Summary
Immunoglobulins emerging from B lymphocytes and capable of recognizing almost all kinds of antigens owing to the extreme diversity of their antigen‐binding portions, known as variable (V) regions, play an important role in immune responses. The exons encoding the V regions are known as V (variable), D (diversity), or J (joining) genes. V, D, J segments exist as multiple copy arrays on the chromosome. The recombination of the V(D)J gene is the key mechanism to produce antibody diversity. The recombinational process, including randomly choosing a pair of V, D, J segments, introducing double‐strand breaks adjacent to each segment, deleting (or inverting in some cases) the intervening DNA and ligating the segments together, is defined as V(D)J recombination, which contributes to surprising immunoglobulin diversity in vertebrate immune systems. To enhance both the ability of immunoglobulins to recognize and bind to foreign antigens and the effector capacities of the expressed antibodies, naive B cells will undergo class switching recombination (CSR) and somatic hypermutation (SHM). However, the genetics mechanisms of V(D)J recombination, CSR and SHM are not clear. In this review, we summarize the major progress in mechanism studies of immunoglobulin V(D)J gene recombination and CSR as well as SHM, and their regulatory mechanisms.
Keywords: B cell, class switch recombination, mechanism, somatic hypermutation, V(D)J recombination
Developments of novel experimental methodologies and analysis techniques have elucidated more molecules and mechanisms concerning the generation and regulation of immunoglobulin. This review summarizes the current state of research concerning the mechanism of immunoglobulin V(D)J recombination, CSR as well as SHM, and their regulatory mechanisms.
Abbreviations
- 3′RR
3′ regulatory region
- 3′Eα
3′ Cα enhancer
- 53BP1
p53‐binding protein 1
- AID
activation‐induced cytidine deaminase
- Alt‐NHEJ
alternative NHEJ
- APE1/2
apurinic/apyrimidinic endonuclease
- ATM
ataxia telangiectasia mutated
- Bach2
BTB and CNC homology 2
- BER
base excision repair
- Blimp‐1
B lymphocyte‐induced maturation protein‐1
- BRCA1
breast cancer 1
- C‐NHEJ
canonic NHEJ
- CSR
class switch recombination
- CTCF
CCCTC‐Binding Factor
- DDX1
DEAD‐box RNA helicase 1
- DNA‐PK
DNA protein kinase
- DNA‐PKcs
DNA‐dependent protein kinase catalytic subunit
- DSB
double‐strand breaks
- Ebf1
early B‐cell factor 1
- eEF1A
eukaryotic elongation factor 1 α
- Erag
enhancer region of RAG1/2
- Exo1
exonuclease 1
- FOXO
Forkhead box
- G4
G‐quadruplexes
- HMG
high‐mobility group
- hs
hypersensitive sites
- IgH
immunoglobulin heavy
- IgL
immunoglobulin light
- Lig
ligase
- LSR
locus suicide recombination
- Mbd4
methyl‐CpG binding domain protein 4
- MDC1
mediator of damage checkpoint protein
- MLH1
MutL homologue 1
- MMR
mismatch repair
- NBD
nonamer binding domain
- NHEJ
non‐homologous end joining
- PARP1
poly‐ADP ribose polymerase 1
- PAXX
paralogue of XRCC4 and XLF
- PCNA
proliferating cell nuclear antigen
- pol
polymerases
- RAP80
receptor‐associated protein 80
- RPA
replication protein A
- RSS
recombination signal sequence
- SHM
somatic hypermutation
- ssDNA
single‐stranded DNA
- XLF
XRCC4‐like factor
- XRCC4
X‐ray cross complementing Group 4
Immunoglobulin structure and genetic encoding
Immunoglobulin is comprised of two immunoglobulin heavy (IgH) chains encoded by the IgH heavy‐chain locus (chromosome 14 in human and 12 in the mouse) and two immunoglobulin light (IgL) chains encoded by either the Igκ (chromosome 2 in human and 6 in the mouse) or Igλ (chromosome 22 in human and 16 in the mouse) light‐chain loci. IgH chains have five major isotypes (Igμ, Igα, Igγ, Igδ and Igε), with four subtypes of IgG (Igγ1, Igγ2, Igγ3 and Igγ4) and two subtypes of IgA (Igα1 and Igα2). The IgL chain has two types (Igκ and Igλ). IgH chains have four (Igγ, Igα and Igδ) or five (Igμ and Igε) domains while IgL chains have two domains. The N‐terminal regions of heavy and light chains, known as variable (V) region, are highly variable in their sequences and are the antigen‐binding portions of the antibody.1 The C‐terminal regions of immunoglobulin, both IgH and IgL, have only a few sequential variations in the same species but different individuals, so are called constant (C) regions.2 Igγ, Igα and Igδ all have three CH whereas both Igμ and Igε have four CH (see Supplementary material, Fig. S1a).3
The V region of IgH chain is encoded by V(variable), D(diversity), J(joining) genes whereas the V region of IgL chain is encoded by V and J segments with the absence of D segments in the light‐chain loci. The C regions of different immunoglobulin isotypes are encoded by different CH exon clusters, which are organized in the order of Cμ, Cδ, Cγ, Cε and Cα in the IgH locus.4 These genes are assembled in the developing lymphocyte by V(D)J recombination to form a complete immunoglobulin (see Supplementary material, Fig. S1b).
V(D)J recombination
The V regions are assembled through V(D)J recombination of VH, DH and JH genes on the heavy chain and VL and JL genes on the light chain. This recombination is initiated by double‐strand breaks (DSB) produced by the Recombination‐activating gene (RAG1–RAG2) recombinase at specific recombination signal sequences (RSS).
Recombination signal sequences
The RSS (see Supplementary material, Fig. S2a) normally consists of a highly conserved heptamer motif (consensus sequence 5′‐CACAGTG‐3′) and a conserved nonamer sequence (consensus sequence 5′‐ACAAAAACC‐3′) separated by a poorly conserved spacer sequence of 12 or 23 nucleotides.3, 5 The heptamer sequence is considered to be the essential recognition element. The first three nucleotides of the heptamer (closest to the coding flank) show the highest sequence conservation, and are critical for recombination, whereas the remaining heptamer positions are much less important. The nonamer sequence is dispensable for recombination, with only a few highly conserved positions.
Both the RSS at the 3′ end of the IgH V fragment and the 5′ end of the J fragment carry a 23‐bp spacer sequence, while the D fragment has a 12‐bp spacer sequence at both the 5′ and 3′ ends of the RSS (see Supplementary material, Fig. S2b).3 According to the 12/23 rule,6 efficient recombination only occurs between RSS with different spacer lengths. Therefore, D can interact with J and V while the V cannot be connected to the J because it does not comply with the 12/23 rule. In addition, proximal sites undergo recombination substantially more frequently than distal ones.6 So the order is first D‐J then V‐DJ. For the light chain, the 3′ end of the V fragment and the 5′ end of the J fragment are reverse complementarily complementary, with a 12‐ or 23‐bp spacer sequence, respectively, which can result in a V‐J linkage.
However, recently, it has been widely proved that the 12/23 rule of genomic recombination is frequently violated under physiological conditions, resulting in unanticipated hybrid recombinations. Non‐12/23 junctions under physiological conditions are thought to be rare, the most common being in VDDJ rearrangements of the IgH locus that occur once per 800 cells. In addition to violating the 12/23 rule, other non‐canonical rearrangements form hybrid signal‐to‐coding junctions and are typically generated in artificial systems, but may also be detected at low frequency in vivo.7
RAG1/RAG2
The V(D)J recombinase consists of two lymphoid‐specific proteins, RAG1 and RAG2, of 1040 and 527 residues, which carry out DNA cleavage. Both RAG proteins are large multi‐domain proteins that consist of core regions and non‐core regions (see Supplementary material, Fig. S3). The catalytic core and DNA‐binding functions are largely contained within RAG‐1.6 The ‘core’ region of RAG1 is required for binding to the nonamer as well as catalysis of cleavage. The non‐core domains of the RAG proteins provide important regulatory functions, but the mechanism is not well understood. However, the role for RAG2 is less clear, which may function to activate RAG1 for sequence‐specific binding and cleavage, and also provide additional DNA‐binding capability.8, 9
The mechanism of V(D)J recombination
Efficient cleavage of a DNA substrate requires only RAG1, RAG2, a divalent metal ion, and high‐mobility group (HMGB1 or HMGB2) proteins. The first step in V(D)J recombination is binding of RAG, probably together with HMGB1 or HMGB2. RAG–RSS interactions have been studied. The RAG complex catalyzes two reactions, nicking and hairpin formation, without dissociation. First, RAG complex binds either a 12‐RSS or a 23‐RSS, resulting in a 12 or 23 signal complex. Then a nicking is introduced precisely at the junction between the coding segment and the RSS. Another type of RSS is captured to form a paired complex, and nicking is also follows.10 In vitro, HMGB1 or HMGB2 have been shown to stimulate the activity of RAG in DNA binding, nicking and hairpin formation, presumably by inducing RSS bending.9, 11 Upon paired complex formation, the free 3′‐hydroxyl released from the nicking step attacks the opposing strand by transesterification in the presence of Mg2+ to create a hairpin coding segment and the cleaved RSS ends. Proteins in the non‐homologous end joining (NHEJ) DNA repair pathway are recruited to the coding ends. The coding ends are opened by NHEJ DNA repair factors and then joined, forming imprecise coding joints that contain added nucleotides9, 12, 13 (Fig. 1a). After V(D)J recombination of IgH and IgL chain genes, the resulting immature naive B cells transcribe the IgH and IgL genes and the transcripts undergo alternative splicing to produce IgD and IgM by fusing the μ and δ exons to the J exon.1
Figure 1.
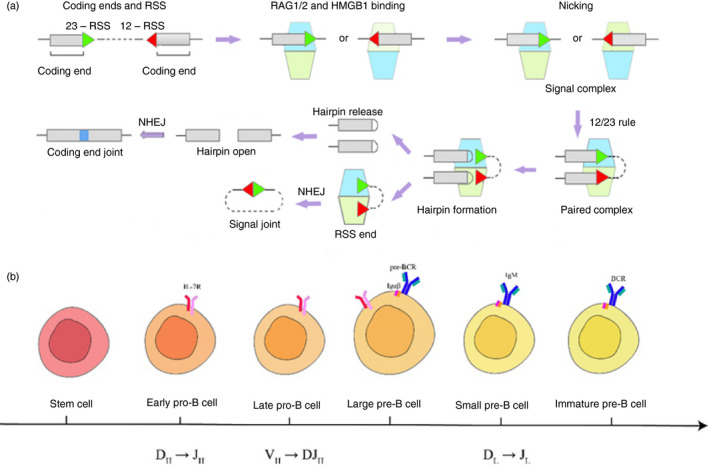
Overview of V(D)J recombination. (a) The RAG1–RAG2 complex is shown as light blue and light green trapezoids. Briefly, RAG binds a single recombination signal sequence (RSS) in the presence of HMGB1 to form a 12 or 23 signal complex. Another RSS is captured to form a paired complex. The DNA is cleaved, generating hairpin coding ends and RSS ends with a 3′ hydroxyl (OH) group in the presence of Mg2+. The hairpin is released and opened and rejoined by non‐homologous end joining (NHEJ), resulting in imprecise coding joints that contain added nucleotides (blue bars). RSS ends are processed through the NHEJ pathway as well, creating a signal joint. (b) B‐cell ontogeny during V(D)J recombination. This figure shows the chronological order of B cells in different stages of development in the bone marrow. B cells progress from stem cells to pro‐B cells, pre‐B cells and immature pre‐B‐cell stages. During this differentiation, V(D)J rearrangement of heavy chains occurs in pro‐B‐cell periods, resulting in the expression of pre‐B‐cell receptors (pre‐BCR), which are composed of Igμ and surrogate light chains (composed of VpreB and λ‐5). After receiving the pre‐BCR signaling, the light chains rearrange in small pre‐B cells, resulting in the expression of a mature BCR (composed by rearranged heavy and light chains).
Non‐homologous end joining is the major DSB rejoining process and occurs in all cell cycle stages, including two competing NHEJ pathways, canonic NHEJ (C‐NHEJ) and alternative NHEJ (Alt‐NHEJ; Fig. 2).
Figure 2.
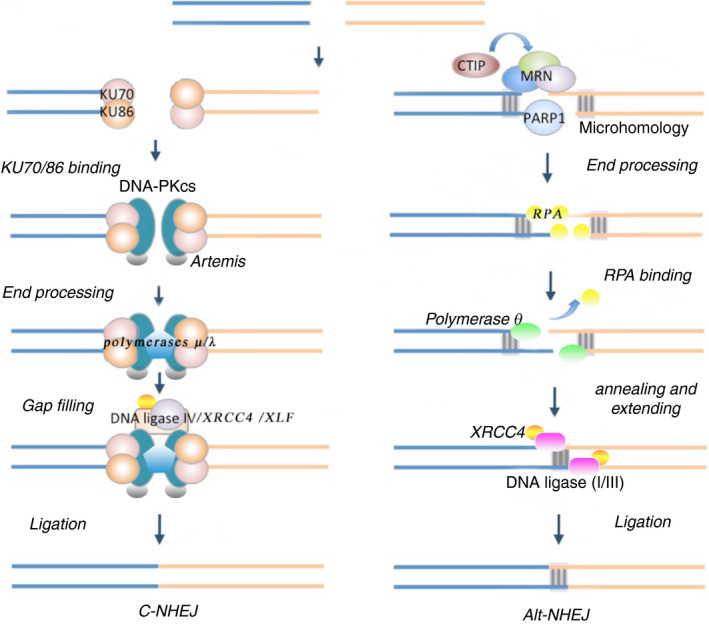
A model for canonical non‐homologous end joining (C‐NHEJ) and alternate NHEJ (Alt‐NHEJ). C‐NHEJ (left) and Alt‐NHEJ (right) are shown. For C‐NHEJ, Ku70/80 binds to the double strand breaks (DSB) first and protects DNA ends from digestion. The Ku:DNA complex recruits DNA‐dependent protein kinase catalytic subunit (DNA‐PKcs) in complex with Artemis, generating DNA‐PK. DNA‐PKcs undergoes autophosphorylation and activates Artemis, which then gains various nuclease activities, ensuring the DSB are compatible by resecting damaged DNA or non‐ligatable end groups. The gap created is filled by Polμ/λ recreated by Ku. DNA‐PK also facilitates recruitment of DNA ligase IV, XRCC4 and XLF to complete the ligation of DSB. For Alt‐NHEJ, PARP1 recognizes the broken DNA ends and recruits Mre11‐Rad50‐Nbs1 (MRN) and CtIP complex to initiate 5′‐3′ DNA resection, creating ssDNA overhangs. Resection exposes microhomology internal to break sites, facilitating spontaneous annealing of ssDNA. The binding of replication protein A (RPA) to the ssDNA overhangs removes secondary structure and prevents annealing of overhangs, which hinders Alt‐NHEJ. Polθ‐helicase acts as an ATP‐dependent annealing helicase that dissociates RPA to promote DNA annealing and stimulate Alt‐NHEJ. The paired ssDNA overhangs are subsequently extended by Po lθ‐polymerase. Finally, end rejoining is carried out by the DNA ligase I or III (Lig I or Lig III)/XRCC1 complex.
The mechanism of C‐NHEJ has been well explained. The main proteins required for C‐NHEJ involve the Ku70/Ku80 heterodimer, the catalytic subunit of the DNA‐dependent protein kinase (DNA‐PKcs), the endonuclease Artemis, the DNA ligase IV (Lig IV), the DNA polymerases μ (pol μ) and λ (pol λ), X‐ray cross complementing Group 4 (XRCC4) and XRCC4‐like factor (XLF).14 Ku70/80 is thought to be the first protein to bind to the DSB, based on its abundance and tight affinity.15 Ku70/80 protects DNA ends from digestion, and the Ku:DNA complex also serves as a node at which the nuclease, polymerases and ligase of C‐NHEJ can attach. There is a Ku:DNA complex at each of the two DNA ends being joined, permitting each DNA end to be modified in preparation for joining. Ku:DNA complex recruits DNA‐PKcs and Artemis, a DNA‐PKcs‐activated endonuclease, generating a DNA protein kinase (DNA‐PK). DNA‐PKcs undergoes autophosphorylation and activates Artemis, which then gains various nuclease activities including 5′‐endonuclease activity, 3′‐endonuclease activity, and hairpin opening activity.15, 16 The nuclease activities ensure that the two ends are compatible by resecting damaged or non‐ligatable end groups at the end of the DSB. The gap created after processing is filled by Polμ/λ recruited by Ku.17 DNA‐PK also facilitates recruitment of a ligation complex, which encompasses LigIV, XRCC4 and XLF. PAXX (paralogue of XRCC4 and XLF) is a recently discovered protein with structural similarity to XRCC4 and XLF and promotes DSB repair by interacting with Ku.18, 19
The role for Alt‐NHEJ in V(D)J recombination was originally thought to be a backup pathway for DSB repair.20 However, emerging evidence has demonstrated that Alt‐NHEJ can also occur in C‐NHEJ‐intact cells.20, 21 The higher affinity of Ku for DSB compared with Poly‐ADP ribose polymerase 1 (PARP1) could explain the predominance of C‐NHEJ. The mechanism of Alt‐NHEJ involves 5′–3′ resection of the DNA ends, annealing of microhomology, fill‐in synthesis and ligation. PARP1 can recognize the broken DNA ends and serves as a scaffold for the recruitment of other DNA damage factors.22 First, Mre11‐Rad50‐Nbs1 and CtIP complex initiates 5′–3′ DNA resection, creating short single‐stranded DNA (ssDNA) overhangs.20 Resection exposes microhomology internal to break sites, which could facilitate spontaneous annealing of ssDNA. The binding of Replication protein A (RPA) to the resulting short ssDNA overhangs removes secondary structure and prevents annealing of overhangs, which hinders Alt‐NHEJ.23, 24 Polθ‐helicase acts as an ATP‐dependent annealing helicase that dissociates RPA to promote DNA annealing and stimulate Alt‐NHEJ.25 The paired ssDNA overhangs are subsequently extended by Polθ. Finally, end rejoining is carried out by the DNA ligase I or III (Lig I or Lig III)/XRCC1 complex in coordination.22, 26, 27
The recombination described above proceeds in such a way that intervening DNA is deleted. In fact, the intervening DNA can also be inverted without deletion. Whether recombination proceeds in a deletional or inversional manner depends on the relative orientation of the two RSS. V and J segments located on the same strand are recombined by deletion, leaving the coding segment on the chromosome and the signal joint on an excised circle of DNA. If V and J segments are on opposite strands, they are joined by inversion between the RSS, generating a signal joint and a coding joint.
Notably, to generate an efficient immune repertoire, V(D)J recombination favors genetic diversity at two levels. The first level corresponds to a diverse rearrangement of V, D and J genes. The second level is characterized by a particular joining mechanism of coding segments characterized by the loss or addition of extra nucleotides. During V(D)J recombination, the terminal deoxynucleotidyl transferase adds random nucleotides (N‐segments) at the V‐D and D‐J junctions in heavy chains of immunoglobulin, thereby significantly contributing to the diversity of the immune repertoire.28
Regulatory mechanism of V(D)J recombination
V(D)J recombination undergoes regulation at multiple levels, including the regulation of RAG1/2 activity, subnuclear reposition, spatial conformation of chromatin, and Vκ recombination, and has some specific regulation mechanisms.
The regulation of RAG1/RAG2 activity
The RAG are important enzymes in V(D)J recombination, so their activity and quantity have an important influence on this process. The RAG proteins can coordinate their activity through autoregulatory properties. The RAG1 N‐terminus is also associated with ubiquitylation‐dependent regulatory processes by acting as an E3 ligase.29 RAG2 is a regulatory cofactor, which promotes RAG1 to bind and cleave DNA. However, with the RAG1 C‐terminus (residues 1009–1040) collaboration, the C‐terminus of RAG2 efficiently autoinhibits RAG activity.30
Phosphorylation events are related to the activity of RAG as well. RAG2 has a conserved serine‐glutamine (SQ) phosphorylation site on 365 to 366. This ataxia telangiectasia mutated (ATM) ‐mediated phosphorylation site of RAG2 has the function of feedback control of cleavage and maintenance of genome stability.31 Thr‐490 is an essential Cyclin‐dependent kinases phosphorylation site in a conserved degradation signal sequence, which links the amount of RAG2 to the cell cycle.32 RAG activity can also be enhanced by the AMP‐activated protein kinase, which directly phosphorylates RAG1 at serine‐528. This phosphorylation event can increase the catalytic activity of the RAG, and so initiates V(D)J recombination.33
In addition, the sensitivity of the gene to RAG cleavage is related to the epigenetics of histone (see Supplementary material, Fig. S4).
The accessibility hypothesis postulates that the initial step of V(D)J recombination must be chromatin changes, which render the different genes of the immunoglobulin loci accessible to the V(D)J recombination machinery. The 3′ proximal region of immunoglobulin loci is activated by enhancer‐mediated recruitment of histone‐modifying enzymes. Consequently, J segments that are adjacent to the Eμ enhancer are characterized by an abundance of active histones like H3K4me2 (trimethylation of histone H3 at lysine 4), H3K4me3 and H3K9ac in pro‐B cells. Through a plant homeodomain finger, the RAG2 proteins are able to specifically bind the active histone mark H3K4me3 in the RSS that are associated with J segments.34 H3K4me3 subsequently changes the conformation of catalytic and substrate‐binding regions of RAG1 and the autoinhibitory domain (residues 352 to 405) in RAG2, increasing substrate affinity and catalytic rate.35, 36 Additionally, SUV39H1, a histone lysine N‐methyltransferase involved in the formation of heterochromatin, is involved in the establishment of H3K9me3 and H4K20me3, the hallmarks of heterochromatin. The formation of heterochromatin is known to influence RAG2, catalyzing the VDJ recombination. Moreover, SUV39H1 has a role in the methylation of RAG2, which changes RAG2 subnuclear localization, and might regulate the chromatin binding of RAG2.37
Apart from these ways to regulate RAG activity, there are also some mechanisms to regulate the amount of RAG. As RAG2 protein is only expressed in the G0‐G1 phase, proteins that control the cell cycle can also influence the amount of RAG. The RNA‐binding proteins ZFP36L1 and ZFP36L2 can suppress mRNAs, helping B‐cell entry into and exit from the cell cycle. Cells that lack ZFP36L1 and ZFP36L2 show a delayed V(D)J recombination.38
Recent studies have shown that Forkhead box O (FOXO) protein has an important regulatory effect on the activity of RAG proteins (see Supplementary material, Fig. S5). Pre‐B‐cell receptors (pre‐BCR) synergize with interleukin‐7 receptor (IL‐7R) signaling pathways, which is found to negatively regulate RAG mRNA and protein levels during the clonal expansion period of pre‐BCR+ cells, but positively regulates the expression of RAG in contrast to small pre‐B‐cell period.39 FOXO protein is a major RAG transcription activator. It can activate the expression of RAG1 and RAG2, as well as up‐regulate the amount of p27, a stabilizer for RAG2.40 Phosphatidyl 3‐kinase and Akt kinase, activated by tonic (foreign antigen‐independent) receptor signaling, can phosphorylate and result in cytoplasmic separation of FOXO protein.41 Moreover, early B‐cell factor 1 (Ebf1), a transcription factor, can either directly regulate the accessibility of specific genes at the loci of IgH and Igλ, or indirectly contributes to V(D)J recombination via the activation of Pax5.42 The expression of Ebf1 is driven by the downstream effector Stat5 of IL‐7R. During pre‐B‐cell proliferation, Ebf1 and its downstream target c‐Myb have a negative influence on RAG transcription, through the negative regulation of FOXO1 binding to the RAG locus. Ebf1 can directly downgrade FOXO1 expression and upgrade zinc finger protein Gfi1b expression.39, 43 Similarly, activation of DNA damage is able to depress RAG expression by ATM‐mediated canonical nuclear factor‐κB transcription factors as well as the loss of ATM‐dependent FOXO1 binding to the enhancer region of RAG1/2 locus (Erag).43, 44
Subnuclear reposition
Evidence from several model systems has indicated that the location of genes, especially their proximity to heterochromatin, may be important for initiating or maintaining silence of a gene, and modulating locus accessibility. There are two repressive compartments, the nuclear periphery and pericentromeric heterochromatin, that are important for propagating the inactive state of genes. It has been revealed that subnuclear location will change during the development of a B cell.45, 46 In progenitors, the IgH locus is anchored at the nuclear periphery via the distal VH genes that are in an accessibility repression stage;47 meanwhile, the 3′ proximal D and J clusters are free, which facilitates D‐J rearrangements in lymphoid progenitors. Both IgH alleles will undergo D‐J recombination, which leads to a pro‐B cell. IgH loci are subsequently relocated from the nuclear periphery to more central positions within the nucleus. This subnuclear repositioning reactivates V genes in preparation for V‐DJ recombination. The two alleles behave differently at this stage of pre‐B cells. Following successful V‐DJ rearrangement of one IgH allele, which gives rise to pre‐BCR, the signaling leads to repositioning of the second, incompletely rearranged IgH allele to repressive pericentromeric heterochromatin,48 whereas the functionally rearranged IgH allele remains in a central position. This feedback inhibition of V‐DJ recombination at the second allele ensures that each B lymphocyte expresses only one monospecific BCR, known as allelic exclusion (see Supplementary material, Fig. S6).49 Mono‐allelic expression seems to take place preferentially on the associated allele.45 However, it has also been reported that activated B cells can transcribe both IgH alleles as well.50 To what extent the two IgH alleles differ remains unclear.
Spatial conformation of chromatin
Chromatin interactions can control the gene assembly of IgH chains. In the pro‐B cell, in which D‐J recombination has finished but V‐DJ recombination is going to take place, contraction will happen in the distal V cluster through looping.48 The localized activity of the RAG1/2 at the proximal chromatin is strictly dependent on locus contraction for subsequent V(D)J recombination. The long‐range contraction of the IgH locus also facilitates V‐DJ rearrangements because of the proximity of both elements. In addition, different V genes can recombine with similar frequency, which is essential for the generation of a highly diverse immunoglobulin repertoire. Little is known about the molecular mechanisms controlling locus contraction and only a few trans‐acting factors like Pax‐5,51 YY152 and CCCTC‐binding factor (CTCF)53, 54 have been implicated in the regulation of contraction.49
Chromatin interactions control the gene assembly of IgH chains. In developing B lymphocytes, V(D)J recombination is also regulated by remote cis‐acting elements. At distant sites of the IgH variable region, the 3′ regulatory region (3′RR) conveys a transcriptional silencing activity on transcription of the immunoglobulin gene, but the repressive activity was switched off after V‐DJ recombination, promoting transcription of immunoglobulin in mature B cells.55
Besides, the highly conserved 11‐zinc finger protein CTCF can mediate long‐range chromatin interactions that influence the organization and function of the mammalian genome. CTCF also affects the complex configuration of chromatin loops that control the functional interactions between elements. CTCF binding site orientations can define the chromatin architecture that supports V(D)J recombination.54
In addition, histone deacetylase 3, a catalytic component of the NCoR/SMRT co‐repressor complexes, is involved in global changes in chromatin structure, which likely have effects on distal V‐DJ recombination.56
Vκ recombination regulation
The mechanisms of Vκ recombination regulation are different from VH. First, the RSS for each V gene is different in quality, which can be quantified as RSS Information Content scores. Theoretically, a higher score indicates a tendency for recombination. In addition, Igκ recombination in pre‐B cells is activated by the binding of some transcription factors like PU.1, E2A, IRF4 and IRF8 at the iEκ and 3′Eκ enhancers. PU.1 binding at the RSS is a key feature to determine whether the Vκ gene will actively recombine,57 and the antisense lncRNA PU.1 is able to inhibit PU.1 mRNA translation by blocking tRNA recruitment.58 In early B‐cell progenitors, two DNA methyltransferases, Dnmt3a and Dnmt3b participate in the methylation of Tcfe2a gene, which encodes E2A. The lack of Dnmt3a and Dnmt3b elevates the level of E2A. Moreover, H3K4 methylation and IKAROS binding at the RSS are important for the promotion of higher recombination frequencies.59, 60
Long‐range genomic interactions, which distantly located genomic regions on the same chromosome undergo, are dynamically controlled at the Igκ light‐chain locus. Yin Yang 1, a widely expressed transcription factor, contributes to long‐range chromatin interactions in the IgH chain. Also, CTCF can regulate the expression of developmentally regulated genes. However, their roles in Igκ locus contraction have not been determined.61, 62 Recent research shows that a B‐cell‐specific enhancer, E88, is a major hub of long‐range chromatin interactions, which connects subtopologically associated domains in the V gene region with a recombination center at the J genes in Igκ. This enhancer ensures a proper folding structure of Igκ locus for V(D)J recombination.63
The primary immunoglobulin repertoire is initially composed of IgM after the V(D)J recombination. The range of reactivity of the primary immunoglobulin repertoire can be further modified by somatic hypermutation (SHM) and class switch recombination (CSR) at the immunoglobulin loci. SHM results in mutations being introduced into the V region of the heavy chain and light chain, altering the affinity of the immunoglobulin for its antigen. Whereas in CSR, the initial heavy‐chain C regions are replaced by another isotype, modifying the effector activity of the immunoglobulin but not its specificity. Both CSR and SHM are initiated by an enzyme called activation‐induced cytidine deaminase (AID), which is expressed specifically in activated B cells. So these two processes do not occur in T‐cell receptor genes.
Somatic hypermutation and class switch recombination
After going through V(D)J recombination, B cells subsequently undergo two genetic modifications, SHM and CSR. The purpose of these alterations, mostly in the germinal center, is to increase the affinity and alter the biological properties of immunoglobulin but with a specificity for the antigen. For some species, like chickens, immunoglobulin gene conversion is used in addition to or instead of SHM.64 The molecular mechanism of SHM, CSR as well as gene conversion, shows many similarities and shares several enzymes. But AID is thought to be the only factor that is absolutely necessary for all these mechanisms. Below, the mechanism of SHM and CSR is described in detail.
AID is a 198‐amino‐acid protein. It is one of 12 members of the APOBEC family of DNA/RNA cytidine deaminases, and shares a conserved catalytic domain with other members. The catalytic domain contains His56, Cys87 and Cys90, which bind Zn2+ and are essential for catalytic activity, and Glu58, which serves as a general acid–base catalyst. The C‐terminal domain is essential for AID to mediate CSR.65 AID is a ssDNA‐specific cytidine deaminase, which converts cytosine bases (C) to uracils (U) in vitro and in vivo but with no activity on dsDNA, RNA or RNA:DNA hybrids. The deamination reaction proceeds via a direct nucleophilic attack at position 4 of the pyrimidine ring of cytosine by Zn2+ coordinated to AID. However, AID is an inefficient enzyme, deaminating only about 3% of the cytidines even at preferred hotspot motifs. How this AID inefficiency effects CSR and SHM remains unclear at present.
Uracils caused by AID can trigger several types of DNA repair including the mismatch repair (MMR) and the base excision repair (BER) pathways. In the MMR, U mismatched with G is detected by the heterodimer MutSα, comprising MSH2 and MSH6.66 MutSα recruits MutLα (MLH1‐PMS2) and exonuclease 1 (Exo1). The endonuclease activity of PMS2 then cleaves the DNA 5′ of the mismatch, creating a DNA nick that serves as a point of entry for the Exo1, which initiates resection from the nick going past the mismatch site and creates an extended patch of ssDNA. Proliferating cell nuclear antigen (PCNA) subsequently recruits Polδ to replicate over the gap and Ligase 1 (Lig 1) finalizes repair. Whereas in BER, U is recognized by uracil‐DNA glycosylase, which removes U from DNA67, leaving an abasic site that is recognized by apurinic/apyrimidinic endonuclease 1/2 (APE1/2). APE1/2 nicks the DNA 5′ of the abasic site. PARP1 is activated and scaffold XRCC1, Polβ and a ligase are recruited. Polβ then removes the 5′ deoxyribose and inserts a single nucleotide, followed by a ligation and so completes the repair.68 Interestingly, both uracil‐DNA glycosylase and MutSα are conserved enzymes that initiate DNA repair with high fidelity through either MMR or BER. Hence, a non‐canonical MMR or BER pathway is needed for transition or transversion mutations in SHM and DSB formation required for CSR (Fig. 3).
Figure 3.
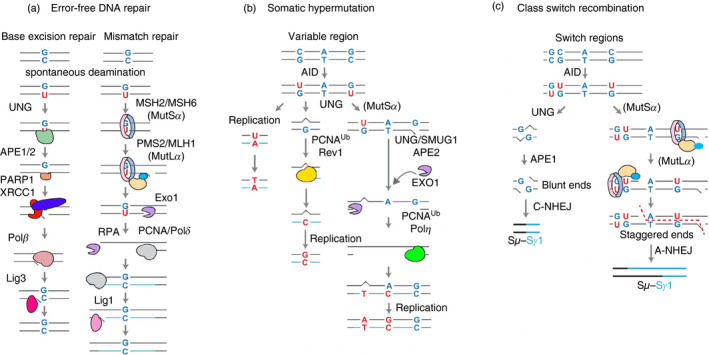
Overview of error‐free basic excision repair (BER) and mismatch repair (MMR), somatic hypermutation (SHM) and class switching recombination (CSR). (a) Error‐free repair pathways for BER (left) and MMR (right). Canonical BER is initiated by uracil‐DNA glycosylase, creating an abasic site that is recognized by APE1/2. APE1/2 nick the DNA 5′ of the abasic site. PARP1 and XRCC1 are activated and recruit proliferating cell nuclear antigen (PCNA) and polymerase β (Polβ). Polβ removes the remaining 5′ deoxyribose and insert a single nucleotide, followed by ligation with Lig3. In MMR, MutSα recognizes the U:G mispair and recruits MutLα. MutLα nicks the DNA 5′ of the mismatch via PMS2. Exo1 creates an ssDNA from the nick going past the mismatch site. PCNA subsequently recruits Polδ to fill over the gap, following ligation by Lig1. (b) During SHM, uracil can act as a template for replication leading to a C–T transition mutation. Alternatively, non‐canonical BER or MMR recruit low‐fidelity polymerases Polη through PCNA ubiquitination (PCNA‐Ub) leads to transition or transversion mutations. (c) For CSR, non‐canonical BER can lead to double strand breaks (DSB) when two uracils in opposite strands are closely spaced. MMR can process distantly spaced uracils, leading to staggered DSB. Blunt DSB are joined by canonical non‐homologous end joining, whereas staggered breaks are repaired by alternative non‐homologous end joining.
Mechanism of SHM
For error‐prone MMR in SHM, APE2 rather than MutLα is recruited to nick the mismatch and provide an entry for the Exo1. Furthermore, low‐fidelity polymerases like Polη instead of Polδ are recruited through PCNA ubiquitination69 to resynthesize the DNA gap and introduce mutation, which is mainly at A:T residues. Whereas in non‐canonical BER during SHM, some abasic sites serve as a non‐informative template. REV, a Y‐family DNA polymerase recruited by PCNA ubiquitination as well, inserts dCMP into the new DNA strand opposite the abasic site. After a further round of DNA replication this can result in a stable transversion mutation at the site of the original C:G base pair.
Mechanism of CSR
CSR is a DNA deletional‐recombination reaction that proceeds through the generation of DSB in the switch (S) region preceding each CH gene and is completed by NHEJ between donor and acceptor S regions, resulting in the replacement of the expressed CH exon cluster.
The S regions are repetitive DNA elements that locate upstream of all the CH genes except Cδ. Mouse S regions are composed of 1–10 kb of tandem repeats that are G‐rich on the non‐template strand. Both the sequence and number of tandem repeats vary among mouse S regions. Sμ, for example, is approximately 3·2 kb long and is comprised of GAGCT pentameric motifs, with the AGCT palindromic sequence representing a canonical RGYW/WRCY sequence. Sγ1 is approximately 10 kb long and has multiple RGYW/WRCY sequences embedded within 49‐bp repeat units. Other repeat motifs found in S regions include GGGGA/T in Sμ, Sγ1, Sγ2b and Sγ3 and GGGCT in Sε and Sα. S regions are the targets for AID that initiates CSR.65, 70
There are a transcriptional intervening (I) promoter and an I exon upstream of all C genes except Cδ. At the onset of CSR, I promoters are selectively activated and produce non‐coding germline transcripts, which initiate at the I exon, proceed through the S region and terminate downstream of the corresponding CH gene.71 AID is a ssDNA‐specific cytidine deaminase, so germline transcripts are thought to separate the two dsDNA and provide access to AID.72 AID binds to the S region by 14‐3‐3 adaptors73 and recruits protein kinase A as well. AID is phosphorylated at Ser38 of the N‐terminal region by protein kinase A, generating a binding site for RPA.74 RPA enhances the deamination activity of AID and transfers C into U. If two U are sufficiently near in opposite strands, a DSB is resulted through the BER pathway. Conversion of two more distal U into a blunt DSB, which is necessary for CSR, requires MMR.75
AID‐initiated DSB in two S regions activate the ATM‐dependent DNA damage response. The Mre11‐Rad50‐Nbs1 complex is first recruited to a DSB, where it recruits the protein kinase ATM. ATM phosphorylates and recruits numerous DNA damage response proteins including H2AX, Mediator of Damage Checkpoint protein (MDC1),76 ubiquitin ligases RNF8 and RNF168, p53‐binding protein 1 (53BP1), breast cancer 1 (BRCA1) and receptor‐associated protein 80 (RAP80).77 H2AX and 53BP1 mediate synapsis of upstream and downstream DSB,78 and 53BP1 can also prevent the rejoining of intra S‐region DSB.79 DSB in different S regions are eventually rejoined by C‐NHEJ or Alt‐NHEJ. Both C‐NHEJ and Alt‐NHEJ pathways are used in CSR.80 DSB generated by the MMR pathway seem to favor Alt‐NHEJ, possibly by exposing microhomologies after Exo processing.75
IgD CSR is restricted to a few B‐cell subsets in specific lymphoid tissues such as mesenteric lymph nodes, peritoneal cavity and mucosa‐associated tissue in both mice and humans.81, 82, 83 A recent study suggested that IgD CSR is initiated by microbiota,83 implying a role of IgD in the regulation of microbial homeostasis.
Locus suicide recombination
Immunoglobulin production relies on the selection of antigen‐specific B cells. This implies not only proliferation and differentiation of those B cells that specifically bind antigen but also elimination of the less efficient or inappropriately activated cells.84 It is important for B cells to undergo such a choice as inappropriate production of useless cells might trigger autoimmunity and inflammation. Locus suicide recombination (LSR) is reported as a variant form of the recombination mechanism to eliminate inappropriate B cells. LSR is characterized by recombination between Sμ and the 3′RR downstream of the C region, deleting the whole IgH constant gene cluster and thereby terminating B‐cell function. Apart from direct junctions from Sμ to 3′RR, sequence analysis of LSR both in vivo and in vitro reveals the presence of junctions of Sγ or Sα regions rather than Sμ with 3′RR, indicating that LSR has taken place in previous CSR in at least 14% of cases.85 Murine 3′RR includes repetitive DNA sequences resembling S regions. In humans, there are two 3′RRs due to the interval duplication of Cα gene on the IgH locus (3′RR1 and 3′RR2, respectively, downstream of Cα1 and Cα2).86 Analyzing the structure of human 3′RR shows their high similarity with the mouse 3′RR and both of them share multiple features with S regions. These ‘Like switch’ regions promote LSR through a process that is similar to CSR: non‐coding germline transcription of Sµ and ‘Like switch’ within the 3′RR, DSB caused by AID, and finally DNA repair, mainly by the NHEJ pathway as in CSR.87 However, major involvement of the A‐NHEJ rather than C‐NHEJ during DSB repair in LSR compared with CSR was recently reported,88 suggesting a mechanism to control the balance between CSR and LSR. LSR is mostly detected during B‐cell activation but remains undetectable in long‐lived memory cells or plasma cells.85 Interestingly, as LSR is a process towards cell death, it remains difficult to evaluate its regulation.85
Regulation of AID
Given that AID plays a significant role in both CSR and SHM, it is a crucial mechanism to regulate CSR and SHM by modulating AID expression, stability and localization.
Epigenetic regulation of AID
Histone modification and miRNA are two significant epigenetic mechanisms that regulate gene expression. In B‐cell activation, stimulatory signals induce several histone‐modifying enzymes to active H3K4me3, H3K9ac and H3K14ac in the promoter regions of AID and remove the repressive H3K27me3 and H3K9me3, changing the chromatin structure or recruiting other modifying factors and so up‐regulating AID. These signals also activate miRNA like mir‐16, mir‐155 and mir‐181b that decrease the expression of AID by binding to and degrading complementary sequences of the mRNA.89
Phosphorylation also modulates AID activity at a post‐translation level. Between 5 and 15% of the AIDs in activated B cells are phosphorylated at serine 38 by protein kinase A, and it has a role in targeting AID to DNA and increasing its activity.90 Moreover, threonine 140 can also be phosphorylated by PKC family members, enhancing AID relatively. Threonine 140 phosphorylation preferentially affects SHM, suggesting that post‐translational modifications may contribute to the choice between CSR and SHM.91 Serine 3 is another phosphorylation point controlled by protein phosphatase 2. Increasing S3 phosphorylation leads to a reduced activity of AID.92
The localization and stability of AID
It is contradictory that AID is predominantly located in the cytoplasm whereas both CSR and SHM occur in nucleus. Further studies have revealed that AID is transported between nucleus and cytoplasm continuously, although the mechanism for this is not clear. Germinal center‐associated nuclear protein, an RNA‐binding protein, is assumed to transport AID into the nucleus in mice.93 Cytoplasmic AID interacts with eukaryotic elongation factor 1α (eEF1A), a translation elongation factor involved in AID cytoplasmic retention.94 Export of AID is mediated by the Ran‐dependent nuclear exportin CRM1 because AID has a powerful nuclear export signal at the C terminus.95
Localization of AID is also associated with its stability. In the nucleus, AID undergoes a ubiquitin‐dependent protein degradation as well as a Reg‐γ‐mediated ubiquitin‐independent degradation.96 Whereas in the cytoplasm, molecular chaperones like Hsp90 and DnaJa1 can inhibit the polyubiquitination and proteasomal degradation of AID, and so increase the stability of AID.97 The complex regulatory mechanism of AID subcellular localization and stability allows proper amounts of AID to access into the genome, protects the genome from continuous mutations, and minimizes the detrimental off‐target effects.
Bach2
BTB and CNC homology 2 (Bach2) is a B‐cell‐specific transcription repressor and is thought to be the common pathway in controlling the expression, protein stability and subcellular location of AID.98 Bach2 up‐regulates AID mainly by suppressing B lymphocyte‐induced maturation protein‐1 (Blimp‐1), a factor that drives plasma differentiation by suppressing mature B‐cell‐associated genes including AID.99 Bcl6 can co‐bind to the cis‐regulatory sequence of the Blimp‐1 gene with Bach2, up‐regulating AID and maintaining the stability of the Bach2.100 However, Bcl6 as well as other suppressors of Blimp‐1 like Pax5 and Spib can be in turn blocked by Blimp‐1 at a transcriptional level, forming a negative feedback regulatory pathway.101 Moreover, Bach2 can also upgrade the expression of AID through a Blimp‐1‐independent pathway.98 Apart from a decreased amount of AID in B cells, Bach2 deficiency also profoundly decreased AID level in the nucleus.98 Bach2‐deficient cells also had reduced expression of the molecular chaperones known to stabilize cytoplasmic AID.102
Other regulatory mechanism of CSR
3′ regulatory region
The 3′RR is located at the most distal 3′ region of the immunoglobulin H chain locus and comprises four hypersensitive sites: hs3a, hs1,2, hs3b and hs4. Each hs is a weak enhancer but they form a strong enhancer synergistically.72, 103 The 3′RR is crucial for conventional CSR,82, 104 but its mechanism remains unclear. Conclusions drawn from knock‐out mice are that the 3′RR plays a role in promoting germline transcription105, 106 and synapsis between S regions in CSR.107 In addition, it has also been reported that 3′RR mainly controls the S acceptor region rather than the Sμ donor region in multiple aspects including alterations of epigenetic marks, germline transcription, R‐loop formation, paused RNA PolII, AID targeting and generation of DSB. Once DSB are generated, the Sμ–Sγ1 junctions are not affected by the 3′RR.108 The complete 3′RR deletion dramatically affects CSR and immunoglobulin secretion for all isotypes.104 But the IgG1 class displays a special status as only Cγ1 transcription and CSR are partly preserved after alteration or even complete deletion of the 3′RR, whereas all other class‐switched immunoglobulin was nearly abolished, suggesting that other elements may support the γ1 transcription.104, 105, 106 Furthermore, it seems that IgD CSR109 and IgA CSR in B1 cells110 are totally independent on the 3′RR. The Sμ–σδ junction is mainly produced by A‐NHEJ rather than C‐NHEJ,109 which markedly differs from IgG, IgA and IgE CSR.
Chromosome looping
The activation of CSR likely needs the signal from BCR, and the secondary signals from CD40 and Toll‐like receptors. Next, the transcription of unrearranged CH genes regulates naive B cells to a specific isotype. The type of cytokines can regulate the direction of CSR by chromosome looping, as different transcription promoters, located upstream of each acceptor S region, are activated by different cytokines. Then specific S‐C loci (like Sε‐Cε loci) are closed to the Eμ‐Sμ‐Cμ and 3′ Cα enhancer (3′Eα) region. Finally, the formation of chromosome looping can induce cell switching to that specific isotype (Fig. 4).111
Figure 4.

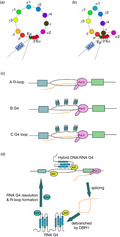
Chromosome looping and R‐loop. (a) This picture shows the structure of the chromosome looping, and each colorful circle representing the specific S‐C loci. Eμ and 3′Eα interact, causing Sμ and the downstream S regions located in a chromosomal loop. (b) After the Igε transcription promoter (located at the upstream of Igε acceptor S region) activated, Sε‐Cε loci are close to the 3′Eα segment and Eμ‐Sμ‐Cμ, inducing the B cell to switch to IgE. (c) Structures that recruit activation‐induced cytidine deaminase (AID): the switch sequence has a high G‐richness on the non‐template chain. During transcription, these G‐rich regions form secondary structures such as the R‐loop, G‐quadruplexes (G4) and G‐loop, which help recruit AIDs in the S region. (d) DDX1 promotes R‐loop formation: RNA polymerase II (Pol II) produces switch transcripts, whose intron contains the G4. After a splicing step, the intron lariat intermediate is debranched by RNA lariat (intron) debranching (DBR1) enzyme. Bound G4–AID complex, DDX1 targets to the S‐region DNA. At last G4 is resolved, and an R‐loop is formed in the S region.
G‐quadruplex
G‐quadruplexes (G4) are secondary structures that are formed in nucleic acids by guanine‐rich sequences. They are constructed around continuous G‐tetrads of Hoogsteen hydrogen‐bond guanine bases.112 G4 DNA structures have been previously reported to be present on the non‐template DNA strand of Sμ and Sγ regions in vitro. Recent studies show that AID deaminates ssDNA more robustly in the context of structured substrates, such as G4. Some studies have shown that G4 formation plays a role in CSR. G4 mediates recruitment and oligomerization of AID, which helps to create mutation clusters and DSB on IgH chain.73 Conversely, G4‐stabilizing agents, like (3,11‐difluoro‐6,8,13‐trimethyl‐8H‐quino(4,3,2‐kl)acridinium), can inhibit CSR and decrease immunoglobulin secretion.113 Moreover, two nucleoside diphosphate kinase (NME) isoforms are novel players in the CSR process. Before CSR activation, NME1 binds to the S region and suppresses CSR. Upon stimulation, NME1 dissociates from activated S region, whereas NME2 binds to G4 in the S region to promote CSR. NME1 and NME2 act as coordinated inhibitory stimulus pairs to modulate CSR.114
R‐loop
R‐loop is a stable intermediate of CSR, which is composed of a DNA:RNA hybrid and the associated non‐template ssDNA. R‐loop forms behind the extended RNA Pol II in the S region and results in ssDNA that may act as a substrate for AID, the ssDNA‐specific cytidine deaminase. In recent years, many studies have found that, compared with the original model, R‐loop is unlikely to play a role in the initial AID‐dependent mutagenesis phase, and it must have other AID‐independent roles in CSR. For example, R‐loop can regulate the DNA replication environment at the IgH locus.115, 116
Many other factors participate in the regulation of CSR through the R‐loop pathway. DEAD‐box RNA helicase 1 (DDX1) is a DEAD‐box RNA helicase required at the IgH locus for CSR in vivo. DDX1 binds to G4 structures, converting them into an S‐region R‐loop. This process targets AID to the S regions, resulting in the promotion of CSR (Fig. 4d),116 which is how DDX1 promotes R‐loop formation.
Other regulatory pathways
Many other molecules have been discovered that regulate CSR. For example, histone methyltransferase and multiple myeloma SET domain can promote AID‐mediated DNA breaks during CSR.117 CD11b, an integrin expressed on the surface of activated B2 cells induces AID expression by the nuclear factor‐κB signaling pathway.118 Estrogen receptors (ER) in activated B cells may control CSR by the direct binding of ER to key regulatory elements in the IgH chain locus, such as Sμ, Eμ and the 3′RR.119 basic leucine zipper transcription factor, ATF‐like promotes the production of class‐switched immunoglobulin by positively regulating the expression of Nfil3 and miR155hg, but negatively regulating Wnt10a.120 The BER enzyme methyl‐CpG binding domain protein 4 (Mbd4) participates in MMR‐directed DNA end processing by interacting with MutL homologue 1 (MLH1).121 The nuclear structural protein NuMA interacts with 53BP1, controlling 53BP1 diffusion and negatively regulating 53BP1 in DSB repair.122 The serine/threonine phosphatase PP4 shows a promoting component of CSR by involvement in sequential recruitment of RPA and NBS1.123
Immunoglobulin‐specific isoform induction
In addition to the mechanisms above, certain conditions can also induce immunoglobulin‐specific isoform production. Isotype switch recombination is controlled by cytokine receptors and their downstream signals. It is widely believed that IgG1 and IgG3 are mainly produced in response to protein antigens, but chronic stimulation with these antigens leads to an increase in the proportion of IgG4. Interleukin‐4 can induce IgE and IgG secretion whereas IL‐10 can induce the production of IgG1 or IgG3. Transforming growth factor‐β is involved in the conversion to IgA, and interferon‐γ can induce the production of IgG2a or IgG3.124, 125
There are many newly discovered inducing factors for IgE with a lower activation threshold, so compared with IgG1, IgE has a higher transcription probability in the early choice.126 Thymic stromal lymphopoietin from damaged epithelial cells following allergen stimulation can activate dendritic cells, triggering naive CD4+ T cells to differentiate into inflammatory T helper type 2 effector cells and secrete the cytokines of IL‐4, IL‐9 and IL‐13, which enhance IgE production. Dendritic cells also activate allergen‐specific T helper type 2 memory cells, helping immunoglobulin switches from IgM to IgE.127 In addition, microRNA‐146a up‐regulates the expression of 14‐3‐3σ , which is a key protein of CSR. This process enhances class switch and secretion of IgE in B cells.128
There have also been some recent discoveries about IgG and IgA CSR. Specific inhibitors of elastase cause cells to secrete higher levels of IgG and IgA in mice, and increase transcription of factors involved in murine B‐cell differentiation, like mRNA for AID, IL‐10, B‐cell‐activating factor of the tumor necrosis factor family (BAFF) and a proliferation‐inducing ligand (APRIL).129 Dectin‐1 agonists, like heat‐killed Saccharomyces cerevisiae, can selectively induce IgG1 class switching in lipopolysaccharide‐activated B cells through reinforcing surface expression and direct stimulation of Dectin‐1.130
Conclusion
Given the importance of the diverse repertoire of immunoglobulin in immune responses, it is reasonable to suppose that sophisticated genetic mechanisms have evolved to generate and regulate these proteins. However, these mechanisms are still not fully elucidated. After more than one century of research, it has been discovered that the generation of immunoglobulin undergoes a series of specific genetic events including V(D)J recombination, CSR and SHM. V(D)J recombination contributes to the generation of surprising immunoglobulin diversity. CSR and SHM can increase the affinity of immunoglobulin to recognize and bind with various antigens. Even though the molecular mechanisms of V(D)J recombination, CSR and SHM have been studied in detail, much remains unclear. For example, how inefficient AID initiates CSR and SHM, how CSR and SHM are well regulated and compete with error‐free DNA repair pathways, and how B cells regulate LSR are still not well understood. Identification of regulatory factors and their function in V(D)J recombination, SHM and CSR, especially those associated with RAG and AID, do require further elucidation.
Disclosures
The authors declare no financial or commercial conflicts of interest.
Supporting information
Figure S1. Immunoglobulin gene locus and IgG structure.
Figure S2. RSS sequence and location.
Figure S3. RAG1/2 structure and function.
Figure S4. Histone modification and contraction of V cluster.
Figure S5. Molecules that regulate the amount of RAG by FOXO.
Figure S6. Subnuclear location of the IgH locus.
Acknowledgements
This work was supported by research grants from the Key Support Projects of the National Natural Science Foundation's Major Research Programme (91642206), The Strategic Priority Research Programme of the Chinese Academy of Sciences, China (XDA16010500), Major International Cooperation Projects of the National Natural Science Foundation (81320108020), The National Natural Science Fund for Distinguished Young Scholars (31625016), The Research Institute Fund of the NHC Key Laboratory of Medical Immunology, Peking University (BMU2018JDJS010), The Non‐profit Central Research Institute Fund of the Chinese Academy of Medical Sciences (2017PT31037), Shenzhen Science and Technology Programme (JCYJ20170413141047772 and JCYJ20180507181659781), CAS Key Research Projects of Frontier Science (QYZDY‐SSW‐SMC027), The CAS Youth Innovation Promotion Association (2018133 and 2016097), and The Shanghai Municipal Science and Technology Major Project (2017SHZDZX01).
References
- 1. Bashford‐Rogers RJM, Smith KGC, Thomas DC. Antibody repertoire analysis in polygenic autoimmune diseases. Immunology 2018; 155:3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boyd SD, Joshi SA. High‐throughput DNA sequencing analysis of antibody repertoires. Microbiol Spectr 2014; 2:345–62. [DOI] [PubMed] [Google Scholar]
- 3. Hwang JK, Alt FW, Yeap LS. Related mechanisms of antibody somatic hypermutation and class switch recombination. Microbiol Spectr 2015; 3:MDNA3‐0037‐2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu Z, Zan H, Pone EJ, Mai T, Casali P. Immunoglobulin class‐switch DNA recombination: induction, targeting and beyond. Nat Rev Immunol 2012; 12:517–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carmona LM, Schatz DG. New insights into the evolutionary origins of the recombination‐activating gene proteins and V(D)J recombination. FEBS J 2017; 284:1590–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim MS, Lapkouski M, Yang W, Gellert M. Crystal structure of the V(D)J recombinase RAG1‐RAG2. Nature 2015; 518(7540):507–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Parkinson NJ, Roddis M, Ferneyhough B, Zhang G, Marsden AJ, Maslau S et al Violation of the 12/23 rule of genomic V(D)J recombination is common in lymphocytes. Genome Res 2015; 25:226–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rodgers KK. Riches in RAGs: Revealing the V(D)J Recombinase through High‐Resolution Structures. Trends Biochem Sci 2017; 42:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ru H, Chambers MG, Fu TM, Tong AB, Liao M, Wu H. Molecular mechanism of V(D)J recombination from synaptic RAG1‐RAG2 complex structures. Cell 2015; 163:1138–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Villartay JP. Congenital defects in V(D)J recombination. Br Med Bull 2015; 114:157–67. [DOI] [PubMed] [Google Scholar]
- 11. Zagelbaum J, Shimazaki N, Esguerra ZA, Watanabe G, Lieber MR, Rothenberg E. Real‐time analysis of RAG complex activity in V(D)J recombination. Proc Natl Acad Sci USA 2016; 113:11853–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Panchakshari RA, Zhang X, Kumar V, Du Z, Wei PC, Kao J et al DNA double‐strand break response factors influence end‐joining features of IgH class switch and general translocation junctions. Proc Natl Acad Sci USA 2018; 115:762–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lescale C, Abramowski V, Bedora‐Faure M, Murigneux V, Vera G, Roth DB et al RAG2 and XLF/Cernunnos interplay reveals a novel role for the RAG complex in DNA repair. Nat Commun 2016; 7:10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maezawa S, Nakano S, Kuniya T, Koiwai O, Koiwai K. Double‐strand break repair based on short‐homology regions is suppressed under terminal deoxynucleotidyltransferase expression, as revealed by a novel vector system for analysing DNA repair by nonhomologous end joining. FEBS Open Bio 2016; 6:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non‐homologous DNA end joining and alternative pathways to double‐strand break repair. Nat Rev Mol Cell Biol 2017; 18:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang HH, Lieber MR. Structure‐Specific nuclease activities of Artemis and the Artemis: DNA‐PKcs complex. Nucleic Acids Res 2016; 44:4991–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pryor JM, Waters CA, Aza A, Asagoshi K, Strom C, Mieczkowski PA et al Essential role for polymerase specialization in cellular nonhomologous end joining. Proc Natl Acad Sci USA 2015; 112:E4537–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tadi SK, Tellier‐Lebegue C, Nemoz C, Drevet P, Audebert S, Roy S et al PAXX is an accessory c‐NHEJ factor that associates with Ku70 and has overlapping functions with XLF. Cell Rep 2016; 17:541–55. [DOI] [PubMed] [Google Scholar]
- 19. Ochi T, Blackford AN, Coates J, Jhujh S, Mehmood S, Tamura N et al PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double‐strand break repair. Science 2015; 347:185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Truong LN, Li Y, Shi LZ, Hwang PY, He J, Wang H et al Microhomology‐mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double‐strand breaks in mammalian cells. Proc Natl Acad Sci USA 2013; 110:7720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M et al IgH class switching and translocations use a robust non‐classical end‐joining pathway. Nature 2007; 449(7161):478–82. [DOI] [PubMed] [Google Scholar]
- 22. Wei H, Yu X. Functions of PARylation in DNA damage repair pathways. Genomics Proteomics Bioinformatics 2016; 14:131–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deng SK, Chen H, Symington LS. Replication protein A prevents promiscuous annealing between short sequence homologies: implications for genome integrity. BioEssays 2015; 37:305–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deng SK, Gibb B, de Almeida MJ, Greene EC, Symington LS. RPA antagonizes microhomology‐mediated repair of DNA double‐strand breaks. Nat Struct Mol Biol 2014; 21:405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mateos‐Gomez PA, Gong F, Nair N, Miller KM, Lazzerini‐Denchi E, Sfeir A. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature 2015; 518:254–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Doksani Y, de Lange T. Telomere‐internal double‐strand breaks are repaired by homologous recombination and PARP1/Lig3‐dependent end‐joining. Cell Rep 2016; 17:1646–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sfeir A, Symington LS. Microhomology‐mediated end joining: a back‐up survival mechanism or dedicated pathway? Trends Biochem Sci 2015; 40:701–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Motea EA, Berdis AJ. Terminal deoxynucleotidyl transferase: the story of a misguided DNA polymerase. Biochim Biophys Acta 2010; 1804:1151–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yurchenko V, Xue Z, Sadofsky M. The RAG1 N‐terminal domain is an E3 ubiquitin ligase. Genes Dev 2003; 17:581–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Teng G, Schatz DG. Regulation and evolution of the RAG recombinase. Adv Immunol 2015; 128:1–39. [DOI] [PubMed] [Google Scholar]
- 31. Hewitt SL, Wong JB, Lee JH, Nishana M, Chen H, Coussens M et al The conserved ATM kinase RAG2‐S365 phosphorylation site limits cleavage events in individual cells independent of any repair defect. Cell Rep 2017; 21:979–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Z, Dordai DI, Lee J, Desiderio S. A conserved degradation signal regulates RAG‐2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity 1996; 5:575–89. [DOI] [PubMed] [Google Scholar]
- 33. Um JH, Brown AL, Singh SK, Chen Y, Gucek M, Lee BS et al Metabolic sensor AMPK directly phosphorylates RAG1 protein and regulates V(D)J recombination. Proc Natl Acad Sci USA 2013; 110:9873–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell 2010; 141:419–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bettridge J, Na CH, Pandey A, Desiderio S. H3K4me3 induces allosteric conformational changes in the DNA‐binding and catalytic regions of the V(D)J recombinase. Proc Natl Acad Sci USA 2017; 114:1904–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ward A, Kumari G, Sen R, Desiderio S. The RAG‐2 inhibitory domain gates accessibility of the V(D)J recombinase to chromatin. Mol Cell Biol 2018; 38:e00159–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kudithipudi S, Schuhmacher MK, Kebede AF, Jeltsch A. The SUV39H1 protein lysine methyltransferase methylates chromatin proteins involved in heterochromatin formation and VDJ recombination. ACS Chem Biol 2017; 12:958–68. [DOI] [PubMed] [Google Scholar]
- 38. Galloway A, Saveliev A, Lukasiak S, Hodson DJ, Bolland D, Balmanno K et al RNA‐binding proteins ZFP36L1 and ZFP36L2 promote cell quiescence. Science 2016; 352:453–9. [DOI] [PubMed] [Google Scholar]
- 39. Timblin GA, Schlissel MS. Ebf1 and c‐Myb repress rag transcription downstream of Stat5 during early B cell development. J Immunol 2013; 191:4676–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Herzog S, Reth M, Jumaa H. Regulation of B‐cell proliferation and differentiation by pre‐B‐cell receptor signalling. Nat Rev Immunol 2009; 9:195–205. [DOI] [PubMed] [Google Scholar]
- 41. Kuo TC, Schlissel MS. Mechanisms controlling expression of the RAG locus during lymphocyte development. Curr Opin Immunol 2009; 21:173–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boller S, Li R, Grosschedl R. Defining B cell chromatin: lessons from EBF1. Trends Genet 2018; 34:257–69. [DOI] [PubMed] [Google Scholar]
- 43. Ochodnicka‐Mackovicova K, Bahjat M, Maas C, van der Veen A, Bloedjes TA, de Bruin AM et al The DNA Damage response regulates RAG1/2 expression in Pre‐B cells through ATM‐FOXO1 signaling. J Immunol 2016; 197:2918–29. [DOI] [PubMed] [Google Scholar]
- 44. Fisher MR, Rivera‐Reyes A, Bloch NB, Schatz DG, Bassing CH. Immature lymphocytes inhibit Rag1 and Rag2 transcription and V(D)J recombination in response to DNA double‐strand breaks. J Immunol 2017; 198:2943–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Skok JA, Brown KE, Azuara V, Caparros ML, Baxter J, Takacs K et al Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat Immunol 2001; 2:848–54. [DOI] [PubMed] [Google Scholar]
- 46. Holwerda SJ, van de Werken HJ, Ribeiro de Almeida C, Bergen IM, de Bruijn MJ, Verstegen MJ et al Allelic exclusion of the immunoglobulin heavy chain locus is independent of its nuclear localization in mature B cells. Nucleic Acids Res 2013; 41:6905–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Powers SE, Mandal M, Matsuda S, Miletic AV, Cato MH, Tanaka A et al Subnuclear cyclin D3 compartments and the coordinated regulation of proliferation and immunoglobulin variable gene repression. J Exp Med 2012; 209:2199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Roldan E, Fuxa M, Chong W, Martinez D, Novatchkova M, Busslinger M et al Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy‐chain gene. Nat Immunol 2005; 6:31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ebert A, Hill L, Busslinger M. Spatial regulation of V‐(D)J recombination at antigen receptor loci. Adv Immunol 2015; 128:93–121. [DOI] [PubMed] [Google Scholar]
- 50. Daly J, Licence S, Nanou A, Morgan G, Martensson IL. Transcription of productive and nonproductive VDJ‐recombined alleles after IgH allelic exclusion. EMBO J 2007; 26:4273–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V‐to‐DJ rearrangements and locus contraction of the immunoglobulin heavy‐chain gene. Genes Dev 2004; 18:411–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu H, Schmidt‐Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H et al Yin Yang 1 is a critical regulator of B‐cell development. Genes Dev 2007; 21:1179–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Merkenschlager M, Odom DT. CTCF and cohesin: linking gene regulatory elements with their targets. Cell 2013; 152:1285–97. [DOI] [PubMed] [Google Scholar]
- 54. Rawat P, Jalan M, Sadhu A, Kanaujia A, Srivastava M. Chromatin domain organization of the TCRb locus and its perturbation by ectopic CTCF binding. Mol Cell Biol 2017; 37:e00557–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Braikia FZ, Conte C, Moutahir M, Denizot Y, Cogne M, Khamlichi AA. Developmental switch in the transcriptional activity of a long‐range regulatory element. Mol Cell Biol 2015; 35:3370–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stengel KR, Barnett KR, Wang J, Liu Q, Hodges E, Hiebert SW et al Deacetylase activity of histone deacetylase 3 is required for productive VDJ recombination and B‐cell development. Proc Natl Acad Sci USA 2017; 114:8608–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kleiman E, Loguercio S, Feeney AJ. Epigenetic enhancer marks and transcription factor binding influence vκ gene rearrangement in Pre‐B cells and pro‐B cells. Front Immunol 2018; 9:2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dahl M, Kristensen LS, Gronbaek K. Long non‐coding RNAs guide the fine‐tuning of gene regulation in B‐cell development and malignancy. Int J Mol Sci 2018; 19:2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Matheson LS, Bolland DJ, Chovanec P, Krueger F, Andrews S, Koohy H et al Local chromatin features including PU.1 and IKAROS binding and H3K4 methylation shape the repertoire of immunoglobulin kappa genes chosen for V(D)J recombination. Front Immunol 2017; 8:1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Manoharan A, Du Roure C, Rolink AG, Matthias P. De novo DNA Methyltransferases Dnmt3a and Dnmt3b regulate the onset of Igκ light chain rearrangement during early B‐cell development. Eur J Immunol 2015; 45:2343–55. [DOI] [PubMed] [Google Scholar]
- 61. Ribeiro de Almeida C, Hendriks RW, Stadhouders R. Dynamic control of long‐range genomic interactions at the immunoglobulin κ light‐chain locus. Adv Immunol 2015; 128:183–271. [DOI] [PubMed] [Google Scholar]
- 62. Dekker J, Misteli T. Long‐range chromatin interactions. Cold Spring Harb Perspect Biol 2015; 7:a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Barajas‐Mora EM, Kleiman E, Xu J, Carrico NC, Lu H, Oltz EM et al A B‐cell‐specific enhancer orchestrates nuclear architecture to generate a diverse antigen receptor repertoire. Mol Cell 2019; 73:48–60.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tang ES, Martin A. Immunoglobulin gene conversion: synthesizing antibody diversification and DNA repair. DNA Repair (Amst) 2007; 6:1557–71. [DOI] [PubMed] [Google Scholar]
- 65. Zan H, Casali P. Regulation of Aicda expression and AID activity. Autoimmunity 2013; 46:83–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jiricny J. Postreplicative mismatch repair. Cold Spring Harb Perspect Biol. 2013; 5:a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Krokan HE, Bjoras M. Base excision repair. Cold Spring Harb Perspect Biol. 2013; 5:a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem 2007; 76:1–22. [DOI] [PubMed] [Google Scholar]
- 69. Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell 2007; 129:665–79. [DOI] [PubMed] [Google Scholar]
- 70. Matthews AJ, Zheng S, DiMenna LJ, Chaudhuri J. Regulation of immunoglobulin class‐switch recombination: choreography of noncoding transcription, targeted DNA deamination, and long‐range DNA repair. Adv Immunol 2014; 122:1–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Feldman S, Wuerffel R, Achour I, Wang L, Carpenter PB, Kenter AL. 53BP1 contributes to Igh locus chromatin topology during class switch recombination. J Immunol 2017; 198:2434–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kim A, Han L, Santiago GE, Verdun RE, Yu K. Class‐switch recombination in the absence of the IgH 3′ regulatory region. J Immunol 2016; 197:2930–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Qiao Q, Wang L, Meng FL, Hwang JK, Alt FW, Wu H. AID recognizes structured DNA for class switch recombination. Mol Cell 2017; 67:361–73.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Basu U, Chaudhuri J, Alpert C, Dutt S, Ranganath S, Li G et al The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature 2005; 438:508–11. [DOI] [PubMed] [Google Scholar]
- 75. Eccleston J, Yan C, Yuan K, Alt FW, Selsing E. Mismatch repair proteins MSH2, MLH1, and EXO1 are important for class‐switch recombination events occurring in B cells that lack nonhomologous end joining. J Immunol 2011; 186:2336–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ghosh R, Das D, Franco S. The role for the DSB response pathway in regulating chromosome translocations. Adv Exp Med Biol 2018; 1044:65–87. [DOI] [PubMed] [Google Scholar]
- 77. Khair L, Guikema JE, Linehan EK, Ucher AJ, Leus NG, Ogilvie C et al ATM increases activation‐induced cytidine deaminase activity at downstream S regions during class‐switch recombination. J Immunol 2014; 192:4887–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Robert I, Gaudot L, Yelamos J, Noll A, Wong HK, Dantzer F et al Robust immunoglobulin class switch recombination and end joining in Parp9‐deficient mice. Eur J Immunol 2017; 47:665–76. [DOI] [PubMed] [Google Scholar]
- 79. Dong J, Panchakshari RA, Zhang T, Zhang Y, Hu J, Volpi SA et al Orientation‐specific joining of AID‐initiated DNA breaks promotes antibody class switching. Nature 2015; 525:134–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Boboila C, Alt FW, Schwer B. Classical and alternative end‐joining pathways for repair of lymphocyte‐specific and general DNA double‐strand breaks. Adv Immunol 2012; 116:1–49. [DOI] [PubMed] [Google Scholar]
- 81. Chen K, Xu W, Wilson M, He B, Miller NW, Bengten E et al Immunoglobulin D enhances immune surveillance by activating antimicrobial, proinflammatory and B cell‐stimulating programs in basophils. Nat Immunol 2009; 10:889–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rouaud P, Saintamand A, Saad F, Carrion C, Lecardeur S, Cogne M et al Elucidation of the enigmatic IgD class‐switch recombination via germline deletion of the IgH 3′ regulatory region. J Exp Med 2014; 211:975–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Choi JH, Wang KW, Zhang D, Zhan X, Wang T, Bu CH et al IgD class switching is initiated by microbiota and limited to mucosa‐associated lymphoid tissue in mice. Proc Natl Acad Sci USA 2017; 114:E1196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mayer CT, Gazumyan A, Kara EE, Gitlin AD, Golijanin J, Viant C et al The microanatomic segregation of selection by apoptosis in the germinal center. Science 2017; 358:eaao2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Dalloul I, Boyer F, Dalloul Z, Pignarre A, Caron G, Fest T et al Locus suicide recombination actively occurs on the functionally rearranged IgH allele in B‐cells from inflamed human lymphoid tissues. PLoS Genet 2019; 15:e1007721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Pinaud E, Aupetit C, Chauveau C, Cogne M. Identification of a homolog of the Cα 3′/hs3 enhancer and of an allelic variant of the 3′IgH/hs1,2 enhancer downstream of the human immunoglobulin α 1 gene. Eur J Immunol 1997; 27:2981–5. [DOI] [PubMed] [Google Scholar]
- 87. Peron S, Laffleur B, Denis‐Lagache N, Cook‐Moreau J, Tinguely A, Delpy L et al AID‐driven deletion causes immunoglobulin heavy chain locus suicide recombination in B cells. Science 2012; 336:931–4. [DOI] [PubMed] [Google Scholar]
- 88. Boutouil H, Boyer F, Cook‐Moreau J, Cogne M, Peron S. IgH locus suicide recombination does not depend on NHEJ in contrast to CSR in B cells. Cell Mol Immunol. 2019; 16:201–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wu H, Deng Y, Feng Y, Long D, Ma K, Wang X et al Epigenetic regulation in B‐cell maturation and its dysregulation in autoimmunity. Cell Mol Immunol 2018; 15:676–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. McBride KM, Gazumyan A, Woo EM, Barreto VM, Robbiani DF, Chait BT et al Regulation of hypermutation by activation‐induced cytidine deaminase phosphorylation. Proc Natl Acad Sci USA 2006; 103:8798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. McBride KM, Gazumyan A, Woo EM, Schwickert TA, Chait BT, Nussenzweig MC. Regulation of class switch recombination and somatic mutation by AID phosphorylation. J Exp Med 2008; 205:2585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Gazumyan A, Timachova K, Yuen G, Siden E, Di Virgilio M, Woo EM et al Amino‐terminal phosphorylation of activation‐induced cytidine deaminase suppresses c‐myc/IgH translocation. Mol Cell Biol 2011; 31:442–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Stavnezer J. Complex regulation and function of activation‐induced cytidine deaminase. Trends Immunol 2011; 32:194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Methot SP, Litzler LC, Trajtenberg F, Zahn A, Robert F, Pelletier J et al Consecutive interactions with HSP90 and eEF1A underlie a functional maturation and storage pathway of AID in the cytoplasm. J Exp Med 2015; 212:581–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1‐dependent nuclear export of activation‐induced deaminase. J Exp Med 2004; 199:1235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Aoufouchi S, Faili A, Zober C, D'Orlando O, Weller S, Weill JC et al Proteasomal degradation restricts the nuclear lifespan of AID. J Exp Med 2008; 205:1357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Uchimura Y, Barton LF, Rada C, Neuberger MS. REG‐γ associates with and modulates the abundance of nuclear activation‐induced deaminase. J Exp Med 2011; 208:2385–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Budzynska PM, Kylaniemi MK, Kallonen T, Soikkeli AI, Nera KP, Lassila O et al Bach2 regulates AID‐mediated immunoglobulin gene conversion and somatic hypermutation in DT40 B cells. Eur J Immunol 2017; 47:993–1001. [DOI] [PubMed] [Google Scholar]
- 99. Ochiai K, Katoh Y, Ikura T, Hoshikawa Y, Noda T, Karasuyama H et al Plasmacytic transcription factor Blimp‐1 is repressed by Bach2 in B cells. J Biol Chem 2006; 281:38226–34. [DOI] [PubMed] [Google Scholar]
- 100. Huang C, Geng H, Boss I, Wang L, Melnick A. Cooperative transcriptional repression by BCL6 and BACH2 in germinal center B‐cell differentiation. Blood 2014; 123:1012–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Shapiro‐Shelef M, Calame K. Regulation of plasma‐cell development. Nat Rev Immunol 2005; 5:230–42. [DOI] [PubMed] [Google Scholar]
- 102. Orthwein A, Zahn A, Methot SP, Godin D, Conticello SG, Terada K et al Optimal functional levels of activation‐induced deaminase specifically require the Hsp40 DnaJa1. EMBO J 2012; 31:679–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Santos JM, Braikia FZ, Oudinet C, Haddad D, Conte C, Dauba A et al Duplication of a germline promoter downstream of the IgH 3′ regulatory region impairs class switch recombination. Sci Rep 2018; 8:9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Vincent‐Fabert C, Fiancette R, Pinaud E, Truffinet V, Cogne N, Cogne M et al Genomic deletion of the whole IgH 3′ regulatory region (hs3a, hs1,2, hs3b, and hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood 2010; 116:1895–8. [DOI] [PubMed] [Google Scholar]
- 105. Lieberson R, Giannini SL, Birshtein BK, Eckhardt LA. An enhancer at the 3′ end of the mouse immunoglobulin heavy chain locus. Nucleic Acids Res 1991; 19:933–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Pinaud E, Khamlichi AA, Le Morvan C, Drouet M, Nalesso V, Le Bert M et al Localization of the 3′ IgH locus elements that effect long‐distance regulation of class switch recombination. Immunity 2001; 15:187–99. [DOI] [PubMed] [Google Scholar]
- 107. Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T et al S‐S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation‐induced deaminase. Immunity 2007; 27:711–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Saintamand A, Rouaud P, Saad F, Rios G, Cogne M, Denizot Y. Elucidation of IgH 3′ region regulatory role during class switch recombination via germline deletion. Nat Commun 2015; 6:7084. [DOI] [PubMed] [Google Scholar]
- 109. Issaoui H, Ghazzaui N, Saintamand A, Denizot Y, Boyer F. IgD class switch recombination is not controlled through the immunoglobulin heavy chain 3′ regulatory region super‐enhancer. Cell Mol Immunol 2017; 14:871–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Issaoui H, Ghazzaui N, Saintamand A, Carrion C, Oblet C, Denizot Y. The IgH 3′ regulatory region super‐enhancer does not control IgA class switch recombination in the B1 lineage. Cell Mol Immunol 2018; 15:289–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Stavnezer J, Schrader CE. IgH chain class switch recombination: mechanism and regulation. J Immunol 2014; 193:5370–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zhang S, Wu Y, Zhang W. G‐quadruplex structures and their interaction diversity with ligands. ChemMedChem 2014; 9:899–911. [DOI] [PubMed] [Google Scholar]
- 113. Dalloul Z, Chenuet P, Dalloul I, Boyer F, Aldigier JC, Laffleur B et al G‐quadruplex DNA targeting alters class‐switch recombination in B cells and attenuates allergic inflammation. J Allergy Clin Immunol 2018; 142:1352–5. [DOI] [PubMed] [Google Scholar]
- 114. Zheng S, Kusnadi A, Choi JE, Vuong BQ, Rhodes D, Chaudhuri J. NME proteins regulate class switch recombination. FEBS Lett 2019; 593:80–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Pavri R. R Loops in the regulation of antibody gene diversification. Genes (Basel) 2017; 8:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ribeiro de Almeida C, Dhir S, Dhir A, Moghaddam AE, Sattentau Q, Meinhart A et al RNA helicase DDX1 converts RNA G‐quadruplex structures into R‐loops to promote IgH class switch recombination. Mol Cell 2018; 70:650–662.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Nguyen HV, Dong J, Panchakshari RA, Kumar V, Alt FW, Bories JC. Histone methyltransferase MMSET promotes AID‐mediated DNA breaks at the donor switch region during class switch recombination. Proc Natl Acad Sci USA 2017; 114:E10560–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Park S, Sim H, Kim HI, Jeong D, Wu G, Cho SY et al CD11b regulates antibody class switching via induction of AID. Mol Immunol 2017; 87:47–59. [DOI] [PubMed] [Google Scholar]
- 119. Jones BG, Penkert RR, Xu B, Fan Y, Neale G, Gearhart PJ et al Binding of estrogen receptors to switch sites and regulatory elements in the immunoglobulin heavy chain locus of activated B cells suggests a direct influence of estrogen on antibody expression. Mol Immunol 2016; 77:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Morman RE, Schweickert PG, Konieczny SF, Taparowsky EJ. BATF regulates the expression of Nfil3, Wnt10a and miR155hg for efficient induction of antibody class switch recombination in mice. Eur J Immunol 2018; 48:1492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Grigera F, Wuerffel R, Kenter AL. MBD4 facilitates immunoglobulin class switch recombination. Mol Cell Biol 2017; 37:e00316–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Salvador Moreno N, Liu J, Haas KM, Parker LL, Chakraborty C, Kron SJ et al The nuclear structural protein NuMA is a negative regulator of 53BP1 in DNA double‐strand break repair. Nucleic Acids Res 2019; 47:2703–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Chen MY, Hsu WC, Hsu SC, Yang YS, Chuang TH, Lin WJ et al PP4 deficiency leads to DNA replication stress that impairs immunoglobulin class switch efficiency. Cell Death Differ 2019; 26:1221–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Briere F, Servet‐Delprat C, Bridon JM, Saint‐Remy JM, Banchereau J. Human interleukin 10 induces naive surface immunoglobulin D+ (sIgD+) B cells to secrete IgG1 and IgG3. J Exp Med 1994; 179:757–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Malisan F, Briere F, Bridon JM, Harindranath N, Mills FC, Max EE et al Interleukin‐10 induces immunoglobulin G isotype switch recombination in human CD40‐activated naive B lymphocytes. J Exp Med 1996; 183:937–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wu YL, Stubbington MJ, Daly M, Teichmann SA, Rada C. Intrinsic transcriptional heterogeneity in B cells controls early class switching to IgE. J Exp Med 2017; 214:183–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Lin SC, Cheng FY, Liu JJ, Ye YL. Expression and regulation of thymic stromal lymphopoietin and thymic stromal lymphopoietin receptor heterocomplex in the innate‐adaptive immunity of pediatric asthma. Int J Mol Sci 2018; 19:1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Li F, Huang Y, Huang YY, Kuang YS, Wei YJ, Xiang L et al MicroRNA‐146a promotes IgE class switch in B cells via upregulating 14‐3‐3σ expression. Mol Immunol 2017; 92:180–9. [DOI] [PubMed] [Google Scholar]
- 129. Attia Z, Rowe JC, Kim E, Varikuti S, Steiner HE, Zaghawa A et al Inhibitors of elastase stimulate murine B lymphocyte differentiation into IgG‐ and IgA‐producing cells. Eur J Immunol 2018; 48:1295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Seo BS, Park HY, Yoon HK, Yoo YC, Lee J, Park SR. Dectin‐1 agonist selectively induces IgG1 class switching by LPS‐activated mouse B cells. Immunol Lett 2016; 178:114–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Immunoglobulin gene locus and IgG structure.
Figure S2. RSS sequence and location.
Figure S3. RAG1/2 structure and function.
Figure S4. Histone modification and contraction of V cluster.
Figure S5. Molecules that regulate the amount of RAG by FOXO.
Figure S6. Subnuclear location of the IgH locus.


