Inherited arrhythmias may be caused by mutations in genes encoding cardiac ion channels and associated proteins (primary electrical disorders), and may occur in the setting of hereditary cardiomyopathies secondary to mutations in cardiac structural proteins.1 Together, they may contribute to up to 15–20% of all sudden cardiac deaths (SCDs).2 Progress in genetic, molecular, and (electro)physiological aspects of inherited arrhythmia disorders has enabled the identification of diagnostic and therapeutic strategies. However, insight into disease mechanisms and arrhythmia triggers is still limited, precluding the development of mechanism-driven therapies. In addition, inherited arrhythmia syndromes are often associated with reduced penetrance and variability in disease expressivity and severity.1 Hence, predicting who is most at risk for SCD remains difficult. In this Spotlight Issue, a series of reviews highlight the current state of the art for the various inherited arrhythmia disorders, review novel targets for risk stratification and therapy, and describe the remaining challenges and future perspectives.
In line with their role in action potential formation, mutations in ion channel genes are well-established causes of inherited arrhythmia syndromes associated with altered depolarization and/or repolarization.3 A wide range of molecularly diverse potassium channels are present in the myocardium, which together maintain the resting membrane potential and mediate action potential repolarization. Mutations in genes encoding these channels can impair their assembly, trafficking, and gating, leading to long QT syndrome types and 2 (LQTS1, LQTS2), short QT syndrome (SQTS), J wave syndromes, and atrial fibrillation. As discussed by Crotti et al.,4 mechanism-based therapies are largely missing for these disorders, despite extensive knowledge on their genetic and molecular basis. In the last decade, transgenic rabbit LQTS/SQTS models (which show more similarities to human cardiac repolarization than for instance mice) have provided valuable mechanistic insight into arrhythmia mechanisms, including the role of dispersion of repolarization, electromechanical remodelling, sympathetic activity, and hormones. Human induced pluripotent stem cell-derived cardiomyocyte (hiPSC-CM) and zebrafish models are furthermore enabling identification and testing of novel therapeutic strategies aimed at allele-specific RNA interference, modulation of ion currents, or rescue of trafficking-deficient channels. In addition to potassium channelopathies, mutations in the SCN5A gene encoding the cardiac sodium channel NaV1.5 also cause a broad spectrum of inherited arrhythmia disorders, including long QT syndrome type 3 (LQTS3), Brugada syndrome (BrS), cardiac conduction disease, atrial fibrillation, and sick sinus syndrome. In their review, Rivaud et al. discuss the multifunctionality of sodium channels and propose a novel classification of NaV1.5 (dys)function.5 Reduced peak sodium current or increased late sodium current (‘direct ionic’ effects) consequent to SCN5A mutations are well-established causes of pro-arrhythmic conduction slowing and repolarization disturbances, respectively. However, cardiac abnormalities are increasingly reported in SCN5A mutation carriers, including cardiac fibrosis, dilated cardiomyopathy, and arrhythmogenic (right ventricular) cardiomyopathy (ACM/ARVC). Such structural remodelling may occur consequent to alterations in intracellular sodium and calcium homeostasis (‘indirect ionic’ effects), or secondary to disrupted interactions of NaV1.5 with partner proteins within the macromolecular complex (‘non-ionic’ effects). Since these non-canonical actions of NaV1.5 may contribute significantly to arrhythmogenesis, they clearly warrant further exploration.
In addition to ion channelopathies, arrhythmias and SCD also occur in the setting of hereditary cardiomyopathies, including hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), arrhythmogenic cardiomyopathy (ACM), and left ventricular non-compaction cardiomyopathy (LVNC). As reviewed by Marian et al.,6 these cardiomyopathies are predominantly caused by mutations in genes encoding sarcomeric proteins (HCM), cytoskeletal proteins (DCM), and desmosomal proteins (ACM). Pro-arrhythmic mechanisms include cardiac hypertrophy and dilatation, myocardial fibrosis, ion channel and connexin remodelling, and intracellular calcium dysregulation. Interestingly, mutations in a number of genes encoding ion channels or transporters, including SCN5A, HCN4, PLN (phospholamban), and RYR2 (ryanodine receptor 2) have been associated with DCM, ACM, and/or LVNC. Conversely, mutations in the LMNA gene encoding lamin A/C lead to cardiac conduction defects and ventricular arrhythmias, and mutations in titin (TTN) to atrial fibrillation. Hence, inherited cardiomyopathies and cardiac arrhythmias are closely interrelated, and may have in part a shared genetic aetiology. This is further exemplified by ACM/ARVC, which is predominantly caused by mutations in desmosomal genes but which was recently also linked to mutations in SCN5A. The pathogenesis of ACM includes intercalated disc remodelling, cardiomyocyte loss, adiposis, and inflammation. As discussed by Van der Voorn et al.,7 novel anti-fibrotic and anti-inflammatory strategies are currently being investigated in available mouse, zebrafish, and hiPSC-CM ACM disease models. Various mechanisms may contribute to arrhythmias in ACM: cardiac structural alterations and fibrosis formation may set the stage for re-entrant based arrhythmias in advanced disease stages, whereas NaV1.5 remodelling and calcium dysregulation may occur in early disease stages and cause arrhythmias prior to overt cardiomyopathic changes. Novel strategies enabling early detection of pathological remodelling and hence risk stratification in ACM patients include (bio)markers of fibrosis and inflammation, desmosomal protein remodelling in buccal mucosa smears, circulating desmoglein-2 autoantibodies, and assessment of cardiac conduction delay through high resolution imaging and sodium channel blocker challenge. Abnormalities in sodium current and calcium homeostasis also contribute to arrhythmogenesis in the setting of HCM, as reviewed by Coppini et al.8 HCM is a major cause of SCD in young individuals (occurring during exercise or at rest), but the factors predisposing to ventricular arrhythmias remain incompletely known and hence risk stratification options are limited. While structural alterations may provide a pro-arrhythmic substrate, a clear association between e.g. cardiac fibrosis and arrhythmia risk is lacking in HCM patients. More recently, the use of mouse, rabbit, and hiPSC-CM models of HCM-related mutations have provided essential mechanistic insight into electrophysiological remodelling on the cardiomyocyte level. Decreased potassium currents, increased L-type calcium current and enhanced late sodium current have been shown to underlie action potential prolongation and EAD formation, attributed at least in part to a sustained CamKII activation. In addition, global dysregulation of intracellular calcium homeostasis occurs in HCM cardiomyocytes, secondary to alterations in L-type calcium current and sarcoplasmic reticulum and NCX function. Diastolic calcium levels are furthermore elevated consequent to altered sarcomere function and set the stage for DAD formation and triggered activity. Pharmacological late sodium current inhibition has been shown to improve calcium handling and reduce pro-arrhythmia in HCM cardiomyocytes, indicating a potential therapeutic strategy.
While genetic studies in the inherited arrhythmia field have traditionally focused on Mendelian disorders, it has become increasingly clear that the genetic basis for these disorders is often far more complex.1 As discussed above, mutations in one gene can lead to multiple disease phenotypes, and conversely one disease can be caused by mutations in multiple genes. In addition, genetic modifiers likely contribute to the reduced penetrance and variable disease severity observed in patients. In some disorders, such as for instance Brugada syndrome (BrS), an oligogenic or polygenic basis has been demonstrated, with multiple common or rare genetic variants in aggregate predisposing to the disease. In their review, Glinge et al. discuss the use of genome-wide association studies (GWAS) for the identification of genetic variants (single nucleotide polymorphisms) that govern interindividual variability in electrocardiographic parameters in the general population.9 Identified variants can be used to explore their possible role in modifying disease susceptibility and severity in inherited disorders such as BrS and LQTS. Ultimately, polygenic risk scores may be generated but their use in clinical risk stratification is still limited. Importantly, functional investigation of identified loci/variants may identify new genes relevant for cardiac electrical function and reveal novel mechanistic insight, but as yet such studies are rarely conducted. In terms of therapy, gene-targeted approaches are increasingly investigated for their potential use in inherited arrhythmia disorders. As reviewed by Bezzerides et al.,10 gene therapy strategies currently being explored include gene replacement (using AAV viral transduction), allele-specific silencing (using a mutation-specific short hairpin RNA or an allele-specific antisense oligonucleotide), modulation of signalling pathways (e.g. CamKII), modulation of splicing (by e.g. oligonucleotide-induced exon skipping), or genome editing by CRISPR/Cas9. So far, the most promising results for gene therapy have been obtained in experimental models of HCM and catecholaminergic polymorphic ventricular tachycardia (CPVT). In Casq2 deficient or mutant mice, AAV9-mediated cardiac expression of wild type CASQ2 prevented catecholamine-induced arrhythmias. Allele-specific silencing of the mutant Ryr2 allele furthermore reduced arrhythmia inducibility in a CPVT mouse model. While promising, such approaches are mutation-specific and hence therapeutic strategies applicable to a broader range of mutations would be more efficient, such as for instance gene delivery of CamKII inhibitory peptides. In HCM mouse and/or hiPSC-CM models, MYBPC3 gene replacement, antisense-based exon skipping, and transgenic expression of a CaMKII inhibitory peptide prevented hypertrophy and improved cardiac function. Despite a number of practical limitations in relation to clinical applicability, gene therapy may ultimately prove promising treatment strategies for certain inherited arrhythmias.
In conclusion, the research field of inherited arrhythmia disorders is fast evolving, aided by the availability of appropriate animal and human disease models, in-depth (electro)physiological and molecular functional studies to unravel arrhythmia mechanisms, compound screens for the identification of novel therapies, genetic studies revealing novel biological pathways and enabling improved risk stratification, and discovery of more efficient gene therapy approaches. Ongoing translational research efforts will no doubt continue to facilitate development of improved strategies for diagnosis, risk stratification, prevention, and treatment of patients with inherited arrhythmias.
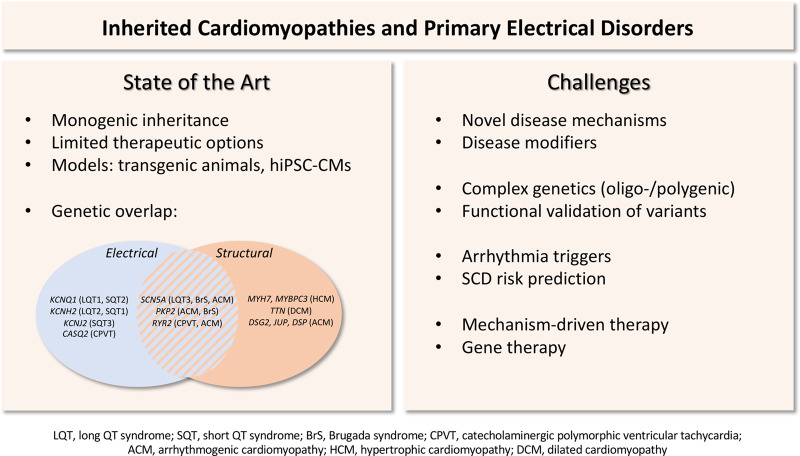
Figure 1.
Inherited cardiomyopathies and primary electrical disorders: state of the art and challenges.
Conflict of interest: none declared.
Funding
This work was supported by a grant from the Netherlands Cardio Vascular Research Initiative (CVON): the Dutch Heart Foundation, Dutch Federation of University Medical Centres, the Netherlands Organization for Health Research and Development and the Royal Netherlands Academy of Sciences (CVON-eDETECT 2015-12, CVON-PREDICT2 2018-30) to C.A.R.
References
- 1. Bezzina CR, Lahrouchi N, Priori SG.. Genetics of sudden cardiac death. Circ Res 2015;116:1919–1936. [DOI] [PubMed] [Google Scholar]
- 2. Chugh SS, Reinier K, Teodorescu C, Evanado A, Kehr E, Al Samara M, Mariani R, Gunson K, Jui J.. Epidemiology of sudden cardiac death: clinical and research implications. Progr Cardiovasc Dis 2008;51:213–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Offerhaus JA, Bezzina CR, Wilde AAM.. Epidemiology of inherited arrhythmias. Nat Rev Cardiol 2020;17:205–215. [DOI] [PubMed] [Google Scholar]
- 4. Crotti L, Odening KE, Sanguinetti MC.. Heritable arrhythmias associated with abnormal function of cardiac potassium channels. Cardiovasc Res 2020;116:1542–1556. [DOI] [PubMed] [Google Scholar]
- 5. Rivaud MR, Delmar M, Remme CA.. Heritable arrhythmia syndromes associated with abnormal cardiac sodium channel function: ionic and non-ionic mechanisms. Cardiovasc Res 2020;116:1557–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marian AJ, Asatryan B, Wehrens XHT. Genetic basis and molecular biology of cardiac arrhythmias in cardiomyopathies. Cardiovasc Res 2020;116:1600–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van der Voorn SM, Te Riele ASJM, Basso C, Calkins H, Remme CA, van Veen TABet al. Arrhythmogenic cardiomyopathy: pathogenesis, pro-arrhythmic remodeling, and novel approaches for risk stratification and therapy. Cardiovasc Res 2020;116:1571–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coppini R, Santini L, Olivotto I, Michael J, Cerbai E. Abnormalities in sodium current and calcium homeostasis as drivers of arrhythmogenesis in hypertrophic cardiomyopathy. Cardiovasc Res 2020;116:1585–1599. [DOI] [PubMed] [Google Scholar]
- 9. Glinge C, Lahrouchi N, Jabbari R, Tfelt-Hansen J, Bezzina CR. Genomic approaches for the identification of novel genes and disease mechanisms in inherited arrhythmia syndromes. Cardiovasc Res 2020;116:1620–1634. [Google Scholar]
- 10. Bezzerides VJ, Prondzynski M, Carrier L, Pu WT.. Gene therapy for inherited arrhythmias. Cardiovasc Res 2020;116:1635–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]