Abstract
Plasma concentration of Cystatin C (CysC) level is a biomarker of glomerular filtration rate in the kidney. We use a Systems Genetics approach to investigate the genetic determinants of plasma CysC concentration. To do so we perform Quantitative Trait Loci (QTL) and expression QTL (eQTL) analysis of 120 Diversity Outbred (DO) female mice, 56 weeks of age. We performed network analysis of kidney gene expression to determine if the gene modules with common functions are associated with kidney biomarkers of chronic kidney diseases. Our data demonstrates that plasma concentrations and kidney mRNA levels of CysC are associated with genetic variation and are transcriptionally coregulated by immune genes. Specifically, Type-I interferon signaling genes are coexpressed with Cst3 mRNA levels and associated with CysC concentrations in plasma. Our findings demonstrate the complex control of CysC by genetic polymorphisms and inflammatory pathways.
Keywords: Quantitative trait loci, Multi parental models, Cystatin C, Kidney biomarkers, Type-I interferon signalling pathway, Multiparent Advanced Generation Inter-Cross (MAGIC), multiparental populations, MPP
The kidney is a complex organ responsible for excretion (Finco 1997), secretion (Davis et al. 1961), reabsorption (Lemann et al. 1970), and activating vitamin D (Fraser and Kodicek 1973). The gold standard for assessing kidney function is the glomerular filtration rate (GFR), but it is difficult to measure with precision. Therefore, GFR is often estimated from circulating creatinine (Ferguson et al. 2015). Creatinine is an amino acid derivative, released by muscle, and freely filtered by the kidney glomerulus (Narayanan and Appleton 1980). However, the level of plasma creatinine is influenced by a number of factors, including: diet, muscle mass, medication, chronic illness, age, sex, and race, limiting its accuracy to represent true GFR (Stevens et al. 2006). An alternative to creatinine is Cystatin C (CysC), often used in research as the basis for estimating glomerular filtration rate. CysC is produced by all mammalian cells, secreted into the blood, filtered through the glomerulus, and catabolized by tubular cells (Inker and Okparavero 2011). Plasma CysC had been found to be unaltered by age, sex, race, and metabolic disorders and was proposed as a clinical biomarker for GFR (Filler et al. 2005; Newman et al. 1995). Several clinical trials reported CysC as a superior marker compared to creatinine (Plebani et al. 1998; Kyhse-Andersen et al. 1994; Harmoinen et al. 1999) while a few others could not show a significant difference between CysC and creatinine (Donadio et al. 2001; Oddoze et al. 2001). Although CysC is not influenced by physiological factors, several recent studies (Köttgen et al. 2010; Köttgen et al. 2009; Hwang et al. 2007) have found associations between SNPs near the Cystatin C gene (Cst3) and plasma CysC levels or CysC-based estimated glomerular filtration rate (eGFRcys). How these genetic variants relate to plasma CysC protein level or its mRNA level remains to be determined.
In the current study, we focus on the genetic determination of CysC protein and mRNA levels using a “Systems Genetics” approach, which is useful for the discovery of genes and pathways associated with a reduced kidney function (Keller et al. 2012). We also perform co-expression analysis and expression QTL (eQTL) analysis to further investigate the control of CysC levels using the Diversity Outbred (DO) population, which are derived from eight founder strains (A/J, C57BL/6J, 129S1/SvImJ, NOD/ShiLtJ, NZO/HiLtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ). Since the DO is a highly recombinant population with immense genetic and phenotypic variation (Svenson et al. 2012; Smallwood et al. 2014), DO mice have been successfully used in studies focused on the genetic response to environmental toxin exposure and diseases (French et al. 2015; Tyler et al. 2017). The DO mice capture the same set of allelic variants as the eight founder strains, and their genetic backgrounds are well-studied, which make them an excellent model for researching the genetic susceptibility to disease. In this study, we focus on the genetic variation associated with CysC gene expression and plasma CysC concentration in DO mice. We then performed co-expression network analysis to identify gene modules associated with CysC and identified two gene modules associated with CysC levels.
Materials and Methods
Study design and sample collection
Female diversity outbred (DO) mice (n = 120; J: DO; G16; stock number 009376) were obtained from the Jackson Laboratory (Bar Harbor, ME) at 4 weeks of age. These mice were used to generate progeny for another study (Didion et al. 2015) and then aged. At 56 weeks of age, all mice were injected intraperitoneally with a volume of sterile isotonic saline equivalent to 10% of their body weight and urine was collected in a chilled metabolic cage system (Hatteras Inc, NC). On the following day, mice were fasted for 4 hr, plasma was collected from the retro-orbital sinus into EDTA-containing microtubes and centrifuged. Mice were euthanized, dissected, and tissue samples were snap frozen in liquid nitrogen. Biological samples were stored at -80° until assayed. All procedures were approved by the IACUC at UNC, Chapel Hill (IACUC Protocol Number 13-103).
Plasma and urine biochemical assays
Plasma CysC was measured by a commercially available quantitative sandwich ELISA kit (R&D Systems, MN, USA) for mice according to the manufacturer’s instructions. In brief, plasma samples were diluted 200-fold and incubated in a 96 well microplate pre-coated with CysC specific antibody and CysC concentration was determined from the color intensity of oxidized Tetramethylbenzidine (TMB) measured at 450 nm. Urinary total protein, creatinine and plasma blood urea nitrogen were measured by COBAS INTEGRA 400 plus analyzer (Roche Diagnostics, Rotkreuz, Switzerland). To measure urinary Na+, samples were diluted 1500-fold with 1.0% v/v trace metal free nitric acid and analyzed by using a Varian VISTA AX CCD Simultaneous Inductively Coupled Plasma Atomic Emission Spectroscopy (Varian, CA, USA). Standards Na+ for Inductively Coupled Plasma Mass Spectrometry (Spex CertiPrep, NJ, USA) was used to make standards ranging from 0.05 ppm to 5.0 ppm. Certified urine controls (Seronorm, Stasjonsveien, Norway) and sample pool controls were used to check the accuracy of the assay.
Kidney mRNA microarray
Total RNA was extracted from about 25 mg of kidney tissue using automated instrumentation (Maxwell 16 Tissue LEV Total RNA Purification Kit, Promega). RNA concentration was measured by fluorometry (Picogreen Life Technologies), and RNA quality was verified using a microfluidics platform (Bioanalyzer, Agilent Technologies). 95 RNA samples were chosen for microarray analysis and 1 sample was run in duplicate as a control. RNA was hybridized to Affymetrix Mouse Gene 2.1 ST 96-Array Plate using a GeneTitan instrument from Affymetrix according to the manufacturer’s protocols. We used the robust multiarray average method (RMA) implemented in the Affymetrix gene expression console with default settings (median polish and sketch-quantile normalization) to estimate the normalized expression levels of transcripts. All probes containing known single nucleotide polymorphisms (SNPs) from the eight founder inbred mouse strains of the DO mouse population were masked (165,204 probes) during normalization by downloading the SNPs from the Sanger sequencing website (http://www.sanger.ac.uk/science/data/mouse-genomes-project) and overlapping them with probe sequences. The data are available at NCBI Gene Expression Omnibus (GEO) database under the accession ID GSE122061.
Genotyping
DNA was extracted and purified from tail samples using Qiagen DNeasy kit (Qiagen, MD, USA) according to the manufacturer’s instructions. Genotyping was performed using the Mega Mouse Universal Genotyping Array (MegaMUGA) by GeneSeek (Neogen, Lansing, MI) (Welsh et al. 2012). The MegaMUGA array is built on the Illumina Infinium platform and contains 77,800 SNP markers that are distributed throughout the genome at an average spacing of 33 Kb. Genotypes of DO mice are accessible through UNC’s Mutant Mouse Resource and Research Centers website (https://www.med.unc.edu/mmrrc/genotypes/). We estimated heritability (h2) from allele probability using a linear mixed model in R package QTL2 version 0.18 (Broman et al. 2019).
QTL mapping
QTL mapping and Genome-wide association (GWAS) analysis were performed using the R package QTL2 (Broman et al. 2019) on the University of California, Davis’ high-performance cluster computing system, which has 6,752 CPUs with 64 GB – 1 TB of RAM on each node. Genotype probability was calculated from the allele calls. The probability of a founder SNP haplotype was calculated from genotype probabilities. QTL mapping was carried out by regressing the phenotypes on the founder haplotypes with an adjustment for kinship matrix as a random effect of the linear mixed effect model in the QTL2 “scan1” and “scan1perm” functions. Kinship matrix is a measure of genetic similarity among individuals used to control for the random polygenic effect in genome scanning. In this study, we calculated a matrix of the proportion of matching alleles per chromosome using the leave out one chromosome at a time (type = “loco”) method in the QTL2 R package. Candidate genes were identified by position based on the Wellcome Trust Sanger mouse genomes database, www.sanger.ac.uk, release 1303 based on genome assembly GRCm38/mm10 (Yalcin et al. 2011). QTL support intervals were defined by the 95% Bayesian Credible Interval (BCI), calculated by normalizing the area under the QTL curve on a given chromosome (Chen and Kendziorski 2007). We used the Best Linear Unbiased Predictors (BLUPs) model (Robinson 1991) in the QTL2 package to estimate the allelic contributions of the 8 founder strains to the significant QTL in which the founder allelic effect was identified using a regression of the phenotype on the founder genotype probabilities at the locus. The mapping statistic reported as a log of the odds ratio (LOD). The significance LOD thresholds at P < 0.05 for each kidney biomarker and gene probeset were determined by performing 1,000 permutations of genome-wide scans by shuffling genotype data in relation to phenotype data to generate a null distribution from the maximum LOD score (Churchill and Doerge 1994). A QTL or eQTL was considered significant when the LOD score for the phenotype is above the permutation LOD threshold at P < 0.05 for that particular phenotype. Instead of estimating a single global LOD threshold from permutation testing of a randomly selected subset of genes, we performed permutation testing for each annotated transcript cluster ID to determine its individual LOD threshold. A single LOD threshold for all transcript cluster IDs allows variation of the significant p-value cut off across transcript cluster ID, leading to possible increases in both Type –I error for some transcript cluster ID and type-II error for others (Figure S1). To accomplish > 23,000,000 genome scans, we utilized a high-performance cluster computing system, which allowed us to perform the parallelization of CPU for computationally intensive permutation genome scans to empirically determine the significance threshold for each annotated transcript cluster ID on the microarray. An eQTL for microarray data were considered “cis” when a transcript cluster ID was located at the same genomic position (within a ± 2Mb interval) of the probe (Bennett et al. 2010). We compared the single empirical LOD threshold-based eQTL results to the individual threshold method and identified 274 transcript cluster ID with Type-II error or false negative. Similarly, the single empirical LOD based analysis incorrectly identified 1,495 non-significant eQTLs as significant (Type-I error or false positive). Therefore, in our study, we found a single empirical LOD threshold-based analysis had 49.5% Type-I error and 9.1% Type-II error (Figure S1C).
Weighted Gene Correlation Network Analysis
Co-occurrence network analysis was performed by using R package WGCNA-Weighted Gene Correlation Network Analysis (Langfelder and Horvath 2008) version 1.66. Gene expression data were available for 95 samples. A total of 23,612 transcript cluster IDs were filtered to 8,045 those were expressed above robust multi-array average (RMA) value of 6 in more than 87.5% of the samples (Aylor et al. 2011). 4 samples were identified as outliers (Figure S2A) and removed, resulting in 91 samples for gene module analysis. A soft threshold approach was used with a power of 4 (based on scales free topology, Figure S2B and S2C) in a WGCNA default unsigned network with dynamic tree cutting (deep split = 2) and a min Module Size = 15 as parameters for the dynamic tree cut function (Langfelder et al. 2007). The module eigengene, defined as the first principle component of a module’s gene expression matrix, was used to calculate the Spearman correlation between a module and kidney biomarkers. Gene network modules were visualized using Cytoscape 3.7.1 (Shannon et al. 2003).
Functional annotation
We performed gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis on each module using Enrichr (Chen et al. 2013) with “GO biological process 2018” and “KEGG 2019 mouse” dataset, respectively, to determine if any gene set in the modules were associated with shared functional annotations or biochemical pathway. For each term in the gene-set library, the rank-based ranking of each of the GO term was derived from running Fisher’s exact test for many random gene sets to compute a mean rank and standard deviation from the expected rank and then z-score was calculated to assess the deviation from the expected rank of the enriched GO term. The combined score for the enriched GO term was computed by multiplying the z-score and -log of the p-value of the Fisher exact test. Gene annotation for tissue-specific expression was performed using several databases including: DAVID (Huang et al. 2009) and BioGPS Mouse Cell Type and Tissue Gene Expression dataset (Wu et al. 2013). The human GWAS data at the CysC locus were obtained from the European population (Pattaro et al. 2016) and was queried for LocusZoom (Pruim et al. 2010) plotting. The effect of SNP and indel variation on protein function was determined by a web-based tool Protein Variation Effect Analyzer -PROVEAN (Choi and Chan 2015). In silico transcription factor (TF) binding site prediction was performed by using CiiiDER (Gearing et al. 2019) with JASPAR 2020 motif database (Khan et al. 2018). The functional relevance of SNPs located within the predicted transcription motif was estimated by using R package for tRAP (Thomas-Chollier et al. 2011).
Quantitative PCR
To validate the kidney genes expression we utilized archived kidney samples from a strain survey of DO/CC Progenitor mice (O’Connor et al. 2014) in which the mice were perfused prior to tissue collection. Total RNA was isolated using MagMAX mirVana Total RNA Isolation Kit (Thermo Fisher, MA, USA) and cDNA was synthesized using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher, MA, USA) according to the manufacturer’s recommendation. The qPCR was performed using the primer listed in the Table S1 using PowerUp SYBR Green Master Mix (Thermo Fisher, MA, USA) on QuantStudio 6 and 7 Flex Real-Time PCR Systems (Thermo Fisher, MA, USA). PCR was run in triplicate and relative gene expression was determined using an efficiency corrected method, and efficiency was determined from a 2-log serial dilutions standard curve made from cDNA pooled from all samples (Bennett et al. 2013) and normalized by GAPDH expression.
Other statistical analyses for the phenotype data
Clinical phenotype data for DO mice were checked for normality. Non-normally distributed data were transformed using log10, Box-Cox power, or rank-normal functions before any statistical analysis. The variables with Shapiro-Wilk “W” value >0.95 were considered as normally distributed. The correlation between CysC and variables indicating kidney biomarkers were determined by Spearman correlation. Regression analysis involving CysC was adjusted for body weight as a confounding factor. Data are presented as means ± SD unless otherwise indicated. Statistical analysis was performed using R 3.6.1 for windows release (R Core 2019). Correlation p-value was adjusted for multiple comparisons using “Benjamini & Hochberg” (Benjamini and Hochberg 1995), and an adjusted p-value <0.05 was considered significant for all analyses unless stated otherwise.
Data availability
Microarray gene expression data are available at GEO with the accession ID GSE122061. Genotypes of DO mice are accessible through UNC’s MMRRC website (https://www.med.unc.edu/mmrrc/genotypes/). Further information is available from the corresponding authors if required. This study was approved by the IACUC at UNC, Chapel Hill (IACUC Protocol Number 13-103). DO mouse kidney RNASeq (DO-RNASeq) data are available at the NCBI’s Gene Expression Omnibus (GEO) with the accession ID GSE121330 and also at Dr. Churchill’s laboratory web site (https://churchilllab.jax.org/qtlviewer/JAC/DOKidney). The C57Bl/6J mouse RNASeq (B6-RNASeq) data (Söllner et al. 2017) is available at https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA375882 and EBI under the Array Express ID: E-MTAB-6081; https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6081/. Supplemental material available at figshare: https://doi.org/10.25387/g3.10307972.
Results
Characteristics of the study DO mice population
The mean body weight of the mice was 32.4 ± 7.4g with a range from 19.0 to 58.5g; a variation of approximately 23% among our DO study population (Table 1). Plasma CysC had similar variation as bodyweight, approximately 25% variation, with a mean concentration of 535.5 ± 137.5 ng/mL and ranging from 260 to 922 ng/ml. Although both body weight and plasma CysC had similar variations within the population of mice they were not significantly correlated with each other (r = 0.028, P = 0.80).
Table 1. Characteristics of the study mice.
| Characteristics | na | Mean ± SD | Median (25th, 75th) | Correlation with Cystatin C | Correlation with Cystatin C (Cst3) mRNAb | ||
|---|---|---|---|---|---|---|---|
| r | p | r | p | ||||
| Body weight, g | 120 | 32.4 ± 7.4 | 31.5 (26.3, 37.3) | −0.014 | 0.88 | 0.028 | 0.80 |
| Plasma cystatin C, ng/mL | 109 | 535.5 ± 137.5 | 515.1 (427.7, 629.5) | 1.000 | NA | 0.500 | 1.22 x 10−06 |
| Blood urea nitrogen, mg/dL | 120 | 14.9 ± 5.0 | 14.3 (11.9, 16.7) | −0.079 | 0.41 | −0.097 | 0.38 |
| Urine pH | 111 | 6.6 ± 0.8 | 6.5 (6.0, 7.2) | −0.062 | 0.54 | −0.085 | 0.44 |
| Urine Osmolality, mOsm/kg of water | 117 | 534.8 ± 201.2 | 491 (400, 625) | 0.181 | 0.062 | 0.219 | 0.046 |
| Urine volume, µL | 120 | 826.4 ± 651.4 | 745.0 (285.0, 1210.0) | −0.126 | 0.19 | −0.007 | 0.95 |
| Urinary protein, mg/L | 83 | 369.4 ± 321.7 | 255.6 (105.2, 570.4) | 0.176 | 0.12 | 0.177 | 0.16 |
| Urinary creatinine, mmol/L | 83 | 0.3 ± 0.2 | 0.3 (0.3, 0.5) | 0.183 | 0.11 | 0.228 | 0.07 |
| Urinary protein: creatinine, mg/mmol | 83 | 2080 ± 6530 | 789 (349, 1400) | 0.121 | 0.29 | 0.112 | 0.37 |
| Urinary total Na, ng | 92 | 2415 ± 1526 | 2060 (1296, 3380) | 0.006 | 0.95 | 0.121 | 0.31 |
| Na excretion rate, ng/h | 92 | 1208 ± 763 | 1030 (648, 1690) | −0.003 | 0.98 | 0.113 | 0.34 |
| Urinary Na: creatinine, ng/mmol | 82 | 10.3 ± 16.7 | 6.8 (4.8, 10.6) | −0.122 | 0.29 | −0.128 | 0.31 |
| Urinary total K, ng | 92 | 1002 ± 698 | 876 (514, 1326) | 0.045 | 0.68 | 0.115 | 0.34 |
| K excretion rate, ng/h | 92 | 501 ± 349 | 438 (257, 663) | 0.020 | 0.85 | 0.078 | 0.52 |
| Urinary K: creatinine, ng/mmol | 82 | 3.7 ± 5.0 | 2.9 (2.1, 3.8) | −0.202 | 0.08 | −0.249 | 0.045 |
Number represents the number of successful mouse phenotype observes depending on the availability of the sample.
Gene expression data were available for 95 mice.
We also measured levels of the following urinary analytes: creatinine, total protein, protein: creatinine ratio, total sodium, sodium excretion rate, sodium: creatinine ratio, total potassium, potassium excretion rate, and potassium: creatinine ratio. Additionally, blood urea nitrogen was measured. We observed a significant variation within urinary creatinine, urinary protein, urinary total protein: creatinine, urinary total sodium, urinary sodium excretion rate, and urinary sodium: creatinine (Table 1). To better assess how these urinary markers are related to renal function, we correlated plasma CysC and cystatin C (Cst3) gene mRNA expression levels with each urinary analyte. Plasma CysC did not show any significant correlation with other blood or urinary kidney biomarkers (Table 1). Cst3 expression level was significantly correlated with CysC (r = 0.498, P = 1.22 × 10−06) and Urinary K: creatinine (r = -0.249, P = 0.045; Table 1).
Quantitative trait locus (QTL) analysis of plasma and urinary traits demonstrate that plasma CysC concentration is under genetic determination in the DO
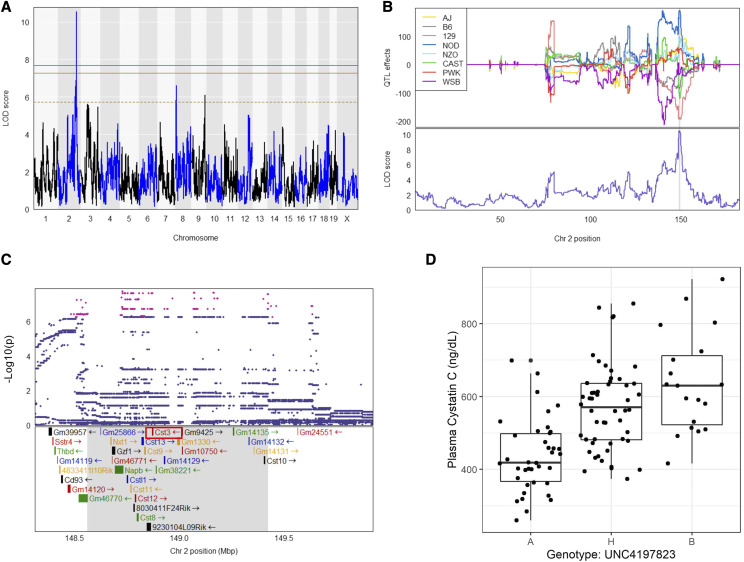
We first assessed the heritability (h2) of plasma CysC which was 47.5% in our DO population, indicating a significant portion, nearly 50%, of the variation in plasma CysC is genetic. To identify if there are specific loci regulating CysC levels, we next performed QTL analysis. We identified a single locus regulating plasma CysC on Chromosome 2 at position 148.7 Mb with a logarithm of the odds ratio (LOD) score of 10.6 which exceeded genome-wide threshold for plasma CysC as determined by permutation testing (Figure 1A). The CysC QTL on Chromosome 2 remained significant even after adjusting for the body weight as an additive covariate (Figure S3). We determined the 95% confidence interval by calculating the Bayesian Credible Interval (BCI), and identified a 1 Mb region of Chr 2, from 148.6-149.4 Mb associated with CysC.
Figure 1.
Plasma cystatin C QTL. (A) Genome scan of plasma CysC level. Red, solid golden, and broken golden lines show permutation-derived (N = 1000) significance thresholds at P < 0.05, P < 0.10, and P < 0.63, respectively. (B) The Best Linear Unbiased Predictors (BLUPs) coefficient plot of eight founder mice strains to the CysC QTL (top). Color represents the eight founder mice strains as indicated. The bottom portion represents the LOD score for plasma CysC on the chromosome 2. (C) Zoomed view of the peak position with an additive SNP model and the known genes in that region. Red dots indicate the significant SNPs at P < 0.05 (D) Genotype by phenotype plot of the top SNP located at the peak LOD for plasma CysC concentration. Genotype: A – AA, B – GG and H – AG. Shaded areas on the figure B and C represent approximate 95% Bayesian credible interval.
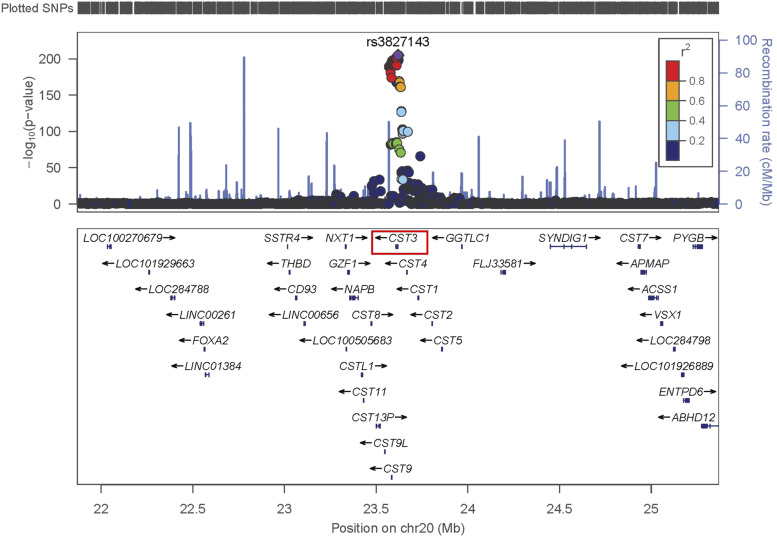
Human GWAS studies have identified several loci associated with kidney disease and related physiological traits. Notably, the locus on Chr 2 for CysC in DO mice corresponds to the homologous locus in humans, on Chromosome 20, which has been replicated several times (Akerblom et al. 2014; Suhre et al. 2017; Köttgen et al. 2010) for measured human kidney function using CysC based estimates of glomerular filtration rate (eGFRcys). This association for human eGFRcys is near the Cst3 gene locus on human Chr 20 between 22 Mb and 25 Mb in a European population (Pattaro et al. 2016) (Figure 2), which is a region of conserved synteny between humans and mice.
Figure 2.
Locuszoom association plot for human eGFR-cys at the Cst3 gene locus on human chromosome 20 between 22 Mb and 25 Mb. The human data were obtained from the European population (Pattaro et al. 2016) and is a region of conserved synteny between humans and mice.
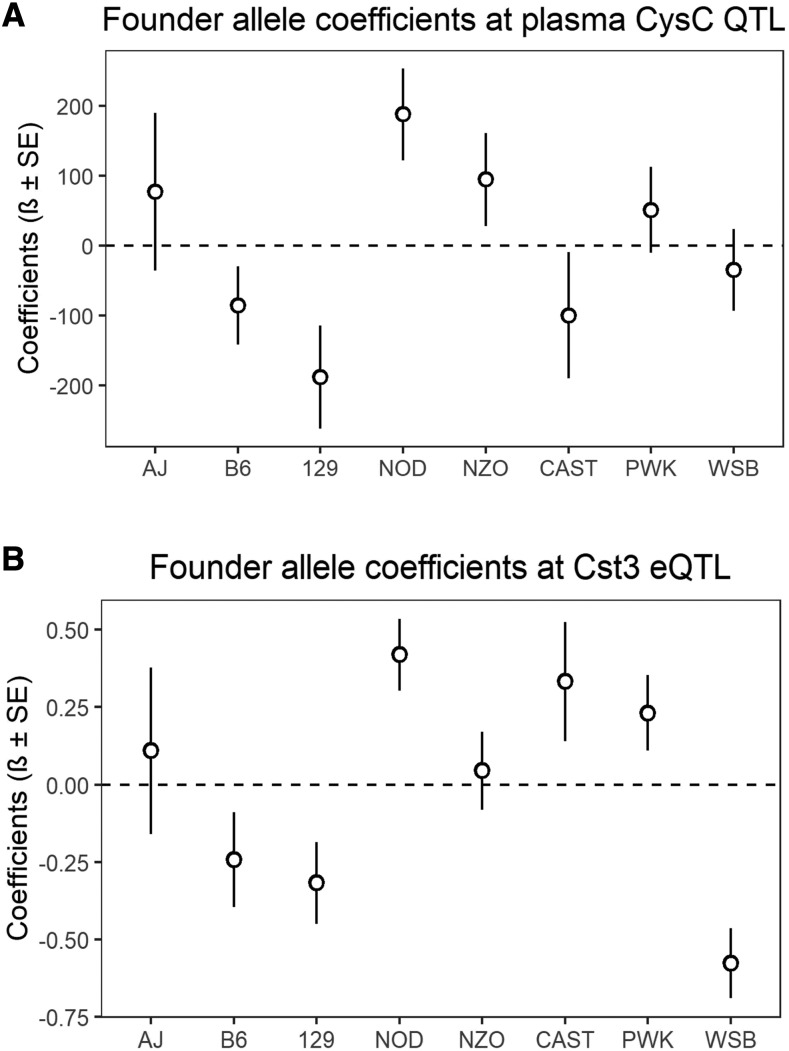
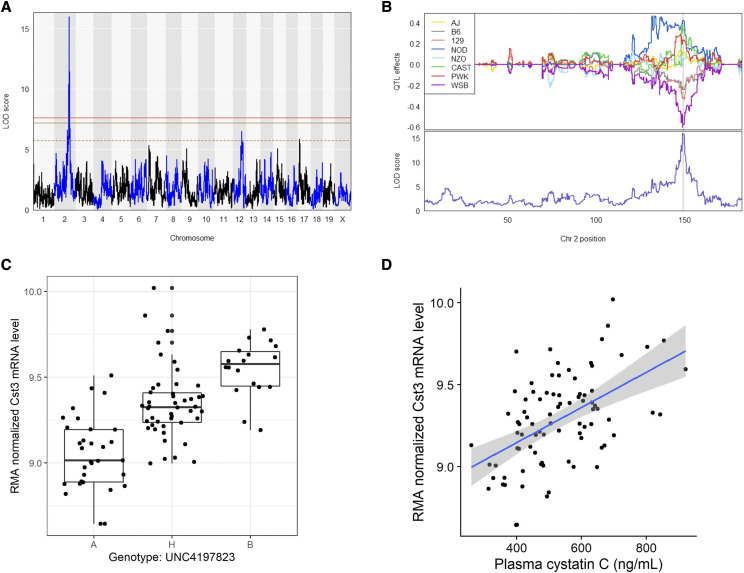
To further characterize this QTL, we estimated the allelic contributions of the 8 founder strains (Figure 1B and Figure 3A) and found that the haplotypes separate into two groups, in which, DO mice containing the 129, CAST, and B6 alleles have lower plasma CysC levels than mice harboring NOD and NZO haplotypes. We then determine the distribution of the founder allele among our DO mouse population and found that about 30.0%, 49.2%, 32.5%, 13.5%, 14.2%, 3.3%, 18.3% and 19.2% mouse are harboring genome from AJ, B6, 129, NOD, NOZ, CAST, PWK and WSB mouse, respectively either as homozygous or heterozygous state at the significant haplotype block of the CysC QTL (Table S2). This locus on the Chr 2 contains 32 pseudogenes and genes (Broman 2019; Yue et al. 2014) including Cst3 (Cystatin C gene) (Figure 1C, Table S3). Among them, gene expression values of 16 genes were available and only Cst3 mRNA expression was found to be significantly correlated (r = 0.52, adjusted p-value =6.57X10−6) with plasma CysC concentration (Table S3). Mice homozygous AA at the marker (UNC4197823, LOD = 10.3) had a lower plasma CysC level compared to heterozygous AG and homozygous GG (Figure 1D). To evaluate what other kidney gene expression levels might be associated with plasma CysC protein level we determined the correlation between plasma CysC level, and all genes expressed in kidney. Plasma CysC significantly correlated with a number of mRNA levels in kidney including Cst3 (cystatin C; r = 0.52, adjusted p-value = 0.0048), Trav16d-dv11 (T cell receptor alpha variable 16D-DV11; r = 0.52, adjusted p-value = 0.0048), Pecam1 (Platelet/endothelial cell adhesion molecule 1; r = 0.50, adjusted p-value = 0.0064), and Zfp768 (zinc finger protein 768; r=-0.45, adjusted p-value = 0.043; Table 2). We also performed QTL analysis of all other phenotypes listed on Table 1, which yielded a number of suggestive loci (Table S4).
Figure 3.
Regression coefficients of the association between genotype marker UNC4197823 located on Chr2 and (A) plasma CysC level or (B) Cst3 gene expression level for eight founder strain determined by BLUP analysis.
Table 2. Genes significantly correlated with plasma Cystatin C, and their eQTLs.
| Gene name | ENTREZID | Gene location | Correlation | eQTL | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Symbol | Chr | Stand | Start (Mb) | r | p | adj.p | SNP Position | LOD | |||
| Chr | Mb | ||||||||||
| T cell receptor alpha variable 16D-DV11 | Trav16d-dv11 | 547329 | Chr14 | + | 54.20 | 0.522 | 2.98x10−7 | 0.005 | |||
| Cystatin C | Cst3 | 13010 | Chr2 | — | 148.87 | 0.517 | 4.11x10−7 | 0.005 | Chr2 | 148.7 | 15.99 |
| Platelet/endothelial cell adhesion molecule 1 | Pecam1 | 18613 | Chr11 | — | 106.65 | 0.502 | 9.67x10−7 | 0.006 | Chr6 | 136.7 | 9.50 |
| Leupaxin | Lpxn | 107321 | Chr19 | + | 12.80 | 0.500 | 1.09x10−6 | 0.006 | |||
| FYVE, RhoGEF and PH domain containing 2 | Fgd2 | 26382 | Chr17 | + | 29.36 | 0.476 | 4.10x10−6 | 0.019 | |||
| Ribonuclease, RNase A family, 6 | Rnase6 | 78416 | Chr14 | + | 51.13 | 0.469 | 5.93x10−6 | 0.023 | |||
| Cytokine receptor-like factor 3 | Crlf3 | 54394 | Chr11 | — | 80.05 | 0.461 | 9.03x10−6 | 0.030 | |||
| Formin binding protein 1 | Fnbp1 | 14269 | Chr2 | — | 31.03 | 0.454 | 1.25x10−5 | 0.037 | |||
| Zinc finger protein 768 | Zfp768 | 233890 | Chr7 | — | 127.34 | −0.449 | 1.62x10−5 | 0.043 | |||
| Protein kinase C, eta | Prkch | 18755 | Chr12 | + | 73.58 | 0.445 | 1.98x10−5 | 0.043 | |||
| Selectin, platelet (p-selectin) ligand | Selplg | 20345 | Chr5 | — | 113.82 | 0.445 | 1.99x10−5 | 0.043 | |||
Genome-wide expression QTL (eQTLs) analysis of Renal Gene Expression indicates CysC mRNA is under genetic regulation
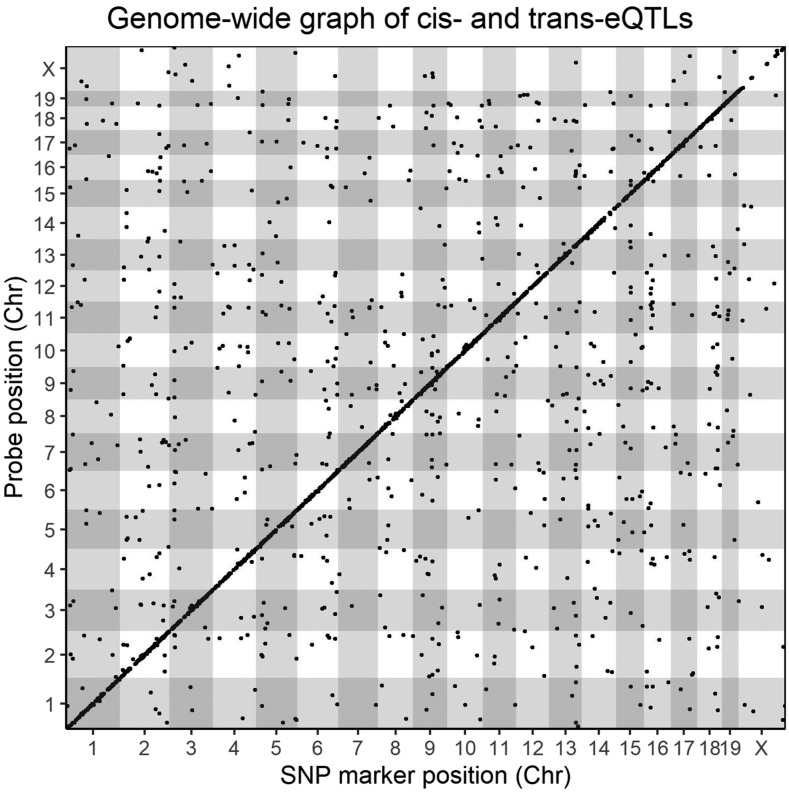
We performed individual QTL analysis for all annotated probesets available on the microarray to identify genes with significant eQTL (Methods). We determined the significant eQTL based on single p-value cut off (P < 0.05), allowing for a unique LOD threshold for each individual probeset as determined by permutation testing (Methods), and observed a total of 3,022 significant eQTL hits (Figure 4) for 2,866 unique transcripts. The average genome-wide LOD threshold at P < 0.05 for all annotated probesets was 7.94 ± 0.67, with a median of 7.79 (25th percentile= 7.69, 75th percentile= 7.93), and a range 6.96– 21.27. Among the total 3,022 statistically significant (P < 0.05) eQTL, a total of 2,004 were cis-eQTL (peak SNP within ±2Mb of the gene start position (Bennett et al. 2010)) and 1,018 were trans-eQTL (Figure 3). A list of all significant and suggestive eQTL and their corresponding LOD threshold are provided in Table S5.
Figure 4.
Locations of kidney eQTLs in the mouse genome. The positions of the significant eQTLs (P < 0.05) are plotted against the locations of the corresponding transcript (y-axis) along with the genome (x-axis). The significance thresholds for each individual probeset were determined by performing 1000 permutations of genome-wide scans by shuffling genotype data in relation to individual gene expression data for every single probeset on the microarray. Cis-eQTLs, occurring within a 4-Mb (±2Mb) genomic window, are located at the diagonal, all other dots represent trans-regulated genes.
We next investigated the expression pattern and genetic determination of genes at the CysC locus. We found that the mRNA levels of Cst3 has a heritability of 58.8% and has a significant cis-eQTL (LOD = 16.0) at the same CysC locus on the Chr 2 (Figure 4A-C) with a similar association with the peak SNP indicating a possible connection between genetic architecture and plasma CysC level through mRNA levels. Indeed, in our study, plasma CysC concentration was associated with Cts3 mRNA level [β (se) = 254.3 (45.6), P < 0.001] and remained significant even after adjusting with body weight [β (se) = 254.3 (46.0), P < 0.001] (Figure 5D). Mouse harboring the WSB, 129, and B6 alleles had lower Cst3 mRNA level compared to mice harboring CAST, PWK, NOD and NZO alleles (Figure 3B).
Figure 5.
eQTL for Plasma Cystatin C (Cst3) mRNA. (A) Genome scan of CysC gene (Cst3) mRNA expression levels. Red, solid golden, and broken golden lines show permutation-derived (N = 1000) significance thresholds at P < 0.05, P < 0.10, and P < 0.63, respectively. (B) BLUP coefficient plot of eight founder mice strains to the Cst3 eQTL (top). Color represents the eight founder mice strain as indicated. The bottom portion represents the LOD score for Cst3 eQTL model on the Chr 2. The shaded area represents approximate 95% Bayesian credible interval. (C) Genotype by phenotype plot of the top SNP located at the peak LOD. Genotype: A – AA, B – GG and H – AG. (D) Correlation between plasma cystatin C and Cst3 mRNA level.
Genetic variant analysis at the CysC locus
To better understand the genetic variation at the Cst3 locus, we compared genetic polymorphism present in the 8 founder mouse genome available on the Sanger Institute’s mouse database (www.sanger.ac.uk) and found that there are several 5′ and 3′ UTR variations, splice region variants, upstream and downstream gene variant as SNPs or insertion or deletion in the DO founders, which may lead to the mRNA abundance. We took an in silico approach to predict transcription factor (TF) binding sites using CiiiDER (Gearing et al. 2019) with JASPAR 2020 motif database (Khan et al. 2018). This analysis predicted 426 potential TFs interacting to 2,657 TF motifs in the Cst3 promotor region (-1500 bps to +500 bps of Cst3 transcription starting position). We next identified which of the sequence variants from the DO founders are located within one of the TF binding sites. The 8 founder strains of the DO contain 31 SNPs located within these predicted transcription motifs. To assess the functional significance of these variants predicted changes in TF binding affinity was also assessed in-silco using the R package tRAP (Thomas-Chollier et al. 2011), which calculates the affinity of transcription factors for a DNA sequences on the basis of a biophysical model and determine which TF is affected the most by a regulatory SNP (Thomas-Chollier et al. 2011). tRAP predicted that 50 transcription motifs alter binding affinity at a level of >20% due to the presence of SNPs. A number of these SNPs including rs225697750, rs27261906, rs244335261, rs220753689, rs236432550, rs220753689, rs387212829, rs27261907, rs581036327, and rs236432550 were distributed differently among alleles arising from WSB and NOD, CAST, or PWK founder strains (Table S6).
We also examined the Cst3 locus for structural variants using publicly available data in addition to variants affecting transcription (www.sanger.ac.uk). Among the 8 founder strains of the DO, two missense SNP variants in the Cst3 gene are present in the CAST allele at position 148872018 bp and 148875196 bp (rs27261909). Both of these specific variants could potentially affect the protein function and the effect of these polymorphisms was assessed using an in-silico prediction tool, Protein Variation Effect Analyzer -PROVEAN (Choi and Chan 2015). However, the both missense variants in the Cst3 gene were not predicted to have a known deleterious structural consequence (Table S7).
Co-expression modules of kidney mRNA showed association Between CysC and biologically related gene sets
Our correlation/regression analysis identified that 24.8% of the variation in plasma CysC protein could be accounted for by differences in the Cst3 gene expression. We hypothesized that additional genes and pathways affecting CysC levels could be identified using Weighted Gene Correlation Network Analysis (WGCNA). WGCNA can identify genetic pathways or modules of genes associated with clinical traits and provide a complementary approach to traditional QTL analysis. We constructed a gene co-expression network from 8,045 expressed genes in the kidney using WGCNA, as described in the method section. Out of these 8,045 genes, 4,166 transcripts formed 25 co-expressed gene modules, which contain a varying number of genes, ranging from 21 to 928 (Figure S4A-C). The remaining transcripts (∼3,900), had reduced topological overlap and were not assigned to a module.
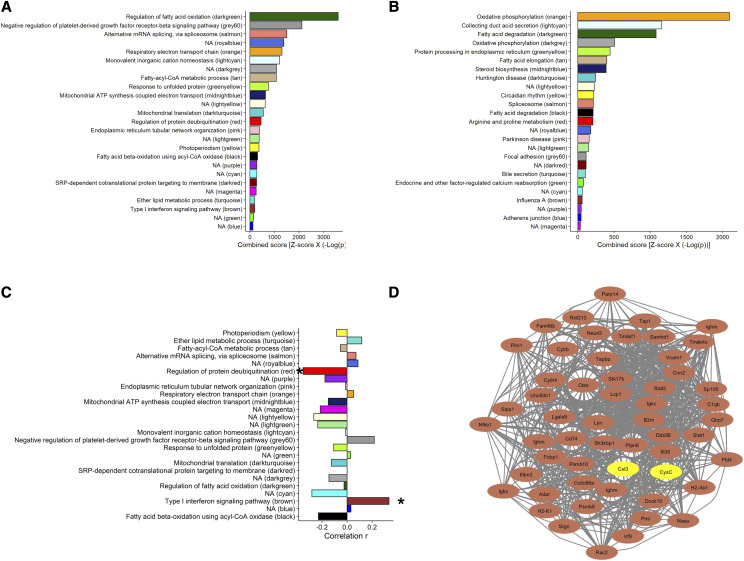
To determine the correlation between gene modules and kidney biomarkers, we calculated the module eigengene (ME), which is the first principal component for the module member gene expression values. The average percent variance explained by the MEs was 27.0 ± 5.3% and ranged from 20.7% (greenyellow) to 39.0% (royalblue) (Table S8). In contrast, ME of the transcripts not assigned to a module explained 3.6% variability. Then we performed gene set enrichment analysis (Chen et al. 2013) for the GO and KEGG terms (Tables S9 and S10, respectively), as described in the method, on each module to determine their shared gene ontology of the biological function. 16 out of 25 modules were significantly enriched with biological process related GO terms (Figure 6A) and 18 were significantly enriched with KEGG pathway terms (Figure 6B).
Figure 6.
Co-expression network analysis of kidney gene. (A) Top GO and (B) top KEGG pathway for each of the gene-module. (C) Correlation coeffiencent of module eigengenes with plasma CysC. “*” < 0.05. (D) Cytoscape network visualization of to 50 hubgene in the brown module and their relationship with Cst3 mRNA level is kidney and plasma CysC concentration. The brown nodes denote hubgenes in the module and the yellow nodes denote Cst3 gene and CysC protein concentration in plasma.
The modules identified in the network were prioritized by their correlation with CysC (Figure 6C). Two modules, brown and red, showed significant positive (r = 0.33, adjusted p-value = 0.027) and negative (r = -0.35, adjusted p-value = 0.027) correlation with plasma CysC, respectively (Figure 6C). This indicates that the brown and red modules explain about 10.9% and 12.3% variation of the plasma CysC levels. The brown module contains 323 genes and is highly enriched for genes related to the immune system, specifically Type-I interferon signaling (GO:0060337, adjusted p-value = 1.05 x10−5, Odd Ratio =10.6, and combined score [Z-score x -log(p)] = 200.0 (Table 3). The 50 most connected transcripts within the brown module (Table S11) showed a strong correlation among themselves, Cst3 gene, and CysC protein levels (Figure S5). We used Cytoscape network to visualize the connectivity among these 50 transcripts in the brown module and their connection with Cst3 mRNA level in kidney and plasma CysC concentration (Figure 6D). Among these 50 transcripts are several well-characterized immune genes, including Signal transducer and activator of transcription 1 (Stat1), Signal transducer and activator of transcription 2 (Stat2), Interferon induced transmembrane protein 3 (Ifitm3), Nuclear antigen sp100 (Sp100), Interferon-induced protein 35 (Ifi35), Sam domain and HD domain, 1 (Samhd1), and Interferon regulatory factor 9 (Irf9).
Table 3. Enrichment analysis of the brown module genes.
| Term | P | adj.P | Odd Ratio | Combined Score a |
|---|---|---|---|---|
| Type I interferon signaling pathway (GO:0060337) | 6.14 x 10−9 | 1.05 x 10−5 | 10.6 | 200.0 |
| Regulation of interleukin-8 secretion (GO:2000482) | 2.70 x 10−5 | 0.01 | 13.6 | 142.9 |
| Positive regulation of response to cytokine stimulus (GO:0060760) | 5.67 x 10−5 | 0.02 | 17.9 | 174.6 |
| Response to interferon-alpha (GO:0035455) | 1.30 x 10−4 | 0.03 | 14.7 | 131.6 |
| Positive regulation of interleukin-6 secretion (GO:2000778) | 1.30 x 10−4 | 0.03 | 14.7 | 131.6 |
| Positive regulation of interferon-alpha production (GO:0032727) | 2.06 x 10−4 | 0.04 | 13.2 | 111.7 |
| Regulation of dendritic cell apoptotic process (GO:2000668) | 4.48 x 10−4 | 0.05 | 18.8 | 144.6 |
| Positive regulation of tumor necrosis factor secretion (GO:1904469) | 6.09 x 10−4 | 0.07 | 17.0 | 126.2 |
| Ribose phosphate metabolic process (GO:0019693) | 8.02 x 10−4 | 0.08 | 15.6 | 111.4 |
| Pentose-phosphate shunt (GO:0006098) | 8.02 x 10−4 | 0.08 | 15.6 | 111.4 |
| Response to chemokine (GO:1990868) | 0.004 | 0.19 | 20.8 | 116.8 |
| Cellular response to chemokine (GO:1990869) | 0.004 | 0.19 | 20.8 | 116.8 |
| Positive regulation of ATP biosynthetic process (GO:2001171) | 0.004 | 0.18 | 20.8 | 116.8 |
| Vesicle fusion with endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membrane (GO:1990668) | 0.004 | 0.18 | 20.8 | 116.8 |
| Antigen processing and presentation of endogenous peptide antigen (GO:0002483) | 0.004 | 0.18 | 20.8 | 116.8 |
| Regulation of blood vessel remodelling (GO:0060312) | 0.004 | 0.18 | 20.8 | 116.8 |
| Regulation of interleukin-12 secretion (GO:2001182) | 0.004 | 0.18 | 20.8 | 116.8 |
| Regulation of interferon-alpha secretion (GO:1902739) | 0.004 | 0.17 | 20.8 | 116.8 |
| Positive regulation of interferon-alpha secretion (GO:1902741) | 0.004 | 0.17 | 20.8 | 116.8 |
| Glomerulus vasculature development (GO:0072012) | 0.004 | 0.17 | 20.8 | 116.8 |
Results shows the top 20 gene ontology (GO) of biological processes based on combined score.
The expected mean rank and standard deviation was calculated from running the Fisher exact test for many random gene sets for each term in the gene-set library and then the z-score was calculated to assess the deviation from the expected rank for the enriched GO term for the genes in the module. Finally, the combined score was computed by multiplying the z-score by the negative log of the p-value from the Fisher exact test.
Additionally, the red module was negatively associated with plasma CysC level (r =-0.340, adjusted p-value = 0.021). The red module contains 195 genes including D-amino acid oxidase (Dao), aldehyde dehydrogenase 2, mitochondrial (Aldh2), 4-hydroxy-2-oxoglutarate aldolase 1 (Hoga1), proline dehydrogenase (Prodh); proline dehydrogenase (oxidase) 2 (Prodh2), aldehyde dehydrogenase family 7, member A1 (Aldh7a1), and aldehyde dehydrogenase 9, subfamily A1 (Aldh9a1). KEGG pathway analysis revealed that the red module is enriched with arginine and proline metabolizing enzymes (adjusted p-value = 1.60 × 10−4, OR = 14.36, and combined Z-score = 207.6) (Table S10). The red module is also significantly enriched with regulation of protein deubiquitination (GO:0090085, adjusted p-value= 0.026) with an OR of 44.0 and combined score of 456.3 (Table S9).
Publicly available kidney gene expression datasets support association Between CysC and Type-I interferon signaling genes
In order to determine if the CysC-brown module association identified in the current study was robust we sought to identify appropriate publicly available datasets to investigate. We identified two gene expression data sets which utilize RNAseq, one utilizing C57BL/6J mice (EBI Array Express: E-MTAB-6081) as previously reported (Söllner et al. 2017) and one utilizing Diversity Outbred mice (GEO: GSE121330). We will refer to these as B6-RNAseq and DO-RNAseq. First, we ensured that the genes named within the manuscript are in fact expressed in the kidney. In supplemental Table S12 we list the expression of each gene mentioned in the manuscript and the two aforementioned datasets and found that they are expressed in the kidney. Additionally, we calculated the average expression for all genes in the datasets and listed this in the table along with the corresponding average expression for all eQTL genes and genes used in the Network analysis and found that they are comparable. Furthermore, we correlated the average expression values of the all common genes in our study, DO-RNAseq, and B6-RNAseq data sets and identified that broadly the genes selected are highly correlated (n = 16,599; r = 0.66; P < 0.001) and (n = 21,261; r = 0.60; P < 0.001), respectively). These data indicate that our measures of gene expression via microarray are representative of RNAseq methods and are reproducibly expressed in the kidney. We repeated this analysis with the 8,045 genes used for WGCNA analysis and these genes were also correlated between our data and renal samples of the DO-RNAseq (n = 7,818; r = 0.58; P < 0.001), and B6-RNAseq datasets (n = 7,825; r = 0.44; P < 0.001).
In addition to these global analyses, we also focused on the expression of brown module transcripts. We performed correlation analysis of the 50 most connected brown module transcripts and Cst3 in the DO-RNAseq dataset. The correlation between these genes and Cst3 was robust and ranged between r = 0.123 and 0.661. 48 of the selected transcripts were significant after multiple comparison testing (Figure S5). Most notable, the immune transcripts Stat1, Stat2, Ifitm3, Sp100, Ifi35, Samhd1 and Irf9 were all significantly correlated with Cst3 which is congruent with the immune system enrichment identified in the brown module. As a final confirmation quantitative PCR assays were performed on archived kidney samples from a strain survey of DO Progenitor mice (O’Connor et al. 2014) in which the mice were perfused prior to tissue collection and these transcripts were expressed in perfused samples (Table S13).
Discussion
Establishing the genetic architecture of kidney biomarkers remains critical to the development of clinical strategies for understanding kidney function. Advanced genetic mapping panels such as the Diversity Outbred (DO) mice provide a tremendous opportunity to examine the genetic determination of kidney function and disease. In this study, we utilize DO mice to dissect the genetic architecture of the renal biomarker CysC which yields 3 key results. The first is that CysC is associated with genetic polymorphism in aged, female DO mice. The second is that eQTL analysis identified a concordant eQTL for CysC mRNA, revealing a positive correlation between Cst3 gene expression and plasma CysC levels. The third is two module of genes co-expressed associated with CysC levels. Each of these are discussed in detail below.
We identified a QTL associated with plasma concentration of CysC on Chromosome 2 at approximately 148 Mb. This locus contains the gene Cst3 whose transcript is ultimately translated into the Cystatin C protein. eQTL analysis identified a significant cis-eQTL for the Cst3 gene at the same locus and there was a positive correlation between Cst3 mRNA and plasma CysC protein levels. Similar to our result, a meta-analysis of human GWAS data found that the estimated GFR based on plasma CysC was associated with SNPs proximal or within the physical location of Cst3 on Chr20 at 23.6 Mb in human (Köttgen et al. 2010; Köttgen et al. 2009; Hwang et al. 2007). The identification of a colocalized eQTL for the Cst3 gene and the correlation between transcript and protein levels in the current study suggest that a portion of the variable concentration of plasma CysC could be transcriptionally mediated. However, the potential causal variant(s) affecting mRNA levels remains to be elucidated. Detailed sequence analysis and utilization of in-silico prediction of the functional consequences of SNPs within the DO founder strains on transcription factor and protein function highlight the tremendous genetic variation contained in the DO at this locus. More than a hundred SNP variants were identified at the Cst3 locus (Keane et al. 2011). Our in-silico analysis predicted 50 SNPs which could possibly affect Cst3 mRNA levels and plasma CysC concentrations. However, these results remain to be confirmed.
The correlation between Cst3 mRNA and plasma CysC suggests complex regulation of CysC beyond a simple relationship between mRNA levels and plasma CysC concentrations as only 24.8% of the variation in plasma CysC protein could be accounted by the variation in the Cst3 gene expression. It is well understood that genetic variants are critically important factors affecting clinical traits, but we acknowledge that many complex traits are affected by a multitude of biological and environmental factors. One approach to address this biological complexity is the use of gene co-expression network analysis. Genes are often coregulated through complex biological pathways and co-expressed gene network analysis allows us to explore modulation of complex traits and disease phenotypes that are not understood by focusing on single genes. Therefore, we also identified a transcriptional networks (pathways) in the kidney associated with plasma CysC concentrations by using WGCNA analysis. Our gene module analysis showed that the plasma CysC level is highly co-expressed with genes involved in Type-I interferon (IFN) signaling pathway genes. Specifically, IFN-stimulated gene factor 3 (ISGF3), comprised of Stat1, Stat2, and Irf9, are strongly associated with CysC levels. To the best of our knowledge, this is the first report of local renal Type-I interferon signaling pathway gene expression being associated with transcriptional adjustment of CysC. The cell surface receptor for Type-I IFN are expressed on most cells, including kidney cells (Secombes and Zou 2017) and resident macrophages (York et al. 2007). When Type-I IFN binds with its receptor on the cell surface, a signaling cascade initiates, causing phosphorylation and activation of STATs (Schreiber 2017). We note that in-vitro experiments using a reporter assay identified a cis element named IRF (interferon regulatory factor)‐Ets composite sequence (IECS) that regulates both cystatin C and cathepsin C expression (Tamura et al. 2005). These data suggest a potential mechanism by which inflammatory signals contribute to plasma CysC levels. We note that our network analysis is non-directional and thus we cannot demonstrate a direct causal effect of IFN signaling and CysC gene expression. Furthermore, the relationship between CysC and Type-I IFN signaling genes is intriguing but complicated by the fact that CysC itself is expressed by monocytes and dendritic cells (El-Sukkari et al. 2003) and may have an important role in innate immune responses (Zi and Xu 2018). An additional study is needed to determine if this module of genes is regulating CysC or perhaps if CysC levels are influencing kidney inflammation. The Cst3 gene expression only explained about 25% variation of the plasma CysC concentration, indicating that in addition to environmental factors, there might be some other genes or cluster of genes that contributing plasma CysC concentration.
Our results indicate that the DO mice can be used to study renal function in the context of varying genetic determination of Cst3 levels, similar to what occurs in humans. Thus, perturbations that affect renal function through surgical or chemical manipulation could be used in DO mice to evaluate their effect on both renal function and CysC levels. Additionally, these studies are critical as studies in model organisms are effective at eliminating the random environmental factors. For example, studies in mice can tightly control the environment which allows the determination of the genetic contribution to the phenotype, independent of the environmental confounders.
The QTL, eQTL and WGCNA analysis identify a number of interesting factors affecting plasma CysC in this relatively small study utilizing DO mice. In addition to the sample size we acknowledge several areas for future investigation. Plasma CysC was measured only once at 56 weeks of age and thus we could not examine aging related changes in circulating CysC levels, which may be critical predictors of disease susceptibility. The DO cohort used in the current study is small (n = 120) and thus we were only able to detect QTL with strong effect size. We did not find QTL for kidney traits beyond plasma CysC and the cis-eQTL for Cst3 which could be a consequence of our study population. Another limitation of our study is that we did not perfuse the kidney with PBS before collection. Thus, we cannot eliminate the possibility that differences in circulating white cells are contributing in part to gene expression differences observed in this study. To address this limitation and to confirm renal expression of the genes discussed in this paper, we compared our data with two different publicly available mouse kidney gene expression datasets. Additionally, the expression of a number of transcripts was confirmed by qPCR in kidney isolated from a set of DO progenitor mice which were perfused before tissue collection. Our expression studies are comprehensive but were performed on kidney tissue not specific cell types and thus provide limited insight into the specific kidney cell types mediating trait associations. Lastly, the results associating IFN signaling pathway gene expression with CysC levels is intriguing, but questions remain related to the temporal aspect of this association and how perturbation of genes in this module affect plasma CysC levels.
In conclusion, this study identified a locus on Chr 2 associated with variation in both plasma CysC concentration and the Cst3 gene expression and a correlation between transcript levels of Cst3 and plasma CysC. In silico sequence analysis highlighted a tremendous genetic variation contained in the DO population at this locus which potentially may affect transcript and protein levels, but a putative causal variant remained to be determined. Network analysis identified potentially novel inflammatory pathway associated with CysC concentration. Future DO mouse investigations are needed to explore the causal relationship between CysC, kidney inflammation and filtration function and their implications for CKD progression and adverse cardiovascular disease outcomes.
Acknowledgments
This research was supported in part by U42OD010924 (FPMV), R01HL128572 (BJB), P30DK056350 (BJB), a pilot grant from the Nutrition Research Institute (BJB), NIDDK K23 DK0099442 (BR), R03 DK114502 (BR), R01HL133169 (HA), R01ES021801 (HA), R01ES025786 (HA), and P01ES022845 (HA), and U.S. EPA Grant RD83544101 (HA). We are grateful to Dr. Gary A. Churchill, The Jackson Laboratory Nathan Shock Center for providing us the DO-RNASeq data (NIH grant P30 AG038070) available at (GEO: GSE121330). We appreciate Michael Vernon and the UNC Functional Genomics Core for help with the microarray studies. This work was supported by USDA/ARS/Western Human Nutrition Research Center project funds (2032-51000-022-00D). We also acknowledge Nikhil Joshi from the University of California, Davis Bioinformatics Core for his help on the analysis and database management.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.10307972.
Communicating editor: D.-J. De Koning
Literature Cited
- Akerblom A., Eriksson N., Wallentin L., Siegbahn A., Barratt B.J. et al. , 2014. Polymorphism of the cystatin C gene in patients with acute coronary syndromes: Results from the PLATelet inhibition and patient Outcomes study. Am Heart J 168: 96–102 e102. [DOI] [PubMed] [Google Scholar]
- Aylor D. L., Valdar W., Foulds-Mathes W., Buus R. J., Verdugo R. A. et al. , 2011. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 21: 1213–1222. 10.1101/gr.111310.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., and Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Bennett B. J., de Aguiar Vallim T. Q., Wang Z., Shih D. M., Meng Y. et al. , 2013. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 17: 49–60. 10.1016/j.cmet.2012.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B. J., Farber C. R., Orozco L., Kang H. M., Ghazalpour A. et al. , 2010. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 20: 281–290. 10.1101/gr.099234.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Gatti D. M., Simecek P., Furlotte N. A., Prins P. et al. , 2019. R/qtl2: software for mapping quantitative trait loci with high-dimensional data and multiparent populations. Genetics 211: 495–502. 10.1534/genetics.118.301595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z. et al. , 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., and Kendziorski C., 2007. A statistical framework for expression quantitative trait loci mapping. Genetics 177: 761–771. 10.1534/genetics.107.071407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., and Chan A. P., 2015. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31: 2745–2747. 10.1093/bioinformatics/btv195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., and Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. O., Carpenter C. C., Ayers C. R., Holman J. E., and Bahn R. C., 1961. Evidence for secretion of an aldosterone-stimulating hormone by the kidney. J. Clin. Invest. 40: 684–696. 10.1172/JCI104301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didion J. P., Morgan A. P., Clayshulte A. M., McMullan R. C., Yadgary L. et al. , 2015. A multi-megabase copy number gain causes maternal transmission ratio distortion on mouse chromosome 2. PLoS Genet. 11: e1004850 10.1371/journal.pgen.1004850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donadio C., Lucchesi A., Ardini M., and Giordani R., 2001. Cystatin C, β2-microglobulin, and retinol-binding protein as indicators of glomerular filtration rate: comparison with plasma creatinine. J. Pharm. Biomed. Anal. 24: 835–842. 10.1016/S0731-7085(00)00550-1 [DOI] [PubMed] [Google Scholar]
- El-Sukkari D., Wilson N. S., Hakansson K., Steptoe R. J., Grubb A. et al. , 2003. The protease inhibitor cystatin C is differentially expressed among dendritic cell populations, but does not control antigen presentation. J. Immunol. 171: 5003–5011. 10.4049/jimmunol.171.10.5003 [DOI] [PubMed] [Google Scholar]
- Ferguson T. W., Komenda P., and Tangri N., 2015. Cystatin C as a biomarker for estimating glomerular filtration rate. Curr. Opin. Nephrol. Hypertens. 24: 295–300. 10.1097/MNH.0000000000000115 [DOI] [PubMed] [Google Scholar]
- Filler G., Bokenkamp A., Hofmann W., Le Bricon T., Martinez-Bru C. et al. , 2005. Cystatin C as a marker of GFR–history, indications, and future research. Clin. Biochem. 38: 1–8. 10.1016/j.clinbiochem.2004.09.025 [DOI] [PubMed] [Google Scholar]
- Finco D. R., 1997. Kidney function, pp. 441–484 in Clinical Biochemistry of Domestic Animals. Elsevier, Amsterdam. [Google Scholar]
- Fraser D., and Kodicek E., 1973. Regulation of 25-hydroxycholecalciferol-1-hydroxylase activity in kidney by parathyroid hormone. Nat. New Biol. 241: 163–166. 10.1038/newbio241163a0 [DOI] [PubMed] [Google Scholar]
- French J. E., Gatti D. M., Morgan D. L., Kissling G. E., Shockley K. R. et al. , 2015. Diversity Outbred Mice Identify Population-Based Exposure Thresholds and Genetic Factors that Influence Benzene-Induced Genotoxicity. Environ. Health Perspect. 123: 237–245. 10.1289/ehp.1408202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearing L. J., Cumming H. E., Chapman R., Finkel A. M., Woodhouse I. B. et al. , 2019. CiiiDER: A tool for predicting and analysing transcription factor binding sites. PLoS One 14: e0215495 10.1371/journal.pone.0215495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmoinen A., Kouri T., Wirta O., Lehtimäki T., Rantalaiho V. et al. , 1999. Evaluation of plasma cystatin C as a marker for glomerular filtration rate in patients with. Clin. Nephrol. 52: 363–373. [PubMed] [Google Scholar]
- Huang D.W., Sherman B. T., and Lempick R. A., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Hwang S. J., Yang Q., Meigs J. B., Pearce E. N., and Fox C. S., 2007. A genome-wide association for kidney function and endocrine-related traits in the NHLBI’s Framingham Heart Study. BMC Med. Genet. 8: S10 10.1186/1471-2350-8-S1-S10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inker L. A., and Okparavero A., 2011. Cystatin C as a marker of glomerular filtration rate: prospects and limitations. Curr. Opin. Nephrol. Hypertens. 20: 631–639. 10.1097/MNH.0b013e32834b8850 [DOI] [PubMed] [Google Scholar]
- Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K. et al. , 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294. 10.1038/nature10413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller B. J., Martini S., Sedor J. R., and Kretzler M., 2012. A systems view of genetics in chronic kidney disease. Kidney Int. 81: 14–21. 10.1038/ki.2011.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Fornes O., Stigliani A., Gheorghe M., Castro-Mondragon J. A. et al. , 2018. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46: D260–D266. 10.1093/nar/gkx1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köttgen A., Glazer N. L., Dehghan A., Hwang S. J., Katz R. et al. , 2009. Multiple loci associated with indices of renal function and chronic kidney disease. Nat. Genet. 41: 712–717. 10.1038/ng.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köttgen A., Pattaro C., Boger C. A., Fuchsberger C., Olden M. et al. , 2010. New loci associated with kidney function and chronic kidney disease. Nat. Genet. 42: 376–384. 10.1038/ng.568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyhse-Andersen J., Schmidt C., Nordin G., Andersson B., Nilsson-Ehle P. et al. , 1994. Serum cystatin C, determined by a rapid, automated particle-enhanced turbidimetric method, is a better marker than serum creatinine for glomerular filtration rate. Clin. Chem. 40: 1921–1926. 10.1093/clinchem/40.10.1921 [DOI] [PubMed] [Google Scholar]
- Langfelder P., and Horvath S., 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Zhang B., and Horvath S., 2007. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24: 719–720. 10.1093/bioinformatics/btm563 [DOI] [PubMed] [Google Scholar]
- Lemann J., Lennon E., Piering W., Prien E., and Ricanati E., 1970. Evidence that glucose ingestion inhibits net renal tubular reabsorption of calcium and magnesium in man. J. Lab. Clin. Med. 75: 578–585. [PubMed] [Google Scholar]
- Narayanan S., and Appleton H. D., 1980. Creatinine: a review. Clin. Chem. 26: 1119–1126. 10.1093/clinchem/26.8.1119 [DOI] [PubMed] [Google Scholar]
- Newman D. J., Thakkar H., Edwards R. G., Wilkie M., White T. et al. , 1995. Serum cystatin C measured by automated immunoassay: a more sensitive marker of changes in GFR than serum creatinine. Kidney Int. 47: 312–318. 10.1038/ki.1995.40 [DOI] [PubMed] [Google Scholar]
- O’Connor A., Quizon P. M., Albright J. E., Lin F. T., and Bennett B. J., 2014. Responsiveness of cardiometabolic-related microbiota to diet is influenced by host genetics. Mamm. Genome 25: 583–599. 10.1007/s00335-014-9540-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddoze C., Morange S., Portugal H., Berland Y., and Dussol B., 2001. Cystatin C is not more sensitive than creatinine for detecting early renal impairment in patients with diabetes. Am. J. Kidney Dis. 38: 310–316. 10.1053/ajkd.2001.26096 [DOI] [PubMed] [Google Scholar]
- Pattaro C., Teumer A., Gorski M., Chu A. Y., Li M. et al. , 2016. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat. Commun. 7: 10023 10.1038/ncomms10023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plebani M., Amico R. D., Mussap M., Montini G., Ruzzante N. et al. , 1998. Is serum cystatin C a sensitive marker of glomerular filtration rate (GFR)? A preliminary study on renal transplant patients. Ren. Fail. 20: 303–309. 10.3109/08860229809045115 [DOI] [PubMed] [Google Scholar]
- Pruim R. J., Welch R. P., Sanna S., Teslovich T. M., Chines P. S. et al. , 2010. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26: 2336–2337. 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core, R., T., 2019 R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Austria, 2019. URL http://www.R-project.org.
- Robinson G. K., 1991. That BLUP is a good thing: the estimation of random effects. Stat. Sci. 6: 15–32. 10.1214/ss/1177011926 [DOI] [Google Scholar]
- Schreiber G., 2017. The molecular basis for differential type I interferon signaling. J. Biol. Chem. 292: 7285–7294. 10.1074/jbc.R116.774562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secombes C. J., and Zou J., 2017. Evolution of Interferons and Interferon Receptors. Front. Immunol. 8: 209 10.3389/fimmu.2017.00209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T. et al. , 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood T. L., Gatti D. M., Quizon P., Weinstock G. M., Jung K. C. et al. , 2014. High-resolution genetic mapping in the diversity outbred mouse population identifies Apobec1 as a candidate gene for atherosclerosis. G3 (Bethesda) 4: 2353–2363 (Bethesda) 10.1534/g3.114.014704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner J. F., Leparc G., Hildebrandt T., Klein H., Thomas L. et al. , 2017. An RNA-Seq atlas of gene expression in mouse and rat normal tissues. Sci. Data 4: 170185 10.1038/sdata.2017.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L. A., Coresh J., Greene T., and Levey A. S., 2006. Assessing kidney function—measured and estimated glomerular filtration rate. N. Engl. J. Med. 354: 2473–2483. 10.1056/NEJMra054415 [DOI] [PubMed] [Google Scholar]
- Suhre K., Arnold M., Bhagwat A. M., Cotton R. J., Engelke R. et al. , 2017. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 8: 14357 10.1038/ncomms14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson K. L., Gatti D. M., Valdar W., Welsh C. E., Cheng R. et al. , 2012. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics 190: 437–447. 10.1534/genetics.111.132597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T., Thotakura P., Tanaka T. S., Ko M. S., and Ozato K., 2005. Identification of target genes and a unique cis element regulated by IRF-8 in developing macrophages. Blood 106: 1938–1947. 10.1182/blood-2005-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M., Hufton A., Heinig M., O’keeffe S., El Masri N. et al. , 2011. Transcription factor binding predictions using TRAP for the analysis of ChIP-seq data and regulatory SNPs. Nat. Protoc. 6: 1860–1869. 10.1038/nprot.2011.409 [DOI] [PubMed] [Google Scholar]
- Tyler A. L., Ji B., Gatti D. M., Munger S. C., Churchill G. A. et al. , 2017. Epistatic Networks Jointly Influence Phenotypes Related to Metabolic Disease and Gene Expression in Diversity Outbred Mice. Genetics 206: 621–639. 10.1534/genetics.116.198051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh C. E., Miller D. R., Manly K. F., Wang J., McMillan L. et al. , 2012. Status and access to the Collaborative Cross population. Mamm. Genome 23: 706–712. 10.1007/s00335-012-9410-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Macleod I., and Su A. I., 2013. BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 41: D561–D565. 10.1093/nar/gks1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B., Wong K., Agam A., Goodson M., Keane T. M. et al. , 2011. Sequence-based characterization of structural variation in the mouse genome. Nature 477: 326–329. 10.1038/nature10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York M. R., Nagai T., Mangini A. J., Lemaire R., van Seventer J. M. et al. , 2007. A macrophage marker, siglec-1, is increased on circulating monocytes in patients with systemic sclerosis and induced by type i interferons and toll-like receptor agonists. Arthritis Rheum. 56: 1010–1020. 10.1002/art.22382 [DOI] [PubMed] [Google Scholar]
- Yue F., Cheng Y., Breschi A., Vierstra J., Wu W. et al. , 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364. 10.1038/nature13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi M., and Xu Y., 2018. Involvement of cystatin C in immunity and apoptosis. Immunol. Lett. 196: 80–90. 10.1016/j.imlet.2018.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Microarray gene expression data are available at GEO with the accession ID GSE122061. Genotypes of DO mice are accessible through UNC’s MMRRC website (https://www.med.unc.edu/mmrrc/genotypes/). Further information is available from the corresponding authors if required. This study was approved by the IACUC at UNC, Chapel Hill (IACUC Protocol Number 13-103). DO mouse kidney RNASeq (DO-RNASeq) data are available at the NCBI’s Gene Expression Omnibus (GEO) with the accession ID GSE121330 and also at Dr. Churchill’s laboratory web site (https://churchilllab.jax.org/qtlviewer/JAC/DOKidney). The C57Bl/6J mouse RNASeq (B6-RNASeq) data (Söllner et al. 2017) is available at https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA375882 and EBI under the Array Express ID: E-MTAB-6081; https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6081/. Supplemental material available at figshare: https://doi.org/10.25387/g3.10307972.