Abstract
COVID-19 is the shocking viral pandemics of this year which affected the health, economy, communications, and all aspects of social activities all over the world. Early diagnosis of this viral disease is very important since it can prevent lots of mortalities and care consumption.
The functional similarities between COVID-19 and COVID-2 in inducing acute respiratory syndrome lightened our mind to find a diagnostic mechanism based on early traces of mitochondrial ROS overproduction as lung cells’ dysfunctions induced by the virus. We designed a simple electrochemical sensor to selectively detect the intensity of ROS in the sputum sample (with a volume of less than 500 μl). Comparing the results of the sensor with clinical diagnostics of more than 140 normal and involved cases resulted in a response calibration with accuracy and sensitivity both 97%. Testing the sensor in more than 4 hospitals shed promising lights in ROS based real-time tracing of COVID-19 from the sputum sample.
Keywords: Viral pandemic, COVID-19, ROS, Sputum, Electrochemical sensor
Highlights
-
•
A novel electrotechnical diagnosis system for fast screening of COVID-19 was developed.
-
•
It has been based on early tracing of reactive oxygen species (ROS) overproduction.
-
•
MWCNTs have a crucial role in selective detection of released ROS induced by the virus.
-
•
These impressive results were achieved while the sensor declared diagnosis in less than 30 s.
1. Introduction
COVID-19 has become the main health challenge of the world since February 2020 (Nourizadeh et al., 2020). Its similar symptoms to SARS (had been found in 2003) such as respiratory syndromes convinced the scientist to name it SARS-CoV-2 (Long et al., 2020), (Shi et al., 2020), (Wu et al., 2020). But it's contagious is much more severe than SARS-CoV-2 (Petrosillo et al., 2020). Due to the reports of WHO, more than 7 million people have been involved in this disease up to June 2020. Moreover, 400000 people were died because of COVID-19 (“Coronavirus Cases: Statistics and Charts,” 2020). The non-controlled pandemic of this infection forces the researchers to develop new assays for early diagnosis or pre-screening to better quarantine the suspicious people.
One of the crucial side effects of COVID viruses in lung host cells is inducing mitochondrial ROS functions to promote viral replications (Cheng et al., 2014) as far it has been reported that cellular reactive oxygen species were markedly increased in SARS-CoV3L pro-expressing cells (Lin et al., 2006).
This would occur because mitochondrial reactive oxygen species were important for SARS-CoV 3a-induced NLRP3 inflammasome activation (Chen et al., 2019).
Dysregulation of inflammatory cytokines may be involved in lung injury and the pathogenesis of SARS-COV in respiratory system with the underlying molecular mechanisms: Nod-like receptor family, pyrin domain-containing 3 (NLRP3) has been reported to be activated by virus infection (Bauernfeind et al., 2011) and release of reactive oxygen species (ROS) from damaged mitochondria (Nakahira et al., 2011), (Shimada et al., 2012) as far activation of the NLRP3 inflammasome showed significant dependency on the generation of ROS (Shimada et al., 2012). The proposed mechanism reported for this phenomenon is that mitophagy/autophagy blockade results in the accumulation of damaged, ROS-generating mitochondria, which activates the NLRP3 inflammasome (Zhou et al., 2011). Hence, all known NLRP3 activators generate ROS which results in the secretion of IL-1β in an NLRP3-ASC-caspase-1-dependent manner in THP-1 human macrophages to fight against the viral disease (Zhou et al., 2010), (Dostert et al., 2008).
One of the most important secreted samples that might contain plenty of viral involved lung epithelium is fresh sputum (Tse et al., 2004). We designed a real-time electrochemical diagnostic system in detecting the reactive oxygen species (ROS) levels in the sputum of candidates for COVID screening either their disease had been confirmed by other assays or not. In this system, the simple integrated sensor whose working electrode covered by functionalized MWCNTs, as a well-known material in biochemical sensing (Zanganeh et al., 2016) was entered into a sputum sample and record the ROS peaks. Although there are some various ROS detection techniques and methodologies such as spectroscopic methods (Electron spin resonance or electron paramagnetic resonance) (Roubaud et al., 1998), fluorescent-dependent methods (Wang and Joseph, 1999), chemiluminescent probes (Stockert and Blázquez-Castro, 2016), etc. were developed, none of them could be applied in in-vivo ROS detection induced by the virus. Some of the reported advantages and limitations of these techniques were presented in Table S1.
By testing more than 140 candidates and comparing the peak results by Clinical judgment standards (CT Scan, CBC test, and RT-PCR) in involving or non-involving to COVID-19, calibration pattern for our sensor was achieved. The calibrated sensor then was tested on more than 30 patients and showed accuracy and sensitivity of 94% and 92% with respect to CT-Scan as the gold standard.
This sensor entered to sputum sample showed the COVID-19 involvement in suspicious cases in less than 30 s. The sensors were dispensed as a non-invasive real-time system in 4 hospitals for doing observational clinical trials (Due to Ethics committee ID: IR.TUMS.VCR.REC.1399.230).
In some respiratory diseases, such as asthma, acute pneumonia, fibrosis patients with chronic Pseudomonas aeruginosa lung infection (Kolbeck et al., 1997), and etc., there is an increase in ROS. It is worth noting that seasonal influenza (might be miss diagnosed instead of COVID-19) induces a reduction in ROS levels of immune-system (Sun and Metzger, 2014) and suppresses NADPH oxidase-dependent bacterial clearance (Shen et al., 2016). It has been shown that reduced production of TNF-α and IL-1β cytokines, which prime in respiratory burst in phagocyte (Gauss et al., 2007), (El-Benna et al., 2008), influenza-infected mice might be responsible for such decreased ROS (Sun and Metzger, 2014). While we observe increased levels of cytokines (cytokine storm) in COVID-19 patients (Vinciguerra et al., 2020) due to COVID-19's pandemic conditions with no ROS based similarity to influenza and the low rate of people with asthma and the rare onset of conventional acute respiratory pneumonia in the warm season, this sensor has a great ability at this stage to warn people suspected of having corona. In this system, we performed a calibration related to the COVID-19 by testing about 170 normal, suspicious, and virus-infected cases. It showed great promising results in the real-time screening of the people during this pandemic.
2. Materials and methods
2.1. Data collection
All patients with suspected COVID-19 provided written informed consent according to an ethically approved protocol by the institutional review board of Tehran University of Medical Science (IR.TUMS.VCR.REC.1399.230) at our central hospitals and assistant laboratories for the use of their samples. We collected and analyzed data on patients with laboratory-confirmed COVID-19 infection by real-time RT-PCR assays and clinical judgment (HR-CT, ESR, CRP, CBC, Lymphopenia, and observational symptoms). Clinical, laboratory, and radiological characteristics and diagnosis and outcomes data were collected from all patients. Fresh sputum samples prepared from 172 patients who had been admitted to the intensive care unit (ICU) and those who had not, were recorded. Our team also communicated with patients or their families to record their epidemiological and symptom data. Outcomes were also compared between patients who were hospitalized in ICU (ranged from 1 to 10 days of ICU care), patients who were hospitalized and didn't need the ICU care, non-hospitalized candidates who were tested by RT-PCR, and/or CT for COVID-19 diagnosis and normal candidates with confirmed non-involvement to COVID-19.
2.2. The electrochemical diagnostic system
The COVID-19 induced ROS detector system includes a known electrochemical ROS/H2O2 system (Patent pub. No: US, 2018/0299401 A1, Pub. Date: Oct. 18, 2018) introduced by the authors. This consists of an integrated portable automatic electrochemical readout board and a sensing disposable sensor as the main diagnostic part of the system. The sensor was fabricated by the growth of Multi-Wall Carbon Nanotubes (MWCNTs) on the tip of steel needles in the conformation of three electrodes, named Working (WE), Counter (CE), and Reference (RE), with a triangular distance of 3 mm from each other. Also, the software was designed based on experimental calibration to analyze the data and diagnose whether the responses are related to the positive or negative detection of COVID-19. This ability of the device provides a free and flexible method for the phlebotomist or physicians to utilize the device in the laboratories or clinics. For electrochemical measurements, the potential was swept in the range from −0.8 to +0.8 V, using a scan rate of 100 mV s−1 as conventional parameters in biological solutions (Shashaani et al., 2016).
2.3. Sampling method
To perform the test, first, the sputum sampling container (Falcon) was given to the candidates to throw their Fresh sputum into the Falcon. But for patients who were hospitalized in ICU, bronchoalveolar lavage (BAL) fluids were taken for tests. After sampling, the head probe sensor was connected to the probe and the needles of the head probe were entered into sputum solution. Then the test was started and just in 30 s showed the ROS levels of sputum with the calibrated response in correlation with the probability of COVID-19 involvement. We also designed and fabricated a special COVID-19 isolation booth for our test (Fig. S1).
2.4. Statistical analysis
The diagnostic results of the different methods were used in this study were presented as means ± standard deviation (Mean ± S.D.). Graph Pad Prism (V. 8.0.1) and SPSS (V. 26) software were used for statistical analysis. To assess the significance of the differences between the negative and positive diagnostic results of different groups, a statistical analysis was performed using a Kruskal-Wallis method followed by Dunn's multiple comparisons test for repeated measurements with the significance assessed at the 1% significance level (P < 0.01).
3. Results and discussions
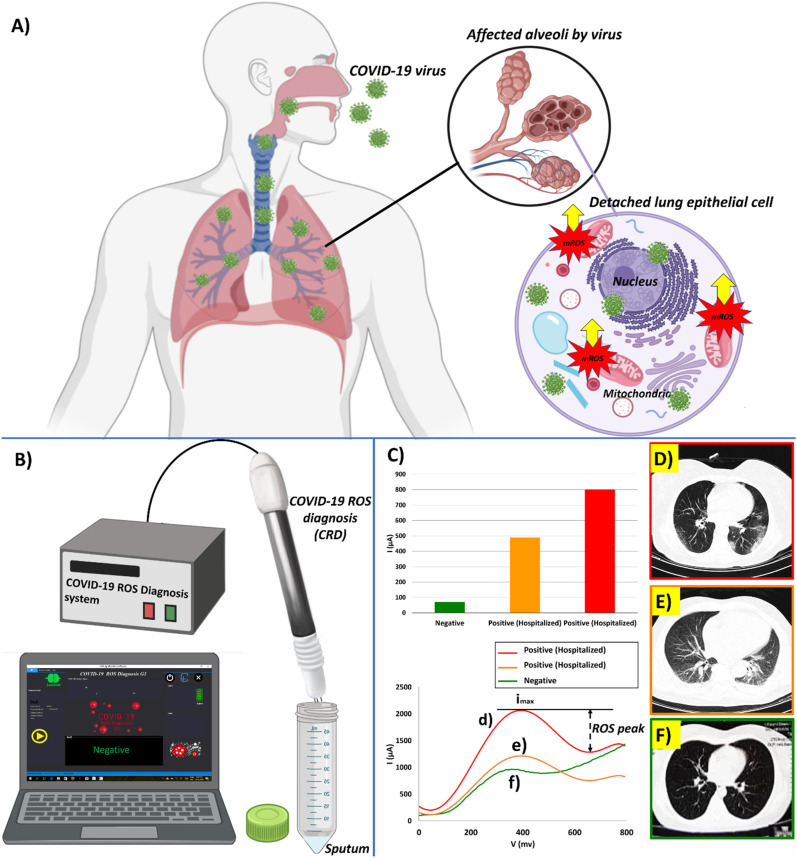
The ROS detector in the sputum sample (RDSS) which could be named as COVID-19 associated ROS diagnosis (CRD) system during this pandemic is a simple electrochemical sensor to detect the ROS levels in the sputum of people candidates for COVID-19 screening due to the production of mitochondrial ROS induced by COVID virus in respiratory epithelial host cells (Fig. 1 A). The system traces the intensity of released ROS levels in sputum by cyclic voltammetry procedure (Fig. 1B). The detection mechanism of CRD has been based on the electrochemical recording of ROS released from viral-infected lung epithelium. The released ROS would interact by the CNT electrodes. The multi-wall carbon nanotube (MWCNT) decorated electrode are well known as a selective electrochemical tracer of the super oxidant such as H2O2/ROS (Gooran, 2015),. The released electric charges due to reaction of ROS molecules on nanotube working electrodes would be transferred through counter electrode (Kumar et al., 2017), (Liu et al., 2018). Hence, CRD would present quantitative response signals in correlation with the viral infection in the sputum sample (Fig. S2). Repeatability tests on some randomly selected fabricated sensors tested on individual sputum of some normal cases and COVID-19 patients were shown in Fig. S3. Well acceptable similarity was observed in responses to the different sensors that had been tested on the same sputum (Fig. S3).
Fig. 1.
(A) Schematic of the COVID viruses side effect in lung host cells by inducing mitochondrial ROS overproduction to promote viral replications, (B) The COVID-19 ROS diagnosis (CRD) system consists of three needle electrodes coated by functionalized multi-wall carbon nanotubes, (C) Selective electrochemical reactions of released ROS on MWCNTs produces cathodic ionic peak. ROS related electrochemical cyclic voltammetry cathodic peaks from the fresh sputum of two different patients were involved to COVID-19 and hospitalized in comparison with a confirmed normal case (D), (E) The intensity of the electrochemical ROS peak currents is correlated with the amount of the viral-induced mitochondrial ROS production found in the sputum. It is meaningfully higher in the sputum sample of the patient ID 34 (I = 800 μA) than patient ID 37 (I = 490 μA) with severe lung affected by COVID-19 viruses. The CT-Scan of the patient ID 34′s lung showed more distinctive hazy patches with gross glassy opacity in both lobes of the lung, (F) Also, a normal candidate with no complaint's cases who were clinically checked by a physician in hospital and confirmed as non-COVID cases showed peak current 71 μA. The CT-Scan of this patient showed blood vessels without any viral involvement effects.
Sensor responses showed meaningful correlation with the CT-Scan results of the patients with hazy patches in their both lung lobes (COVID positive patients) showed significantly high levels of ROS cyclic voltammetry (CV) peaks (more than 490 μA) with respect candidate with normal lung (70 μA) (Fig. 1C).
The detailed information of 142 candidates who were known cases of positive and negative COVID-19 confirmed by clinical judgment (HR-CT-Scan, ESR, CRP, CBC, Lymphopenia, and observational symptoms) and RT-PCR assays, were presented in Table 1 . HR-CT was done only for suspicious patients, but other tests were done for both suspicious and non-suspicious cases.
Table 1.
Baseline characteristics and symptoms of 172 patients who were investigated in this study.
| Characteristics and symptoms | All Patients (n = 172) | ICU care (n = 25) | hospitalized without need to ICU care (n = 36) | Non-hospitalized candidates which was checked by RT-PCR or CT-Scan (n = 75) | Normal candidates with non-involvement to COVID-19 (n = 36) |
|---|---|---|---|---|---|
| Characteristics | |||||
| Age | 46.3 (21–76) | 53.7 (41–65) | 47 (21–76) | 45.3 (24–58) | 39.1 (22–60) |
| Sex | |||||
| Female | 67 (39%) | 8 (32%) | 12 (33%) | 32 (43%) | 15 (42%) |
| Male | 105 (61%) | 17 (68%) | 24 (67%) | 43 (57%) | 21 (58%) |
| Race | White | White | White | White | White |
| Current Smoking | 28 (16%) | 0 | 2 (6%) | 19 (25%) | 7 (19%) |
| Any comorbidity | |||||
| Diabetes | 30 (17%) | 9 (36%) | 7 (19%) | 12 (16%) | 2 (6%) |
| Hypertension | 14 (8%) | 6 (24%) | 3 (8%) | 5 (7%) | 0 |
| pulmonary disease | 2 (1%) | 2 (8%) | 0 | 0 | 0 |
| Cardiovascular disease | 11 (6%) | 5 (20%) | 3 (8%) | 2 (3%) | 1 (3%) |
| Chronic liver disease | 6 (3%) | 1 (4%) | 2 (6%) | 3 (4%) | 0 |
| Hypothyroidism | 21 (12%) | 3 (12%) | 2 (6%) | 13 (17%) | 3 (8%) |
| Hyperthyroidism | 7 (4%) | 1 (4%) | 0 | 6 (8%) | 0 |
| Immune deficiency disease | 1 (1%) | 1 (4%) | 0 | 0 | 0 |
| Symptoms | |||||
| Fever | 107 (62%) | 25 (100%) | 34 (94%) | 42 (56%) | 6 (17%) |
| Cough | 84 (49%) | 23 (92%) | 31 (86%) | 25 (34%) | 5 (14%) |
| Myalgia or fatigue | 75 (44%) | 22 (88%) | 29 (81%) | 22 (29%) | 2 (6%) |
| Dyspnoea | 72 (42%) | 24 (96%) | 28 (78%) | 18 (24%) | 2 (6%) |
| Diarrhoea | 4 (2%) | 0 | 2 (6%) | 2 (3%) | 0 |
| Vomit | 11 (6%) | 2 (8%) | 2 (6%) | 7 (9%) | 0 |
| Headache | 47 (27%) | 5 (20%) | 12 (33%) | 27 (36%) | 3 (8%) |
| Lack of appetite | 80 (47%) | 22 (88%) | 31 (86%) | 25 (33%) | 2 (6%) |
| Vertigo | 6 (3%) | 2 (8%) | 4 (11%) | 0 | 0 |
| Chest pain | 14 (8%) | 4 (16%) | 7 (19%) | 3 (4%) | 0 |
| Sore throat | 36 (21%) | 1 (4%) | 5 (14%) | 25 (33%) | 5 (14%) |
| Haemoptysis | 2 (1%) | 1 (4%) | 1 (3%) | 0 | 0 |
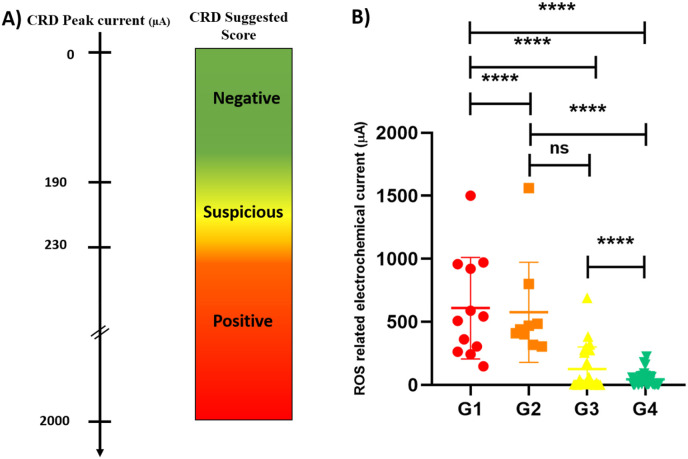
Meaningful results were achieved in a way that a calibration pattern was provided between the responses of our sensor and the clinical state of the patient. 92% of patients with severe symptoms who were hospitalized in ICU (ranged from 1 to 10 days of ICU care) showed peak currents between 230 and 1500 μA, 100% of the patients with moderate symptoms who were hospitalized with no need for ICU showed current peaks between 315 and 1560 μA (Fig. 1d,e). 83% of negative COVID cases due to RT-PCR results showed peak currents lower than 180 μA (Fig. 1f). 94% of normal candidates with no complaint cases (include some nurses and healthy people) who were clinically checked by physicians in hospitals and confirmed as non-COVID cases showed peak currents lower than 190 μA. Among all of the cases whose COVID-19 were confirmed by CBC and CT-Scan results (as a clinical gold standard), 95% showed peak currents higher than 230μA by CRD. Hence, we assumed peak currents higher than 230 μA as the positive score of CRD. Moreover, among non-hospitalized COVID free cases who were clinically checked by physicians and diagnosed as normal cases, 94% showed CRD peaks lower than 200 μA. Among the patients whose COVID-free diagnosis was just carried out by RT-PCR, 84% showed CRD current peaks lower than 200 μA. In this regard, a calibrated diagnostic cut-off based on CBC/CT-Scan gold standard results were defined for CRD scores: current peaks lower than 190 μA were scored as COVID-Free cases, between 190 and 230 μA were scored as suspicious cases who were recommended to do CT-Scan and current peaks higher than 230 μA were scored as COVID-Positive cases (Table 1, Fig. 2 , and Section S1).
Fig. 2.
(A) The calibration table of CRD current peak of 142 candidates who were known cases of positive and negative COVID-19 confirmed by clinical judgment (HR-CT, ESR, CRP, CBC, Lymphopenia, and observational symptoms), RT-PCR assays. Based on the mentioned study, the validated diagnostic ranges of the CRD for positive, suspicious, and negative ranges were obtained as higher than 230 μA, 190–230 μA, and lower than 190 μA, respectively. (B) The comparative diagnostic results of each group are expressed as mean ± SD and analyzed using a one-way ANOVA method followed by Tukey's multiple comparisons test. The p-value amount of each group was shown in the figure. Differences in mean current peak responses between involvement and non-involvement patients to COVID-19 were highly significant (G1 vs. G4: p < 0.0001, G2 vs. G4: p < 0.0001), NS: non-significant. G1, G2, G3, and G4 refers to the patients who were hospitalized in ICU (n = 25), hospitalized without need to ICU care (n = 36), Non-hospitalized candidates which were checked by RT-PCR (n = 45), and normal candidates with confirmed non-involvement to COVID-19 (n = 36), respectively.
In the next step, the donated sputum from 30 patients (collected from 4 individual medical centers) who were recommended to do CT-Scan by the physicians (due to their blood tests and physical symptoms), were tested and scored by CRD (Table 1).
CRD diagnosed 13/30 of the patients' positive COVID-19 cases which 12/13 of those cases were confirmed by CT-Scan images. Just one case was scored as suspicious, which was not confirmed as positive COVID-19 by CT-Scan. 17/30 of the cases were negatively scored by CRD among which just one case showed COVID positive CT-Scan result. So, discrepancies between the results of CRD and clinical judgment on those 30 patients were less than 7% with false negatives of less than 3% (Table 1, Table 2 and Section S2).
Table 2.
Comparative diagnostic results of the CRD system on, A) 142 candidates who were known cases of positive and negative COVID-19 confirmed by clinical judgment (HR-CT, ESR, CRP, CBC, Lymphopenia and observational symptoms) and RT-PCR assays, B) Donated sputum from 30 patients who were recommended to do CT-Scan by the physicians (due to their blood tests and physical symptoms).
| Diagnosis results | A (n = 142) | B (n = 30) |
|---|---|---|
| TP | 61 | 12 |
| TN | 72 | 16 |
| FP | 7 | 1 |
| FN | 2 | 1 |
| Accuracy | 97% | 94% |
| Sensitivity | 97% | 92% |
| Specificity | 91% | 94% |
| Selectivity | 88% | 87% |
Also, two COVID-free patients with oral disease and Purulent sore throat that may induce ROS in sputum samples were checked by CRD which showed current peaks as low as 70 μA (Patient ID 6 and 22 in Table S5). Hence, non-COVID based ROS accumulated in sputum samples might not perturb the results of CRD for COVID detection.
4. Conclusion
In summary, advanced electronic biosensors could also be helpful in the early-stage screening of COVID-19. Here an electrochemical ROS detector in the sputum sample was applied for real-time screening of people might be suspicious to COVID-19. More than 97% of true positive patients were detected while the sensor declares the diagnosis in less than 30 s.
It can be used as a power full assistant in the fast screening of the patients who need further medical examination during this pandemic and may be used in the future to reduce the number of cases that must be undergone CT-Scan for COVID-19 diagnosis.
Availability of data and materials
The authors declare that all the sources of data supporting the findings of this study are available within the article and its supplementary information files and from the corresponding author upon reasonable request. The system was fabricated and produced under the financial supports of Nano Hesgarsazan Salamat Arya Co. and Iran NANOFUND institution (USA Patent Prov. No: IP 9902). Hence, the financial and intellectual rights of the CRD product are for the company.
CRediT authorship contribution statement
Zohreh Sadat Miripour: Data curation, Formal analysis, Methodology, Investigation. Ramin Sarrami-Forooshani: Methodology, Validation, Investigation. Hassan Sanati: Investigation, Visualization. Jalil Makarem: Methodology, Validation, Investigation. Morteza Sanei Taheri: Methodology, Validation, Investigation. Fatemeh Shojaeian: Investigation, Visualization. Aida Hasanzadeh Eskafi: Investigation, Visualization. Fereshteh Abbasvandi: Methodology, Validation, Investigation. Naser Namdar: Software, Methodology, Validation. Hadi Ghafari: Methodology, Visualization. Parisa Aghaee: Formal analysis, Methodology, Visualization. Ashkan Zandi: Validation. Mahsa Faramarzpour: Visualization. Meisam Hoseinyazdi: Methodology, Validation, Investigation. Mahtab Tayebi: Methodology, Validation, Investigation. Mohammad Abdolahad: Conceptualization, Formal analysis, Investigation, Methodology, Supervision, Project administration, Writing - original draft, Writing - review & editing.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
We thank Professor Saeed Sarkar, Hani Shashaani and Mehdi Zeighami for their great support on this project, and Dr. Sahraian from Tehran University of Medical Sciences his helpful collaboration in facilitating clinical procedures.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2020.112435.
Source of funding
This research was supported by Iran NANOFUND financial institution, P.O. Box 1533984611, Tehran, Iran.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Bartok Eva, Bauernfeind Franz, Ablasser Andrea, Kim Sarah, Schmid-Burgk Jonathan, Cavlar Taner, Hornung Veit. Inflammasomes: current understanding and open questions. Cell. Mol. Life Sci. 2011;68(5):765–783. doi: 10.1007/s00018-010-0567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.-Y., Moriyama M., Chang M.-F., Ichinohe T. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M.-L., Weng S.-F., Kuo C.-H., Ho H.-Y. Enterovirus 71 induces mitochondrial reactive oxygen species generation that is required for efficient replication. PloS One. 2014;9 doi: 10.1371/journal.pone.0113234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus Cases Statistics and Charts. 2020. https://www.worldometers.info/coronavirus/?zarsrc=130
- Dostert C., Pétrilli V., Van Bruggen R., Steele C., Mossman B.T., Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science (80-.) 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Benna J., Dang P.M.-C., Gougerot-Pocidalo M.-A. Seminars in Immunopathology. Springer; 2008. Priming of the neutrophil NADPH oxidase activation: role of p47phox phosphorylation and NOX2 mobilization to the plasma membrane; pp. 279–289. [DOI] [PubMed] [Google Scholar]
- Gauss K.A., Nelson‐Overton L.K., Siemsen D.W., Gao Y., DeLeo F.R., Quinn M.T. Role of NF‐κB in transcriptional regulation of the phagocyte NADPH oxidase by tumor necrosis factor‐α. J. Leukoc. Biol. 2007;82:729–741. doi: 10.1189/jlb.1206735. [DOI] [PubMed] [Google Scholar]
- Gooran Anal. Chem. 2015;87(12):5989–5996. doi: 10.1021/acs.analchem.5b00059. [DOI] [PubMed] [Google Scholar]
- Kolbeck R.C., She Z.-W., Callahan L.A., Nosek A.N.D.T.M. Increased superoxide production during fatigue in the perfused rat diaphragm. Am. J. Respir. Crit. Care Med. 1997;156:140–145. doi: 10.1164/ajrccm.156.1.9610041. [DOI] [PubMed] [Google Scholar]
- Kumar A., Hsu L.H.-H., Kavanagh P., Barrière F., Lens P.N.L., Lapinsonnière L., Schröder U., Jiang X., Leech D. The ins and outs of microorganism–electrode electron transfer reactions. Nat. Rev. Chem. 2017;1:1–13. [Google Scholar]
- Lin C.-W., Lin K.-H., Hsieh T.-H., Shiu S.-Y., Li J.-Y. Severe acute respiratory syndrome coronavirus 3C-like protease-induced apoptosis. FEMS Immunol. Med. Microbiol. 2006;46:375–380. doi: 10.1111/j.1574-695X.2006.00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Shi L., Gu J.-D. Microbial electrocatalysis: redox mediators responsible for extracellular electron transfer. Biotechnol. Adv. 2018;36:1815–1827. doi: 10.1016/j.biotechadv.2018.07.001. [DOI] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020:1–4. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Nakahira K., Haspel J.A., Rathinam V.A.K., Lee S.-J., Dolinay T., Lam H.C., Englert J.A., Rabinovitch M., Cernadas M., Kim H.P. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011;12:222. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourizadeh M., Rasaee M.J., Moin M., Allergy J. Asthma Immunol; Iran: 2020. COVID-19 Pandemic: a Big Challenge in Iran and the World. [DOI] [PubMed] [Google Scholar]
- Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin. Microbiol. Infect. 2020;26(6):729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubaud V., Sankarapandi S., Kuppusamy P., Tordo P., Zweier J.L. Quantitative measurement of superoxide generation and oxygen consumption from leukocytes using electron paramagnetic resonance spectroscopy. Anal. Biochem. 1998;257:210–217. doi: 10.1006/abio.1997.2542. [DOI] [PubMed] [Google Scholar]
- Shashaani H., Faramarzpour M., Hassanpour M., Namdar N., Alikhani A., Abdolahad M. Silicon nanowire based biosensing platform for electrochemical sensing of Mebendazole drug activity on breast cancer cells. Biosens. Bioelectron. 2016;85:363–370. doi: 10.1016/j.bios.2016.04.081. [DOI] [PubMed] [Google Scholar]
- Shen F., Tang X., Cheng W., Wang Y., Wang C., Shi X., An Y., Zhang Q., Liu M., Liu B. Fosfomycin enhances phagocyte-mediated killing of Staphylococcus aureus by extracellular traps and reactive oxygen species. Sci. Rep. 2016;6:19262. doi: 10.1038/srep19262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., Bucci E., Piacentini M., Ippolito G., Melino G. 2020. COVID-19 Infection: the Perspectives on Immune Responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K., Crother T.R., Karlin J., Dagvadorj J., Chiba N., Chen S., Ramanujan V.K., Wolf A.J., Vergnes L., Ojcius D.M. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockert J.C., Blázquez-Castro A. Establishing the subcellular localization of photodynamically-induced ROS using 3, 3′-diaminobenzidine: a methodological proposal, with a proof-of-concept demonstration. Methods. 2016;109:175–179. doi: 10.1016/j.ymeth.2016.04.031. [DOI] [PubMed] [Google Scholar]
- Sun K., Metzger D. 2014. Influenza Infection Suppresses NADPH Oxidase-dependent Phagocytic Bacterial Clearance and Enhances Susceptibility to Secondary MRSA Infection. (MPF2P. 811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse G.M.K., Hui P.K., Ma T.K.F., Lo A.W.I., To K.F., Chan W.Y., Chow L.T.C., Ng H.K. Sputum cytology of patients with severe acute respiratory syndrome (SARS) J. Clin. Pathol. 2004;57:256–259. doi: 10.1136/jcp.2003.012948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra M., Romiti S., Greco E. Atherosclerosis as pathogenetic substrate for sars-Cov2 ‘“Cytokine storm. ’. 2020 doi: 10.3390/jcm9072095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Joseph J.A. Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radic. Biol. Med. 1999;27:612–616. doi: 10.1016/s0891-5849(99)00107-0. [DOI] [PubMed] [Google Scholar]
- Wu J.T., Leung K., Bushman M., Kishore N., Niehus R., de Salazar P.M., Cowling B.J., Lipsitch M., Leung G.M. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat. Med. 2020;26:506–510. doi: 10.1038/s41591-020-0822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanganeh S., Khodadadei F., Tafti S.R., Abdolahad M. Folic acid functionalized vertically aligned carbon nanotube (FA-VACNT) electrodes for cancer sensing applications. J. Mater. Sci. Technol. 2016;32:617–625. [Google Scholar]
- Zhou R., Tardivel A., Thorens B., Choi I., Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat. Immunol. 2010;11:136. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- Zhou R., Yazdi A.S., Menu P., Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all the sources of data supporting the findings of this study are available within the article and its supplementary information files and from the corresponding author upon reasonable request. The system was fabricated and produced under the financial supports of Nano Hesgarsazan Salamat Arya Co. and Iran NANOFUND institution (USA Patent Prov. No: IP 9902). Hence, the financial and intellectual rights of the CRD product are for the company.