and then inoculated with rMDV-Cas9-gLTR1/6 or the parental MDV (rMDV) 7 days later. The body weights (A), bursa/body (B), and thymus/body (C) weight ratios were monitored after MDV inoculation. The REV proviral DNA loads in blood samples were detected by quantitative real-time PCR (D). (E) 1-day-old chicks were first inoculated with rMDV-Cas9-gLTR1/6 or the parental MDV and then challenged with REV at 8 days of age. The REV proviral DNA loads in blood samples were detected by quantitative real-time PCR following the REV challenge. ns, no significant difference; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Table 1.
Detection of Viremia and Viral Shedding in Chickens Pre-challenged with REV and Inoculated with rMDV-Cas9-gLTR1/6 Latera
| Group | REV Viremiab |
REV Sheddingc |
||||||
|---|---|---|---|---|---|---|---|---|
| 7d | 14 | 21 | 28 | 7 | 14 | 21 | 28 | |
| REV+rMDV-Cas-gLTR1/6 | 7/10 | 4/10 | 3/10 | 3/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| REV+rMDV | 8/10 | 8/10 | 6/10 | 7/10 | 2/10 | 3/10 | 0/10 | 0/10 |
| Mock | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
Chickens were pre-challenged with REV at 1 day of age before inoculation with rMDV-Cas9-gLTR1/6 or the WT parental MDV (rMDV) at 8 days of age. Plasmas and vaginal swabs were collected after MDV inoculation for REV viremia and shedding detection.
Ratios of REV viremic birds to total detected birds.
Ratios of REV shedding birds to total detected birds.
Days after MDV inoculation.
In animal experiment 2, we first inoculated 1-day-old chicks with the recombinant MDV or the parental MDV, and then challenged them with REV at 8 days of age. Due to the age resistance of chickens against REV, we did not observe the runting-stunting syndrome and bursa/thymus atrophy, as well as viral shedding in both groups of chickens after the REV challenges (Figure S1; Table 2). However, the viremia rate and the proviral DNA loads were notably reduced in chickens inoculated with rMDV-Cas9-gLTR1/6, compared to the parental MDV-inoculated group (Table 2; Figure 5E). These results indicated that the duplex anti-LTR gRNAs-Cas9 delivered by MDV vector could also immunize chickens against REV infection.
Table 2.
Detection of REV Viremia and Viral Shedding in Chickens Inoculated with rMDV-Cas9-gLTR1/6 and Challenged with REV Latera
| Group | REV Viremiab |
REV Sheddingc |
||||||
|---|---|---|---|---|---|---|---|---|
| 7d | 14 | 21 | 28 | 7 | 14 | 21 | 28 | |
| rMDV-Cas-gLTR1/6+REV | 2/10 | 1/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| rMDV+REV | 6/10 | 4/10 | 1/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| Mock | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
Chickens were inoculated with rMDV-Cas9-gLTR1/6 or the WT parental MDV (rMDV) at 1 day of age, and challenge with REV at 8 days of age. Plasmas and vaginal swabs were collected after REV challenge for viremia and viral shedding detection.
Ratios of REV viremic birds to total detected birds.
Ratios of REV shedding birds to total detected birds.
Days after REV challenge.
Discussion
Poultry accounts for about 30% of total meat production worldwide and is an important component of global food security and agricultural economies. RE, an oncogenic and runting disease in multiple avian hosts, causes severe damages to the poultry industry.2,9 REV infection causes severe immunosuppression, resulting in poor immune responses to vaccines against other avian viruses, such as avian influenza virus that can infect humans.42,43 As an avian retrovirus, REV reverse transcribes its single-stranded RNA genome into double-stranded DNA and the resulting viral DNA is then integrated into the host genome as a latent provirus;44 these features have created an obstacle in effective vaccine development. The CRISPR-Cas9 technology is widely used in the fields of virus infection, genetic diseases, and cancer.45, 46, 47 Here, we found that LTR-directed gRNA-Cas9 eradicates the REV proviral genome and immunizes target cells against REV infection; the CRISPR-Cas9 components were efficiently delivered to chickens using a recombinant herpesvirus vector. Our results suggest that CRISPR-Cas9 can be engineered to provide an efficacious preventive and therapeutic approach against retrovirus infection in chickens.
We designed a panel of sgRNA targeting the REV LTR and structural regions. In general, targeting the LTR-R region has a higher impact on REV expression, which is consistent with the critical role of LTR for retrovirus expressional regulations by the transcriptional machinery. Previous studies demonstrated that a combination of two gRNAs can diminish the chances of HIV-1 escaping gRNA targeting.48,49 In this study, we confirmed that using a combination of two antiviral gRNAs targeting the REV LTR enhances the inhibition of REV expression in host cells, compared to single gRNA. With this duplex gRNAs-Cas9 system, we successfully excised the REV pro-viral DNA in the infected cells, thereby efficiently disrupting REV expression. The CRISPR-Cas9-mediated inhibition of REV infection might be attributed to Cas9, which either directly targets the viral genomic DNA reverse transcribed from the viral RNA before integration into the host genome or disrupts the pro-viral DNA already integrated in the host genome. In this study, we optimized the CRISPR-Cas9 system to improve its gene-editing efficiency in chicken cells. We used a hybrid CMV enhancer/chicken β-actin promoter to control Cas9 expression which was shown to be more effective than other promoters in chicken cells.40 Moreover, we enhanced Cas9 expression in chicken cells by codon optimizing the spCas9 gene for chicken usage. Additionally, the Cas9 and sgRNA expression cassettes were inserted into the US2 site in MDV genome, which resulted in high expression of foreign genes without affecting the viral replication.
The CRISPR-Cas9 system can be readily delivered into cells in vitro; however, the large size of Cas9 limits efficient delivery of CRISPR-Cas9 in vivo.28 Lentivirus or adenovirus vectors are currently employed for delivery of CRISPR-Cas9 components.50, 51, 52 However, there are drawbacks to lentivirus or adenovirus delivery systems, as the capacity of these viral vectors is relatively small, resulting in a low titer of viruses carrying the CRISPR-Cas9 insert.28 Moreover, one cannot guarantee that the viral payload goes to the same cells infected by the targeted virus in vivo. Here, we utilized MDV as the vector to deliver the REV-targeted CRISPR-Cas9 system into chickens. Specifically, an attenuated MDV vaccine strain was selected, which is widely used in chickens for the prevention of Marek’s disease.39 As a herpesvirus, MDV has a large genome, and several regions are nonessential for viral replication, rendering it suitable for the insertion of foreign genes.39 The MDV vaccine virus can be inoculated into 1-day-old field chicks to establish an early immunity. More importantly, MDV and REV have the same tropism for lymphocytes in vivo,1,30 and previous studies demonstrated that REV LTR can be readily integrated into the MDV genome during REV and MDV co-infection in chickens, indicating that the recombinant MDV carrying the CRISPR-Cas9 system can efficiently transduce REV target cells in vivo.34,36,38
While REV is highly prevalent worldwide, there is no commercial vaccine currently available.2,11,12 REV control typically depends on the elimination of infected chickens; however, with the widespread distribution of this virus in chicken flocks, and the lack of organization in the poultry industry, controlling and eradicating REV is likely to be a very difficult and high-cost process. In our study, we demonstrated that the recombinant MDV expressing anti-REV CRISPR-Cas9 notably diminished REV replication in REV pre-infected chickens and drastically reduced the viremia and runting-stunting syndrome caused by REV early infection. Moreover, the recombinant MDV also immunized chickens against REV infection. These results suggested that the MDV-delivered CRISPR-Cas9 may provide a therapeutic approach to eliminate viral reservoirs from REV-positive chickens and be used as a novel and effective gene-editing-based vaccine against REV infection in healthy chickens. In addition, as we used an attenuated MDV vaccine strain as the delivery vector, the recombinant MDV carrying anti-REV CRISPR-Cas9 system can potentially serve as a bivalent vaccine for the prevention of MDV and REV co-infection.
To our knowledge, this is the first study to demonstrate the effective excision of proviral DNA of an avian retrovirus in chickens using multiplex gRNAs and Cas9 delivered by an all-in-one herpesvirus vector. The MDV-delivered CRISPR-Cas9 efficiently reduced REV replication in vivo and immunized chickens against REV infection. This work contributes to earlier in vitro studies that demonstrated the effective CRISPR-Cas9-mediated excision of proviral DNA from infected cells, supporting the use of this technology as a robust antiviral strategy in vivo. The strategy of using a herpesvirus vector for the delivery of CRISPR-Cas9 perhaps provides a means to eradicate and vaccinate against other pathogenic viruses in mammals and humans.
Materials and Methods
Animals and Ethics Statement
The SPF chickens and fertilized SPF chicken eggs were purchased from the State Resource Center of Laboratory Animal for Poultry (Harbin, China). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of China.53 The use of SPF eggs and chickens and the animal experiments were approved by the Animal Ethics Committee of Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences, and were performed in accordance with animal ethics guidelines and approved protocols (SYXK [Hei] 2017-009).
Virus, Cells, and Antibodies
REV strain HLJR0901 (GenBank: GQ415646) was isolated from a REV-infected layer in Heilongjiang, China. CEFs were prepared using 10-day-old SPF chicken embryos and cultured in Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies, Grand Island, NY, USA) containing 5% fetal bovine serum (FBS, Sigma-Aldrich, St. Louis, MO, USA). Chicken fibroblast cell line DF-1 (ATCC CRL-12203) was cultured in DMEM supplemented with 10% FBS. The REV-transformed cell line AVOL-2 was cultured in RPMI 1640 medium containing 10% FBS, 10% tryptose phosphate broth, 0.1% 2-mercaptoethanol, and 1% sodium pyruvate. Anti-gp90 and anti-p30 antibodies were prepared in our laboratory; anti-CRISPR-Cas9 antibody was obtained from Abcam (Shanghai, China).
gRNA Design and Plasmid Construction
The gRNAs targeting REV HLJR0901 were designed with CRISPR sgRNA designing software (https://www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design) and cloned into pGEM-T vector (Promega) under control of the human U6 polymerase III promoter from pX459-v2 (Addgene plasmid # 62988). To enhance Cas9 expression in chicken cells, we codon optimized the spCas9 gene for chicken usage and cloned into the pCAGGS vector under control of the Pec promoter (CMV enhancer/chicken β-actin promoter) with a C-terminal nuclear localization sequence (NLS). The pCAGGS-Cas9-EGFP plasmid was constructed by inserting the IRES2-EGFP fragment from the pIRES2-EGFP vector (Clontech, Mountain View, CA) into pCAGGS-Cas9 between the Cas9-NLS gene and the poly(A) sequence. A gateway entry vector pENTR-MCS was constructed by replacing the gus gene in the pENTR-gus plasmid with a multiclonal site sequence. Then, an all-in-one plasmid pENTR-Cas9-gLTR1/6 expressing a pair of sgRNA (gLTR1 and gLTR6) together with Cas9 was constructed by inserting the gLTR1, gLTR6, and Cas9 expression cassettes into pENTR-MCS between the attL1 and attL2 sequences. Plasmid pENTR-Cas9-EGFP-gLTR1/6 expressing gLTR1 and gLTR6 together with Cas9-IRES2-EGFP was constructed with the similar way.
Transfection and Flow Cytometry
DF-1 cells and CEFs were transfected with plasmids expressing Cas9 and sgRNAs together with the REV-EGFP reporter using the TransIT-X2 dynamic delivery system (Mirus, Madison, WI, USA) according to the manufacturer’s instructions. For flow cytometry analyses, DF-1 cells were trypsinized, washed with PBS, and analyzed using a flow cytometer (Cytomics TM FC 500, Beckman Coulter, Brea, CA, USA). REV-transformed AVOL-2 cells were transfected with pENTR-Cas9-EGFP-gLTR1/6 using electroporation. The EGFP-positive AVOL-2 cells were collected using a SONY SH800S cell sorter (Sony Biotechnology).
PCR and Sequencing
Genomic DNA was purified using a DNeasy tissue kit (QIAGEN, Shanghai, China). The LTR locus was amplified using the following primers LTR-F: 5′-AAT GTG GGA GGG AGC TCC GGG GGA ATG TGG GA-3′ and LTR-R: 5′-CCC CCA AAT GTT GTA CCG AAA TAC TAC G-3′. The products were resolved in 2% (wt/vol) agarose gel. The bands of interest were gel-purified and TA cloned into the pMD18-T vector (TaKaRa, Dalian, China), and the individual clones were sequenced.
Provirus Copy Number Detection
Genomic DNA extracted from AVOL-2 cells or the blood of REV-infected chickens was analyzed by quantitative real-time PCR for measuring REV proviral loads. The quantitative real-time PCR analyses were carried out using LightCycler480 (Roche, Mannheim, Germany) as described previously.54 In each run, a series of dilutions of the plasmid standard were included along with the DNA samples. The quantitation data, in terms of Cp values, were determined using the second derivative method of the LightCycler software (Roche, Mannheim, Germany). All controls and treated samples were examined in triplicate in the same plate.
REV Replication Analyses
To determine REV replication in cells treated with or without the REV-targeting CRISPR-Cas9 system, we infected CEFs in 60-mm-plates with 104 50% tissue culture infective dose (TCID50) of REV HLJR0901. Infected cell cultures were harvested at 1, 3, and 5 days post-infection, and the titer of infectious progeny was determined with TCID50 per milliliter using the Reed-Muench formula directed by an immunofluorescence assay with anti-gp90 antibodies. The mean values and standard deviations (SDs) of the data obtained from three independent experiments were calculated.
Western Blotting
The expression of Cas9 and REV viral proteins gp90 and p30 were detected by western blotting analyses with mouse anti-Cas9, anti-gp90, and anti-p30 antibodies. For western blotting, whole-cell lysates were obtained by lysing cells in NP-40 lysis buffer (Beyotime, Beijing, China). The proteins were separated by electrophoresis on 12% SDS-polyacrylamide gels, transferred onto nitrocellulose membranes, and incubated with the indicated primary antibodies and IRDye 680RD goat anti-mouse antibodies (LICOR, Lincoln, NE, USA). Images were acquired with the Odyssey infrared imaging system (LICOR, Lincoln, NE, USA).
Immunofluorescence Assay
The REV replication in cells and Cas9 expression from recombinant MDVs were detected by immunofluorescence assays. The REV-infected cells were detected with anti-gp90 antibodies at 1, 3, and 5 days post-infection, and the cells infected with recombinant MDVs were detected with anti-Cas9 antibodies at 5 days post-infection. Non-infected cells were used as a negative control. For the immunofluorescence assay, cells were washed once with PBS and fixed with ethanol for 20 min at room temperature. The fixed cells were incubated with the indicated primary antibodies and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (IgG) antibody (Sigma, St. Louis, MO, USA). After washing five times with PBS, the cells were examined by fluorescence microscopy.
Rescue of Recombinant MDV Delivering REV-Targeting CRISPR-Cas9
Five fosmid clones that contain genomic sequences spanning the entire genome of the MDV vaccine strain 814 were constructed in our preliminary studies. To simplify the insertion of foreign genes into MDV genome, we modified fosmid 814E by inserting a dual selection marker encoding a kanamycin resistance gene (KanR) and a ccdB gene flanked by attR1 and attR2 sequences into the US2 gene of MDV with the Counter-Selection BAC modification kit (Gene Bridges, Heidelberg, Germany). To insert the sgRNA and Cas9 expression cassettes into MDV genome, we mixed the entry plasmid pENTR-Cas9-gLTR1/6 expressing a pair of sgRNAs (gLTR1 and gLTR6) and Cas9 with the modified fosmid 814EUS2-KanccdB and treated with LR Clonase II enzyme (Invitrogen, Shanghai, China). The mixtures were transformed into Escherichia coli EPI300-T1. Only fosmids with the KanR-ccdB marker replaced by the gLTR1/6 and Cas9 cassettes could replicate and be selected in EPI300-T1.
To rescue the recombinant MDV, we used the five fosmid combinations with or without gLTR1/6 and Cas9 insertions that covered the entire MDV genome to transfect CEFs in 60-mm-dishes following the calcium phosphate procedure.55 The cytopathic effects-positive samples were harvested and characterized. To verify that the gLTR1/6 and Cas9 cassettes were inserted into the MDV genome at the desired sites, we analyzed the viral genomic DNA by PCR and sequencing. The Cas9 and gRNA expression was detected by immunofluorescence and RT-PCR, respectively.
Animal Experiments
In animal experiment 1, 75 1-day-old chicks were randomly divided into three groups with 25 chicks in each group. Group 1 and group 2 were challenged intraperitoneally with 104 TCID50 of REV HLJR0901 strain at 1 day of age and inoculated with rMDV-Cas9-gLTR1/6 or the parental MDV (rMDV) at 8 days of age, respectively. Chicks in group 3 were challenged and inoculated with DMEM as the mock controls. These chickens were monitored daily for signs of illness after REV challenge. At 7, 14, 21, and 28 days post-inoculation of recombinant MDV, five chickens were randomly selected from each group, weighed, humanely killed, and necropsied. After necropsy, the bursa and thymus samples were weighed for the calculation of the bursa/thymus-to-body weight ratios. The blood samples were aseptically collected in heparinized tubes at weekly intervals after MDV inoculation. The blood genomic DNA was extracted using the Blood DNA Kit (Omega, Norcross, GA, USA) and detected by real-time PCR assay.54 REV viremia was detected in CEFs using the plasma samples, through an immunofluorescence assay with anti-gp90 antibodies. In addition, vaginal swabs were collected from 10 birds on each sampling day and examined for the presence of REV by inoculation of cell cultures and detection with immunofluorescence assays.
In animal experiment 2, 75 1-day-old chicks were randomly divided into three groups. Chickens in group 1 and group 2 were first inoculated with rMDV-Cas9-gLTR1/6 or the WT parental MDV (rMDV) at 1 day of age, and then challenged with REV HLJR0901 strain at 8 days of age. Group 3 was inoculated and challenged with DMEM as mock controls. At 7, 14, 21, and 28 days post-REV challenge, five chickens were randomly selected from each group and examined as in animal experiment 1. Plasmas and vaginal swabs were collected after the REV challenge for REV viremia and viral shedding detection.
Statistical Analysis
All data were presented as the mean ± SD. One-way ANOVA was employed to evaluate the statistical differences among groups using SPSS 17.0 software (SPSS, Chicago, IL, USA). Statistical significance was set at p < 0.05 for all tests.
Author Contributions
K.L., L.G., Y.Y., V.N., and X.W. conceived the project and designed the study. K.L. and Y.L. performed the experiments and wrote the paper. Z.X., Yu Zhang, C.L, Yanping Zhang, Y.G., X.Q., and H.C. participated in critical analysis of the data. All authors reviewed and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This research was supported by grants from National Natural Science Foundation of China (31970162, 31600127, and 31761133002); China Postdoctoral Science Foundation (2015M580157); Heilongjiang Natural Science Foundation of China (QC2016042); Heilongjiang Provincial Natural Science Foundation of China (TD2019C003); and China Agricultural Research System (CARS-41-G15).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.06.009.
Contributor Information
Li Gao, Email: gaoli@caas.cn.
Xiaomei Wang, Email: wangxiaomei@caas.cn.
Supplemental Information
References
- 1.Bohls R.L., Linares J.A., Gross S.L., Ferro P.J., Silvy N.J., Collisson E.W. Phylogenetic analyses indicate little variation among reticuloendotheliosis viruses infecting avian species, including the endangered Attwater’s prairie chicken. Virus Res. 2006;119:187–194. doi: 10.1016/j.virusres.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Woźniakowski G., Frant M., Mamczur A. Avian Reticuloendotheliosis in Chickens - An Update on Disease Occurrence and Clinical Course. J. Vet. Res. (Pulawy) 2018;62:257–260. doi: 10.2478/jvetres-2018-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buscaglia C. Mixed infections of Marek’s disease and reticuloendotheliosis viruses in layer flocks in Argentina. Avian Dis. 2013;57(2, Suppl):569–571. doi: 10.1637/10398-100112-Case.1. [DOI] [PubMed] [Google Scholar]
- 4.Sun G.R., Zhang Y.P., Zhou L.Y., Lv H.C., Zhang F., Li K., Gao Y.L., Qi X.L., Cui H.Y., Wang Y.Q. Co-infection with Marek’s disease virus and reticuloendotheliosis virus increases illness severity and reduces Marek’s disease vaccine efficacy. Viruses. 2017;9:E158. doi: 10.3390/v9060158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y., Cui S., Cui Z., Chang S., Zhao P. Genome analysis and pathogenicity of reticuloendotheliosis virus isolated from a contaminated vaccine seed against infectious bursal disease virus: first report in China. J. Gen. Virol. 2016;97:2809–2815. doi: 10.1099/jgv.0.000588. [DOI] [PubMed] [Google Scholar]
- 6.Witter R.L., Salter D.W. Vertical transmission of reticuloendotheliosis virus in breeder turkeys. Avian Dis. 1989;33:226–235. [PubMed] [Google Scholar]
- 7.Fadly A.M., Witter R.L. Comparative evaluation of in vitro and in vivo assays for the detection of reticuloendotheliosis virus as a contaminant in a live virus vaccine of poultry. Avian Dis. 1997;41:695–701. [PubMed] [Google Scholar]
- 8.Liu Q., Zhao J., Su J., Pu J., Zhang G., Liu J. Full genome sequences of two reticuloendotheliosis viruses contaminating commercial vaccines. Avian Dis. 2009;53:341–346. doi: 10.1637/8579-010609-Reg.1. [DOI] [PubMed] [Google Scholar]
- 9.Cheng Z., Shi Y., Zhang L., Zhu G., Diao X., Cui Z. Occurrence of reticuloendotheliosis in Chinese partridge. J. Vet. Med. Sci. 2007;69:1295–1298. doi: 10.1292/jvms.69.1295. [DOI] [PubMed] [Google Scholar]
- 10.Qin L.T., Gao Y.L., Pan W., Deng X.Y., Sun F.F., Li K. Investigation of co-infection of ALV-J with REV, MDV, CAV in layer chicken flocks in some regions of China. Chin. J. Prev. Vet. Med. 2010;32:90–93. [Google Scholar]
- 11.Deng X.Y., Qi X.L., Gao Y.L., Qin L.T., Gao L., Wu G. Molecular characteristics of gp90 gene of 14 reticuloendotheliosis viruses isolated in China. Agric. Sci. Technol. 2011;12:1954–1957. [Google Scholar]
- 12.Peng Z., Ma C.T., Yan D., Wu Z.C., Cui Z.Z. Serological survey of the reticuloendotheliosis virus infection in China native chicken flocks. Pak. Vet. J. 2012;32:621–623. [Google Scholar]
- 13.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 16.Lin S.R., Yang H.C., Kuo Y.T., Liu C.J., Yang T.Y., Sung K.C., Lin Y.Y., Wang H.Y., Wang C.C., Shen Y.C. The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo. Mol. Ther. Nucleic Acids. 2014;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seeger C., Sohn J.A. Targeting Hepatitis B Virus With CRISPR/Cas9. Mol. Ther. Nucleic Acids. 2014;3:e216. doi: 10.1038/mtna.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy E.M., Kornepati A.V., Goldstein M., Bogerd H.P., Poling B.C., Whisnant A.W., Kastan M.B., Cullen B.R. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J. Virol. 2014;88:11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhen S., Hua L., Takahashi Y., Narita S., Liu Y.H., Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem. Biophys. Res. Commun. 2014;450:1422–1426. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 20.Wang J., Quake S.R. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc. Natl. Acad. Sci. USA. 2014;111:13157–13162. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuen K.S., Chan C.P., Wong N.M., Ho C.H., Ho T.H., Lei T., Deng W., Tsao S.W., Chen H., Kok K.H., Jin D.Y. CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human cells. J. Gen. Virol. 2015;96:626–636. doi: 10.1099/jgv.0.000012. [DOI] [PubMed] [Google Scholar]
- 22.Dampier W., Nonnemacher M.R., Sullivan N.T., Jacobson J.M., Wigdahl B. HIV excision utilizing CRISPR/Cas9 technology: attacking the proviral quasispecies in reservoirs to achieve a cure. MOJ Immunol. 2014;1:00022. doi: 10.15406/moji.2014.01.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebina H., Misawa N., Kanemura Y., Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu W., Kaminski R., Yang F., Zhang Y., Cosentino L., Li F., Luo B., Alvarez-Carbonell D., Garcia-Mesa Y., Karn J. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc. Natl. Acad. Sci. USA. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C., Guan X., Du T., Jin W., Wu B., Liu Y., Wang P., Hu B., Griffin G.E., Shattock R.J., Hu Q. Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. J. Gen. Virol. 2015;96:2381–2393. doi: 10.1099/vir.0.000139. [DOI] [PubMed] [Google Scholar]
- 26.Liao H.K., Gu Y., Diaz A., Marlett J., Takahashi Y., Li M., Suzuki K., Xu R., Hishida T., Chang C.J. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat. Commun. 2015;6:6413. doi: 10.1038/ncomms7413. [DOI] [PubMed] [Google Scholar]
- 27.Zhu W., Lei R., Le Duff Y., Li J., Guo F., Wainberg M.A., Liang C. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology. 2015;12:22. doi: 10.1186/s12977-015-0150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lino C.A., Harper J.C., Carney J.P., Timlin J.A. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25:1234–1257. doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marek J. Multiple Nervenentzündung (Polyneuritis) bei Hühnern. Dtsch. Tierarztl. Wochenschr. 1907;15:417–421. [Google Scholar]
- 30.Venugopal K. Marek’s disease: an update on oncogenic mechanisms and control. Res. Vet. Sci. 2000;69:17–23. doi: 10.1053/rvsc.2000.0396. [DOI] [PubMed] [Google Scholar]
- 31.Bao K.Y., Zhang Y.P., Zheng H.W., Lv H.C., Gao Y.L., Wang J.F., Gao H.L., Qi X.L., Cui H.Y., Wang Y.Q. Isolation and full-genome sequence of two reticuloendotheliosis virus strains from mixed infections with Marek’s disease virus in China. Virus Genes. 2015;50:418–424. doi: 10.1007/s11262-015-1191-z. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y.P., Bao K.Y., Sun G.R., Lv H.C., Cui H.Y., Gao Y.L., Wang X.M., Liu C.J. Characterization of a Gallid herpesvirus 2 strain with novel reticuloendotheliosis virus long terminal repeat inserts. Virus Genes. 2017;53:386–391. doi: 10.1007/s11262-017-1427-1. [DOI] [PubMed] [Google Scholar]
- 33.Su S., Cui N., Cui Z., Zhao P., Li Y., Ding J., Dong X. Complete genome sequence of a recombinant Marek’s disease virus field strain with one reticuloendotheliosis virus long terminal repeat insert. J. Virol. 2012;86:13818–13819. doi: 10.1128/JVI.02583-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui Z., Zhuang G., Xu X., Sun A., Su S. Molecular and biological characterization of a Marek’s disease virus field strain with reticuloendotheliosis virus LTR insert. Virus Genes. 2010;40:236–243. doi: 10.1007/s11262-009-0437-z. [DOI] [PubMed] [Google Scholar]
- 35.Bülow V.V. Immunological effects of reticuloendotheliosis virus as potential contaminant of Marek’s disease vaccines. Avian Pathol. 1977;6:383–393. doi: 10.1080/03079457708418247. [DOI] [PubMed] [Google Scholar]
- 36.Kost R., Jones D., Isfort R., Witter R., Kung H.J. Retrovirus insertion into herpesvirus: characterization of a Marek’s disease virus harboring a solo LTR. Virology. 1993;192:161–169. doi: 10.1006/viro.1993.1018. [DOI] [PubMed] [Google Scholar]
- 37.Witter R.L., Li D., Jones D., Lee L.F., Kung H.J. Retroviral insertional mutagenesis of a herpesvirus: a Marek’s disease virus mutant attenuated for oncogenicity but not for immunosuppression or in vivo replication. Avian Dis. 1997;41:407–421. [PubMed] [Google Scholar]
- 38.Sun A.J., Xu X.Y., Petherbridge L., Zhao Y.G., Nair V., Cui Z.Z. Functional evaluation of the role of reticuloendotheliosis virus long terminal repeat (LTR) integrated into the genome of a field strain of Marek’s disease virus. Virology. 2010;397:270–276. doi: 10.1016/j.virol.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 39.Li K., Liu Y., Liu C., Gao L., Zhang Y., Cui H., Gao Y., Qi X., Zhong L., Wang X. Recombinant Marek’s disease virus type 1 provides full protection against very virulent Marek’s and infectious bursal disease viruses in chickens. Sci. Rep. 2016;6:39263. doi: 10.1038/srep39263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li K., Liu Y., Liu C., Gao L., Zhang Y., Gao Y., Cui H., Qi X., Zhong L., Wang X. Effects of different promoters on the protective efficacy of recombinant Marek’s disease virus type 1 expressing the VP2 gene of infectious bursal disease virus. Vaccine. 2016;34:5744–5750. doi: 10.1016/j.vaccine.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 41.Li K., Liu Y., Liu C., Gao L., Gao Y., Zhang Y., Cui H., Qi X., Zhong L., Wang X. Evaluation of two strains of Marek’s disease virus serotype 1 for the development of recombinant vaccines against very virulent infectious bursal disease virus. Antiviral Res. 2017;139:153–160. doi: 10.1016/j.antiviral.2016.11.024. [DOI] [PubMed] [Google Scholar]
- 42.Kawamura H., Wakabayashi T., Yamaguchi S., Taniguchi T., Takayanagi N. Inoculation experiment of Marek’s disease vaccine contaminated with a reticuloendotheliosis virus. Natl. Inst. Anim. Health Q. (Tokyo) 1976;16:135–140. [PubMed] [Google Scholar]
- 43.Yoshida I., Sakata M., Fujita K., Noguchi T., Yuasa N. Modification of low virulent Newcastle disease virus infection in chickens infected with reticuloendotheliosis virus. Natl. Inst. Anim. Health Q. (Tokyo) 1981;21:1–6. [PubMed] [Google Scholar]
- 44.Rice N.R., Bonner T.I., Gilden R.V. Nucleic acid homology between avian and mammalian type C viruses: relatedness of reticuloendotheliosis virus cdna to cloned proviral DNA of the endogenous Colobus virus CPC-1. Virology. 1981;114:286–290. doi: 10.1016/0042-6822(81)90279-8. [DOI] [PubMed] [Google Scholar]
- 45.Zhang F., Wen Y., Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum. Mol. Genet. 2014;23:R40–R46. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- 46.Yin H., Xue W., Anderson D.G. CRISPR-Cas: a tool for cancer research and therapeutics. Nat. Rev. Clin. Oncol. 2019;16:281–295. doi: 10.1038/s41571-019-0166-8. [DOI] [PubMed] [Google Scholar]
- 47.Huang Z., Tomitaka A., Raymond A., Nair M. Current application of CRISPR/Cas9 gene-editing technique to eradication of HIV/AIDS. Gene Ther. 2017;24:377–384. doi: 10.1038/gt.2017.35. [DOI] [PubMed] [Google Scholar]
- 48.Lebbink R.J., de Jong D.C., Wolters F., Kruse E.M., van Ham P.M., Wiertz E.J., Nijhuis M. A combinational CRISPR/Cas9 gene-editing approach can halt HIV replication and prevent viral escape. Sci. Rep. 2017;7:41968. doi: 10.1038/srep41968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang G., Zhao N., Berkhout B., Das A.T. A Combinatorial CRISPR-Cas9 Attack on HIV-1 DNA Extinguishes All Infectious Provirus in Infected T Cell Cultures. Cell Rep. 2016;17:2819–2826. doi: 10.1016/j.celrep.2016.11.057. [DOI] [PubMed] [Google Scholar]
- 50.Heckl D., Kowalczyk M.S., Yudovich D., Belizaire R., Puram R.V., McConkey M.E., Thielke A., Aster J.C., Regev A., Ebert B.L. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schiwon M., Ehrke-Schulz E., Oswald A., Bergmann T., Michler T., Protzer U., Ehrhardt A. One-vector system for multiplexed CRISPR/Cas9 against hepatitis B virus cccDNA utilizing high-capacity adenoviral vectors. Mol. Ther. Nucleic Acids. 2018;12:242–253. doi: 10.1016/j.omtn.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding Q., Strong A., Patel K.M., Ng S.L., Gosis B.S., Regan S.N., Cowan C.A., Rader D.J., Musunuru K. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ. Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ministry of Science and Technology of China . Central People’s Government of the People’s Republic of China, Beijing; 2017. Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of China.http://www.gov.cn/gongbao/content/2017/content_5219148.htm [Google Scholar]
- 54.Li K., Gao H., Gao L., Qi X., Qin L., Gao Y., Xu Y., Wang X. Development of TaqMan real-time PCR assay for detection and quantitation of reticuloendotheliosis virus. J. Virol. Methods. 2012;179:402–408. doi: 10.1016/j.jviromet.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 55.Morgan R.W., Cantello J.L., McDermott C.H. Transfection of chicken embryo fibroblasts with Marek’s disease virus DNA. Avian Dis. 1990;34:345–351. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.