Abstract
The oncogenic transcription factor FOXM1 has been previously shown to play a critical role in carcinogenesis by inducing cellular proliferation in multiple cancer types. A small molecule compound, RCM-1, has been recently identified from high throughput screen as an inhibitor of FOXM1 in vitro and in mouse model of allergen-mediated lung inflammation. In the present study, we examined anti-tumor activities of RCM-1 using tumor models. Treatment with RCM-1 inhibited tumor cell proliferation as evidenced by increased cell cycle duration. Confocal imaging of RCM-1-treated tumor cells indicated that delay in cellular proliferation was concordant with inhibition of FOXM1 nuclear localization in these cells. RCM-1 reduced the formation and growth of tumor cell colonies in the colony formation assay. In animal models, RCM-1 treatment inhibited growth of mouse rhabdomyosarcoma Rd76–9, melanoma B16-F10 and human H2122 lung adenocarcinoma. RCM-1 decreased FOXM1 protein in the tumors, reduced tumor cell proliferation and increased tumor cell apoptosis. RCM-1 decreased protein levels and nuclear localization of β-catenin, and inhibited protein-protein interaction between β-catenin and FOXM1 in cultured tumor cells and in vivo. Altogether, our study provides important evidence of anti-tumor potential of the small molecule compound RCM-1, suggesting that RCM-1 can be a promising candidate for anti-cancer therapy.
Introduction
Robert Costa Memorial drug-1 (RCM-1) is a small molecule compound that has been recently identified by high throughput screening as a potent inhibitor of FOXM1 transcription factor (1). The RCM-1 compound inhibited the nuclear localization of FOXM1 protein, increased its ubiquitination and caused degradation of FOXM1 in proteasomes (1). While in mouse asthma models RCM-1 effectively inhibited FOXM1 without altering expression of other transcription factors or causing toxicity, the efficacy of the RCM-1 compound in cancer models remains unknown. FOXM1, a member of the Winged Helix or Forkhead Box (Fox) family of transcription factors, has been implicated in activation of cellular proliferation in multiple cell types, making it a promising target for anticancer therapy (2–6). Unlike other FOX proteins, FOXM1 is expressed in highly proliferative cells but is inhibited during cellular differentiation. FOXM1 is increased in various progenitors and in regenerating adult tissues (7–9). FOXM1 is overexpressed in a variety of human tumors and is used as biomarker for multiple types of cancer (4,10). We have previously demonstrated that FOXM1 overexpression increases lung tumor growth in transgenic mice, while lung epithelial-specific deletion of FOXM1 inhibits pulmonary tumorigenesis (11–14). FOXM1 has been reported to regulate progression of carcinogenesis and its increased expression is correlated with poor prognoses in patients with prostate cancer (15–17), lung cancers (5), glioblastoma (18), rhabdomyosarcoma (19), melanoma (20), colorectal cancer, breast cancer (9), liver cancer (21), pancreatic cancer and gastric cancers (22,23). FOXM1 was also implicated in chemo- and radio-resistance in various cancer types (22,24–26).
In addition to its prognostic significance, the exclusive expression of FOXM1 in the cancer tissue makes it an excellent cancer drug target (6). Inhibition of FOXM1 transcriptional activity led to significant inhibition of carcinogenesis and rendered the tumors responsive to standard chemotherapeutic drugs and radio-therapy (13,16,27,28). Pharmacological treatment with antibiotics and proteasome inhibitors, such as bortezomib, sialomycin, thiostrepton, MG115 and MG132, decreased FOXM1 expression in cancer cell lines and in vivo (27,29). However, while proteasomal inhibitors efficiently inhibit FOXM1, they affect multiple signaling pathways and cannot be viewed as specific inhibitors of FOXM1. Development of specific pharmacological inhibitors of FOXM1 represents a considerable clinical value. However, pharmacological targeting of transcription factors has been difficult due to the lack of enzymatic activity (30). This issue accounts for the lack of advanced target-specific inhibitors of previously characterized transcription factors.
We have recently reported the use of high throughput screening to identify FOXM1-inibiting small-molecule compound, RCM-1 (1). RCM-1 efficiently and specifically inhibited the nuclear localization of FOXM1 protein, causing its ubiquitination and degradation by proteasomes (1). The current study was designed to examine the efficacy of RCM-1 in the inhibition of carcinogenesis in different tumor models.
Materials and Methods
Cell lines and reagents
The cell lines, mouse melanoma B16-F10, human lung adenocarcinoma A549 and H2122, mouse mammary carcinoma 4T1 and mouse prostate cancer MyC-CaP, were obtained from American Type Culture Collection (ATCC, Manassas, VA). Rd76–9 rhabdomyosarcoma were the kind gift from Tim Cripe. KPC-2 cells were a kind gift from Matthew Flick. The compound RCM-1 (2-{[2-oxo-2-(thiophen-2-yl) ethyl]sulfanyl} −4,6-di(thiophen-2-yl)pyridine-3-carbonitrile) was synthesized by Vitas-M Laboratory (95% purity). The structure of RCM-1 has been published already in our previous manuscript (1).
Mouse models
Mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). All animal studies were approved by Animal Care and Use Committee (IACUC) of Cincinnati Children’s Research Foundation. For rhabdomyosarcoma model, 1×106 Rd76–9 cells were injected intramuscularly in the flanks of C56Bl/6J mice. For melanoma model, 1×106 B16-F10 cells were injected subcutaneously into C56Bl/6J mice. For lung adenocarcinoma model, 1×106 H2122 cells were injected subcutaneously into Nod-Scid-Gamma (NSG) mice. For mammary carcinoma model, 1×106 4T1 cells were inoculated into the fat pad of Balb/C mice. The tumor bearing mice were randomly divided into two groups (n=5–8) and treated with equal volumes of vehicle (DMSO) or RCM1. RCM1 was dissolved in DMSO and delivered intraperitoneally (IP) in a small volume of 40 μl at a dose of 20 mg/kg of body weight (mg/kg b.w.). 20 mg is equal to 47.1 μM of the compound. The tumor volume was measured using a digital caliper. Serum samples were collected from DMSO- or RCM-1- treated animals for liver enzyme profiling.
Growth curve analysis
Tumor cells (2×104 cells per well) were seeded in triplicates in 6-well plates and treated with 20 μM RCM-1 or equal volumes of DMSO. Automated cell counter (Countess II FL, ThermoFisher Scientific) was used to count the total number of viable cells at 24, 48 and 72 h. Trypan blue was used to exclude dead cells. Experiments were performed in triplicates.
EdU, BrdU incorporation assays
Vehicle control and RCM-1 treated tumor cells were incubated with EdU or BrdU, and immunofluorescence staining for EdU, BrdU, PH3 and Ki67 was performed as previously described (31,32).
Phase-contrast live cell imaging
DMSO or RCM-1 treated Rd76–9, B16-F10 and MyC-CaP cells were imaged using Leica DMI 6000b inverted microscope (Leica). Four fields per well were photographed every 5 min for 2–3 days as previously described (31). Mitotic duration was measured as the average time between nuclear envelope breakdown and anaphase onset. Cell cycle duration was measured as the average time interval between consecutive mitoses (n=100 cells) (31).
Immunohistochemistry, immunofluorescence and confocal imaging
Lung tissue sections were stained using anti-FOXM-1, anti-Ki-67, anti-PH3 (Santa Cruz) and anti-Cleaved Caspase-3 (Abcam) antibodies, as described previously (33). Tumor cells growing on coverslips were treated with 20 μM of RCM-1 for 24 h, fixed and stained with antibodies against FOXM1, FOXA1, β-catenin, Ki-67 and α-tubulin (Santa Cruz) as previously described (32).
Colony formation assay
Colony formation assay was performed as previously described (13). 2×103 tumor cells per well were seeded in 6-well plates and treated with 1, 5, 10 and 20 μM of RCM-1. The colonies were fixed at day 7, stained with crystal violet and the numbers of colonies containing ≥50 cells were counted. To study the effect of RCM-1 on colony formation, the tumor cells were treated with RCM-1 for three days (Day 3). RCM-1 was removed and growth of colonies was assessed after another three days (Day 6). To study the effect of RCM-1 on the growth of pre-existing colonies, RCM-1 treatment was started at Day 3 of culturing tumor cells. The growth of colonies was assessed three days later (Day 6).
Wound healing assay
The in vitro wound healing assay was performed as described previously (34). Briefly, 2×105 cells were seeded in 6-well plates and grown to a confluency of 80–90%. A wound was created in the middle of the well and cells were treated with RCM-1 (5 and 10 μM) for 12 to 18 h. Carl Zeiss inverted microscope and ImageJ 1.46r software were used to analyze wound closure.
siRNA-mediated knockdown of FOXM1
The siRNA transfection experiments were performed as described previously (35). Cells were transfected with siRNAs for human or mouse FOXM1 genes (Sigma-Aldrich) using Lipofectamine™ 2000 reagent (Invitrogen). Scrambled siRNA was used as a negative control.
Real time PCR
RNA Stat-60 RNA Extraction Reagent (Amsbio) was used to isolate total RNA from tumor cells. Taqman gene expression assays for FOXM1, β-catenin, Cyclin D1, and β-actin were purchased from Applied Biosystems. qRT-PCR was performed using the StepOnePlus Real-Time PCR system (Applied Biosystems) as previously described (36).
Western blot and co-immunoprecipitation (co-IP) analysis
Protein extracts were prepared from tumor cells using RIPA buffer as described (37). Cytoplasmic and nuclear extracts were prepared using NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (ThermoFisher Scientific). Western blot analysis was done as previously described (33). The following antibodies were used: FOXM1, β-catenin, Cyclin D1, Histone H3 (Santa Cruz) and β-actin (Millipore). For co-IP, 3T3 fibroblasts were treated with DMSO or RCM-1 for 48 h. Protein extracts were isolated either from cells incubated for an hour with 1 μM Dithiobis-succinimidyl propionate (DSP) to cross link protein-protein complexes or from DSP-untreated cells (no cross linking). IP reactions were performed using Universal Magnetic CoIP kit (Active Motif). For cell-free coIP reactions, whole cell lysate of RD76–9 cells was prepared and incubated with 40 μM RCM-1 and FOXM1 antibody overnight. The protein extracts from cells without RCM-1 treatment and IgG controls were used as positive and negative control respectively. Immunoprecipitation reactions were performed using Universal Magnetic Co-IP Kit (Active Motif). Rd76–9 whole cell lysate was used as input control for both groups.
UPLC-MS/MS analysis for serum RCM-1 concentration
Serum concentration of RCM-1 was measured after protein precipitation. RCM-1 were quantified by using UPLC coupled with electrospray tandem mass spectrometry (ESI-MS/MS) on an Acquity Quattro Premier (Waters Corp) with multiple reaction ion monitoring (MRM) in positive ion mode. RCM-1 was retained and eluted from a reversed-phase Supelco® LC18-DB column (Sigma-Aldrich). Chromatography was performed with a gradient mobile phase of solvent A/solvent B at a flow rate 200 μL/min at 25°C. RCM-1 was detected and quantified by monitoring the positive ion MRM transition m/z 425→313 generated by fragmentation, and the structural analog internal standard was detected from the corresponding positive ion MRM transition m/z 447→147. The concentrations of RCM-1 were determined from the peak area ratio of each ion relative to the internal standard and by interpolation of this area ratio against calibration curves plotted for known concentrations of standard.
Statistical analysis
Student’s t test, Fisher’s exact test and one-way analysis of variance (ANOVA) were used to determine statistical significance. P values <0.05 were considered significant. Values were shown as mean±SD or mean±SE. For the mitotic and cell cycle duration experiment, p-values were obtained using GraphPad Prism version 6 (GraphPad, San Diego, CA, USA). Data were tested for parametric vs. non-parametric distribution using D’Agostino-Pearson omnibus normality test. As data followed a non-parametric distribution, Mann-Whitney test was applied.
Results
RCM-1 inhibits cancer cell proliferation by increasing the mitotic and cell cycle duration in vitro
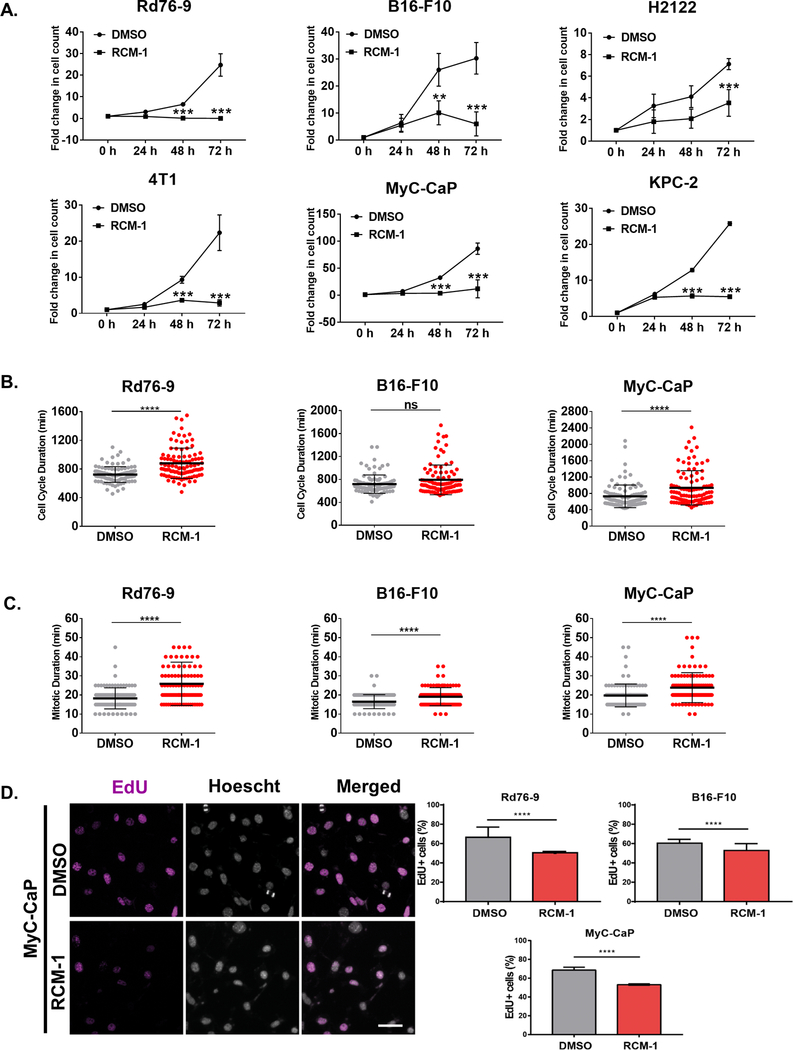
To assess the effect of RCM-1 on the proliferation of cancer cells in vitro, we performed growth curve analyses. RCM-1 significantly inhibited the growth of several tumor cell lines, including mouse rhabdomyosarcoma Rd76–9, melanoma B16-F10, human lung adenocarcinoma H2122, breast carcinoma 4T1, prostate adenocarcinoma MyC-CaP and pancreatic adenocarcinoma KPC-2 cells (Fig. 1A). The number of dead cells was not changed (Supplemental Fig. S1A). Since the inhibition of cell growth in RCM-1-treated cell cultures can be due to delayed progression of cell cycle, we performed 3-day phase-contrast live-cell imaging to follow individual cell behavior in Rd76–9, B16-F10 and MyC-CaP cell cultures treated with RCM-1 or vehicle (Supplemental Movies 1–6). When measuring cell cycle duration, the interval between anaphase onset of one mother cell and the anaphase onset of each daughter cell (n=100), we found a delay in Rd76–9 and MyC-Cap cell cultures treated with RCM-1 versus untreated controls (Fig. 1B; Supplemental Movies 1–6). We also measured mitotic duration as the interval between nuclear envelope breakdown and anaphase onset. RCM-1 significantly delayed the completion of mitotic division in Rd76–9, B16-F10 and MyC-CaP cell lines (Fig. 1C, Supplemental Fig. S1B and Supplemental movies 1–6). Consistent with these results, RCM-1 decreased the number of tumor cells undergoing mitosis as determined by immunostaining for phospho-histone H3 (Supplemental Fig. S2A–B). Next, we performed the EdU incorporation assay to determine the number of cells undergoing S phase during EdU labeling period. EdU incorporation was decreased in Rd76–9, B16-F10 and MyC-Cap cells after RCM-1 treatment (Fig. 1D). Immunostaining for Ki-67 and BrdU were also reduced in RCM-1-treated cells (Supplemental Fig. S2A). Altogether, RCM-1 inhibits tumor cell proliferation in vitro by decreasing DNA replication and mitosis.
Figure 1. RCM-1 inhibits cancer cell proliferation and increases the mitotic and cell cycle duration in vitro.
(A) RCM-1 inhibits tumor cell growth. Growth curves for RCM-1 treated Rd76–9 rhabdomyosarcoma, B16-F10 melanoma, H2122 lung adenocarcinoma, 4T1 mammary carcinoma, MyC-CaP prostate carcinoma and KPC-2 pancreatic carcinoma cells were determined in culture by counting alive cells. Cells treated with vehicle (DMSO) were used as controls. Graphs show fold change in the number of cells compared to 0 h time point and represent data from three independent experiments. (B) RCM-1 inhibits cell cycle duration. Live images of tumor cells were acquired an epifluorescence microscope every 5 min for 3 days. Cell cycle duration was measured as the interval between anaphase onset of the mother cell and its daughter cells. Graphs represent average cell cycle duration of DMSO- and RCM-1-treated cells (n=100). Values are shown as mean±SD from three independent experiments. (C) RCM-1 inhibits mitotic duration. Live images of tumor cells were acquired every 5 min for 3 days. Mitotic duration was measured as the interval between nuclear envelope breakdown and anaphase onset. Graphs represent average mitotic duration of DMSO- and RCM-1-treated cells (n=100). (D) RCM-1 inhibits DNA replication. Rd76–9, B16-F10 and MyC-CaP tumor cells were incubated with EdU. Left panels show representative images of EdU incorporation in DMSO- and RCM-1-treated MyC-CaP cells after 24 h (EdU staining, magenta; DNA staining, grey). Right panels show the percentage of EdU-positive cells after DMSO- and RCM-1 treatment (n>500). ns p>0.05, *p < 0.05, **p < 0.01 and ***p < 0.001 and ****p≤0.0001.
RCM-1 inhibits FOXM1 in tumor cells
Published studies demonstrated that treatment with RCM-1 compound decreased FOXM1 protein in airway epithelial cells in vitro and in vivo (1). To determine whether RCM-1 inhibits FOXM1 in tumor cells, we examined FOXM1 abundance and nuclear localization by immunostaining. FOXM1 staining was detected in the nuclei of Rd76–9, B16-F10, MyC-CaP, KPC-2, A549 and 4T1 tumor cells (Supplemental Fig. S3A–D and S4A–B). Treatment with RCM-1 decreased the nuclear staining of FOXM1 in these tumor cells (Supplemental Fig. S3A–D and S4A–B), which is consistent with the published studies (1). Nuclear expression of FOXA1, a related transcription factor from FOX family, was not altered after RCM-1 treatment (Supplemental Fig. S3A–D and S4A–B).
RCM-1 decreases tumorigenicity and migration of tumor cells in vitro
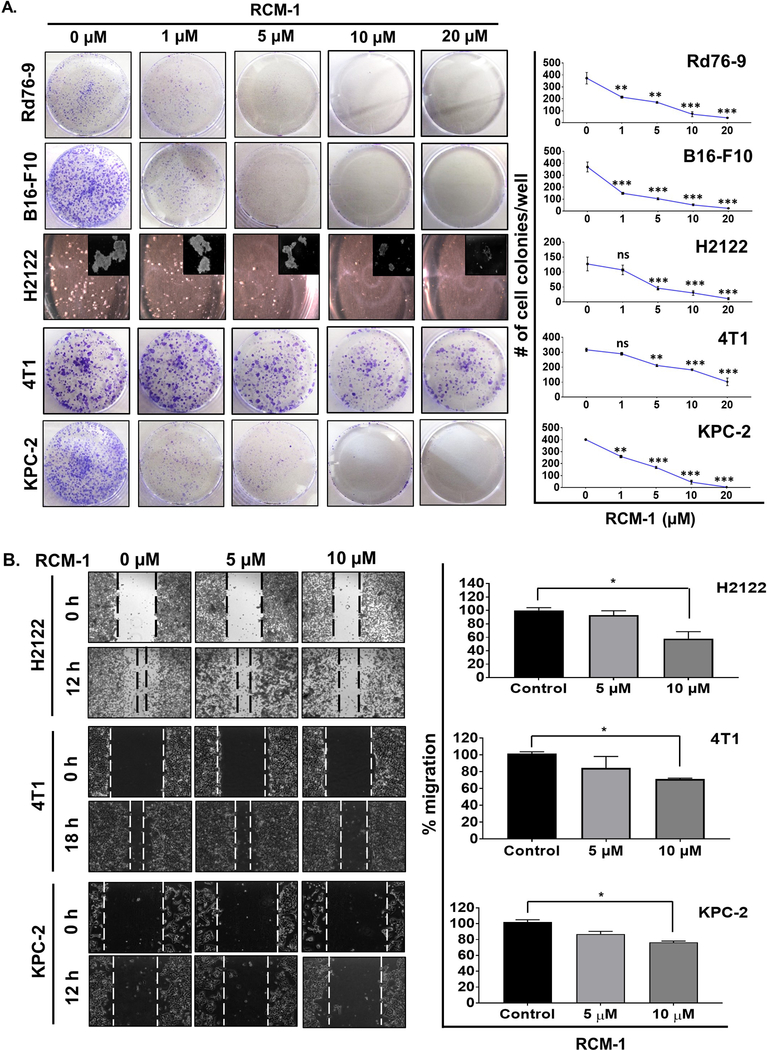
The ability of tumor cells to form colonies is a measure of tumorigenicity in vitro. Effect of RCM-1 on the in vitro colony forming potential of different tumor cells was determined using anchorage-dependent colony formation assay. Dose-dependent inhibition of colony formation was observed in Rd76–9, B16-F10, H2122, 4T1 and KPC-2 tumor cells; as low as 5 μM RCM-1 effectively inhibited colony formation (Fig. 2A). RCM-1 also inhibited the colony formation in A549 and LLC lung adenocarcinoma cells (Supplemental Fig. S5A). Next, we used scratch assay to examine the effects of RCM-1 on tumor cell migration. RCM-1 significantly inhibited the migration of H2122, 4T1, KPC-2 and A549 cells (Fig. 2B and Supplemental Fig. S5B). Altogether, these data demonstrate that RCM-1 inhibits the ability of tumor cells to form colonies and migrate in vitro.
Figure 2. RCM-1 decreases tumorigenicity and migration of tumor cells in vitro.
(A) Continuous treatment with RCM-1 decreases the number of colonies formed in cultured tumor cells. Rd76–9, B16-F10, H2122, 4T1 and KPC-2 tumor cells were treated with either DMSO or different concentrations of RCM-1 for 7 seven days. Graphs represent average number of colonies (mean±SE) that were obtained from three independent experiments. (B) RCM-1 inhibits migration of tumor cells in vitro. Data is presented as percent migration in RCM-1-treated groups compared to DMSO-treated control (mean±SE). *p < 0.05, **p < 0.01 and ***p < 0.001.
RCM-1 inhibits colony-formation and reduces the growth of pre-existing tumor cell colonies in vitro
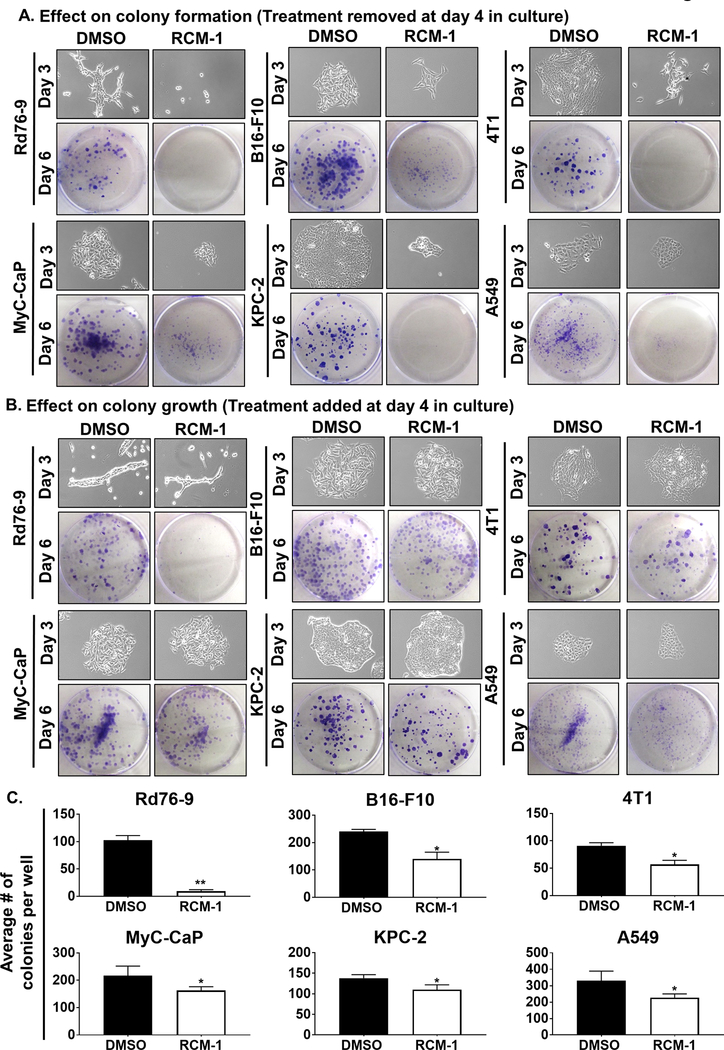
Tumor relapse in patients following the completion of anti-cancer treatment remains an important concern for oncologists (38). We assessed the effect of removal of RCM-1 treatment after a short exposure by colony formation assay. Even short exposure to RCM-1 was sufficient to decrease frequency and sizes of the cell colonies formed in Rd76–9, B16-F10, 4T1, MyC-CaP, KPC-2 and A549 tumor cell lines (Fig. 3A). Thus, RCM-1-treated cells cannot regain the ability to grow and form cell colonies even after the removal of RCM-1. We also analyzed the effect of RCM-1 on pre-existing cancer colonies. We found that RCM-1 inhibits further growth of pre-existing colonies in vitro. The number of cell colonies in the RCM-1-treated cultures were significantly reduced compared to vehicle-treated cell cultures (Fig. 3B–C). Thus, RCM-1 effectively suppressed the colony formation and reduced the growth of pre-existing cell colonies in vitro.
Figure 3. RCM-1 inhibits initial colony-formation and reduces the growth of pre-existing tumor cell colonies in vitro.
(A) Treatment of tumor cells with RCM-1 for 3 days followed by RCM-1 removal prevented further growth of tumor cell colonies. Upper panels show representative images of colonies at the day of RCM-1 removal (Day 3). Lower panels show crystal violet stained colonies three days after RCM-1 removal (Day 6). DMSO was used as a vehicle control. Images are representative of three independent experiments. (B) RCM-1 inhibited growth of pre-existing tumor colonies. Upper panels show individual colonies before RCM-1 treatment (Day 3). Lower panels show crystal violet stained colonies of tumor cells after three days of treatment either with DMSO or RCM-1 (Day 6). Images are representative of three independent experiments. (C) Quantification of RCM-1 effect on the pre-existing tumor colonies. Graphs represent average number of tumor cell colonies per well after three days of RCM-1 treatment (Day 6) compared to DMSO-treated control (mean±SE). *p < 0.05, **p < 0.01 and ***p < 0.001.
RCM-1 inhibits β-catenin in tumor cells
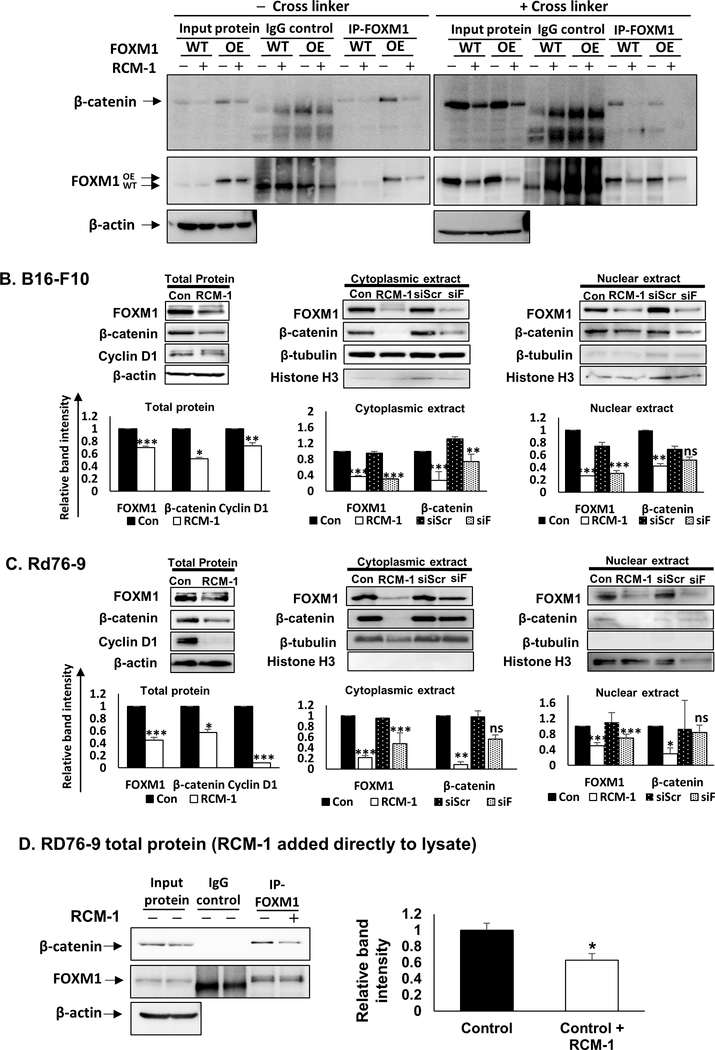
Similar to FOXM1, β-catenin is an important transcriptional regulator of carcinogenesis which promotes cancer growth & metastasis and maintains stem cell characteristics (39,40). Persistent activation of Wnt/β-catenin signaling has been implicated in a variety of human cancers (40). Since FOXM1 directly binds to β-catenin to promote glioma growth and metastasis (18), we examined the effect of RCM-1 on FOXM1-β-catenin protein-protein interactions. Consistent with published studies (18), FOXM1-β-catenin protein-protein interactions were detected by co-immunoprecipitation (CoIP) using total cell lysates (Fig. 4A). Treatment with RCM-1 decreased FOXM1 protein binding to β-catenin (Fig. 4A). Furthermore, RCM-1 decreased the protein abundance of β-catenin and FOXM1 in both nuclear and cytoplasmic fractions (Fig. 4B–C and Supplemental Fig. S6A). The FoxM1 and β-catenin mRNAs were not affected by RCM-1 treatment, indicating that RCM-1 does not inhibit these genes at transcriptional level (Supplemental Fig. S6B). These findings are consistent with previous studies highlighting the importance of FOXM1-β-catenin interactions for β-catenin protein stability (18). Consistent with decreased β-catenin protein levels, its downstream target Cyclin D1 was reduced in RCM-1-treated cells (Fig. 4B–C and Supplemental Fig. S6A). RCM-1 was more efficient in inhibiting FOXM1 and β-catenin compared to FoxM1 siRNA (Fig. 4B–C and Supplemental Fig. S6A). Consistent with CoIP and Western blots, RCM-1 inhibited nuclear localization of β-catenin in the tested cell lines (Supplemental Fig. S6C and S7A–D). Importantly, FOXM1-β-catenin protein complex was directly affected by RCM-1, since addition of RCM-1 to the protein lysate from the control tumor cells decreased FOXM1-β-catenin protein-protein interactions (Fig. 4D). Thus, RCM-1 decreased protein abundance of FOXM1 and β-catenin and disrupted FOXM1-β-catenin protein-protein interactions in cell nuclei.
Figure 4. RCM-1 inhibits FOXM1/ β-catenin protein-protein interaction.
(A) RCM-1 decreased FOXM1 protein binding to β-catenin protein in control (WT) and FOXM1 overexpressing (OE) 3T3 fibroblasts. Protein lysates were prepared without cross-linking (– cross linker, left panel) or with cross-linking peptide DSP (+cross linker) to stabilize protein-protein complexes. β-actin was used as loading control. Blots are representative of two independent experiments. (B) RCM-1 decreased protein levels of FOXM1, β-catenin and Cyclin D1 in B16-F10 melanoma cells (left panels). Cytoplasmic (middle panels) and nuclear (right panels) protein extracts of RCM-1-treated tumor cells were compared to siFOXM1-depleted cells. DMSO-treated (Con) or shScrambled (shScr) cells were used as controls. β-actin, α/β-tubulin, and Histone H3 were used as loading controls for whole cell lysates, cytoplasmic-, and nuclear extracts, respectively. Blots are representative of three independent experiments. (C) RCM-1 decreased protein levels of FOXM1, β-catenin and Cyclin D1 in Rd76–9 rhabdomyosarcoma cells (left panels). Cytoplasmic (middle panels) and nuclear (right panels) protein extracts of RCM-1-treated cells were compared to siFOXM1-transfected cells. DMSO-treated (Con) or shScrambled (shScr) cells were used as controls. β-actin, α/β-tubulin, and Histone H3 were used as loading controls for whole cell lysates, cytoplasmic-, and nuclear extracts, respectively. Blots are representative of three independent experiments. (D) RCM-1 decreases FOXM1 binding to β-catenin protein. Total protein lysate of Rd76–9 tumor cells were incubated with RCM-1 overnight and used for immunoprecipitation with either FOXM1 or IgG antibodies. β-actin was used as loading control. Blots are representative of two independent experiments. Graphs represent relative band intensities (mean±SE). *p < 0.05, **p < 0.01 and ***p < 0.001 compared to DMSO-treated control.
RCM-1 inhibits tumor growth in animal models
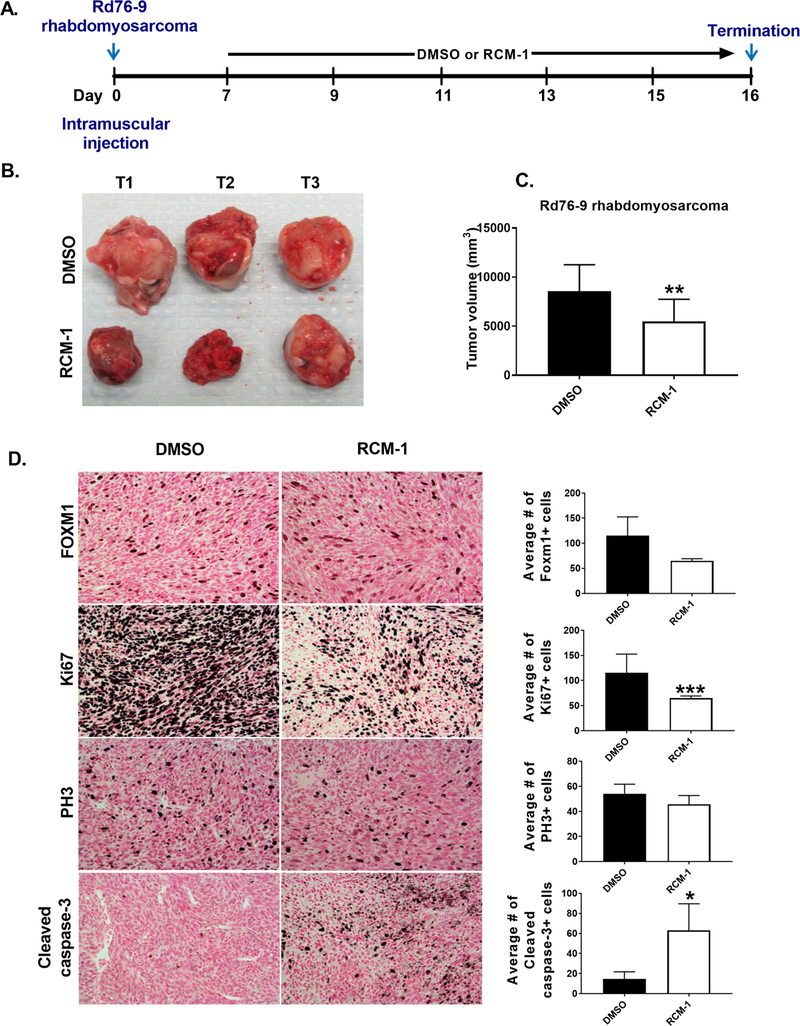
Since RCM-1 decreased the ability of tumor cells to form colonies in vitro (Fig. 2A and 3A–B), we examined the efficacy of RCM-1 in the mouse tumor models in vivo. Rhabdomyosarcoma Rd76–9 cells were injected into the muscle of syngeneic mice, and seven days later, RCM-1 was given to tumor-bearing mice (Fig. 5A). RCM-1 reduced the Rd76–9 tumor size compared to the vehicle-treated control group (Fig. 5B). The volumes of RCM-1-treated rhabdomyosarcoma tumors were significantly decreased (Fig. 5C). FOXM1 staining in cell nuclei was decreased and the number of FOXM1-positive tumor cells was also reduced considerably in RCM-1-treated tumors, consistent with efficient inhibition of FOXM1 by RCM-1 (Fig. 5D). To determine whether reduced tumor sizes in RCM-1-treated mice were due to decreased tumor cell proliferation, immunostaining for Ki-67 and PH3 was performed. The number of Ki-67- and PH3- positive cells were decreased in RCM-1 treated tumors compared to vehicle-treated tumors (Fig. 5D). Furthermore, the number of tumor cells positive for cleaved caspase-3 was significantly increased in RCM-1-treated tumors (Fig. 5D), indicating an increase in tumor cell apoptosis. Thus, RCM-1 decreased tumor growth, reduced tumor cell proliferation and increased apoptosis in mice bearing Rd76–9 rhabdomyosarcomas.
Figure 5. RCM-1 treatment reduces growth of orthotopic Rd76–9 rhabdomyosarcoma in mice.
(A) Schematic diagram of experimental protocol. (B-C) RCM-1 treatment decreased Rd76–9 tumor growth in mice. DMSO-treated animals were used as control. Data presented as average tumor volume (mean±SD). (D) RCM-1 decreased tumor cell proliferation and increased apoptosis in Rd76–9 tumors. RCM-1-treated tumors showed reduced FOXM1, Ki67 and PH3 staining. Apoptotic cleaved caspase-3 staining was increased in the RCM-1-treated tumors. Graphs in right panels show average numbers of FOXM1-, Ki67-, PH3- and Cleaved caspase-3- positive cells (mean±SD). Staining-positive cells were counted in 6 random microscopic fields. Magnification: x200. *p < 0.05, **p < 0.01 and ***p < 0.001 compared to DMSO-treated control.
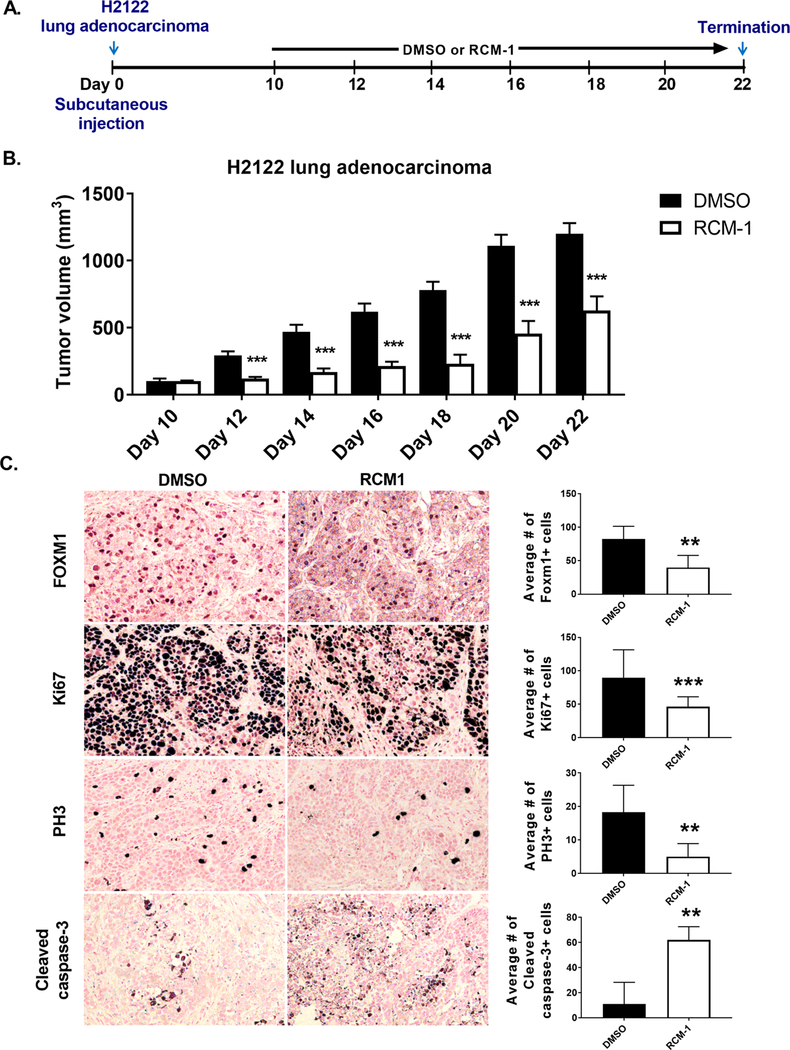
To demonstrate that the effect of RCM-1 is not limited to the rhabdomyosarcoma Rd76–9 tumor model, we also used mouse melanoma B16-F10, human lung adenocarcinoma H2122 and mouse breast carcinoma 4T1 tumor models. RCM-1 significantly inhibited tumor growth in H2122 lung adenocarcinoma and B16-F10 melanoma consistent with loss of FOXM1 and β-catenin in cell nuclei (Fig. 6A–C and Supplemental Fig. S8A–C, Supplemental Fig. S9A), coinciding with diminished tumor cell proliferation and increased apoptosis of tumor cells (Fig. 6A–C and Supplemental Fig. S8A–C). Treatment with RCM-1 did not change the growth of orthotopic 4T1 breast tumors significantly (Supplemental Fig. S9B–D).
Figure 6. RCM-1 treatment reduces growth of human H2122 lung adenocarcinomas in mice.
(A) Schematic representation of experimental protocol. (B) RCM-1 decreases H2122 tumor growth in vivo. Graph shows average tumor volume (mean±SD). DMSO-treated animals were used as control. (C) RCM-1 decreases tumor cell proliferation, shown with Ki67 and PH3 staining and increases apoptosis, shown with cleaved caspase staining. Right panels show average numbers of FOXM1-, Ki67-, PH3- and Cleaved caspase-3-positive cells. Positive cells were counted in 6 random microscopic fields (mean±SD). Magnification: x200. *p < 0.05, **p < 0.01 and ***p < 0.001 compared to DMSO-treated control.
To measure the RCM-1 concentration in blood serum, the UPLC-MS/MS analysis was used (Supplemental Fig. S10A–B). RCM-1 was detected in blood serum of animals within 15 min after a single i.p. injection (Supplemental Fig. S10C–D, Supplemental Table-1). Serum concentrations of RCM-1 were maintained at the same levels for 2 h (Supplemental Fig. S10C–D, Supplemental Table-1). Multiple injections of RCM-1 did not affect body weights (Supplemental Fig. S11A) or concentrations of liver enzymes ALT and ALK (Supplemental Fig. S11B). Albumin, bilirubin, total protein, and Gamma-glutamyltransferase (GGT) were unchanged in blood serum of RCM-1-treated mice (Supplemental Fig. S11B), indicating that RCM-1 does not cause liver toxicity. Consistent with our in vitro findings, RCM-1 effectively inhibited FOXM1 and β-catenin in tumor-bearing mice without visible toxicity even with prolonged treatment. Altogether, RCM-1 reduced FOXM1 and β-catenin, decreased tumor cell proliferation and growth in rhabdomyosarcoma, melanoma and lung adenocarcinoma tumors in vivo.
Discussion
Previous studies demonstrated that FOXM1 is critical for embryonic development and organ regeneration following tissue injury (4). While FOXM1 is decreased after tissue repair, aberrantly high levels of FOXM1 are common for many different types of cancers. Overexpression of FOXM1 increases the initiation, growth, and metastasis in different types of tumors in orthotopic and transgenic mouse models as well as human xenografts (5,11–13,15,28). Diverse and extensive involvement of FOXM1 in the process of carcinogenesis of different tissue origins suggests that FOXM1 inhibition can be utilized for anticancer therapy. Our recent study identified the small molecule FOXM1 inhibitor RCM-1 in a high-throughput screen (1). RCM-1 inhibited nuclear localization and increased ubiquitination of FOXM1 in cultured epithelial cells and in the mouse model of goblet cell metaplasia (1). In the present study, RCM-1 inhibited tumor growth, decreased tumor cell proliferation, and increased apoptosis in the mouse Rd76–9 rhabdomyosarcoma, B16-F10 melanoma and human H2122 lung cancer models. Our results suggest that RCM-1-modulated suppression of tumor growth is mediated by inhibition of FOXM1.
FOXM1 is known to function as a typical proliferation-specific transcription factor. It stimulates cellular proliferation by promoting entry into S-phase and M-phase of the cell cycle and controls proper execution of mitotic cell division (41). RCM-1-mediated inhibition of FOXM1 expression was associated with the inhibition of cell cycle progression. Targeted inhibition of proteins involved in the regulation of the cell cycle has been linked to delays in the duration of mitosis in cancer cells (42). Similar to previous studies with siRNA inhibition of FOXM1 (43,44), RCM-1 increased the duration of mitotic division and overall length of the cell cycle and inhibited DNA replication in tumor cells. However, we cannot rule out the possibility that RCM-1 affects other molecular targets in addition to FOXM1.
The cancer stem cell (tumor initiating cell) hypothesis identifies in vitro clonogenesis as the indication of tumorigenic abilities (33). FOXM1 has been implicated in the maintenance of the cancer stem cell state (9), and FOXM1 silencing resulted in reduced tumorigenicity in glioblastoma (40). Consistent with these studies, RCM-1-mediated inhibition of FOXM1 led to suppression of clonogenicity of cancer cells in vitro. Furthermore, FOXM1 promoted cancer cell migration and invasion, and genetic deletion of FOXM1 inhibited invasion and metastasis of lung, liver, prostate, colon cancers in vivo (13), 17, (21) (16,45). Consistent with these studies, we observed inhibition of tumor cell migration in RCM-1 treated cells, which suggests the prospective anti-metastatic potential of this small molecule compound.
FOXM1 was implicated in tumor recurrence in human non-small cell lung and oral cancers (46,47). We found that cancer cells, when briefly exposed to RCM-1, have reduced cell growth and clonogenicity, indicating the potential of this molecule in delaying or preventing tumor relapse in cancer patients. In addition, the ability of RCM-1 to inhibit pre-existing tumor colonies suggests that the compound has therapeutic potential during cancer progression. Interestingly, interactions of FOXM1 with β-catenin, a critical regulator of the canonical Wnt signaling pathway, were reduced in RCM-1-treated tumor cells in vitro and in vivo. Wnt signaling through β-catenin was implicated in tumor relapses, metastasis and tumor cell proliferation (48). FOXM1 directly binds to β-catenin protein, stabilizing β-catenin in cell nuclei and promoting glioma tumorigenesis (18). Our data suggest that RCM-1 directly disrupts FOXM1-β-catenin protein complex, decreasing β-catenin protein stability in tumor cells. Interestingly, FOXM1 was reported to induce β-catenin gene expression in cultured endothelial cells and lung injury models (49). Based on our findings, it is unlikely that RCM-1 inhibits β-catenin gene expression in neoplastic cells since β-catenin mRNA was not affected by RCM-1 in all the tumor types. Therefore, diminished β-catenin protein levels were not a consequence of decreased β-catenin gene expression in RCM-1-treated tumor cells. RCM-1 directly or indirectly disrupted the protein-protein interactions between FOXM1 and β-catenin. Since FOXM1/β-catenin interactions are critical for stability of both proteins, decreased amounts of FOXM1 and β-catenin in RCM-1-treated cells can be a consequence of disruption of FOXM1/β-catenin complexes. Altogether, RCM-1 disrupts FOXM1-β-catenin interactions, which can contribute to diminished tumorigenesis in RCM-1-treated mice.
In summary, the small molecule compound RCM-1 effectively inhibits FOXM1 and β-catenin in vitro and in vivo, delaying tumor cell proliferation, increasing apoptosis, and reducing tumor growth. Anticancer effects of RCM-1 are not limited to one tumor type, suggesting a potential for broad use of this small molecule FOXM1 inhibitor in anticancer therapy.
Supplementary Material
Acknowledgements
This work was supported by NIH grants R01HL132849 (T.V. Kalin), R56HL126660 (T.V. Kalin), RO1HL84151 (V.V. Kalinichenko) and RO1HL123490 (V.V. Kalinichenko).
Footnotes
The authors declare no potential conflicts of interest.
REFERENCES
- 1.Sun L, Ren X, Wang IC, Pradhan A, Zhang Y, Flood HM, et al. The FOXM1 inhibitor RCM-1 suppresses goblet cell metaplasia and prevents IL-13 and STAT6 signaling in allergen-exposed mice. Sci Signal 2017;10(475). [DOI] [PubMed] [Google Scholar]
- 2.Balli D, Ren X, Chou FS, Cross E, Zhang Y, Kalinichenko VV, et al. Foxm1 transcription factor is required for macrophage migration during lung inflammation and tumor formation. Oncogene 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balli D, Zhang Y, Snyder J, Kalinichenko VV, Kalin TV. Endothelial cell-specific deletion of transcription factor FoxM1 increases urethane-induced lung carcinogenesis. Cancer Res 2011;71(1):40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalin TV, Ustiyan V, Kalinichenko VV. Multiple faces of FoxM1 transcription factor. Cell Cycle 2011;10(3):396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milewski D, Balli D, Ustiyan V, Le T, Dienemann H, Warth A, et al. FOXM1 activates AGR2 and causes progression of lung adenomas into invasive mucinous adenocarcinomas. PLoS Genet 2017;13(12):e1007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalinichenko VV, Kalin TV. Is there potential to target FOXM1 for ‘undruggable’ lung cancers? Expert Opin Ther Targets 2015;19(7):865–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang X, Dai Z, Cai L, Sun K, Cho J, Albertine KH, et al. Endothelial p110gammaPI3K Mediates Endothelial Regeneration and Vascular Repair After Inflammatory Vascular Injury. Circulation 2016;133(11):1093–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ustiyan V, Zhang Y, Perl AK, Whitsett JA, Kalin TV, Kalinichenko VV. beta-catenin and Kras/Foxm1 signaling pathway are critical to restrict Sox9 in basal cells during pulmonary branching morphogenesis. Dev Dyn 2016;245(5):590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang N, Wang C, Wang Z, Zona S, Lin SX, Wang X, et al. FOXM1 recruits nuclear Aurora kinase A to participate in a positive feedback loop essential for the self-renewal of breast cancer stem cells. Oncogene 2017;36(24):3428–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nandi D, Cheema PS, Jaiswal N, Nag A. FoxM1: Repurposing an oncogene as a biomarker. Semin Cancer Biol 2017. [DOI] [PubMed] [Google Scholar]
- 11.Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV, et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res 2006;66(4):2153–61. [DOI] [PubMed] [Google Scholar]
- 12.Wang IC, Meliton L, Tretiakova M, Costa RH, Kalinichenko VV, Kalin TV. Transgenic expression of the forkhead box M1 transcription factor induces formation of lung tumors. Oncogene 2008;27(30):4137–49. [DOI] [PubMed] [Google Scholar]
- 13.Wang IC, Meliton L, Ren X, Zhang Y, Balli D, Snyder J, et al. Deletion of Forkhead Box M1 transcription factor from respiratory epithelial cells inhibits pulmonary tumorigenesis. PLoS One 2009;4(8):e6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang IC, Ustiyan V, Zhang Y, Cai Y, Kalin TV, Kalinichenko VV. Foxm1 transcription factor is required for the initiation of lung tumorigenesis by oncogenic Kras(G12D.). Oncogene 2014;33(46):5391–6. [DOI] [PubMed] [Google Scholar]
- 15.Kalin TV, Wang IC, Ackerson TJ, Major ML, Detrisac CJ, Kalinichenko VV, et al. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res 2006;66(3):1712–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai Y, Balli D, Ustiyan V, Fulford L, Hiller A, Misetic V, et al. Foxm1 expression in prostate epithelial cells is essential for prostate carcinogenesis. J Biol Chem 2013;288(31):22527–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aytes A, Mitrofanova A, Lefebvre C, Alvarez MJ, Castillo-Martin M, Zheng T, et al. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell 2014;25(5):638–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, et al. FoxM1 promotes beta-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell 2011;20(4):427–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuda M, Kohashi K, Yamada Y, Maekawa A, Kinoshita Y, Nakatsura T, et al. FOXM1 expression in rhabdomyosarcoma: a novel prognostic factor and therapeutic target. Tumour Biol 2016;37(4):5213–23. [DOI] [PubMed] [Google Scholar]
- 20.Kruiswijk F, Hasenfuss SC, Sivapatham R, Baar MP, Putavet D, Naipal KA, et al. Targeted inhibition of metastatic melanoma through interference with Pin1-FOXM1 signaling. Oncogene 2016;35(17):2166–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gusarova GA, Wang IC, Major ML, Kalinichenko VV, Ackerson T, Petrovic V, et al. A cell-penetrating ARF peptide inhibitor of FoxM1 in mouse hepatocellular carcinoma treatment. J Clin Invest 2007;117:99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui J, Xia T, Xie D, Gao Y, Jia Z, Wei D, et al. HGF/Met and FOXM1 form a positive feedback loop and render pancreatic cancer cells resistance to Met inhibition and aggressive phenotypes. Oncogene 2016;35(36):4708–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang C, Qiu Z, Wang L, Peng Z, Jia Z, Logsdon CD, et al. A novel FoxM1-caveolin signaling pathway promotes pancreatic cancer invasion and metastasis. Cancer Res 2012;72(3):655–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nestal de Moraes G, Delbue D, Silva KL, Robaina MC, Khongkow P, Gomes AR, et al. FOXM1 targets XIAP and Survivin to modulate breast cancer survival and chemoresistance. Cell Signal 2015;27(12):2496–505. [DOI] [PubMed] [Google Scholar]
- 25.Tassi RA, Todeschini P, Siegel ER, Calza S, Cappella P, Ardighieri L, et al. FOXM1 expression is significantly associated with chemotherapy resistance and adverse prognosis in non-serous epithelial ovarian cancer patients. J Exp Clin Cancer Res 2017;36(1):63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y, Kim KH, Kim DG, Cho HJ, Kim Y, Rheey J, et al. FoxM1 Promotes Stemness and Radio-Resistance of Glioblastoma by Regulating the Master Stem Cell Regulator Sox2. PLoS One 2015;10(10):e0137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwok JM, Myatt SS, Marson CM, Coombes RC, Constantinidou D, Lam EW. Thiostrepton selectively targets breast cancer cells through inhibition of forkhead box M1 expression. Mol Cancer Ther 2008;7(7):2022–32. [DOI] [PubMed] [Google Scholar]
- 28.Koo C-Y, Muir KW, Lam EWF. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta 2012;1819(1):28–37. [DOI] [PubMed] [Google Scholar]
- 29.Gartel AL. A new target for proteasome inhibitors: FoxM1. Expert Opin Investig Drugs 2010;19(2):235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagenbuchner J, Ausserlechner MJ. Targeting transcription factors by small compounds--Current strategies and future implications. Biochem Pharmacol 2016;107:1–13. [DOI] [PubMed] [Google Scholar]
- 31.Macedo JC, Vaz S, Bakker B, Ribeiro R, Bakker PL, Escandell JM, et al. FoxM1 repression during human aging leads to mitotic decline and aneuploidy-driven full senescence. Nat Commun 2018;9(1):2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Black M, Milewski D, Le T, Ren X, Xu Y, Kalinichenko VV, et al. FOXF1 Inhibits Pulmonary Fibrosis by Preventing CDH2-CDH11 Cadherin Switch in Myofibroblasts. Cell Rep 2018;23(2):442–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shukla S, Khan S, Kumar S, Sinha S, Farhan M, Bora HK, et al. Cucurbitacin B Alters the Expression of Tumor-Related Genes by Epigenetic Modifications in NSCLC and Inhibits NNK-Induced Lung Tumorigenesis. Cancer Prev Res (Phila) 2015;8(6):552–62. [DOI] [PubMed] [Google Scholar]
- 34.Milewski D, Pradhan A, Wang X, Cai Y, Le T, Turpin B, et al. FoxF1 and FoxF2 transcription factors synergistically promote rhabdomyosarcoma carcinogenesis by repressing transcription of p21(Cip1) CDK inhibitor. Oncogene 2017;36(6):850–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai Y, Bolte C, Le T, Goda C, Xu Y, Kalin TV, et al. FOXF1 maintains endothelial barrier function and prevents edema after lung injury. Sci Signal 2016;9(424):ra40. [DOI] [PubMed] [Google Scholar]
- 36.Kalin TV, Meliton L, Meliton AY, Zhu X, Whitsett JA, Kalinichenko VV. Pulmonary mastocytosis and enhanced lung inflammation in mice heterozygous null for the Foxf1 gene. Am J Respir Cell Mol Biol 2008;39(4):390–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia H, Ren X, Bolte CS, Ustiyan V, Zhang Y, Shah TA, et al. Foxm1 regulates resolution of hyperoxic lung injury in newborns. Am J Respir Cell Mol Biol 2015;52(5):611–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warren JL, Yabroff KR. Challenges and opportunities in measuring cancer recurrence in the United States. J Natl Cancer Inst 2015;107(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lucero OM, Dawson DW, Moon RT, Chien AJ. A re-evaluation of the “oncogenic” nature of Wnt/beta-catenin signaling in melanoma and other cancers. Curr Oncol Rep 2010;12(5):314–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017;31(4):591–606 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol 2005;25(24):10875–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koo CY, Giacomini C, Reyes-Corral M, Olmos Y, Tavares IA, Marson CM, et al. Targeting TAO Kinases Using a New Inhibitor Compound Delays Mitosis and Induces Mitotic Cell Death in Centrosome Amplified Breast Cancer Cells. Mol Cancer Ther 2017;16(11):2410–21. [DOI] [PubMed] [Google Scholar]
- 43.Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev 2004;18(7):830–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang I-C, Chen Y-J, Hughes D, Petrovic V, Major ML, Park HJ, et al. Forkhead Box M1 Regulates the Transcriptional Network of Genes Essential for Mitotic Progression and Genes Encoding the SCF (Skp2-Cks1) Ubiquitin Ligase. Mol Cell Biol 2005;25(24):10875–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Qiu W, Liu B, Yao R, Liu S, Yao Y, et al. Forkhead box transcription factor 1 expression in gastric cancer: FOXM1 is a poor prognostic factor and mediates resistance to docetaxel. J Transl Med 2013;11(1):204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mermod M, Hiou-Feige A, Bovay E, Roh V, Sponarova J, Bongiovanni M, et al. Mouse model of postsurgical primary tumor recurrence and regional lymph node metastasis progression in HPV-related head and neck cancer. Int J Cancer 2018;142(12):2518–28. [DOI] [PubMed] [Google Scholar]
- 47.Xu N, Wu SD, Wang H, Wang Q, Bai CX. Involvement of FoxM1 in non-small cell lung cancer recurrence. Asian Pac J Cancer Prev 2012;13(9):4739–43. [DOI] [PubMed] [Google Scholar]
- 48.Islam F, Gopalan V, Smith RA, Lam AK. Translational potential of cancer stem cells: A review of the detection of cancer stem cells and their roles in cancer recurrence and cancer treatment. Exp Cell Res 2015;335(1):135–47. [DOI] [PubMed] [Google Scholar]
- 49.Mirza MK, Sun Y, Zhao YD, Potula HH, Frey RS, Vogel SM, et al. FoxM1 regulates re-annealing of endothelial adherens junctions through transcriptional control of beta-catenin expression. J Exp Med 2010;207(8):1675–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.