Abstract
Actin-binding protein Anillin plays a pivotal role in regulating cytokinesis during the cell cycle, and involves in tumorigenesis and progress. However, the exact regulation mechanism of Anillin in human hepatocellular carcinoma (HCC) remains largely unknown. In this study, we examined and verified the anomalous high expression of Anillin in both HCC patients' specimens and HCC cell lines. High expression of Anillin is associated with dismal clinicopathologic features of HCC patients and poor prognosis. We conducted loss-of and gain-of function studies in HCC Hep3B cells. Anillin presented a significantly facilitating effect on cell proliferation in vitro and induced remarkable tumor growth in vivo. We found that the over-expression of Anillin was driven by a potential axis of miR-138/SOX4. Transcription factor SOX4 presented a high expression profile positive correlated with Anillin, and ChIP assay validated the interaction between SOX4 and the specific sequence of the promoter region of Anillin gene. While, we verified miR-138 as an upstream regulator of SOX4, which is abrogated in HCC cells and exerts degenerating effect on SOX4 mRNA. In our conclusion, Anillin facilitates the cell proliferation and enhances tumor growth of HCC, and is modulated by miR-138/SOX4 axis which regulates the transcriptional activity of Anillin. Findings above demonstrate us a probable axis for HCC diagnosis and treatment.
Summary of the main point
Anillin facilitates the cell proliferation and enhances tumor growth in HCC. The transcriptional activity of Anillin is modulated by miR-138/SOX4 axis. Findings above demonstrate us a probable axis for HCC diagnosis and treatment.
Introduction
As one of the devastating human malignancies, hepatocellular carcinoma (HCC) ranks the sixth most common tumors and causes the second cancer related mortality worldwide in counting over 750,000 people died from it per year [1,2]. Despite of the innovative methods and improvements on HCC prevention, diagnosis and treatment strategies, the multi-focal lesions when diagnosed, with metastasis and high rate of recurrence lead to dismal outcome of HCC patients generally [3].
Cytokinesis is the final step of cell mitosis that generates two daughter cells from one parental cell [4]. Proper cytokinesis ensures the stabilization of genome and the cell proliferation regularly, while the defection of cytokinesis could induce tumorigenesis in different ways of chromosomal instability [5]. According to recent reports, anomalous hyperactivity of cytokinesis is contributing to enhance the proliferation of HCC cells, which promotes the progression of HCC [6]. Thus, interference with cytokinesis provides the researchers a probable strategy against HCC tumor progression [7]. However, there involves quite a lot of medium and regulators in the process of mitosis and cytokinesis, and it is a rigorous challenge to discover a proper gene among them as an effective and safety target.
Anillin is an actin-binding protein works as kind of critical scaffold intracellular, organizing and maintaining the actomyosin contractile rings necessary for cytokinesis [8]. Structurally and functionally, the N-terminus of Anillin binds to myosin and F-actin, while the C-terminus of Anillin respectively binds to RhoA through its anillin homology (AH) domain and recruit of anillin to the equatorial membrane through its pleckstrin homology (PH) [9]. The above understanding of Anillin illustrates that Anillin plays a role as the hub of the mid-zone membrane regulation and of the cytokinesis modulation [10]. Knock-down of Anillin will lead to exact failure of cytokinesis, and might be one of the innovative approaches contributing for anti-tumorigenesis and progression [11]. However, the specific regulation upstream Anillin is still obscure, and we believe that the intensive understanding of the corresponding mechanism will contribute to the research and clinical translational study on HCC.
In this study, we validated the high expression of Anillin in both HCC tumor samples and HCC cell lines. Analysis of the clinicopathologic features illustrated the significant correlation between increase in Anillin from tumor tissues and unsatisfying clinical parameters, including larger tumor dimension, advanced TNM stages, microsatellite formation occurrence and liver cirrhosis. The conduction of loss-of function and gain-of function study either in vivo or in vitro demonstrates that high Anillin facilitates HCC cell proliferation, and promotes tumor growth in xenograft mouse models. On basis of this, we further explored the upstream transcription factor of Anillin, which induces transcriptional activity of Anillin gene in HCC cells. Combining with the prediction and validation, SRY-Box Transcription Factor 4 (SOX4) was screened out as a positive transcription factor effectively activating Anillin transcription. Intriguingly, we noticed microRNA-138 (miR-138), which was lowly expressed in HCC cells, presents a negative regulation on SOX4, abrogating the regulation of SOX4 on Anillin. Our findings indicate that Anillin is regulated via miR-138/SOX4 axis in HCC cells, and exerts pivotal roles in enhancing HCC tumor growth. Targeting Anillin and miR-138/SOX4 is hopeful to provide innovative strategy for HCC treatment.
Materials and methods
Cell culture
Three HCC cell lines (Huh7, HepG2 and Hep3B) used in this study were purchased, and the normal human hepatic cell L02 was used as control (Shanghai Institutes for Biological Sciences, Chinese Academy of Science, Shanghai, China). All Cells in our study, were cultured in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS), incubated at the environment temperature of 37 °C, with 100 μg/ml streptomycin and 100 U/ml Penicillin in a humidified cell and an atmosphere of 5% CO2. Specifically, for the transfected cells, medium mixed with G418 (Santa Cruz Biotechnology, Inc; 400 μg/ml) was used for selection.
Pathological specimens
Eighty-seven tumor specimens paired with the adjacent non-cancerous tissues were collected from the radical resection samples from the HCC patients without preoperative treatment during 2014–2017, at the Department of Hepatobiliary Surgery, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. Clinicopathologic features of the patients including gender, age, tumor size, number of lesions, grades et.al were collected. Informed consent was obtained and the study was approved by the Ethics Committee of Ruijin Hospital, Shanghai Jiaotong University School of Medicine.
RT-qPCR assay, Western blot analysis and immunohistochemistry assay
Tissue and cell RNA isolation for the Real-time quantitative polymerase chain reaction (RT-qPCR) were carried out following the instruction of TRIzol reagent (Invitrogen, USA). The first-strand cDNA was synthesized using High-Capacity cDNA Reverse Transcription Kit (ABI, USA). RT-primers for the target mRNAs were synthesized by Sangon Biotech Company (Shanghai, China) (Suppl. Table. 1). The RT-qPCR was conducted according to the TaqMan Gene Expression Assays protocol (ABI, USA).
Antibodies against Anillin and SOX4 and the other related proteins were purchased (Abcam, USA) for the using of the Western blot analysis and immunohistochemistry assay. The methods for the assays above were performed as the same protocol described in we previously research [12]. Especially, the protein levels obtained from IHC were assigned to two experienced pathologists and examined in blind, then set into two groups as staining intensity graded subjectively: no to low staining (0–1+) and moderate to high staining (2 + ~3+).
Plasmid construction and transfection
Hep3B cells in exponential phase were prepared and transfected with shRNA suppressing Anillin through pGU6/Neo vectors (GenePharma, Shanghai, China) along with the construction of the control ones. Transfected cells were selected using medium mixed with G418 (Santa Cruz Biotechnology, Inc; 400 μg/ml). Hep3B cells over-expressing miR-138 (Hep3B/miR-138) was constructed through the mimic method similar to the description in our previous study, and the negative control were set as NigmiR. The lentiviral vector pWPXL (Addgene, Cambridge, USA) was introduced (pWPXL-SOX4) for upregulating SOX4 in cells or reversing the SOX4 abrogation induced by over-expressing miR-138, and the negative control were set as pWPXL-Null. Similar method in using lentivirus was applied in re-introducing Anillin into Hep3B cells.
Cell proliferation assay and cell cycle analysis
The treated Hep3B cells (1 × 106) were cultured in 96-well microtiter plates in triplicate and incubated for 5 days with an atmosphere of 5% CO2 and 37 °C. Microplate computer software (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was applied measuring the OD following the Cell Counting Kit-8 (CCK-8) assay kit protocol (Dojindo, Tokyo, Japan). The cell proliferation curves were plotted.
Cells were treated in steps with ethanol fixation, RNase A treatment and propidium iodide staining. Flow cytometry detection was conducted by using FACSCalibur (Becton-Dickinson, Franklin Lakes, NJ, USA) for quantifying cell populations at the G0/G1, S and G2/M phases, and ModFit software e(Becton-Dickinson) was used. The debris and fixation artifacts of the cells were xcluded.
Cell apoptosis analysis
Cell apoptosis rate was calculated by using PE-Annexin V Apoptosis Detection Kit I (BD Pharmingen, USA) following the product instructions. Stable transfected cells were resuspended in 1× Binding Buffer in the concentration of 1 × 106 cells/ml. 5 μl of FITC and 5 μl of PI were added into 100 μl of the cell suspension, followed by a 15 min incubation in darkness, added with 400 μl × Binding Buffer. The analysis of apoptosis rate was conducted by flow cytometry (Becton Dickinson, USA). Both Annexin V-FITC-positive and PI-negative cells were considered as the apoptosis cells.
Orthotopic transplantation of mouse liver
Four- to five-week-old BALB/c nude male mice (Institute of Zoology Chinese Academy of Sciences) were subscribed and housed in the Animal Laboratory Unit, Shanghai Jiao Tong University School of Medicine, China, within the pathogen-free environment. Experiment on animal models was performed in accordance with the guidelines of the Shanghai Medical Experimental Animal Care Commission. HCC cells were suspended in 25 μl serum-free DMEM mixed with 25 μl Matrigel (1:1, v/v) every 1 × 106 cell. Cells above were orthotopic ally injected into hepatic lobes of each mouse. All mice were sacrificed in 6 weeks post-injection. And the weight of all the xenografted livers was measured. For histological analysis, liver and lung from mice were collected, stained with hematoxylin and eosin (HE), and detected by two experienced pathologists independently.
Dual-luciferase reporter assay
Prediction by using online tool of microcosm (http://mirecords.biolead.org) indicated miR-138 as a potential upstream regulator of SOX4 mRNA via directly interaction. A 202 bp sequence containing putative binding site of miR-138 was selected from the 3′-UTR of SOX mRNA, set along with the mutative sequence (Suppl. Table 2). Either of the designed sequences was cloned into the pMIR-REPORT luciferase vector containing firefly luciferase, and the pRL-TK vector containing renilla luciferase was used as control (Promega, Madison, WI, USA). The two vectors above were co-transfected into Hep3B cells overexpressing miR-138 and the control ones. The luciferase activity was measured through Dual-Glo Luciferase assay system (Promega) 48 h later after the transfection.
On the other hand, the Dual-luciferase reporter assay was also used for detecting the influence of miR-138 on regulating the transcription factor activity of SOX4. In this study, pGL-3 vectors (Promega, USA) were used to construct pGL3-Anillin, which contains the promoter sequence of Anillin gene. And pGL3-basic was set for negative control. These vectors were transfected into Hep3B cells, either treated by miR-138 mimics or treated by the control. And the transcriptional factor activity of SOX4 was detected through the Dual-luciferase reporter method.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was applied to investigate the binding interaction between the promoter region of Anillin genes and the transcription factor SOX4. A total of 5 × 106 cells were cultured in each 10 cm dish and subjected to the protocol of ChIP by using ChIP-ITTM Kit (Active Motif). Chromatin was immune-precipitated with 2 μg of either SOX4 antibody (Abcam,USA) or IgG as the negative control. The extracted DNA followed was then analyzed through PCR and RT-qPCR by introducing the relative primers (Suppl. Table 1) for amplification of the fragment including the promoter sequence of Anillin gene.
Statistical analysis
Statistical analysis was carried out by using SPSS 20.0. P-values were calculated using an unpaired Student's t-test and Fisher's exact test. Differences were considered statistically significant at P-values <0.05.
Results
Anillin is anomalously expressed in HCC tissues and cells
Before initiating the exploration in tissues and cell lines, the expression profile of Anillin in pan-cancer and multiple human cancer cell lines was respective investigated through analyzing the Cancer Genome Atlas (TCGA) database and Cancer Cell Line encyclopedia (CCLE) database. Basically, Anillin shows a common high expression in most of the solid human malignancies, including HCC, compared with the normal tissues; Consistently, Anillin presents high expression level in various cancer cell lines, including 29 HCC cell lines (Suppl. Fig. 1).
According to the findings above, we further investigated the exact expression status of Anillin in 87 paired HCC tissues along with their adjacent liver tissues. The cases were divided into two groups on basis of the expressing intensity of Anillin, Anillin high expressed group and low expressed group. For the tumor tissues, we observed 67.8% (59/87) of the cases showing highly expressed Anillin, and the rest 32.2% (28/87) ones with lower Anillin expression. On contrast, in non-cancerous tissues high expression of Anillin was observed only in a small portion of the cases, counted 25.3% (22/87); And the other 74.7% (65/87) cases present extremely low Anillin expression compared with the tumor tissues (Fig. 1A, B).
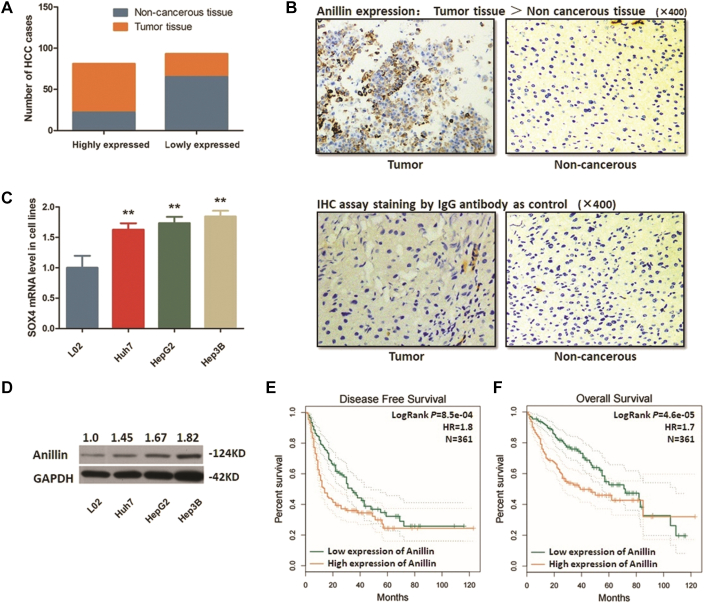
Fig. 1.
Anillin is anomalously expressed in HCC tissues and cells.
A. Statistic of number of cases with higher or lower expression of Anillin in HCC specimens. Anillin was up-regulated in most of the tumor tissues (59/87), and was expressed at a lower level in most of the adjacent non-cancerous tissues (22/87) (P < 0.01). B. Representative graph of immunohistochemistry analysis (400×) of the HCC cases. Specimens stained IgG anti-body were regarded as control. Anillin expression in tumor specimens was significantly higher than in adjacent non-cancerous tissues. C. RT-qPCR assay demonstrated a significantly high expression of Anillin mRNA in three HCC cell lines than the control L02 cells (**P < 0.01). D. The Western blot analysis demonstrated an obviously high expression of Anillin protein in three HCC cell lines than the control L02 cells (P < 0.01). E. Disease free survival (DFS) analysis according to HCC patients' follow-up information was presented by Kaplan-Meier plot. High Anillin expression in HCC tissue is correlated poor OS (361 cases, P = 8.5e-04). F. Overall survival (OS) analysis according to HCC patients' follow-up information was presented by Kaplan-Meier plot. High Anillin expression in HCC tissue is correlated poor OS (361 cases, P = 4.6e-05).
In consistent with the observation in tumor tissues, we selected 3 HCC cell lines (Huh7, HepG2 and Hep3B) for detection compared with the control L02 cells, and found that Anillin was remarkably highly expressed in all of the three HCC cells, at both mRNA and protein stages. Fig. 1C, D shows the results obtained from RT-qPCR assay and the Western blot analysis.
High expression of Anillin correlates with dismal clinicopathological features of the HCC patients
We reviewed and analyzed the disease free survival (DFS) and the over-all survival (OS) datasets of a 361 cases cohort of the HCC patients. As shown in Fig. 1E, F, higher Anillin expression is significantly correlated with poor prognosis and high mortality, and the DFS rate (logRank P = 8.5e-04, HR = 1.8) and OS rate (logRank P = 4.6e-04, HR = 1.7) were remarkably dismal in the patients with high Anillin levels.
The correlationship between Anillin and the clinicopathological features of those 87 HCC patients was calculated as Table 1 shown. We observed no significant correlation between Anillin and the age, gender, Alpha-fetoprotein (AFP) level or virus control status. Yet, a significant positive trend towards larger tumor size, advanced TNM stages, more incidence of tumor microsatellite formation, venous invasion and liver cirrhosis was discovered in patients with highly expressed Anillin in tumor tissues.
Table 1.
Correlation between Anillin transcript and clinicopathologic features in 87 HCC specimens.
Anillin transcript level associated with clinicopathologic features in 87 HCC patients, including age, gender, tumor size, tumor stage (AJCC), tumor encapsulation, tumor microsatellite formation, vein invasion, HBsAg status, AFP level, and liver cirrhosis. Statistically significance was assessed by Fish's exact text.
| Clinicopathologic parameters | Anillin expression |
P* | |
|---|---|---|---|
| Low (n = 28) | High (n = 59) | ||
| Age (years) | |||
| ≤50 | 13 | 33 | 0.492 |
| >50 | 15 | 26 | |
| Gender | |||
| Male | 18 | 28 | 0.172 |
| Female | 10 | 31 | |
| Diameter (cm) | |||
| ≤5 | 19 | 24 | 0.0225 |
| >5 | 9 | 35 | |
| TNM stage | |||
| I–II | 16 | 16 | 0.0164 |
| III–IV | 12 | 41 | |
| Tumor encapsulation | |||
| Absent | 15 | 21 | 0.162 |
| Present | 13 | 38 | |
| Tumor microsatellite formation | |||
| Absent | 16 | 21 | 0.067 |
| Present | 12 | 38 | |
| Venous invasion | |||
| No | 14 | 14 | 0.026 |
| Yes | 14 | 45 | |
| HBsAg | |||
| Negative | 4 | 7 | 0.741 |
| Positve | 24 | 52 | |
| AFP (ng/ml) | |||
| ≤400 | 9 | 11 | 0.181 |
| >400 | 19 | 48 | |
| Cirrhosis | |||
| Absent | 7 | 3 | 0.011 |
| Present | 21 | 56 | |
Knock-down Anillin blocks cell proliferation of Hep3B cells, induces cell apoptosis and suppresses tumor growth in orthotopic xenograft mouse model
Among the three HCC cell lines, Hep3B cell shows the highest Anillin expression level. Thus, we knock-down Anillin in Hep3B cells for the loss-of function study by using pGU6/Neo vector containing shRNA. The significant decrease of Anillin was validated as Fig. 2A shown.
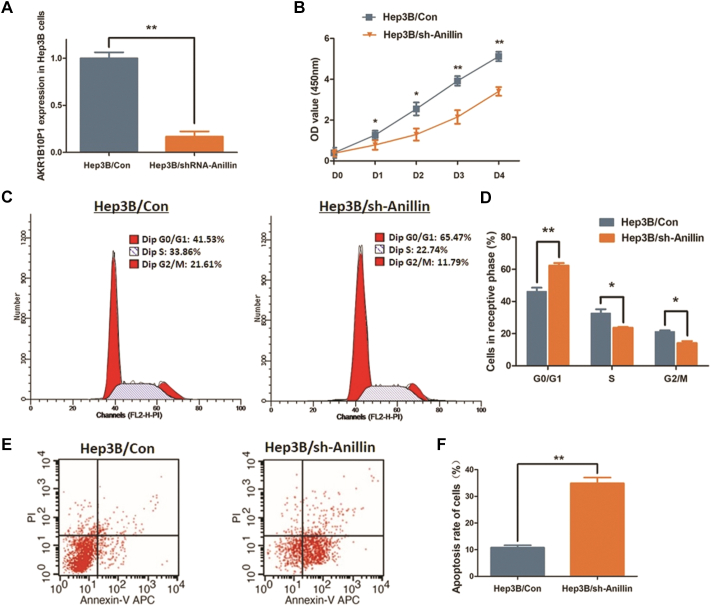
Fig. 2.
Knock-down Anillin blocks cell proliferation of Hep3B cells and induces cell apoptosis.
A. Anillin was knocked-down in Hep3B cells via shRNA transfection. RT-qPCR assay indicated a significant decrease of Anillin expression in the treated cells (**P < 0.01). B. CCK8 assay was applied for investigating the cell proliferation ability. Cell proliferation in Hep3b cells was significantly impaired after Anillin knock-down (*P < 0.05, **P < 0.01). C. The representative histograms describe the cell cycle profiles of Hep3B cells by using flow cytometry. D. The cell cycle of Hep3B cells was arrested in G0/G1 phase after Anillin knock-down. The results are means of three independent experiments±SD. (*P < 0.05, **P < 0.01). E. The representative histograms describing cell apoptosis profile in Hep3B cells through flow cytometry. F. The apoptosis rate of Hep3B cells was significantly increased from 10.79% to 34.89% (**P < 0.01) through knocking-down AKR1B10P1. The results are means of three independent experiments ±SD. (**P < 0.01).
As expected, knock-down Anillin impaired the proliferation of Hep3B cells, which was verified through CCK8 assay. Respective, the P-value was <0.05 for days 1–2, and <0.01 for days 3–4 (Fig. 2B). The percentage of Hep3B cells maintaining at G0/G1 phase was increased from 46.19% to 62.28% (P < 0.01). Simultaneously, the S phase cells and the G2/M phase cells were respectively declined from 32.63% to 23.62% (P < 0.05) and from 21.15% to 14.09% (P < 0.05) (Fig. 2C, D).
Simultaneously, the flow cytometric analysis was conducted to learn the influence of Anillin on cell apoptosis. In Fig. 2E, F, the graphics show an effective induction of cell apoptosis in Hep3B cells through knock-down Anillin, with the apoptosis rate raised from 10.79% to 34.89% in average (P < 0.01).
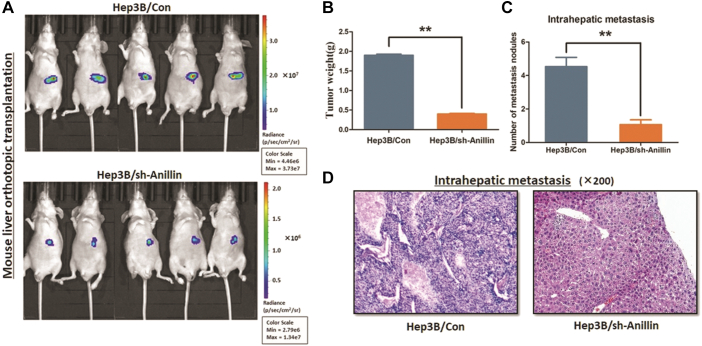
Xenograft mouse models were studied by injecting Hep3B cells treated with Anillin knock-down or the control ones. The quantification of the orthotopic transplanted masses in liver were measured 6 weeks post-orthotopic, and the result demonstrated smaller tumor masses generated in mouse livers when Anillin was knocked-down (Fig. 3A, C). Meanwhile, the examination under HE staining indicated that by knocking-down Anillin, less intrahepatic metastasis was generated in the mice models than those control ones (Fig. 3B, D).
Fig. 3.
Knock-down Anillin suppresses HCC growth in orthotopic xenograft mouse model.
Mouse liver orthotopic transplantation model was constructed in four- to five-week-old male BALB/c nude mice. A. The orthotopic tumor growth was significantly suppressed in the mice treated with Anillin knock-down Hep3B cells. B. Anillin knock-down Hep3b cells induced significant decrease of the xenograft tumor weight in vivo (**P < 0.01). C–D. The xenograft tumor tissue was checked under HE staining examination. AKR1B10P1 depleted Hep3B cells induced less intrahepatic metastasis lesion and lung metastasis in mice compared with the control Hep3B cells (**P < 0.01).
These findings indicate that Anillin exerts pivotal effects on tumor growth both in vitro and in vivo, and this gene might be a probable target for intensive research and investigation on its upstream regulation.
Anillin gene transcription is activated by SOX4 in HCC cells
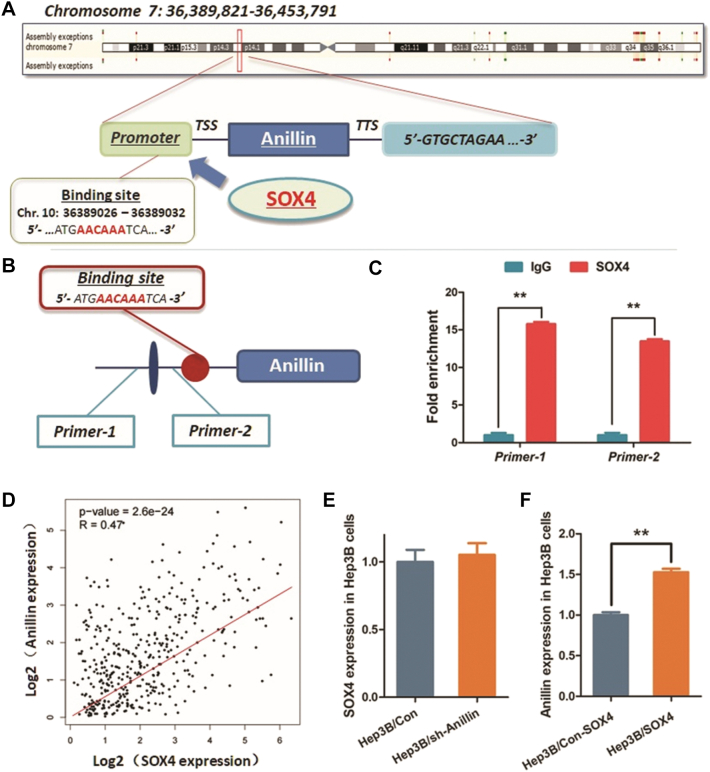
We collected a 3000 bp fragment of the promoter sequence in Anillin gene for predicting the probable binding site for transcription factor activating Anillin gene transcription. According the results comprehensively evaluated through both Database of Human Transcription Factor Targets (http://bioinfo.life.hust.edu.cn/hTFtarget#!/) and Gene-Cloud of Biotechnology Information (GCBI, https://www.gcbi.com.cn), we noticed that SOX4 is a potential transcription factor binding to Anillin gene promoter region at the site with a specific binding sequence (5′-AACAAA-3′, Chr. 7: 36389026 to 36,389,032) (Fig. 4A). The exact interaction between SOX4 and Anillin promoter region was then validated by carrying out ChIP assay (Fig. 4B, C).
Fig. 4.
Anillin gene transcription is activated by SOX4 in HCC cells.
A. SOX4 is a potential transcription factor binds to the promoter region of Anillin gene (5′-AACAAA-3′, Chr. 7: 36389026 to 36389032). B–C. ChIP assay was conducted to investigate the interaction between SOX4 and the promoter region of Anillin gene. IgG was used as the negative control. D. Analysis on the data from TCGA liver cancer database demonstrates a significant positive relationship between SOX4 and Anillin expression (P = 2.6e-24). E. No significant expression change of SOX4 was observed in Hep3B cells treated by knocking down Anillin. F. Up-regulation of SOX4 in these Hep3B cells through pWPXL-SOX4 lentivirus vector increased the mRNA expression of Anillin (**P < 0.01).
The expression profile of SOX4 was verified up-regulated in either HCC tissues or cell lines (Suppl. Fig. 2). According the findings above, we further investigated the exact transcriptional activating effect of SOX4 on Anillin. As Fig. 4D shown, the data obtained from TCGA liver cancer database demonstrate a significant positive relationship between SOX4 and Anillin expression. Simultaneously, in this study, we did not observe the significant expression change of SOX4 in Hep3B cells treated by knocking down Anillin. However, up-regulation of SOX4 in these Hep3B cells through pWPXL-SOX4 lentivirus vector increased the mRNA expression of Anillin (Fig. 4E, F). Results above indicate that the SOX4 directly activates the transcription of Anillin gene in HCC process.
MiR-138 negatively regulates SOX4 expression in HCC cells
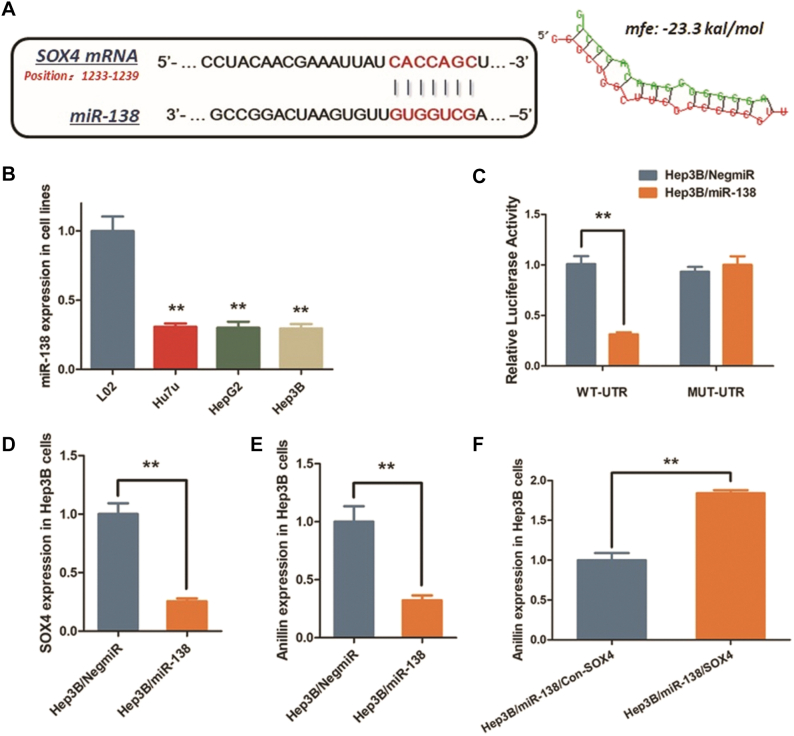
By using microcosm online software (https://www.microcosm.com/), we predicted and noticed miR-138 as a post-transcriptional regulator upstream SOX4, and miR-138 was supposed to directly interact with the 3′-untranslated region (3′-UTR) of SOX4 mRNA (Fig. 5A). Considering the up-regulation of SOX4 in HCC, we further measured the expression of miR-138 in HCC cells. As we observed, miR-138 expression was tremendously decreased in all of the three HCC cell lines, compared with the control L02 cells (Fig. 5B). The vectors containing a 202 bp 3′-UTR fragment of SOX4 mRNA (WT-UTR) and the control luciferase vectors containing the mutated sequence (MUT-UTR) were constructed. And then the Dual-luciferase reporter assay was conducted. We utilized mimics for cell transfection and up-regulated miR-138 in Hep3B cells. Consistent with our prediction, miR-138 up-regulation in Hep3B cells (Hep3B/miR-138) significantly declined the luciferase signal of SOX4/pMIR/WT, compared with the negative control (Hep3B/NigmiR). The signal suppressing effect induced by miR-138 was abrogated in Hep3B cells transfected with mutated binding sequence, along with a sequential decrease of SOX4 mRNA expression through over-expressing miR-138 (Fig. 5C). Results above demonstrate that miR-138 directly binds to SOX4 mRNA and degenerates the target transcript.
Fig. 5.
MiR-138 negatively regulates SOX4 expression in Hep3B cells via binding the 3′-UTR of SOX4 mRNA.
A. MiR-138 was predicted as the up-stream regulator of SOX4 in Hep3B cells by interacting with the 3′-UTR of SOX4 mRNA. The minimum free energy (Mfe) hybridization is calculated as: −23.3kal/mol. B. MiR-138 expression was decreased in Hep3B cell lines compared with L02 cells (**P < 0.01). C. The direct interaction between SOX4 and miR-138 was detected by Dual-luciferase reporter assay. Ectopic expression of miR-138 in Hep3B cells (Hep3B/miR-138) significantly declined the luciferase signal of wildtype binding sequence, compared with the negative control (Hep3B/NigmiR). The signal suppression induced by miR-138 was defected in Hep3B cells transfected with mutated binding sequence (**P < 0.01). D–E. RT-qPCR assay demonstrated that both SOX4 and Anillin mRNA expression in Hep3B cells transfected with miR-138 mimics was significantly decreased (**P < 0.01). F. The decrease of Anillin induced by miR-138 was reversed in Hep3B cells when SOX4 was re-introduced as a rescuing treatment (**P < 0.01).
MiR-138/SOX4 axis modulates Anillin in HCC cells
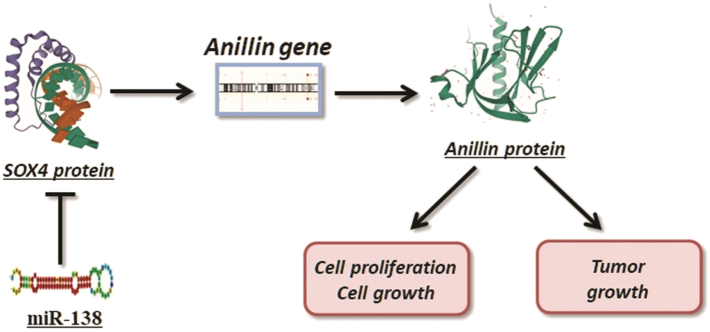
Since miR-138 was involved in SOX4 mRNA degeneration, we supposed that Anillin is regulated through the axis of miR-138/SOX4 in HCC cells. Both SOX4 and Anillin mRNA expression in Hep3B cells transfected with miR-138 mimics was significantly decreased (Fig. 5D, E). Further, we conducted the rescue experiment by re-introducing SOX4 into Hep3B cells. As supposed, the decrease of Anillin induced by up-regulation of miR-138 was consequentially reversed in Hep3B cells when SOX4 was re-introduced (Fig. 5F). Thus, we concluded the regulating axis of miR-138/SOX4/Anillin involved with HCC tumor growth for alternative therapeutic targets. And Fig. 6 briefly describes the regulation process of this axis in HCC cells.
Fig. 6.
MiR-138/SOX4 axis modulates Anillin in HCC cells.
The summary of the regulational axis of miR-138/SOX4/Anillin involved with HCC tumor growth for alternative therapeutic targets.
Discussion
HCC shows extremely aggressive characteristics in tumor growth, invasiveness and metastasis, and leads to poor outcome of the patients along with high recurrence and mortality [13,14]. As acknowledged, cell proliferation is one of the vital indicators for measuring and evaluating the cell viability, which reflects the specific ability of growth, adaption and tumor development in human malignancies [15,16]. However, the intensive research on the mechanism promoting HCC cell growth and tumor development remains largely unknown.
Concerning with the cell proliferation process, cytokinesis is essential for cell division and complete mitosis [17]. Theoretically, deliberate induction to fail cytokinesis impairs the cell proliferation in tumor progress, which might be a potential approach for tumor control. While, cautious consideration should be taken for the probable detrimental impact on cytokinesis failure of normal tissues, and there has no sufficient evidence supporting the safety and universal effects of this assumed treatment strategy yet.
Notably, as the largest digestive organ in human being composed of tremendous amount of functional cells and undertaking complex physiological functions, liver provides unique micro-environment and characteristics as a hopeful target organ to practice the therapeutic treatment concerning with cytokinesis modulation. Accumulating evidence has revealed that cytokinesis failure is one of the common cellular events incidents in normal hepatocytes, and results in the generation of polyploid hepatocytes [18,19]. According to the published literatures, polyploid cells were respectively counted up to 90% and 40% hepatocytes of mouse and human liver cells [20]. And recently, Zhu et al. [21,22] suggested that the polyploid hepatocytes generated from cytokinesis failure could reduce HCC development in mouse models without adverse reactions including degeneration of either cell functions and liver regeneration. Information above prompts the possibility to interfere HCC progression and development via regulating cytokinesis.
The process of cytokinesis involves multiple regulators and effectors. In briefly, cytokinesis results in the cell division from one individual parental cell into two daughter cells, under the modulation of constricting force generated by the contractile ring. The contractile ring is structured with filamentous actin and motor protein myosin. And Anillin, a phylogenetically conserved protein, is the essential component that plays the scaffold protein role and links the actomyosin ring to the membrane and RhoA, which is a pivotal effector directing the localization of the contractile actomyosin ring [23]. In other words, dysfunction of Anillin could impair the efficient integration of RhoA functions and leads to failure of cytokinesis [24].
Considering that hepatocytes are unique functional units perfectly adapted the cell status of polyploidy induced by cytokinesis failure of no obvious adverse effects, we suppose that specific organ targeting treatment for hepatocyte would be an innovative strategy to interfere liver tumor development. According to the review of published literatures, we found that Anillin has been reported as an effective driver of cell proliferation in multiple human malignancies, including breast, ovarian, kidney, colorectal cancer, and even liver cancer [25,26]. Thus, we supposed that Anillin might be the potential target for clinical practical using.
In our study, we found that Anillin exactly presents a remarkable up-regulated expression in HCC tissues and cell lines. We collected the follow-up information and the clinicopathologic parameters of the patients, and found a significant positive correlationship between high Anillin expression in tumor tissues and poor DFS and OS rate. While, patients with anomalous up-regulation of Anillin in tumor were declined to have larger tumor size, higher serum Alpha-fetoprotein generated by the tumor cells, microsatellite formation occurrence and liver cirrhosis, which are all decisive indicators for advanced TNM stages of HCC. Impressively, these findings strongly prompt Anillin as an indicator of HCC tumor growth.
Then, we conducted the loss-of and Gain-of function experiment to investigate the bio-functional changes via modulating Anillin. Obviously, knock-down Anillin significantly impaired the cell proliferation of HCC cells, arresting a large portion of the Hep3B cells in the G0/G1 stage of cell cycle. Apoptosis rate of Hep3B cells was remarkably raised when Anillin was knocked down, which indicated an anti-apoptosis function of Anillin in HCC cells. The xenograft further illustrated that loss of Anillin in Hep3B cells led to smaller tumor generation and delayed tumor growth. Moreover, the metastasis lesions in mice models treated with Anillin knocked-down cells were counted significantly less than the control ones. Thus, we confidently supposed that Anillin acts as an enhancer of HCC development.
Considering the promoting effect of Anillin on HCC growth, we wondered the up-streaming modulation of Anillin in HCC cells. As described, miR-138/SOX4 axis was pointed out in our study that presents a critical regulating function in Anillin expression. The first step led to discover this axis was to investigate the transcriptional regulation of Anillin gene. According to the learning of the sequence intercepted from the promoter of Anillin gene, quit a lot of transcription factors potentially functioning in HCC cells were predicted out. We mining the datasets for discovering the most hopeful candidates that have to share the similar expression profile with Anillin and could exactly bind to the promoter region of Anillin gene. SOX4 was then being noticed since its extremely significant positive expression relationship along with Anillin.
SOX4 is known as an essential transcription factor involves in the regulation of cell stemness, differentiation and progenitors development [27]. Consistent with the exclusive functions above, SOX4 participates in multiple pathways concerning with tumorigenesis and progress, including Wnt, TGF-β and PI3K signaling, and has been identified as one of the oncogenes amplified in different cancers like breast cancer, ovarian cancer and lung cancer [28,29]. Report on the exact interaction or relationship between SOX4 and Anillin is limited. In this study, we confirmed the significant positive correlation between SOX4 and Anillin expression in Hep3B cells again. And intriguingly, SOX4 definitely did not show any significant change after Anillin knock-down. And the ChIP assay provided the evidence of the direct interaction between SOX4 and the particular sequence located at the promoter of Anillin gene mentioned above. All the findings and the later rescue experiment then demonstrate an activating effect of the transcription factor SOX4 on the HCC growth enhancing Anillin gene.
Lastly, by combining with the prediction through online tools and the following Dual-luciferase reporter assay, we had screened miR-138 out as a noticeable up-streaming regulator of SOX4, which could directly bind to the 3′-UTR of SOX4 mRNA and degenerate the transcripts. As known, miR-138 is a tumor suppressor through post-transcriptional reported in different human malignancies, such as bladder cancer, gastric cancer, colorectal cancer [[30], [31], [32]]. It can degenerate the targeting mRNA by directly binding to the 3′-UTR and results in decrease of targeting gene expression. In our study, we investigated the exact expression of miR-138 in HCC cells, and observed an extremely low expression of miR-138. To our observation, the up-regulation of miR-138 in Hep3B consequentially induced an obvious decrease of Anillin, and the rescue experiment by re-introducing SOX4, the potential transcription factor of Anillin, successfully reverted the abrogation of Anillin impacted by miR-138. Therefore, this finding strongly supported the modulating effect of miR-138/SOX4 axis on Anillin.
Comprehensively, Anillin enhances HCC cell proliferation and tumor growth. And we discovered an axis of miR-138/SOX4 regulating the transcriptional activity of Anillin through a post-transcription way in classic miRNA-mRNA degeneration way. Moreover, the study provides us a lot for further investigation and discussion, which is limited so far in understanding. For instance, the regulation upstream miR-138 is still unclear, and the effect of Anillin on HCC cell division remains to be intensively illustrated. In summary, our findings above hopefully provide us innovative strategies to control HCC development and new targets for therapeutic treatment.
The following are the supplementary data related to this article.
The primers for RT-qPCR assay.
Selected sequence of the predicted miR-138 binding site of SOX4 mRNA 3′-UTR, along with the relative mutated sequence.
Supplementary figures
Abbreviations
- HCC
hepatocellular carcinoma
- miRNA
microRNA
- miR-138
microRNA-138
- SOX4
Sex-determining region Y-related high-mobility group box 4
- RT-qPCR
quantitative real-time polymerase chain reaction
- shRNA
short hairpin RNA
- ChIP
Chromatin immunoprecipitation
- TCGA
Cancer Genome Atlas database
- CCLE
Cancer Cell Line encyclopedia database
- DFS
Disease free survival
- OS
Over-all survival
Declaration of competing interest
No potential conflicts of interest were disclosed.
Acknowledgements
The authors thank Xinping Ren, Lei Dong, Zhiyuan Wu for providing valuable technical supports and assistance.
This study was kindly supported by grants from the following: National Natural Science Foundation of China (No. 81602544); Shanghai Pujiang Talent Project (No. 18PJD029); Research Physician Project from Shanghai Jiao Tong University School of Medicine (20191901).
References
- 1.Vilgrain V., Pereira H., Assenat E., Guiu B., Ilonca A.D., Pageaux G.P., Sibert A., Bouattour M., Lebtahi R., Allaham W., Barraud H., Laurent V., Mathias E., Bronowicki J.P., Tasu J.P., Perdrisot R., Silvain C., Gerolami R., Mundler O., Seitz J.F., Vidal V., Aube C., Oberti F., Couturier O., Brenot-Rossi I., Raoul J.L., Sarran A., Costentin C., Itti E., Luciani A., Adam R., Lewin M., Samuel D., Ronot M., Dinut A., Castera L., Chatellier G., Group ST Efficacy and safety of selective internal radiotherapy with yttrium-90 resin microspheres compared with sorafenib in locally advanced and inoperable hepatocellular carcinoma (SARAH): an open-label randomised controlled phase 3 trial. Lancet Oncol. 2017;18:1624–1636. doi: 10.1016/S1470-2045(17)30683-6. [DOI] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Pinyol R., Montal R., Bassaganyas L., Sia D., Takayama T., Chau G.Y., Mazzaferro V., Roayaie S., Lee H.C., Kokudo N., Zhang Z., Torrecilla S., Moeini A., Rodriguez-Carunchio L., Gane E., Verslype C., Croitoru A.E., Cillo U., de la Mata M., Lupo L., Strasser S., Park J.W., Camps J., Sole M., Thung S.N., Villanueva A., Pena C., Meinhardt G., Bruix J., Llovet J.M. Molecular predictors of prevention of recurrence in HCC with sorafenib as adjuvant treatment and prognostic factors in the phase 3 STORM trial. Gut. 2019;68:1065–1075. doi: 10.1136/gutjnl-2018-316408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potapova T.A., Zhu J., Li R. Aneuploidy and chromosomal instability: a vicious cycle driving cellular evolution and cancer genome chaos. Cancer Metastasis Rev. 2013;32:377–389. doi: 10.1007/s10555-013-9436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGranahan N., Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168:613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 6.Yang X.M., Cao X.Y., He P., Li J., Feng M.X., Zhang Y.L., Zhang X.L., Wang Y.H., Yang Q., Zhu L., Nie H.Z., Jiang S.H., Tian G.A., Zhang X.X., Liu Q., Ji J., Zhu X., Xia Q., Zhang Z.G. Overexpression of Rac GTPase activating protein 1 contributes to proliferation of cancer cells by reducing hippo signaling to promote cytokinesis. Gastroenterology. 2018;155:1233–1249. doi: 10.1053/j.gastro.2018.07.010. (e1222) [DOI] [PubMed] [Google Scholar]
- 7.Desdouets C., Avila M.A. Inhibiting cytokinesis in the liver: a new way to reduce tumor development. Gastroenterology. 2018;154:1229–1231. doi: 10.1053/j.gastro.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Chougule A.B., Hastert M.C., Thomas J.H. Drak is required for actomyosin organization during Drosophila cellularization. G3 (Bethesda) 2016;6:819–828. doi: 10.1534/g3.115.026401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirashima T., Tanaka R., Yamaguchi M., Yoshida H. The ABD on the nascent polypeptide and PH domain are required for the precise Anillin localization in Drosophila syncytial blastoderm. Sci. Rep. 2018;8:12910. doi: 10.1038/s41598-018-31106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norden C., Mendoza M., Dobbelaere J., Kotwaliwale C.V., Biggins S., Barral Y. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell. 2006;125:85–98. doi: 10.1016/j.cell.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 11.Jananji S., Risi C., Lindamulage I.K.S., Picard L.P., Van Sciver R., Laflamme G., Albaghjati A., Hickson G.R.X., Kwok B.H., Galkin V.E. Multimodal and polymorphic interactions between Anillin and actin: their implications for cytokinesis. J. Mol. Biol. 2017;429:715–731. doi: 10.1016/j.jmb.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 12.Wang J., Zhou Y., Fei X., Chen X., Chen Y. Biostatistics mining associated method identifies AKR1B10 enhancing hepatocellular carcinoma cell growth and degenerated by miR-383-5p. Sci. Rep. 2018;8:11094. doi: 10.1038/s41598-018-29271-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong M.C., Jiang J.Y., Goggins W.B., Liang M., Fang Y., Fung F.D., Leung C., Wang H.H., Wong G.L., Wong V.W., Chan H.L. International incidence and mortality trends of liver cancer: a global profile. Sci. Rep. 2017;7:45846. doi: 10.1038/srep45846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Llovet J.M., Zucman-Rossi J., Pikarsky E., Sangro B., Schwartz M., Sherman M., Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 15.Goodlad R.A. Quantification of epithelial cell proliferation, cell dynamics, and cell kinetics in vivo. Wiley Interdiscip. Rev. Dev. Biol. 2017;6 doi: 10.1002/wdev.274. (10.1002) [DOI] [PubMed] [Google Scholar]
- 16.Wood C.E., Hukkanen R.R., Sura R., Jacobson-Kram D., Nolte T., Odin M., Cohen S.M. Scientific and Regulatory Policy Committee (SRPC) review: interpretation and use of cell proliferation data in Cancer risk assessment. Toxicol. Pathol. 2015;43:760–775. doi: 10.1177/0192623315576005. [DOI] [PubMed] [Google Scholar]
- 17.Nakayama Y., Soeda S., Ikeuchi M., Kakae K., Yamaguchi N. Cytokinesis failure leading to chromosome instability in v-Src-induced oncogenesis. Int. J. Mol. Sci. 2017;18:811–823. doi: 10.3390/ijms18040811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hognas G., Tuomi S., Veltel S., Mattila E., Murumagi A., Edgren H., Kallioniemi O., Ivaska J. Cytokinesis failure due to derailed integrin traffic induces aneuploidy and oncogenic transformation in vitro and in vivo. Oncogene. 2012;31:3597–3606. doi: 10.1038/onc.2011.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv L., Zhang T., Yi Q., Huang Y., Wang Z., Hou H., Zhang H., Zheng W., Hao Q., Guo Z., Cooke H.J., Shi Q. Tetraploid cells from cytokinesis failure induce aneuploidy and spontaneous transformation of mouse ovarian surface epithelial cells. Cell Cycle. 2012;11:2864–2875. doi: 10.4161/cc.21196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bou-Nader M., Caruso S., Donne R., Celton-Morizur S., Calderaro J., Gentric G., Cadoux M., L'Hermitte A., Klein C., Guilbert T., Albuquerque M., Couchy G., Paradis V., Couty J.P., Zucman-Rossi J., Desdouets C. Polyploidy spectrum: a new marker in HCC classification. Gut. 2020;69:355–364. doi: 10.1136/gutjnl-2018-318021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Y.H., Zhang S., Zhu M., Lu T., Chen K., Wen Z., Wang S., Xiao G., Luo D., Jia Y., Li L., MacConmara M., Hoshida Y., Singal A., Yopp A., Wang T., Zhu H. Mice with increased numbers of polyploid hepatocytes maintain regenerative capacity but develop fewer tumors following chronic liver injury. Gastroenterology. 2020;158:1698–1712. doi: 10.1053/j.gastro.2020.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S., Zhou K., Luo X., Li L., Tu H.C., Sehgal A., Nguyen L.H., Zhang Y., Gopal P., Tarlow B.D., Siegwart D.J., Zhu H. The polyploid state plays a tumor-suppressive role in the liver. Dev. Cell. 2018;44:447–459. doi: 10.1016/j.devcel.2018.01.010. (e445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basant A., Lekomtsev S., Tse Y.C., Zhang D., Longhini K.M., Petronczki M., Glotzer M. Aurora B kinase promotes cytokinesis by inducing centralspindlin oligomers that associate with the plasma membrane. Dev. Cell. 2015;33:204–215. doi: 10.1016/j.devcel.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H., Johnson J.M., Lera R.F., Brahma S., Burkard M.E. Anillin phosphorylation controls timely membrane association and successful cytokinesis. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall P.A., Todd C.B., Hyland P.L., McDade S.S., Grabsch H., Dattani M., Hillan K.J., Russell S.E. The septin-binding protein anillin is overexpressed in diverse human tumors. Clin. Cancer Res. 2005;11:6780–6786. doi: 10.1158/1078-0432.CCR-05-0997. [DOI] [PubMed] [Google Scholar]
- 26.Hall G., Lane B.M., Khan K., Pediaditakis I., Xiao J., Wu G., Wang L., Kovalik M.E., Chryst-Stangl M., Davis E.E., Spurney R.F., Gbadegesin R.A. The human FSGS-causing ANLN R431C mutation induces dysregulated PI3K/AKT/mTOR/Rac1 signaling in podocytes. J. Am. Soc. Nephrol. 2018;29:2110–2122. doi: 10.1681/ASN.2017121338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.She Z.Y., Yang W.X. SOX family transcription factors involved in diverse cellular events during development. Eur. J. Cell Biol. 2015;94:547–563. doi: 10.1016/j.ejcb.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J.F., Wyczalkowski M.A., Leiserson M.D.M., Miller C.A., Welch J.S., Walter M.J., Wendl M.C., Ley T.J., Wilson R.K., Raphael B.J., Ding L. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreno C.S. SOX4: the unappreciated oncogene. Semin. Cancer Biol. 2019;18:30145–30147. doi: 10.1016/j.semcancer.2019.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y., Pan Z.G., Shu L., Li Q.J. Podocalyxin-like, targeted by miR-138, promotes colorectal cancer cell proliferation, migration, invasion and EMT. Eur. Rev. Med. Pharmacol. Sci. 2018;22:8664–8674. doi: 10.26355/eurrev_201812_16631. [DOI] [PubMed] [Google Scholar]
- 31.Pang L., Li B., Zheng B., Niu L., Ge L. miR-138 inhibits gastric cancer growth by suppressing SOX4. Oncol. Rep. 2017;38:1295–1302. doi: 10.3892/or.2017.5745. [DOI] [PubMed] [Google Scholar]
- 32.Blanca A., Sanchez-Gonzalez A., Requena M.J., Carrasco-Valiente J., Gomez-Gomez E., Cheng L., Cimadamore A., Montironi R., Lopez-Beltran A. Expression of miR-100 and miR-138 as prognostic biomarkers in non-muscle-invasive bladder cancer. APMIS. 2019;127:545–553. doi: 10.1111/apm.12973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The primers for RT-qPCR assay.
Selected sequence of the predicted miR-138 binding site of SOX4 mRNA 3′-UTR, along with the relative mutated sequence.
Supplementary figures