Abstract
Genetic background commonly modifies the effects of mutations. We discovered that worms mutant for the canonical rol-1 gene, identified by Brenner in 1974, do not roll in the genetic background of the wild strain CB4856. Using linkage mapping, association analysis and gene editing, we determined that N2 carries an insertion in the collagen gene col-182 that acts as a recessive enhancer of rol-1 rolling. From population and comparative genomics, we infer the insertion is derived in N2 and related laboratory lines, likely arising during the domestication of Caenorhabditis elegans, and breaking a conserved protein. The ancestral version of col-182 also modifies the phenotypes of four other classical cuticle mutant alleles, and the effects of natural genetic variation on worm shape and locomotion. These results underscore the importance of genetic background and the serendipity of Brenner’s choice of strain.
Keywords: collagen, cuticle, genetic interaction, genetic background
Since Morgan’s first white-eyed fly, forward genetics has been one of our most powerful tools for discovering biological mechanisms. In 1974, Sydney Brenner introduced geneticists to C. elegans, an experimental organism with properties ideal for probing the molecular basis of development and neurobiology (Brenner 1974). Brenner began by isolating mutants with conspicuous effects under the evocative nomenclature of Dumpy, Squat, Long, Blistered, and Roller phenotypes, including a single allele of rol-1. This mutation causes helical twisting of the adult worm’s cuticle, which manifests most obviously as sinusoidal motion along the short axis of locomoting animals and consequent gyration on the uniform surface of an agar plate.
rol-1, and several other genes from Brenner’s first screen, opened the door not only to linkage mapping in C. elegans but also to decades of productive work on the worm cuticle. The cuticle is a complex structure, made primarily of cross-linked collagens generated anew with each larval molt (Page and Johnstone 2007). It plays an integral structural role as both barrier and morphological scaffold for muscle attachment. Epistasis analysis of collagens and collagen-modifying enzymes represents a landmark example of the power of transmission genetics to reveal molecular and developmental mechanisms (Higgins and Hirsh 1977; Cox et al. 1980; Kramer and Johnson 1993; McMahon et al. 2003).
Brenner’s original screen, and the vast majority of subsequent research in C. elegans, took place in the genetic context of the inbred reference strain, N2. Over the past decade, researchers have discovered that N2 evolved during its adaptation to laboratory conditions, and that wild isolates of C. elegans differ from the lab strain in diverse and substantive ways (Hodgkin and Doniach 1997; de Bono and Bargmann 1998; McGrath et al. 2009; Duveau and Félix 2012; Andersen et al. 2014; Sterken et al. 2015; Large et al. 2016; Gimond et al. 2019). Critically, the effects of mutations are often modified by genetic background. This kind of background dependence both complicates experimental analyses and underlies important genetic phenomena such as variable penetrance of Mendelian diseases in humans (Summers 1996; Scriver and Waters 1999; Dipple and McCabe 2000; Gibson and Dworkin 2004; Paaby and Rockman 2014; Paaby and Gibson 2016).
While using a rol-1 allele as a visible marker for genetic mapping experiments, we discovered that its rolling phenotype is substantially suppressed by a wild strain background. We mapped the major-effect locus responsible for this suppression, finding that N2 carries a derived insertion in col-182, a collagen gene with no known mutational effects. The ancestral allele of this collagen, found in all wild isolates of C. elegans and highly conserved among Caenorhabditis species, also modifies the effects of other canonical cuticle mutants, including those with Blistered and Squat phenotypes. These results underscore the importance of genetic background and the serendipity of Brenner’s choice of strain.
Materials and Methods
Strains
All experiments were carried out at 20° with NGM-agarose plates and OP50-1 E. coli for food, unless otherwise noted.
We used the following strains: BE8: sqt-3(sc8) V, BE13: sqt-1(sc13) II, BE22: rol-1(sc22) II, BE44: dpy-8(sc44) X, BE93: dpy-2(e8) II, BE108: sqt-2(e108) II, CB61: dpy-5(e61) I, CB91: rol-1(e91) II, CB224: dpy-11(e224) V, CB768: bli-2(e768) II, CB769: bli-1(e769) II, CB1166: dpy-4(e1166) IV, CB2070: bli-1(e935) rol-1(e91) II, CB4856: Hawaiian wild type, COP1834: col-182(knu732) X, EG7993: oxTi412 [eft-3p::TdTomato::H2B] X, EG8951: oxTi1015 [eft-3p::GFP::NLS + NeoR] X, N2: laboratory wild type, QG2797: rol-1(e91) II; ajIR6 [X, CB4856 > N2] X, QG2798: rol-1(e91) II; oxTi412 [eft-3p::TdTomato::H2B] oxTi1015 [eft-3p::GFP::NLS + NeoR] X, QG2804: mIs12 rol-1(e91) II, QG2952: bli-1(e769) II; col-182(knu732) X, QG2954: dpy-11(e224) V; col-182(knu732) X, QG2953: rol-1(e91) II; col-182(knu732) X, QG2955: dpy-4(e1166) IV; col-182(knu732) X, QG2956: bli-1(e935) rol-1(e91) II; col-182(knu732) X, QG2957: rol-1(sc22) II; col-182(knu732) X, QG2958: dpy-2(e8) II; col-182(knu732) X, QG2960: dpy-5(e61) I; col-182(knu732) X, QG2961: sqt-2(e108) II; col-182(knu732) X, QG2962: bli-2(e768) II; col-182(knu732) X, QG3070: sqt-3(sc8) V; col-182(knu732) X, QG3072: sqt-1(sc13) II; col-182(knu732) X, QG3074: dpy-10(cn64) II; col-182(knu732) X, QG3076: dpy-8(sc44) col-182(knu732) X, SP419: unc-4(e120) rol-1(e91) II, TN64: dpy-10(cn64) II, and WE5241: ajIR6 [X, CB4856 > N2] X.
COP1834 col-182(knu732) X was generated by Knudra Biosciences (now InVivo Biosystems), using the CRISPR/Cas9 homology-directed repair protocol of Paix et al. (2015). We refer to this synthetic ancestral allele in the main text as col-182anc. In the N2 background, Cas9 was directed to sites on each side of the target insertion with sgRNA sequences CTGGATATAGTTGTTCCCGG and CAAGGAGAGATCGGACGCGA, and repair was directed by the DNA oligo AGGACGGATGCCGCGTCTGCCCACCAGGACCACCGGGTA- CTACCGGTTACCCAGGTCAGCCTGGCCCCCAGGGTCTCC- CAGGTAACCAAGGAGACCAAGGTGAAATTGGTCGCGAA- GGATCACCAGGAGCCCAGGGAGCTCCAGGAG. This oligo precisely excises the 8-bp insertion sequence from the N2 col-182 gene and simultaneously alters 16 additional sites, all silent codon positions, to prevent recutting by Cas9.
We verified the presence of the rol-1(e91) mutation in these strains by Sanger sequencing using primers RolF3: CAAATTCGACAAAGCGACAA and RolR3: GAGCATCGTAAGGCTGGAAA. We verified the presence of the col-182(knu732) mutation by Sanger sequencing using primers X12.636F: TAGGCAAACTTGCTGCACAC and X12.636R: GAGACAGGCTGGAAATGAGC.
Observation of segregation distortion
As described in Kaur and Rockman (2014), we crossed wild isolate CB4856 and a strain carrying unc-4(e120) rol-1(e91) II in the N2 background. hermaphrodites were singled and Rol nonUnc adults were isolated and genotyped by Illumina GoldenGate Assay. Genotyping was performed by the DNA Sequencing and Genomics Core Facility of the University of Utah. Allele frequencies are in File S2.
Complementation crosses
We used visible markers to generate animals that are homozygous rol-1 on chromosome II and heterozygous on the X chromosome, with one X chromosome from N2 and the second from the strain of interest (SOI). If the SOI carries the dominant suppressor of rol-1, then these animals are expected to show suppressed rolling behavior.
Specifically, we crossed mIs12[GFP] rol-1 II males to SOI hermaphrodites to generate mIs12 rol-1 / + + II; SOI X males. These we crossed to unc-4 rol-1 II hermaphrodites. For each SOI, we tested six GFP-positive nonUnc hermaphrodite progeny of this cross for rolling behavior in the second day of adulthood. These animals are mostly unc-4 + rol-1 / + mIs12 rol-1 II; SOI/N2 X. A small fraction of the phenotyped animals could be rol-1 heterozygotes due to rare recombination events between mIs12 and rol-1 in the males. In addition, each tested animal is heterozygous (N2/SOI) for a random fraction (expectation 1/2) of the autosomes except for chromosome II.
We used this approach to test 5 recombinant inbred advanced intercross lines (QX33, QX43, QX126, QX150, and QX202). We also tested 8 wild isolates by this approach (CB4856, PB306, EG4347, QX1211, JU319 [CeNDR isotype JU311], JU1088, PX179, and JU400 [CeNDR isotype JU394]), along with N2 and LJS1, which are laboratory-adapted strains.
Fine mapping with recombinants
To fine-map the suppressor, we crossed rol-1(e91) II; CB4856 X and rol-1(e91) II; oxTi1015 oxTi412 X. The latter strain carries integrated single-copy transgenes on the X expressing fluorescent proteins, tdTomato::H2B from X:11.049 Mb and GFP::NLS from X:13.480 Mb [Wormbuilder; Frøkjær-Jensen et al. (2014)]. Both strains carry N2-derived autosomes. We homozygosed X chromosomes that were recombinant between the transgenes, scored their rolling behavior, and genotyped them at SNP markers in the mapping interval. Recombinant strain AM_GNR9 placed the suppressor to the right of SNP WBVar00083599 at X:12,579,121, and recombinant AM_GNR13 placed it to the left of SNP WBVar1602269 at X:12,662,633.
Genotyping primers used for mapping were: WBVar01981458 indel, TGGGTAAACATCGGCTCCAT and TGTTCTGCACGGGAAAAGAT; WBVar00083599 SNP, CGACATCCAAAGTTTTTGAGACT and GAGAAAGTGTTATGGGCATGG; WBVar01602269 SNP, CGTGTGTTTCCGTTGTGAAT and TTCAGTGTTCATCGCAATCTG.
Association mapping
Variant data for 330 C. elegans isotypes were downloaded as the 20180527 CeNDR release soft-filtered vcf. We tested for a match between variants in the recombination-mapping interval (X:12,579,122 - X:12,662,632) and rol-1 suppression phenotype in the panel of N2, CB4856, LSJ1, and the seven other tested wild isolates.
Gene model
Sequence and annotation data for the Caenorhabditis genus were downloaded from the Caenorhabditis Genomes Project. RNAseq data from young adults of C. elegans wild isolates were downloaded from the NCBI SRA: CB4856 PRJNA437313 (Zamanian et al. 2018), AB1 and ED3040 PRJNA288824 (Vu et al. 2015). Reads were mapped to a 100 Kb region of the N2 WS220 genome centered on col-182 with bwa mem version 0.7.17 (Li and Durbin 2010), assembled with Trinity version 2.3.2 in genome-guided mode (Grabherr et al. 2011), and homologous transcripts were extracted by blast version 2.9 (Altschul et al. 1990) against col-182 orthologs across the genus. Coding and protein sequences were aligned with mafft version 7.3.10 in L-INS-i mode (Katoh et al. 2005), and homology and gene structures were plotted using R packages (Paradis and Schliep 2019), (Yu et al. 2017), (Pagès et al. 2019), and (Yin et al. 2012). Species with only computationally predicted annotations (C. kamaaina, C. becei, C. panamensis) and two species (C. quiockensis and C. sulstoni) with extremely long predicted ortholog sequences potentially deriving from annotation errors were excluded. The CGP C. remanei col-182 protein sequence contains a large deletion that is not present in the WormBase version, and we opted to use the latter. Collagen triplet stability scores shown in Figure 2B were obtained from (Persikov et al. 2005), and ignore higher-order interactions. Gene models and coding sequence alignments are in Files S10 and S11.
Epistasis analysis of visible mutants
For each tested mutation, we grew the mutant line and col-182(knu732) double mutant in parallel at low population densities for several generations and performed synchronous egg lays to generate animals for phenotyping. For most strains we observed these synchronized animals three days later as young adults. For sqt-2(sc108), because the heterozygous phenotype (Rol) is different from the homozygous Squat phenotype (Sqt), we also scored sqt-2/+ animals. For rol-1 bli-1 and rol-1 bli-1; col-182 animals, we followed 200 of each genotype through the third day of adulthood. Videos of rol-1 worms (CB91, QG2979, QG2957, BE22, QG2953) were taken on day four of adulthood. Ten worms were picked to fresh seeded plates and imaged for 8 min at 12 frames per minute. Videos of sqt-3 worms (BE8, QG3070) were taken on day two of adulthood. Twenty worms were picked to fresh seeded plates and imaged for five minutes at 12 frames per minute. Video samples are in Files S3-S9.
Quantitative locomotion analysis
We generated quantitative phenotype data in three genetic backgrounds (wild-type, rol-1(sc22), and sqt-3(sc108)), each with the ancestral and derived col-182 allele. Conditions and worm tracking have been described previously (Noble et al. 2017; Mallard et al. 2019). Lines (N2, COP1834, BE8, BE22, QG2957, QG3070) were each split to duplicate lineages, bleached, and grown under common conditions on HT115 bacteria in 90mm plates at 20° for two generations before assay, with each generation starting from around 500 L1 larvae that had been starvation synchronized in M9 buffer for 18 hr. Young adults were imaged in random order, on food, during day three post-L1, and again on two further generations (treated as above) for a total of six replicate plates per genotype. Worms were tracked for 8 min using the Multi-Worm Tracker (Swierczek et al. 2011), the final 4 of which were analyzed after subsampling to 4 Hz, and 11-point skeletons and outlines from Choreography were parsed to generate summary track statistics based on size and movement. More precisely, we used Choreography-defined measurements of length, width, area, speed, acceleration, angular momentum (turning rate), mean body curvature, kink (the maximum ratio of angles between head/tail and body), and the length of continuous runs of Forward, Backward or Still motion (“bias”). Raw data are in File S12.
For each worm track we took the median and variance of each metric, as a whole and split by bias state. Exploratory behavior was quantified as the area and circularity ( ) of the track convex hull (Pebesma and Bivand 2005), averaged first across 30 sec intervals for each of the longest 100 tracks from each plate, then across tracks. Traits were log transformed where strongly non-normal (an improvement in Shapiro-Wilk -log10 p-value > 6), and effects of assay block, defined by lineage and assay day, were removed by linear regression. Processed data are in File S13.
We show univariate and multivariate (classical multidimensional scaling on the mean centered and scaled Euclidean distance matrix of plate means, base R ) analysis. In Figure 3A-B we used multivariate analysis of a subset of traits selected using sparse linear discriminant analysis (Clemmensen et al. 2011). From 25 traits (repeatability < 0.5, thinned to reduce maximum colinearity to < 0.5), we selected the five metrics most associated with suppression of rol-1(sc22) or sqt-3(sc8), separately, using plate means. Traits retained for sqt-3 (BE8 vs. QG3070, COP1834, N2), ordered by absolute loading on the discriminant function, were log transformed curvature (S), width (F), kink, width (S) variance, and acceleration (F). Traits retained for rol-1 (BE22 vs. QG2957, COP1834, N2) were log transformed circularity, velocity variance, width (F), kink (F), and run length (F) variance. R code for this analysis is in File S14.
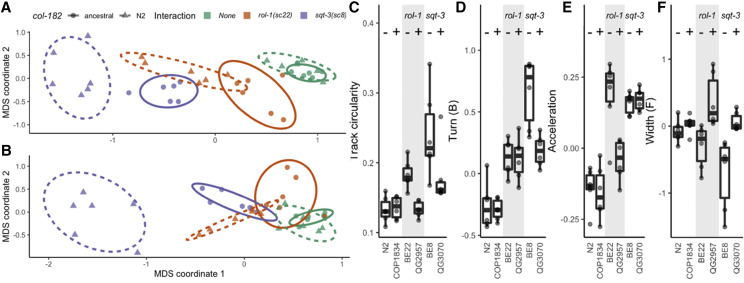
Figure 3.
Ancestral col-182 suppresses rol-1 and sqt-3 alleles. A-B. Multidimensional scaling of locomotion and size traits selected by sparse discriminant analysis that maximize suppression of rol-1 (A) or sqt-3 (B) by ancestral col-182. Point shapes show col-182 genotype (ancestral or N2), bounded by ellipses giving the 95% confidence interval under a multivariate t-distribution (solid lines for ancestral col-182), colors show genetic interaction (none in the N2 background, or alleles of rol-1 and sqt-3). Each point is the grand mean of tracks from around 500 young adult worms per replicate plate measured over three consecutive generations for each genotype, assayed for N2 (green triangles) and COP1834 (ancestral col-182 in the N2 background; green circles), BE22 (N2 col-182; rol-1(sc22); orange triangles) and QG2957 (ancestral col-182; rol-1(sc22); orange circles), and BE8 (N2 col-182; sqt-3(sc8); purple triangles) and QG3070 (ancestral col-182; sqt-3(sc8); purple circles). C-F. Univariate comparisons show variable effects across backgrounds. col-182 genotype is indicated with – (N2 allele) and + (ancestral) symbols. Complete suppression of track circularity is seen for rol-1(sc22) (C), and of worm width in the forward state for sqt-3(sc8) (F), however partial (or no) suppression is the most common outcome. Raw and processed data in Files S12 and S13, and code in File S14.
Epistasis analysis in the CeMEE
To test for potential modifying effects of col-182 on natural variation segregating in the C. elegans multiparent experimental evolution (CeMEE) panel we genotyped the N2 indel from existing sequence data (Noble et al. 2019) using bcftools (Li 2011) after indel realignment (DePristo et al. 2011), obtaining calls for 365 recombinant inbred lines (RILs; from populations A6140, CA[1-3]50 and GA[1,2,4]50) for which locomotion has been measured on NGM (Mallard et al. 2019). Two lines were excluded as multivariate outliers based on Mahalanobis distance. Genotypes were marker set 1 from Noble et al. (2019). The N2 col-182 allele is at a frequency of 16.5% in these lines, providing sufficient power to detect pairwise interactions conditional on joint allele frequency. In total, 167,187 diallelic SNPs where all four genotype classes were present at a minimum frequency of 10, excluding any uncertain imputations, were tested.
To test for col-182-by-genotype interactions we fit nested bivariate linear models for three pairs of partially correlated traits: length and width (forward state, log transformed), the rol-1 and sqt-3 suppression discriminant functions that are linear combinations of five traits (see Quantitative locomotion analysis), and the single traits with the highest loading in each discriminant function, curvature and track circularity (log transformed). Trait values were best linear unbiased predictions (BLUPs) extracted from linear mixed effects models (R package ) fit to replicate observations, with fixed effects of population replicate. Significance testing followed the univariate approach in Noble et al. (2017). In brief, we first tested for genetic effects by likelihood ratio (Pillai’s trace statistic) for a full model with additive and interaction effects of col-182 genotype and focal marker genotype, against a null model (intercept only). Genome-wide significance was declared against a null distribution of >1000 test statistics generated by permuting lines within populations and retaining the minimum observed p-value. We used a relative permissive false discovery rate (FDR) of 0.2. Quantile-quantile plots showed statistics were well calibrated for length/width and the Rol/Sqt discriminant functions at , but strongly deflated for curvature/circularity. Inflation was evident for length/width at , independent of linkage disequilibrium, consistent with additional polygenic interactions. For loci with significant genetic effects, interaction significance was then assessed at a nominal threshold of by parametric bootstrap against the additive model (Bůžková et al. 2011), resampling responses jointly among lines 5000 times. Genotype and phenotype data, and R code for this analysis are in Files S15, S16.
Gene expression analysis
We extracted data for 199 recombinant inbred advanced intercross lines (RIAILs) from Rockman et al. (2010), after excluding data from lines with annotation issues (Zych et al. 2017). We performed structured nonparametric trait mapping as in Rockman and Kruglyak (2009) for abundance of 15,617 transcripts whose genes are more than a megabase from col-182 (to exclude local linkages for genes near col-182 that have their own -acting variants). We retained traits with genome- and experiment-wide significant linkage peaks (LOD > 4.3, 5% FDR) within 1 RIAIL-effective cM of col-182 (approximately 400 Kb). The nine significantly linked transcripts were tested for functional enrichment using the WormBase Enrichment Analysis Suite (Angeles-Albores et al. 2018).
Data availability
All quantitative data and code to reproduce our analyses and main figures are available from FigShare and https://github.com/lukemn/cuticle. Strains are available upon request. File S1 details all supplemental files, File S2 contains genotyping data from Figure 1, Files S3-S9 contain short videos of mutant and suppressed adult hermaphrodites on plates, File S10 contains gene models from Figure 2A, File S11 contains coding sequence alignments for Figure 2B, Files S12 and S13 contain raw and processed Multi-Worm tracker data for Figure 3 with associated R code in File S14. File S15 contains genotypes and phenotypes used for detecting genetic interactions between col-182 and SNPs in the CeMEE (Figure 4), with associated R code in File S16. Supplemental material available at figshare: https://doi.org/10.25387/g3.12293138.
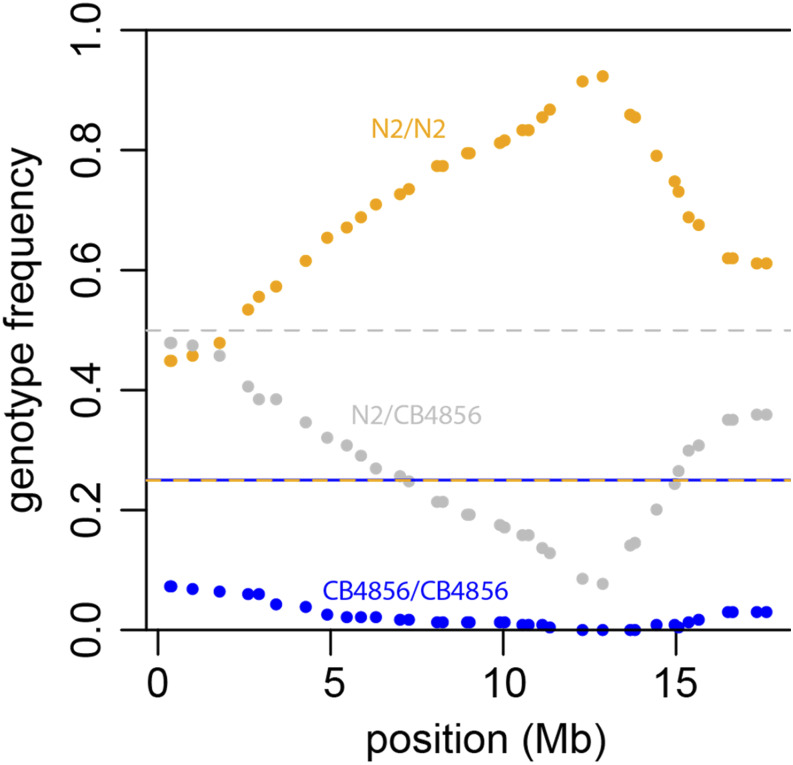
Figure 1.
Allele frequencies along the X chromosome in Rol nonUnc s from a cross of CB4856 and N2-background unc-4(e120) rol-1(e91) II. Dashed lines show the Mendelian expectations for heterozygotes (1/2) and homozygotes (1/4 each).
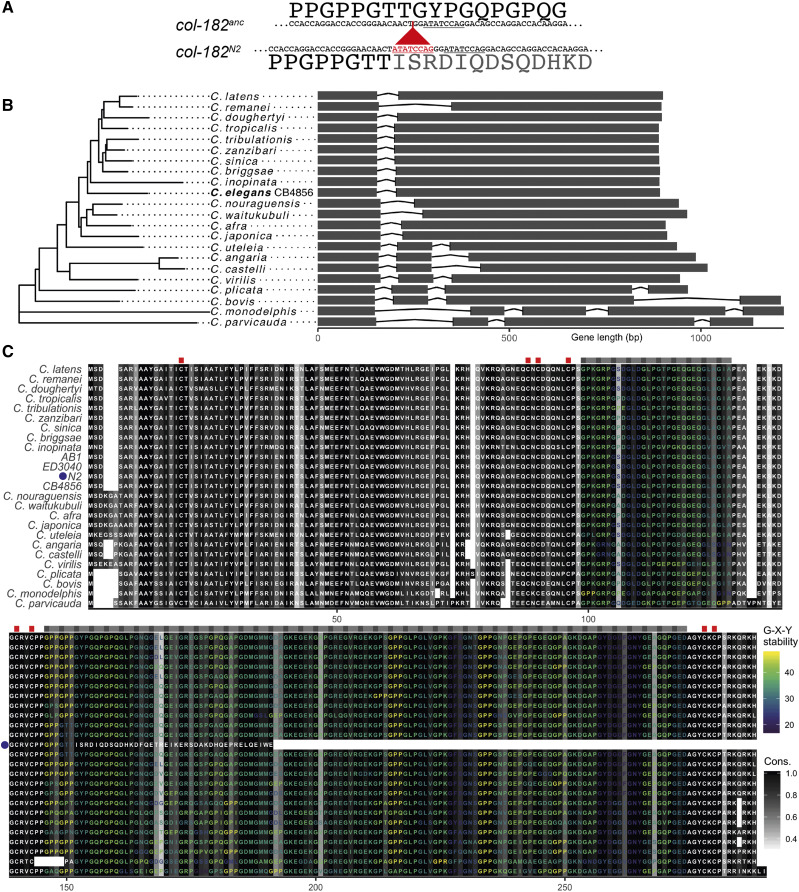
Figure 2.
A. N2 carries a derived insertion of 8 bp (red) relative to the ancestral allele. The insertion duplicates an 8-bp sequence present in the ancestor (underlined). B. col-182 gene tree and model for C. elegans CB4856 and other Caenorhabditis species. The x-axis shows distance from the start codon for each genome. C. Protein alignment, including predicted sequences for C. elegans wild isolates CB4856, AB1 and ED3040 assembled from young adult RNAseq data, and the frame-shifted N2 translation (marked with a blue dot). Gray-scale shading represents site conservation (% identity), and labels are colored by predicted collagen triplet stability (melting temperature) for runs of >1 G-X-Y repeats conserved across all sequences other than N2. The positions of conserved triplets are indicated above the alignment by gray boxes, and the positions of conserved cysteine residues potentially involved in inter-strand disulphide bridges are shown as red boxes.
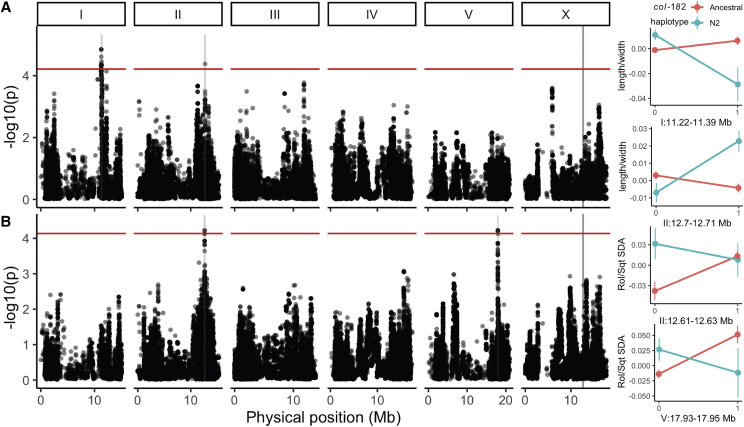
Figure 4.
col-182 modifies the effects of natural genetic variation on worm movement and shape. Genome-wide statistics are shown for two bivariate response models of col-182 indel SNP genotype, worm length/width (A) and Rol/Sqt sparse discriminant functions (B), using diallelic SNPs segregating in 363 recombinant inbred lines of the C. elegans multiparent experimental evolution (CeMEE) panel. Statistics are from a likelihood ratio test for a full additive and interaction linear model against a null model of no genetic effects. Permutation thresholds for genome-wide significance (FDR = 0.2) are shown in red, light shaded regions show 1.5 LOD drop QTL intervals (expanded to a minimum of 300 Kb for visibility), and the location of col-182 on the X chromosome is indicated by a dark gray line. Effect plots at right show genotype class means and standard errors for QTL with significant interactions (parametric bootstrap against additive models, ). Trait values shown are the first principal component for length/width, and the Sqt discriminant function, which explains most of the interaction in the Rol/Sqt bivariate model. Reference-based genotype is on the x-axis. Genotype and phenotype data in File S15, code in File S16.
Results
CB4856 carries a dominant suppressor of rol-1
During a study of recombination patterns on C. elegans chromosome II (Kaur and Rockman 2014), we crossed unc-4(e120) rol-1(e91) II worms (N2 background) with wild isolate strain CB4856, isolated in Hawaii in 1972 (Hodgkin and Doniach 1997). After allowing hermaphrodites to reproduce by selfing, we isolated Rol nonUnc s, which are homozygous for the N2 rol-1 allele on chromosome II but should show Mendelian segregation of other chromosomes [ the peel-1 zeel-1 incompatibility on chromosome I (Seidel et al. 2008)]. We noticed strong segregation distortion on the X chromosome among these Rol nonUnc s, which we had genotyped at 37 SNP markers (Figure 1, File S2). Distortion favored the N2 background, with a peak around 13 Mb where zero of 234 worms were homozygous for the CB4856 genotype (expectation 1/4 = 58.5). At the peak, 216 (92%) worms were N2 homozygotes and the remaining 18 (8%) were heterozygotes.
We hypothesized that the CB4856 X chromosome carries a dominant suppressor of rol-1. We crossed the rol-1(e91) allele into an X-chromosome substitution strain, which carries the CB4856 X chromosome but is otherwise N2, and confirmed that the resulting rol-1(e91) II CB4856 X strain is strongly, but incompletely, suppressed for rolling. The worms retain a slight helical twist and can be distinguished from wildtype N2 and CB4856, but the dramatic rolling and circling behaviors of rol-1(e91) are absent (Videos: CB91 rol-1(e91); N2 X vs. QG2797 rol-1(e91); CB4856 X in Files S3 and S4). Suppression of rol-1 is also observed in X chromosome heterozygotes, consistent with a dominant suppressor on the CB4856 X and explaining the pattern of segregation distortion we observed in the unc-4 rol-1 crosses.
Suppression often reveals interactions between genes in physical association or in specific developmental pathways, but can also reflect altered transcription, splicing, or translation of a mutant gene. These informational suppressors can be allele specific, masking only particular kinds of missense or nonsense or splice site variants, for example (Hodgkin 2005). There are two rol-1 alleles with known effects on phenotype at present, and we found that the second, sc22, is also suppressed by the CB4856 X chromosome. Although the sc22 molecular lesion is unknown, e91 and sc22 mutants show distinct phenotypic profiles, including temperature sensitivity (Cox et al. 1980), and we concluded that allele-specific interaction was thus unlikely.
The suppressor maps to an indel polymorphism in col-182
We performed complementation testing with a panel of N2-CB4856 recombinant inbred advanced intercross lines (Rockman and Kruglyak 2009) with breakpoints near the peak allele frequency distortion, taking advantage of the dominant mode of action of the CB4856 allele. Using this approach, we tested 5 RIAILs, scoring based on Rolling, with boundaries defined by RIAIL QX43, which carries the suppressor, and RIAIL QX126, which does not. These strains place the suppressor to the right of SNP WBVar00083496 at X:12,364,484 and to the left of indel WBVar01981458 at X:12,699,819 (WS272 coordinates).
Next we used integrated single-copy fluorescent marker transgenes (Frøkjær-Jensen et al. 2014) to select for N2/CB4856 recombinants in the interval, in N2 autosomal genetic backgrounds. These data localized the causative locus to the interval between 12.579 and 12.662 Mb.
Complementation testing with seven additional wild isolates found that all exhibit suppression of rol-1. Laboratory strain LSJ1, which shares a laboratory ancestor with N2 but was cultured separately since 1963, does not exhibit suppression. Among all variants segregating in these strains in the mapped interval, only one exhibits perfect cosegregation with rol-1 suppression: an 8-base pair (bp) deletion in the gene col-182 (WBVar01928355). Like rol-1, col-182 is one of the 181 collagen genes in the C. elegans genome (Teuscher et al. 2019).
N2 carries a derived insertion mutation in col-182
Among 330 genome-sequenced C. elegans isotypes from around the world (CeNDR freeze 20180527), the 8-bp deletion is present in every strain except for the lab strains N2, LSJ1, and ECA252. These three strains are all derived from a single isolate of C. elegans sampled by L. N. Staniland in 1951 (McGrath et al. 2009; Weber et al. 2010; Sterken et al. 2015; Cook et al. 2017). The deletion state is also found in orthologs of all other examined Caenorhabditis species (Figure 2). This strongly suggests the reference allele is a derived insertion, one that arose either in the wild, and happened to be sampled by Staniland in 1951, or in the lab sometime before 1963 [likely before 1958 (Gimond et al. 2019)]. The eight basepair sequence, ATATCCAG, is also present in the ancestral allele, two bases downstream of the insertion site, so that it now occurs twice in N2 (Figure 2A).
C. elegans collagens contain two blocks of Gly-X-Y repeats, flanked and separated by short cysteine-containing domains involved in interchain disulphide bonds (Page and Johnstone 2007). Until recently, col-182 was annotated as a three-exon, two-intron gene encoding a near-canonical collagen protein, missing only the middle cysteine domain (Teuscher et al. 2019). However, new RNAseq data suggests that the earlier gene model was erroneous, and that the N2 transcript has only one intron (WormBase WS274). With this RNAseq-supported gene model, the 8-bp insertion causes a frameshift that eliminates the entire second Gly-X-Y domain and final cysteine domain (Figure 2B). As a consequence, col-182 has been reclassified as a pseudogene. The ancestral version of the gene, preserved in CB4856 and all other wild isolates, encodes a perfect canonical cuticular collagen. The C-terminal Gly-X-Y domain includes 43 consecutive Gly-X-Ys, tying it with four other genes for the longest such collagenous stretch among C. elegans collagens [with col-72, col-75, col-104, and col-113, none of which have known mutant phenotypes; (Teuscher et al. 2019)].
To test whether col-182 is the rol-1 suppressor and whether the ancestral allele functions via the predicted isoform, we commissioned a strain that carries the ancestral splicing and reading frame in an otherwise N2 background (see strains). CRISPR-Cas9 conversion removed the 8-bp insertion and altered 16 additional base pairs, each a synonymous third codon position change in the predicted ancestral reading frame. We then confirmed that this ancestral allele col-182anc suppressed rol-1, both the e91 and sc22 alleles (videos: CB91 rol-1(e91) vs. QG2953 rol-1(e91); col-182anc, and BE22 rol-1(sc22) vs. QG2957 rol-1(sc22); col-182anc, Files S3 and S5-S7). We generated quantitative data for the sc22 allele (selected arbitrarily) by tracking young adult hermaphrodites and extracting an array of statistics describing locomotion, size, and shape (see Quantitative locomotion analysis). Reduction of this data by multidimensional scaling into two orthogonal axes provided quantitative confirmation that the ancestral col-182anc allele suppresses some of these phenotypes in the rol-1 background (Figure 3A). By univariate analysis, double mutants are indistinguishable from wild-type N2 for a metric that captures circling locomotion (Figure 3C), but weak to no suppression is the predominant outcome across the correlated set of single traits (Figure 3D-F).
col-182 interacts with other collagen mutants
rol-1 is part of a complex network of collagens and collagen-modifying enzymes that interact genetically, often in quite complex ways (Cox et al. 1980; Kusch and Edgar 1986). We therefore expected that the ancestral col-182 might modify rol-1’s epistatic relationships with other genes, and might itself interact with other cuticle-specifying genes.
Blistered mutants, described along with rol-1 in Brenner’s original screen, develop fluid-filled blisters along the adult cuticle (Brenner 1974). In the N2 background, rol-1 suppresses the blister phenotype of bli-1 mutants (Cox et al. 1980). We found that rol-1 bli-1; col-182 triple mutants show the expected suppression of rolling but are also unblistered, indicating that col-182 suppresses rol-1’s Rol phenotype but not its suppression of bli-1. Moreover, col-182 modified bli-1’s phenotype by itself; blisters were largely suppressed in bli-1; col-182 double mutant hermaphrodites. The penetrance of the blister phenotype was reduced and the individual blisters were spatially restricted to the head region (Table 1). Conversely, col-182 enhances the blister phenotype of bli-2 worms; bli-2 young adults had small blisters, typically restricted to their heads, but bli-2; col-182 blisters were larger, sometimes extending the whole length of the worm (Table 1). Like col-182 and rol-1, bli-1 and bli-2 encode collagens.
Table 1. Epistasis analysis for Blister phenotype. Each cell shows the counts of mutants/total worms. “Head-only” and “Full-body” count the number of worms with blisters that showed blisters either restricted to the head region or extending the entire length of the animal. Asterisks report p-values from Fisher’s Exact Test for a difference between the single and double mutant strains for each bli gene and phenotype. ns, not significant, * P < 0.01, ** P < 10-6.
| Genotype | Blisters | Head-only | Full-body |
|---|---|---|---|
| 33/36 | 3/33 | 0/33 | |
| bli-1; col-182anc | |||
| bli-2 | 39/40 | 24/39 | 0/39 |
| bli-2; col-182anc | 30/30ns |
Cox et al. (1980) identified alleles in several genes that give rise to left-handed rollers, like rol-1(e91). Although we observed no gross phenotypic effect of col-182 on dpy-8(sc44), dpy-10(cn64), or sqt-1(sc13), we observed partial suppression of rolling in sqt-3(sc8), which we quantified by worm tracking (Figure 3, and see videos: BE8 sqt-3(sc8) vs. QG3070 sqt-3(sc8) col-182anc, Files S8,S9). Suppression was qualitatively and quantitatively distinct to that of rol-1; near complete for the single measure of worm width, but again highly variable and generally weak for other measures of locomotion and morphology (Figure 3B-F).
sqt-2(sc108) exhibits right-handed rolling as a heterozygote in the N2 background, and did so as well in the col-182anc background. However, sqt-2 heterozygotes showed slowed development in the N2 col-182 background, while the ancestral allele suppressed the developmental delays. Finally, alleles of several additional genes involved in cuticle development – collagens dpy-2(e8), dpy-4(e1166) and dpy-5(e61), and thioredoxin dpy-11(e224) – showed no gross phenotypic modification in the col-182anc background.
col-182 modifies effects of natural variation on worm shape and locomotion
Collagens are known to influence body size (Brenner 1974; Fernando et al. 2011; Madaan et al. 2018), and our locomotion analysis identified specific axes of worm size, posture and locomotion modified by col-182 in two genetic backgrounds (Figure 3 B-F). We next sought to test more broadly for interactions between col-182 and natural genetic variation for these traits in the C. elegans Multiparent Experimental Evolution (CeMEE) panel, a collection of recombinant inbred lines derived from the pooled standing genetic diversity of 14 wild isolates and two N2-related strains (Teotónio et al. 2012; Noble et al. 2017, 2019).
We genotyped the N2 insertion in RILs sampled from an ancestral laboratory-adapted population, A6140, and from six populations derived from A6140 that evolved under varying mating system and environment (Noble et al. 2017). Using Multi-Worm Tracker data for 363 lines, we fit bivariate linear models for three sets of correlated traits (length and width, body curvature and track circularity, and the Rol/Sqt discriminant functions from Figure 3B) to test for interaction effects. Univariate tests showed that col-182 genotype had no effect on the means of these population-centered traits ( by likelihood ratio test).
We detected four loci with significant genetic effects at a per-model false discovery rate of 20% (Figure 4). Two QTL were detected for length/width with clear genetic interactions ( by bootstrap against the additive model): the first, on chromosome I, fell within the central recombination rate domain (1.5 LOD drop interval around 170 Kb); the second, on chromosome II, was contained by a single very large N2 protein coding gene, tbc-17, with several missense variants, a splice-donor change, and heterozygous SNP calls suggestive of copy number variation segregating in the CeMEE founder haplotypes (Cook et al. 2017). tbc-17 encodes a highly conserved predicted Rab family GTPase activator which, based on homology, may be involved in intracellular trafficking, a process critically important for collagen secretion from hypodermal cells (Roberts et al. 2003; Ackema et al. 2013). Two QTL were detected for the Rol/Sqt discriminant functions: one on chromosome II (interaction bootstrap ; 18 Kb interval) contained nine N2 annotated protein-coding genes of unknown function, mostly of the nematode-specific peptide group E family; the second, on chromosome V (interaction ; 25 Kb interval), was a specific interaction with MY16 haplotypes, spanning predicted ubiquitin protease usp-50 partially, and dpy-21 fully, along with eight non-coding RNAs. dpy-21 is a non-essential, non-condensin subunit of the dosage compensation (DC) complex (Meyer and Casson 1986), with additional DC-independent roles in gene regulation (Webster et al. 2013), and loss-of-function mutants show an enrichment in dysregulation of genes involved in the cuticle (Kramer et al. 2015).
col-182 does not have systemic effects on gene expression
Several of the alleles that arose during C. elegans domestication have large and systemic effects on C. elegans biology. These include npr-1 (de Bono and Bargmann 1998; McGrath et al. 2009; Andersen et al. 2014; Zhao et al. 2018), nath-10 (Duveau and Félix 2012), nurf-1 (Large et al. 2016), and Y17G9B.8 (Rockman et al. 2010; Burga et al. 2018). We therefore investigated whether col-182 is linked to systemic effects on gene expression in adult hermaphrodites, using a published dataset of gene expression in 199 N2/CB4856 recombinant inbred advanced intercross lines [RIAILs; (Rockman et al. 2010)]. The RIAILs provide much higher genotypic replication than a typical pairwise contrast of strains, as each col-182 allele is homozygous in approximately half the RIAILs, but effects that map to col-182 may be due to nearby variants in other genes. As expected, col-182 abundance shows strong linkage to its own location. Nine other genes show significant linkage to the col-182 region, including two glutathione S-transferases and two cytochrome P450 enzymes (Table S1). However, none are known to be involved in cuticle development and collectively they show no enrichment for any particular tissue. Thus the col-182 mutation in N2 appears to have limited effects on gene expression in young adult hermaphrodites, at least under ordinary laboratory conditions.
Discussion
The structural complexity of the nematode cuticle is reflected in its developmental and genetic regulation, and its environmental (temperature) and genetic sensitivity. Around 4% of the worm genome is dedicated to expressing, processing, and assembling the collagens, cuticulins, glycoproteins and other components of the multilayered extracellular matrix (Teuscher et al. 2019). Yet, of 173 predicted cuticular collagen genes, phenotypes from extensive mutagenesis screens have been detected for just 21 (Page and Johnstone 2007). To this number we can now add col-182, though we have also shown that even this select list might well have been shorter had Brenner not adopted N2 as the C. elegans reference genetic background.
In the absence of molecular and structural data, the precise role of col-182 in the worm cuticle and its mode of interaction with other collagens remains obscure. The derived N2 insertion represents an evolved enhancer of rol-1 rolling, and the ancestral col-182 modifies to a variable extent the phenotypes from other classical mutant alleles of bli-1, bli-2, sqt-2 and sqt-3, but not obviously those of dpy-2, -4, -5, -8, -10 and -11, or sqt-1.
The expression of cuticular genes during worm development offers no clear insight. Of the tested genes, only rol-1, for which suppression by ancestral col-182 was strongest, shows strong stage specificity, being around 30-fold enriched in L4 (i.e., when the adult cuticle is manufactured; Figure S1). But bli-1, bli-2 and rol-1 show generally similar patterns and levels of transcriptional activity over the life-cycle, with very low expression in embryonic and early larval stages. The Blistered phenotype is thought to be due to defects in struts linking basal and cortical layers of the adult cuticle, and of six Blistered mutants, three are enzymes rather than structural components.
sqt-1, -2 and -3 are all highly expressed collagens that interact genetically, with similar stage specificity from L2 onward (Figure S1). sqt-3 is unique among collagens in its essentiality (Priess and Hirsh 1986; van der Keyl et al. 1994; Novelli et al. 2006), and is strongly expressed in the embryo as well. sqt-1 also interacts genetically with bli-1 and bli-2, and 22 other genes (Cox et al. 1980; Kusch and Edgar 1986; Kramer and Johnson 1993; Kramer 1994; Westlund et al. 1997; Nyström et al. 2002; Byrne et al. 2007; Shephard et al. 2011; Cai et al. 2011), yet we saw no obvious modification of the left rolling phenotype of sqt-1(sc13) mutants in the ancestral col-182 background. This may be explained by the extreme specificity of allelic interactions among collagen mutants, and sqt mutants in particular. The sqt-1(sc13) allele tested is a recessive C-terminal C>Y substitution, altering cross-linking (Kramer and Johnson 1993; Yang and Kramer 1999), while sqt-2(sc108) is an N-terminal R>C substitution of unknown structural effect. Alleles of sqt-1 vary markedly in their type, severity, temperature sensitivity, and degree of dominance of phenotypes, as well as inter- and intragenic interaction effects; from near wild-type for a null allele, to left or right rolling, abnormal hermaphrodite tail or male rays, or variation in body length. Enrichment in CeMEE interaction statistics for worm length/width over a small region spanning sqt-1 () provides some fuel for speculation that allele-specific interactions with col-182 may exist.
Lastly, no grossly visible interactions were seen for collagens involved in annuli formation and shape dpy-2, -5, -8, -10 (McMahon et al. 2003). In sum, we surmise that col-182 likely plays a role, apparently redundant under laboratory conditions, in one or both of the strut-anchored adult cuticular layers. The oft-touted genetic simplicity of C. elegans breaks down somewhat when considering the cuticle, and targeted biochemical and structural analysis, together with epistasis analysis encompassing natural genetic variation, will be required to clarify the precise role of col-182 and the majority of other collagens with no known function in the N2 background.
Effects of genetic background are ubiquitous in complex genetic systems wherever they are carefully considered. Studies mixing natural with domesticated genetic variation have amply shown the importance of genetic interaction on the phenotypic outcome of allelic effects in C. elegans (Seidel et al. 2008; McGrath et al. 2009; Bendesky et al. 2012; Duveau and Félix 2012; Gaertner et al. 2012; Andersen et al. 2014; Glater et al. 2014; Greene et al. 2016; Ben-David et al. 2017; Bernstein et al. 2018; Zhao et al. 2018). This extends to classical mutations of the cuticle, some of the first mutants isolated in C. elegans and core components of the worm geneticist’s toolkit.
Acknowledgments
This work was supported by the National Science Foundation (DDIG 1210762 to TK), the National Institutes of Health (R01GM121828 to MVR), the NYU Dean’s Undergraduate Research Fund (AM), and the European Commission Sklodowska-Curie Fellowship (H2020-MSCA-IF-2017-798083 to LMN). For sharing data and facilities, we thank Henrique Teotónio. For strains, we thank Erik Andersen and the Caenorhabditis Genetics Center, funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). We thank Jia Shen, John Yuen, Arielle Martel, James Hong, and Ambika Natesan for assistance with experiments, and Zhenhao Guo and the Chalfie lab for helpful comments on the preprint.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.12293138.
Communicating editor: D. Fay
Literature Cited
- Ackema K. B., Sauder U., Solinger J. A., and Spang A., 2013. The ArfGEF GBF-1 Is Required for ER Structure, Secretion and Endocytic Transport in C. elegans. PLoS One 8: e67076 10.1371/journal.pone.0067076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Andersen E. C., Bloom J. S., Gerke J. P., and Kruglyak L., 2014. A variant in the neuropeptide receptor npr-1 is a major determinant of Caenorhabditis elegans growth and physiology. PLoS Genet. 10: e1004156 10.1371/journal.pgen.1004156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeles-Albores D., Lee R. Y. N., Chan J., and Sternberg P. W., 2018. Two new functions in the WormBase Enrichment Suite. microPublication Biology. 10.17912/W25Q2N [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David E., Burga A., and Kruglyak L., 2017. A maternal-effect selfish genetic element in Caenorhabditis elegans. Science 356: 1051–1055. 10.1126/science.aan0621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendesky A., Pitts J., Rockman M. V., Chen W. C., Tan M.-W. et al. , 2012. Long-range regulatory polymorphisms affecting a GABA receptor constitute a quantitative trait locus (QTL) for social behavior in Caenorhabditis elegans. PLoS Genet. 8: e1003157 10.1371/journal.pgen.1003157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein M. R., Zdraljevic S., Andersen E. C., and Rockman M. V., 2019. Tightlylinked antagonistic-effect loci underlie polygenic demographic variation in C. elegans. Evolution Letters. 3: 462–473. 10.1002/evl3.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The Genetics of Caenorhabditis Elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burga A., Ben-David E., Vergara T. L., Boocock J., and Kruglyak L., 2019. Fast genetic mapping of complex traits in C. elegans using millions of individuals in bulk. bioRxiv 428870; doi: 10.1101/428870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne A. B., Weirauch M. T., Wong V., Koeva M., Dixon S. J. et al. , 2007. A global analysis of genetic interactions in Caenorhabditis elegans. J. Biol. 6: 8 10.1186/jbiol58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bůžková P., Lumley T., and Rice K., 2011. Permutation and parametric bootstrap tests for gene-gene and gene-environment interactions. Ann. Hum. Genet. 75: 36–45. 10.1111/j.1469-1809.2010.00572.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Phong B. L., Fisher A. L., and Wang Z., 2011. Regulation of Fertility, Survival, and Cuticle Collagen Function by the Caenorhabditis elegans eaf-1 and ell-1 Genes. J. Biol. Chem. 286: 35915–35921. 10.1074/jbc.M111.270454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmensen L., Witten D., Hastie T., and Ersbøll B., 2011. Sparse Discriminant Analysis. Technometrics 53: 406–413. 10.1198/TECH.2011.08118 [DOI] [Google Scholar]
- Cook D. E., Zdraljevic S., Roberts J. P., and Andersen E. C., 2017. CeNDR, the Caenorhabditis elegans natural diversity resource. Nucleic Acids Res. 45: D650–D657. 10.1093/nar/gkw893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G. N., Laufer J. S., Kusch M., and Edgar R. S., 1980. Genetic and Phenotypic Characterization of Roller Mutants of Caenorhabditis Elegans. Genetics 95: 317–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bono M., and Bargmann C. I., 1998. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell 94: 679–689. 10.1016/S0092-8674(00)81609-8 [DOI] [PubMed] [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R. et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipple K. M., and McCabe E. R., 2000. Phenotypes of patients with “simple” Mendelian disorders are complex traits: thresholds, modifiers, and systems dynamics. Am. J. Hum. Genet. 66: 1729–1735. 10.1086/302938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duveau F., and Félix M.-A., 2012. Role of pleiotropy in the evolution of a cryptic developmental variation in Caenorhabditis elegans. PLoS Biol. 10: e1001230 10.1371/journal.pbio.1001230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando T., Flibotte S., Xiong S., Yin J., Yzeiraj E. et al. , 2011. C. elegans ADAMTS ADT-2 regulates body size by modulating TGFβ signaling and cuticle collagen organization. Dev. Biol. 352: 92–103. 10.1016/j.ydbio.2011.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Sarov M., Taylor J., Flibotte S. et al. , 2014. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat. Methods 11: 529–534. 10.1038/nmeth.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertner B. E., Parmenter M. D., Rockman M. V., Kruglyak L., and Phillips P. C., 2012. More than the sum of its parts: a complex epistatic network underlies natural variation in thermal preference behavior in Caenorhabditis elegans. Genetics 192: 1533–1542. 10.1534/genetics.112.142877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G., and Dworkin I., 2004. Uncovering cryptic genetic variation. Nat. Rev. Genet. 5: 681–690. 10.1038/nrg1426 [DOI] [PubMed] [Google Scholar]
- Gimond C., Vielle A., Silva-Soares N., Zdraljevic S., McGrath P. T. et al. , 2019. Natural Variation and Genetic Determinants of Caenorhabditis elegans Sperm Size. Genetics 213: 615–632. 10.1534/genetics.119.302462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glater E. E., Rockman M. V., and Bargmann C. I., 2014. Multigenic natural variation underlies Caenorhabditis elegans olfactory preference for the bacterial pathogen Serratia marcescens. G3 (Bethesda) 4: 265–276. 10.1534/g3.113.008649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A. et al. , 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene J. S., Brown M., Dobosiewicz M., Ishida I. G., Macosko E. Z. et al. , 2016. Balancing selection shapes density-dependent foraging behaviour. Nature 539: 254–258. 10.1038/nature19848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins B. J., and Hirsh D., 1977. Roller mutants of the nematode Caenorhabditis elegans. Molecular and General Genetics MGG 150: 63–72. 10.1007/BF02425326 [DOI] [PubMed] [Google Scholar]
- Hodgkin J., 2005. Genetic suppression. WormBook ed. The C. elegans Research Community, WormBook, 10.1895/wormbook.1.59.1, http://www.wormbook.org. [DOI] [Google Scholar]
- Hodgkin J., and Doniach T., 1997. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics 146: 149–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Kuma K.-i., Toh H., and Miyata T., 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33: 511–518. 10.1093/nar/gki198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur T., and Rockman M. V., 2014. Crossover heterogeneity in the absence of hotspots in Caenorhabditis elegans. Genetics 196: 137–148. 10.1534/genetics.113.158857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer J. M., 1994. Genetic Analysis of Extracellular Matrix in C. Elegans. Annu. Rev. Genet. 28: 95–116. 10.1146/annurev.ge.28.120194.000523 [DOI] [PubMed] [Google Scholar]
- Kramer J. M., and Johnson J. J., 1993. Analysis of Mutations in the Sqt-1 and Rol-6 Collagen Genes of Caenorhabditis Elegans. Genetics 135: 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M., Kranz A.-L., Su A., Winterkorn L. H., Albritton S. E. et al. , 2015. Developmental dynamics of X–Chromosome dosage compensation by the DCC and H4K20me1 in C. elegans. PLoS Genet. 11: e1005698 10.1371/journal.pgen.1005698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch M., and Edgar R. S., 1986. Genetic Studies of Unusual Loci That Affect Body Shape of the Nematode Caenorhabditis Elegans and May Code for Cuticle Structural Proteins. Genetics 113: 621–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Large E. E., Xu W., Zhao Y., Brady S. C., Long L. et al. , 2016. Selection on a Subunit of the NURF Chromatin Remodeler Modifies Life History Traits in a Domesticated Strain of Caenorhabditis elegans. PLoS Genet. 12: e1006219 10.1371/journal.pgen.1006219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R., 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaan U., Yzeiraj E., Meade M., Clark J. F., Rushlow C. A. et al. , 2018. BMP Signaling Determines Body Size via Transcriptional Regulation of Collagen Genes in Caenorhabditis elegans. Genetics 210: 1355–1367. 10.1534/genetics.118.301631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard F., Noble L., Guzella T., Afonso B., Baer C. F. et al. , 2019. Selection and drift determine phenotypic stasis despite genetic divergence. bioRxiv. doi: 10.1101/778282 (Preprint posted September 23, 2019). [Google Scholar]
- McGrath P. T., Rockman M. V., Zimmer M., Jang H., Macosko E. Z. et al. , 2009. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron 61: 692–699. 10.1016/j.neuron.2009.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon L., Muriel J. M., Roberts B., Quinn M., and Johnstone I. L., 2003. Two Sets of Interacting Collagens Form Functionally Distinct Substructures within a Caenorhabditis elegans Extracellular Matrix. Mol. Biol. Cell 14: 1366–1378. 10.1091/mbc.e02-08-0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B. J., and Casson L. P., 1986. Caenorhabditis elegans compensates for the difference in X chromosome dosage between the sexes by regulating transcript levels. Cell 47: 871–881. 10.1016/0092-8674(86)90802-0 [DOI] [PubMed] [Google Scholar]
- Noble L. M., Chelo I., Guzella T., Afonso B., Riccardi D. D. et al. , 2017. Polygenicity and epistasis underlie fitness-proximal traits in the Caenorhabditis elegans multiparental experimental evolution (CeMEE) panel. Genetics 207:1663–1685. 10.1534/genetics.117.300406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble L. M., Rockman M. V., and Teotónio H., 2019. Gene-level quantitative trait mapping in an expanded C. elegans multiparent experimental evolution panel. bioRxiv. (Preprint posted March 26, 2019). 10.1101/589432 [DOI] [Google Scholar]
- Novelli J., Page A. P., and Hodgkin J., 2006. The C Terminus of Collagen SQT-3 Has Complex and Essential Functions in Nematode Collagen Assembly. Genetics 172: 2253–2267. 10.1534/genetics.105.053637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyström J., Shen Z.-Z., Aili M., Flemming A. J., Leroi A. et al. , 2002. Increased or Decreased Levels of Caenorhabditis elegans lon-3, a Gene Encoding a Collagen, Cause Reciprocal Changes in Body Length. Genetics 161: 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby A. B., and Gibson G., 2016. Cryptic Genetic Variation in Evolutionary Developmental Genetics. Biology (Basel) 5: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby A. B., and Rockman M. V., 2014. Cryptic genetic variation: evolution’s hidden substrate. Nat. Rev. Genet. 15: 247–258. 10.1038/nrg3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A., and Johnstone I. L., 2007. The cuticle. WormBook ed. The C. elegans Research Community, WormBook, 10.1895/wormbook.1.138.1, http://www.wormbook.org. [DOI] [Google Scholar]
- Pagès, H., P. Aboyoun, R. Gentleman, and S. DebRoy, 2019 Biostrings: Efficient manipulation of biological strings. R package version 2.50.2.
- Paix A., Folkmann A., Rasoloson D., and Seydoux G., 2015. High Efficiency, Homology-Directed Genome Editing in Caenorhabditis elegans Using CRISPR-Cas9 Ribonucleoprotein Complexes. Genetics 201: 47–54. 10.1534/genetics.115.179382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., and Schliep K., 2019. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35: 526–528. 10.1093/bioinformatics/bty633 [DOI] [PubMed] [Google Scholar]
- Pebesma E. J., and Bivand R. S., 2005. Classes and methods for spatial data in R. R News 5: 9–13. [Google Scholar]
- Persikov A. V., Ramshaw J. A. M., and Brodsky B., 2005. Prediction of Collagen Stability from Amino Acid Sequence. J. Biol. Chem. 280: 19343–19349. 10.1074/jbc.M501657200 [DOI] [PubMed] [Google Scholar]
- Priess J. R., and Hirsh D. I., 1986. Caenorhabditis elegans morphogenesis: the role of the cytoskeleton in elongation of the embryo. Dev. Biol. 117: 156–173. 10.1016/0012-1606(86)90358-1 [DOI] [PubMed] [Google Scholar]
- Roberts B., Clucas C., and Johnstone I. L., 2003. Loss of SEC-23 in Caenorhabditis elegans Causes Defects in Oogenesis, Morphogenesis, and Extracellular Matrix Secretion. Mol. Biol. Cell 14: 4414–4426. 10.1091/mbc.e03-03-0162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., and Kruglyak L., 2009. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 5: e1000419 10.1371/journal.pgen.1000419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., Skrovanek S. S., and Kruglyak L., 2010. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science 330: 372–376. 10.1126/science.1194208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scriver C. R., and Waters P. J., 1999. Monogenic traits are not simple: lessons from phenylketonuria. Trends Genet. 15: 267–272. 10.1016/S0168-9525(99)01761-8 [DOI] [PubMed] [Google Scholar]
- Seidel H. S., Rockman M. V., and Kruglyak L., 2008. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science 319: 589–594. 10.1126/science.1151107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shephard F., Adenle A. A., Jacobson L. A., and Szewczyk N. J., 2011. Identification and functional clustering of genes regulating muscle protein degradation from amongst the known C. elegans muscle mutants. PLoS One 6: e24686 10.1371/journal.pone.0024686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterken M. G., Snoek L. B., Kammenga J. E., and Andersen E. C., 2015. The laboratory domestication of Caenorhabditis elegans. Trends Genet. 31: 224–231. 10.1016/j.tig.2015.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers K. M., 1996. Relationship between genotype and phenotype in monogenic diseases: Relevance to polygenic diseases. Hum. Mutat. 7: 283–293. [DOI] [PubMed] [Google Scholar]
- Swierczek N. A., Giles A. C., Rankin C. H., and Kerr R. A., 2011. High-throughput behavioral analysis in C. elegans. Nat. Methods 8: 592–598. 10.1038/nmeth.1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotónio H., Carvalho S., Manoel D., Roque M., and Chelo I. M., 2012. Evolution of outcrossing in experimental populations of Caenorhabditis elegans. PLoS One 7: e35811 10.1371/journal.pone.0035811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuscher A. C., Jongsma E., Davis M. N., Statzer C., Gebauer J. M. et al. , 2019. The in-silico characterization of the Caenorhabditis elegans matrisome and proposal of a novel collagen classification. Matrix Biology Plus 1: 100001 10.1016/j.mbplus.2018.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Keyl H., Kim H., Espey R., Oke C. V., and Edwards M. K., 1994. Caenorhabditis elegans sqt-3 mutants have mutations in the col-1 collagen gene. Dev. Dyn. 201: 86–94. 10.1002/aja.1002010109 [DOI] [PubMed] [Google Scholar]
- Vu V., Verster A. J., Schertzberg M., Chuluunbaatar T., Spensley M. et al. , 2015. Natural Variation in Gene Expression Modulates the Severity of Mutant Phenotypes. Cell 162: 391–402. 10.1016/j.cell.2015.06.037 [DOI] [PubMed] [Google Scholar]
- Weber K. P., De S., Kozarewa I., Turner D. J., Babu M. M. et al. , 2010. Whole Genome Sequencing Highlights Genetic Changes Associated with Laboratory Domestication of C. elegans. PLoS One 5: e13922 10.1371/journal.pone.0013922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster C. M., Wu L., Douglas D., and Soukas A. A., 2013. A non-canonical role for the C. elegans dosage compensation complex in growth and metabolic regulation downstream of TOR complex 2. Development 140: 3601–3612. 10.1242/dev.094292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlund B., Berry L. W., and Schedl T., 1997. Regulation of Germline Proliferation in Caenorhabditis elegans, pp. 43–80 in Advances in Developmental Biology (1992), Vol. 5, edited by Wassarman P. M. Cambridge, MA: Academic Press; 10.1016/S1566-3116(08)60035-3 [DOI] [Google Scholar]
- Yang J., and Kramer J. M., 1999. Proteolytic Processing of Caenorhabditis elegans SQT-1 Cuticle Collagen Is Inhibited in Right Roller Mutants whereas Cross-linking Is Inhibited in Left Roller Mutants. J. Biol. Chem. 274: 32744–32749. 10.1074/jbc.274.46.32744 [DOI] [PubMed] [Google Scholar]
- Yin T., Cook D., and Lawrence M., 2012. ggbio: an R package for extending the grammar of graphics for genomic data. Genome Biol. 13: R77 10.1186/gb-2012-13-8-r77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Smith D. K., Zhu H., Guan Y., and Lam T. T.-Y., 2017. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8: 28–36. 10.1111/2041-210X.12628 [DOI] [Google Scholar]
- Zamanian M., Cook D. E., Zdraljevic S., Brady S. C., Lee D. et al. , 2018. Discovery of genomic intervals that underlie nematode responses to benzimidazoles. PLoS Negl. Trop. Dis. 12: e0006368 10.1371/journal.pntd.0006368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Long L., Xu W., Campbell R. F., Large E. E. et al. , 2018. Changes to social feeding behaviors are not sufficient for fitness gains of the Caenorhabditis elegans N2 reference strain. eLife 7: e38675 10.7554/eLife.38675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zych K., Snoek B. L., Elvin M., Rodriguez M., Velde K. J. V. et al. , 2017. reGenotyper: Detecting mislabeled samples in genetic data. PLoS One 12: e0171324 10.1371/journal.pone.0171324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All quantitative data and code to reproduce our analyses and main figures are available from FigShare and https://github.com/lukemn/cuticle. Strains are available upon request. File S1 details all supplemental files, File S2 contains genotyping data from Figure 1, Files S3-S9 contain short videos of mutant and suppressed adult hermaphrodites on plates, File S10 contains gene models from Figure 2A, File S11 contains coding sequence alignments for Figure 2B, Files S12 and S13 contain raw and processed Multi-Worm tracker data for Figure 3 with associated R code in File S14. File S15 contains genotypes and phenotypes used for detecting genetic interactions between col-182 and SNPs in the CeMEE (Figure 4), with associated R code in File S16. Supplemental material available at figshare: https://doi.org/10.25387/g3.12293138.
Figure 1.
Allele frequencies along the X chromosome in Rol nonUnc s from a cross of CB4856 and N2-background unc-4(e120) rol-1(e91) II. Dashed lines show the Mendelian expectations for heterozygotes (1/2) and homozygotes (1/4 each).
Figure 2.
A. N2 carries a derived insertion of 8 bp (red) relative to the ancestral allele. The insertion duplicates an 8-bp sequence present in the ancestor (underlined). B. col-182 gene tree and model for C. elegans CB4856 and other Caenorhabditis species. The x-axis shows distance from the start codon for each genome. C. Protein alignment, including predicted sequences for C. elegans wild isolates CB4856, AB1 and ED3040 assembled from young adult RNAseq data, and the frame-shifted N2 translation (marked with a blue dot). Gray-scale shading represents site conservation (% identity), and labels are colored by predicted collagen triplet stability (melting temperature) for runs of >1 G-X-Y repeats conserved across all sequences other than N2. The positions of conserved triplets are indicated above the alignment by gray boxes, and the positions of conserved cysteine residues potentially involved in inter-strand disulphide bridges are shown as red boxes.
Figure 4.
col-182 modifies the effects of natural genetic variation on worm movement and shape. Genome-wide statistics are shown for two bivariate response models of col-182 indel SNP genotype, worm length/width (A) and Rol/Sqt sparse discriminant functions (B), using diallelic SNPs segregating in 363 recombinant inbred lines of the C. elegans multiparent experimental evolution (CeMEE) panel. Statistics are from a likelihood ratio test for a full additive and interaction linear model against a null model of no genetic effects. Permutation thresholds for genome-wide significance (FDR = 0.2) are shown in red, light shaded regions show 1.5 LOD drop QTL intervals (expanded to a minimum of 300 Kb for visibility), and the location of col-182 on the X chromosome is indicated by a dark gray line. Effect plots at right show genotype class means and standard errors for QTL with significant interactions (parametric bootstrap against additive models, ). Trait values shown are the first principal component for length/width, and the Sqt discriminant function, which explains most of the interaction in the Rol/Sqt bivariate model. Reference-based genotype is on the x-axis. Genotype and phenotype data in File S15, code in File S16.