Summary
During mammalian meiotic prophase I, programmed DNA double-strand breaks are repaired by non-crossover or crossover events, the latter predominantly occurring via the class I crossover pathway and requiring the cyclin N-terminal domain-containing 1(CNTD1) protein. Using an epitope-tagged Cntd1 allele, we detect a short isoform of CNTD1 in vivo that lacks a predicted N-terminal cyclin domain and does not bind cyclin-dependent kinases. Instead, we find that the short-form CNTD1 variant associates with components of the replication factor C (RFC) machinery to facilitate crossover formation, and with the E2 ubiquitin conjugating enzyme, CDC34, to regulate ubiquitylation and subsequent degradation of the WEE1 kinase, thereby modulating cell-cycle progression. We propose that these interactions facilitate a role for CNTD1 as a stop-go regulator during prophase I, ensuring accurate and complete crossover formation before allowing metaphase progression and the first meiotic division.
Keywords: meiosis, recombination, CNTD1, RFC, CDC34, WEE1, CDK1, CDK2, MutLγ, ubiquitylation
Graphical Abstract

Highlights
-
•
CNTD1 associates with sites of crossing over in meiosis, co-localizing with MutLγ
-
•
In the testis, CNTD1 does not interact with CDKs or with known crossover regulators
-
•
CNTD1 regulates crossing over via interactions with the replication factor C complex
-
•
CNTD1 regulates cell-cycle progression via interactions with the SCF complex
The cyclin family member, CNTD1, is critical for establishing crossovers during meiosis. The predominant CNTD1 isoform in mouse spermatocytes lacks features that enable interaction with CDKs. Gray et al. find that CNTD1 coordinates crossover regulation with cell-cycle progression through interactions with the replication factor C and SKP1-Cullin-F-box ubiquitin ligase complexes.
Introduction
Meiosis is a specialized cell division consisting of one round of DNA replication followed by two rounds of chromosome segregation. During meiosis I, homologous chromosomes must pair, synapse, and form crossovers to enable accurate segregation at the first meiotic division (MI). During the second meiotic division (MII), the paired sister chromatids segregate, resulting in the formation of haploid gametes (Gray and Cohen, 2016).
Crossing over is achieved through meiotic recombination, which is dependent on, and required for, accurate synapsis. Synapsis is achieved by the formation of the synaptonemal complex (SC), whose assembly and status defines the five sub-stages of prophase I. Meiotic recombination is initiated by the formation of programmed DNA double-strand breaks (DSBs) throughout the genome in early leptonema (Baudat et al., 2000; Keeney et al., 1997, 1999, Robert et al., 2016a, 2016b; Romanienko and Camerini-Otero, 2000). In the mouse, 200–300 DSBs are generated and processed through common intermediate repair steps to yield either non-crossover or crossover events (Gray and Cohen, 2016). While the majority of DSBs (∼90% in the mouse) repair as non-crossovers in zygonema and early pachynema, the crossovers that form during pachynema establish the inter-homolog tethers that allow for their correct segregation at MI (Cole et al., 2014). Thus, the correct timing, frequency, and distribution of crossovers ensures that at least one crossover per pair is generated (the obligate crossover) (Jones, 1984), that crossovers are maintained at the expense of non-crossovers (crossover homeostasis) (Cole et al., 2012; Martini et al., 2006), and that the formation of one crossover prevents the formation of nearby crossovers (interference) (Jones and Franklin, 2006). How these rules are enforced within the realm of DSB repair and shape crossover and non-crossover decisions remains unclear.
In the mouse, crossovers can form by at least two mechanisms. The class I (ZMM) pathway is responsible for the majority of crossovers and uses the DNA mismatch repair endonuclease MutLγ, a heterodimer of MLH1 and MLH3 (Edelmann et al., 1996; Lipkin et al., 2002). MLH1 and MLH3 co-localize along chromosome cores at a frequency of ∼23 foci at pachynema (Baker et al., 1996), reflecting 90%–95% of the final crossover count in male meiosis (Lipkin et al., 2002; Svetlanov et al., 2008). The distribution of MutLγ foci during pachynema in mouse spermatocytes is interference dependent and satisfies the requirement for the obligate crossover (Broman et al., 2002). Loss of either component of MutLγ results in infertility due to the loss of most crossovers, leading to arrest before metaphase I in males and meiosis I non-disjunction in females (Baker et al., 1996; Edelmann et al., 1996; Kolas et al., 2005; Lipkin et al., 2002). The remaining 5%–10% crossovers are generated by the class II MUS81-EME1-dependent pathway. These class II crossovers are interference independent, but cannot account for all of the class I-independent crossovers since the mutation of both Mlh3 and Mus81 leads to the persistence of 1–3 chiasmata (Holloway et al., 2008).
One of the major questions in mammalian meiosis concerns how crossovers are selected from the initial pool of 200–300 DSB repair intermediates. Initially, a subset (∼150) of these repair intermediates accrue the MutSγ heterodimer of MSH4 and MSH5 (Edelmann et al., 1999; Kneitz et al., 2000), an event called crossover licensing. Of these, only 23–26 MutSγ sites subsequently become loaded with MutLγ to form class I crossovers, while the remaining sites are repaired either through the class II crossover pathway or via the formation of non-crossovers (Cole et al., 2014; Holloway et al., 2008; Milano et al., 2019). The mechanism by which MutSγ becomes further selected by the accrual of MutLγ has been called crossover designation, leading to the idea that crossover homeostasis is imposed sequentially by the association of these pro-crossover MutS/MutL proteins (Cole et al., 2012; Hunter, 2015; Moens et al., 2002).
Recent studies have revealed a number of regulatory molecules that aid in crossover designation and that are essential for class I crossovers, including crossover site-associated-1 (COSA-1) in Caenorhabditis elegans (Yokoo et al., 2012) and its mammalian ortholog cyclin N-terminal domain-containing-1 (CNTD1) (Holloway et al., 2014). Loss of COSA-1 in worms results in a failure to accumulate MSH-5 at DSB repair intermediates and the loss of all crossovers (Yokoo et al., 2012). The loss of CNTD1 in the mouse results in similar meiotic failure characterized by persistently elevated early crossover factors through pachynema and failure to load crossover designation factors such as MutLγ, the crossover site-associated cyclin-dependent kinase-2 (CDK2), and the putative ubiquitin E3 ligase HEI10 (Holloway et al., 2014). In the present study, we sought to further elucidate the function of CNTD1 in driving crossover formation. We generated a dual epitope-tagged allele of Cntd1 (Cntd1 FH ) to facilitate the tracking of CNTD1 protein in mouse spermatocytes and to enable the analysis of the CNTD1 interactome. We find that CNTD1 localizes to sites that are loaded with MutLγ, but surprisingly, the predominant form of CNTD1 in spermatocytes lacks the ability to interact with known meiotic CDKs or crossover factors. Instead, CNTD1 drives crossover designation and cell-cycle progression through distinct interactions with key regulatory complexes involved in facilitating the activity of MutLγ (namely replication factor C [RFC]) and those involved in modulating cell-cycle progression through the ubiquitylation of critical cell-cycle regulators (namely CDC34-containing SKP1-Cullin-Fbox [SCF] complex).
Results
Epitope Tagging of CNTD1 to Create a Cntd1FLAG-HA Allele Reveals a Short-Form CNTD1
We used CRISPR-Cas9 to generate a dual C-terminal FLAG-hemagglutinin (HA) epitope tagged allele (Figure S2A), called Cntd1 FH. The dual tag was placed at the 3′ end of the protein, preceding the stop codon. Cntd1 FH/FH male mice are almost indistinguishable from wild-type littermates (Figure S1).
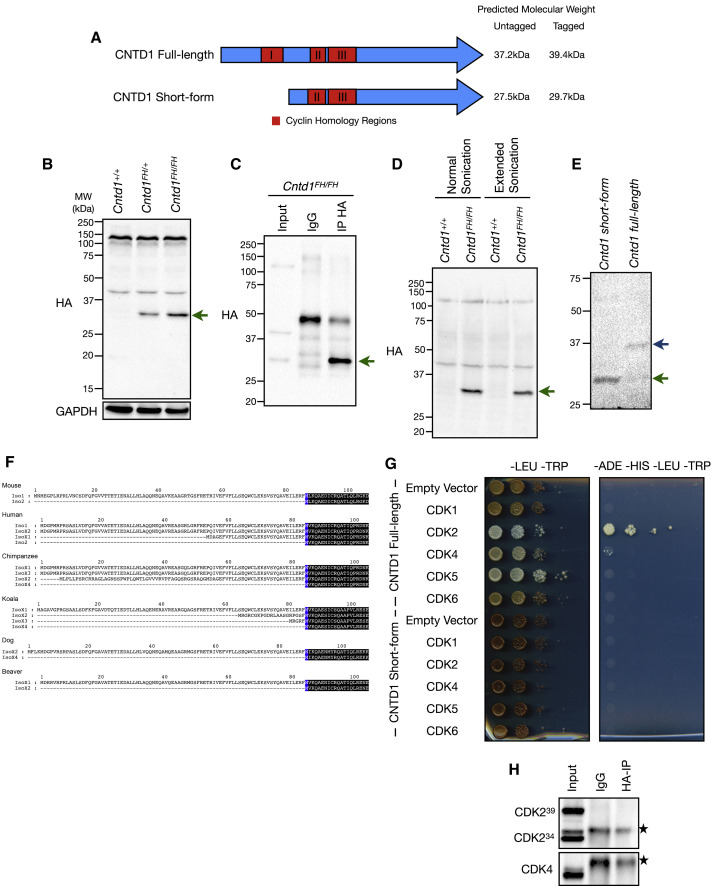
Annotation of the Cntd1 genomic locus describes a 7-exon gene encoding a 334-amino acid protein with a predicted molecular weight of ∼40 kDa for the full-length, tagged form (Figures 1 A and S2A) (NCBI: NM_026562). Western blotting (WB) of whole testis extracts from Cntd1 +/+, Cntd1 FH/+, and Cntd1 FH/FH adult matched littermates demonstrated the presence of the protein specifically in mice bearing the Cntd1 FH allele, but revealed a smaller-than-expected band at ∼30 kDa, present only in the testis (Figures 1B and S2B, arrow). Prior characterization of the Cntd1 locus described the use of a start codon near the beginning of exon 3 (NCBI: DS033671), which would produce a 27.5-kDa endogenous protein and a 29.7-kDa tagged protein, matching the size we observe by WB (Figures 1A, 1B, and S2A). We call this protein product CNTD1 short form. Since a non-specific band around the predicted size (40 kDa) for CNTD1FH is observed, we undertook immunoprecipitation (IP) followed by WB using two independent anti-HA antibodies to determine whether a form of Cntd1 FH exists that is obscured by this non-specific band. We observe the enrichment of the smaller 30-kDa band in the IP fraction (Figure 1C, arrow), loss of the non-specific band at ∼40 kDa, and no additional specific bands (Figure 1C).
Figure 1.
CNTD1FH Protein Is Smaller Than Predicted, Lacking an N-Terminal Cyclin Homology Required for CDK Interaction
(A) Schematic of annotated full-length and short-form CNTD1, including predicted cyclin homology regions (red).
(B) WB using antibodies against the HA epitope and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) from testis lysate of Cntd1+/+, Cntd1FH/+, and Cntd1FH/FH mice. The green arrow indicates the CNTD1-specific band.
(C) WB using the rabbit HA antibody against immunoprecipitated testis lysate from Cntd1FH/FH mice using rat anti-HA antibody. The green arrow indicates the specific enriched band.
(D) WB using the anti-HA antibody against testis lysate from Cntd1+/+ and Cntd1FH/FH mice following normal and extended sonication. The green arrow indicates the specific band.
(E) WB using the anti-HA antibody against yeast lysate expressing Cntd1 short-form and full-length Cntd1 from PADH1.
(F) N-terminal splice variants of mammalian CNTD1 homologs. Blue coloring denotes the conserved methionine of the short form in each homolog, with the immediate downstream amino acid sequences that follow colored black. In the mouse, Iso1 and Iso2 are equivalent to the annotated full-length CNTD1 and CNTD1 short form, respectively.
(G) Yeast two-hybrid with full-length CNTD1 and CNTD1 short form (bait) with CDKs expressed (prey), grown on control (-LEU-TRP) and selective plates (-ADE-HIS-LEU-TRP).
(H) WB using antibodies against CDK2 and CDK4 from anti-HA IP of Cntd1FH/FH testis lysate. The stars indicate the non-specific bands.
We considered the possibility that standard sonication for protein extraction may not sufficiently liberate proteins associated with dense chromatin (T. Tran and J. Schimenti, personal communication). To determine whether this may explain the absence of a native 40-kDa CNTD1 isoform, we undertook an extended sonication technique, but we were unable to identify additional bands by WB (Figure 1D). We also considered whether the CNTD1 full-length protein could migrate smaller than predicted by WB. However, when we expressed constructs encoding the FLAG-HA-tagged full-length and short forms of CNTD1, we detected both forms of the protein by WB using an anti-HA antibody (Figure 1E). The CNTD1 short form migrates at the expected 30 kDa size. Thus, anti-HA antibodies can detect both full-length and short-form CNTD1, but they detect only the latter in mouse spermatocyte extracts, leading us to conclude that the major CNTD1 variant found in mouse testis is the short-form 30-kDa protein.
Comparative Analysis of CNTD1 Orthologs Reveals N-Terminal Variability
Next, we examined the sequences of other CNTD1 orthologs. Sequence alignment shows strong conservation of CNTD1 across a diverse set of species, with a few important caveats. First, we identified an internal methionine that is absolutely conserved throughout the entire family (Figure S3, blue). This methionine corresponds to the alternative start codon that produces the short form of CNTD1 in the mouse (Figures 1F and S3). Second, we observe greater variability in the N-terminal portion of the protein upstream of this methionine (Figure 1F). This manifests as splice variants within species that exhibit multiple annotated isoforms (Figure 1F). Splicing always preferentially modifies the N terminus, leading to truncations that, at a minimum, constitute the short form of the protein. The koala ortholog exemplifies this pattern of variability (Figure 1F). It is important to note that one species, the American alligator, has only a single short isoform described that begins at the alternative methionine start site (Figure S3, bottom row). These in silico findings underscore the evolutionary conservation of the CNTD1 short form and suggest its broader biological importance.
The CNTD1 N Terminus Mediates Interactions with Cyclin-Dependent Kinases
CNTD1 has predicted homology to cyclin proteins across 3 regions: amino acids 57–88, 117–135, and 140–180 (Figure 1A). Truncation of the N terminus in the CNTD1 short form removes the first 85 amino acids, encompassing the majority of cyclin homology region 1 (Figure 1A). Using yeast two-hybrid analysis, we investigated whether the CNTD1 short form would associate with the CDKs that predominate in mouse spermatocytes, CDKs 1, 2, 4, 5, and 6 (Adhikari et al., 2012; Ashley et al., 2001; Barrière et al., 2007; Berthet et al., 2003; Session et al., 2001). The full-length mouse CNTD1 interacts with CDK2 and CDK4 in yeast two-hybrid assays (Figure 1G, top half). The CNTD1 short form, however, fails to interact with any of the tested CDKs (Figure 1G, bottom half). To determine whether CDK2 and CDK4 interactions with CNTD1 exist in vivo, we immunoprecipitated CNTD1FH from mouse testis using the anti-HA antibody and probed with antibodies against CDK2 and CDK4. IP with anti-HA antibodies failed to enrich for either CDK2 or CDK4 (Figure 1H), which is consistent with the conclusion that the CNTD1 short form is the predominant isoform in mouse and lacks the ability to interact with key prophase I CDKs.
CNTD1FH Forms Discrete Foci in Pachytene Spermatocytes
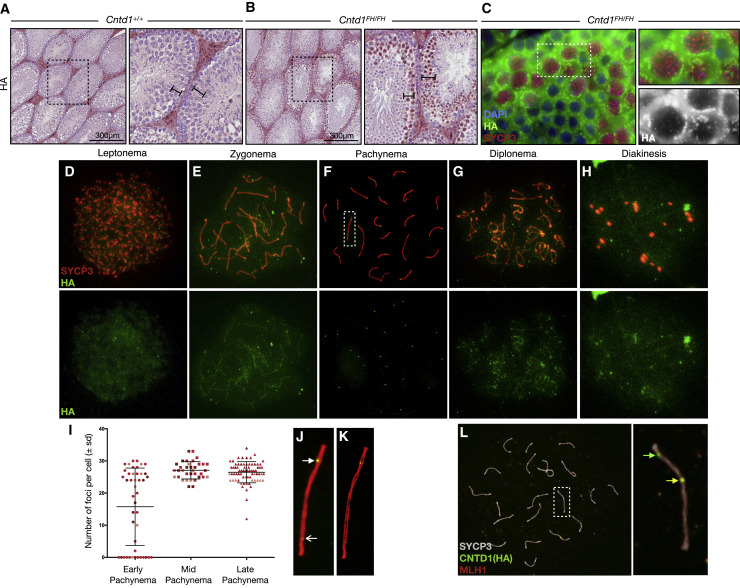
We next undertook immunohistochemistry on testis sections from Cntd1 +/+ and Cntd1 FH/FH littermates. In testis sections from Cntd1 FH/FH adults, we observed staining in primary spermatocytes using antibodies against the HA tag (Figures 2 A, 2B, S4A, and S4B). The localization of CNTD1 is restricted to prophase I cells, but appears to shuttle between the nucleus and cytoplasm in a stage-specific manner (Figures 2B and S4B). Cytoplasmic localization is observed when staining intensity is increased by prolonged exposure to 3,3′-diaminobenzidine (DAB) exposure (Figure S4B, square arrows), but staining remains consistently heaviest in the nucleus (Figure S4B, arrows). No HA signal was observed in testis sections from wild-type littermates (Figures 2A and S4A).
Figure 2.
CNTD1FH Localizes to Spermatocytes and Forms Discrete Foci at Pachytene
(A and B) Immunohistochemistry using anti-HA antibody on formalin-fixed testis sections of Cntd1+/+(A) and Cntd1FH/FH (B) mice, imaged at 200×. Bar, 300 μm.
(C) Immunofluorescence staining using anti-HA (green) and anti-SYCP3 (red) antibodies on formalin-fixed testis sections of Cntd1+/+ and Cntd1FH/FH mice, imaged at 200×. Right-hand panels are enlargements of the dotted-outline box. Lower right-hand box shows the anti-HA staining (white) with cytoplasmic staining and discreet foci.
(D–H) Immunofluorescence staining on spread meiotic spermatocyte preparations from Cntd1FH/FH mice using antibodies against the HA epitope (green) and SYCP3 (red). Leptonema (D), zygonema (E), pachynema (F), diplonema (G), and diakinesis (H) staged spermatocytes defined by SYCP3 morphology.
(I) Quantification of HA foci (means ± SDs) in pachytene spermatocytes of Cntd1FH/FH mice.
(J) Enlarged image of dotted-outline box in (F). The closed arrowhead indicates strong intensity focus; the open arrowhead indicates low intensity focus.
(K) Structural illumination microscopy rendering of (J).
(L) Representative immunofluorescent staining of pachynema spermatocyte from Cntd1FH/FH testis using antibodies against MLH1 (red), HA (green), and SYCP3 (white). The right panel is the enlarged insert of the dotted-outline box. Colocalization of HA and MLH1 is identified by the yellow arrow; the green arrow indicates HA-only foci.
We next used immunofluorescence staining on fixed testis sections and chromosome spread preparations. Using the same anti-HA antibody that we used in WB, which detected only the CNTD1FH short orm, we observed discrete foci of CNTD1FH at pachynema, distributed along the cores of the SC in testis sections and spread meiotic spermatocytes (Figures 2C–2L). For chromosome spreads, we used colocalization with the SC protein SYCP3 to assess the prophase I stage, providing a temporal profile of CNTD1FH through prophase I. In leptonema, we observed diffuse nuclear staining of CNTD1FH, which accumulates along the SC in zygonema, with some foci observed along the SC (Figures 2D and 2E). Throughout pachynema, the diffuse nuclear staining of CNTD1FH is no longer observed, being replaced by discrete foci along the SC at a frequency reminiscent of MutLγ numbers (Figures 2F and J–L; Baker et al., 1996; Lipkin et al., 2002). In early pachynema, localization of CNTD1FH is observed in two distinct patterns: one population of cells has yet to accumulate any CNTD1FH while a second population of cells shows increasing focus frequency (Figure 2I). By mid-pachynema, the focus frequency for CNTD1FH is more homogeneous, at 27 ± 2.7, and persisting at 26.5 ± 3.3 in late pachynema (Figure 2I). By diplonema, the diffuse nuclear staining pattern resumes, but clear foci of HA signal remain associated with the SC (Figures 2G and 2H), and is lost by diakinesis.
From mid-pachynema onward, the number of HA-marked CNTD1 foci is consistently higher than focus frequencies for MLH1 and MLH3 (Baker et al., 1996; Lipkin et al., 2002; Figures 2I and S1F). Dual staining of CNTD1FH and MLH1 reveals the frequent presence of CNTD1FH foci that do not co-localize with MLH1 foci but not vice versa (Figure 2L). However, while we observe colocalization of CNTD1FH and MLH1, given the spatial distance over which fluorescent molecules excite, this only indicates proximity but does not imply direct interaction.
In addition to the temporospatial pattern of CNTD1FH protein signal, we observed 2 distinct intensity patterns for CNTD1FH foci during pachynema: the majority of HA-stained foci are bright and robust, while ∼1–5 foci per cell are of weaker intensity (Figure 2J). Increasing the imaging exposure time does not reveal additional faint foci that are not included in our quantitation nor do we observe any non-specific HA+ signal on chromosome spreads from wild-type males (Figures S4C–S4F).
STA-PUT Protein Profiling
To define the temporal dynamics of CNTD1 appearance through prophase I relative to other key meiotic and cell-cycle regulators, an improved gravitational cell separation (STA-PUT) strategy was devised to allow the recovery of all stages of prophase I in a single procedure (Bellvé, 1993; Bellvé et al., 1977; Romrell et al., 1976). Meiotic chromosome spreads were prepared from 46 fractions and scored based upon SC morphology and presence of the phosphorylated histone variant γH2AFX (Gray and Cohen, 2016). The purity of STA-PUT-enriched fractions for each germ cell stage reached 67% leptonema, 45% zygonema, 89% pachynema, 79% diplonema, and 98% sperm across different fractions (Figure 3 A). WB performed against proteins from the STA-PUT cell fractions resulted in a dynamic profile of protein levels using a battery of antibodies against meiosis regulators (Figure 3A), all compared to the dynamic expression of CNTD1 as detected by the HA tag. Detailed protein dynamics are delineated below, describing the data in Figure 3A.
Figure 3.
STA-PUT Allows Isolation of Prophase I Stage-Specific Fractions Revealing Dynamic Protein Expression
(A) STA-PUT assay performed on Cntd1FH/FH testis single-cell suspensions. Top: quantification of STA-PUT cell fraction composition based on a minimum of 200 cells from spread preparations stained for SYCP3 and γH2AFX. WBs performed using the described antibodies against each STA-PUT fraction. Each WB row is a composite of four individual WBs performed and imaged under identical conditions.
(B) Model depicting the 2 complexes identified as interacting partners with CNTD1 in our MS analysis: RFC (left) and SCF (right). The hypothesized interaction between RFC-PCNA and MutLγ is based upon in vitro experiments. The direct interactions with CNTD1 are depicted by black arrows.
CNTD1FH Expression (HA)
The peak HA signal (for CNTD1FH) is observed within fractions enriched for leptonema through pachynema, which is coincident with the staining observed in spread testis spermatocytes. As with our previous WBs (Figure 1), a single band of 30 kDa is observed in our isolated purified cell populations.
Cell-Cycle Regulation
We observe the expression of CDK1 primarily in fractions enriched for leptotene through pachytene spermatocytes. A form of CDK1 with increased electrophoretic mobility is observed in pachynema. CDK2 has 2 characterized isoforms in the mouse; a 34-kDa short form (CDK234) lacking 48 amino acids compared with the longer 39-kDa (CDK239) isoform generated by alternate splicing (Ellenrieder et al., 2001). The abundance and modifications of the two isoforms differs across STA-PUT-enriched fractions.
The expression of CDK4 is observed in fractions enriched with leptotene, zygotene, pachytene, and sperm cells. CDK4 staining also shows an additional higher-molecular-weight-modified band that appears from leptonema until pachynema, in line with previously published localization patterns for CDK2 and CDK4 (Ashley et al., 2001).
Meiotic Proteins
Expression of the early RING domain crossover factor RNF212 is observed coincident with the enrichment of leptotene cells and spans fractions enriched for leptonema though early pachynema. The appearance of the later-stage crossover protein HEI10 occurs from zygonema onward and persists in pachytene-enriched fractions. The expression of RNF212 and HEI10 fits the temporal progression of crossover sites gaining first, RNF212, followed by HEI10 observed in spread meiotic spermatocytes (Qiao et al., 2014). In addition, Cntd1 mutants load RNF212 but fail to load HEI10 (Holloway et al., 2014). The appearance of the CNTD1FH signal relative to RNF212 and HEI10 fits the temporal observations defined by chromosome spreads, with RNF212 observed earliest, followed by CNTD1FH, and then finally HEI10 (Qiao et al., 2014; Reynolds et al., 2013).
MLH3 protein accumulation is observed from leptotene- through pachytene-enriched fractions. MLH3 antibody detects two different bands of similar molecular weight, indicating possible protein modification. Two isoforms of SYCP3 exist in the mouse, with the longer form a consequence of N-terminal extension (Alsheimer et al., 2010). Both isoforms of SYCP3 are present in leptotene- through diplotene-enriched fractions. The large form is predominant in the sperm- and leptotene-enriched fractions and the short form is predominant in the diplotene-enriched fractions. Both forms of SYCP3 appear to have multiple bands migrating around the similar molecular weight, indicating protein modification. The central element SYCP1 forms between synapsed chromosomes during zygotene and pachytene, and appears in STA-PUT fractions enriched in zygotene through pachytene spermatocytes.
DNA Repair Factors
Following DSB formation, phosphorylation of H2AFX on serine 139 (referred to as γH2AFX) occurs, leading to the recruitment of repair factors and chromatin remodeling. On meiotic spread chromosomes, γH2AFX is observed throughout leptotene as DSBs occur and persists during DSB repair. During zygotene and pachytene γH2AFX appears around the XY chromosomes, forming the sex body. Detection of γH2AFX in the STA-PUT fractions is consistent with these cytological observations. γH2AFX appears as multiple bands throughout leptonema and becomes a more discrete band in pachytene-enriched fractions. These observations suggest that early, DSB-associated γH2AFX may undergo different modifications than that of the XY-associated protein.
Following DSB formation and early processing, RecA homologs RAD51 and DMC1 undertake strand invasion to repair from homologous DNA templates. Consistent with RAD51 staining on chromosome spreads, we observe RAD51 in leptotene through pachytene-enriched fractions, with levels decreasing as the fractions start to contain diplotene cells.
Mass Spectrometry (MS) of CNTD1FH-Enriched STA-PUT Fractions Reveals Interactions with RFC and SCF Complexes
Stage-matched prophase I cell fractions from Cntd1 +/+ and Cntd1 FH/FH testes were used for IP against the HA epitope, followed by MS to identify interacting proteins (green box in Figure S5). A total of 588 proteins (Table S1) were enriched in the CNTD1FH sample compared with Cntd1 +/+. A total of 181 proteins were identified by ≥2 peptides. Given the localization pattern and function in crossover formation, we did not identify any proteins important for class I crossovers (MSH4, MSH5, MLH1, MLH3, RNF212, HEI10, or CDK2), nor did we identify any CDKs, consistent with the predominant expression of Cntd1 short form in mouse testis. Instead, most CNTD1-interacting proteins fall into Gene Ontology groups involved in cellular and metabolic processes (Figure S6A). Of these, we prioritized our continued analysis based on protein interactors that may explain crossover designation, cell-cycle progression, and the recent understanding of the role of post-translational modifications in driving these functions during meiosis (Ahuja et al., 2017; Chen et al., 2015; Reynolds et al., 2013). Based on these criteria, two specific complexes were of particular interest (Figure 3B). First, CNTD1FH interacts with components of the RFC complex, which in somatic cells acts in the recruitment/activation of the DNA mismatch repair pathway (Bowen and Kolodner, 2017; Constantin et al., 2005; Kadyrov et al., 2006). Second, CNTD1FH interacts with many proteins belonging to the ubiquitylation pathway (Figures 3B and S6B), including components of the SKP1-Cullin-Fbox (SCF; reviewed in Lee and Diehl, 2014), namely the E2 conjugating enzyme CDC34 (Figures 3B and S6B).
RFC Complex
RFC is a pentameric complex, consisting of RFC1–5, which loads proliferating cell nuclear antigen (PCNA) onto DNA during DNA replication (Figure 3B; Majka and Burgers, 2004). RFC and PCNA have been implicated in the activation of MutL endonucleases (Bowen and Kolodner, 2017; Constantin et al., 2005; Kadyrov et al., 2006; Smith et al., 2015). Our MS data revealed CNTD1 interaction with RFC3 and RFC4 (Table S1), which was confirmed by anti-HA-IP followed by WB using anti-RFC3 and RFC4 antibodies (Figure 4 A). WB analysis on whole-testis lysate from Cntd1 +/+ and Cntd1 −/− mice demonstrated no difference in the total protein levels of RFC3, RFC4, or PCNA (Figure 4B). In STA-PUT fractions from Cntd1 +/+ testes, we detected RFC3 and RFC4 protein in fractions enriched for leptonema through pachynema cells, while PCNA persists until diplonema (Figure 4C). In STA-PUT fractions from Cntd1 −/− testes, RFC3 and RFC4 protein levels were decreased in lanes representing mostly pachytene cells, compared to similar lanes in WBs from STA-PUT-sorted Cntd1 +/+ fractions (Figure 5 A).
Figure 4.
CNTD1 Mutation Leads to a Decrease in CDC34 Levels Compared to Wild Type
(A and B) WB using antibodies against CNTD1FH-interacting proteins from immunoprecipitated material using anti-HA antibody from Cntd1FH/FH mice (A) and from testis lysate (B) of Cntd1FH/FH, Cntd1FH/−, and Cntd1−/− mice.
(C) STA-PUT assay performed on Cntd1FH/FH testis single-cell suspensions showing expression of CNTD1FH interacting factors and downstream interactors relative to prophase I stage. Top panel: quantification of STA-PUT cell fraction composition based on a minimum of 200 cells from spread preparations stained with SYCP3 and γγH2AFX antibodies. WBs were performed using the described antibodies against each STA-PUT fraction.
(D) WB using antibodies against CDC34 and GAPDH on lysate from whole testis, Wild-type, Cntd1−/−, and Mlh1−/− mice.
(E) WB against predicted downstream CDC34 targets and GAPDH control.
(F) WEE1 inhibitory phosphorylation on CDK1 and CDK2.
Figure 5.
CNTD1 Mutation Causes a Decrease in Nuclear RFC4 Signal and Persistent Increased Nuclear Signal of PCNA in Prophase I Cells
(A) STA-PUT assay performed on Cntd1−/− testis single-cell suspension from 12 adult mice showing expression of RFC3, RFC4, PCNA, and CDC34 relative to prophase I stage. Top panel: quantification of STA-PUT cell fraction composition based on a minimum of 200 cells from spread preparations stained with SYCP3 and γH2AFX antibodies. WBs were performed using the described antibodies against each STA-PUT fraction. Each WB row is an amalgamation of 4 individual WBs performed and imaged under identical conditions.
(B–G) Immunofluorescence staining of pachynema staged spread meiotic spermatocytes from Cntd1FH/FH (B, D, and F) and Cntd1−/− (C, E, and G) mice using RFC3 (B and C), RFC4 (D and E), and PCNA (F and G) antibodies.
(H and I) Immunofluorescence staining using antibodies against RFC4 (green) and SYCP3 (red) on formalin-fixed testis sections of Cntd1FH/FH (H) and Cntd1−/− (I) mice, imaged at 200×. The white bars indicate the prophase I cells. The arrowheads indicate spermatogonia and the arrows indicate spermatocytes. The right-hand panels show representative images of the prophase I stages from enlarged sections of (H) and (I), respectively, staged by SYCP3 morphology.
(J and K) Immunofluorescence staining using antibodies against PCNA (green) and SYCP3 (red) on formalin-fixed testis sections of Cntd1FH/FH (J) and Cntd1−/− (K) mice, imaged at 200×. The white bars indicate the prophase I cells. The arrowheads indicate spermatogonia and the arrows indicate spermatocytes. The right-hand panels show representative images of the prophase I stages from enlarged sections of (J) and (K), respectively, staged by SYCP3 morphology.
Immunofluorescence staining on spread meiotic chromosomes using antibodies against RFC3 and RFC4 revealed faint nuclear staining and focus formation along regions of the synaptonemal complex in pachynema. In pachytene spermatocytes from Cntd1 −/− males, the overall nuclear staining of RFC3 and RFC4 was reduced, as was the specific number of RFC3/4 foci along autosomal synaptonemal complexes compared with that observed in Cntd1 FH/FH cells (Figures 5B–5E). By contrast, PCNA immunofluorescence staining in pachytene spermatocytes from Cntd1 FH/FH males revealed diffuse nuclear staining in addition to discrete foci of PCNA along chromosome cores, some reminiscent of the patterning observed when staining for class I crossover proteins (Figure 5F). By contrast, PCNA staining in pachytene spermatocytes from Cntd1 −/− males was more intense across the entire nucleus, including the signal observed along the synaptonemal complex (Figure 5G).
Immunolocalization of RFC4 protein in testis sections revealed a robust RFC4 signal within the nuclei of spermatogonia and primary spermatocytes in wild-type testes (Figure 5H), with a steadily increasing signal for RFC4 as prophase I progresses (Figure 5B). In testis sections from Cntd1 −/− males, we observe persistent, stronger staining of RFC4 in spermatogonia compared with Cntd1 FH/FH males (Figure 5I), but decreased staining in the primary spermatocytes at all stages of prophase I relative to that observed in Cntd1 +/+ spermatocytes (Figures 5H and 5I, right-hand gray panels, and S7).
PCNA staining of Cntd1 +/+ testis revealed nuclear staining in occasional spermatogonia and all of the primary spermatocytes (Figure 5J). Similar staining is observed in primary spermatocytes in Cntd1 −/− males, with an increased intensity of PCNA signal due to the higher proportion of spermatocytes compared to that observed in adult Cntd1 +/+ testis (Figures 5K and S7). However, the staining intensity in individual sub-stages of prophase I appears persistently more robust in spermatocytes from Cntd1 −/− males than in wild type (Figures 5J and 5K, right-hand gray panels, S7). These results demonstrate the robust localization of RFC and PCNA in late-stage primary spermatocytes, distinct from their expected staining in cells undergoing DNA replication (e.g., proliferating spermatogonia). In this regard, our data suggest that RFC and PCNA may play a role in DSB repair during meiosis that is analogous to their role in DNA repair in somatic cells in regard to MutL complexes (Majka and Burgers, 2004).
CDC34 E2 Conjugating Enzyme
The SCF complex is the most widely characterized of the superfamily of Cullin/RING ubiquitin ligases (CRLs) (Sarikas et al., 2011). The SCF core complex works together with E1 (activating) and E2 (conjugating) enzymes to define ubiquitylation substrates (Figure 3B; Reitsma et al., 2017). Our MS data revealed that CNTD1 interacts with 22 components of the ubiquitylation/de-ubiquitylation family (Figure S6B; Table S1), including the E2 conjugating enzyme CDC34.
The distribution of CDC34 protein across STA-PUT fractions from Cntd1 +/+ males primarily revealed protein in fractions enriched with round spermatids, sperm, leptotene, and zygotene cell types (Figure 4C, fractions 32–42). IP-WB confirmed the interactions between CNTD1FH and CDC34 (Figure 4A), while WB from whole-testis lysates revealed that CDC34 levels in Cntd1 −/− mutants were drastically decreased compared with those of Cntd1 FH/FH animals (Figure 4B). Because of this interaction, we asked whether CDC34 levels were similarly altered in crossover mutants for genes acting downstream of CNTD1. CDC34 protein levels were reduced in Mlh1 −/− mice to a similar extent as those observed in Cntd1 −/− mice (Figure 4D). The WB profile of CDC34 protein across STA-PUT fractions from Cntd1 −/− males, where prophase I cells are present but sperm is absent, showed a similar drastic reduction in CDC34 protein (Figure 5K). Therefore, the lack of CDC34 protein is a consequence of a failure to form a competent crossover complex and subsequent lack of later-stage cells.
Immunofluorescence staining on testis sections revealed the most prominent CDC34 signal in round and elongating spermatids of Cntd1 FH/FH males. Cntd1 −/− males, lacking these post-meiotic cell types, failed to show significant CDC34 staining (Figures 6A and 6B).
Figure 6.
CDC34 Levels Are Decreased and Downstream Targets Are Mislocalized in CNTD1 Mutants
(A and B) Immunofluorescence using antibodies against CDC34 (green) and SYCP3 (red) on formalin-fixed testis sections of Cntd1FH/FH (A) and Cntd1−/− (B) mice, imaged at 200×. The white bars indicate the prophase I cells. The right-hand gray panels show representative images of the prophase I stages from enlarged sections of (A) and (B), respectively, staged by the morphology of the synaptonemal complex from SYCP3 staining.
(C–J) Immunofluorescence using antibodies against WEE1 (C and D) and its targets, including pan-tyrosine15 (E and F), CDK1 (G and H), and CDK2 (I and J), on formalin-fixed testis sections of Cntd1FH/FH (left column panels C, E, G, and I) and Cntd1−/− (right column panels D, F, H, and J), imaged at 200×. The white bars indicate the prophase I cells; the arrow highlights the spermatocytes. The right-hand gray panels show representative images of the prophase I stages from enlarged sections, staged by SYCP3 morphology.
Targets of CDC34 Are Drivers of Cell-Cycle Progression and Crossover Regulation during Prophase I
CDC34 targets the WEE1 kinase for degradation in mitosis (Michael and Newport, 1998). WEE1 functions as a mitotic inhibitor (Michael and Newport, 1998; Morgan, 1995; Sadowski et al., 2007; Sandoval et al., 2015; Smith et al., 2007), specifically inhibiting the CDK1 component of maturation-promoting factor (MPF) to prevent premature metaphase I entry (Dunphy, 1994; Morgan, 1995; Nurse, 1990; Figure 4F). WEE1 degradation thus allows MPF activation to facilitate cell-cycle progression. Accordingly, WEE1 staining in fixed testis sections revealed strong cytoplasmic and diffuse faint nuclear localization in prophase I spermatocytes from Cntd1 FH/FH males, with the nuclear localization disappearing by diplonema (Figure 6C).
Given the lack of post-spermatocyte cells in Cntd1 −/− males and the decrease in CDC34 protein in these animals, we hypothesized that the regulation of WEE1 was defective in Cntd1 −/− males. WEE1 was decreased in protein extracts from testes of Cntd1 −/− males compared with Cntd1 FH/FH males, whereas CDK1 and CDK2 protein levels were not significantly altered (Figure 4E). The nuclear localization of WEE1, however, is observed throughout prophase I and remains elevated at diplonema in spermatocytes of Cntd1 −/− males rather than diminishing as in wild-type testes (Figure 6D).
WEE1 inhibits CDK1 and CDK2 activity by phosphorylation on tyrosine 15 (Gould and Nurse, 1989; Gu et al., 1992; Krek et al., 1992; Morla et al., 1989), but phospho-specific antibodies against tyrosine 15 of CDK1 and CDK2 did not reveal any significant change in whole-testis lysate from Cntd1 −/− males compared to Cntd1 FH/FH animals (Figure 4E). Therefore, to assess prophase I stage-specific changes in WEE1 phosphorylation, we stained testis sections from Cntd1 FH/FH and Cntd1 −/− males using a pan-CDK phospho-tyrosine 15 antibody (Figures 6E and 6F). We observed strong cytoplasmic staining of early prophase I Cntd1 FH/FH spermatocytes that is lost by diplonema (Figure 6E). By contrast, we observed the retention of the cytoplasmic pan-CDK phospho-tyrosine 15 signal throughout prophase I in testis sections of Cntd1 −/− males (Figure 6F).
CDK1 and CDK2 staining in Cntd1 FH/FH cells appear to be dynamic between the cytoplasm and nucleus throughout prophase I (Figures 6G and 6I). In early prophase I, CDK1 and CDK2 are distributed between both cellular compartments, but by diplonema show strong nuclear staining. By contrast, staining of CDK1 and CDK2 in testis sections from Cntd1 −/− males reveals much less-intense nuclear signal with only faint cytoplasmic staining and no localization in the nucleus by diplonema (Figures 6H and 6J). These data suggest that SCF is misregulated in Cntd1 −/− males relative to Cntd1 FH/FH, leading to decreased CDC34 levels. This in turn results in increased WEE1+ cells within the seminiferous epithelium of Cntd1 −/− males coupled with a decrease in CDK1 and CDK2 staining in the nucleus of late prophase I cells and a persistence of inhibited CDKs in the cytoplasm (Figures 6G–6L). We postulate that the insufficient degradation of WEE1 arising from the loss of CNTD1 and CDC34 directly contributes to the failure of cell-cycle progression.
In Vitro Inhibition of WEE1 Facilitates Progression into Metaphase I and Is a Prerequisite for Spindle Assembly Checkpoint (SAC) Activation
Following prophase I, cells progress to metaphase I, the stage at which the SAC monitors correct microtubule attachment before entry into anaphase I. We postulated that spermatocytes from Cntd1 −/− males fail to progress past prophase I to the metaphase-to-anaphase transition, and thus do not activate the SAC. To test this, we cultured prophase I cells from Cntd1 +/+ and Cntd1 −/− males for short periods of time in the presence of adavosertib (MK-1775) (0.25–10 μg/mL), a WEE1 kinase inhibitor, and/or nocodazole (1–80 μg/mL), a microtubule polymerization inhibitor. As a readout for drug activity and metaphase entry/progression, we used WB analysis to monitor the levels of the anaphase I inhibitor MAD2L2 and inhibitory phospho-tyrosine 15 on CDK1 and CDK2 for SAC and WEE1 activity, respectively.
Cells from Cntd1 −/− males showed reduced CDK1 and CDK2 inhibitory phosphorylation when treated with adavosertib alone, while cells treated with nocodazole alone failed to show any increase in the levels of MAD2L2 (Figures 7A and 7B). Only when both drugs were applied to these cells was the inhibition of anaphase I progression observed, reflected by an increase in MAD2L2 protein levels (Figures 7A and 7B). Accordingly, whole-testis lysates from Cntd1 −/− males showed a decrease in MAD2L2 protein levels compared to those of wild-type males (Figure 7C), which is indicative of a reduced number of cells completing prophase I.
Figure 7.
In Vitro Short-Term Cultures Reveal a WEE1-Dependent Meiotic Crossover Checkpoint for Progression into Metaphase I
(A) WB using antibodies against WEE1 inhibitory phosphorylation targets CDK1-pY15 and CDK2-pY15 and anaphase inhibitor MAD2L2 following the treatment of cultured spermatocytes from Cntd1+/+and Cntd1−/− males with adavosertib, nocodazole, or both drugs. The triangles indicate increasing drug concentrations (see Method Details).
(B) Analysis of protein signal from WB in (A) divided by GAPDH signal, relative to the V.O. (vehicle only) control.
(C) WB against MAD2L2 in whole testis lysates from Cntd1+/+and Cntd1−/− males.
(D) Inhibitory phosphorylation of CDK1 and CDK2 by WEE1, which itself is degraded in vivo by CDC34 or inhibited by adavosertib treatment. WEE1 degradation/inhibition allows the activation of the MPF and progression into metaphase I. The spindle assembly checkpoint (SAC) monitors the correct attachment of microtubules and prevents the progression from metaphase I to anaphase I. Nocodazole inhibits microtubule formation and promotes/maintains the SAC.
(E) Model of CNTD1 function based on results presented in the present article.
To our surprise, cultured spermatocytes from wild-type males showed a similar requirement for both adavosertib and nocodazole treatment to elicit a MAD2L2 response (Figures 7A and 7B). These data indicate that the inhibition of WEE1 is required for progression into metaphase and that the treatment of wild-type spermatocytes with adavosertib results in the massive and rapid induction of metaphase I entry, thus permitting progression through to anaphase I. Without adavosertib, too few cells naturally progress into metaphase to be detected by MAD2L2 protein induction (Figure 7D).
Discussion
Crossovers are essential for the accurate segregation of homologous chromosomes during meiosis I. The present article shows that CNTD1FH foci are found in a similar distribution and frequency to those of MutLγ, often co-localizing with this complex. Approximately 1–5 MLH1-independent foci exist per cell, reminiscent of the number of crossovers expected to form through MutLγ-independent mechanisms (Holloway et al., 2008). Thus, we propose that CNTD1 localizes to all nascent crossover sites, not only those destined to form MutLγ-dependent crossovers. We do not find any discrete CNTD1FH signal before pachynema, suggesting that CNTD1 accumulates at a time when class I and II crossovers are becoming designated.
Using an epitope-tagged allele of Cntd1, we demonstrate that a short form of CNTD1, arising from the translation of an internal start codon in exon 3, predominates in the mouse testis. Two lines of evidence support this conclusion: (1) migration of CNTD1-FH on WB and the corroborating localization of the tagged protein on meiotic chromosome cores in prophase I (Figures 1B–1E and 2C–2L) and (2) multiple anti-HA antibodies can detect both full-length and short forms of the protein when expressed in yeast, and they migrate relative to their predicted size, indicating that if a full-length protein did exist in spermatocytes, we would be able to detect it. The CNTD1 short-form protein lacks the first cyclin homology domain and is incapable of associating directly with any known CDKs. Our MS and IP-WB data, however, show that the CNTD1 short form can interact with the regulators of CDK activity, thereby providing an indirect mechanism by which progression through the cell cycle can be modulated following appropriate crossing over. We cannot exclude the possibility that the CNTD1 short form maintains some cyclin-like functions since it still contains two cyclin homology domains with predicted structural similarity to cyclin A. In fact, these cyclin domains may be critical for protein translocation between the cytoplasm and the nucleus at defined stages during prophase I, as observed in CNTD1 short-form staining (Figure S4) and as has previously been shown for cyclin domain-containing proteins (Kong et al., 2000). We also cannot rule out that the full-length version of CNTD1 exists in low, undetectable levels or that it is more predominant in females. In testis from male mice, however, we find only a single band migrating at a size that corresponds with the epitope-tagged short form of CNTD1, while the same antibodies used on chromosome spread preparations localize exclusively to sites that correspond with nascent crossovers and mostly correspond to MutLγ localization.
Although the internal methionine start site used to generate the short-form mouse CNTD1 is absolutely conserved, the CNTD1 N terminus shows variability across and within species, with many organisms showing multiple isoforms in this region. These differences may reflect divergent CNTD1 functions, with some homologs retaining the ability to maintain direct CDK interactions and act as canonical cyclins, while others must exert their effects indirectly.
Our MS identified interactions between CNTD1 and components of the RFC complex. While RFC and PCNA are required for the activity of human and yeast MutLα in vitro (Constantin et al., 2005; Kadyrov et al., 2006; Smith et al., 2015), yeast MutLγ displays activity independent of these proteins (Manhart and Alani, 2016). However, more recent studies (currently in preprint form) have demonstrated that RFC and PCNA activate mammalian MutLγ (Cannavo et al., 2020; Kulkarni et al., 2020). While no information exists to describe the interaction between RFC and PCNA complexes in mammalian germ cells in vivo, it is likely, given the observations from mitotic, biochemical, and in vitro assays, that these complexes interact, potentially in a DNA substrate-specific manner. Moreover, our previous studies have demonstrated the importance of PCNA in meiotic prophase I in the mouse (Roa et al., 2008). In the present work, we observe a decreased cellular signal of RFC4 but a stronger nuclear staining pattern of PCNA throughout prophase I in testis sections from Cntd1 mutants. On meiotic spread preparations, PCNA localization along the SC increases in the absence CNTD1, coincident with the decreased localization of RFC3/4. Previous characterizations of Cntd1 GT/GT male mice revealed normal prophase I progression before crossover formation and normal DSB formation and repair kinetics with no persistent DSBs observed (Holloway et al., 2014). The progression of spermatocytes through to pachynema of prophase I in testis from Cntd1 −/− animals (Figures 5 and 6) indicates that the kinetics of early DSB induction and repair are largely normal. The persistent strong nuclear and SC staining of PCNA that we observe within Cntd1 null mutants is therefore not a consequence of the failure to undergo appropriate early DNA repair events. This is further supported by diakinesis spreads that show decreased bivalent formation in Cntd1 −/− animals, but not fragmented chromosomes forming as a result of unrepaired DSBs. Based on these observations, we propose that the persistent PCNA signal (and not RFC) is not related to ongoing DNA synthesis associated with meiotic recombination in Cntd1 −/− animals, but is instead a prelude to active crossover formation, with CNTD1-RFC-PCNA being required to localize proteins at the correct location and time to achieve class I crossovers. New observations from the Cejka and Hunter labs support our observations and our conclusion that PCNA-RFC interactions facilitate MutLγ-stimulated crossover formation (Cannavo et al., 2020; Kulkarni et al., 2020).
CNTD1 interacts with several components of the SCF ubiquitylation complex (Figure S6B), including the E2 conjugating enzyme CDC34. Although we have not identified the partner E3 ubiquitin ligase(s) that functions with CDC34, previous findings in Xenopus identified the cell-cycle kinase WEE1 as a specific target of CDC34 (Michael and Newport, 1998). WEE1 exerts inhibitory effects through the phosphorylation of CDK1 and CDK2, and its removal enables mitotic cell-cycle progression (Gu et al., 1992; Morgan, 1995; Welburn et al., 2007). We find a similar mechanism functioning during meiotic prophase I in the male mouse. In this context, WEE1 phosphorylation of CDK1 maintains MPF in an inactive state and prevents cyclin B-CDK1-induced progression to metaphase. Cntd1 null mutants show a decreased CDC34 signal, a retention of WEE1 in the nucleus throughout prophase I, continued inhibitory phosphorylation of its targets within the cytoplasm, and a consequent failure to localize CDK1 and CDK2 within the nucleus of late-stage prophase I cells. However, the total WEE1 signal from the testis of Cntd1 −/− animals showed a decrease compared with that of Cntd1 FH/FH animals. While we cannot fully explain this difference, the lack of post-meiosis I cells in the Cntd1 −/− animals or the difference in total protein found in the cytoplasmic versus nuclear compartments may account for the difference in total protein. These observations strongly argue that CNTD1 acts upstream of WEE1 through association with CDC34 and relieves the inhibitory effects of WEE1 phosphorylation, thereby coordinating the timing of cell-cycle progression with proper crossover formation.
To clarify the connection between CNTD1 and cell-cycle progression, we devised a strategy to investigate how CNTD1-mediated WEE1 inhibition could perturb the completion of prophase I. To test this in an in vitro short-term culture system, we used the small-molecule WEE1 inhibitor adavosertib in conjunction with the metaphase-to-anaphase inhibitor nocodazole. Only with combined treatment using both drugs were we able to observe the inhibition of metaphase I progression, indicative of an induced SAC, and such a result was observed in spermatocytes from both wild-type and Cntd1 mutant males. These results demonstrate a requirement for WEE1 degradation/inhibition to allow metaphase I entry and thus to facilitate progression to the metaphase-anaphase boundary (Figure 7D).
Collectively, our work defines two modes of action for CNTD1 in meiosis: simultaneously stimulating crossover formation through association with RFC while concomitantly regulating cell-cycle progression through interactions with CDC34 and subsequent ubiquitylation of WEE1 (Figure 7E). While these activities are distinct, the localization of both CNTD1FH and CDK2 at crossover sites, together with the fact that CDK1 and CDK2 are both downstream targets of the SCF complex, support the integrated regulation by CNTD1 of the processes governing crossover formation and cell-cycle progression. Despite the persistent colocalization of CNTD1 and CDK2 in recombination foci throughout the ∼7 days of mouse pachynema, we cannot find any evidence of a physical interaction between these two proteins in vivo. Instead, our findings show that CNTD1 interacts with key players of crossover formation and cell-cycle progression and that the loss of CNTD1 leads to the misregulated localization of these factors. We propose that CNTD1 functions to coordinate these processes, indirectly affecting the activity of cell-cycle regulators and implicating CNTD1 as a stop-go regulator mechanism, monitoring crossover formation akin to a crossover-specific checkpoint, which may also be activated by the cellular machinery downstream of CNTD1. CNTD1 interactions with RFC in the context of the RAD17-RFC complex, an established cell-cycle checkpoint mediator (Ohashi and Tsurimoto, 2017), could provide additional crosstalk and control of these pathways (Figure 7E). While our findings have immediate implications for the timing and control of prophase I in the male mouse, the dual nature of CNTD1 pathway interactions that we have uncovered here and the potential of variable splicing at the N terminus implicate CNTD1 as a regulatory nexus throughout the animal kingdom.
Limitations of Study
The present article describing our elucidation of CNTD1 function was submitted for consideration in November 2019. Reviewers’ comments were received in February 2020, but we were unable to fully address some of the points due to the outbreak of coronavirus disease 2019 (COVID-19). While we addressed the vast majority of points raised by the reviewers, due to our inability to undertake additional experiments, we have identified three limitations to our study. First, while we have identified a short-form CNTD1 protein by WB, a reviewer suggested a modified protein extraction technique to identify whether an additional CNTD1 form could be observed. Second, while we have shown interactions of CNTD1 with components of the RFC complex and CDC34 by unbiased (MS) and biased (IP-WB) methods, because of the importance of these interactions, a reviewer asked for further confirmation of interaction by other biochemical methods. Finally, reviewers asked us to build on data in Figure 4 and data within Figure 7 with the inclusion of drug treatment on an additional meiotic crossover mutant (Mlh1 −/−). Such studies will be pursued once our laboratories reopen.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CDC34 | Proteintech | Cat# 10964-2-AP; RRID: AB_2077925 |
| CDK1 | Millipore | Cat# MAB8878; RRID: AB_2074771 |
| CDK1-pY15 | Abcam | Cat# ab47594; RRID: AB_869073 |
| CDK2 | Santa Cruz | Cat# sc-163; RRID: AB_631215 |
| CDK2-pY15 | Abcam | Cat# ab76146; RRID: AB_1310069 |
| CDK4 | Cell Signaling | Cat# 12790S; RRID: AB_2631166 |
| Cyclin B1 | Cell Signaling | Cat# 4138S; RRID: AB_2072132 |
| GAPDH | Invitrogen | Cat# MA5-15738-HRP; RRID: AB_2537659 |
| HA (Rabbit) | Cell Signaling | Cat# 3724S; RRID: AB_1549585 |
| HA (Rat) | Roche | Cat# 11867423001; RRID: AB_390918 |
| HEI10 | Abcam | Cat# ab71977; RRID: AB_1310045 |
| MLH1 | BD Biosciences | Cat# 550838; RRID: AB_2297859 |
| MLH3 | Custom made (Holloway et al., 2008) | N/A |
| MAD2L2 | Proteintech | Cat# 12683-1-AP; RRID: AB_2139530 |
| Pan CDK p-Y15 | Abcam | Cat# ab133463; RRID: AB_2811262 |
| PCNA | Proteintech | Cat# 10205-2-AP; RRID: AB_2160330 |
| RAD51 | Millipore | Cat# PC130-100UL; RRID: AB_569857 |
| RFC3 | Proteintech | Cat# 11814-1-AP; RRID: AB_2178470 |
| RFC4 | Proteintech | Cat# 10806-1-AP; RRID: AB_2178477 |
| RNF212 | Novus | Cat# NBP1-83471; RRID: AB_11030667 |
| SYCP1 | Abcam | aCat# b15090; RRID: AB_301636 |
| SYCP3 | Custom made (Kolas et al., 2005) | N/A |
| WEE1 | Novus | Cat# NBP1-33506; RRID: AB_2215876 |
| γH2AFX | Millipore | Cat# 05-636; RRID: AB_309864 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Adavosertib | MedChemExpress | Cat# HY-10993 |
| Nocodazole | Sigma Aldrich | Cat# SML1665-1ML |
| Experimental Models: Organisms/Strains | ||
| M. musculus C57BL/6J | The Jackson Laboratory | 000664 |
| S. cerevisiae Y2H Gold | Takarabio | 630498 |
| Oligonucleotides | ||
| CNTD1-FH crRNA: ccgcuuccucuaacacguga | This Paper | N/A |
| CNTD1-FH HDR Donor: cctgggccacag cagcctgttccccacaaggcagccagagctctga ggactgctgccgctgccgcttcctctaacacg GACTACAAAGACGATGACGACAAGcttT ACCCATACGATGTTCCAGATTACGCTt gagggtggctgacccgcactgccttgtcactgg acctctttccttgtttacttttatactcaggg |
This Paper | N/A |
| CNTD1-FH Sequencing Primer, Forward: gcgcacgagtgtttgtgcact | This Paper | N/A |
| CNTD1-FH Sequencing Primer, Reverse: ccagtgacaaggcagtgcgggtcagcc | This Paper | N/A |
| CNTD1 Mutant crRNA: UAGUGACUUUCA GUUCGGAG |
This Paper | N/A |
| CNTD1 Wildtype and Mutant Primer, Forward: CCAAGGTGTGGCAGAAGATTC | This Paper | N/A |
| CNTD1 Wildtype Primer, Reverse: ctgtcaattccaggccagcc | This Paper | N/A |
| CNTD1 Mutant Primer, Reverse: AGCCCTTGGAGGAGCCC | This Paper | N/A |
| Recombinant DNA | ||
| pGADT7 AD | Clontech | 630442 |
| pGBKT7 DNA-BD | Clontech | 630443 |
| pX330 | Addgene | 42230 |
| Software and Algorithms | ||
| Adobe Creative Cloud | Adobe | https://www.adobe.com/uk/creativecloud.html |
| Fiji-ImageJ | (Schneider et al., 2012) | https://imagej.nih.gov/ij/ |
| GraphPad Prism 6.0 | GraphPad | https://www.graphpad.com |
| Image Lab 6.0 | Bio-Rad | https://www.bio-rad.com/en-us/product/image-lab-software?ID=KRE6P5E8Z |
| Zen 2.0 | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Paula Cohen (paula.cohen@cornell.edu)
Materials Availability
Plasmids and mouse lines generated within this study are available upon request from the Lead Contact, Paula Cohen (paula.cohen@cornell.edu)
Data and Code availability
This study did not generate any code. Mass spectrometry dataset generated in this study is found within Table S1.
Experimental Model and Subject Details
Mouse strains
All mouse alleles were maintained on a C57BL6/J background, with at least six successive backcrosses. CRISPR/Cas9 genome edited mice were created initially on a FVB x C57B6 background before undergoing backcrossing. In all cases, analysis was performed on adult male mice between the ages of 10 and 14 weeks. All mice were maintained under strictly defined conditions of constant temperature and light:day cycles, with food and water ad libitum. Animal handling and procedures were performed following approval by the Cornell Institutional Animal Care and Use Committee, under protocol 2004-0063.
Transgenic mice generation
Cntd1-FLAG-HA: CRISPR/Cas9 was used to insert the FLAG-HA epitope at the C terminus of the Cntd1 locus. The RNA guide was generated by cloning the Cntd1 specific sequence (GCCGCTTCCTCTAACACGTGA) in between the BbsI restriction sites of pX330, courtesy of Feng Zhang (Addgene #42230). Once integrated, the region including the gRNA scaffold was PCR amplified to include the T7 promoter. The PCR fragment was used to in an in vitro transcription reaction using the Ambion MEGAshortscript T7 Transcription Kit (AM1354). Single-stranded DNA homology donor containing 78 nucleotides of homology upstream of the guide sequence, FLAG and HA epitopes separated by a leucine (generating a HindIII restriction site) and 66 nucleotides of homology downstream of the Cntd1 stop codon was generated by IDT. gRNA, DNA donor and Cas9 mRNA was injected into 146 C57BL/6J x FVB F1 zygotes. 119 two-cell embryos were surgically implanted in 5 female recipients leading to the birth of 46 founder mice. PCR screening identified 7 founders with DNA integrated at the locus, 3 of which were the correct size. Sequencing revealed one founder mouse having the tags integrated in the correct frame. The founder mouse was backcrossed with C57BL/6J for four generations before being used for experiments described.
Cntd1 null allele
CRISPR/Cas9 was used to generate a DNA double-strand break at the Cntd1 locus and founder mice were screened for non-homologous end joining events generating a frameshift mutation. The gRNA was generated using long PCR primers incorporating the T7 promoter, targeting sequence and Cas9 scaffold followed by PCR and in vitro transcription (see above). Founders were screened for mutations by PCR of the Cntd1 locus followed by sequencing. One founder carried an incorporation of 324 nucleotides of chromosome 12 at the targeted locus leading to disruption of the Cntd1open reading frame.
Yeast strain
Y2H Gold yeast strain (detailed in Key Resources Table) was used for yeast two-hybrid experiments and protein expression.
Method Details
Plasmid construction
Yeast two-hybrid
Testis cDNA was generated from Trizol extracted RNA using the Invitrogen SuperScript® III First-Strand Synthesis System. CDKs 1, 2, 4, 5 and 6 were PCR amplified to include NdeI and BamHI restriction sites upstream and downstream respectively. The CDKs were cloned into Clontech yeast two-hybrid prey vector, pGADT7, using the Roche Rapid DNA Dephos & Ligation Kit. Annotated full-length CNTD1 and CNTD1 short-form sequences were PCR amplified to include NcoI and BamHI restriction sites upstream and downstream respectively. The CNTD1 sequences were cloned into the Clontech yeast two-hybrid bait vector pGBKT7.
CNTD1 expression constructs
The yeast CEN/ARS region from pRS414 was PCR amplified, introducing restriction sites SpeI upstream and EcoRV downstream. This fragment was cloned into pFA6a-HphMX. The promoter of ADH1 from pGADT7 along with FLAG-HA epitope tagged CNTD1 full-length and short-forms from mouse testis cDNA was PCR amplified and cloned into the previously constructed vector using Gibson Assembly.
Cntd1 Orthology Sequence Alignment
Sequence alignments carried out using Clustal Omega. NCBI database IDs for annotated isoform sequences are as follows: Mus musculus (mouse), Iso1: NP_080838.1, Iso2: EDL01034.1; Homo sapiens (human), Iso1: NP_775749.2, IsoX2: XP_006721753.1, IsoX1: XP_005257100.2, Iso2: NP_001317151.1; Pan troglodytes (chimpanzee), IsoX1: XP_009430406, IsoX3, XP_016787256.1, IsoX2: XP_016787255.1, IsoX4: XP_024206054.1; Phascolarctos cinereus (koala), IsoX1: XP_020828270.1, IsoX2: XP_020828271.1, IsoX3: XP_020828272.1, IsoX4: XP_020828273.1; Canis familiaris (dog), IsoX2: XP_022278673.1, IsoX4: XP_022278675.1; Castor canadensis (American beaver), IsoX1: XP_020020731.1, IsoX2: XP_020020732.1.
Yeast two-hybrid assay
The yeast two-hybrid assay was performed as described by the manufacturer, Clontech. Briefly, combinations of bait and prey plasmids were transformed into the yeast two-hybrid reporter strain Y2HGold and plated onto –LEU –TRP selection plates. Positive colonies were re-streaked and tested for plasmids using PCR. Single colonies were inoculated into –LEU –TRP media and grown overnight. Colonies were normalized to an OD600 or 1.0 and four, 10-fold dilutions made. 10μl drops were placed on selection plates and quadruple dropout plates, which were incubated at 30°C for 3 days.
Chromosome analysis and immunofluorescence
Chromosome spreads were made as previously described in Holloway et al. (2014). Briefly, decapsulated testis tubules were incubated in hypertonic elution buffer (30 mM Tris pH7.2, 50 mM sucrose, 17 mM trisodium dehydrate, 5 mM EDTA, 0.5 mM DTT, 0.1 mM PMSF, pH8.2-8.4) for one hour. Small sections of testis tubule were dissected in 100mM sucrose and spread onto 1% Paraformaldehyde, 0.15% Triton X coated slides and incubated in a humid chamber for 2.5hrs at room temperature. Slides were dried for 30minutes, washed in 1x PBS, 0.4% Photoflo (Kodak 1464510) and either stored at −80°C or stained. For staining, slides were washed in 1xPBS, 0.4% Photoflo for 10 minutes, followed by a 10 minute wash in 1x PBS, 0.1% Triton X and finally a 10 minute wash in 10% antibody dilution buffer (3% BSA, 10% Goat Serum, 0.0125% Triton X, 1 x PBS) 1x PBS. Antibodies used in this study are described in Table S1. Primary antibodies were diluted in 100% antibody dilution buffer, placed as a bubble on parafilm within a humid chamber, and the surface of the slide spread on the parafilm allowing the antibody to spread across the surface of the slide. Slides were incubated at 4°C overnight. Slides were washed in 1xPBS, 0.4% Photoflo for 10 minutes, followed by a 10 minute wash in 1x PBS, 0.1% Triton X and finally a 10 minute wash in 10% antibody dilution buffer. Secondary antibodies were diluted as the primary antibodies and spread in a similar fashion. Slides were incubated at 37°C for one hour. Slides were washed in 1xPBS, 0.4% Photoflo for 10 minutes, three times. Finally slides were left to dry, mounted using DAPI/antifade mix and either imaged or stored at 4°C for later imaging. Slides were imaged on a Zeiss Axiophot with Zen 2.0 software.
Histology and immunofluorescence
Adult testes were dissected and incubated in either 10% neutral buffered formalin (for IHC) or Bouin’s fixative (for hematoxylin and eosin [H&E] staining) for 8hrs to overnight. Fixed testis was then washed 4 x in 70% ethanol, embed in paraffin and 5 μM sections mounted on slides. For H&E staining, slides were rehydrated in safeclear followed by decreasing amounts of ethanol. Slides were then stained with hematoxylin followed by eosin then gradually dehydrated by incubation in increasing concentrations of ethanol. Finally, slides were mounted using permount and imaged on an Aperio Scanscope.
For immunofluorescence, slides were rehydrated as previously mentioned and then boiled in sodium citrate buffer (10mM Sodium Citrate, 0.05% Tween 20, pH 6.0) for 20 minutes. Following subsequent cooling, slides were blocked in blocking buffer (1 x PBST, 1% BSA, 3% Goat Serum) for an hour and primary antibody dilutions incubated on the sections overnight at 4°C. Slides were then washed, and incubated with fluorescence-conjugated secondary antibodies for one hour at 37°C, washed and then mounted using a DAPI/Antifade mix. Slides were imaged on a Zeiss Axiophot with Zen 2.0 software. Antibodies used in this study are described in the Key Resources Table.
Immunohistochemical staining of testis sections was similar to that described for immunofluorescent staining, except for the use of secondary antibodies conjugated to horseradish peroxidase (HRP; Jackson Immunochemicals). HRP detection was achieved using the Pierce DAB substrate Kit (ThermoFisher Scientific) according to the manufacturers detailed protocol.
Sperm counts
Caudal epididymides of adult mice was transferred and dissected into sperm count media (4% BSA in 1 x PBS). Sample was incubated at 30°C for 30 minutes allowing sperm to swim out into the media. 1:10 dilutions were made in 10% neutral buffered formalin and stored at 4°C until counting on a haemocytometer.
Spermatocyte diakinesis spread preparations
Spread diakinesis preparations were made as described in Holloway et al. (2014). Briefly, testis cells were liberated by manual dissection of tubules in 0.5% KCl. Multiple mixing and slow centrifugation followed by fixation in 30% methanol: 10% acetic acid: 0.05% chloroform. Cells were finally fixed in 30% methanol: 10% acetic acid and then pipette onto heated slides. Finally slides were stained in using Giemsa and imaged. Slides were imaged on a Zeiss Axiophot with Zen 2.0 software.
Gravitational cell separation (STA-PUT)
Cells were separated using a protocol. The number of adult mice (8-12 weeks, age matched) used in each STA-PUT are as follows: Figures 3 and 4 and S5: 8 mice of the respective genotypes, Figure 5: 12 mice. Testis extracts were dissected into approximately 30 mL of 1 x Krebs buffer (Sigma K3753) supplemented with amino acids (GIBCO 11130-051, Sigma M7145) and glutaMAX (GIBCO 35050-061) in a Petri dish lid. Extract was incubated in a shaking water bath at 34°C, 150rpm in 10 mL of 2 mg/ml Collagenase (Sigma C5138) in 1 x Krebs for 15 minutes. Following three rounds of centrifugation and washing in 20 mL 1 x Krebs, the testis extract was re-suspended in 20 mL of 2.5mg/ml Trypsin (Sigma T0303), 200μg/ml DNase (Sigma DN25) in 1 x Krebs and incubated in a shaking water bath at 34°C, 150rpm for 15 minutes. Extract was then centrifuged and washed three times in 20 mL 1 x Krebs, followed by re-suspension in 20 mL 0.5% BSA (Sigma A7906). Extract was then loaded into the STA-PUT apparatus along with 275 mL 4% BSA in 1 x Krebs, 275 mL 2% BSA in 1 x Krebs and 100 mL 0.5% BSA in 1 x Krebs. Following loading of the BSA into the separating chamber along with the extract, the sample was allowed to sediment for 2 hr. Following sedimentation, cell fractions of 13 mL were collected, centrifuged at 2,000rpm for 5 minutes and washed in 200 μl 1 x PBS. For immunofluorescence analysis of cell types 5 μl of each fraction was re-suspended in 50mM sucrose and incubated at room temperature for 20 minutes. 30μl drops of 1% Paraformaldehyde (EMS 19200) were placed on each well of an 8 well slide and the 50mM sucrose cell mix were added to each well. Slides were incubated in a humid chamber overnight, followed by drying, washing and staining against proteins defining stages of prophase I, in our case SYCP3 and γH2AX. The remaining extract was centrifuged at 2,000 rpm for 5 minutes and re-suspended in 1 x PBS Lysis buffer (1 x PBS, 0.01% NP-40, 5% Glycerol, 150mM NaCl, 1 x Roche cOmplete), sonicated and stored at −20°C for downstream applications.
Protein extraction
Decapsulated testis extract was re-suspended into 1 x PBS Lysis buffer (1 x PBS, 0.01% NP-40, 5% Glycerol, 150mM NaCl, 1 x Roche cOmplete) and sonicated for 20 s at 22% amplitude in cycles of 0.4 s on and 0.2 s off.
SDS-PAGE and Western Blotting
Protein samples were separated by SDS-PAGE on gels varying in percentage from 6%–14% and transferred to methanol activated PVDF membranes using a Biorad Mini Trans-Blot Cell. Membranes were incubated in 5% BSA, 1 x TBST for 30minutes to 2 hours at room temperature while rotating at 60rpm. Membranes were incubated overnight in primary antibodies in 1 x TBST. Membranes were washed three times in 1 x TBST and subsequently incubated for one hour in secondary antibodies in 1 x TBST. Finally membranes were washed three times in 1 x TBST, developed using the ECL reagent and imaged using a Biorad ChemiDoc imager. Antibodies used in this study are described in The Key Resources Table.
Colloidal Coomassie Staining
Protein extracts separated on SDS-PAGE gels were fixed in 40% methanol, 10% acetic acid for 30 minutes. Gels were stained using the Invitrogen Colloidal Coomassie staining kit (LC6025) in 20% methanol, 20% stainer A, 5% stainer B and incubated at room temperature overnight while slowly rocking. Gels were subsequently washed in double distilled water and imaged.
Image acquisition
Imaging was performed using a Zeiss Axiophot Z1 microscope attached to a cooled charge-coupled device (CCD) Black and White Camera (Zeiss McM). Images were captured and pseudo-colored using ZEN 2 software (Carl Zeiss AG, Oberkochen. Germany). Higher resolution images were acquired using an ELYRA 3D-Structured Illumination Super resolution Microscopy (3D-SIM) from Carl Zeiss with ZEN Black software (Carl Zeiss AG, Oberkochen. Germany). Images are shown as maximum intensity projections of z stack images. To reconstruct high-resolution images, raw images were computationally processed by ZEN Black. Channel alignment was used to correct for chromatic shift. The brightness and contrast of images were adjusted using ImageJ (National Institutes of Health, USA).
Mass spectrometry and protein identification
Mass spectrometry was performed in the Cornell University Proteomics and Mass Spectrometry facility. The nanoLC-MS/MS analysis was carried out using an Orbitrap Fusion (Thermo-Fisher Scientific, San Jose, CA) mass spectrometer equipped with a nanospray Flex Ion Source using high energy collision dissociation (HCD) and coupled with the UltiMate3000 RSLCnano (Dionex, Sunnyvale, CA). Each reconstituted sample was injected onto a PepMap C-18 RP nano trap column (3 μm, 100 μm × 20 mm, Dionex) with nanoViper Fittings at 20 μL/min flow rate for on-line desalting and then separated on a PepMap C-18 RP nano column (3 μm, 75 μm x 25 cm), and eluted in a 120 min gradient of 5% to 35% acetonitrile (ACN) in 0.1% formic acid at 300 nL/min. The instrument was operated in data-dependent acquisition (DDA) mode using FT mass analyzer for one survey MS scan for selecting precursor ions followed by 3 s “Top Speed” data-dependent HCD-MS/MS scans in Orbitrap analyzer for precursor peptides with 2-7 charged ions above a threshold ion count of 10,000 with normalized collision energy of 38.5%. For label-free protein analysis, one MS survey scan was followed by 3 s “Top Speed” data-dependent CID ion trap MS/MS scans with normalized collision energy of 30%. Dynamic exclusion parameters were set at 1 within 45 s exclusion duration with ± 10 ppm exclusion mass width. All data are acquired under Xcalibur 3.0 operation software and Orbitrap Fusion Tune 2.0 (Thermo-Fisher Scientific).
All MS and MS/MS raw spectra from each experiment were processed and searched using the Sequest HT search engine within the Proteome Discoverer 2.2 (PD2.2, Thermo). The default search settings used for relative protein quantitation and protein identification in PD2.2 searching software were: two mis-cleavage for full trypsin with fixed carbamidomethyl modification of cysteine and oxidation of methionine and demaidation of asparagine and glutamine and acetylation on N-terminal of protein were used as variable modifications. Identified peptides were filtered for maximum 1% false discovery rate (FDR) using the Percolator algorithm in PD 2.2. The relative label free quantification method within Proteome Discoverer 2.2 software was used to calculate the protein abundances. The intensity values of peptides, which were summed from the intensities values of the number of peptide spectrum matches (PSMs), were summed to represent the abundance of the proteins. Protein ratios are calculated based on pairwise ratio, where the median of all possible pairwise ratios calculated between replicates of all connected peptides.
Short-term testis drug culture
Cultures were performed as described elsewhere (Brieño-Enríquez et al., 2016; Wiltshire et al., 1995), with some modifications. Testis extracts were dissected into 1 x PBS and incubated in a shaking water bath at 34°C, 150rpm in 2 mg/ml Collagenase (Sigma C5138) in 1 x PBS for 15 minutes. Following multiple rounds of centrifugation and washing in 1 x PBS, the testis extract was re-suspended in 2.5 mg/ml Trypsin (Sigma T0303), 200 μg/ml DNase (Sigma DN25) in 1 x PBS and incubated in a shaking water bath at 34°C, 150rpm for 15 minutes. Extract was then centrifuged and washed five times in 4 mL spermatocyte culture medium (SCM) (GIBCO DMEM without red phenol (21063-029), fetal calf serum (GIBCO 10082139), penicillin-streptomycin 1003 (GIBCO 15140-122); lactic acid (Sigma L13750, NaHCO3 9s8761) and sodium pyruvate 1003 (Sigma 11360-070). Cells were re-suspended in 600μl of SCM and placed in 12-well treated culture dishes (Corning #3513). Adavosertib (MedChemExpress HY-10993) was diluted in 10% DMSO and added at 0.25, 0.5, 1, 2, 5 and 10 μg/ml. Nocodazole (Sigma SML1665-1ML) was diluted in 10% DMSO and added at 1, 5, 10, 20, 50 and 80 μg/ml. Cultures were incubated at 37°C for 6 hours followed by protein extraction.
Quantification and Statistical Analyses
Statistical analyses were performed using GraphPad Prism version 6.00 for Macintosh and Microsoft Excel. Signal intensity of prophase I staged cells from testis tubules (Figure S7) was performed by outlining a minimum of 6 cells per stage and calculating the average intensity of each channel using ImageJ. Specific analyses are described within the text and the corresponding figures. Mean values are all presented ± standard deviation (s.d.) and alpha value was established at 0.05. All statistical analyses performed using two-sided tests.
Acknowledgments
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development (NICHD) of the National Institutes of Health under award no. R01HD041012 to P.E.C. Salary for S.G. was funded in part by a NICHD award to S.G. (K99HD092618). J.S.C. is a Meinig Family Investigator in the Life Sciences. The transgenic mice creation was supported in part by Empire State Stem Cell Fund contract no. C024174. Imaging data were acquired through the Cornell University Biotechnology Resource Center, with NSF1428922 funding for the shared Zeiss Elyra super-resolution microscope. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Lynn Dong for providing assistance with immunohistochemistry, Dr. Sheng Zhang and his team for performing the MS studies, and Rob Munroe and Chris Abratte of the Cornell Transgenic Mouse Core Facility for generation of new mouse lines. We also thank John Schimenti and Tina Tran for helpful conversations and Eric Alani, Michael Lichten, and Kathryn Grive for critical review of this manuscript.
Author Contributions
S.G. and P.E.C. conceived and designed the experiments and analyzed the data; S.G. conducted the experiments; E.R.S. and J.S.C. performed the computational alignment and analyzed the data; S.G., J.S.C., and P.E.C wrote the paper.
Declaration of Interests
The authors declare no competing interests.
Published: July 7, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107858.
Contributor Information
Stephen Gray, Email: stephen.gray@nottingham.ac.uk.
Paula E. Cohen, Email: paula.cohen@cornell.edu.
Supplemental Information
References
- Adhikari D., Zheng W., Shen Y., Gorre N., Ning Y., Halet G., Kaldis P., Liu K. Cdk1, but not Cdk2, is the sole Cdk that is essential and sufficient to drive resumption of meiosis in mouse oocytes. Hum. Mol. Genet. 2012;21:2476–2484. doi: 10.1093/hmg/dds061. [DOI] [PubMed] [Google Scholar]
- Ahuja J.S., Sandhu R., Mainpal R., Lawson C., Henley H., Hunt P.A., Yanowitz J.L., Börner G.V. Control of meiotic pairing and recombination by chromosomally tethered 26S proteasome. Science. 2017;355:408–411. doi: 10.1126/science.aaf4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsheimer M., Baier A., Schramm S., Schütz W., Benavente R. Synaptonemal complex protein SYCP3 exists in two isoforms showing different conservation in mammalian evolution. Cytogenet. Genome Res. 2010;128:162–168. doi: 10.1159/000303341. [DOI] [PubMed] [Google Scholar]
- Ashley T., Walpita D., de Rooij D.G. Localization of two mammalian cyclin dependent kinases during mammalian meiosis. J. Cell Sci. 2001;114:685–693. doi: 10.1242/jcs.114.4.685. [DOI] [PubMed] [Google Scholar]
- Baker S.M., Plug A.W., Prolla T.A., Bronner C.E., Harris A.C., Yao X., Christie D.M., Monell C., Arnheim N., Bradley A. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- Barrière C., Santamaría D., Cerqueira A., Galán J., Martín A., Ortega S., Malumbres M., Dubus P., Barbacid M. Mice thrive without Cdk4 and Cdk2. Mol. Oncol. 2007;1:72–83. doi: 10.1016/j.molonc.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F., Manova K., Yuen J.P., Jasin M., Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- Bellvé A.R. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 1993;225:84–113. doi: 10.1016/0076-6879(93)25009-q. [DOI] [PubMed] [Google Scholar]
- Bellvé A.R., Millette C.F., Bhatnagar Y.M., O’Brien D.A. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J. Histochem. Cytochem. 1977;25:480–494. doi: 10.1177/25.7.893996. [DOI] [PubMed] [Google Scholar]
- Berthet C., Aleem E., Coppola V., Tessarollo L., Kaldis P. Cdk2 knockout mice are viable. Curr. Biol. 2003;13:1775–1785. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- Bowen N., Kolodner R.D. Reconstitution of Saccharomyces cerevisiae DNA polymerase ε-dependent mismatch repair with purified proteins. Proc. Natl. Acad. Sci. USA. 2017;114:3607–3612. doi: 10.1073/pnas.1701753114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieño-Enríquez M.A., Moak S.L., Toledo M., Filter J.J., Gray S., Barbero J.L., Cohen P.E., Holloway J.K. Cohesin Removal along the Chromosome Arms during the First Meiotic Division Depends on a NEK1-PP1γ-WAPL Axis in the Mouse. Cell Rep. 2016;17:977–986. doi: 10.1016/j.celrep.2016.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K.W., Rowe L.B., Churchill G.A., Paigen K. Crossover interference in the mouse. Genetics. 2002;160:1123–1131. doi: 10.1093/genetics/160.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E., Sanchez A., Anand R., Ranjha L., Hugener J., Adam C., Acharya A., Weyland N., Aran-Guiu X., Charbonnier J.-B. Regulation of the MLH1-MLH3 endonuclease in meiosis. bioRxiv. 2020 doi: 10.1101/2020.02.12.946293. [DOI] [PubMed] [Google Scholar]
- Chen X., Suhandynata R.T., Sandhu R., Rockmill B., Mohibullah N., Niu H., Liang J., Lo H.-C., Miller D.E., Zhou H. Phosphorylation of the Synaptonemal Complex Protein Zip1 Regulates the Crossover/Noncrossover Decision during Yeast Meiosis. PLOS Biol. 2015;13:e1002329. doi: 10.1371/journal.pbio.1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole F., Kauppi L., Lange J., Roig I., Wang R., Keeney S., Jasin M. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat. Cell Biol. 2012;14:424–430. doi: 10.1038/ncb2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole F., Baudat F., Grey C., Keeney S., de Massy B., Jasin M. Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nat. Genet. 2014;46:1072–1080. doi: 10.1038/ng.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin N., Dzantiev L., Kadyrov F.A., Modrich P. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J. Biol. Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy W.G. The decision to enter mitosis. Trends Cell Biol. 1994;4:202–207. doi: 10.1016/0962-8924(94)90142-2. [DOI] [PubMed] [Google Scholar]
- Edelmann W., Cohen P.E., Kane M., Lau K., Morrow B., Bennett S., Umar A., Kunkel T., Cattoretti G., Chaganti R. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- Edelmann W., Cohen P.E., Kneitz B., Winand N., Lia M., Heyer J., Kolodner R., Pollard J.W., Kucherlapati R. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 1999;21:123–127. doi: 10.1038/5075. [DOI] [PubMed] [Google Scholar]
- Ellenrieder C., Bartosch B., Lee G.Y., Murphy M., Sweeney C., Hergersberg M., Carrington M., Jaussi R., Hunt T. The long form of CDK2 arises via alternative splicing and forms an active protein kinase with cyclins A and E. DNA Cell Biol. 2001;20:413–423. doi: 10.1089/104454901750361479. [DOI] [PubMed] [Google Scholar]
- Gould K.L., Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- Gray S., Cohen P.E. Control of meiotic crossovers: from double-strand break formation to designation. Annu. Rev. Genet. 2016;50:175–210. doi: 10.1146/annurev-genet-120215-035111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Rosenblatt J., Morgan D.O. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J. 1992;11:3995–4005. doi: 10.1002/j.1460-2075.1992.tb05493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway J.K., Booth J., Edelmann W., McGowan C.H., Cohen P.E. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLOS Genet. 2008;4:e1000186. doi: 10.1371/journal.pgen.1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway J.K., Sun X., Yokoo R., Villeneuve A.M., Cohen P.E. Mammalian CNTD1 is critical for meiotic crossover maturation and deselection of excess precrossover sites. J. Cell Biol. 2014;205:633–641. doi: 10.1083/jcb.201401122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 2015;7:a016618. doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G.H. The control of chiasma distribution. Symp. Soc. Exp. Biol. 1984;38:293–320. [PubMed] [Google Scholar]
- Jones G.H., Franklin F.C.H. Meiotic crossing-over: obligation and interference. Cell. 2006;126:246–248. doi: 10.1016/j.cell.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Kadyrov F.A., Dzantiev L., Constantin N., Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Keeney S., Giroux C.N., Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Keeney S., Baudat F., Angeles M., Zhou Z.H., Copeland N.G., Jenkins N.A., Manova K., Jasin M. A mouse homolog of the Saccharomyces cerevisiae meiotic recombination DNA transesterase Spo11p. Genomics. 1999;61:170–182. doi: 10.1006/geno.1999.5956. [DOI] [PubMed] [Google Scholar]
- Kneitz B., Cohen P.E., Avdievich E., Zhu L., Kane M.F., Hou H., Jr., Kolodner R.D., Kucherlapati R., Pollard J.W., Edelmann W. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 2000;14:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Kolas N.K., Svetlanov A., Lenzi M.L., Macaluso F.P., Lipkin S.M., Liskay R.M., Greally J., Edelmann W., Cohen P.E. Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J. Cell Biol. 2005;171:447–458. doi: 10.1083/jcb.200506170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong M., Barnes E.A., Ollendorff V., Donoghue D.J. Cyclin F regulates the nuclear localization of cyclin B1 through a cyclin-cyclin interaction. EMBO J. 2000;19:1378–1388. doi: 10.1093/emboj/19.6.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek W., Marks J., Schmitz N., Nigg E.A., Simanis V. Vertebrate p34cdc2 phosphorylation site mutants: effects upon cell cycle progression in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 1992;102:43–53. doi: 10.1242/jcs.102.1.43. [DOI] [PubMed] [Google Scholar]
- Kulkarni D.S., Owens S., Honda M., Ito M., Yang Y., Corrigan M.W., Chen L., Quan A.L., Hunter N. PCNA activates the MutLγ endonuclease to promote meiotic crossing over. bioRxiv. 2020 doi: 10.1101/2020.02.12.946020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.K., Diehl J.A. SCFs in the new millennium. Oncogene. 2014;33:2011–2018. doi: 10.1038/onc.2013.144. [DOI] [PubMed] [Google Scholar]
- Lipkin S.M., Moens P.B., Wang V., Lenzi M., Shanmugarajah D., Gilgeous A., Thomas J., Cheng J., Touchman J.W., Green E.D. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 2002;31:385–390. doi: 10.1038/ng931. [DOI] [PubMed] [Google Scholar]
- Majka J., Burgers P.M.J. The PCNA-RFC families of DNA clamps and clamp loaders. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:227–260. doi: 10.1016/S0079-6603(04)78006-X. [DOI] [PubMed] [Google Scholar]
- Manhart C.M., Alani E. Roles for mismatch repair family proteins in promoting meiotic crossing over. DNA Repair (Amst.) 2016;38:84–93. doi: 10.1016/j.dnarep.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E., Diaz R.L., Hunter N., Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126:285–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael W.M., Newport J. Coupling of mitosis to the completion of S phase through Cdc34-mediated degradation of Wee1. Science. 1998;282:1886–1889. doi: 10.1126/science.282.5395.1886. [DOI] [PubMed] [Google Scholar]
- Milano C.R., Holloway J.K., Zhang Y., Jin B., Smith C., Bergman A., Edelmann W., Cohen P.E. Mutation of the ATPase Domain of MutS Homolog-5 (MSH5) Reveals a Requirement for a Functional MutSγ Complex for All Crossovers in Mammalian Meiosis. G3 (Bethesda) 2019;9:1839–1850. doi: 10.1534/g3.119.400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens P.B., Kolas N.K., Tarsounas M., Marcon E., Cohen P.E., Spyropoulos B. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell Sci. 2002;115:1611–1622. doi: 10.1242/jcs.115.8.1611. [DOI] [PubMed] [Google Scholar]
- Morgan D.O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- Morla A.O., Draetta G., Beach D., Wang J.Y. Reversible tyrosine phosphorylation of cdc2: dephosphorylation accompanies activation during entry into mitosis. Cell. 1989;58:193–203. doi: 10.1016/0092-8674(89)90415-7. [DOI] [PubMed] [Google Scholar]
- Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- Ohashi E., Tsurimoto T. Functions of Multiple Clamp and Clamp-Loader Complexes in Eukaryotic DNA Replication. Adv. Exp. Med. Biol. 2017;1042:135–162. doi: 10.1007/978-981-10-6955-0_7. [DOI] [PubMed] [Google Scholar]
- Qiao H., Prasada Rao H.B.D., Yang Y., Fong J.H., Cloutier J.M., Deacon D.C., Nagel K.E., Swartz R.K., Strong E., Holloway J.K. Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat. Genet. 2014;46:194–199. doi: 10.1038/ng.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitsma J.M., Liu X., Reichermeier K.M., Moradian A., Sweredoski M.J., Hess S., Deshaies R.J. Composition and regulation of the cellular repertoire of SCF ubiquitin ligases. Cell. 2017;171:1326–1339.e14. doi: 10.1016/j.cell.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A., Qiao H., Yang Y., Chen J.K., Jackson N., Biswas K., Holloway J.K., Baudat F., de Massy B., Wang J. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 2013;45:269–278. doi: 10.1038/ng.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa S., Avdievich E., Peled J.U., Maccarthy T., Werling U., Kuang F.L., Kan R., Zhao C., Bergman A., Cohen P.E. Ubiquitylated PCNA plays a role in somatic hypermutation and class-switch recombination and is required for meiotic progression. Proc. Natl. Acad. Sci. USA. 2008;105:16248–16253. doi: 10.1073/pnas.0808182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert T., Nore A., Brun C., Maffre C., Crimi B., Bourbon H.M., de Massy B. The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science. 2016;351:943–949. doi: 10.1126/science.aad5309. [DOI] [PubMed] [Google Scholar]
- Robert T., Vrielynck N., Mézard C., de Massy B., Grelon M. A new light on the meiotic DSB catalytic complex. Semin. Cell Dev. Biol. 2016;54:165–176. doi: 10.1016/j.semcdb.2016.02.025. [DOI] [PubMed] [Google Scholar]
- Romanienko P.J., Camerini-Otero R.D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- Romrell L.J., Bellvé A.R., Fawcett D.W. Separation of mouse spermatogenic cells by sedimentation velocity. A morphological characterization. Dev. Biol. 1976;49:119–131. doi: 10.1016/0012-1606(76)90262-1. [DOI] [PubMed] [Google Scholar]
- Sadowski M., Mawson A., Baker R., Sarcevic B. Cdc34 C-terminal tail phosphorylation regulates Skp1/cullin/F-box (SCF)-mediated ubiquitination and cell cycle progression. Biochem. J. 2007;405:569–581. doi: 10.1042/BJ20061812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval D., Hill S., Ziemba A., Lewis S., Kuhlman B., Kleiger G. Ubiquitin-conjugating enzyme Cdc34 and ubiquitin ligase Skp1-cullin-F-box ligase (SCF) interact through multiple conformations. J. Biol. Chem. 2015;290:1106–1118. doi: 10.1074/jbc.M114.615559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarikas A., Hartmann T., Pan Z.-Q. The cullin protein family. Genome Biol. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Session D.R., Fautsch M.P., Avula R., Jones W.R., Nehra A., Wieben E.D. Cyclin-dependent kinase 5 is expressed in both Sertoli cells and metaphase spermatocytes. Fertil. Steril. 2001;75:669–673. doi: 10.1016/s0015-0282(00)01794-5. [DOI] [PubMed] [Google Scholar]
- Smith A., Simanski S., Fallahi M., Ayad N.G. Redundant ubiquitin ligase activities regulate wee1 degradation and mitotic entry. Cell Cycle. 2007;6:2795–2799. doi: 10.4161/cc.6.22.4919. [DOI] [PubMed] [Google Scholar]
- Smith C.E., Bowen N., Graham W.J., 5th, Goellner E.M., Srivatsan A., Kolodner R.D. Activation of Saccharomyces cerevisiae Mlh1-Pms1 Endonuclease in a Reconstituted Mismatch Repair System. J. Biol. Chem. 2015;290:21580–21590. doi: 10.1074/jbc.M115.662189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetlanov A., Baudat F., Cohen P.E., de Massy B. Distinct functions of MLH3 at recombination hot spots in the mouse. Genetics. 2008;178:1937–1945. doi: 10.1534/genetics.107.084798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J.P.I., Tucker J.A., Johnson T., Lindert L., Morgan M., Willis A., Noble M.E.M., Endicott J.A. How tyrosine 15 phosphorylation inhibits the activity of cyclin-dependent kinase 2-cyclin A. J. Biol. Chem. 2007;282:3173–3181. doi: 10.1074/jbc.M609151200. [DOI] [PubMed] [Google Scholar]
- Wiltshire T., Park C., Caldwell K.A., Handel M.A. Induced premature G2/M-phase transition in pachytene spermatocytes includes events unique to meiosis. Dev. Biol. 1995;169:557–567. doi: 10.1006/dbio.1995.1169. [DOI] [PubMed] [Google Scholar]
- Yokoo R., Zawadzki K.A., Nabeshima K., Drake M., Arur S., Villeneuve A.M. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell. 2012;149:75–87. doi: 10.1016/j.cell.2012.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any code. Mass spectrometry dataset generated in this study is found within Table S1.