Abstract
Pleiotropy, the concept that a single gene controls multiple distinct traits, is prevalent in most organisms and has broad implications for medicine and agriculture. The identification of the molecular mechanisms underlying pleiotropy has the power to reveal previously unknown biological connections between seemingly unrelated traits. Additionally, the discovery of pleiotropic genes increases our understanding of both genetic and phenotypic complexity by characterizing novel gene functions. Quantitative trait locus (QTL) mapping has been used to identify several pleiotropic regions in many organisms. However, gene knockout studies are needed to eliminate the possibility of tightly linked, non-pleiotropic loci. Here, we use a panel of 296 recombinant inbred advanced intercross lines of Caenorhabditis elegans and a high-throughput fitness assay to identify a single large-effect QTL on the center of chromosome V associated with variation in responses to eight chemotherapeutics. We validate this QTL with near-isogenic lines and pair genome-wide gene expression data with drug response traits to perform mediation analysis, leading to the identification of a pleiotropic candidate gene, scb-1, for some of the eight chemotherapeutics. Using deletion strains created by genome editing, we show that scb-1, which was previously implicated in response to bleomycin, also underlies responses to other double-strand DNA break-inducing chemotherapeutics. This finding provides new evidence for the role of scb-1 in the nematode drug response and highlights the power of mediation analysis to identify causal genes.
Keywords: C. elegans, QTL, pleiotropy, chemotherapeutics, mediation
Pleiotropy refers to the well established notion that a single gene or genetic variant affects multiple distinct traits (Paaby and Rockman 2013), and the discovery of pleiotropic genes can provide meaningful insights into the molecular mechanisms of these traits (Tyler et al. 2016). It has become easier to identify pleiotropic genes with the advent of reverse-genetic screens and quantitative trait locus (QTL) mapping (Paaby and Rockman 2013). For example, pleiotropic QTL for diverse growth and fitness traits have been identified in organisms such as yeast (Cubillos et al. 2011; Jerison et al. 2017; Peltier et al. 2019), Arabidopsis (McKay et al. 2003; El‐Assal et al. 2004; Fusari et al. 2017), Drosophila (Brown et al. 2013; McGuigan et al. 2014), and mice (White et al. 2013; Leamy et al. 2014; Lin et al. 2014). These studies have led to important questions in the field of evolutionary genetics regarding the ‘cost of complexity’ (Fisher; Orr 2000), as a single mutation might be beneficial for one trait and harmful for another (Wagner and Zhang 2011). Furthermore, human association studies have identified pleiotropic variants associated with different diseases (Sivakumaran et al. 2011; Pavlides et al. 2016; Chesmore et al. 2018), highlighting both the ubiquity and importance of certain immune-related genes and oncogenes across unrelated diseases (Borrello et al. 2008; Gratten and Visscher 2016). Perhaps the strongest evidence of pleiotropy exists for molecular phenotypes. Large-scale expression QTL (eQTL) mapping studies have identified single regulatory variants that control expression and likely the functions of hundreds of genes at once, opening a window into the mechanisms for how traits are controlled (Keurentjes et al. 2007; Breitling et al. 2008; Rockman et al. 2010; Albert and Kruglyak 2015; Hasin-Brumshtein et al. 2016; Albert et al. 2018).
The nematode Caenorhabditis elegans provides a tractable metazoan model to identify and study pleiotropic QTL (Paaby and Rockman 2013). A large panel of recombinant inbred advanced intercross lines (RIAILs) derived from two divergent strains, N2 and CB4856 (Rockman and Kruglyak 2009; Andersen et al. 2015), has been leveraged in several linkage mapping analyses (Li et al. 2006; Gutteling et al. 2007b, 2007a; Kammenga et al. 2007; Seidel et al. 2008, 2011; Reddy et al. 2009; McGrath et al. 2009; Doroszuk et al. 2009; Viñuela et al. 2010; Rockman et al. 2010; Bendesky et al. 2011, 2012; Bendesky and Bargmann 2011; Rodriguez et al. 2012; Andersen et al. 2014; Glater et al. 2014; Snoek et al. 2014; Balla et al. 2015; Schmid et al. 2015; Singh et al. 2016; Zdraljevic et al. 2017, 2019; Lee et al. 2017; Zamanian et al. 2018a; Evans et al. 2018; Brady et al. 2019). Quantitative genetic analysis using these panels and a high-throughput phenotyping assay (Andersen et al. 2015) has facilitated the discovery of numerous QTL (Zamanian et al. 2018b), several quantitative trait genes (QTG) (Brady et al. 2019) and quantitative trait nucleotides (QTN) (Zdraljevic et al. 2017, 2019) underlying fitness-related traits in the nematode. Additionally, three pleiotropic genomic regions were recently found to influence responses to a diverse group of toxins (Evans et al. 2018). However, overlapping genomic regions might not represent true pleiotropy but could demonstrate the co-existence of tightly linked loci (Paaby and Rockman 2013).
Here, we use linkage mapping to identify a single overlapping QTL on chromosome V that influences the responses to eight chemotherapeutic compounds. We show that these drug-response QTL also overlap with an expression QTL hotspot that contains the gene scb-1, previously implicated in bleomycin response (Brady et al. 2019). Although the exact mechanism of scb-1 is yet unknown, it is hypothesized to act in response to stress (Riedel et al. 2013) and has weak homology to a viral hydrolase (Kelley et al. 2015; Zhang et al. 2018). Together, these data suggest that the importance of scb-1 expression might extend beyond bleomycin response. We validated the QTL using near-isogenic lines (NILs) and performed mediation analysis to predict that scb-1 expression explains the observed QTL for four of the eight drugs. Finally, we directly tested the effect of scb-1 loss of function on chemotherapeutic responses. We discovered that expression of scb-1 underlies differential responses to several chemotherapeutics that cause double-strand DNA breaks, not just bleomycin. This discovery of pleiotropy helps to further define the role of scb-1 by expanding its known functions and provides insights into the molecular mechanisms underlying the nematode drug response.
Materials and Methods
Strains
Animals were grown at 20° on modified nematode growth media (NGMA) containing 1% agar and 0.7% agarose to prevent burrowing and fed OP50 (Ghosh et al. 2012). The two parental strains, the canonical laboratory strain, N2, and the wild isolate from Hawaii, CB4856, were used to generate all recombinant lines. 208 recombinant inbred advanced intercross lines (RIAILs) generated previously by Rockman et al. (Rockman and Kruglyak 2009) (set 1 RIAILs) were phenotyped for expression QTL mapping (detailed below). A second set of 296 RIAILs generated previously by Andersen et al. (Andersen et al. 2015) (set 2 RIAILs) was used more extensively for drug phenotyping and linkage mapping. The set 2 RIAILs were used for linkage mapping because they addressed the three main disadvantages of the set 1 RIAILs detailed previously (Andersen et al. 2015), namely a structured population, the laboratory-derived variant in npr-1 (Sterken et al. 2015), and the peel-1 zeel-1 incompatibility (Seidel et al. 2008, 2011). Because of these limitations, the set 2 RIAILs were generated using QX1430 and CB4856. QX1430 is from the N2 strain background but contains a transposon insertion in peel-1 and the CB4856 npr-1 allele introgressed on chromosome X (Andersen et al. 2015). Near-isogenic lines (NILs) were generated by backcrossing a selected RIAIL for several generations to the parent strain (N2 or CB4856) (Brady et al. 2019) using PCR amplicons for insertion-deletion (indels) variants to track the introgressed region. NILs were whole-genome sequenced to verify introgressions were only in the targeted genomic intervals. CRISPR-Cas9-mediated deletions of scb-1 were described previously (Brady et al. 2019). All strains are available upon request or from the C. elegans Natural Diversity Resource (Cook et al. 2017). Primers used to generate ECA1114 can be found in the Supplemental Information.
High-throughput fitness assays for linkage mapping
For dose responses and RIAIL phenotyping, we used a high-throughput fitness assay described previously (Andersen et al. 2015). In summary, populations of each strain were passaged and amplified on NGMA plates for four generations. In the fifth generation, gravid adults were bleach-synchronized and 25-50 embryos from each strain were aliquoted into 96-well microtiter plates at a final volume of 50 µL K medium (Boyd et al. 2012). The following day, arrested L1s were fed HB101 bacterial lysate (Pennsylvania State University Shared Fermentation Facility, State College, PA; (García-González et al. 2017)) at a final concentration of 5 mg/mL in K medium and were grown to the L4 larval stage for 48 hr at 20° with constant shaking. Three L4 larvae were sorted into new 96-well microtiter plates containing 10 mg/mL HB101 bacterial lysate, 50 µM kanamycin, and either diluent (1% water or 1% DMSO) or drug dissolved in the diluent using a large-particle flow cytometer (COPAS BIOSORT, Union Biometrica; Holliston, MA). Sorted animals were grown for 96 hr at 20° with constant shaking. The next generation of animals and the parents were treated with sodium azide (50 mM in 1X M9) to straighten their bodies for more accurate length measurements. Animal length (median.TOF), optical density integrated over animal length (median.EXT), and brood size (norm.n) were quantified for each well using the COPAS BIOSORT. Nematodes get longer (animal length) and become thicker and more complex (optical density) over developmental time. Because animal length and optical density are highly correlated, we calculated a fourth trait (median.norm.EXT) that normalizes optical density by animal length (median.EXT / median.TOF). Phenotypic measurements collected by the BIOSORT were processed and analyzed using the R package easysorter (Shimko and Andersen 2014) as described previously (Brady et al. 2019). Differences among strains within the control conditions were controlled by subtracting the mean control-condition value from each drug-condition replicate for each strain using a linear model (drug_phenotype ∼ mean_control_phenotype). In this way, we are addressing only the differences among strains that were caused by the drug condition and the variance in the control condition does not affect the variance in the drug condition. For plotting purposes, these residual values (negative and positive residuals) were normalized from 0 to 1 where 0 refers to the smallest residual phenotypic value in that condition and 1 refers to the largest.
Dose-response assays
Four genetically divergent strains (N2, CB4856, JU258, and DL238) were treated with increasing concentrations of each of the eight drugs using the high-throughput fitness assay described above. The dose of each drug that provided a reproducible drug-specific effect that maximizes between-strain variation while minimizing within-strain variation across the four traits was selected for the linkage mapping experiments. The chosen concentrations are as follows: 100 µM amsacrine hydrochloride (Fisher Scientific, #A277720MG) in DMSO, 50 µM bleomycin sulfate (Fisher, #50-148-546) in water, 2 µM bortezomib (VWR, #AAJ60378-MA) in DMSO, 250 µM carmustine (Sigma, #1096724-75MG) in DMSO, 500 µM cisplatin (Sigma, #479306-1G) in K media, 500 µM etoposide (Sigma, #E1383) in DMSO, 500 µM puromycin dihydrochloride (VWR, #62111-170) in water, and 150 µM silver nitrate (Sigma-Aldrich, #S6506-5G) in water.
Linkage mapping
Set 1 and set 2 RIAILs were phenotyped in each of the eight drugs and controls using the high-throughput fitness assay described above. Linkage mapping was performed on each of the drug and gene expression traits using the R package linkagemapping (https://github.com/AndersenLab/linkagemapping) as described previously (Brady et al. 2019). The cross object derived from the whole-genome sequencing of the RIAILs containing 13,003 SNPs was loaded using the function load_cross_obj(“N2xCB4856cross_full”). The RIAIL phenotypes were merged into the cross object using the merge_pheno function with the argument set = 1 for expression QTL mapping and set = 2 for drug phenotype mapping. A forward search (fsearch function) adapted from the R/qtl package (Broman et al. 2003) was used to calculate the logarithm of the odds (LOD) scores for each genetic marker and each trait as -n(ln(1-R2)/2ln(10)) where R is the Pearson correlation coefficient between the RIAIL genotypes at the marker and trait phenotypes (Bloom et al. 2013). A 5% genome-wide error rate was calculated by permuting the RIAIL phenotypes 1000 times. The marker with the highest LOD score above the significance threshold was selected as the QTL then integrated into the model as a cofactor and mapping was repeated iteratively until no further significant QTL were identified. Finally, the annotate_lods function was used to calculate the effect size of each QTL and determine 95% confidence intervals defined by a 1.5 LOD drop from the peak marker using the argument cutoff = proximal.
Modified high-throughput fitness assay for NIL validation
NILs and scb-1 deletion strains were tested using a modified version of the high-throughput fitness assay detailed above. Strains were propagated for two generations, bleach-synchronized in three independent replicates, and titered at a concentration of 25-50 embryos per well of a 96-well microtiter plate. The following day, arrested L1s were fed HB101 bacterial lysate at a final concentration of 5 mg/mL with either diluent or drug. After 48 hr of growth at 20° with constant shaking, nematodes were treated with sodium azide (5 mM in water) prior to analysis of animal length and optical density using the COPAS BIOSORT. As only one generation of growth is observed, brood size was not calculated. A single trait (median.EXT) was chosen to represent animal growth generally, as the trait is defined by integrating optical density over length. Because of the modified timing of the drug delivery, lower drug concentrations were needed to recapitulate the previously observed phenotypic effect. The selected doses are as follows: 12.5 µM amsacrine in DMSO, 12.5 µM bleomycin in water, 2 µM bortezomib in DMSO, 250 µM carmustine in DMSO, 125 µM cisplatin in K media, 62.5 µM etoposide in DMSO, 300 µM puromycin in water, and 100 µM silver in water.
Expression QTL analysis
Microarray data for gene expression using 15,888 probes were previously collected from synchronized young adult populations of 208 set 1 RIAILs (Rockman et al. 2010). Expression data were corrected for dye effects and probes with variants were removed (Andersen et al. 2014). Linkage mapping was performed as described above for the remaining 14,107 probes, and a significance threshold was determined using a permutation-based False Discovery Rate (FDR). FDR was calculated as the ratio of the average number of genes across 10 permutations expected by chance to show a maximum LOD score greater than a particular threshold vs. the number of genes observed in the real data with a maximum LOD score greater than that threshold. We calculated the FDR for a range of thresholds from 2 to 10, with increasing steps of 0.01, and set the threshold so that the calculated FDR was less than 5%.
Local eQTL were defined as linkages whose peak LOD scores were within 1 Mb of the starting position of the probe (Rockman et al. 2010). eQTL hotspots were identified by dividing the genome into 5 cM bins and counting the number of distant eQTL that mapped to each bin. Significance was determined as bins with more eQTL than the Bonferroni-corrected 99th percentile of a Poisson distribution with a mean of 3.91 QTL (total QTL / total bins) (Brem et al. 2002; Rockman et al. 2010; Evans et al. 2018). We identified nine eQTL hotspots (II, IVL, IVC, IVR, VL, VC, VR, XL, and XC). To avoid false positives, we increased the LOD threshold for QTL to be counted in the hotspot analysis to a LOD > 5 or LOD > 6. At a LOD > 5, six of the nine eQTL hotspots persist (IVL, IVR, VC, VR, XL, and XC), and at a LOD > 6, three persist (IVL, IVR, and XL). We further looked for spurious eQTL hotspots in ten permuted datasets. At a LOD > 5, we identified four hotspots, and at a LOD > 6, we identified one hotspot.
Mediation analysis
A total of 159 set 1 RIAILs were phenotyped in each of the eight drugs and controls using the standard high-throughput fitness assay described above. Mediation scores were calculated with bootstrapping using the mediate function from the mediation R package (version 4.4.7) (Tingley et al. 2014) for each QTL identified from the set 1 RIAILs and all 49 probes (including scb-1, A_12_P104350) that mapped to the chromosome V eQTL hotspot using the following models:
| (1) |
| (2) |
The output of the mediate function can be summarized as follows: the total effect of genotype on phenotype, ignoring expression (tau.coef); the direct effect of genotype on phenotype, while holding expression constant (z0); the estimated effect of expression on phenotype (d0); the proportion of the total effect that can be explained by expression data (n0). This mediation proportion (n0) can be a useful way to identify the impact of gene expression on the overall phenotype. However, cases of inconsistent mediation (where the direct effect is either smaller than or in the opposite direction of the indirect mediation effect) render this measurement uninterpretable with values greater than one or less than zero (MacKinnon et al. 2007). We used the estimated effect of expression on phenotype (z0) as the final mediation score for this reason. Because the effect size can be positive or negative, mediation scores range from -1 to 1, and we evaluated the absolute value of mediation estimates to compare across traits. Each mediation estimate generated a p-value, indicating confidence in the estimate, derived from bootstrapping with 1000 simulations. The likelihood of scb-1 mediating a given QTL effect was calculated relative to the other 48 probes with an eQTL in the region (Table S1). Traits in which scb-1 was at or above the 90th percentile of this distribution were prioritized over other traits.
Statistical analysis
Broad-sense heritability was calculated from the dose response phenotypes using the lmer function in the lme4 R package (Bates et al. 2014) with the formula phenotype ∼1 + (1|strain) for each dose. For the NIL and scb-1 deletion high-throughput assays, statistical significance of phenotypic differences between each strain pair was tested using the TukeyHSD function (R Core Team 2017) on an ANOVA model with the formula phenotype ∼ strain to assess differences between strains in the control-regressed phenotype data.
Data availability
File S1 contains the results of the original dose response high-throughput fitness assay. File S2 contains the residual phenotypic values for all 159 set 1 RIAILs, 296 set 2 RIAILs, and parent strains (N2 and CB4856) in response to all eight chemotherapeutics. File S3 contains the linkage mapping results for the set 2 RIAILs for all 32 drug-response traits tested in the high-throughput fitness assay. File S4 is a VCF that reports the genotype of ECA1114. File S5 contains the simplified genotype of all the NILs in the study. File S6 contains the raw pruned phenotypes for the NIL dose response with the modified high-throughput fitness assay. File S7 contains the pairwise statistical significance for all strains and high-throughput assays. File S8 contains the microarray expression data for 14,107 probes from Rockman et al. 2010. File S9 contains the linkage mapping results for the expression data obtained with the set 1 RIAILs. File S10 contains the location of each eQTL hotspot and a list of genes with an eQTL in each hotspot. File S11 contains the linkage mapping results from the set 1 RIAILs for all 32 drug-response traits tested in the high-throughput fitness assay. File S12 contains the pairwise mediation estimates for all 32 drug-response traits and all 49 probes. File S13 contains the raw pruned phenotypes for the scb-1 deletion modified high-throughput fitness assay. The datasets and code for generating figures can be found at https://github.com/AndersenLab/scb1_mediation_manuscript. Supplemental material available at figshare: https://doi.org/10.25387/g3.12250091.
Results
Natural variation on chromosome V underlies differences in responses to several chemotherapeutics
We measured C. elegans development and chemotherapeutic sensitivity as a function of animal length (median.TOF), optical density (median.EXT), and brood size (norm.n) with a high-throughput assay developed using the COPAS BIOSORT (see Methods) (Andersen et al. 2015; Zdraljevic et al. 2017, 2019; Evans et al. 2018; Brady et al. 2019). Animal length and optical density (animal thickness and composition) are both measures of nematode development, and brood size is a measure of nematode reproduction (Andersen et al. 2015). Because optical density is calculated as a function of length and these traits are related, a fourth trait that captures the optical density normalized by length (median.norm.EXT) was also included. We exposed four genetically divergent strains (N2, CB4856, JU258, and DL238) to increasing doses of eight chemotherapeutic compounds. Five of these compounds (bleomycin, carmustine, etoposide, amsacrine, and cisplatin) are known to cause double-strand DNA breaks and/or inhibit DNA synthesis (Dorr 1992; Ketron et al. 2012; Dasari and Tchounwou 2014; Montecucco et al. 2015; Nikolova et al. 2017). The remaining three compounds either inhibit protein synthesis (puromycin) (Azzam and Algranati 1973), inhibit the proteosome and subsequent protein degradation (bortezomib) (Piperdi et al. 2011), or cause cellular toxicity in a poorly defined way (silver nitrate) (Kaplan et al. 2016) (Table 1). In the presence of each drug, nematodes were generally shorter, less optically dense, and produced smaller broods compared to non-treated nematodes (Figure S1, File S1). We observed significant phenotypic variation among strains and identified a substantial heritable genetic component for most traits (average H2 = 0.52 +/− 0.53).
Table 1. Main mechanism of action for eight chemotherapeutic drugs.
| Drug | Drug class | Mechanism of action |
|---|---|---|
| Amsacrine | Topoisomerase inhibitors | DNA intercalation and inhibition of topoisomerase II, causing DNA double-strand breaks, cell cycle arrest, and cell death |
| Bleomycin | Antitumor antibiotic | Forms complexes with iron that reduce molecular oxygen to form free radicals which in turn cause DNA single- and double-strand breaks |
| Bortezomib | Proteosome inhibitors | Reversibly inhibits the 26S proteosome and inhibits nuclear factor (NF)-kappaB causing disruption of various cell signaling pathways, cell cycle arrest, and cell death. |
| Carmustine | Alkylating agents | Alkylates and cross-links DNA causing cell cycle arrest and cell death |
| Cisplatin | Alkylating agents | Alkylates and cross-links DNA causing cell cycle arrest and cell death |
| Etoposide | Topoisomerase inhibitors | Binds to and inhibits topoisomerase II causing an increase of DNA single- and double-strand breaks, cell cycle arrest, and cell death |
| Puromycin | Aminonucleoside antibiotic | Acts as analog of 3′ terminal end of aminoacyl-tRNA and incorporates itself into growing polypeptide chain causing premature termination and inhibition of protein synthesis |
| Silver | NA | Multi-faceted induction of apoptosis |
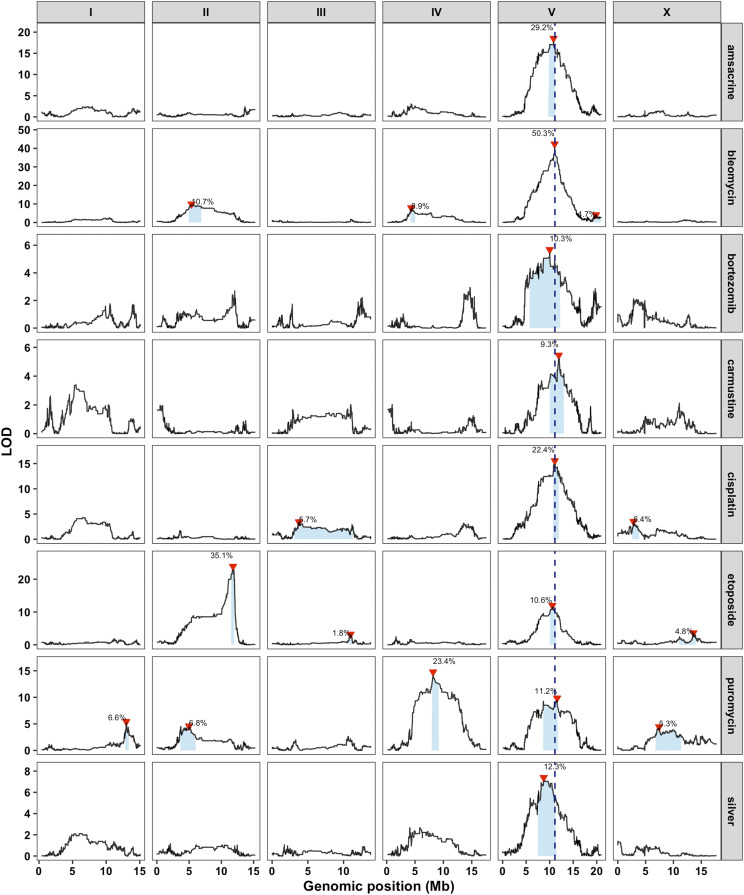
We exposed a panel of 296 RIAILs (set 2 RIAILs, see Methods) to all eight chemotherapeutics at a selected concentration that both maximizes among-strain and minimizes within-strain phenotypic variation (File S2). Linkage mapping for all four traits for each of the eight drugs (total of 32 traits) identified 79 QTL from 31 traits (one trait had no significant QTL), several of which have been identified previously (Zdraljevic et al. 2017; Evans et al. 2018; Brady et al. 2019) (File S3, Figure S2). Strikingly, a QTL on the center of chromosome V was linked to variation in responses to all eight compounds (Figure 1). In all cases, the CB4856 allele on chromosome V is associated with greater resistance to the drug than the N2 allele (Figure S2, File S2, File S3). We previously identified this genomic interval as a QTL hotspot, defined as a region heavily enriched for toxin-response QTL (Evans et al. 2018). Because several of the chemotherapeutics share a similar mechanism of action, a single pleiotropic gene might underlie the observed QTL for multiple drugs.
Figure 1.
A large-effect QTL on the center of chromosome V underlies responses to several chemotherapeutics. Linkage mapping results with the set 2 RIAILs for a representative trait for each drug are shown (amascrine: median.norm.EXT, bleomycin median.TOF:, bortezomib: median.TOF, carmustine: norm.n, cisplatin: median.TOF, etoposide: norm.n, puromycin: median.TOF, silver: median.TOF). Genomic position (x-axis) is plotted against the logarithm of the odds (LOD) score (y-axis) for 13,003 genomic markers. Each significant QTL is indicated by a red triangle at the peak marker, and a blue rectangle shows the 95% confidence interval around the peak marker. The percentage of the total variance in the RIAIL population that can be explained by each QTL is shown above the QTL. The dotted vertical line represents the genomic position of scb-1.
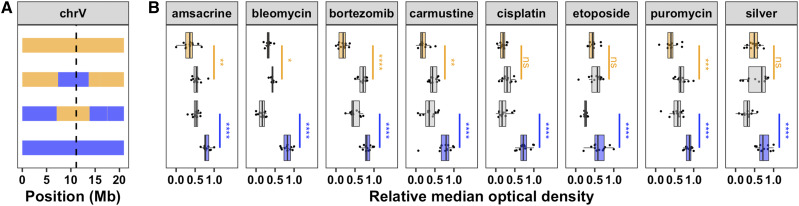
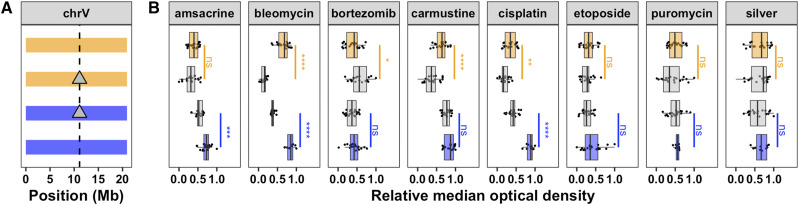
In order to isolate and validate the effect of this QTL, we constructed reciprocal near-isogenic lines (NILs) by introgressing a genomic region on chromosome V from the resistant CB4856 strain into the sensitive N2 background and vice versa (File S4, File S5). We used a modified high-throughput assay (see Methods) to measure length and optical density of a population of animals grown in the presence of the drug for 48 hr (from larval stages L1 to L4). In this modified assay, less drug was required to observe the same phenotypic effect as before (Figure S3, File S6). Statistical significance was calculated in a pairwise manner for each strain (see Methods; File S7). For all eight chemotherapeutics tested, the strain with the N2 introgression was significantly more sensitive than its CB4856 parent and/or the strain with the CB4856 introgression was significantly more resistant than its N2 parent (Figure 2, File S6, File S7). These data confirm that one or more genetic variant(s) within this region on chromosome V cause increased drug sensitivities in N2.
Figure 2.
Near-isogenic lines validate the chromosome V QTL. (A) NIL genotypes on chromosome V are shown, colored orange (N2) and blue (CB4856). From top to bottom, strains are N2, ECA232, ECA1114, and CB4856. The dotted vertical line represents the location of scb-1. (B) NIL phenotypes in eight chemotherapeutics (12.5 µM amsacrine, 12.5 µM bleomycin, 2 µM bortezomib, 250 µM carmustine, 125 µM cisplatin, 62.5 µM etoposide, 300 µM puromycin, and 100 µM silver) are plotted as Tukey box plots with strain (y-axis) by relative median optical density (median.EXT, x-axis). Statistical significance was calculated for each strain pair (File S7). Significance of each strain compared to its parental strain (ECA232 to N2 and ECA1114 to CB4856) is shown above each strain pair and colored by the parent strain against which it was tested (ns = non-significant (p-value > 0.05); *, **, ***, and **** = significant (p-value < 0.05, 0.01, 0.001, or 0.0001, respectively).
Expression QTL mapping identifies a hotspot on the center of chromosome V
Genetic variation can affect a phenotype most commonly through either modifications of the amino acid sequence that lead to altered protein function (or even loss of function) or changes in the expression level of the protein. In the latter case, measuring the intermediate phenotype (gene expression) can be useful in elucidating the mechanism by which genetic variation causes phenotypic variation. More specifically, cases with overlap between expression QTL (eQTL) and drug-response QTL suggest that a common variant could underlie both traits and provide evidence in support of causality for the candidate gene in question (Huang et al. 2015; Sasaki et al. 2018).
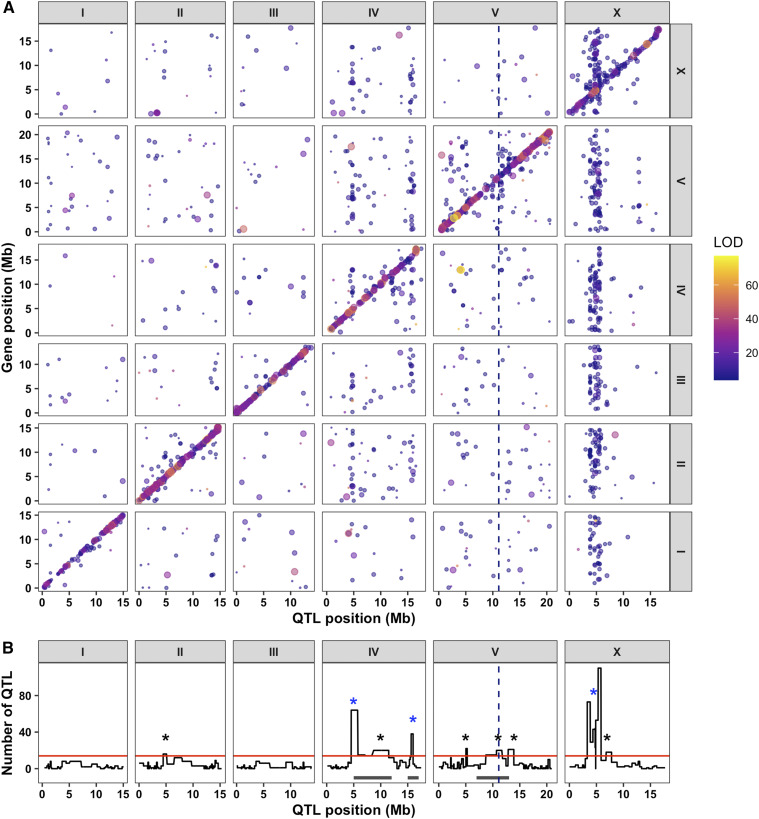
To identify such cases of overlap between expression QTL and the drug-response QTL on chromosome V, we need genome-wide expression data for the RIAILs. In a previous study, expression of 15,888 probes were measured using microarrays for a panel of 208 RIAILs (set 1 RIAILs, see Methods) between N2 and CB4856 (Rockman and Kruglyak 2009) (File S8). This study used the variation in gene expression as a phenotypic trait to identify eQTL using linkage mapping with 1,455 variants (Rockman et al. 2010). They identified 2,309 eQTL and three regions with significantly clustered distant eQTL (eQTL hotspots), suggesting that these regions are pleiotropic, wherein one or more variant(s) are affecting expression of multiple genes. We recently performed whole-genome sequencing for these strains and identified 13,003 informative variants (Brady et al. 2019). Using this new set of variants, we re-analyzed the eQTL mapping by performing linkage mapping analysis for a selected 14,107 of the 15,888 probes without genetic variation in CB4856 (Andersen et al. 2014). We identified 2,540 eQTL associated with variation in expression of 2,196 genes (Figure 3A, File S9). These eQTL have relatively large effect sizes compared to the drug-response QTL. On average, each eQTL explains 23% of the phenotypic variance in gene expression among the RIAIL population. Half of the eQTL (50.2%; 1,276) mapped within 1 Mb of the gene whose expression was measured and were classified as local (see Methods) (Albert and Kruglyak 2015). The other half (49.7%; 1,264) were found distant from their respective gene, and over a third (37%; 940) were found on different chromosomes entirely. In general, eQTL effect sizes increased, max LOD scores decreased, and confidence intervals became smaller compared to the original mapping results (File S9). These differences and the additional eQTL observed between this analysis and the original are possibly caused by the integration of new genetic markers. Additionally, we found several differences in methodology between the current approach and the previous one. These differences include ignoring the population structure of the set 1 RIAILs, adding the forward-search marker-regression linkage mapping, and altering the linkage mapping method itself (see Methods, (Rockman et al. 2010)).
Figure 3.
Expression QTL mapping identifies several hotspots. (A) The genomic locations of the eQTL peaks derived from linkage mapping using the set 1 RIAILs (x-axis) are plotted against the genomic locations of the probe (y-axis). The size of the point corresponds to the effect size of the QTL. eQTL are colored by the LOD score, increasing from purple to pink to yellow. The diagonal band represents local eQTL, and vertical bands represent eQTL hotspots. (B) Quantification of eQTL hotspots identified by overlapping distant eQTL. The number of distant eQTL (y-axis) in each 5 cM bin across the genome (x-axis) is shown. Bins above the red line are significant and marked with an asterisk. The bins with the blue asterisks are most significant and have been identified in a previous analysis. The dotted vertical line represents the genomic position of scb-1. Gray rectangles below the plot represent locations of the drug-response QTL hotspots previously identified.
We noticed regions of the genome that appeared to be enriched for distant eQTL. We identified eQTL hotspots in a similar manner to the previous study (see Methods) and found a total of nine eQTL hotspots (Figure 3B, File S10). Six of the nine eQTL hotspots withstood more stringent filtering methods (see Methods), and three (left of chromosome IV, right of chromosome IV, and left of chromosome X) were the most significant. These three hotspots also overlap with the most significant eQTL hotspots in the previous study (Rockman et al. 2010). Notably, three of the eQTL hotspots (center of chromosome IV, right of chromosome IV, and center of chromosome V) overlap with the previously identified drug-response QTL hotspots on chromosomes IV and V (Figure 3B) (Evans et al. 2018). The overlap of these eQTL and drug-response QTL hotspots could provide strong candidate genes whose expression underlies the differences in nematode drug responses generally. Expression of one gene of interest, scb-1, has been previously implicated in response to bleomycin (Brady et al. 2019) and resides within the eQTL hotspot region on the center of chromosome V (File S10, Table S1). Although the exact mechanism of how scb-1 responds to bleomycin is unknown, its putative hydrolase activity (Kelley et al. 2015; Zhang et al. 2018; Brady et al. 2019) suggests that it might act to break down chemotherapeutic compounds. These data suggest that variation in expression of scb-1 and responses to these eight chemotherapeutics (including bleomycin) could be mechanistically linked through the metabolic breakdown of chemotherapeutic drugs.
Mediation analysis suggests that scb-1 expression plays a role in responses to several chemotherapeutics
Mediation analysis seeks to explain the relationship between an independent and a dependent variable by including a third intermediary variable. We can use mediation analysis to understand how certain genetic variants on chromosome V (independent variable) affect drug responses (dependent variable) through differential gene expression of genes within the eQTL hotspot (mediator variable) (Figure S4). We measured brood size, animal length, and optical density in response to all eight chemotherapeutics in the set 1 RIAILs and performed linkage mapping for these traits (File S2, File S11, Figure S5). Although the power to detect QTL with these strains is lower than in our original mapping set (set 2 RIAILs; see Methods) (Andersen et al. 2015), we still identified overlapping QTL on chromosome V for half of the drugs tested (bleomycin, cisplatin, silver, and amsacrine) (Figure S5, File S11).
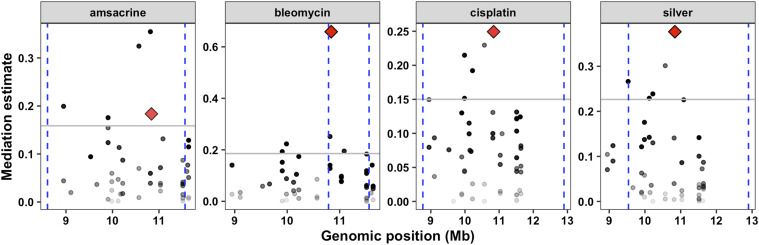
We calculated the effect that variation in expression of scb-1 had on drug-response traits compared to the other 48 genes with an eQTL in the chromosome V eQTL hotspot using mediation analysis (see Methods). We estimated that the effect of expression variation of scb-1 on bleomycin response is 0.65 (set 1 RIAILs, Figure 4, Figure S6, File S12). Moreover, out of all 49 genes with an eQTL in the region (Table S1), scb-1 was a clear mediation score outlier. All of the remaining three chemotherapeutics with a QTL on the center of chromosome V in the set 1 RIAIL mapping showed moderate evidence of scb-1 mediation, with scb-1 falling well above the 90th percentile of mediation estimates for all genes with an eQTL in this region (Figure 4, Figure S6, File S12). We further performed this mediation analysis on all 32 drug-response traits, regardless of the presence of a QTL in the set 1 RIAIL panel (Figure S6, File S12). Etoposide and puromycin also showed evidence of scb-1 mediation. This in silico approach indicated that expression of scb-1 might be an intermediate link between genetic variation on chromosome V and responses to several of the chemotherapeutic drugs tested.
Figure 4.
Mediation analysis for the eQTL hotspot on the center of chromosome V. Mediation estimates calculated as the indirect effect that differences in expression of each gene plays in the overall phenotype (y-axis) are plotted against the genomic position of the eQTL (x-axis) on chromosome V for 49 probes (including scb-1 (red diamond)) that map to the chromosome V eQTL hotspot (set 1 RIAILs). A representative trait for each drug from the set 1 linkage mapping analysis are shown: amascrine (median.EXT), bleomycin (median.EXT), cisplatin (median.TOF), and silver (median.norm.EXT). The 90th percentile of the distribution of mediation estimates for each trait are represented by the horizontal gray lines. The confidence intervals for the QTL (set 1 RIAILs) are shown with the vertical blue dotted lines. The confidence of the estimate increases (p-value decreases) as points become less transparent.
Expression of scb-1 affects responses to several chemotherapeutics that cause double-strand DNA breaks
To empirically test whether scb-1 expression modulates the chromosome V QTL effect for each drug, we used the modified high-throughput assay (see Methods) to expose two independently derived strains with scb-1 deletions (Brady et al. 2019) to each chemotherapeutic (Figure 5, Figure S7, File S13). Statistical significance was calculated in a pairwise manner for each strain (see Methods; File S7). Because RIAILs with the CB4856 allele on chromosome V express higher levels of scb-1 than RIAILs with the N2 allele (File S8, File S9), we expect that loss of scb-1 will cause increased drug sensitivity in the CB4856 background but might not have an effect in the N2 background. We validated the results of Brady et al. and confirmed that ablated scb-1 expression causes hyper-sensitivity to bleomycin in both N2 and CB4856 (Figure 5, Figure S7, Figure S8 File S7, File S13). We also observed similarly increased sensitivity to cisplatin with scb-1 deletions in both backgrounds. Furthermore, removing scb-1 shows moderately increased sensitivity in the CB4856 background for amsacrine and in the N2 background for carmustine. The remaining four drugs did not show a significantly different phenotype between the parental N2 and CB4856 strains, suggesting these traits might be less reproducible or that scb-1 variation does not underlie these drug differences. Overall, these results provide evidence for the pleiotropic effect of scb-1, which appears to mediate responses to at least four of the eight chemotherapeutic drugs.
Figure 5.
Testing the role of scb-1 in drug responses. (A) Strain genotypes on chromosome V are shown, colored orange (N2) and blue (CB4856). From top to bottom, strains are N2, ECA1132, ECA1134, and CB4856. Deletion of scb-1 is indicated by a gray triangle. The dotted vertical line represents the location of scb-1. (B) Phenotypes of strains in eight chemotherapeutics (12.5 µM amsacrine, 12.5 µM bleomycin, 2 µM bortezomib, 250 µM carmustine, 125 µM cisplatin, 62.5 µM etoposide, 300 µM puromycin, and 100 µM silver) are plotted as Tukey box plots with strain (y-axis) by relative median optical density (median.EXT, x-axis). Statistical significance was calculated for each strain pair (File S7). Significance of each deletion strain compared to its parental strain (ECA1132 to N2 and ECA1134 to CB4856) is shown above each strain pair and colored by the parent strain against which it was tested (ns = non-significant (p-value > 0.05); *, **, ***, and **** = significant (p-value < 0.05, 0.01, 0.001, or 0.0001, respectively).
Discussion
In this study, we identified overlapping QTL on the center of chromosome V that influence sensitivities to eight chemotherapeutic drugs. Because five of these drugs are known to cause double-strand DNA breaks, we hypothesized that this genomic region might be pleiotropic – a single shared genetic variant affects the responses to each drug. Because this variant might affect drug responses by regulating gene expression levels, we looked for the co-existence of drug-response QTL and expression QTL on chromosome V. We identified 2,540 eQTL and nine eQTL hotspots, including a region on the center of chromosome V. We calculated the mediation effect of all 49 genes with an eQTL that maps to this hotspot region and identified scb-1 as a candidate gene whose expression influences the responses to several chemotherapeutics. We used CRISPR-Cas9-mediated scb-1 deletion strains to empirically validate the role of scb-1 in the chemotherapeutic response. In addition to bleomycin (Brady et al. 2019), we discovered that responses to cisplatin, amsacrine, and carmustine are affected by scb-1 expression. In this study, we found evidence that several overlapping QTL are representative of pleiotropy at the gene level and further elucidated the function of scb-1 as a potential response to double-strand DNA break stress.
Mediation of drug-response QTL using gene expression to identify causal genes
Mediation analysis often suggests potential candidate genes that underlie different traits (Huang et al. 2015; Sasaki et al. 2018) and could be applied to drug responses. Using C. elegans strains and high-throughput assays, we can rapidly validate hypotheses generated by mediation analysis. Three of the eight chemotherapeutics that map to an overlapping drug-response QTL and were potentially mediated by scb-1 were validated using targeted deletion strains.
Although mediation analysis provided moderate evidence that expression of scb-1 could also play a role in sensitivity to etoposide and puromycin, we observed no experimental evidence of this relationship. Additionally, we have evidence that expression of scb-1 might mediate response to carmustine. However, mediation analysis disagrees. The discrepancy between the mediation analysis and validation of causality using targeted deletion strains could be partially explained by one of several possibilities. First, different traits were measured in each experiment. The mediation analysis used traits measured over 96 hr of growth in drug conditions spanning two generations, but the causality test used traits measured over 48 hr of growth in drug conditions within one generation. Second, the precision of our mediation estimates was likely reduced by the poor quality drug traits for the set 1 RIAIL panel (Andersen et al. 2015). Indeed, bortezomib, carmustine, etoposide, and puromycin did not map to the center of chromosome V using the set 1 RIAILs (Figure S5). Expression data for the set 2 RIAIL panel would likely generate more accurate mediation estimates, especially if data were collected using RNA sequencing to avoid the inherent reference bias of microarray data (Zhao et al. 2014). Third, our mediation analysis was performed using expression data collected in control conditions and phenotype data collected in drug conditions. This analysis will only provide evidence of mediation if the baseline expression differences affect an individual’s response to the drug. Collecting expression data from drug-treated nematodes could help us learn more about how gene expression varies in response to treatment with the chemotherapeutic. Finally, as we only directly assessed the complete loss of scb-1 in drug sensitivity, it is still possible that reduction of function (or change in function) caused by a single nucleotide variant or other structural variation in CB4856 could validate the role of scb-1 in responses to these drugs.
This study demonstrates the power of pairing genome-wide linkage mapping of gene expression and drug response data using simple colocalization as well as more complex mediation analysis techniques. In addition to providing a resource for candidate gene prioritization within a QTL interval, mediation analysis can help to identify the mechanism by which genetic variation causes phenotypic differences. This type of approach could be even more powerful using genome-wide association (GWA) where the lower linkage disequilibrium between variants also has smaller confidence intervals in some genomic regions. Smaller intervals have fewer spurious overlapping eQTL, which could help to narrow the list of candidate genes. Although mediation analysis is only effective if a change in expression is observed and might not be useful for identifying effects from protein-coding variation, many current studies show that the majority of genetic variants associated with complex traits lie in regulatory regions (Hindorff et al. 2009). Whole-genome expression analysis could provide the missing link to the identification of causal genes underlying complex traits.
New evidence for the pleiotropic function of scb-1
We identified eight chemotherapeutics with a QTL that mapped to a genomic region defined as a QTL hotspot on the center of chromosome V (Evans et al. 2018). Multiple genes in close proximity, each regulating an aspect of cellular growth and fitness, might underlie each QTL independently. Alternatively, genetic variation within a single gene might regulate responses to multiple (or all) of the eight drugs tested, particularly if the gene is involved in drug transport or metabolism or if the drug mechanisms of action were shared (e.g., repair of double-strand DNA breaks). Expression of scb-1, a gene previously implicated in modulating responses to bleomycin, was found to reduce sensitivity to half of the drugs tested. This pleiotropic effect of scb-1 provides new evidence for the function of the gene and possible molecular mechanisms underlying nematode drug responses. It is hypothesized that SCB-1 might function as a hydrolase that metabolizes compounds like bleomycin (Brady et al. 2019) or somehow plays a role in the nematode stress response (Riedel et al. 2013). Both hypotheses are consistent with our data, explaining why nematodes with low expression of scb-1 are highly sensitive to the compound. Furthermore, all four of these chemotherapeutics, whose responses are mediated by expression of scb-1, are known to cause double-strand DNA breaks (Dorr 1992; Ketron et al. 2012; Dasari and Tchounwou 2014; Nikolova et al. 2017). Although the results for bortezomib, puromycin, and silver were inconclusive, we found no clear evidence that expression of scb-1 dictates their responses. Together, these data suggest a potential role for scb-1 specifically in response to stress induced by double-strand DNA breaks. However, the lack of sensitivity in etoposide, which also causes double-strand DNA breaks (Montecucco et al. 2015), indicates that this response might be more complex.
The exact variant that causes the differential expression of scb-1 is still unknown. Importantly, scb-1 lies within an eQTL hotspot region where it is hypothesized that genetic variation at a single locus might regulate expression of the 49 genes with an eQTL in this region. It is possible that the same causal variant that regulates expression of scb-1 could also underlie the QTL for the remaining four chemotherapeutics through differential expression of other genes. For example, mediation analysis for both bortezomib and etoposide indicated that expression variation of a dehydrogenase (D1054.8) may underlie their differential responses (File S12). Alternatively, the causal variants underlying these drug-response QTL might be distinct but physically linked in the genome. This result would suggest a cluster of genes essential for the nematode drug response. Overall, our study highlights the power of using mediation analysis to connect gene expression to organismal traits and describes a novel function for the pleiotropic gene scb-1.
Acknowledgments
We would like to thank members of the Andersen Lab for helpful comments on the manuscript. This work was supported by an American Cancer Society Research Scholar Grant (127313-RSG-15-135-01-DD) to E.C.A. Additionally, K.S.E. received support from the NSF-Simons Center for Quantitative Biology at Northwestern University (awards Simons Foundation/SFARI 597491-RWC and the National Science Foundation 1764421).
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.12250091.
Communicating editor: M. Rockman
Literature Cited
- Albert F. W., Bloom J. S., Siegel J., Day L., and Kruglyak L., 2018. Genetics of trans-regulatory variation in gene expression. eLife 7 10.7554/eLife.35471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert F. W., and Kruglyak L., 2015. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 16: 197–212. 10.1038/nrg3891 [DOI] [PubMed] [Google Scholar]
- Andersen E. C., Bloom J. S., Gerke J. P., and Kruglyak L., 2014. A variant in the neuropeptide receptor npr-1 is a major determinant of Caenorhabditis elegans growth and physiology. PLoS Genet. 10: e1004156 10.1371/journal.pgen.1004156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen E. C., Shimko T. C., Crissman J. R., Ghosh R., Bloom J. S. et al. , 2015. A Powerful New Quantitative Genetics Platform, Combining Caenorhabditis elegans High-Throughput Fitness Assays with a Large Collection of Recombinant Strains. G3 (Bethesda) 5: 911–920. 10.1534/g3.115.017178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam M. E., and Algranati I. D., 1973. Mechanism of puromycin action: fate of ribosomes after release of nascent protein chains from polysomes. Proc. Natl. Acad. Sci. USA 70: 3866–3869. 10.1073/pnas.70.12.3866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla K. M., Andersen E. C., Kruglyak L., and Troemel E. R., 2015. A wild C. elegans strain has enhanced epithelial immunity to a natural microsporidian parasite. PLoS Pathog. 11: e1004583 10.1371/journal.ppat.1004583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Mächler M., Bolker B., and Walker S., 2014. Fitting Linear Mixed-Effects Models using lme4. arXiv. 10.18637/jss.v067.i01 [DOI]
- Bendesky A., and Bargmann C. I., 2011. Genetic contributions to behavioural diversity at the gene-environment interface. Nat. Rev. Genet. 12: 809–820. 10.1038/nrg3065 [DOI] [PubMed] [Google Scholar]
- Bendesky A., Pitts J., Rockman M. V., Chen W. C., Tan M.-W. et al. , 2012. Long-range regulatory polymorphisms affecting a GABA receptor constitute a quantitative trait locus (QTL) for social behavior in Caenorhabditis elegans. PLoS Genet. 8: e1003157 10.1371/journal.pgen.1003157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendesky A., Tsunozaki M., Rockman M. V., Kruglyak L., and Bargmann C. I., 2011. Catecholamine receptor polymorphisms affect decision-making in C. elegans. Nature 472: 313–318. 10.1038/nature09821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. S., Ehrenreich I. M., Loo W. T., Lite T.-L. V., and Kruglyak L., 2013. Finding the sources of missing heritability in a yeast cross. Nature 494: 234–237. 10.1038/nature11867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrello M. G., Degl’Innocenti D., and Pierotti M. A., 2008. Inflammation and cancer: the oncogene-driven connection. Cancer Lett. 267: 262–270. 10.1016/j.canlet.2008.03.060 [DOI] [PubMed] [Google Scholar]
- Boyd W. A., Smith M. V., and Freedman J. H., 2012. Caenorhabditis elegans as a model in developmental toxicology. Methods Mol. Biol. 889: 15–24. 10.1007/978-1-61779-867-2_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S. C., Zdraljevic S., Bisaga K. W., Tanny R. E., Cook D. E. et al. , 2019. A Novel Gene Underlies Bleomycin-Response Variation in Caenorhabditis elegans. Genetics 212: 1453–1468. 10.1534/genetics.119.302286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R., Li Y., Tesson B. M., Fu J., Wu C. et al. , 2008. Genetical genomics: spotlight on QTL hotspots. PLoS Genet. 4: e1000232 10.1371/journal.pgen.1000232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R. B., Yvert G., Clinton R., and Kruglyak L., 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755. 10.1126/science.1069516 [DOI] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., and Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. 10.1093/bioinformatics/btg112 [DOI] [PubMed] [Google Scholar]
- Brown E. B., Layne J. E., Zhu C., Jegga A. G., and Rollmann S. M., 2013. Genome-wide association mapping of natural variation in odour-guided behaviour in Drosophila. Genes Brain Behav. 12: 503–515. 10.1111/gbb.12048 [DOI] [PubMed] [Google Scholar]
- Chesmore K., Bartlett J., and Williams S. M., 2018. The ubiquity of pleiotropy in human disease. Hum. Genet. 137: 39–44. 10.1007/s00439-017-1854-z [DOI] [PubMed] [Google Scholar]
- Cook D. E., Zdraljevic S., Roberts J. P., and Andersen E. C., 2017. CeNDR, the Caenorhabditis elegans natural diversity resource. Nucleic Acids Res. 45: D650–D657. 10.1093/nar/gkw893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos F. A., Billi E., Zörgö E., Parts L., Fargier P. et al. , 2011. Assessing the complex architecture of polygenic traits in diverged yeast populations. Mol. Ecol. 20: 1401–1413. 10.1111/j.1365-294X.2011.05005.x [DOI] [PubMed] [Google Scholar]
- Dasari S., and Tchounwou P. B., 2014. Cisplatin in cancer therapy: molecular mechanisms of action. Eur. J. Pharmacol. 740: 364–378. 10.1016/j.ejphar.2014.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroszuk A., Snoek L. B., Fradin E., Riksen J., and Kammenga J., 2009. A genome-wide library of CB4856/N2 introgression lines of Caenorhabditis elegans. Nucleic Acids Res. 37: e110 10.1093/nar/gkp528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr R. T., 1992. Bleomycin pharmacology: mechanism of action and resistance, and clinical pharmacokinetics. Semin. Oncol. 19: 3–8. [PubMed] [Google Scholar]
- El‐Assal S. E. D., Alonso‐Blanco C., Hanhart C. J., and Koornneef M., 2004. Pleiotropic Effects of the Arabidopsis Cryptochrome 2 Allelic Variation Underlie Fruit Trait‐Related QTL. Plant Biol. 6: 370–374. 10.1055/s-2004-820890 [DOI] [PubMed] [Google Scholar]
- Evans K. S., Brady S. C., Bloom J. S., Tanny R. E., Cook D. E. et al. , 2018. Shared Genomic Regions Underlie Natural Variation in Diverse Toxin Responses. Genetics 210: 1509–1525. 10.1534/genetics.118.301311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A.1930 The genetical theory of natural selection. The Clarendon Press.
- Fusari C. M., Kooke R., Lauxmann M. A., Annunziata M. G., Enke B. et al. , 2017. Genome-Wide Association Mapping Reveals That Specific and Pleiotropic Regulatory Mechanisms Fine-Tune Central Metabolism and Growth in Arabidopsis. Plant Cell 29: 2349–2373. 10.1105/tpc.17.00232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-González A. P., Ritter A. D., Shrestha S., Andersen E. C., Yilmaz L. S. et al. , 2017. Bacterial Metabolism Affects the C. elegans Response to Cancer Chemotherapeutics. Cell 169: 431–441.e8. 10.1016/j.cell.2017.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R., Andersen E. C., Shapiro J. A., Gerke J. P., and Kruglyak L., 2012. Natural variation in a chloride channel subunit confers avermectin resistance in C. elegans. Science 335: 574–578. 10.1126/science.1214318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glater E. E., Rockman M. V., and Bargmann C. I., 2014. Multigenic natural variation underlies Caenorhabditis elegans olfactory preference for the bacterial pathogen Serratia marcescens. G3 (Bethesda) 4: 265–276. 10.1534/g3.113.008649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratten J., and Visscher P. M., 2016. Genetic pleiotropy in complex traits and diseases: implications for genomic medicine. Genome Med. 8: 78 10.1186/s13073-016-0332-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteling E. W., Doroszuk A., Riksen J. A. G., Prokop Z., Reszka J. et al. , 2007a Environmental influence on the genetic correlations between life-history traits in Caenorhabditis elegans. Heredity 98: 206–213. 10.1038/sj.hdy.6800929 [DOI] [PubMed] [Google Scholar]
- Gutteling E. W., Riksen J. A. G., Bakker J., and Kammenga J. E., 2007b Mapping phenotypic plasticity and genotype-environment interactions affecting life-history traits in Caenorhabditis elegans. Heredity 98: 28–37. 10.1038/sj.hdy.6800894 [DOI] [PubMed] [Google Scholar]
- Hasin-Brumshtein Y., Khan A. H., Hormozdiari F., Pan C., Parks B. W. et al. , 2016. Hypothalamic transcriptomes of 99 mouse strains reveal trans eQTL hotspots, splicing QTLs and novel non-coding genes. eLife 5 10.7554/eLife.15614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff L. A., Sethupathy P., Junkins H. A., Ramos E. M., Mehta J. P. et al. , 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 106: 9362–9367. 10.1073/pnas.0903103106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-T., Liang L., Moffatt M. F., Cookson W. O. C. M., and Lin X., 2015. iGWAS: Integrative Genome-Wide Association Studies of Genetic and Genomic Data for Disease Susceptibility Using Mediation Analysis. Genet. Epidemiol. 39: 347–356. 10.1002/gepi.21905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerison E. R., Kryazhimskiy S., Mitchell J. K., Bloom J. S., Kruglyak L. et al. , 2017. Genetic variation in adaptability and pleiotropy in budding yeast. eLife 6 10.7554/eLife.27167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammenga J. E., Doroszuk A., Riksen J. A. G., Hazendonk E., Spiridon L. et al. , 2007. A Caenorhabditis elegans wild type defies the temperature-size rule owing to a single nucleotide polymorphism in tra-3. PLoS Genet. 3: e34 10.1371/journal.pgen.0030034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan A., Akalin Ciftci G., and Kutlu H. M., 2016. Cytotoxic, anti-proliferative and apoptotic effects of silver nitrate against H-ras transformed 5RP7. Cytotechnology 68: 1727–1735. 10.1007/s10616-015-9922-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. E., 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10: 845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketron A. C., Denny W. A., Graves D. E., and Osheroff N., 2012. Amsacrine as a topoisomerase II poison: importance of drug-DNA interactions. Biochemistry 51: 1730–1739. 10.1021/bi201159b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes J. J. B., Fu J., Terpstra I. R., Garcia J. M., van den Ackerveken G. et al. , 2007. Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc. Natl. Acad. Sci. USA 104: 1708–1713. 10.1073/pnas.0610429104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamy L. J., Elo K., Nielsen M. K., Thorn S. R., Valdar W. et al. , 2014. Quantitative trait loci for energy balance traits in an advanced intercross line derived from mice divergently selected for heat loss. PeerJ 2: e392 10.7717/peerj.392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Yang H., Kim J., Brady S., Zdraljevic S. et al. , 2017. The genetic basis of natural variation in a phoretic behavior. Nat. Commun. 8: 273 10.1038/s41467-017-00386-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Alvarez O. A., Gutteling E. W., Tijsterman M., Fu J. et al. , 2006. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2: e222 10.1371/journal.pgen.0020222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. H. S., Chen J., Ziman B., Marshall S., Maizel J. et al. , 2014. Endostatin and kidney fibrosis in aging: a case for antagonistic pleiotropy? Am. J. Physiol. Heart Circ. Physiol. 306: H1692–H1699. 10.1152/ajpheart.00064.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon D. P., Fairchild A. J., and Fritz M. S., 2007. Mediation analysis. Annu. Rev. Psychol. 58: 593–614. 10.1146/annurev.psych.58.110405.085542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath P. T., Rockman M. V., Zimmer M., Jang H., Macosko E. Z. et al. , 2009. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron 61: 692–699. 10.1016/j.neuron.2009.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuigan K., Collet J. M., McGraw E. A., Ye Y. H., Allen S. L. et al. , 2014. The nature and extent of mutational pleiotropy in gene expression of male Drosophila serrata. Genetics 196: 911–921. 10.1534/genetics.114.161232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay J. K., Richards J. H., and Mitchell-Olds T., 2003. Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Mol. Ecol. 12: 1137–1151. 10.1046/j.1365-294X.2003.01833.x [DOI] [PubMed] [Google Scholar]
- Montecucco A., Zanetta F., and Biamonti G., 2015. Molecular mechanisms of etoposide. EXCLI J. 14: 95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova T., Roos W. P., Krämer O. H., Strik H. M., and Kaina B., 2017. Chloroethylating nitrosoureas in cancer therapy: DNA damage, repair and cell death signaling. Biochim. Biophys. Acta 1868: 29–39. [DOI] [PubMed] [Google Scholar]
- Orr H. A., 2000. Adaptation and the cost of complexity. Evolution 54: 13–20. 10.1111/j.0014-3820.2000.tb00002.x [DOI] [PubMed] [Google Scholar]
- Paaby A. B., and Rockman M. V., 2013. The many faces of pleiotropy. Trends Genet. 29: 66–73. 10.1016/j.tig.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlides J. M. W., Zhu Z., Gratten J., McRae A. F., Wray N. R. et al. , 2016. Predicting gene targets from integrative analyses of summary data from GWAS and eQTL studies for 28 human complex traits. Genome Med. 8: 84 10.1186/s13073-016-0338-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier E., Friedrich A., Schacherer J., and Marullo P., 2019. Quantitative Trait Nucleotides Impacting the Technological Performances of Industrial Saccharomyces cerevisiae Strains. Front. Genet. 10: 683 10.3389/fgene.2019.00683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperdi B., Ling Y.-H., Liebes L., Muggia F., and Perez-Soler R., 2011. Bortezomib: understanding the mechanism of action. Mol. Cancer Ther. 10: 2029–2030. 10.1158/1535-7163.MCT-11-0745 [DOI] [PubMed] [Google Scholar]
- R Core Team , 2017. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
- Reddy K. C., Andersen E. C., Kruglyak L., and Kim D. H., 2009. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science 323: 382–384. 10.1126/science.1166527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel C. G., Dowen R. H., Lourenco G. F., Kirienko N. V., Heimbucher T. et al. , 2013. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat. Cell Biol. 15: 491–501. 10.1038/ncb2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., and Kruglyak L., 2009. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 5: e1000419 10.1371/journal.pgen.1000419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., Skrovanek S. S., and Kruglyak L., 2010. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science 330: 372–376. 10.1126/science.1194208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M., Snoek L. B., Riksen J. A. G., Bevers R. P., and Kammenga J. E., 2012. Genetic variation for stress-response hormesis in C. elegans lifespan. Exp. Gerontol. 47: 581–587. 10.1016/j.exger.2012.05.005 [DOI] [PubMed] [Google Scholar]
- Sasaki E., Frommlet F., and Nordborg M., 2018. GWAS with Heterogeneous Data: Estimating the Fraction of Phenotypic Variation Mediated by Gene Expression Data. G3 (Bethesda) 8: 3059–3068. 10.1534/g3.118.200571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid T., Snoek L. B., Fröhli E., van der Bent M. L., Kammenga J. et al. , 2015. Systemic Regulation of RAS/MAPK Signaling by the Serotonin Metabolite 5-HIAA. PLoS Genet. 11: e1005236 10.1371/journal.pgen.1005236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., Ailion M., Li J., van Oudenaarden A., Rockman M. V. et al. , 2011. A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLoS Biol. 9: e1001115 10.1371/journal.pbio.1001115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., Rockman M. V., and Kruglyak L., 2008. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science 319: 589–594. 10.1126/science.1151107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimko T. C., and Andersen E. C., 2014. COPASutils: an R package for reading, processing, and visualizing data from COPAS large-particle flow cytometers. PLoS One 9: e111090 10.1371/journal.pone.0111090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K. D., Roschitzki B., Snoek L. B., Grossmann J., Zheng X. et al. , 2016. Natural Genetic Variation Influences Protein Abundances in C. elegans Developmental Signalling Pathways. PLoS One 11: e0149418 10.1371/journal.pone.0149418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumaran S., Agakov F., Theodoratou E., Prendergast J. G., Zgaga L. et al. , 2011. Abundant pleiotropy in human complex diseases and traits. Am. J. Hum. Genet. 89: 607–618. 10.1016/j.ajhg.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek L. B., Orbidans H. E., Stastna J. J., Aartse A., Rodriguez M. et al. , 2014. Widespread genomic incompatibilities in Caenorhabditis elegans. G3 (Bethesda) 4: 1813–1823. 10.1534/g3.114.013151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterken M. G., Snoek L. B., Kammenga J. E., and Andersen E. C., 2015. The laboratory domestication of Caenorhabditis elegans. Trends Genet. 31: 224–231. 10.1016/j.tig.2015.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingley D., Yamamoto T., Hirose K., Keele L., and Imai K., 2014. mediation: R Package for Causal Mediation Analysis. Journal of Statistical Software. Articles 59: 1–38. [Google Scholar]
- Tyler A. L., Crawford D. C., and Pendergrass S. A., 2016. The detection and characterization of pleiotropy: discovery, progress, and promise. Brief. Bioinform. 17: 13–22. 10.1093/bib/bbv050 [DOI] [PubMed] [Google Scholar]
- Viñuela A., Snoek L. B., Riksen J. A. G., and Kammenga J. E., 2010. Genome-wide gene expression regulation as a function of genotype and age in C. elegans. Genome Res. 20: 929–937. 10.1101/gr.102160.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G. P., and Zhang J., 2011. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat. Rev. Genet. 12: 204–213. 10.1038/nrg2949 [DOI] [PubMed] [Google Scholar]
- White J. K., Gerdin A.-K., Karp N. A., Ryder E., Buljan M. et al. , 2013. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154: 452–464. 10.1016/j.cell.2013.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian M., Cook D. E., Zdraljevic S., Brady S. C., Lee D. et al. , 2018a Discovery of genomic intervals that underlie nematode responses to benzimidazoles. PLoS Negl. Trop. Dis. 12: e0006368 10.1371/journal.pntd.0006368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian M., Cook D. E., Zdraljevic S., Brady S. C., Lee D. et al. , 2018b Discovery of genomic intervals that underlie nematode responses to benzimidazoles. PLoS Negl. Trop. Dis. 12: e0006368 10.1371/journal.pntd.0006368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdraljevic S., Fox B. W., Strand C., Panda O., Tenjo F. J. et al. , 2019. Natural variation in C. elegans arsenic toxicity is explained by differences in branched chain amino acid metabolism. eLife 8 10.7554/eLife.40260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdraljevic S., Strand C., Seidel H. S., Cook D. E., Doench J. G. et al. , 2017. Natural variation in a single amino acid substitution underlies physiological responses to topoisomerase II poisons. PLoS Genet. 13: e1006891 10.1371/journal.pgen.1006891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Li L., Yan L., Ming Z., Jia Z. et al. , 2018. Structural and Biochemical Characterization of Endoribonuclease Nsp15 Encoded by Middle East Respiratory Syndrome Coronavirus. J. Virol. 92 10.1128/JVI.00893-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Fung-Leung W.-P., Bittner A., Ngo K., and Liu X., 2014. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One 9: e78644 10.1371/journal.pone.0078644 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
File S1 contains the results of the original dose response high-throughput fitness assay. File S2 contains the residual phenotypic values for all 159 set 1 RIAILs, 296 set 2 RIAILs, and parent strains (N2 and CB4856) in response to all eight chemotherapeutics. File S3 contains the linkage mapping results for the set 2 RIAILs for all 32 drug-response traits tested in the high-throughput fitness assay. File S4 is a VCF that reports the genotype of ECA1114. File S5 contains the simplified genotype of all the NILs in the study. File S6 contains the raw pruned phenotypes for the NIL dose response with the modified high-throughput fitness assay. File S7 contains the pairwise statistical significance for all strains and high-throughput assays. File S8 contains the microarray expression data for 14,107 probes from Rockman et al. 2010. File S9 contains the linkage mapping results for the expression data obtained with the set 1 RIAILs. File S10 contains the location of each eQTL hotspot and a list of genes with an eQTL in each hotspot. File S11 contains the linkage mapping results from the set 1 RIAILs for all 32 drug-response traits tested in the high-throughput fitness assay. File S12 contains the pairwise mediation estimates for all 32 drug-response traits and all 49 probes. File S13 contains the raw pruned phenotypes for the scb-1 deletion modified high-throughput fitness assay. The datasets and code for generating figures can be found at https://github.com/AndersenLab/scb1_mediation_manuscript. Supplemental material available at figshare: https://doi.org/10.25387/g3.12250091.