Abstract
SULT2B1b (SULT2B) is a prostate-expressed hydroxysteroid sulfotransferase, which may regulate intracrine androgen homeostasis by mediating 3β-sulfation of dehydroepiandrosterone (DHEA), the precursor for 5α-dihydrotestosterone (DHT) biosynthesis. The aldo-keto reductase (AKR)1C3 regulates androgen receptor (AR) activity in castration-resistant prostate cancer (CRPC) by promoting tumor tissue androgen biosynthesis from adrenal DHEA and also by functioning as an AR-selective coactivator. Herein we report that SULT2B-depleted CRPC cells, arising from stable RNA interference or gene knockout (KO), are markedly upregulated for AKR1C3, activated for ERK1/2 survival signal, and induced for epithelial-to-mesenchymal (EMT)-like changes. EMT was evident from increased mesenchymal proteins and elevated EMT-inducing transcription factors SNAI1 and TWIST1 in immunoblot and single-cell mass cytometry analyses. SULT2B KO cells showed greater motility and invasion in vitro; growth escalation in xenograft study; and enhanced metastatic potential predicted on the basis of decreased cell stiffness and adhesion revealed from atomic force microscopy analysis. While AR and androgen levels were unchanged, AR activity was elevated, since PSA and FKBP5 mRNA induction by DHT-activated AR was several-fold higher in SULT2B-silenced cells. AKR1C3 silencing prevented ERK1/2 activation and SNAI1 induction in SULT2B-depleted cells. SULT2B was undetectable in nearly all CRPC metastases from 50 autopsy cases. Primary tumors showed variable and Gleason score (GS)-independent SULT2B levels. CRPC metastases lacking SULT2B expressed AKR1C3. Since AKR1C3 is frequently elevated in advanced prostate cancer, the inhibitory influence of SULT2B on AKR1C3 upregulation, ERK1/2 activation, EMT-like induction, and on cell motility and invasiveness may be clinically significant. Pathways regulating the inhibitory SULT2B-AKR1C3 axis may inform new avenue(s) for targeting SULT2B-deficient prostate cancer.
Keywords: SULT2B1b sulfotransferase, AKR1C3 aldo-keto reductase, androgen receptor activity, epithelial-to-mesenchymal transition, ERK1/2 survival signaling, prostate cancer
Reactivation of androgen receptor (AR) signaling drives the lethal progression of prostate cancer. On gaining resistance to peripheral castration from androgen deprivation therapy, rapid advance of the castration-recurrent disease on second-line inhibitions of the androgen-AR axis is aided by intratumoral changes such as AR gene amplification and increased expression, upregulation of AR’s transcriptional program, and elevated intratumoral expression/activity of the androgen biosynthetic machinery (1, 2). Adrenal DHEA (dehydroepiandrosterone), the precursor for androgen biosynthesis, transits through circulation as a sulfate conjugate and participates in peripheral tissue steroidogenesis as a desulfated steroid. SULT2B1b (SULT2B hereafter), a 3β-hydroxysteroid sulfotransferase with DHEA-sulfating activity, and steroid sulfatase (STS) with sulfate deconjugation activity regulate DHEA/DHEA sulfate homeostasis (3). De novo DHT biosynthesis entails multistep metabolic conversions of DHEA by the steroidogenic enzymes 3β-HSD1, SRD5α isozymes, and the aldo-keto reductase (AKR)1C3—the last one catalyzing a final reductive step (4, 5). AKR1C3 also has AR-coactivating function (6).
SULT2B is a 43-kDa cytoplasmic sulfotransferase, which mediates sulfation of DHEA, cholesterol, and selected oxysterols (3, 7, 8). Apart from the prostate, SULT2B is present in several other tissues, including at high abundance in skin and intestine. SULT2B and the neuron-expressed isoform SULT2B1a arise from 6 exons of the SULT2B1 gene through utilization of separate transcription start sites and alternate splicing of exon-1 to exon-2 (9). SULT2A1, a third member of the SULT2 subfamily, converts adrenal DHEA to a hydrophilic sulfate, which facilitates DHEA transit in circulation and its subsequent import into the prostate aided by organic anion uptake transporters (3, 10).
Association of SULT2B with prostate cancer was identified in our earlier study with a limited set of clinical primary prostate specimens (11). Immunostaining of tissue microarrays showed that SULT2B expression in primary prostate cancer is variable—ranging from relatively strong expression to significant reduction or undetectable. Calcitriol, a hormonally active vitamin D3, which is known to display antiprostate cancer activity in multiple cell and mouse models, can induce SULT2B in prostate cancer cells through a vitamin D receptor binding element in the upstream SULT2B1 promoter (11). SULT2B induction by calcitriol alludes to a potential role of this sulfotransferase in the regulation of prostate cancer.
Herein we report that SULT2B-ablated castration-resistant prostate cancer (CRPC) cells manifest increased aggressive traits such as enhanced motility and invasion; epithelial-to-mesenchymal (EMT)-like changes such as elevated EMT markers; and escalation of xenograft growth. Reduced stiffness and adhesion of SULT2B knocked out CRPC cells, detected by atomic force microscopy (AFM), and linked loss of SULT2B to enhanced metastatic potential of the cells. Loss of SULT2B led to AKR1C3 upregulation and activation of ERK1/2 Map kinase, while AKR1C3 silencing in SULT2B-depleted cells prevented ERK activation and induction of the EMT factor SNAI1. AR and 5α-DHT levels did not change, whereas AR activity increased markedly. SULT2B was undetectable by immunohistochemistry (IHC) in nearly all CRPC metastases from multiple distant sites of 50 autopsy cases. Notably, AKR1C3, which is frequently elevated in advanced prostate cancer, was found to be expressed in the same specimens that lack SULT2B. Upregulation of AKR1C3, heightened AR and ERK1/2 activities, EMT-like induction, and enhanced invasive activity observed in SULT2B-depleted CRPC cells, as well as SULT2B-negative status of clinical CRPC metastases, suggest that pathways regulating the inhibitory SULT2B–AKR1C3 axis may inform new avenue(s) of prostate cancer inhibition.
Materials and Methods
Stable depletion of SULT2B in CRPC cells
C4-2 and C4-2B CRPC cells (ViroMed, Hopkins, MN), which are derived from castration-sensitive LNCaP cells (12), were made SULT2B deficient by stable gene KO in C4-2 cells or stable knockdown (KD) by RNA interference in C4-2B cells. The mycoplasma-free status of cells was verified routinely.
Stable KO lines under puromycin selection were developed using a CRISPR/Cas9 gene KO kit (sc-404529, Santa Cruz Biotech, Dallas, TX). The kit contains 3 U6 promoter-driven CRISPR/Cas9 coexpressing plasmids—each containing 20-nucleotide SULT2B1-specific guide sequences and a homology directed repair plasmid (SC-404529-HDR). Plasmid DNAs were transfected into C4-2 cells using the UltraCruz transfection reagent (sc-395739). Guide RNA sequences are TGTCGTCGTCCCGCACATCT, sense (sc-404529A); GGATCCGCTCCGTG-CCCATC, sense (sc-404529B); TGTCCCACAAGCCCGGGATC, sense (sc-404529C).
For stable SULT2B KD, C4-2B cells were infected with SULT2B1 MISSION® shRNA lentiviral particles (SHCLNV-NM_004605, Sigma-Aldrich) and puromycin-resistant clonal lines were isolated. Viral particles expressing shRNAs (from pLKO-puro) that target separate regions of SULT2B1 mRNAs (TRCN0000333182; TRCN0000333183; TRCN0000333184; TRCN0000035350; TRCN00000-35351) were used individually for separate transductions. For the nontargeted (NT) control, we used SHC002V MISSION nonmammalian shRNA transduction particles.
DHEA sulfotransferase activity in cells was assayed from DHEA and DHEA sulfate levels in cells ± DHEA (2.5 μM) incubated for 1 hour in charcoal-stripped serum. Washed cell pellets were processed as before (13) and assayed for DHEA and DHEA sulfate by LC-MS/MS (liquid chromatography tandem mass spectroscopy). On organic extraction of cell homogenates, DHEA was recovered from the organic phase and DHEA sulfate from the aqueous phase. Internal standards were as follows: for DHEA, 5-androsten-3b-ol-17-one 2,3,4 13C (13C3-DHEA); and for DHEA-sulfate, 5-androsten-3b-ol-17-one-3 sulfate sodium salt 2,2,3,4,4-d5 (DHEAS-d5). DHEAS-d5 co-eluted with DHEAS at ~4.37 minutes of retention time. Data were analyzed using MultiQuant 3.0 software.
Mass cytometry by time-of-flight
A Helios mass cytometer (Fluidigm, South San Francisco, CA) at institutional Bio Analytic Single Cell Core was used. Antibodies for metal conjugation (Fluidigm) are Vimentin, mouse monoclonal immunoglobulin (Ig)G1, NBP1-92687; RRID:AB_11017879 (Novus Biologicals, Centenneal, CO) (14); N-cadherin, polyclonal sheep IgG, AF6426; RRID:AB_10718850 (R&D Systems, Minneapolis, MN) (15); SNAI1, rabbit polyclonal, SAB2108482; RRID:AB_2818978 (Sigma) (16); TWIST1, monoclonal clone #927403, MAB6230; RRID:AB_2818958 (R&D Systems) (17); E-cadherin, polyclonal goat IgG, AF748; RRID:AB_355568 (R&D Systems) (18); EpCAM, AF960; RRID:AB_355745 (R&D Systems) (19). Signals of samples were normalized using mass cytometry by time-of-flight (CyTOF) software (version 6.7.1014, Fluidigm). CyTOF raw data were gated for live cells using Cytobank software (Cytobank, Inc). Filtered data were analyzed in R using Cytofkit, which is an integrated analysis pipeline including functions of data processing, clustering (PhenoGraph), and visualization (t-SNE) (20, 21). For each sample, 10 000 live cells from its fetal calf serum file were analyzed. Heatmaps were derived using MultiExperiment Viewer software (22) and Box Plots using ggplot function in R. Other details are in the online repository (23).
AFM
Adhesion, elastic modulus and deformability of individual cells were analyzed by AFM as before (24, 25). Cells in 55-mm culture dishes, cultivated to 20% confluence, were examined individually with SCANASYST-AIR (Bruker) probes using a Nanoscope Catalyst AFM (Bruker, Billerica, MA). For quantifications, we used the Sneddon model (26), which approximates the mechanics of conical tip interactions with an object. The rules by Sokolov (27) were used to calculate the elastic modulus, assuming high heterogeneity of cell surface properties (brush and rigidity). Mechanical properties were quantified for adhesion, elastic modulus, and deformation using Nanoscope Analysis software, v1.7. Mean values of Young’s modulus, deformation, and adhesion for individual cells were calculated when normal distribution was observed. In the remaining cases, a mode was applied. Averaged data for a cell line are shown as a violin plot. A horizontal line is the mean value of data for individual cells from 3 biological replicates. Details on principal component analysis and other AFM-related information are in the online repository (23).
RNA interference of AKR1C3 and SNAI1
AKR1C3 was silenced in SULT2B KD cells by transfecting AKR1C3-selective siRNA pool (SMARTpool siGENOME Human AKR1C3 (8644), cat #M-008116-02, Dharmacon/Horizon Discovery) using Lipofectamine™3000 (ThermoFisher). Cells in RPMI-1640 containing 5% FBS (no antibiotics) were incubated for 48 hours with the siRNA–lipofectamine complex in the Opti-MEM™ reduced serum medium at an optimized siRNA/lipofectamine ratio, and cell lysates in radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors cocktail (ThermoFisher) were used for Western blotting. A NT siRNA pool was the negative control (siGENOME Non-Targeting siRNA Pool #2 (D-001206-14), Dharmacon).
SNAI1 shRNAs were delivered to SULT2B KD cells via lentivirus transduction. Virus particles expressing SNAI1 shRNAs and scramble shRNAs from the pLKO.1 vector were packaged in 293T cells as described (28). Media from infected 293T cells containing virus particles were provided by Dr. Myron Ignatius (UTHSA, San Antonio). The SNAI1 shRNA of sequence CCAAGGATCTCCAGGCTCGAA, used in our study, efficiently silenced SNAI1 in a previous study (28). The scramble shRNA plasmid, a gift from David Sabatini (29), was from Addgene (plasmid # 1864; http://n2t.net/addgene:1864). For transduction, cells in a 10-cm dish at 80% confluence were placed in 3-mL of fresh medium (RPMI-1640 + 5% FBS, no antibiotics) containing protamine at 20 μg/mL and incubated for 48 hours with 3 mL of medium from infected 293T cells. Lysates of harvested cells were prepared in RIPA buffer containing protease inhibitors.
Western blot, qRT-PCR
For western blotting, cell lysates were prepared in RIPA buffer containing protease inhibitors as before (11). Antibody specifications were the following: from Santa Cruz Biotech: rabbit polyclonal antibody to AR (1:1000) (N-20, sc816; RRID:AB_1563391) (30); Ku86 (1:3000) (H-300, sc9034; RRID:AB_2218743) (31); GAPDH (1:2000) (sc25778; RRID: AB_10167668) (32); RPS (1:3000), mouse monoclonal (C-8, sc74459; RRID:AB_1129205) (33); EpCAM (1:1000), mouse monoclonal (C-10, sc25308; RRID:AB_627531) (34); and E-cadherin (1:1000), mouse monoclonal (sc-21791; RRID: AB_626777) (35). Novus Biologicals (Centennial, CO): vimentin (1:1000), mouse monoclonal (NBP1-92687; RRID: AB_11017879) (14). Sigma-Aldrich: AKR1C3 (1:500), mouse monoclonal (NP6.G6.A6, A6229; RRID: AB_476751) (36). Bio-Rad: AKR1C2 (1:1000), mouse monoclonal (VMA00346; RRID:AB_2818998) (37). Cell signaling Technology: phospho-ERK1/2 (1:500), rabbit polyclonal (9101; RRID: AB_331646) (38); ERK1/2 (1:2000), rabbit monoclonal (9102; RRID:AB_330744) (39). R&D systems: SNAI1 (1:500), goat polyclonal (AF 3639; RRID: AB_2191738) (40). SULT2B antibody, rabbit polyclonal used at 1:1000, was developed in-house (11). To avoid signal loss due to reprobing of stripped membranes, membranes cut in horizontal segments were probed for test and internal control antigens. Size marker positions in gels determined molecular weights of signals. Western blots were quantified using Image J.
Total mRNAs were isolated using TRIzol reagent (ThermoFisher). PrimePCR™SYBR®Green Assay kit was used for qRT-PCR (BioRad, Hercules, CA). BioRad validated primers were used for: SULT2B1, human: UniqueAssayID: qHsaCID0020968 (Cat # 10025636); AKR1C3, human: UniqueAssayID: qHsaCED0038268 (Cat # 10025636); AKR1C2, human: UniqueAssayID: qHsaCED0056434 (Cat # 10025636). Primers for human Ubiquitin B (UBB) and GAPDH mRNAs were supplied by IDT (Coralville, IA). UBB: GGTCCTGCGTCTGAGAGGT, forward; GGCCTTCAGATTTTCGATGGT, reverse. GAPDH: ACCCACTCCTCCACCTTTG, forward; CTCTTGTGCTCTTGCTGGG, reverse.
Cell invasion, migration, and AKR activity
Invasion was assayed in BioCoat Matrigel invasion chambers containing PET membrane inserts and a 24-well culture plate as the lower chamber (Corning). At 72 hours post culture, cells attached to the lower part of the membrane were stained by crystal violet and counted under a microscope in 3 fields (Nikon Eclipse Ti microscope). Averaged counts quantified invasion. Cell motility was quantified using a similar workflow, except that membranes did not have the Matrigel coating; inserts were not rehydrated; and cells were incubated for 24 hours. An AKR Assay Kit (BioVisions, CA) was used to measure total AKR activity.
Xenograft, TMA, IHC
Xenografts were produced subcutaneously in nude mice (6-week-old male) as before (41). Measurement by a digital caliper was on a Mon/Wed/Fri schedule after the tumor reached ~200 mm3 using the following formula: volume = ½ (length × width2), where length is the maximum longitudinal diameter, and width is the maximum transverse diameter. Tumor volume for a mouse at each measurement was calculated relative to its tumor volume at day 1 of measurement. An approved animal protocol was used.
TMAs containing 155 CRPC metastases (73 visceral; 82 bone metastases) from 50 autopsy cases (up to 4 sites per case) were collected at the University of Washington under informed consent at tissue acquisition necropsy within 6 hours of death, and specimens were confirmed histologically. TMA-55 construction has been described previously (42). Bone metastases were decalcified in 10% formic acid. IHC for AKR1C3 was performed at the University of Washington using goat polyclonal anti-AKR1C3 antibody (cat # AB27491, Abcam) (13). IHC staining for SULT2B was done using rabbit polyclonal antibody to SULT2B (11). Primary prostate cancer specimens were from the GU tissue bank at University of Texas Health San Antonio. Institutional Review Board approved protocols were used.
Statistics
In box plots, the differences among subgroups were tested with Duncan’s New Multiple Range Test using the R duncan.test function. AFM data are shown as violin plots with each point representing a single cell. Violin shapes represent kernel smooth data distribution estimated with a Scott bandwidth. A horizontal line corresponds to the mean value and whiskers to 1× standard deviations of data for individual cells from 3 biological replicates. P values were based on Student’s t test using OriginPro 2017. Principal component analysis was performed after data normalization. The association between parameters was tested using Statistica and OriginPro 2017 (OriginLab).
The significance of mRNA induction was calculated by analysis of variance (ANOVA) and Tukey’s post hoc test using Prism 5 (Graphpad Software Inc.). Each point in the xenograft growth curve is the mean of normalized tumor volume ± standard error of the mean. Significance for growth differences between 2 groups at each measurement point was calculated by the t-test assuming unequal variance. Repeated measures ANOVA determined the statistical significance of growth escalation.
Results
SULT2B-ablated KO and KD cells
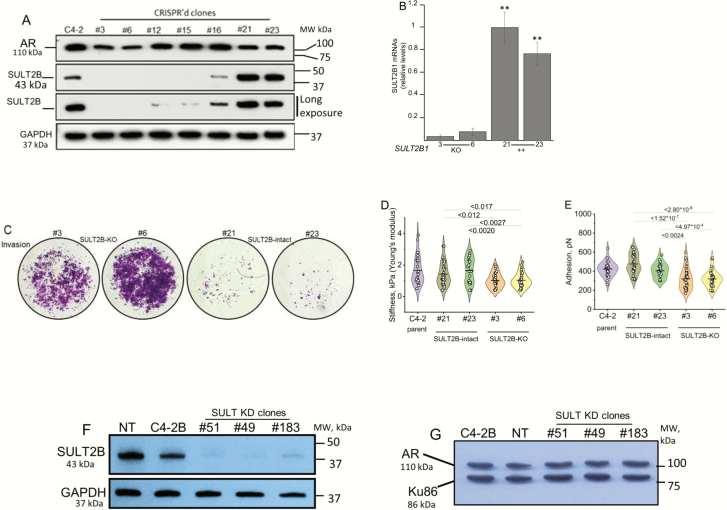
Functional consequences of stable SULT2B depletion in CRPC cells were examined in a C4-2 cell KO model created by SULT2B1 gene disruption (Fig. 1A), and in a C4-2B KD model created by SULT2B1 mRNA silencing with small hairpin RNAs (Fig. 1F). Puromycin-resistant CRISPR’d clones #3 and #6 are SULT2B negative; SULT2B in clones #12, #15, and #16 is barely detectable; SULT2B (43 kDa) is at relatively high levels in clones #21 and #23 (Fig. 1A). SULT2B1 mRNAs are at the background level in KO cells (Fig. 1B). AR (110 kDa) levels are similar for KO and non-KO clones (Fig. 1A).
Figure 1.
AR levels, invasion phenotype and mechanical changes of SULT2B-ablated CRPC cells. (A) SULT2B and AR levels in CRISPR’d clones and parent C4-2 cells. (B) SULT2B1 mRNAs in CRISPR’d clones. Bar charts show average ± SEM of three biological replicates in duplicate qRT-PCR assay. **P < .001. (C). Matrigel invasion of KO and non-KO cells in Transwell chamber assay. Photomicrographs representative of two biological replicates, showing Matrigel-invaded cells, stained with crystal violet at 72 hours post culture. (D,E) AFM analysis of KO, non-KO and parent C4-2 cells. (D) Young’s modulus measuring cell stiffness (in units of Pascal, Pa); (E) AFM tip to cell bonding in units of Newton force N, which measures cell adhesion. Each point is a mode of parameter values measured over a single cell area. Three biological replicates per cell line were analyzed. Number of live cells analyzed for each cell line: 25 (parent C4-2); 31 (clone #21); 23 (clone #23); 30 (clone #3); 31 (clone #6). Horizontal lines are mean values and whiskers are standard deviations. (F,G). SULT2B (F) and AR (G) levels in SULT2B KD cells. GAPDH (a cytosolic protein) and Ku86 (a nuclear protein) are internal controls. Size markers verified molecular weights of immunoblotted proteins.
Mass spectrometry confirmed a lack of DHEA-sulfating activity of KO clones #3 and #6. Figure S-1 in the online repository (23) shows LC-MS/MS chromatograms. In Figure S-1, chromatograms for #21 (left) and #6 (right) ± DHEA show that upon incubation with DHEA, clone #21, not #6, generated the DHEAS peak, which co-eluted with the reference (23). The reference peak appeared for both clones #21 and #6 ± DHEA. An unlabeled DHEAS peak at 4.38 minutes appeared in DHEA-incubated clone-21 only (23). Quantification of peak heights showed that clones #21 and #23 converted DHEA to DHEA sulfate, measuring 2.49 pg, 2.12 pg (#21) and 2.57 pg, 4.06 pg (#23) per mg cell pellet in a duplicate assay (Table 1).
Table 1.
DHEAS levels in cell pellets, assayed by LC-MS/MS. SULT2B-KO and non-KO cells were incubated with 2.5 µM DHEA, 37°C for 2 hours. Harvested cell pellets were analyzed as described in (23).
| SULT2B-KO | SULT2B-positive | |||||||
|---|---|---|---|---|---|---|---|---|
| Clone #3 | Clone #6 | Clone #21 | Clone #23 | |||||
| Vehicle | +DHEA | Vehicle | +DHEA | Vehicle | +DHEA | Vehicle | +DHEA | |
| DHEA | 0.08 | 273.41 | 1.26 | 46.85 | 0.13 | 118.84 | 0.22 | 92.92 |
| (pg/mg) | 0.08 | 390.31 | 0.11 | 49.32 | 0.65 | 99.49 | 0.14 | 155.39 |
| DHEAS | 0.09 | 0.1 | 0.11 | 0.08 | 0.12 | 2.49 | 0.13 | 2.57 |
| (pg/mg) | 0.08 | 0.1 | 0.11 | 0.14 | 0.13 | 2.12 | 0.14 | 4.06 |
KO cells are more invasive in a transwell chamber assay (Fig. 1C) (23). At 72 hours post culture, more KO cells invaded through Matrigel-coated membranes than control cells, (Fig. 1C). Clone #6 KO cells are more invasive than clone #3 KO cells, evident from photomicrographs (Fig. 1C) and from quantified data for invasion activity (23). Proliferation rates between KO and non-KO cells did not differ at 72 hours post culture, indicating that increased KO cell proliferation did not account for the higher number of KO cells that crossed the Matrigel-coated membrane. The motility of KO cells at 24 hours post culture was greater (23). Enhanced invasiveness was also detected for SULT2B KD cells (described below and shown in the online repository (23)).
Mechanical changes in KO cells informed by AFM analysis further suggested that loss of SULT2B is associated with enhanced cellular invasion/metastatic potential. KO cells (clones #3, #6) are less adhesive, based on adhesion force, and less stiff, based on Young’s modulus, than SULT2B-positive clones #21, #23, and C4-2 parent cells (Fig. 1D and 1E). AFM infers cell adhesiveness from the force required to lift the tip of an AFM probe from the cell, approximating the ease with which a cell is detached from a neighbor cell or extracellular matrix, and cell stiffness is defined by its elastic modulus, expressed quantitatively as Young’s modulus (24, 25). Reduced adhesion is expected to weaken cell–cell or cell–extracellular matrix interaction, and reduced stiffness can make cells more pliable. Deformation was similar for KO and non-KO cells (23). Cell softness is quantified by deformation, which reflects the depth of cell surface indentation by a preset force without cell fracture. Principal component analysis of adhesion, Young’s modulus and deformation for 140 cells selected from KO, non-KO, and parent cells showed that KO cells clustered within a single ellipse as 1 distinct group of softer and less adhesive cells. Due to similar mechanical properties, non-KO and parent cells clustered within the same 2 ellipses (23).
As a second model, SULT2B KD lines were examined for functional consequences of SULT2B depletion. Clones #51, #49, and #183 are robustly silenced for SULT2B, while SULT2B expression is maintained in cells expressing nontargeting shRNAs (NT line) (Fig. 1F). AR levels are similar in KD and non-KD cells (Fig. 1G). Similar to KO cells, KD clones are markedly more invasive than NT cells (23). KD clones #49 and #51 appear to be more aggressive than the KD clone #183.
Mesenchyme-like phenotypes of SULT2B-ablated CRPC cells
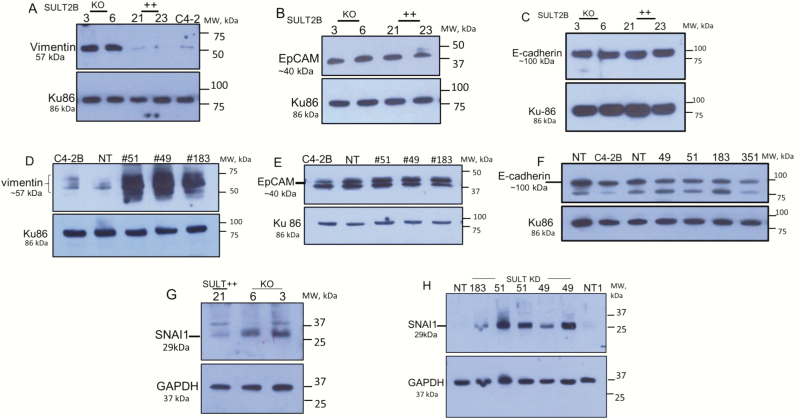
We explored whether increased invasiveness and motility of SULT2B KO cells are associated with an enhanced mesenchymal trait. The 57-kDa vimentin, an intermediate filament protein and mesenchymal cell marker, is elevated in KO clones #3 and #6, while vimentin expression in SULT2B-positive clones #21 and #23 and in parent C4-2 cells is negligible (Fig. 2A). Vimentin in SULT2B KD clones is also strongly induced compared with NT and parent C4-2B cells (Fig. 2D). SNAI1, an EMT-activating transcription factor, was induced in KO and KD cells (Fig. 2G and 2H), providing further evidence for EMT-like changes associated with loss of SULT2B. Since mesenchymal phenotypes drive metastatic progression of epithelial carcinomas (43, 44), enhanced invasion and motility, and elevated vimentin and SNAI1 for KO and KD cells suggest that SULT2B interferes with molecular changes that drive EMT-like transition and progression to more aggressive characteristics of C4-2 and C4-2B carcinoma cells.
Figure 2.
Vimentin, EpCAM, E-cadherin and SNAI1 levels in SULT2B KO and KD cells. (A–C,G) KO/non-KO cells. (D–F,H) KD/NT cells. Results are representative of at least 2 biological replicates.
EpCAM, an epithelial protein, however, was not reduced in KO and KD cells (Fig. 2B and 2E). Although epithelial to mesenchymal transition (EMT) frequently shows decreased expression of EpCAM (43), a high EpCAM level in SULT2B-ablated cells is consistent with the report that EpCAM levels are elevated in clinical cases of metastatic prostate cancer (45). E-cadherin, another epithelial cell marker, is present at high levels in SULT2B KO and KD cells (Fig. 2C and 2F), which is in keeping with augmented E-cadherin levels detected in metastatic prostate cancer (46). Continued E-cadherin expression in SULT2B-ablated cells, which display mesenchyme-like traits, is consistent with the migration characteristic of invading carcinomas, namely that they move as a group of cells connected via homophilic E-cadherin interaction at adherens junctions (44, 47, 48).
Mass cytometry confirmed EMT-like activation
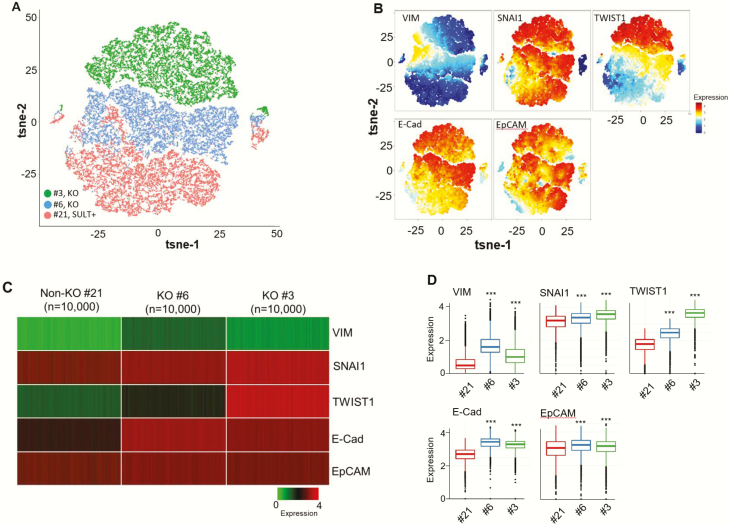
Single-cell CyTOF provided further evidence for EMT-like changes in SULT2B KO C4-2 cells (Fig. 3) and in SULT2B KD C4-2B cells (23). For the KO/non-KO panel, 200 000 individual cells were examined for the expression of vimentin, EMT-inducing transcription factors SNAI1, TWIST1, and epithelial proteins EpCAM and E-cadherin (Fig. 3). From CyTOF data, cells manifesting similar phenotypes were visualized with the t-distributed stochastic neighbor embedding (t-SNE) algorithm. These dimensional reduction approaches position similar cells in nearby points in 2 indices (tSNE1 and tSNE2) after calculation, while dissimilar cells remain far apart. Three distinct clusters were generated from #21, #6, and #3 clones, along with a limited overlap for cells from clone #21 and clone #6 (Fig. 3A). Expression levels of queried proteins in the 3 clusters are shown (Fig. 3B). For each clone, heatmaps of 10 000 randomly selected cells for queried proteins are presented (Fig. 3C). Box plots for mass cytometry data revealed significant upregulation of vimentin, SNAI1, and TWIST 1 in KO clones at P < .0001 (Fig. 3D). Vimentin and SNAI1 induction in KO cells in CyTOF analysis is consistent with immunoblot results (Fig. 2A and 2G). The increase in EpCAM and E-cadherin in KO clones (at P < .001) is also consistent with Western blot data (Fig. 2B and 2C).
Figure 3.
Single-cell mass cytometry of EMT-related proteins in SULT2B-KO and -non-KO C4-2 cells. (A) Visualization of clustered KO/non-KO clones from CyTOF data using t-SNE algorithm. (B) Levels of vimentin, SNAI1, TWIST1, E-cadherin and EpCAM in clustered KO (#3, #6) and non-KO (#21) cells. (C) Heatmaps of proteins queried in panel B for 10 000 randomly selected cells from each clone. (D) Box plots of mass cytometry data. Middle, top and bottom lines of each box signify median, 75th percentile, and 25th percentile of data, respectively. Black dots below or above the boxes are outliers. Protein levels in KO clones differed from the non-KO clone with high significance, showing P < .0001 for vimentin, SNAI1, TWIST1, and P < .001 for E-cadherin, EpCAM. Similar mass cytometry results were obtained with 2 different batches of cells.
SULT2B KD cells showed elevated mesenchymal markers N-cadherin and vimentin, and the EMT factors SNAI1 and TWIST1 in mass cytometry analysis, indicating mesenchyme-like traits of the KD cells (23). The cluster of NT cells significantly overlapped with clusters of KD clones #49 and #51. The overlap is likely due to incomplete SULT2B depletion in KD cells. For clone #51, upregulation of all queried proteins was statistically significant (P < .001). For clone #49, however, N-cadherin and vimentin expression was more variable among individual cells so that their elevated expression is nonsignificant. Nonetheless, increased vimentin in clone #49 was detected by western blotting (Fig. 2D). Also, ~8-fold higher vimentin levels for clones #51 and #49 than NT were observed by mass spectrometry in a separate study. SNAI1 induction revealed from the CyTOF analysis of KD cells is consistent with the elevated SNAI1 level in KD cells shown by western blot analysis (Fig. 2H). EpCAM was elevated for clone #51, while reduced for clone #49 compared with the control NT clone (23). Elevated E-cadherin in KD clones #51 and #49 is consistent with increased E-cadherin levels in KO clones in mass cytometry (Fig. 3D). Complementary analysis by CyTOF and Western blotting strengthens the conclusion that SULT2B deficiency in KO and KD cells promotes EMT-like changes in CRPC carcinomas.
AKR1C3 upregulation and enhanced AR activity
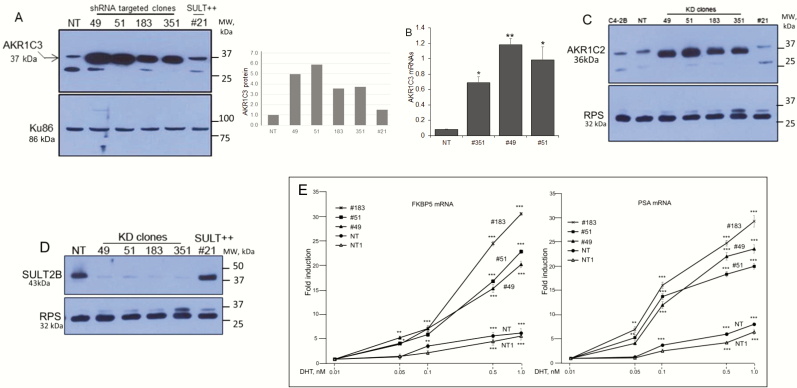
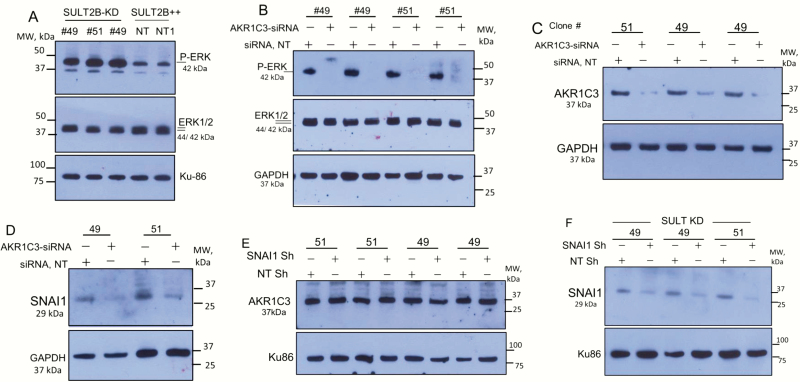
AKR1C3 is markedly upregulated in SULT2B KD cells (Fig. 4A). AKR1C3 was detected as an ~37-kDa protein using a mouse monoclonal antibody (NP6.G6.A6, cat# A6229, Sigma), which is AKR1C3-selective (49) without cross reactivity with AKR1C1, AKR1C2, and AKR1C4. Quantified western blot signals (Fig. 4A, right panel) showed 5- to 6-fold AKR1C3 induction. This fold induction may be an underestimate due to saturation of signals on the X-ray film. Elevated AKR1C3 mRNAs in KD cells (Fig. 4B) indicates that a transcriptional mechanism plays a role in increased AKR1C3 expression. AKR1C2, an AKR closely related to AKR1C3, was upregulated as well in KD cells at protein (Fig.4C) and mRNA (23) levels. It is likely that parallel induction of AKR1C3, which promotes testosterone and 5α-DHT synthesis, and AKR1C2, which metabolizes 5α-DHT to the AR-inert 3α-androstane-diol metabolite (50), at least partly accounts for a lack of increase in testosterone and 5α-DHT levels in SULT2B KO cells relative to non-KO cells in the LC-MS/MS assay. SULT2B is markedly silenced in the same KD cells that show strong upregulation of AKR1C3 and AKR1C2 (Fig. 4D). AKR1C3 elevation (> 2-fold) in KO cells is shown elsewhere (23). Total AKR activity, measured with an assay kit that detects both AKR1C3 and AKR1C2, is higher in KD cells (23).
Figure 4.
Elevated AKR1C3, AKR1C2 expression and enhanced AR activity in SULT2B KD cells. (A) AKR1C3 in multiple clones of KD cells, nontargeted (NT) cells, and SULT2B-expressing CRISPR’d clone #21 cells. Panel at right shows quantified AKR1C3 signals in the Western blot. (B) AKR1C3 mRNAs in KD/non-KD cells. Data are from three biological replicates, assayed in duplicate. *P < .05; **P < .01. (C) AKR1C2 levels. D) SULT2B levels for clones analyzed in panels A, B and C. RPS (ribosomal protein S6, 32 kDa) is a control. (E) FKBP5 and PSA mRNA induction by DHT-activated AR. Induction is relative to the mRNA level at 0.01nM DHT (noninducing dose). NT and NT1 are independent stocks of the NT clone. Data shows average ± SEM from 3 biological replicates, assayed in duplicate by qRT-PCR and normalized to GAPDH and ubiquitin B mRNAs. Significance: *P < .05; **P < .01; ***P < .001.
AR activity increased markedly in KD cells (Fig. 4E). Thus, AR-mediated induction of FKBP5 and PSA mRNAs at 1 nM DHT was 20- to 30-fold in KD clones (#51, #49, and #183) compared with no more than 5- to 7-fold induction in non-KD cells at the same DHT dose. Of note, Fig. 1E shows similar AR levels in KD and non-KD cells. VCaP cells, which arose from vertebral metastasis of clinical prostate cancer, showed high AKR1C3, while SULT2B was barely detectable (23). FKBP5 mRNA induction in VCaP cells reached ≥80-fold upon stimulation by DHT at 0.5 nM or 1 nM (23). Since AKR1C3 can exhibit an AR-selective coactivator function (6), elevated AR activity in SULT2B KD cells may be in part due to enhanced AR coactivation by upregulated AKR1C3.
ERK1/2 activation in SULT2B KD cells and its dependence on AKR1C3
ERK1/2 Map kinase, which promotes cell survival, is strongly activated in SULT2B KD cells, evident from markedly increased phospho-ERK levels (Fig. 5A). Phenotypes of the KD cells such as ERK1/2 activation (Fig. 5A) and SNAI1 induction (Fig. 2H) are AKR1C3 dependent, since AKR1C3 silencing abolished ERK1/2 activation (Fig. 5B) and SNAI1 induction (Fig. 5D). Efficient AKR1C3 silencing by specific siRNAs is shown (Fig. 5C).
Figure 5.
ERK1/2 activation in SULT2B-KD cells, and the role of AKR1C3 in ERK activation and SNAI1 induction. (A) Phospho-ERK1/2 in KD clones (#49, #51) and non-KD clone. Clone #49 and NT/NT1 non-targeting clone were assayed in biological replicates. (B) Loss of ERK1/2 activation in AKR1C3-silenced SULT2B KD cells. Clones #49 and #51 were assayed in biological replicates. (C) AKR1C3 silencing by siRNAs in SULT2B KD clones. (D) Reduced SNAI1 expression in AKR1C3-silenced KD clones. E) AKR1C3 levels in SNAI1 knockdown and SNAI1 intact SULT2B KD cells. (F) SNAI1 levels in SULT2B KD cells treated with SNAI1 shRNA lentivirus or scramble shRNA lentivirus.
SNAI1 silencing, on the other hand, did not alter AKR1C3 levels in SULT2B KD cells (Fig. 5E), indicating that SNAI1 by itself is not involved in AKR1C3 upregulation. SNAI1 was silenced by lentivirus-expressed SNAI1-specific shRNAs (Fig. 5F). Whether other EMT factors such as SNAI2/Slug or members of the TWIST or ZEB family are involved in AKR1C3 upregulation in SULT2B-depleted cells remains to be determined.
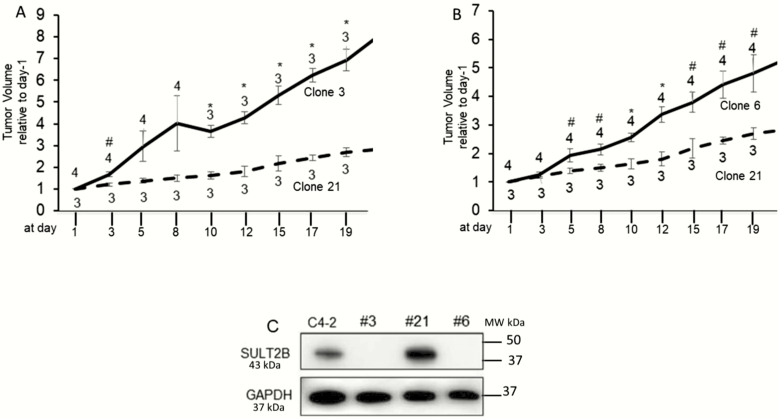
Growth escalation of KO cells in xenograft study
SULT2B-KO clones (#3 and #6) grew faster than the SULT2B-positive clone #21 in xenograft (Fig. 6A and 6B). Tumors were measured 3× per week, and measurement started when xenograft volume reached ~200 mm3. The tumor volume at each data point for an individual mouse was normalized to the tumor volume of the same mouse at the start of measurement (day 1). Statistically significant differences in growth patterns for clone 3 versus clone 21 and clone 6 versus clone 21 were observed. SULT2B status of the cells was reconfirmed prior to inoculation into mice (Fig. 6C).
Figure 6.
Xenograft tumor growth. (A,B) Growth rates for KO clone #3 vs non-KO clone #21 (A) and KO clone #6 vs non-KO clone #21 (B). Measurement started (designated as day-1) when tumor reached ~200 mm3. Each point in the plots is the mean ± SEM for normalized tumor volumes from 3 or 4 mice, as indicated. The tumor volume of a mouse at each time point was normalized to the volume at day-1 for the same mouse. (C) SULT2B levels of cells used for inoculation. Significance in the difference of tumor volumes between two group of mice at each time point was calculated by t-test assuming unequal variance. *P < .01; #P < .05.
Collectively, Figs 1–6 and the material in the online repository (23) led us to conclude that loss of SULT2B promotes prostate cancer progression due to changes that result in EMT-like phenotypes, increased invasion and motility of cells in vitro, and faster tumor growth in xenograft assay.
Loss of SULT2B and elevated AKR1C3 in clinical CRPC metastases
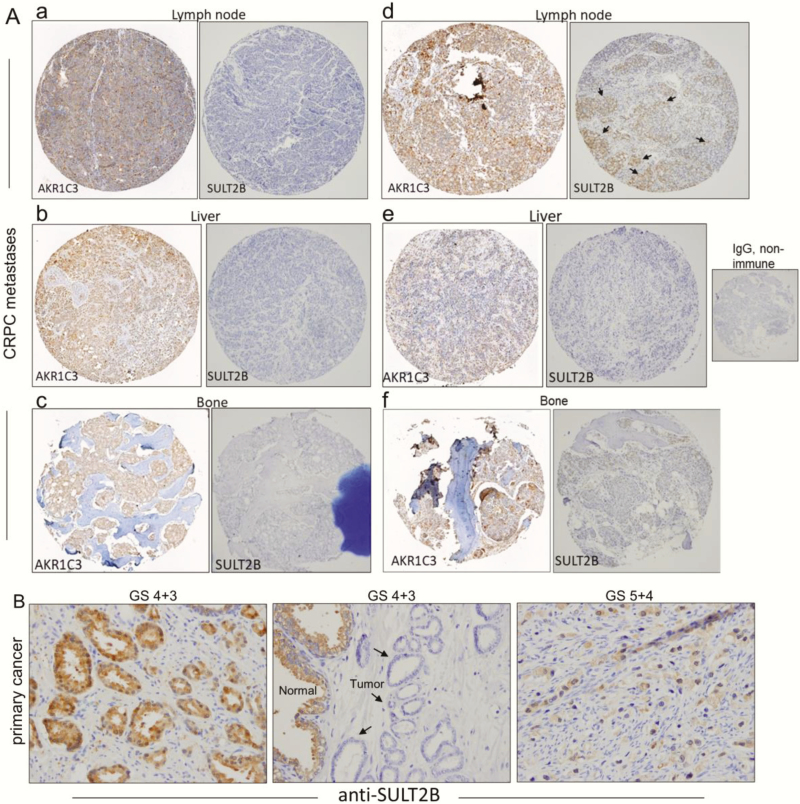
In IHC analysis of tissue microarray (TMA), SULT2B was undetectable in nearly all distant CRPC metastases. Representative photomicrographs of SULT2B-negative specimens (Fig. 7A, panels a–c and e and f) and an example of limited SULT2B staining (marked by arrows, panel d) are shown for TMA-55. TMA-55 contains 155 specimens from 50 autopsy cases, with up to 4 metastatic sites per single case (42). Importantly, specimens, which are SULT2B-negative, showed significant AKR1C3 expression (Fig. 7A, panels a-c and e and f). Strong AKR1C3 positivity is also evident for the CRPC tissue that showed limited SULT2B staining (Fig. 7A, panel d). IHC of AKR1C3 on these specimens was done on a second TMA. No signal was detected in the presence of nonimmune IgG (Fig. 7A). Conclusive evidence for a reciprocal relation between AKR1C3 and SULT2B expression in a tissue context awaits concurrent IHC probing of serial sections of the same TMA for AKR1C3 and SULT2B and quantification of staining intensities.
Figure 7.
Loss of SULT2B in AKR1C3-positive clinical CRPC metastases and variable SULT2B status of primary prostate cancer. (A) IHC of lymph node, liver and bone metastases of CRPC. Each panel with 2 tissue cores from the same specimen shows staining for AKR1C3 (left) and SULT2B (right). For panels (a–c and e,f), specimens are SULT2B-negative, while AKR1C3 expression is relatively strong. In panel d, the AKR1C3-positive CRPC showed limited SULT2B staining (marked by arrows). The non-immune IgG negative control is shown. (B) SULT2B in primary prostate tumor. Middle panel: SULT2B-negative tumor epithelia (shown by arrows) for a GS 7 specimen. Right panel: poorly differentiated GS 9 tumor showing SULT2B-positive status. Left panel: A second GS 7 specimen showing strong SULT2B staining in tumor acini.
Previously, we detected an overall significant reduction of SULT2B in primary prostate cancer samples from a small cohort (benign tissue, n = 29; malignant tissue, n = 12) (11). However, for individual cases, variable SULT2B expression—from strong positive to significant reduction and undetectable—was detected without correlation with GS (11). We confirm these results in Fig. 7B, which shows that tumor cells in specimens at GS 7 (4 + 3) and GS 9 (5 + 4) are SULT2B positive, but for a third specimen at GS 7 (4 + 3), tumor acini are SULT2B negative.
Discussion
The present study provides the first evidence that SULT2B1b (SULT2B), a DHEA and cholesterol/oxysterol sulfating enzyme, exerts inhibitory influences on CRPC cells, since SULT2B ablation led to (1) enhanced motility and invasion in vitro; (2) growth escalation in xenograft study; (3) EMT-like activation; (4) reduced stiffness and adhesion, which is indicative of increased invasion and metastasis potential of the cells; and (5) upregulation of AKR1C3, heightened AR activity, and activated ERK1/2 survival signaling. AKR1C3 silencing prevented activation of ERK1/2 and induction of the EMT factor SNAI1 in SULT2B KD cells. AR or DHT levels did not change in the KD cells—the latter likely due to parallel upregulation of AKR1C2, a DHT-metabolizing AKR, and AKR1C3, which promotes DHT biosynthesis. VCaP cells originating from vertebral metastasis of clinical prostate cancer showed high AKR1C3 levels and high AR activity, but barely detectable SULT2B. CRPC metastases from autopsy cases are mostly SULT2B negative barring a few samples, which showed limited SULT2B staining, whereas specimens lacking SUT2B showed strong AKR1C3-positive status. SULT2B levels in primary prostate cancer are variable and independent of GS.
EMT, which promotes embryonic morphogenesis and wound healing, also regulates cancer progression and metastasis (43, 44). EMT-like changes in SULT2B KO and KD cells, revealed from elevated mesenchymal proteins such as vimentin and N-cadherin, and increased levels of the EMT-inducing transcription factors SNAI1 and TWIST1, are consistent with the faster migration and enhanced invasion activity of these cells. EMT induction in our cell models is also consistent with the report that overexpression of STS led to EMT in PC-3 prostate cancer cells, and DHEA, a product of the sulfatase action of STS on DHEA-sulfate, induced EMT (51).
E-cadherin and EpCAM, which are normally epithelial cell-expressed proteins, are upregulated in SULT2B-ablated cells when assayed by mass cytometry at single-cell resolution. Quantified western blot data showed limited reduction of EpCAM and no significant change for E-cadherin. The lack of significant reduction of E-cadherin and EpCAM is consistent with reports that local and metastatic prostate adenocarcinomas express high levels of these normally epithelium-expressed proteins (45, 46), and circulating tumor cells (CTCs) from patients with advanced prostate cancer can be captured with an anti-EpCAM antibody (52). Also, in xenograft study, EpCAM silencing enhanced prostate tumor sensitivity to radiation and chemotherapy, inactivated the PI3K/AKT/mTORC1 axis, and prolonged survival of host mice (53). Elevated E-cadherin in aggressive prostate tumor xenografts and in clinical CRPC metastases has been reported (46). Persistence of E-cadherin in SULT2B KO and KD cells is likely to be important for cell migration, since invading carcinomas are known to infiltrate surrounding tissue as multicell clusters with cell–cell association via E-cadherin interaction at adherens junctions (44, 47, 48). Since EMT-inducing transcription factors of the SNAI/SNAIL, ZEB, and TWIST families are normally negative regulators of E-cadherin (44), a high E-cadherin level in SULT2B-deficient cells suggests an uncoupling of this negative regulatory program.
AFM study further linked SULT2B loss to an increased invasion and metastasis potential of CRPC cells. The KO cells are less stiff and less adhesive. Since cells showing reduced adhesion are more easily detached from a local environment and less stiff cells are more pliable (24, 25, 54), these mechanical changes can promote escape of detached KO cells and their entry into the bloodstream. Of note, CTCs from CRPC patients were reported to be less stiff and less adhesive but more deformable than CTCs from castration-sensitive patients (24), and in xenograft study CTCs from more aggressive prostate cancer showed lower stiffness and adhesion (25). The deformation index did not change for SULT2B KO cells, possibly due to their partial EMT state.
Our results that ERK1/2 Map kinase activation and SNAI1 induction in SULT2B-depleted cells is dependent on AKR1C3 (Fig. 5A, 5B, and 5D) are in agreement with an earlier report that ERK1/2 along with EMT was activated in AKR1C3-overexpressed PC3 prostate cancer cells (55). Notably, elevated AKR1C3 significantly associates with increased levels of EMT markers in clinical prostate cancer (55). In view of the prostaglandin F synthase activity AKR1C3 (56), we speculate that ERK1/2 activation in SULT2B-depleted cells is at least partly due to prostaglandin F-2α (PGF-2α)-induced signaling, which is known to activate ERK1/2 (50, 57). PGF-2α binding to the cognate G protein coupled receptor (FP receptor) leads to phospholipase C activation and release of the IP3 second messenger, which stimulates downstream events that culminate in ERK1/2 activation (50, 57). In preliminary assay, we detected increased PGF-2α levels in spent media of SULT2B KD cells. A causal link of PGF-2α levels to ERK activation and enhanced aggressive traits of SULT2B KD cells remains to be explored.
The repressive influence of SULT2B on AKR1C3 expression, shown in our cell models, is potentially of clinical significance since ERK1/2 is activated in patients during progression of primary prostate cancer to metastatic CRPC (58) and AKR1C3, which is elevated in advanced prostate cancer (59), is expressed in CRPC metastases that lack SULT2B, as shown in Fig. 7A.
Elevated AR activity may partly account for the growth escalation of xenografts derived from cells lacking SULT2B since AR stimulates prostate cancer cell proliferation (60). AKR1C3, as an AR-selective coactivator, may have played a role in enhancing AR activity in SULT2B-silenced cells despite no changes in AR and androgen levels. Whether enhanced AR activity contributed to EMT-like changes and increased aggressive traits of SULT2B-silenced cells is uncertain, since a role for androgen-induced AR signaling in the migration and invasion of prostate cancer cells is complex. In one study, depending upon the level of AR, DHT either increased or reduced EMT and invasion activity of prostate cancer cells (61), while another study showed that siRNA-driven AR reduction or antagonist-directed AR inactivation in C4-2B CRPC cells inhibited cell migration/invasion and caused reduction of several EMT markers (62). We also note the paradoxical results of a report, which showed that SULT2B silencing by siRNAs led to enhanced susceptibility of LNCaP androgen-sensitive prostate cancer cells to TNFα-induced cell death, despite induction of NF-κB-regulated antiapoptotic genes in single-cell transcriptome analysis (63). TNFα, however, induced apoptosis of SULT2B-silenced C4-2 CRPC cells in the absence of NF-κB activation (63). The significance of these results in the context of stable silencing of SULT2B needs to be evaluated. Also, how TNFα sensitivity might respond to elevated AKR1C3 in cells stably silenced for SULT2B remains an important query.
SULT2B expression is variable in primary PC—ranging from strongly positive to significant reduction or completely undetectable (Fig. 7B and (11)). SULT2B activity for DHEA sulfation is inhibited by abiraterone, a clinically active androgen synthesis blocker (64). Of note, abiraterone and ADT combination is favorably viewed for first-line inhibition of castration-sensitive metastatic prostate cancer due to overall survival advantage of patients (65). Deleterious consequences of reduced SULT2B levels on prostate cancer growth and aggressive traits, revealed from our study in cell and xenograft models, suggest that the SULT2B status of hormone naïve prostate cancer can be a relevant consideration before implementing abiraterone/ADT combination therapy. Finally, in view of the known association of elevated AKR1C3 with advanced prostate cancer, absence of SULT2B in AKR1C3-positive specimens may be clinically significant.
In summary, we show that SULT2B, which catalyzes 3β-sulfation of DHEA, cholesterol, and oxysterols, interferes with AKR1C3 upregulation and CRPC cell progression to increased aggressive traits such as enhanced AR activity, activation of ERK1/2, induction of an EMT-like state, and acceleration of xenograft tumor growth. Loss of SULT2B in CRPC metastases that are AKR1C3-positive highlights the possibility that the repressive influence of SULT2B on AKR1C3, shown in the present in vitro study, is clinically relevant. Insights into regulatory networks driving the SULT2B-AKR1C3 axis may identify a new avenue for prostate cancer inhibition.
Acknowledgment
We thank Dr. Myron Ignatius (UTHSA) for SNAI1 shRNA lentivirus and Sherry Dodds for her interest and help with use of Photoshop. Technical help from Jodie Cropper, Caleb Killer, Jason Pizzini, and from support staff of BASiC on mass cytometry is acknowledged. Part of this study constitutes the dissertation by Sulgi Park for a PhD at Department of Microbiology and Immunology, Pusan National University School of Medicine, South Korea. Ms. Park conducted research in the laboratory of B. Chatterjee as a Visiting Student.
Glossary
Abbreviations
- AFM
atomic force microscopy
- AKR
aldo-keto reductase
- AR
androgen receptor
- CRPC
castration-resistant prostate cancer
- CTC
circulating tumor cell
- CyTOF
mass cytometry by time-of-flight
- DHEA
dehydroepiandrosterone
- DHT
5α-dihydrotestosterone
- EMT
epithelial-to-mesenchymal
- Ig
immunoglobulin
- IHC
immunohistochemistry
- KD
knockdown
- KO
knockout
- LC-MS/MS
liquid chromatography tandem mass spectroscopy
- STS
steroid sulfatase
Financial Support: The study was funded by DoD-W81XWH-14-1-0606, PI: BC; Department of Veterans Affairs Merit-Review (1I01BX000280, PI: BC); Department of Veterans Affairs Res Career Scientist (IK6 BX004207, PI: BC); Morrison Trust (PI: BC). Core services through NIH-P30-CA054174 & NIH/U54-CA113001(UTHSA); CPRIT grants RP150600 (BASiC); RP160732 (Computation); NIH/CTSA-UL1-TR002645 (Statistics). CRPC tissue acquisition, TMA slides, LC-MS/MS were supported through Pacific Northwest Prostate Cancer SPORE (NIH-P50 CA97186, PI: Peter Nelson). Support from NIH-P01-CA163227 (PI, S.P. Balk) and DoD-W81XWH-12-1-0208 (PI, EAM) is acknowledged.
Author Contributions: Data acquisition, analysis: S.P., C.-S.S., S.J., C.-L.L., C.-M.W., B.T.M., P.A.O., M.E.G. Statistical and computational analysis: Y.C., S.P., C.-L.L., P.A.O. Material support: C.M., A.M.M., E.A.M. Conception, design, supervision: B.C.; Scientific critique and input: E.A.M. Manuscript writing: B.C
Additional Information
Disclosure Summary: The authors declare no conflicts of interest.
References
- 1. Sartor O, de Bono JS. Metastatic prostate cancer. N Engl J Med. 2018;378(17):1653–1654. [DOI] [PubMed] [Google Scholar]
- 2. Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15(12):701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mueller JW, Gilligan LC, Idkowiak J, Arlt W, Foster PA. The regulation of steroid action by sulfation and desulfation. Endocr Rev. 2015;36(5):526–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dai C, Heemers H, Sharifi N. Androgen signaling in prostate cancer. Cold Spring Harb Perspect Med. 2017;7: pii: a030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mohler JL. A brief history of intracrine androgen metabolism by castration-recurrent prostate cancer. Am J Clin Exp Urol. 2018;6(2):101–106. [PMC free article] [PubMed] [Google Scholar]
- 6. Yepuru M, Wu Z, Kulkarni A, et al. Steroidogenic enzyme AKR1C3 is a novel androgen receptor-selective coactivator that promotes prostate cancer growth. Clin Cancer Res. 2013;19(20):5613–5625. [DOI] [PubMed] [Google Scholar]
- 7. Strott CA. Sulfonation and molecular action. Endocr Rev. 2002;23(5):703–732. [DOI] [PubMed] [Google Scholar]
- 8. Falany CN, Rohn-Glowacki KJ. SULT2B1: unique properties and characteristics of a hydroxysteroid sulfotransferase family. Drug Metab Rev. 2013;45(4):388–400. [DOI] [PubMed] [Google Scholar]
- 9. Her C, Wood TC, Eichler EE, et al. Human hydroxysteroid sulfotransferase SULT2B1: two enzymes encoded by a single chromosome 19 gene. Genomics. 1998;53(3):284–295. [DOI] [PubMed] [Google Scholar]
- 10. Green SM, Kaipainen A, Bullock K, et al. Role of OATP transporters in steroid uptake by prostate cancer cells in vivo. Prostate Cancer Prostatic Dis. 2017;20(1):20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Seo YK, Mirkheshti N, Song CS, et al. Sulfotransferase SULT2B1b: Induction by vitamin D receptor, loss of expression in prostate cancer. Mol Endocrinol. 2013;27: 925–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thalmann GN, Anezinis PE, Chang SM, et al. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer Res. 1994;54(10):2577–2581. [PubMed] [Google Scholar]
- 13. Mostaghel EA, Zhang A, Hernandez S, et al. Contribution of adrenal glands to intratumor androgens and growth of castration-resistant prostate cancer. Clin Cancer Res. 2019;25(1):426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. RRID:AB_11017879, https://scicrunch.org/resolver/AB_11017879. Accessed December 2019.
- 15. RRID:AB_10718850, https://scicrunch.org/resolver/AB_10718850. Accessed December 2019.
- 16. RRID:AB_2818978, https://scicrunch.org/resolver/AB_2818978. Accessed December 2019.
- 17. RRID:AB_2818958, https://scicrunch.org/resolver/AB_2818958. Accessed December 2019.
- 18. RRID:AB_355568, https://scicrunch.org/resolver/AB_355568. Accessed December 2019.
- 19. RRID:AB_355745, https://scicrunch.org/resolver/AB_355745. Accessed December 2019.
- 20. Chen H, Lau MC, Wong MT, Newell EW, Poidinger M, Chen J. Cytofkit: a bioconductor package for an integrated mass cytometry data analysis pipeline. Plos Comput Biol. 2016;12(9):e1005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van Der Maaten L, Hinton G. Visualizing data using t-SNE. J Machine Learning Res . 2008;9:2579–2625. [Google Scholar]
- 22. Howe EA, Sinha R, Schlauch D, Quackenbush J. RNA-Seq analysis in MeV. Bioinformatics. 2011;27(22):3209–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park S, Song C-S, Lin C-L, et al. Supplemental Figures and Methods from: Inhibitory interplay of SULT2B1b sulfotransferase with AKR1C3 aldo-keto reductase in prostate cancer. Deposited September 22, 2019.https://figshare.com/s/5e71c08931aafc35fecb. [DOI] [PMC free article] [PubMed]
- 24. Osmulski P, Mahalingam D, Gaczynska ME, et al. Nanomechanical biomarkers of single circulating tumor cells for detection of castration resistant prostate cancer. Prostate. 2014;74(13): 1297–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang G, Osmulski PA, Bouamar H, et al. TGF-β signal rewiring sustains epithelial-mesenchymal transition of circulating tumor cells in prostate cancer xenograft hosts. Oncotarget. 2016;7(47):77124–77137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoffman BD, Crocker JC. Cell mechanics: dissecting the physical responses of cells to force. Annu Rev Biomed Eng. 2009;11:259–288. [DOI] [PubMed] [Google Scholar]
- 27. Iyer S, Gaikwad RM, Subba-Rao V, Woodworth CD, Sokolov I. Atomic force microscopy detects differences in the surface brush of normal and cancerous cells. Nat Nanotechnol. 2009;4(6):389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ignatius MS, Hayes MN, Lobbardi R, et al. The NOTCH1/SNAIL1/MEF2C pathway regulates growth and self-renewal in embryonal rhabdomyosarcoma. Cell Reports 2017;19(11):2304–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307(5712):1098–1101. [DOI] [PubMed] [Google Scholar]
- 30. RRID: AB_156339, https://scicrunch.org/resolver/AB_156339
- 31. RRID:AB_2218743, https://scicrunch.org/resolver/AB_2218743
- 32. RRID: AB_10167668, https://scicrunch.org/resolver/AB_10167668
- 33. RRID:AB_1129205, https://scicrunch.org/resolver/AB_1129205
- 34. RRID:AB_627531, https://scicrunch.org/resolver/AB_627531
- 35. RRID: AB_626777, https://scicrunch.org/resolver/AB_626777
- 36. RRID: AB_476751, https://scicrunch.org/resolver/AB_476751
- 37. RRID:AB_2818998, https://scicrunch.org/resolver/AB_2818998
- 38. RRID: AB_331646, https://scicrunch.org/resolver/AB_2818998
- 39. RRID:AB_330744, https://scicrunch.org/resolver/AB_330744
- 40. RRID: AB_2191738, https://scicrunch.org/resolver/AB_2191738
- 41. Mirkheshti N, Park S, Jiang S, et al. Dual targeting of androgen receptor and mTORC1 by salinomycin in prostate cancer. Oncotarget. 2016;7(38):62240–62254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang X, Coleman IM, Brown LG, et al. SRRM4 expression and the loss of REST activity may promote the emergence of the neuroendocrine phenotype in castration-resistant prostate cancer. Clin Cancer Res. 2015;21(20):4698–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thompson EW, Nagaraj SH. Transition states that allow cancer to spread. Nature. 2018;556(7702):442–444. [DOI] [PubMed] [Google Scholar]
- 44. Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18(2):128–134. [DOI] [PubMed] [Google Scholar]
- 45. Massoner P, Thomm T, Mack B, et al. EpCAM is overexpressed in local and metastatic prostate cancer, suppressed by chemotherapy and modulated by MET-associated miRNA-200c/205. Br J Cancer. 2014;111(5):955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Putzke AP, Ventura AP, Bailey AM, et al. Metastatic progression of prostate cancer and e-cadherin regulation by zeb1 and SRC family kinases. Am J Pathol. 2011;179(1):400–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Friedl P, Locker J, Sahai E, Segall JE. Classifying collective cancer cell invasion. Nat Cell Biol. 2012;14(8):777–783. [DOI] [PubMed] [Google Scholar]
- 48. Rubtsova SN, Zhitnyak IY, Gloushankova NA. A novel role of E-Cadherin-based adherens junctions in neoplastic cell dissemination. Plos One. 2015;10(7):e0133578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lin HK, Steckelbroeck S, Fung KM, Jones AN, Penning TM. Characterization of a monoclonal antibody for human aldo-keto reductase AKR1C3 (type 2 3alpha-hydroxysteroid dehydrogenase/type 5 17beta-hydroxysteroid dehydrogenase); immunohistochemical detection in breast and prostate. Steroids. 2004;69(13-14):795–801. [DOI] [PubMed] [Google Scholar]
- 50. Penning TM, Byrns MC. Steroid hormone transforming aldo-keto reductases and cancer. Ann N Y Acad Sci. 2009;1155:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shin S, Im HJ, Kwon YJ, et al. Human steroid sulfatase induces Wnt/β-catenin signaling and epithelial-mesenchymal transition by upregulating Twist1 and HIF-1α in human prostate and cervical cancer cells. Oncotarget 2017;8:61604–61617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Diamond E, Lee GY, Akhtar NH, et al. Isolation and characterization of circulating tumor cells in prostate cancer. Front Oncol. 2012;2:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ni J, Cozzi P, Beretov J, et al. Epithelial cell adhesion molecule (EpCAM) is involved in prostate cancer chemotherapy/radiotherapy response in vivo. BMC Cancer. 2018;18(1):1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stroka KM, Konstantopoulos K. Physical biology in cancer. 4. Physical cues guide tumor cell adhesion and migration. Am J Physiol Cell Physiol. 2014;306(2):C98–C109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang B, Gu Y, Hui K, et al. AKR1C3, a crucial androgenic enzyme in prostate cancer, promotes epithelial-mesenchymal transition and metastasis through activating ERK signaling. Urol Oncol. 2018;36(10):472.e11–472.e20. [DOI] [PubMed] [Google Scholar]
- 56. Penning TM. AKR1C3 (type 5 17β-hydroxysteroid dehydrogenase/prostaglandin F synthase): roles in malignancy and endocrine disorders. Mol Cell Endocrinol. 2019;489:82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sales KJ, Milne SA, Williams ALW, Anderson RA, Jabbour HN. Expression, localization, and signaling of prostaglandin F2α receptor in human endometrial adenocarcinoma: regulation of proliferation by activation of epidermal growth factor receptor and mitogen-activated protein kinase signaling pathways. J Clin Endocrinol Metab. 2004;89:986–993. [DOI] [PubMed] [Google Scholar]
- 58. Nickols NG, Nazarian R, Zhao SG, et al. MEK-ERK signaling is a therapeutic target in metastatic castration resistant prostate cancer. Prostate Cancer Prostatic Dis. 2019;22(4):531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mitsiades N, Sung CC, Schultz N, et al. Distinct patterns of dysregulated expression of enzymes involved in androgen synthesis and metabolism in metastatic prostate cancer tumors. Cancer Res. 2012;72(23):6142–6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Balk SP, Knudsen KE. AR, the cell cycle, and prostate cancer. Nucl Recept Signal. 2008;6:e001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhu ML, Kyprianou N. Role of androgens and the androgen receptor in epithelial-mesenchymal transition and invasion of prostate cancer cells. Faseb J. 2010;24(3):769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lin CY, Jan YJ, Kuo LK, et al. Elevation of androgen receptor promotes prostate cancer metastasis by induction of epithelial-mesenchymal transition and reduction of KAT5. Cancer Sci. 2018;109(11):3564–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vickman RE, Yang J, Lanman NA, et al. Cholesterol sulfotransferase SULT2B1b modulates sensitivity to death receptor ligand TNF alpha in castration resistant prostate cancer. Mol Cancer Res. 2019; doi: 10.1158/1541-7786.MCR-18-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yip CKY, Bansal S, Wong SY, Lau AJ. Identification of galeterone and abiraterone as inhibitors of dehydroepiandrosterone sulfonation catalyzed by human hepatic cytosol, SULT2A1, SULT2B1b, and SULT1E1. Drug Metab Dispos. 2018;46(4):470–482. [DOI] [PubMed] [Google Scholar]
- 65. Logothetis CJ. Improved outcomes in men with advanced prostate cancer. N Engl J Med. 2017;377(4):388–390. [DOI] [PubMed] [Google Scholar]