Abstract
Methods that enable rapid, sensitive and specific analyses of nucleic acid sequences have positive effects on precise disease diagnostics and effective clinical treatments by providing direct insight into clinically relevant genetic information. Thus far, many CRISPR/Cas systems have been repurposed for diagnostic functions and are revolutionizing the accessibility of robust diagnostic tools due to their high flexibility, sensitivity and specificity. As RNA-guided targeted recognition effectors, Cas9 variants have been utilized for a variety of diagnostic applications, including biosensing assays, imaging assays and target enrichment for next-generation sequencing (NGS), thereby enabling the development of flexible and cost-effective tests. In addition, the ensuing discovery of Cas proteins (Cas12 and Cas13) with collateral cleavage activities has facilitated the development of numerous diagnostic tools for rapid and portable detection, and these tools have great potential for point-of-care settings. However, representative reviews proposed on this topic are mainly confined to classical biosensing applications; thus, a comprehensive and systematic description of this fast-developing field is required. In this review, based on the detection principle, we provide a detailed classification and comprehensive discussion of recent works that harness these CRISPR-based diagnostic tools from a new perspective. Furthermore, current challenges and future perspectives of CRISPR-based diagnostics are outlined.
Keywords: CRISPR, Nucleic acid detection, Biosensing assays, Imaging assays, DNA sequencing
1. Introduction
Effective identification of the presence of specific nucleic acid targets, as well as their sequence alterations, is vital for the accurate diagnosis and appropriate management of cancer, infections and genetic diseases. For example, ALK (anaplastic lymphoma kinase) gene rearrangement testing has been recommended for patients with nonsquamous non-small-cell lung cancer (NSCLC) and is a prerequisite for targeted therapy with crizotinib (Ettinger et al., 2018). The most effective method for early identification of coronavirus disease (COVID-19) has been established by examining the pathogenic sequences in respiratory tract samples, and such methods are critical for reducing the spread of disease or managing individual patients (Adhikari et al., 2020). Implementation of prenatal screening and diagnosis by analyzing genomic abnormalities in circulating fetal DNA and amniotic fluid cells is the surest way to reduce birth defects (Gray and Wilkins-Haug 2018; Malan et al., 2018). Therefore, nucleic acid detection that incorporates molecular-level information is critical for the effective management of disease. Among numerous factors that can facilitate the detection of target nucleic acids, proper detection methods are of prime importance because a high-performance diagnostic test is a necessary prerequisite for achieving reliable and timely results. Generally, the ideal nucleic acid detection method should be sensitive, specific, rapid, portable, cost-effective and easy to use.
Currently, the most widely used nucleic acid detection methods in clinical practice include quantitative real-time PCR (qPCR), fluorescence in situ hybridization (FISH) and next-generation sequencing (NGS), which have reshaped the landscape of diagnosis by allowing massive molecular-level information to be used during routine testing. Although considerable improvements have been made over the past several decades, these methods still have limitations. The qPCR methodology is the main force in current clinical laboratories and allow for quantitative and precise disease diagnostics (Arya et al., 2005; Bustin and Mueller, 2005). However, the requirement for sophisticated thermal cyclers, specialized expertise and well-established laboratory settings have weakened the generalizability of qPCR, especially in resource-limited areas where the health-care system is usually fragile. FISH analyses offer a single-cell assay that can examine the copy number, amplification, and gene rearrangements that impact targeted therapy implementation (Ettinger et al., 2018; Gradishar et al., 2018); however, the prolonged harsh heat treatment required for FISH probe hybridization makes this tool time-consuming and induces poor cell morphology, which leads to the risk of losing spatial structure information. Moreover, the toxic hazards of formamide used for dsDNA denaturation also pose a significant challenge (Levsky and Singer, 2003). NGS has opened the door to large-scale parallel sequencing capabilities, which represents a prominent trend toward personalized medicine based on an individual's genomic data (Blumenthal et al., 2016; van Dijk et al., 2014). Nevertheless, NGS is limited by many issues, such as inefficient targeted enrichment, which is responsible for elevated cost and reduced sensitivity (Ballester et al., 2016; Mamanova et al., 2010). In addition, the complex and lengthy procedures associated with this method might prevent its use in applications that require rapid results. Therefore, current approaches have not yet function as a master key for all scenarios, and new technologies are needed for optimized strategies and updated solutions. As newly emerging multifunctional toolboxes, clustered regularly interspaced short palindromic repeats (CRISPR) systems have been harnessed beyond their traditional applications of gene editing. In the past five years, CRISPR systems have been repurposed to enable the development of robust diagnostic tools for detecting nucleic acids due to their intrinsic sensitivity, specificity, flexibility and simplicity. Recent reports employing these CRISPR toolboxes have demonstrated their prospects for redefining methods of detecting nucleic acids, thus demonstrating their similarities to other revolutionary technologies, such as PCR, in their infancy.
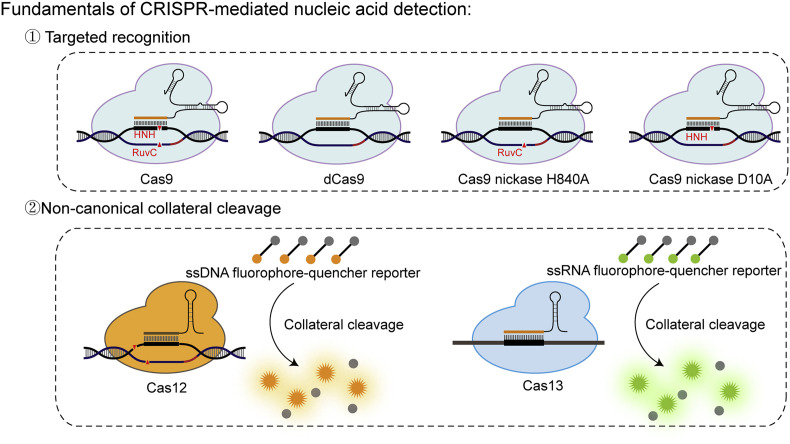
CRISPR systems function as the adaptive immune systems of bacteria and archaea against invasion elements, such as viruses or plasmids. According to the adaptive immune mechanism, invader-derived short fragments (namely, protospacers) along with their repeat flanking sequences are integrated into the CRISPR arrays to establish genetic memory, and these DNA elements are subsequently transcribed and processed into mature crRNAs that can be assembled with CRISPR-related (Cas) proteins to recognize and cleave targets complementary to guide sequences (namely, spacers) of crRNAs (Fig. 1 ). Distinct from restriction enzymes and traditional gene editing tools, such as transcription activator-like effector nucleases (TALENs) and zinc-finger nucleases (ZFNs), the RNA-guided cleavage of the Cas effector enables more flexible utilization of this tool through a simple redesign of spacer sequences for different targets and does not require the modification of the Cas effector.
Fig. 1.
Fundamentals of CRISPR-mediated nucleic acid detection. There are two distinct principles underlying CRISPR diagnostics: RNA-guided targeted recognition (up panel), and target-recognition-triggered collateral cleavage (down panel).
Depending on the architecture of effector integration, CRISPR systems can be divided into two classes: class 1 systems function via multi-effector cascades, and class 2 systems rely on single-effector proteins (Makarova et al., 2015). Possessing remarkable simplicity and operability, CRISPR effectors from class 2 systems are particularly sought after for a variety of purposes, and CRISPR/Cas9 is highly popular (Shmakov et al., 2017). The Cas9 effector guided by its single-guide RNA (sgRNA) specifically recognizes DNA targets that harbor a cognate 20-bp protospacer with a downstream protospacer-adjacent motif (PAM) to induce R-loop formation and double-strand breaks via HNH and RuvC cleavage of spacer-targeted strands and nontargeted strands, respectively (Fig. 1) (Gasiunas et al., 2012; Nishimasu et al., 2014). Notably, the introduction of D10A or H840A silencing mutations into the RuvC or HNH domain results in a programmable Cas9 nickase (Cas9n) that can generate a nick at the corresponding strand of the target, whereas simultaneous inactivation of both domains will lead to a nuclease-deficient Cas9 (dCas9) protein while maintaining targeted reorganization capacity (Fig. 1) (Cong et al., 2013; Jinek et al., 2012). The abovementioned Cas9 variants were first employed as flexible and versatile tools for gene editing and transcriptional regulation (Cong et al., 2013; Mali et al., 2013b; Qi et al., 2013) and later applied to in vitro molecular diagnosis, in which recognition or manipulation by these effectors functions as a critical trigger for subsequent detection procedures (Li et al., 2019b). Due to their intrinsic programmability, flexibility, specificity, ultrasensitivity, and high efficiency (Hsu et al., 2013; Jiang et al., 2015; Josephs et al., 2016; Mekler et al., 2016), Cas9-mediated tools, have offered new opportunities for the development of superior diagnostic utilizations, including facile biosensing assays (Li et al., 2019b), rapid in situ labeling and efficient target enrichment for sequencing analysis, based on their RNA-guided targeted recognition and cleavage activities.
Two other single-component class 2 Cas effectors, Cas12 and Cas13, display RNA-guided targeted recognition and cleavage of double-stranded DNA (dsDNA) and RNA, respectively. However, upon selectively targeting cognate sequences, Cas12 and Cas13 effectors undergo a conformational change and display collateral cleavage of nearby single-stranded DNA (ssDNA) and RNA, respectively (Chen et al., 2018; East-Seletsky et al., 2016) (Fig. 1). This non-canonical collateral cleavage activity of Cas12 and Cas13 effectors radically deviates from the canonical target recognition and cleavage activity of the Cas9 proteins that turn to an inactive state after targeted cleavage. Due to these unexpected activities, both Cas12 and Cas13 effectors can function as elegant signal amplifiers in detecting nucleic acids by translating the presence of specific sequences into multiturnover nuclease activities, thereby resulting in strongly elevated analytical sensitivity. Moreover, this transduction process can be completed rapidly under mild conditions, which is highly desirable for user-friendly utilization in both clinical and field settings.
Collectively, these new applications of CRISPR systems potentially represent ideal candidates for high-performance diagnostic tools. In this review, we discuss recent representative works exploiting CRISPR-based diagnostic tools for human disease-related nucleic acid detection across a range of application fields, including biosensing assays, imaging assays and target enrichment for NGS. We broadly classify these methodologies into two categories based on detection principles: canonical targeted recognition and non-canonical collateral cleavage. We employ this classification because although CRISPR-based diagnostic tools function diversely, their essences are materializations of these two distinct CRISPR fundamentals that lead to different use, performances and features. Therefore, we intend to follow this essential distinction as a clue to discuss the recent advances, current challenges and possible future directions on this topic in a clear and straightforward manner.
2. CRISPR targeted recognition has facilitated a variety of diagnostic tools
The RNA-guided target recognition activities, which are derived mainly from Cas9 variants, have enabled orthogonal tools of high innovation for pathogen and disease detection, thus providing vital complements to methodologies currently used in clinical practice. In this study, we classify these tools into three categories based on the application field: biosensing assays, imaging assays and target enrichment for NGS.
2.1. Biosensing assays
Programmable target recognition activities have suited Cas9 variants for a variety of biosensing assays that primarily depend on the following three mechanisms: 1) RNA-guided DNA cleavage of Cas9, in which input targets are digested to initiate detection or output sequences are sheared as an endpoint readout; 2) RNA-guided DNA binding of dCas9, which transports enzymes or fluorescent or chromogenic compounds to desired positions for detection purposes; and 3) RNA-guided DNA nicking of Cas9n, which generates nick sites for the initiation of strand displacement reactions. In this study, we classify these methods into three categories based on the types of Cas effectors: Cas9, dCas9 and Cas9n. A comparison of different Cas9 variant-based strategies for detecting nucleic acids is listed in Table 1 .
Table 1.
Biosensing assays based on Cas9 variants.
| Classification | Method | Analyte | Amplification | Isothe-rmality | readout | Single-base resolution | Sensitivity | Multi-plexing | Quanti-fication | Time | ref |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cas9-based biosensing | NASBACC | RNA | NASBA | Y | Colorimetry | Y | fM | N | N | 2–6h | Pardee et al. (2016) |
| CAS-EXPAR | ssDNA mRNA | EXPAR | Y | Fluorescence | Y | Sub-aM | N | N | ≤ 1h | Huang et al. (2018) | |
| RACE | miRNA | RCA | Y | Fluorescence | Y | fM | Y | N | 3–4h | Wang et al. (2020b) | |
| CASLFA | DNA | RPA or PCR | Y | Lateral flow | N | 150–200 copies/reaction | N | N | ≤ 1h | Wang et al. (2020c) | |
| dCas9-based biosensing | dCas9/sgRNA-SG I based MRSA detection | DNA | NA | Y | Fluorescence | N | 10 CFU/ml | N | N | ≤30min | Guk et al. (2017) |
| PC reporter | DNA | PCR | N | Fluorescence | N | Single copy | N | N | 10min after PCR | Zhang et al. (2017) | |
| RCH | miRNA | RCA | Y | Colorimetry | Y | fM | N | Y | ˂ 4h | Qiu et al. (2018) | |
| CRISPR–Chip | DNA | NA | Y | electrochemistry | N | fM | N | Y | 45 min | Hajian et al. (2019) | |
| Nanoelectrokinetic platform | cfDNA | NA | Y | Fluorescence | N | pM | N | N | ˂ 1h | Lee et al. (2018) | |
| Cas9n-based biosensing | CRISDA | DNA | KF-mediated amplification | Y | Fluorescence | Y | aM | N | Y | 2–3h | Zhou et al. (2018) |
| Cas9nAR | DNA | KF-mediated amplification | Y | Fluorescence | Y | 2 copies/20 μL reaction | N | Y | ˂ 1h | Wang et al. (2019) |
2.1.1. Cas9-mediated biosensing assays
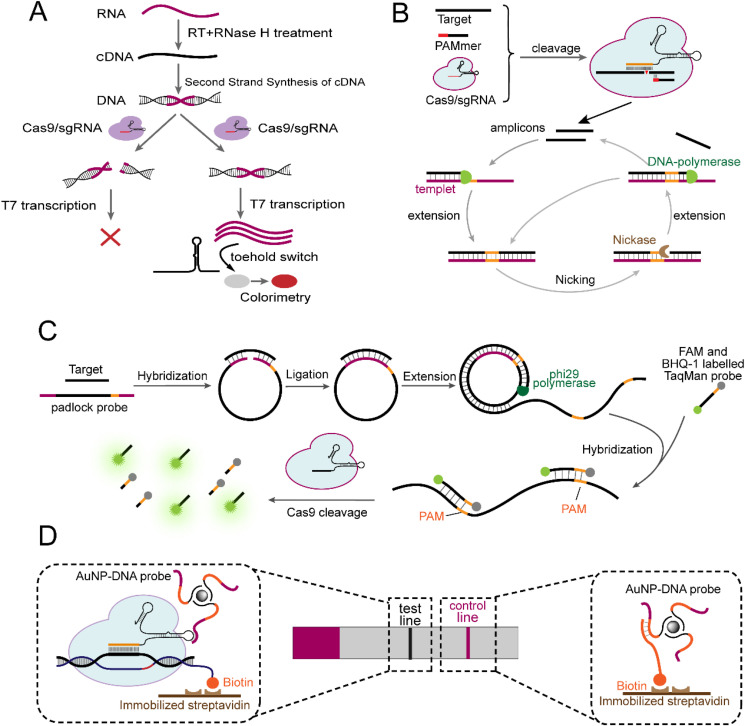
Well-confirmed theories suggest that CRISPR/Cas9 specifically scans its targets in a strict PAM-dependent manner and then creates a double-strand break at the third base upstream of PAM (Knight et al., 2015). Thus, CRISPR/Cas9 can be used to distinguish single base mismatches located in the PAM region. For example, Pardee and colleagues proposed a Cas9-based Zika variant genotyping method with single-base resolution, termed NASBACC, in which the Zika RNA sequences were preamplified by nucleic acid sequence-based amplification (NASBA) (Pardee et al., 2016). The resulting amplicons were applied to trigger the toehold switch of specially designed RNA hairpins and untie the LacZ ribosome binding site (RBS) and start codon motif (AUG) that were otherwise folded and inactivated, thereby initiating enzyme LacZ translation and colorimetric readout. In the NASBA step (Fig. 2 A), RNA amplification was accomplished via three procedures operating in series, during which dsDNA products were intermediates. In the presence of the PAM sequence and an adjacent target site, dsDNA intermediates could be cleaved by the Cas9/sgRNA complex, resulting in a blocked NASBA reaction and thus no endpoint single output. When a mismatch disrupted the PAM, the dsDNA intermediates remained intact such that subsequent amplification and detection can be continued. In this format, NASBACC can accurately genotype and distinguish between African and American Zika variants within several hours, which was the first time Cas9 has been used in nucleic acid biosensing assays.
Fig. 2.
Cas9-mediated biosensing assays. (A) Cas9-based NASBACC assay that can distinguish single-base mismatches located in the PAM region. (B) CAS-EXPAR for ultrasensitive detection of single-stranded targets. (C) RACE for miRNA detection. (D) CASLFA is a Cas9-based lateral flow assay that enables fast and field-deployed detection.
In subsequent research, CRISPR/Cas9, incorporated with varieties of isothermal amplification technologies, including exponential amplification reaction (EXPAR) and rolling circle amplification (RCA), was developed as a highly sensitive and specific nucleic acid detector. Combining CRISPR/Cas9 cleavage with EXPAR, Huang and colleagues proposed a single-stranded nucleic acid biosensing method termed CAS-EXPAR (Huang et al., 2018). Under the assistance of a PAM-presenting oligonucleotide (PAMmer), ssDNA or RNA was cleaved at a specific site to release short fragments that functioned as primers to initiate the EXPAR process, resulting in abundant ds- and ssDNA amplicons (Fig. 2B). The robust exponential amplification capability of EXPAR imparted outstanding analytical sensitivity to CAS-EXPAR to 0.82 aM. In terms of specificity, CAS-EXPAR can discriminate a single-base mismatch located exactly at the cleavage site because the mismatch would hinder primer extension of EXPAR, thereby eliminating readout. In addition, CAS-EXPAR has successfully been applied to Listeria monocytogenes mRNA detection. However, regarding miRNA detection, challenges remained due to their small size at ~19–23 nucleotides (Dong et al., 2013), which clearly impedes the design of sgRNAs. Recently, a Cas9-based strategy for multiple miRNA detection (termed RACE) was developed by integrating the flexibility of Cas9 cleavage with the specificity of the padlock probe (Wang et al., 2020b). With this strategy, miRNA functioned as a ligation template for cyclization of the padlock probe and initiated the RCA reaction to generate ssDNA amplicons containing hundreds or thousands of tandem repeats that subsequently annealed with FAM- and BHQ-1-labeled TaqMan probes, resulting in double-strand substrates for Cas9 cleavage and the ensuing release of the fluorescence signal (Fig. 2C). By simultaneously adding orthogonal sets of padlock probes, sgRNAs and TaqMan probes cognate with miRNAs of interest into a single reaction, RACE has achieved multiple detection of up to three homologous miRNAs (miR-21, miR-221, miR-222).
Compared with the abovementioned biosensing platforms, which require tedious enzymatic steps or sophisticated equipment, the CASLFA method, which is a visual, fast and field-deployed strategy, has been designed by integrating Cas9 detection into a lateral flow format (Wang et al., 2020c). In this scheme (Fig. 2D), dsDNA targets were first amplified using biotinylated primers by recombinase polymerase amplification (RPA) or PCR, thereby generating biotinylated amplicons that could be specifically recognized by Cas9/sgRNA to form ternary complexes. Upon trickling onto the lateral flow device, these ternary complexes flowed laterally and hybridized with the AuNP-DNA probe, an Au affinity-labeled oligonucleotide capable of binding to the loop region of sgRNA, which resulted in the formation of quadruple complexes that were captured by the precoated streptavidin at the test line, while excess AuNP-DNA probes spread forward and finally accumulated at the control line via the immobilized capture probes, collectively providing a chromogenic indication for the presence or absence of DNA analyte. Performing EGFR gene detection as a proof of concept, CASLFA has exhibited the ability to enable rapid (entire test completed within 1 h) detection over a wide temperature range of 20–37 °C with minimal equipment. Although this method is easy to use, the performance is not reduced because CASLFA detected microbe genomic DNA with a sensitivity reaching 150–200 copies/reaction and distinguished L. monocytogenes from orthogonal foodborne pathogens with few cross-reactions. Therefore, this technology offers an attractive option for rapid nucleic acid detection in a simple format, which is particularly relevant for point-of-care testing (POCT) and nonlaboratory environments.
2.1.2. dCas9-mediated biosensing assays
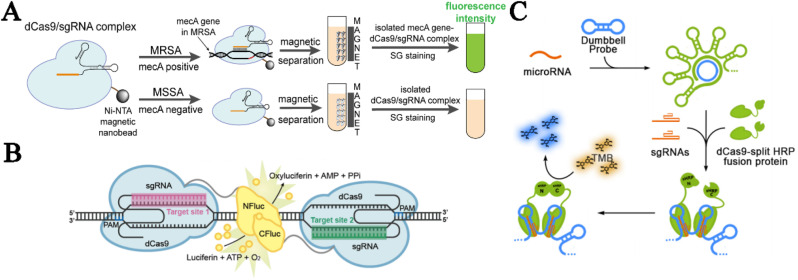
The dCas9 system, a programable DNA-binding tool that can be repurposed only by changing the spacer sequence of sgRNA, functions as an anchor that delivers various effectors, such as fluorescence and enzymes, to desired loci or as a catcher that captures target DNA for further manipulation and transduction, thus providing orthogonal tools for nucleic acid detection. One of the typical examples is a dCas9/sgRNA-SG I-based strategy proposed by Guk's group for methicillin-resistant Staphylococcus aureus (MRSA) detection (Guk et al., 2017). In this strategy, the dCas9/sgRNA complex was repurposed to bind the mecA gene, a MRSA-specific gene responsible for methicillin resistance, and then pulled down the genomic DNA that was subsequently visualized using SYBR green I (SG) staining (Fig. 3 A). This strategy eliminates nucleic acid separation steps by directly using cell lysates, indicating its superiority to traditional methods in which DNA extraction and purification are required before PCR amplification (Hagen et al., 2005). Verified by testing a series of clinical isolates, this strategy enabled rapid detection within 30 min with a limit of detection (LOD) down to 10 CFU/ml.
Fig. 3.
dCas9-mediated biosensing assays. (A) dCas9/sgRNA-SG I-based strategy for MRSA detection. (B) Paired-dCas9 (PC) reporter system for dsDNA detection. (Reprinted with permission from Zhang et al. (2017). Copyright, 2017; American Chemical Society). (C) RCA-CRISPR-split-HRP (RCH) system for miRNA detection. (Reprinted with permission from Qiu et al. (2018). Copyright, 2018; American Chemical Society).
Although dCas9-based DNA binding offers unprecedented superiority, the off-target nature of CRISPR/dCas9 is a potential drawback that might lead to a reduced signal-to-noise ratio (SNR) or even false positive results (Anderson et al., 2018; Tycko et al., 2016). To enhance the specificity of dCas9 detection, a dimerization-dependent strategy that was previously used for ZFNs, TALENs and CRISPR-mediated precise gene editing (Joung and Sander, 2013; Tsai et al., 2014; Urnov et al., 2010), has been introduced. For example, Zhang and colleagues proposed a paired-dCas9 (PC) reporter system to selectively detect dsDNA (Zhang et al., 2017). In this system, an RNA-guided luciferase was constructed by fusing a pair of dCas9 proteins with the N- and C-terminal half of the firefly luciferase enzyme, and dimerization was required for wild-type luciferase activity. Therefore, the fusion dimers were reconstituted and catalyzed luminescence for fluorescent readout only when there were two proximate protospacers spaced at a certain length apart along the target sequence (Fig. 3B). This PC reporter system has high specificity because its output is premised on strict sequence and spatial restrictions. Another embodiment of this principle of dimerization is in the development of the RCA-CRISPR-split-HRP (RCH) method, a novel miRNA detection platform that used, in this case, the split-horseradish peroxidase (HRP) system (Qiu et al., 2018). In this platform, miRNAs acted as primers to initiate RCA using the dumbbell-shaped DNA ring as templates, which enabled discrimination between miRNA orthologs with similar sequences due to the high specificity of the toehold-exchange reaction underlying the hybridization of miRNA primers with dumbbell-shaped templates (Fig. 3C). Compared with the traditional RCA, amplicons of toehold-initiated RCA consisted of hundreds of tandem DNA hairpins, and the double-strand stem regions could recruit dCas9 protein pairs that carried split-HRP, resulting in the generation of reconstituted HRP activity that could initiate the TMB color reaction as a highly specific second-stage amplification. Using let-7a as a model of detection, this platform exhibited a single-base specificity and femtomolar sensitivity attributed to the double enhancement from both the toehold-initiated RCA and proximate reconstitution of the split-HRP system.
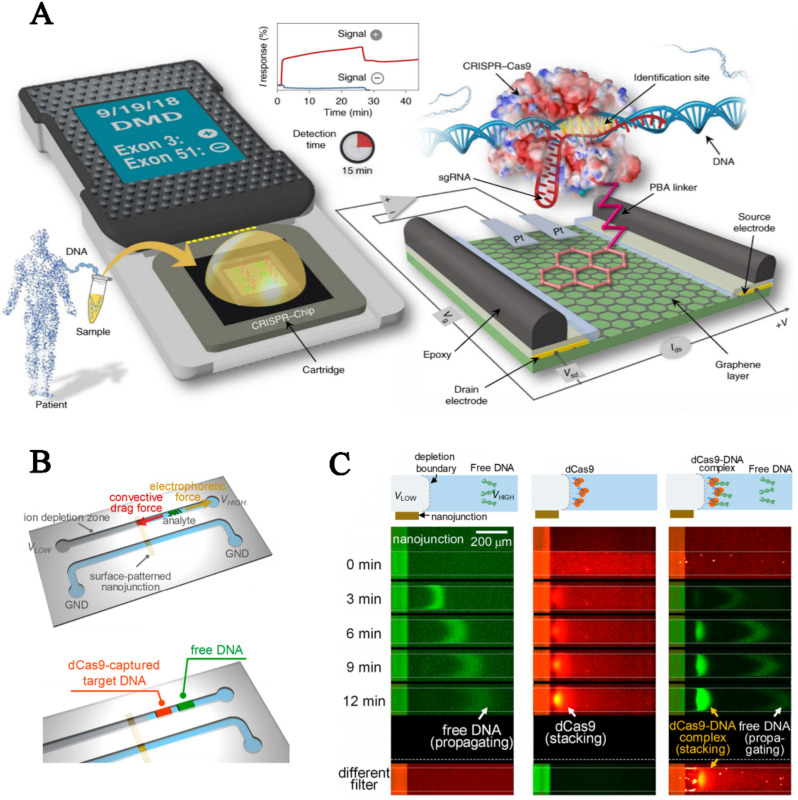
In addition to enzyme-based schemes, dCas9-mediated detections also involve multidisciplinary fundamentals, such as electrochemistry, hydrodynamics, and microfluidics, which has facilitated the development of amplification-free nucleic acid tests. For example, combined with the highly sensitive graphene-based field-effect transistor (gFET) sensor, a dCas9-based handheld device, termed CRISPR–Chip, has been fabricated for simple, rapid and selective on-chip electrical assays of unamplified target genes within intact genomes (Hajian et al., 2019). In this device, the dCas9/sgRNA complex was immobilized on the graphene surface and functioned as a current channel between gate electrodes (Fig. 4 A). Upon application of the genomic DNA, the anchored dCas9/sgRNA complexes selectively captured those harboring cognate protospacers, which led to the accumulation of negatively charged materials on the graphene channel and resultant generation of Donnan potential, which finally facilitated sensitive modulation of the transistor conductivity, thereby creating real-time electrical readout. As a well-designed electrochemical platform, this scheme enables rapid readout within 15 min and high sensitivity as low as 1.7 fM without preamplification. Notably, when integrated with an appropriate nucleic acid isolation protocol, CRISPR–Chip has indisputable potential for POCT of genetic abnormalities, especially large fragment deletions or insertions, because it does not require bulky instruments or multistep enzymatic reactions but rather only a handheld all-in-one analyzer and reaction buffer.
Fig. 4.
dCas9-based amplification-free nucleic acid detection. (A) dCas9-based electrochemical detection using a handheld device. (Reprinted with permission from Hajian et al. (2019). Copyright, 2019, Springer Nature). (B) Schematic of dCas9-based nanoelectrokinetic platform in which analyte migrates along the microchannel driven by electrophoretic force and convective drag force, (C) resulting in concentration and separation of DNA targets from free DNA in a visual format. (Reprinted with permission from Lee et al. (2018). Copyright, 2018; American Chemical Society).
Recently, Lee and colleagues reported a nanoelectrokinetic platform (Lee et al., 2018) for cfDNA detection based on micro/nanofluidic devices in which the analyte migrated along the microchannel driven by two opposite-directed forces, namely, electrophoretic force and convective drag force, derived from voltage configurations and ion concentration polarization (ICP), respectively (Fig. 4B). If the electrophoretic force outstripped the convective drag force, then the negatively charged DNAs migrated toward the bulk electrolyte; if not, they moved in the reverse direction and accumulated at the boundary layer of the nanojunction owing to the ICP phenomenon. The reorganization of DNA targets by dCas9/sgRNA complexes significantly reduced their electrophoretic mobility to a level lower than the defined critical mobility due to the relatively large molecule size of dCas9/sgRNA/DNA ternary complexes, resulting in migration and accumulation toward the nanojunction, whereas untargeted DNA species moved in the opposite direction, toward the bulk reservoir (Fig. 4C). In this format, the concentration and separation of dCas9/sgRNA-captured target DNA from free DNA were simultaneously achieved, thereby enabling amplification-free detection of target DNA in a direct and optical manner.
2.1.3. Cas9n-mediated biosensing assays
RNA-guided Cas9 nickase, which was previously exploited for precise gene editing (Cho et al., 2014; Mali et al., 2013a; Ran et al., 2013), has expanded the accessibility to molecular diagnostics by taking full advantage of the well-characterized intermediate status formed during Cas9n-mediated DNA binding and nicking. Functionally analogous to the Cas9 effector, the Cas9n/sgRNA complex unwinds helices to create an R-loop consisting of a DNA-RNA hybrid and a single-strand DNA that could be cleaved by nickase activity and thereby released from the target-accommodating groove of the Cas9n protein, generating free 3′ termini available for further manipulation (Jinek et al., 2014; Jore et al., 2011) (Fig. 1). Moreover, as a programmable nickase, the dissociation of the Cas9n effector from its target will leave a nick at the desired site, which is the ready substrate for strand extension and displacement reaction. These intriguing and delicate mechanisms form the basis of Cas9n-mediated nucleic acid detection.
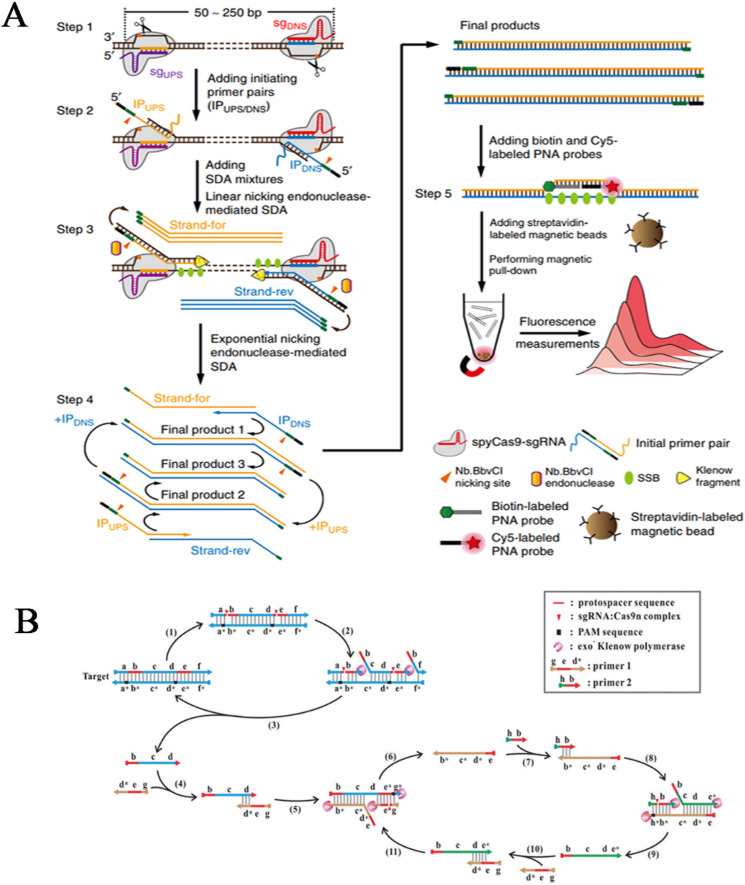
As a typical Cas9n detection scheme, the CRISPR–Cas9-triggered nicking endonuclease-mediated strand displacement amplification (CRISDA) (Zhou et al., 2018) developed by Zhou's team for isothermal amplification and detection of dsDNA is highly innovative (Fig. 5 A). A pair of Cas9n/sgRNA complexes with RuvC activity was designed to recognize the opposite strands of the target-flanking regions in a PAM-out orientation, thereby generating a pair of exposed single-strand DNA with free 3′-hydroxyl termini. Subsequently, a specially designed initiating primer (IP) pair carrying overhanging 5′ Nb.BbvCI nickase recognition motifs were applied to hybridization with exposed single-strand DNA at each border of the target. Catalyzed by exo-Klenow polymerase, the 3′ ends of the annealed primers were extended along the target toward the opposite Cas9n binding site, while the 5′ ends were filled in to create a complete Nb.BbvCI nicking site from which a strand displacement reaction was initiated, collectively resulting in the constant production of replaced single strands. These strands again annealed with IP pairs and initiated new rounds of extension, nicking, and strand displacement repeatedly, eventually generating abundant dsDNA amplicons. To extract target-specific amplicons from noise, a pair of modified PNA probes were designed to invade adjacent regions at the middle of targets, thereby enabling magnetic pull-down and fluorescence measurements. Reproducible results from varied samples have revealed an attomolar sensitivity attributed to Cas9n-mediated exponential amplification and PNA-mediated amplicon enrichment. Encouragingly, CRISDA can efficiently differentiate targets from complex human genome noises due to the triple specificity entrusted by the requirements of two Cas9 recognition sites flanking the target and a PNA complementary region located in the middle. However, CRISDA discriminates only single-nucleotide mutations in the PAM regions and the first nucleotide in the seed regions, thereby resulting in obvious sequence limitation.
Fig. 5.
Cas9n-mediated biosensing assays. (A) CRISDA scheme for dsDNA detection. (Reprinted with permission from Zhou et al. (2018). Copyright, 2018; Springer Nature). (B) Cas9nAR scheme for dsDNA detection. (Reprinted with permission from Wang et al. (2019). Copyright, 2019; Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim).
Although the above CRISDA scheme suggests an innovatively designed strategy for isothermal amplification and ultrasensitive detection, the involvement of complex reagents would be an obstacle. Alternatively, Wangs group proposed a Cas9n-based amplification reaction (Cas9nAR) (Wang et al., 2019) using a pair of Cas9n/sgRNA complexes and homologous primers. As illustrated in Fig. 5B, ssDNA products were displaced from a pair of target-flanking nicks by the collaborative reaction of Cas9n/sgRNA and exo-Klenow polymerase. By annealing with specially designed primers containing 5′-CCN, ssDNA products initiated circuits of extension, nicking and displacement, resulting in constant synthesis of dsDNA amplicons that could be monitored in real time by SYBR Green I staining. This platform has achieved single-molecule sensitivity due to the efficient exponential amplification mechanism and single-nucleotide specificity because of the intrinsic properties of Cas9 effectors.
2.2. Imaging assays
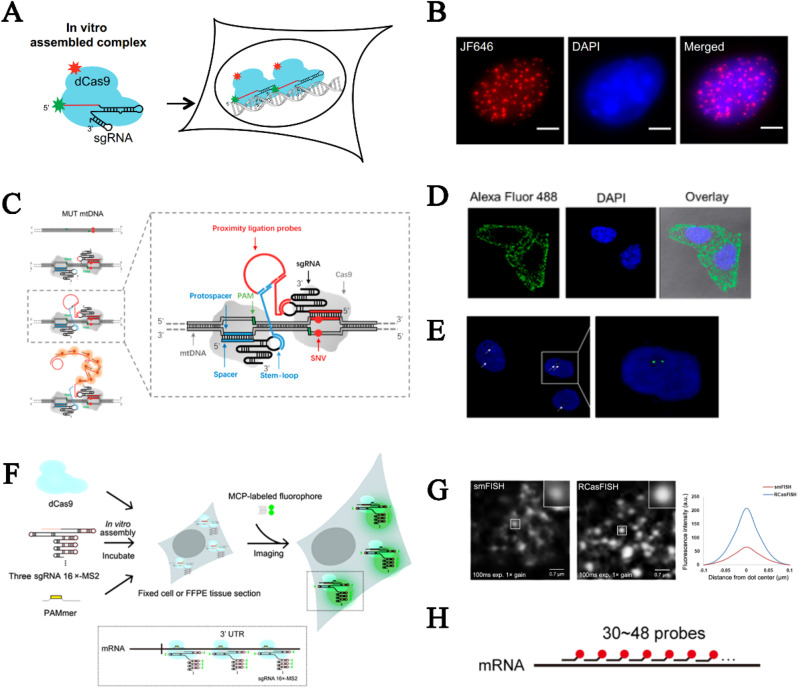
As a single-cell assay for directly visualizing spatial organizations of genetic elements in situ, DNA FISH offers indispensable insight into the mutations associated with the diagnosis and management of genetic disorders and tumors, such as prenatal diagnosis of aneuploidy for high-risk pregnancy (Practice Bulletin, 2016a, 2016b) and determination of HER2 tumor status for targeted therapy of breast cancer (Gradishar et al., 2018; Wolff et al., 2018). Nonetheless, as a paramount procedure of the FISH assay, thermal cycles of denaturation and annealing are applied to fixed cells for probe hybridization, which is time-consuming and could compromise the cell morphology and DNA structure (Levsky and Singer, 2003). FISH is also limited by the hazards of formamide's carcinogenicity and reproductive toxicity and the high cost of the probes. Thus, additional efforts should be made to develop superior approaches. CRISPR-Cas9 currently holds great promise for rapid, cost-effective and facile FISH schemes, as illustrated by the CASFISH approach (Fig. 6 A) from Deng's group (Deng et al., 2015). In this pioneering work, the fluorescently labeled dCas9/sgRNA complex functioned as a specific and efficient enzymatic probe that tiled repeated sequences in telomeres, pericentromeres or coding genes (Fig. 6B). Deviating from the traditional approach, CASFISH expedited in situ labeling without global DNA denaturation under mild conditions because of the high efficiency and stable affinity of dCas9/sgRNA to homologous dsDNA (Table 2 ). However, due to the promiscuous off-target binding and nonspecific adherence from the diversity of applied dCas9/sgRNA complexes, CASFISH have failed to efficiently visualize nonrepetitive elements of strong clinical significance, thereby leading to limited application.
Fig. 6.
FISH imaging based on Cas9 variants. (A) CASFISH enabled rapid in situ imaging by using enzymatic probe tiling repeated sequences in (B) telomere regions (Deng et al., 2015). (C) Schematic of CasPLA for (D) mitochondrial DNA and (E) genomic DNA imaging. (Reprinted with permission from Zhang et al. (2018). Copyright, 2018; American Chemical Society). (F) Schematic of RCasFISH for (G) single-molecular mRNA imaging with fluorescent dots that are brighter than that of (H) smFISH. Distance from dot center: the distance between pixels on the equator of the fluorescent signal dot and the equator center. (Reprinted with permission from Wang et al. (2020a). Copyright, 2020; American Chemical Society).
Table 2.
FISH schemes based on Cas9 variants.
| Method | CASFISH | CasPLA | RCasFISH |
|---|---|---|---|
| Target | highly repetitive sequences | mitochondrial DNA | mRNA |
| Time required after fixation | ~15–60 min | 8–9h | 1.5h |
| Denaturation treatment | N | N | N |
| Single-base resolution | N | Y | N |
| Single molecular imaging | N | Y | Y |
| Efficiency | ≥94% | ~25% | ≥85% |
Clearly, an efficient signal amplification system should be introduced instead of dCas9 tiling for nonrepetitive sequence visualization. To this end, a mechanism that guarantees analytical specificity must be coupled to prevent the background signal from being amplified along with the on-target signal; otherwise, any minor off-target binding might cause significant disturbance. Against this backdrop, the proximity ligation assay (Fredriksson et al., 2002; Soderberg et al., 2006), a highly specific technology previously employed for examining the subcellular localization of protein-protein interactions, has been combined with Cas9/sgRNA for mitochondrial DNA in situ labeling. In this so-called CasPLA method (Zhang et al., 2018), oligo probe pairs guided into close proximity by two adjacently located Cas9/sgRNA complexes served as templates for enzymatic circulation of linear oligonucleotides (Fig. 6C, Table 2). The formed DNA ring, in turn, acted as a template to initiate the RCA reaction, resulting in localized signals of fluorescent spots from target elements (Fig. 6D). It is worth noting that CasPLA has single-base resolution because it requires two indispensable Cas9-mediated target-binding events that are of low tolerance to mismatches in the seed region of sgRNA. In theory, any genetic elements carrying a pair of PAMs in proximity can be visualized and counted with such a scheme, but only approximately 60% of potentially accessible genomic DNA targets are detected (Fig. 6E), which is probably limited by inefficient DNA ring circulation and inaccessible nuclear targets (Nilsson et al., 1994). Therefore, future optimization of the efficiency of genomic DNA imaging is still urgently need to expand the scope of imageable elements by Cas9.
In early studies, FISH examinations of the intracellular distribution of single mRNA transcripts methodologically facilitated a comprehensive understanding of the relevant pathological processes and clinical diagnosis (Femino et al., 1998; Maamar et al., 2007; Raj et al., 2008; Wang et al., 2012). In a typical single-molecular RNA FISH method (Fig. 6H), fluorophore-labeled oligo probe pools tiling the target region are intricately designed as a trade-off between analytical sensitivity and specificity (Raj et al., 2008), which might contribute to the high cost and requirements of laborious optimization. As a specific and efficient enzymatic probe, the RNA-guided dCas9 effector has offered a necessary alternative for easily mapping RNAs within fixed cells and tissues. In this so-called RCasFISH method (Wang et al., 2020a), the sgRNA scaffold harboring 16 MS2-binding motifs was reconstructed so that each dCas9/sgRNA complex can recruit 28 fluorescently labeled MS2 coat proteins (MCPs) (Fig. 6F). Single mRNA transcript was visualized by using three reconstructed dCas9/sgRNA complexes that were distributed within a length of ~200 nt, achieving brighter fluorescent dots than that of smFISH in which 30–48 singly labeled oligonucleotide probes were used to tile a length of 1000-2000 nt length (Raj et al., 2008) (Fig. 6G, Table 2). Notably, three adjacently targeted sgRNAs are sufficient for efficient imaging of individual transcripts due to the specificity of the CRISPR-dCas9 system and signal amplification capacity of the MS2 system, thereby relatively simplifying the scheme design of this method.
2.3. Target enrichment for NGS
Performing the maximum extraction and enhancement of target signal from promiscuous noise is the primary objective across a broad range of applications, especially for NGS technology (Ballester et al., 2016; Mamanova et al., 2010). Observations that the vast majority of clinically significant genomic alterations reside in highly limited regions make targeted sequencing preferable options to whole-genome sequencing (WGS) for increased sequencing depth and decreased cost and complexity in the context of clinical diagnosis (Mamanova et al., 2010; Sun et al., 2015), such as the molecular diagnosis of cancer, wherein sequencing against a panel of relevant genes with high accuracy is performed to identify low-frequency mutations (Surrey et al., 2019), thereby highlighting the need for extracting targets from unwanted species. Technologies for specific target enrichment are highly vital for increased recovery rates, maximized read usability and reduced DNA input (Mamanova et al., 2010). Current methods for target enrichment are primarily based on either PCR or hybridization capture. Each method has advantages but also limitations: PCR is often limited by inevitable false mutation calls derived from the polymerase amplification process and its compromised capacity for multiplexing in a single reaction (Kebschull and Zador, 2015), while hybridization capture suffers from low recovery, especially for small fragments (Samorodnitsky et al., 2015). Recent reports capitalizing on CRISPR-based strategies have displayed their potential to offer a sensible solution. In this section, we discuss the CRISPR-based target enrichment strategies for NGS. Compared with nucleic acid detection platforms described in “biosensing assays” section, methods outlined in this section function as auxiliary tools to improve the analytical performance of NGS, thereby indirectly facilitating nucleic acid detection.
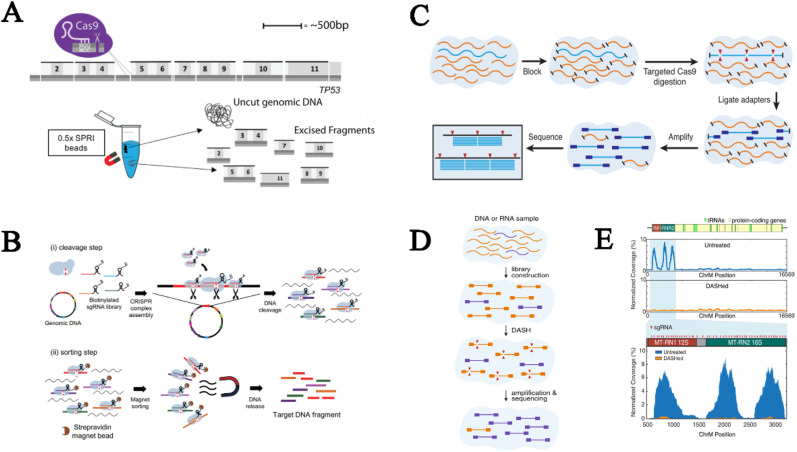
For example, Nachmanson and colleagues proposed a controllable targeted fragmentation strategy termed CRISPR-DS (Nachmanson et al., 2018) wherein the Cas9 system was programmed to evenly cleave the area of interest and generate fragments of homogenous length that could be isolated by a simple SPRI-bead-mediated size selection (Fig. 7 A), resulting in a targeted enrichment of 2000–50000-fold (Table 3 ). The similar size of excised fragments has efficiently reduced PCR amplification bias during library preparation and fueled uniform sequencing coverage and depth and maximized read usability while lowered DNA input, which was particularly suitable for situations where limited source materials were used. Based on a similar principle, Lee's group reported a CRISPR-Cap (Lee et al., 2019) platform in which the Cas9 effector, as well as the constructed sgRNA library, functioned as not only a programmable scissor but also an anchor to densely tile the target region followed by pull-down using streptavidin magnet beads (Fig. 7B). The highly intensive sgRNA tiling with increments of approximately 20 bp contributed to uniform enrichment, which endowed CRISPR-Cap with the ability to estimate allele frequency and gene copy number. Notably, the elimination of hybridization capture and PCR amplification (attributed to the large amount of DNA input during library preparation) remarkably shortened the reaction time over parallel methods (Table 3).
Fig. 7.
Cas9-based target enrichment strategies for NGS. (A) CRISPR-DS for enrichment of small genomic regions (Nachmanson et al., 2018). (B) CRISPR-Cap (Lee et al., 2019). (C) FLASH platform for multiple analysis of AMR (Quan et al., 2019). (D) DASH facilitates metagenomic sequencing (Gu et al., 2016).
Table 3.
Application of CRISPR-Cas systems in sequencing analysis.
| CRISPR-DS | CRISPR-Cap | DASH | FLASH | |
|---|---|---|---|---|
| Application | enrichment of small target regions for targeted NGS | enrichment of small target regions for targeted NGS | depletion of host-derived species for metagenomic NGS | target enrichment for multiple AMR gene sequencing |
| Required time for library preparation library preparation | 3 days | Within 1 day | Within 1 day | Within 1 day |
| Fold enrichment | 2000–50000a | Several hundreda | ~3–10b | 293b |
| DNA input (ng) | 10–500 | ≥100, optimum at 1000 | essentially any amount of input | 0.1 |
Calculated by dividing the percentage of on-target ratio from sequencing data over percentage of target region in genome.
Calculated by dividing sequencing coverage of the targets from enriched sample over that from unenriched sample.
Metagenomic sequencing might suffer from a compromised sensitivity due to the pronouncedly lower-level nucleic acids of infectious agents than that of hosts by several orders of magnitude, thereby justifying the efficient deletion of high-abundance host materials and enrichment of low-abundance pathogen sequences to generate sufficient numbers of pathogenic reads (O'Flaherty et al., 2018). Against this backdrop, Gu and colleagues reported the Depletion of Abundant Sequences by Hybridization (DASH) (Gu et al., 2016) method for deleting high-abundance human-derived fragments from the metagenomic sequencing library. In this work, the CRISPR/Cas9 system was repurposed to selectively digest human 12S and 16S mitochondrial rRNA genes (Fig. 7D), which are the highly abundant sequences in cerebrospinal fluid (CSF)-derived RNA samples, thereby ensuring their depletion in the final sequencing library. After DASH enrichment, the 12S and 16S rRNA were reduced by approximately 10-fold and 3-fold, respectively, while the coverage of representative pathogenic genes increased by approximately 3–10-fold (Fig. 7E, Table 3), which could be translated into higher usability of sequencing space and sensitivity, as well as lower sequencing depth required for nonhuman sequences. These characteristics are critical for the early identification of infectious diseases in a cost-effective manner.
Later, the same group published a novel targeted enrichment system termed FLASH (Quan et al., 2019) for highly multiplexed analyses of antimicrobial resistance (AMR) sequences that rely on Cas9 cleavage against low-abundance targets. The sequences of interest were fragmented and then blocked by phosphatase treatment followed by Cas9/sgRNA digestion of AMR genes (Fig. 7C). The resultant AMR-derived fragments were then available for ligation of sequencing adapters and ensuing amplification, leading to enrichment in the final library. To achieve a high level of multiplexing, a sgRNA collection against all clinically relevant AMR-related genes listed in the CARD and ResFinder databases was computationally defined and optimized, and a subset of which were used in pilot experiments and revealed an enrichment of 293-fold, thereby reducing the required amount of input (down to 100 pg) without compromising the sensitivity and on-target ratio.
It is worth noting that NGS is not the only application area for CRISPR/Cas9-based target enrichment, and the strategy leveraging Cas9-based cleavage to isolate intact large fragments (200 kb target containing BRCA1 gene) from the genome for nanopore sequencing has achieved a targeted enrichment of 237-fold (Gabrieli et al., 2018).
3. CRISPR collateral cleavage has enabled ultrasensitive and portable diagnostic tools
The utility of RNA-guided target-recognition-triggered collateral cleavage as an amplifier for the presence of nucleic acid targets has enabled numerous ultrasensitive and portable approaches that potentially hold significant value for rapid and field-deployed tests. We classify these approaches into two categories according to the types of Cas proteins used: Cas13-based approaches and Cas12-based approaches. A comparison of Cas13- and Cas12-based strategies for nucleic acid detection is listed in Table 4 .
Table 4.
Representative nucleic acid detection strategies based on CRISPR collateral cleavage.
| Classification | Method | Analyte | Amplif-ication | Isothe-rmality | readout | Single-base resolution | Sensitivity | Multi-plexing | Quanti-fication | Time | Porta-bility | ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cas13-based methods | SHERLOCK | RNA/DNA | RPA | Y | Fluorescence | Y | aM | N | N | 2–5h | Y | Gootenberg et al. (2017) |
| SHERLOCKv2 | RNA/DNA | RPA | Y | Fluorescence Colorimetry Lateral flow | Y | zM | Y | Y | 0.5–3h | Y | Gootenberg et al. (2018) | |
| HUDSON + SHERLOCK | RNA/DNA | RPA | Y | Fluorescence Colorimetry Lateral flow | Y | aM | Y | N | ˂ 2h | Y | Myhrvold et al. (2018) | |
| LbuCas13a-based miRNA detection | miRNA | NA | Y | Fluorescence | Y | aM | N | Y | ˂ 30min | N | Shan et al. (2019) | |
| Microfluidic electrochemical biosensor | miRNA | NA | Y | Electrochemistry | Y | pM | N | Y | ˂4h | Y | Bruch et al. (2019) | |
| Automated microfluidic device | RNA | NA | Y | Fluorescence | N | 20 pfu/mL | Y | Y | 5 min | Y | Qin et al. (2019) | |
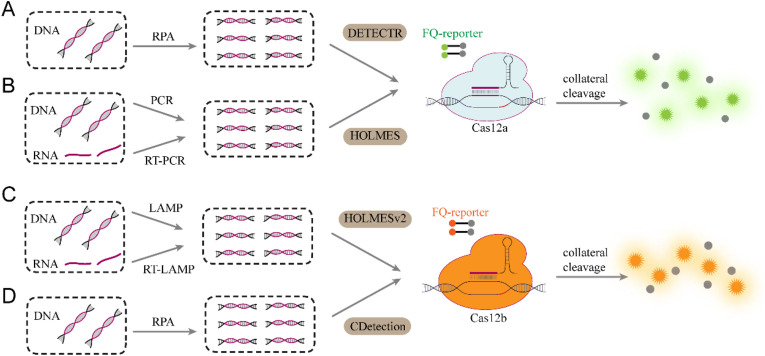
| Cas12-based methods | DETECTR | DNA | RPA | Y | Fluorescence | N | aM | N | N | ~ 1h | Y | Chen et al. (2018) |
| HOLMES | DNA RNA | PCR | N | Fluorescence | Y | aM | N | N | ~ 1h | N | Li et al. (2018) | |
| E-CRISPR | DNA | NA | Y | electrochemistry | Y | pM | N | Y | ~1h | Y | Dai et al. (2019) | |
| hpDNA-based electrochemical biosensor | DNA | NA | Y | Electrochemistry | N | pM | N | Y | ~1h | Y | Zhang et al. (2020) | |
| MAV-chip | DNA | NA | Y | Volumetric Bar-Chart Chip | Y | pM | Y | Y | ~1h | Y | Shao et al. (2019) | |
| HOLMESv2 | DNA RNA | LAMP | Y | Fluorescence | Y | aM | N | Y | ~1h | Y | Li et al. (2019a) | |
| CDetection | DNA | RPA | Y | Fluorescence | Y | 0.1aM | N | N | ~3h | Y | Teng et al. (2019) | |
| CIA based LFB | DNA | LAMP | Y | Lateral flow | N | Single copy | N | N | ˂1h | Y | Mukama et al. (2020a) |
3.1. Cas13-based approaches
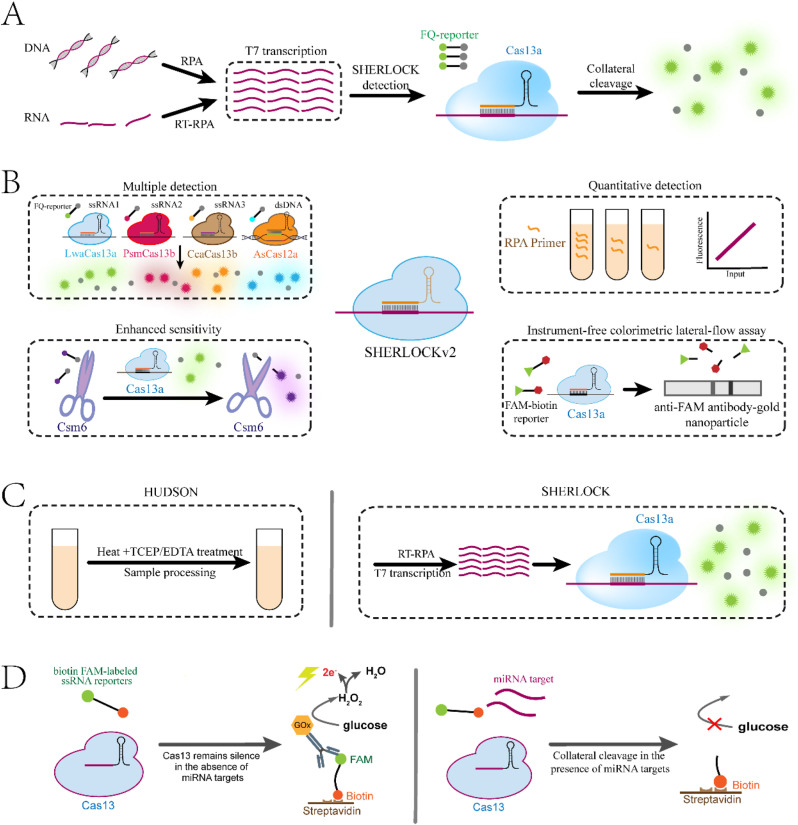
Compared with most orthologs of Class 2 CRISPR systems characterized to date, the Cas13 effector acts as an RNA-guided ribonuclease (RNase) instead of deoxyribonuclease (DNase), which relies on the catalytic activities of its Prokaryotes Nucleotide-binding (HEPN) domains (Shmakov et al., 2017). Within a series of Cas13 proteins, Cas13a (previously known as C2c2) is the first putative subtype and the first developed biosensor for nucleic acid detection. Cas13a is guided by the corresponding crRNA and can be programmed to degrade single-stranded RNA (ssRNA) carrying complementary protospacers. Surprisingly, upon binding to ssRNA targets, Cas13a protein is activated and converted into a nonspecific endoRNase with collateral cleavage activity against nearby nontarget ssRNA at a rate of more than 104 turnovers per target-recognition event (Abudayyeh et al., 2016; East-Seletsky et al., 2016). Such non-canonical activity can serve as an elegant amplifier for the presence of a specific nucleic acid analyte, which notably deviates from the conventional principle of CRISPR-based tools, thereby providing the opportunity to start a brand-new prospect for nucleic acid detection. In 2016, as an initial attempt, East-Seletsky and colleagues used LbuCas13a to detect bacteriophage λ RNA and endogenous β-actin mRNA (East-Seletsky et al., 2016). In this context, Cas13a collateral cleavage was activated by ssRNA targets to degrade the fluorophore quencher-labeled ssRNA (ssRNA-FQ) reporter, which increased the fluorescence intensity and achieved rapid and feasible detection with LOD down to 1–10 pM.
In 2017, Gootenberg and colleagues announced the first comprehensive research characterizing the Cas13a-based diagnostic tool and released “SHERLOCK” (Gootenberg et al., 2017). In this pioneering work, isothermal amplification and in vitro transcription engaged in series into a single reaction along with highly specific LwCas13a detection to achieve a novel solution-based nucleic acid biosensing assay (Fig. 8 A). Specifically, RPA or RT-RPA was used as a preamplification followed by T7 in vitro transcription, which produces enormous RNA amplicons that subsequently triggered Cas13 collateral cleavage of ssRNA-FQ reporters to generate a fluorescent readout. Since these three compatible processes, namely, RPA and T7 transcription and Cas13a detection, imply the process of signal amplification and enable efficient reactions under mild conditions, SHERLOCK is actually a triple signal amplification system with isothermal capacity and a short time course. These superiorities have facilitated both RNA and DNA detection at attomolar concentrations within 1–3 h at 37 °C (Table 4). In addition, SHERLOCK has been used for effective genotyping of virus strains and single nucleotide polymorphisms (SNPs) by mutating single bases in the spacer sequences. This proposed high specificity is guaranteed by the combination of CRISPR recognition and RPA primer annealing. Notably, lyophilized Cas13a systems and paper-based detection have been developed for portable deployment at a cost as low as $0.61/test while maintaining attomolar sensitivity, potentially indicating a robust, cost-effective and cold-chain independent diagnostic for massive manufacturing and distribution. Later, the same team launched an extended version, named SHERLOCKv2 (Gootenberg et al., 2018), which added four-point advancements (Fig. 8B): quadruplex detection by pooling orthogonal Cas effectors that perform unique preferred cleavages together with corresponding FQ reporters; quantification across a wide range of concentrations down to the attomolar level by controlling the progress of exponential preamplification through adjusting the input of RPA primers; increased sensitivity offered by the cascaded activation of Csm6 nuclease activity triggered by Cas13a collateral cleavage; and instrumentation-free colorimetric lateral-flow assays that enabled direct readout with the naked eye and sensitive detection with the LOD down to 2 aM within 90 min. Collectively, SHERLOCK and advanced SHERLOCKv2 present versatile, rapid, portable and cost-effective nucleic acid detection with attomolar sensitivity and single-base specificity, which are highly desired for POCT, epidemic monitoring and pathogen detection, especially in underdeveloped areas where complex infrastructure may not be readily accessible.
Fig. 8.
Nucleic acid detection based on Cas13 collateral cleavage. (A) Schematic of SHERLOCK detection. (B) Four-point advancements of SHERLOCKv2 detection. (C) HUDSON provides a complete workflow from sample to answer for SHERLOCK and SHERLOCKv2 detection. (D) Cas13-based microfluidic electrochemical device for miRNA detection.
In light of the notable boom in Cas13-based diagnostics, the establishment of a complete workflow from sample to answer, which could be paramount to practicable deployment, particularly in field settings, is highly valuable. To provide a feasible solution, Myhrvold and colleagues proposed a viral detection pipeline, known as HUDSON (Myhrvold et al., 2018), by which the viral genome could be released directly from bodily fluids and protected from enzymatic degradation and thus could be ready for direct mixture with Cas13 detection reagents, thereby bridging raw samples and the SHERLOCK assay without nucleic acid purification steps. Various bodily fluids—including urine, whole blood, plasma, serum, and saliva—were compatible with this procedure of heat and TCEP/EDTA treatment within 10–25 min while maintaining the test capacity of SHERLOCK, thereby minimizing instrument requirements for Cas13-based diagnostic workflow (Fig. 8C). To date, these three consecutively published Cas13-based platforms (SHERLOCK, SHERLOCKv2 and HUDSON) indicate the prospects of developing a simple, sensitive, specific, cost-effective and portable diagnosis method for wide use that extends from epidemic monitoring to tumor diagnosis and from clinical routine to field deployment.
Notably, although SHERLOCK has shown excellence in the detection of pathogens, human genomic DNA and cfDNA, no attempt has been made to detect miRNAs because their small size poses a challenge for amplification (Dong et al., 2013). To solve this problem, Shan and colleagues employed the initial reaction rate (measured by the ratio of fluorescence intensity change before reaching the plateau to the corresponding time frame) as the readout instead of fluorescence intensity, and they achieved a LOD down to 4.5 aM without preamplification (Shan et al., 2019). However, the real-time PCR system was integral for the generation and calculation of the initial reaction rate, and this step was less flexible and might add to the complexity of this method. To manufacture a portable and easy-to-use platform, another amplification-free miRNA detection scheme using a microfluidic electrochemical device that consists of an immobilization area and an electrochemical cell is proposed (Bruch et al., 2019). In this scheme (Fig. 8D), biotin FAM-labeled ssRNA reporters were used as linkers anchoring GOx (glucose oxidase)-conjugated anti-FAM antibodies onto the streptavidin-coated microfluidic chip. These immobilized GOx could catalyze the generation of H2O2 and thus triggered a current peak that was positively correlated with GOx activity and H2O2 production. However, in the presence of miRNA targets, the collateral cleavage activities of Cas13a were activated and thus destroyed ssRNA linkers, resulting in the elimination of GOx immobilization and H2O2 production, as well as the output signals. This microfluidic electrochemical sensor enabled picomolar sensitivity without preamplification and achieved good correlation with qRT-PCR, as validated by parallel detection of serum samples from both patients and healthy controls. However, multiple detection still poses a challenge. To this end, an automated and multiplexing microfluidic chip has been fabricated by integrating 24 detection channels with a custom designed fluorometer (Qin et al., 2019). This device has enabled Ebola Virus detection with high sensitivity and can screen 24 samples within 30 min. With further integration of a sample processing module, this device might have higher value in POCT.
3.2. Cas12-based approaches
Cas12, an RNA-guided endonuclease, is the second most important gene editing tool besides of Cas9 (Pickar-Oliver and Gersbach, 2019). The discovery of its collateral cleavage activities has enabled numerous portable diagnostic tools. Distinct from Cas13, the Cas12a-crRNA complex unleashes nonspecific single-strand DNase activity at a catalytic rate of ~1250 turnovers per second upon binding to guide-complementary dsDNA activators, which underlies the use of Cas12a as a biosensor and amplifier for nucleic acid assays (Fig. 9 A, Fig. 9B). Among these schemes, DETECTR (Chen et al., 2018) is a typical representative, receiving almost as much attention as “SHERLOCK”. In this method, RPA was introduced as a preamplification process (Fig. 9A) similar to SHERLOCK, such that the DNA substrate could be detected at concentrations as low as 1 aM. Using carefully designed crRNAs, DETECTR could accurately genotype human papillomavirus (HPV) types 16 and 18 that bear 6 base pair differences between target sequences. Despite sharing common features with SHERLOCK, such as rapid results, remarkable sensitivity, specificity and isothermal capacity, DETECTR may be more inclined to maintain stability in complex clinical settings than SHERLOCK because RNA reporters of SHERLOCK are susceptible to degradation by ubiquitous RNases while the ssDNA reporters are not and thus are more convenient for DNA detection because of the elimination of in vitro transcription processes. This pioneering work has opened the door for the application of the CRISPR-cas12a system to nucleic acid detection fields; however, it did not elucidate whether DETECTR could be employed for SNP and RNA testing, which were highly desirable for broad diagnostic applicability. HOLMES (Fig. 9B), another Cas12a-based nucleic acid detection platform using PCR amplification as a substitute for RPA and published almost at the same time as DETECTR, has been proposed with the ability to detect SNP, even if the site resides at the PAM-distal end (Li et al., 2018). Such a high specificity is provided by the utility of truncated crRNAs (16-nt and 17-nt guide sequences) that can enhance the cleavage specificity of the CRISPR-Cas12a system (Lei et al., 2017). Using specially designed primers to introduce PAM sequences during PCR preamplification, HOLMES can, in theory, detect any sequence in a PAM-independent manner. In addition, HOLMES enables RNA detection via the addition of reverse transcription steps that convert RNA targets into cDNA before preamplification and detection. Notably, although HOLMES theoretically has fewer sequence restrictions and wider usage, it is still not an isothermal test and requires multiple steps. An enhanced version, namely, HOLMESv2, will be discussed in the third paragraph of this part.
Fig. 9.
Classical schemes of Cas12-based diagnostic tools. (A) DETECTR for rapid DNA detection. (B) HOLMES enables both DNA and RNA detection. (C) HOLMESv2 enables isothermal detection by integrating AacCas12b system with LAMP. (D) CDetection enables ultrasensitive DNA detection by integrating AaCas12b system with RPA.
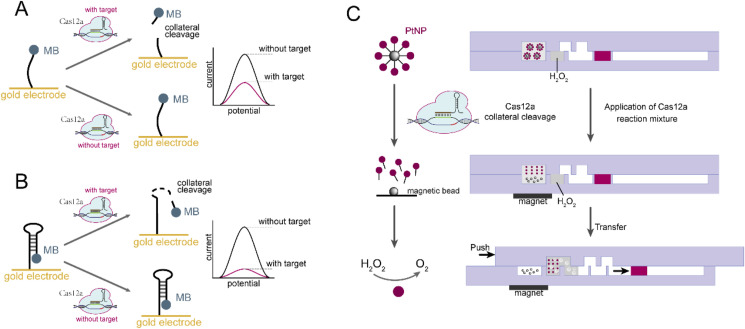
Both DETECTR and HOLMES use fluorescence as a signal transduction tool, and prior to this, nucleic acid amplification is used to increase sensitivity. Many other signal transductions, including electrochemical reporters and nanoparticle systems, are inherently highly sensitive and have been integrated into Cas12a-based nucleic acid detection, thus facilitating more flexible modification and deployment. Recently, capitalizing on the cost-efficiency, sensitivity and portable nature of electrochemical transducers, an electrochemical biosensing tool, named E-CRISPR (Dai et al., 2019), has been proposed for the extended application of Cas12a diagnostics. In this method (Fig. 10 A), modified ssDNA, designed with a methylene blue (MB) tag at the 3′ terminus as the redox label and a thiol group at the 5′ terminus for covalent immobilization onto the gold electrode, was employed as the electrochemical reporter. In the presence of targets, collateral cleavage activity of Cas12a was activated to cleave the ssDNA reporters, thereby unleashing the MB tag from the gold electrode surface and resulting in a peak current. Therefore, the electrical signal could be monitored as a specific proxy for the presence of DNA targets. In this format, E-CRISPR could directly detect unamplified HPV16 DNA at concentrations as low as 50 pM, which was evidently lower than the previously published nanomolar LOD of CRISPR-based nucleic acid detection without preamplification (Gootenberg et al. 2017, 2018). Later, Zhang and his colleagues presented another version of the electrochemical system by replacing linear ssDNA reporters with interfacial hairpin DNA (hpDNA) reporters (Zhang et al., 2020). This stem-loop structure not only brought the MB tag closer to the electrode, thereby increasing the electron transfer rate constant, but also fully exposed the single-strand loop region of hairpin DNA to interference with Cas12a cleavage, collectively leading to an improved detection sensitivity (Fig. 10B). In addition to electrochemistry technology, microfluidics and nanotechnology have also been integrated with the Cas12a platform, and a magnet-assisted V-chip (MAV-chip) has been fabricated for multiple detection and quantification of SNVs (Shao et al., 2019). In this work (Fig. 10C), catalase/platinum nanoparticles (PtNPs), conjugated with magnetic beads by the ssDNA linker, was introduced as a signal transducer, which could be degraded by DNA-target-triggered collateral cleavage activity of Cas12a to release PtNPs. By applying the reaction system onto the volumetric bar-chart chip and using a magnet to pull down magnetic beads, PtNPs were separated and then slid into the hydrogen peroxide reaction wells to generate oxygen that could push red ink forward. In the absence of the target, PtNPs were pulled down together with magnetic beads, therefore, oxygen would not be generated and ink advancement would not occur. Therefore, the distance of ink movement could be measured to quantify of the DNA targets. Employing a multichannel setting and well-designed crRNA, this method enabled the multiple detection of cancer-related SNPs with allelic fractions as low as 0.01% without amplification.
Fig. 10.
Cas12a-based electrochemistry and microfluidics tools for diagnostic applications. (A) Electrochemical detection of DNA targets using linear ssDNA reporters. (B) Electrochemical detection of DNA targets using hairpin ssDNA reporters. (C) Multiple detection and quantification of SNVs based on magnet-assisted V-chip.
The discovery of more orthogonal Cas12 effectors exhibiting collateral cleavage has led to the possibilities of transforming the existing platforms and enabling potentially superior alternatives. Li and colleagues, for example, have developed a one-step HOLMESv2 (Li et al., 2019a) method based on the thermophilic AacCas12b protein. The thermal stability of AacCas12b offers HOLMESv2 compatibility with RT-LAMP to directly amplify and subsequently detect target RNA isothermally at 55 °C, and it omits the intervening reverse transcription step and thereby provides an elegant solution for simple RNA detection with the Cas12b system (Fig. 9C). In addition, HOLMESv2 has been proven to have the ability to accurately quantify DNA targets, as well as their methylation modification, collectively providing opportunities to technologically expand their scope of detection to diverse utilities. However, this method also has some disadvantages, such as high reaction temperature, sequence limitation for SNP detection, and potentially compromised sensitivity. Recently, an AaCas12b-mediated CDetection (Teng et al., 2019) method capable of remarkably sensitive dsDNA detection with single-base specificity was developed (Fig. 9D). Although the observation that AaCas12b (Teng et al., 2018) performs DNA-recognition-triggered collateral cleavage draws obvious parallels to that of Cas12a orthologs, the high cleavage activity of the AaCas12b effector has endowed CDetection with elevated signal amplification capability and improved sensitivity. Concretely, CDetection presented a minimum LOD of 0.1 aM, which was revealed by tests performed on HPV dsDNA diluted with human genomic DNA. Furthermore, combined with tgRNA, a programmed gRNA that harbors a mismatch in the spacer sequence that enables screening of targets with a single-nucleotide resolution, CDetection could clearly discriminate between 3232A > G SNP sites and wild-type alleles with allelic fractions as low as 1%. These data have demonstrated the unique potential of Cas12b, indicating prospects for robust and versatile nucleic acid detection methodologies. It is noteworthy that recent reports integrating Cas12b detection with lateral-flow biosensors have displayed their potential for providing ultrasensitive and specific DNA detection with minimal equipment (Mukama et al. 2020a, 2020b), further suggesting the significant value of Cas12b effectors for diagnostic applications.
In addition to the Cas12 and Cas13 systems, the miniature Cas14 effector also has similar collateral cleavage and the Cas14-DETECTR (Harrington et al., 2018) platform has been established for high-fidelity single-nucleotide polymorphism genotyping. However, the collateral cleavage of Cas14 is triggered by ssDNA-recognition events; therefore, an extra step generating ssDNA from dsDNA targets is required before the detection procedure. This special property of Cas14 might add to the complexity of its applications; therefore, its advantages in diagnostic applications have not been clearly presented to date.
4. Conclusions and future perspectives
Genomic information is becoming increasingly vital for personalized medicine, infectious disease prevention and many other medical implementations. At present, based on approaches that have been reported, CRISPR is providing an opportunity to facilitate superior alternatives or improvements by allowing comprehensive insight into different types of genetic alterations in a more efficient and convenient format. In this review, we have discussed the development of CRISPR-based nucleic acid detection by highlighting representative works. The most essential distinctions between these methods are the intrinsic properties of the different Cas effectors that they use, resulting in different schemes and deployments. As the inherent characteristic of a gene editing tool, CRISPR targeted recognition activity (mainly derived from Cas9 variants) has enabled a wide range of applications, including biosensing assays, imaging assays and target enrichment for NGS. The discovery of Cas effectors with unique collateral cleavage activities has suggested elegant and efficient signal amplification systems for ultrasensitive and portable nucleic acid detection. Despite having different uses, these strategies share several characteristics, such as being fast, isothermal, and flexible, because almost all CRISPR systems can be programmed for repurposed applications and function in a short period, such as minutes, under mild conditions, which is the most prominent advantage of CRISPR-based nucleic acid detection tools.
Although remarkable advances have been made, CRISPR-based diagnostic tools still face a number of challenges. (1) Improved specificity is inevitably required for almost all detection methods, which is particularly concerning in the context of CRISPR-based detection due to the inherent off-target effect. For example, LshC2c2 cannot recognize base-pair mismatches in the spacer-terminal region (Abudayyeh et al., 2016), Cas9-mediated cleavage is highly tolerant to mismatches outside the PAM-proximal 5–12 bp seed regions (Wu et al., 2014), and dCas9 off-target binding is even more chaotic (Kuscu et al., 2014). Off-target recognition may weaken the analysis specificity and might cause false positive signals. Although reports have revealed that the off-target cleavage efficiency in vivo is correlated to the Cas9 expression level (O'Geen et al., 2015), it is not feasible to obtain improved specificity by modulating Cas9 inputs due to the lack of adequate in vitro data and the risk of compromised analytical sensitivity. Therefore, future work should focus on high-fidelity nucleic acid detection. In the past few years, Cas9 variants with reduced off-target cleavage, such as SpCas9-HF1 (Kleinstiver et al., 2016), eSpCas9(1.1) (Slaymaker et al., 2016) and HypaCas9 (Chen et al., 2017), have been developed, thereby providing potential solutions. Structural modulations of sgRNA have also enabled precise targeting (Chavez et al., 2018; Kocak et al., 2019). In addition, scheme designs integrating technologies with inherent high specificity, such as toehold-mediated strand displacement (TMSD) (Zhang and Seelig, 2011) and padlock probes, may further suppress the generation of off-target signals. (2) The requirement of PAM or PFS for CRISPR targeted recognition has contributed to sequence limitation, resulting in a reduced range of sequences that can be detected. For methods such as the PC reporter system (Zhang et al., 2017) and CasPLA (Zhang et al., 2018) that require two adjacent PAMs, their realization of high specificity is at the expense of wide applicability. To expand the scope of detection, Cas variants with altered PAM specificities have been generated (Shmakov et al., 2017), such as SpG (Walton et al., 2020), which recognizes NGN PAMs; SpRY (Walton et al., 2020), which recognizes NRN > NYN PAMs; xCas9 (Hu et al., 2018), which recognizes NG/GAA/GAT PAMs; ScCas9 (Chatterjee et al., 2018), which recognizes NNG PAMs; and AsCas12a variants (Kleinstiver et al., 2019), which recognizes VTTV/TTTT/TTCN/TATV PAMs. Such a variety of PAM specificities has provided the opportunity to remove sequence constraints from CRISPR-based diagnostic tools. (3) The portability conferred by minimized equipment requirements is the noticeable highlight of methods based on CRISPR collateral cleavage activities that possess strong prospects for field deployment, especially for stemming epidemic outbreaks in resource-limited regions. However, most of these methods currently have low throughput; therefore, the workload and possible cross-contamination between samples are still a challenge when dealing with large-scale tests, such as COVID-19 outbreaks. In this context, a multidisciplinary scheme that integrates CRISPR detection with engineering, microelectronics, and miniaturization might increase detection throughput and automation. Since HUDSON has provided biotechnological support (Myhrvold et al., 2018), a multichannel all-in-one handheld device powered by tiny batteries to achieve the whole process from sample to answer should be fabricated to enable real CRISPR POCT. (4) CRISPR-mediated in situ imaging technology has already achieved single-base analysis at the single-cell level, but genomic DNA imaging, which is a real concern, still suffers from inefficiency and failure to meet the standards required for practicable applications. Various measures, including the application of miniaturized Cas9 variants (Kim et al., 2017), optimization of cell pretreatment procedures, and reduction of required reaction steps might be helpful for increasing labeling efficiency.
Although CRISPR diagnostic tools have exhibited clear signs of progress, there are few commercially available kits or devices in clinical use due to the complexity of translating emerging technologies into practice. However, with the emergence of newly developed applications and discovery of more putative Cas orthologs, CRISPR technology has been developed to facilitate numerous diagnostic solutions that can serve human health and thus presents intriguing potential to impact patients, clinicians, and clinical laboratories.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China [81974319]; the National Key R&D Program of China [2018YFE0201604]; and Beijing Dongcheng District Outstanding Talents Team Program [2019DCT-M-01].
References
- Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B., Shmakov S., Makarova K.S., Semenova E., Minakhin L., Severinov K., Regev A., Lander E.S., Koonin E.V., Zhang F. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353(6299):aaf5573. doi: 10.1126/science.aaf5573. (New York, N.Y.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari S.P., Meng S., Wu Y.J., Mao Y.P., Ye R.X., Wang Q.Z., Sun C., Sylvia S., Rozelle S., Raat H., Zhou H. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect. Dis. Poverty. 2020;9(1):29. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K.R., Haeussler M., Watanabe C., Janakiraman V., Lund J., Modrusan Z., Stinson J., Bei Q., Buechler A., Yu C., Thamminana S.R., Tam L., Sowick M.A., Alcantar T., O'Neil N., Li J., Ta L., Lima L., Roose-Girma M., Rairdan X., Durinck S., Warming S. CRISPR off-target analysis in genetically engineered rats and mice. Nat. Methods. 2018;15(7):512–514. doi: 10.1038/s41592-018-0011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya M., Shergill I.S., Williamson M., Gommersall L., Arya N., Patel H.R. Basic principles of real-time quantitative PCR. Expert Rev. Mol. Diagn. 2005;5(2):209–219. doi: 10.1586/14737159.5.2.209. [DOI] [PubMed] [Google Scholar]
- Ballester L.Y., Luthra R., Kanagal-Shamanna R., Singh R.R. Advances in clinical next-generation sequencing: target enrichment and sequencing technologies. Expert Rev. Mol. Diagn. 2016;16(3):357–372. doi: 10.1586/14737159.2016.1133298. [DOI] [PubMed] [Google Scholar]
- Blumenthal G.M., Mansfield E., Pazdur R. Next-generation sequencing in oncology in the era of precision medicine. JAMA oncol. 2016;2(1):13–14. doi: 10.1001/jamaoncol.2015.4503. [DOI] [PubMed] [Google Scholar]
- Bruch R., Baaske J., Chatelle C., Meirich M., Madlener S., Weber W., Dincer C., Urban G.A. CRISPR/Cas13a-Powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv. Mater.(Deerfield Beach, Fla.) 2019;31(51) doi: 10.1002/adma.201905311. [DOI] [PubMed] [Google Scholar]
- Bustin S.A., Mueller R. Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin. Sci. 2005;109(4):365–379. doi: 10.1042/CS20050086. (London, England : 1979) [DOI] [PubMed] [Google Scholar]
- Chatterjee P., Jakimo N., Jacobson J.M. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci. Adv. 2018;4(10) doi: 10.1126/sciadv.aau0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A., Pruitt B.W., Tuttle M., Shapiro R.S., Cecchi R.J., Winston J., Turczyk B.M., Tung M., Collins J.J., Church G.M. Precise Cas9 targeting enables genomic mutation prevention. Proc. Natl. Acad. Sci. U.S.A. 2018;115(14):3669–3673. doi: 10.1073/pnas.1718148115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.S., Dagdas Y.S., Kleinstiver B.P., Welch M.M., Sousa A.A., Harrington L.B., Sternberg S.H., Joung J.K., Yildiz A., Doudna J.A. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550(7676):407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science (New York, N.Y.) 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S., Kim J.S. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24(1):132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science (New York, N.Y.) 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y., Somoza R.A., Wang L., Welter J.F., Li Y., Caplan A.I., Liu C.C. Exploring the trans-cleavage activity of CRISPR-cas12a (cpf1) for the development of a universal electrochemical biosensor. Angew. Chem. 2019;58(48):17399–17405. doi: 10.1002/anie.201910772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Shi X., Tjian R., Lionnet T., Singer R.H. CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc. Natl. Acad. Sci. U.S.A. 2015;112(38):11870–11875. doi: 10.1073/pnas.1515692112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Lei J., Ding L., Wen Y., Ju H., Zhang X. MicroRNA: function, detection, and bioanalysis. Chem. Rev. 2013;113(8):6207–6233. doi: 10.1021/cr300362f. [DOI] [PubMed] [Google Scholar]
- East-Seletsky A., O'Connell M.R., Knight S.C., Burstein D., Cate J.H., Tjian R., Doudna J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538(7624):270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger D.S., Aisner D.L., Wood D.E., Akerley W., Bauman J., Chang J.Y., Chirieac L.R., D'Amico T.A., Dilling T.J., Dobelbower M., Govindan R., Gubens M.A., Hennon M., Horn L., Lackner R.P., Lanuti M., Leal T.A., Lilenbaum R., Lin J., Loo B.W., Martins R., Otterson G.A., Patel S.P., Reckamp K., Riely G.J., Schild S.E., Shapiro T.A., Stevenson J., Swanson S.J., Tauer K., Yang S.C., Gregory K., Hughes M. NCCN guidelines insights: non-small cell lung cancer, version 5.2018. J. Natl. Compr. Canc. Netw. : J. Natl. Compr. Canc. Netw. 2018;16(7):807–821. doi: 10.6004/jnccn.2018.0062. [DOI] [PubMed] [Google Scholar]
- Femino A.M., Fay F.S., Fogarty K., Singer R.H. Visualization of single RNA transcripts in situ. Science (New York, N.Y.) 1998;280(5363):585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gustafsdottir S.M., Ostman A., Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002;20(5):473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- Gabrieli T., Sharim H., Fridman D., Arbib N., Michaeli Y., Ebenstein Y. Selective nanopore sequencing of human BRCA1 by Cas9-assisted targeting of chromosome segments (CATCH) Nucleic Acids Res. 2018;46(14):e87. doi: 10.1093/nar/gky411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2012;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., Myhrvold C., Bhattacharyya R.P., Livny J., Regev A., Koonin E.V., Hung D.T., Sabeti P.C., Collins J.J., Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. (New York, N.Y.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444. doi: 10.1126/science.aaq0179. (New York, N.Y.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradishar W.J., Anderson B.O., Balassanian R., Blair S.L., Burstein H.J., Cyr A., Elias A.D., Farrar W.B., Forero A., Giordano S.H., Goetz M.P., Goldstein L.J., Isakoff S.J., Lyons J., Marcom P.K., Mayer I.A., McCormick B., Moran M.S., O'Regan R.M., Patel S.A., Pierce L.J., Reed E.C., Salerno K.E., Schwartzberg L.S., Sitapati A., Smith K.L., Smith M.L., Soliman H., Somlo G., Telli M.L., Ward J.H., Kumar R., Shead D.A. Breast cancer, version 4.2017, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc. Netw. : J. Natl. Compr. Canc. Netw. 2018;16(3):310–320. doi: 10.6004/jnccn.2018.0012. [DOI] [PubMed] [Google Scholar]
- Gray K.J., Wilkins-Haug L.E. Have we done our last amniocentesis? Updates on cell-free DNA for Down syndrome screening. Pediatr. Radiol. 2018;48(4):461–470. doi: 10.1007/s00247-017-3958-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Crawford E.D., O'Donovan B.D., Wilson M.R., Chow E.D., Retallack H., DeRisi J.L. Depletion of Abundant Sequences by Hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016;17:41. doi: 10.1186/s13059-016-0904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guk K., Keem J.O., Hwang S.G., Kim H., Kang T., Lim E.K., Jung J. A facile, rapid and sensitive detection of MRSA using a CRISPR-mediated DNA FISH method, antibody-like dCas9/sgRNA complex. Biosens. Bioelectron. 2017;95:67–71. doi: 10.1016/j.bios.2017.04.016. [DOI] [PubMed] [Google Scholar]
- Hagen R.M., Seegmuller I., Navai J., Kappstein I., Lehn N., Miethke T. Development of a real-time PCR assay for rapid identification of methicillin-resistant Staphylococcus aureus from clinical samples. Int. J. Med. Microbiol. : IJMM. 2005;295(2):77–86. doi: 10.1016/j.ijmm.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Hajian R., Balderston S., Tran T., deBoer T., Etienne J., Sandhu M., Wauford N.A., Chung J.Y., Nokes J., Athaiya M., Paredes J., Peytavi R., Goldsmith B., Murthy N., Conboy I.M., Aran K. Detection of unamplified target genes via CRISPR-Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019;3(6):427–437. doi: 10.1038/s41551-019-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L.B., Burstein D., Chen J.S., Paez-Espino D., Ma E., Witte I.P., Cofsky J.C., Kyrpides N.C., Banfield J.F., Doudna J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science (New York, N.Y.) 2018;362(6416):839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., Cradick T.J., Marraffini L.A., Bao G., Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J.H., Miller S.M., Geurts M.H., Tang W., Chen L., Sun N., Zeina C.M., Gao X., Rees H.A., Lin Z., Liu D.R. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556(7699):57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Zhou X., Wang H., Xing D. Clustered regularly interspaced short palindromic repeats/cas9 triggered isothermal amplification for site-specific nucleic acid detection. Anal. Chem. 2018;90(3):2193–2200. doi: 10.1021/acs.analchem.7b04542. [DOI] [PubMed] [Google Scholar]
- Jiang F., Zhou K., Ma L., Gressel S., Doudna J.A. STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348(6242):1477–1481. doi: 10.1126/science.aab1452. (New York, N.Y.) [DOI] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, N.Y.) 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Jiang F., Taylor D.W., Sternberg S.H., Kaya E., Ma E., Anders C., Hauer M., Zhou K., Lin S., Kaplan M., Iavarone A.T., Charpentier E., Nogales E., Doudna J.A. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science (New York, N.Y.) 2014;343(6176):1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore M.M., Lundgren M., van Duijn E., Bultema J.B., Westra E.R., Waghmare S.P., Wiedenheft B., Pul U., Wurm R., Wagner R., Beijer M.R., Barendregt A., Zhou K., Snijders A.P., Dickman M.J., Doudna J.A., Boekema E.J., Heck A.J., van der Oost J., Brouns S.J. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011;18(5):529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- Josephs E.A., Kocak D.D., Fitzgibbon C.J., McMenemy J., Gersbach C.A., Marszalek P.E. Structure and specificity of the RNA-guided endonuclease Cas9 during DNA interrogation, target binding and cleavage. Nucleic Acids Res. 2016;44(5):2474. doi: 10.1093/nar/gkv1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J.K., Sander J.D. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013;14(1):49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebschull J.M., Zador A.M. Sources of PCR-induced distortions in high-throughput sequencing data sets. Nucleic Acids Res. 2015;43(21):e143. doi: 10.1093/nar/gkv717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Koo T., Park S.W., Kim D., Kim K., Cho H.Y., Song D.W., Lee K.J., Jung M.H., Kim S., Kim J.H., Kim J.H., Kim J.S. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017;8:14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B.P., Sousa A.A., Walton R.T., Tak Y.E., Hsu J.Y., Clement K., Welch M.M., Horng J.E., Malagon-Lopez J., Scarfo I., Maus M.V., Pinello L., Aryee M.J., Joung J.K. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019;37(3):276–282. doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S.C., Xie L., Deng W., Guglielmi B., Witkowsky L.B., Bosanac L., Zhang E.T., El Beheiry M., Masson J.B., Dahan M., Liu Z., Doudna J.A., Tjian R. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 2015;350(6262):823–826. doi: 10.1126/science.aac6572. (New York, N.Y.) [DOI] [PubMed] [Google Scholar]
- Kocak D.D., Josephs E.A., Bhandarkar V., Adkar S.S., Kwon J.B., Gersbach C.A. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat. Biotechnol. 2019;37(6):657–666. doi: 10.1038/s41587-019-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C., Arslan S., Singh R., Thorpe J., Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 2014;32(7):677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- Lee H., Choi J., Jeong E., Baek S., Kim H.C., Chae J.H., Koh Y., Seo S.W., Kim J.S., Kim S.J. dCas9-mediated nanoelectrokinetic direct detection of target gene for liquid biopsy. Nano Lett. 2018;18(12):7642–7650. doi: 10.1021/acs.nanolett.8b03224. [DOI] [PubMed] [Google Scholar]
- Lee J., Lim H., Jang H., Hwang B., Lee J.H., Cho J., Lee J.H., Bang D. CRISPR-Cap: multiplexed double-stranded DNA enrichment based on the CRISPR system. Nucleic Acids Res. 2019;47(1):e1. doi: 10.1093/nar/gky820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei C., Li S.Y., Liu J.K., Zheng X., Zhao G.P., Wang J. The CCTL (Cpf1-assisted Cutting and Taq DNA ligase-assisted Ligation) method for efficient editing of large DNA constructs in vitro. Nucleic Acids Res. 2017;45(9):e74. doi: 10.1093/nar/gkx018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levsky J.M., Singer R.H. Fluorescence in situ hybridization: past, present and future. J. Cell Sci. 2003;116(Pt 14):2833–2838. doi: 10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- Li S.Y., Cheng Q.X., Wang J.M., Li X.Y., Zhang Z.L., Gao S., Cao R.B., Zhao G.P., Wang J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018;4:20. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li S., Wu N., Wu J., Wang G., Zhao G., Wang J. HOLMESv2: a CRISPR-cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 2019;8(10):2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- Li Y., Li S., Wang J., Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37(7):730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- Maamar H., Raj A., Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science (New York, N.Y.) 2007;317(5837):526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H., Horvath P., Moineau S., Mojica F.J., Terns R.M., Terns M.P., White M.F., Yakunin A.F., Garrett R.A., van der Oost J., Backofen R., Koonin E.V. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015;13(11):722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malan V., Bussieres L., Winer N., Jais J.P., Baptiste A., Le Lorc'h M., Elie C., O'Gorman N., Fries N., Houfflin-Debarge V., Sentilhes L., Vekemans M., Ville Y., Salomon L.J. Effect of cell-free DNA screening vs direct invasive diagnosis on miscarriage rates in women with pregnancies at high risk of trisomy 21: a randomized clinical trial. Jama. 2018;320(6):557–565. doi: 10.1001/jama.2018.9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31(9):833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. (New York, N.Y.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamanova L., Coffey A.J., Scott C.E., Kozarewa I., Turner E.H., Kumar A., Howard E., Shendure J., Turner D.J. Target-enrichment strategies for next-generation sequencing. Nat. Methods. 2010;7(2):111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- Mekler V., Minakhin L., Semenova E., Kuznedelov K., Severinov K. Kinetics of the CRISPR-Cas9 effector complex assembly and the role of 3'-terminal segment of guide RNA. Nucleic Acids Res. 2016;44(6):2837–2845. doi: 10.1093/nar/gkw138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukama O., Wu J., Li Z., Liang Q., Yi Z., Lu X., Liu Y., Liu Y., Hussain M., Makafe G.G., Liu J., Xu N., Zeng L. An ultrasensitive and specific point-of-care CRISPR/Cas12 based lateral flow biosensor for the rapid detection of nucleic acids. Biosens. Bioelectron. 2020;159:112143. doi: 10.1016/j.bios.2020.112143. [DOI] [PubMed] [Google Scholar]
- Mukama O., Yuan T., He Z., Li Z., Habimana J.d.D., Hussain M., Li W., Yi Z., Liang Q., Zeng L. A high fidelity CRISPR/Cas12a based lateral flow biosensor for the detection of HPV16 and HPV18. Sensor. Actuator. B Chem. 2020;316:128119. [Google Scholar]
- Myhrvold C., Freije C.A., Gootenberg J.S., Abudayyeh O.O., Metsky H.C., Durbin A.F., Kellner M.J., Tan A.L., Paul L.M., Parham L.A., Garcia K.F., Barnes K.G., Chak B., Mondini A., Nogueira M.L., Isern S., Michael S.F., Lorenzana I., Yozwiak N.L., MacInnis B.L., Bosch I., Gehrke L., Zhang F., Sabeti P.C. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360(6387):444–448. doi: 10.1126/science.aas8836. (New York, N.Y.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachmanson D., Lian S., Schmidt E.K., Hipp M.J., Baker K.T., Zhang Y., Tretiakova M., Loubet-Senear K., Kohrn B.F., Salk J.J., Kennedy S.R., Risques R.A. Targeted genome fragmentation with CRISPR/Cas9 enables fast and efficient enrichment of small genomic regions and ultra-accurate sequencing with low DNA input (CRISPR-DS) Genome Res. 2018;28(10):1589–1599. doi: 10.1101/gr.235291.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M., Malmgren H., Samiotaki M., Kwiatkowski M., Chowdhary B.P., Landegren U. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science (New York, N.Y.) 1994;265(5181):2085–2088. doi: 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Flaherty B.M., Li Y., Tao Y., Paden C.R., Queen K., Zhang J., Dinwiddie D.L., Gross S.M., Schroth G.P., Tong S. Comprehensive viral enrichment enables sensitive respiratory virus genomic identification and analysis by next generation sequencing. Genome Res. 2018;28(6):869–877. doi: 10.1101/gr.226316.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Geen H., Henry I.M., Bhakta M.S., Meckler J.F., Segal D.J. A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Res. 2015;43(6):3389–3404. doi: 10.1093/nar/gkv137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee K., Green A.A., Takahashi M.K., Braff D., Lambert G., Lee J.W., Ferrante T., Ma D., Donghia N., Fan M., Daringer N.M., Bosch I., Dudley D.M., O'Connor D.H., Gehrke L., Collins J.J. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165(5):1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- Pickar-Oliver A., Gersbach C.A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019;20(8):490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Practice bulletin No. 162: prenatal diagnostic testing for genetic disorders. Obstet. Gynecol. 2016;127(5):e108–122. doi: 10.1097/AOG.0000000000001405. [DOI] [PubMed] [Google Scholar]
- Practice bulletin No. 163: screening for fetal aneuploidy. Obstet. Gynecol. 2016;127(5):e123–137. doi: 10.1097/AOG.0000000000001406. [DOI] [PubMed] [Google Scholar]
- Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P., Park M., Alfson K.J., Tamhankar M., Carrion R., Patterson J.L., Griffiths A., He Q., Yildiz A., Mathies R., Du K. Rapid and fully microfluidic Ebola virus detection with CRISPR-cas13a. ACS Sens. 2019;4(4):1048–1054. doi: 10.1021/acssensors.9b00239. [DOI] [PubMed] [Google Scholar]
- Qiu X.Y., Zhu L.Y., Zhu C.S., Ma J.X., Hou T., Wu X.M., Xie S.S., Min L., Tan D.A., Zhang D.Y., Zhu L. Highly effective and low-cost MicroRNA detection with CRISPR-cas9. ACS Synth. Biol. 2018;7(3):807–813. doi: 10.1021/acssynbio.7b00446. [DOI] [PubMed] [Google Scholar]
- Quan J., Langelier C., Kuchta A., Batson J., Teyssier N., Lyden A., Caldera S., McGeever A., Dimitrov B., King R., Wilheim J., Murphy M., Ares L.P., Travisano K.A., Sit R., Amato R., Mumbengegwi D.R., Smith J.L., Bennett A., Gosling R., Mourani P.M., Calfee C.S., Neff N.F., Chow E.D., Kim P.S., Greenhouse B., DeRisi J.L., Crawford E.D. FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Res. 2019;47(14):e83. doi: 10.1093/nar/gkz418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008;5(10):877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samorodnitsky E., Jewell B.M., Hagopian R., Miya J., Wing M.R., Lyon E., Damodaran S., Bhatt D., Reeser J.W., Datta J., Roychowdhury S. Evaluation of hybridization capture versus amplicon-based methods for whole-exome sequencing. Hum. Mutat. 2015;36(9):903–914. doi: 10.1002/humu.22825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y., Zhou X., Huang R., Xing D. High-fidelity and rapid quantification of miRNA combining crRNA programmability and CRISPR/Cas13a trans-cleavage activity. Anal. Chem. 2019;91(8):5278–5285. doi: 10.1021/acs.analchem.9b00073. [DOI] [PubMed] [Google Scholar]
- Shao N., Han X., Song Y., Zhang P., Qin L. CRISPR-Cas12a coupled with platinum nanoreporter for visual quantification of SNVs on a volumetric bar-chart chip. Anal. Chem. 2019;91(19):12384–12391. doi: 10.1021/acs.analchem.9b02925. [DOI] [PubMed] [Google Scholar]
- Shmakov S., Smargon A., Scott D., Cox D., Pyzocha N., Yan W., Abudayyeh O.O., Gootenberg J.S., Makarova K.S., Wolf Y.I., Severinov K., Zhang F., Koonin E.V. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017;15(3):169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science (New York, N.Y.) 2016;351(6268):84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg O., Gullberg M., Jarvius M., Ridderstrale K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., Landegren U. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006;3(12):995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Sun Y., Ruivenkamp C.A., Hoffer M.J., Vrijenhoek T., Kriek M., van Asperen C.J., den Dunnen J.T., Santen G.W. Next-generation diagnostics: gene panel, exome, or whole genome? Hum. Mutat. 2015;36(6):648–655. doi: 10.1002/humu.22783. [DOI] [PubMed] [Google Scholar]
- Surrey L.F., MacFarland S.P., Chang F., Cao K., Rathi K.S., Akgumus G.T., Gallo D., Lin F., Gleason A., Raman P., Aplenc R., Bagatell R., Minturn J., Mosse Y., Santi M., Tasian S.K., Waanders A.J., Sarmady M., Maris J.M., Hunger S.P., Li M.M. Clinical utility of custom-designed NGS panel testing in pediatric tumors. Genome Med. 2019;11(1):32. doi: 10.1186/s13073-019-0644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F., Cui T., Feng G., Guo L., Xu K., Gao Q., Li T., Li J., Zhou Q., Li W. Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov. 2018;4:63. doi: 10.1038/s41421-018-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F., Guo L., Cui T., Wang X.G., Xu K., Gao Q., Zhou Q., Li W. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biol. 2019;20(1):132. doi: 10.1186/s13059-019-1742-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S.Q., Wyvekens N., Khayter C., Foden J.A., Thapar V., Reyon D., Goodwin M.J., Aryee M.J., Joung J.K. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014;32(6):569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko J., Myer V.E., Hsu P.D. Methods for optimizing CRISPR-cas9 genome editing specificity. Mol. Cell. 2016;63(3):355–370. doi: 10.1016/j.molcel.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov F.D., Rebar E.J., Holmes M.C., Zhang H.S., Gregory P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11(9):636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- van Dijk E.L., Auger H., Jaszczyszyn Y., Thermes C. Ten years of next-generation sequencing technology. Trends Genet. : TIG (Trends Genet.) 2014;30(9):418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Walton R.T., Christie K.A., Whittaker M.N., Kleinstiver B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368(6488):290–296. doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Flanagan J., Su N., Wang L.C., Bui S., Nielson A., Wu X., Vo H.T., Ma X.J., Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. : J. Mod. Dynam. 2012;14(1):22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Liu Y., Sun H.H., Yin B.C., Ye B.C. An RNA-guided Cas9 nickase-based method for universal isothermal DNA amplification. Angew. Chem. 2019;58(16):5382–5386. doi: 10.1002/anie.201901292. [DOI] [PubMed] [Google Scholar]
- Wang M., Chen K., Wu Q., Peng R., Zhang R., Li J. RCasFISH: CRISPR/dCas9-Mediated in situ imaging of mRNA transcripts in fixed cells and tissues. Anal. Chem. 2020;92(3):2468–2475. doi: 10.1021/acs.analchem.9b03797. [DOI] [PubMed] [Google Scholar]
- Wang R., Zhao X., Chen X., Qiu X., Qing G., Zhang H., Zhang L., Hu X., He Z., Zhong D., Wang Y., Luo Y. Rolling circular amplification (RCA)-Assisted CRISPR/Cas9 cleavage (RACE) for highly specific detection of multiple extracellular vesicle MicroRNAs. Anal. Chem. 2020;92(2):2176–2185. doi: 10.1021/acs.analchem.9b04814. [DOI] [PubMed] [Google Scholar]
- Wang X., Xiong E., Tian T., Cheng M., Lin W., Wang H., Zhang G., Sun J., Zhou X. Clustered regularly interspaced short palindromic repeats/cas9-mediated lateral flow nucleic acid assay. ACS Nano. 2020;14(2):2497–2508. doi: 10.1021/acsnano.0c00022. [DOI] [PubMed] [Google Scholar]
- Wolff A.C., Hammond M.E.H., Allison K.H., Harvey B.E., Mangu P.B., Bartlett J.M.S., Bilous M., Ellis I.O., Fitzgibbons P., Hanna W., Jenkins R.B., Press M.F., Spears P.A., Vance G.H., Viale G., McShane L.M., Dowsett M. Human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of American pathologists clinical practice guideline focused update. J. Clin. Oncol. : Off. J. Am. Soc. Clin. Oncol. 2018;36(20):2105–2122. doi: 10.1200/JCO.2018.77.8738. [DOI] [PubMed] [Google Scholar]
- Wu X., Scott D.A., Kriz A.J., Chiu A.C., Hsu P.D., Dadon D.B., Cheng A.W., Trevino A.E., Konermann S., Chen S., Jaenisch R., Zhang F., Sharp P.A. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 2014;32(7):670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.Y., Seelig G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 2011;3(2):103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Qian L., Wei W., Wang Y., Wang B., Lin P., Liu W., Xu L., Li X., Liu D., Cheng S., Li J., Ye Y., Li H., Zhang X., Dong Y., Zhao X., Liu C., Zhang H.M., Ouyang Q., Lou C. Paired design of dCas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains. ACS Synth. Biol. 2017;6(2):211–216. doi: 10.1021/acssynbio.6b00215. [DOI] [PubMed] [Google Scholar]
- Zhang K., Deng R., Teng X., Li Y., Sun Y., Ren X., Li J. Direct visualization of single-nucleotide variation in mtDNA using a CRISPR/Cas9-Mediated proximity ligation assay. J. Am. Chem. Soc. 2018;140(36):11293–11301. doi: 10.1021/jacs.8b05309. [DOI] [PubMed] [Google Scholar]
- Zhang D., Yan Y., Que H., Yang T., Cheng X., Ding S., Zhang X., Cheng W. CRISPR/Cas12a-Mediated interfacial cleaving of hairpin DNA reporter for electrochemical nucleic acid sensing. ACS Sens. 2020;5(2):557–562. doi: 10.1021/acssensors.9b02461. [DOI] [PubMed] [Google Scholar]
- Zhou W., Hu L., Ying L., Zhao Z., Chu P.K., Yu X.F. A CRISPR-Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat. Commun. 2018;9(1):5012. doi: 10.1038/s41467-018-07324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]