Abstract
Social interactions can influence the expression and underlying genetic basis of many traits. Yet, empirical investigations of indirect genetic effects (IGEs) and genotype-by-genotype epistasis—quantitative genetics parameters representing the role of genetic variation in a focal individual and its interacting partners in producing the observed trait values—are still scarce. While it is commonly observed that an individual's traits are influenced by the traits of interacting conspecifics, representing social plasticity, studying this social plasticity and its quantitative-genetic basis is notoriously challenging. These challenges are compounded when individuals interact in groups, rather than (simpler) dyads. Here, we investigate the genetic architecture of social plasticity for exploratory behaviour, one of the most intensively studied behaviours in recent decades. Using genotypes of Drosophila simulans, we measured genotypes both alone, and in social groups representing a mix of two genotypes. We found that females adjusted their exploratory behaviour based on the behaviour of others in the group, representing social plasticity. However, the direction of this plasticity depended on the identity of group members: focal individuals were more likely to emerge from a refuge if group members who were the same genotype as the focal remained inside for longer. By contrast, focal individuals were less likely to emerge from a refuge if partner-genotype group members remained inside for longer. Exploratory behaviour also depended on the identities of both genotypes that composed the group. Together, these findings demonstrate genotype-by-genotype epistasis for exploratory behaviour both within and among groups.
Keywords: social plasticity, quantitative genetics, indirect genetic effects, Drosophila
1. Introduction
Interacting in groups can influence nearly every aspect of an animal's life, and especially its behaviour. Even ‘non-social’ behaviours, such as foraging, can be influenced by social interactions [1,2], representing social plasticity. Indeed, the fitness benefits of social interactions often include coordinating behaviours within a social group [3].
Understanding the quantitative-genetic basis of this social plasticity is critical for forming inferences about how behaviour and its plasticity may evolve. Interacting traits, i.e. those that may be modified by social interactions, can have a unique genetic basis, because the ‘environment’ that influences the expression of the trait is other individuals with their own genotypes. The indirect effect of one individual's genotype on an interacting individual's traits is termed indirect genetic effects (IGEs; [4]). Such effects are described as ‘indirect’ because the alleles that influence the focal individual's traits are expressed not in the focal individual itself, but in the interacting partner: the partner's genotype influences the partner's phenotype, which in turn influences the traits of a focal individual through social plasticity. By contrast, direct genetic effects (DGEs) describe the effects of an individual's own genotype on its own traits.
Furthermore, genotypes can differ in the extent to which their behaviour is modified by IGEs, representing genotype-by-genotype epistasis [5]. For example, in mosquitofish, juveniles reared with other individuals of their own genetically controlled colour morph have poorer body condition than juveniles reared with social partners of the other morph [5]. Thus, under genotype-by-genotype epistasis, the effect of an interacting individual on a focal individual's traits depends on both (or all) participants' genotypes [6]. At the behavioural level, this interdependence between social partners is exactly what we expect, because the idea of social plasticity itself is that characteristics of group-mates influence a focal individual's behaviour, and vice versa [1,7,8]. Furthermore, differences among individuals in plasticity, including social plasticity, are widespread [1,9–11])
In particular, the phenotypic and/or genetic similarity between interacting individuals is expected to be a critical factor structuring social plasticity [12,13], with effects on behaviour, fitness and population genetics parameters [14–17]. For example, phenotypic similarity among individuals in fish shoals plays an important role in anti-predator defence [14,18], and assortative mating by behaviour(s) can influence the success of parental care [19].
One of the central challenges of studying IGEs is that the IGE framework is based on dyadic interactions among individuals, yet social plasticity often occurs in a group context [20–22]. A common workaround for this challenge is to summarize the behaviour of a focal individual's social partners, for example, by averaging the behaviour of everyone in the group [23,24]. This approach is expected to be most successful when the focal individual responds similarly to the behaviour of different group members, i.e. when genotype-by-genotype epistasis is absent. Applying this existing theory about the effects of social partner phenotypic and genetic similarity to formulate predications about IGEs in groups may be useful for two key reasons. First, many groups in nature are composed of individuals with variable relatedness and phenotypic similarity. Groups may include multiple families, for example, or individuals from multiple clutches or broods who were reared in the same local area [20]. Furthermore, because the property of ‘similarity’ is relative to the identities of both interactants, differential social plasticity based on genetic similarity might represent a key source of genotype-by-genotype epistasis. In other words, individuals' behaviours may depend on their own genotype (DGEs), their social partners’ genotypes (IGEs) and the genetic and phenotypic similarity between interacting partners (genotype-by-genotype epistasis)—which is inherently an emergent property of all interacting individuals.
Here, we investigate the social dynamics and underlying quantitative-genetic basis of exploratory behaviour. Exploratory behaviour is one of the most intensively studied behaviours in recent years [25], likely because exploring a new environment is a fundamental step in many ecologically and evolutionarily relevant processes such as foraging, encountering predators [26], patch use [27] and even parental care [28]. Several studies have documented that exploratory behaviour can influence the formation of social bonds [29,30] and that variation in exploratory behaviour within groups can improve the efficiency of collective behaviours [31]. Reciprocally, exploratory behaviour itself can depend on the social environment [32]. Yet, we still know little about how group composition influences social plasticity in exploratory behaviour [29,30], and we know even less about the underlying quantitative-genetic basis of this social plasticity [2]. This gap in knowledge means that the causes of variation in exploratory behaviour, and the potential for this behaviour to evolve, are still uncertain.
To start to fill this gap in the knowledge, we measured genetic variation in exploratory behaviour and its social plasticity. We first established genetic differences in exploratory behaviour in our sample (Experiment 1) and then, for a subset of genotypes, studied their exploratory behaviour in a group setting (Experiment 2). By manipulating and replicating the genetic composition of social groups, we were able to evaluate the role of group-mates' genotypes and behaviours in shaping a focal individual's exploratory behaviour. Specifically, we investigated genotype-by-genotype epistasis at two levels: within groups and among groups. Within groups, we tested whether individuals adjusted their behaviour differently in response to the behaviour of same-genotype versus partner-genotype group members. For example, a particular social partner's genotype and phenotype may have a strong effect on similar focal individuals, but only a weak effect on dissimilar individuals. Among groups, we tested whether different combinations of genotypes produced different effects on the behaviour of each genotype.
2. Material and methods
(a). Study system: Drosophila simulans
Drosophila simulans is a cosmopolitan vinegar fly that feeds on rotting fruit. Drosophila simulans and its familiar sister species, D. melanogaster, are sympatric in many parts of their ranges and share similar ecologies (with important, but subtle, differences; [33,34]). Because flies live on rotting fruit, they must move among fruits relatively frequently as their current habitat degrades in quality (i.e. rots) and new habitats (fallen fruits) become available. Therefore, the decision to leave a current location and explore a novel food patch is an ecologically relevant choice for fruit flies. This exploratory behaviour is particularly important for females, who must meet the metabolic demands of egg production while also choosing suitable oviposition substrates for their progeny [35–37]. Because this link between exploratory behaviour and fitness is better understood for females than males, we studied female D. simulans exploratory behaviour in the current experiments.
(b). Genotypes
Isofemale lines (hereafter ‘genotypes’) of D. simulans were kindly provided by Dr Daniel Matute. Each genotype represents the descendants of a single female whose offspring were subsequently inbred for many generations. Therefore, individuals from the same genotype are much more genetically similar to each other than to individuals from other genotypes [38]. Specifically, our genotypes were LNP 15-001, LNP 15-062 and LNP 15-063, all collected from Zambia; LZV 15-003, collected from the Lower Zambeze Valley; MD 221, MD 223 and MD 242, all collected from Madagascar; NS-05 and NS -39, collected from Nairobi; and NMB-014 and NMB-024, collected from Namibia ([38,39]; J Coughlan 2019, personal communication). These genotypes represent a sample of the natural genetic diversity of D. simulans in Africa and Madagascar.
(c). Rearing and individual marking
Individuals used in all experiments resulted from crossing a virgin female with a male of the same genotype and collecting the virgin female F1 offspring.
Flies studied in groups (Experiment 2) were marked individually. After collection, each individual female was anaesthetized with carbon dioxide, and a small dot of paint was dabbed on the fly's pronotum. In previous studies, this technique has not influenced flies' behaviour [21,40,41]. After painting, each female was isolated in a vial filled with plain fly food and given 72 h to recover before trials began.
(d). Measuring exploratory behaviour
To measure exploratory behaviour, we used the ‘emergence from a refuge’ paradigm similar to studies of exploratory behaviour in many other animals [25]. A single female (Experiment 1) or a group of females (Experiment 2) was gently aspirated into a 1 ml pipette tip. The large end of the pipette tip was covered with mesh to prevent the flies from escaping. The small end of the pipette tip was inserted into a small hole in an arena containing fly food. The arena was composed of two Petri dish bottoms taped together, with the floor of the arena covered in fly food (a photograph of the arena is available in the electronic supplementary material). Therefore, each fly had the choice between (i) remaining in the pipette tip or (ii) emerging out from the small end of the pipette tip into the arena containing food. No food was present in the pipette tip. This approach mimics the situation in the wild in which flies (especially females) hide on a relatively safe leaf or other structure before descending onto a novel food patch [40,42].
We measured the time, in seconds, at which each fly emerged from the pipette tip into the arena (emergence time) during the 5 h trial. If a fly did not emerge within 5 h, her emergence time was measured as 5+ h. We accounted for this censoring in our analysis (see below).
(e). Experiment 1: establishing genotypic differences in emergence behaviour in solo flies
To initially identify genetic differences in exploratory behaviour in D. simulans, and to choose genotypes for social group analysis, we measured the exploratory behaviour of flies individually. Seven to 14 females of each of the 11 genotypes were measured (mean number of replicates per genotype: 10), for a total of 111 flies measured over 555 h of experiments.
Each fly was continuously observed for 5 h, with an observer noting when each individual emerged to the nearest second. These trials were not filmed. Note that these data are also a small component of a separate manuscript, which addresses different questions [43].
(f). Choosing genotypes for social group experiments
We chose four genotypes—genotypes LNP-63, MD-242, NM-14 and NS-39—for social group experiments (Experiment 2, below). These genotypes correspond to the slowest, fastest, 9th fastest and 3rd fastest emergers, respectively, in Experiment 1 (for detailed results, see below). Therefore, these genotypes span a range of exploratory behaviour found in our sample. In addition, we ensured that the four genotypes were originally collected from different sites to avoid the possibility of cryptic genetic relatedness among the genotypes [44].
Note that, because Experiments 1 and 2 were conducted in separate blocks, and using slightly different methods, they are not directly comparable—i.e. the goal Experiment 1 was to choose genotypes for Experiment 2.
(g). Experiment 2: measuring emergence behaviour in groups
To investigate IGEs and genotype-by-genotype epistasis for exploratory behaviour, we measured the emergence times of individuals when in groups of different genotypic composition. Social groups were made up of six individuals total, three individuals of one genotype and three individuals of a different genotype. Given the four genotypes used, all possible combinations of two different genotypes were tested.
Therefore, from each individual's perspective, their group was composed of two individuals who were very similar to them (i.e. individuals from the same genotype) and three individuals who were not (i.e. individuals from a different genotype originally collected from a different location). This design allowed us to estimate the effects of group members' behaviour on the exploratory behaviour of each focal individual. More specifically, we modelled how the exploratory behaviour of same-genotype group members (i.e. the behaviour of group members from the same genotype as the focal) and the exploratory behaviour and genotype of partner-genotype group members (i.e. group members from the other genotype) were associated with variation in the focal individual's exploratory behaviour.
Social groups were formed by aspirating six individual flies from their isolated vials into a single 1 ml pipette tip, with the same set-up as above. As soon as all six flies and the pipette tip were in place, we began filming. Each group was filmed using an apple iPod touch (camera information: f/2.4, 3.3 mm) positioned above the pipette tip and arena. Filming continued for 5 h. Each genotype–genotype combination was replicated four times, for a total of 24 social groups representing 144 total flies measured on 120 h of video.
After the trial was complete, videos were analysed to identify the time, to the nearest second, at which each fly first emerged from the pipette tip into the arena (emergence time).
(i). Analysis methods: approach
All analyses were conducted in R v. 3.6.0 [45]. Code to reproduce these analyses is available as an electronic supplementary material. To model how long it took for flies (individually and in groups) to emerge from the pipette tip, we used mixed-effect proportional hazards models implemented in the coxme package in R [46]. The Cox proportional hazards models are appropriate for time-to-event data because they can be used to understand how fixed and random predictor variables modify an underlying hazard function [47]. These models take the form
where λ(t) is a hazard function representing information about the probability that an event will occur during some time Δt, given that the event has not occurred already. λ0(t) represents the baseline hazard function, β is the vector of fixed-effects coefficients and b is the vector of random effects coefficients [46]. Here, this approach allowed us to test how genotype and social group behaviours influenced the flies' ‘hazard’ of our event of interest: leaving the pipette tip. This method is particularly useful for censored data; our data are right-censored, because many flies (25% of flies in Experiment 1, 27% in Experiment 2) took longer than 5 h to emerge. We modelled censoring by including a variable, ‘status’, that indicated whether each measurement was censored or not. Initial models showed no evidence for non-proportional hazards (Experiment 1: global p = 0.747; Experiment 2: global p = 0.689), indicating that our datasets met the assumption of the Cox proportional hazards models.
For continuous fixed predictors, effects are measured as hazard ratios. These indicate the change in the instantaneous risk of the event occurring if the fixed effect was increased by one unit [48,49]. A hazard ratio of 1 would indicate that the risk is unaffected by the predictor variable. Our fixed effects, described below, are measures of the exploratory behaviour of other group members. Thus, our hazard ratios indicate the change in the focal individual's risk of emerging if their group-mates emerged 1 s faster.
For random effects, variance ‘explained’ values can be obtained as
where σa is the random effect variance of interest; σe is a measure of error variance (e.g. unaccounted differences among individuals or groups); and c is the proportion of censored values in the dataset [50,51].
(ii). Inference
In all analyses, we used likelihood-ratio tests to test the significance of random effects. This test compares the goodness of fits of two nested models, one including the random effect parameter of interest, and one without the parameter [52]. The final models included all the fixed effects noted, plus all random effects found to significantly improve the fit of the model. To evaluate the significance of fixed effects in the final models, we used type III analysis of deviance tests implemented in the car package [53].
For Experiment 2, each measurement of emergence time was tested in two models: the full model that included all data, and one of the single-genotype follow-up models (described below). Therefore, all p-values were doubled; we report these Bonferroni-corrected p-values.
(h). Experiment 1 analysis
(i). Identifying genetic variation in exploratory behaviour
We fit a model where the response variable was each individual's exploratory behaviour in Experiment 1. We included random intercepts for each genotype, allowing us to incorporate estimates of variation in behaviour among individuals who were the same genotype, and across different genotypes. We included a random effect indicating which flies were tested on the same day to account for unavoidable differences in the environment on different days. Finally, we included random intercepts for each individual to enable estimates of ‘variance explained’ by genotype. No fixed effects were included.
(i). Experiment 2 analysis
(i). Overview
In Experiment 2, our goals were to identify how an individual's time to emerge was influenced by (i) its genotype, (ii) the other genotype present in its group (i.e. IGEs), and (iii) the behaviour of the other members of its group. To do so, we fit a model where the response variable was a single individual's exploratory behaviour. As fixed effects, we included the behaviour of other group members, explained in more detail below. As random effects, we included the genotype of the focal fly, and the partner genotype. These correspond to DGEs and IGEs. This approach thus represents a combination of ‘trait-based’ and ‘variance-based’ analysis approaches [54]. Trait-based approaches consider how variation in traits of social partners, which may have a genetic basis, influence the trait(s) of a focal individual. Variance-based approaches consider how social partners of different genotypes influence the traits of a focal individual. Therefore, trait-based approaches represent specific hypotheses about how social interactions influence behaviour, while variance-based approaches provide information about the overall effect of interacting with variable social partners on the focal individual's behaviour. Because we had information about the genotype and exploratory behaviour of all individuals in our experiment, we could combine these approaches [21].
Finally, because our unit of replication was at the group level, we included a random effect to account for our repeated measurements of each group. In other words, each of the six group members were treated as a subsample of their group. Differences among groups may occur due to attributes of the group (i.e. which genotypes were present), random variability among individuals, unavoidable micro-environmental variation among arenas, etc.
In this full model, we did not directly estimate among-group genotype-by-genotype epistasis, because coxme does not support interactions among random effects. To remedy this, we conducted single-genotype follow-up models to elucidate more information about the factors affecting behaviour of each genotype, explained in more detail below.
(ii). Social environment characteristics
For each fly, we computed two measures of the behaviour of its social group. One measure was the median emergence time of the fly's group-mates who were not the same genotype as the focal fly—which we refer to as ‘partner-genotype’ behaviour. The second measure was the median emergence time of the other two flies in the group who were the same genotype as the focal fly—which we refer to as ‘same-genotype’ behaviour. Importantly, the focal fly's own emergence time was not part of either measure. Medians were chosen rather than other measures of central tendency because the data were highly non-normal. These measures were included in the model as fixed effects, allowing us to investigate how the behaviour of group members influenced the behaviour of the focal fly.
(iii). Direct and indirect genetic effects
To estimate DGEs and IGEs, we included random intercepts for each fly's own genotype. We also included a random effect identifying the other genotype present in the group.
Finally, we examined the potential that genotypes differed in the ways that their behaviour was influenced by the behaviour of group members, i.e. within-group genotype-by-genotype epistasis. We specified an interaction between same-genotype behaviour and focal individual genotype, which would indicate that genotypes differ in how their behaviour is influenced by variation in the behaviour of same-genotype social partners. We also used the same approach for partner-genotype behaviour. We tested models in which the intercept and slope for each genotype were assumed to be uncorrelated, as well as models that explicitly estimated this correlation. As noted above, we retained random effects that significantly improved the fit of the model as indicated by likelihood-ratio tests.
(iv). Single-genotype follow-up model
The full model (above) indicated interesting interactions between direct genetic effects and social effects. We next fit four individual models, each one modelling the behaviour of a single focal genotype (as in [23]). For each data subset, we examined the effects of partner genotype, median emergence time of individuals that were the same genotype as the focal fly and median emergence time of individuals of the partner genotype. Among-group genotype-by-genotype epistasis would be evident if the effect of partner genotype on focal individual behaviour differed among focal genotypes, as indicated by the magnitude and/or significance of the parameter corresponding to partner genotype.
3. Results
(a). Experiment 1
(i). Genetic variation in exploratory behaviour
We found support for genetic variation in emergence time when flies were tested alone (, p = 0.00061). The variance estimate for genotype was 0.494, and the unaccounted among-individual variance was 0.182. In this experiment, 32% of observations were censored. Therefore, we estimate that genotypic differences account for about 23% of the observed variance in behaviour (as described above). Note that this value cannot be interpreted as a heritability estimate, because the genotypes were collected from different populations.
(b). Experiment 2: full model
(i). Direct and indirect genetic effects, and variability among groups, contribute to variation in emergence time
In the full model for Experiment 2, we found support for genetic differences in emergence time (, p < 0.0001), as well as effects of the partner genotype, i.e. IGEs (, p = 0.0007). As expected, the random effect corresponding to differences among groups substantially improved the fit of the model (, p = 0.0008; table 1).
Table 1.
Effects, test statistics and interpretation of parameters included in the full model for Experiment 2.
| parameter | estimate and significance testing |
interpretation | |||
|---|---|---|---|---|---|
| fixed effects | parameter estimate | hazard ratio | χ2 | adjusted p | |
| median emergence time of same-genotype group-mates | −2.57 × 10−5 | 0.999959 | 2.266 | 0.391 | no main effect of plasticity in response to behaviour of same-genotype group-mates |
| median emergence time of partner-genotype group-mates | −4.13 × 10−5 | 0.999974 | 6.5128 | 0.02142 | social plasticity in response to behaviour of partner-genotype group-mates |
| random effects | variance | % explained | χ2 | adjusted p | |
| trial ID | 0.000355 | n.a. | 12.62 | 0.0008 | different groups are different |
| focal genotype | 0.089 | 6.1% | 16.38 | <0.0001 | direct genetic effect |
| partner genotype | 0.02246 | 1.6% | 12.78 | 0.0007 | indirect genetic effect |
| focal genotype × median emergence time of same-genotype group-mates | 8.99 × 10−11 | <1% | 12.916 | 0.0006 | genetic variation in social plasticity |
The variance estimate for genotype was 0.089, the variance estimate for partner genotype was 0.02246 and the unaccounted among-group variance was 0.000355. In this experiment, 28% of observations were censored. Therefore, we estimate that genotypic differences account for about 6.1% of the observed variance in behaviour (using the equation noted above), and about 1.6% of variation in behaviour could be attributed to which partner genotype was present. Again, these estimates do not necessarily correspond to heritability estimates, because of the small number of genotypes studied and the fact that they were not a random sample.
(ii). Exploratory behaviour of group members influences individual emergence time
In the full model for Experiment 2, the behaviour of partner-genotype individuals influenced focal emergence time (, p = 0.02142; table 1). The parameter estimate was negative (parameter estimate = −0.000041) corresponding to a hazard ratio below 1 (hazard ratio estimate = 0.9999587). These estimates indicate that, if a focal fly was in a group in which partner-genotype individuals took a long time to emerge (i.e. higher values of median partner-genotype emergence time), the focal fly's instantaneous ‘risk’ of emerging from the pipette tip was lower, compared to focal flies in groups where partner-genotype individuals emerged quickly. For example, a 1 h (3600 s) increase in the median emergence time of partner-genotype individuals should change the risk of focal individuals emerging by a factor of 0.862 (calculated as exp(3600 * parameter estimate); [49]).
By contrast, an overall effect of the behaviour of same-genotype individuals on focal fly emergence time was not statistically supported (, p = 0.391; table 1).
(iii). Genotypes differ in how their behaviour is influenced by same-genotype group members
We found no support for an interaction between partner-genotype behaviour and focal individual genotype (, p = 0.2644). This result indicates that the effects of the emergence behaviour of partner-genotype individuals on focal individual behaviour were similar across all genotypes.
By contrast, we found support for an interaction between same-genotype emergence behaviour and focal individual genotype (, p = 0.00065). Coupled with the lack of support for a main effect of same-genotype emergence behaviour on focal individual emergence behaviour (see above), this result indicates that different genotypes modified their behaviour differently in response to the behaviour of their same-genotype group members.
Relaxing the assumption of no correlation between genotype-specific intercepts and slopes did not improve the fit of the model (, p = 1) indicating that genotype-specific responses to the behaviour of same-genotype social group members were not predictable based on the typical emergence behaviour of the focal genotype.
(c). Experiment 2: single-genotype models
(i). Genetic variation in responses to the behaviour of social partners
The full model indicated that genotypes differ in how they respond to the median emergence behaviour of same-genotype individuals. Single-genotype models further revealed that the direction of this social plasticity was the same for all genotypes, but the magnitude varied (genotype NM-14: hazard ratio = 1.0007474, , p < 0.0001; genotype LNP-63: hazard ratio = 1.0006768, , p < 0.0001; genotype MD-242: hazard ratio = 1.0004040, , p = 0.0002374; genotype NS-39: hazard ratio = 1.0002980, , p < 0.0001). Hazard ratios were all greater than 1, indicating that focal flies whose same-genotype group-mates took longer to emerge (i.e. high values of median emergence time for same-genotype group-mates) had a higher instantaneous risk of emerging. For example, a 1 h (3600 s) increase in the median emergence time of same-genotype individuals would be expected to change the risk of focal individuals emerging by a factor of between 2.92 and 14.73, depending on the focal individual's genotype. These results confirm that genotypes differ in the extent to which individuals are influenced by same-genotype social group members' behaviour.
(ii). Among-group genotype-by-genotype epistasis
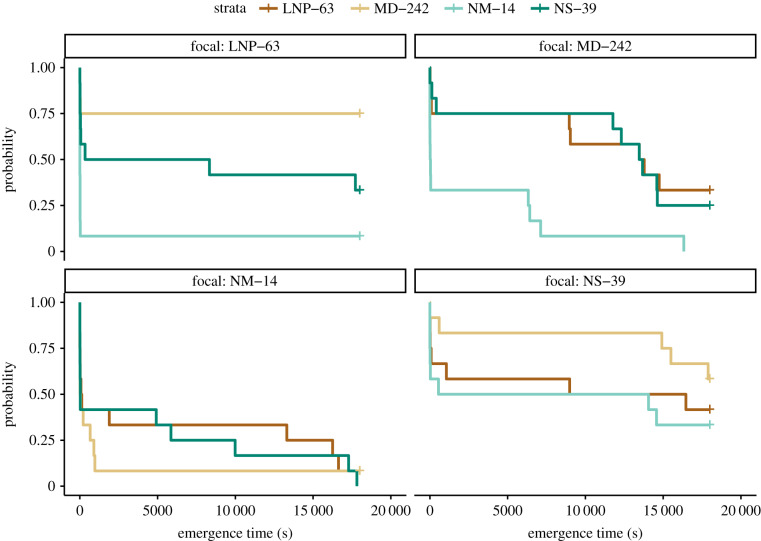
Single-genotype models found statistical support for IGEs only for one of the four genotypes (figure 1; genotype NM-14: , p = 1; genotype LNP-63: , p < 0.0001; genotype MD-242: , p = 1; genotype NS-39: , p = 1). This result indicates that the emergence behaviour of this one genotype (LNP-63) depends on which partner genotype is present in its group.
Figure 1.
Among-group genotype-by-genotype epistasis. Each of the four plots represents one focal genotype, noted in the title directly above. Data are from Experiment 2. The y-axis of the plot represents the probability that a focal fly will emerge from the pipette tip at a particular time, denoted on the y-axis. Different-coloured lines within each plot represent the behaviour of the focal genotype in the presence of different partner genotypes. The partner genotypes are colour-coded as noted at the very top of the plot. Differences among coloured lines on the same plot mean that the focal genotype showed different exploratory behaviour depending on which partner genotype was present in its social group. N = 24 groups, 144 total individuals. (Online version in colour.)
4. Discussion
Identifying the quantitative-genetic basis of social interactions is essential for understanding the ways that interactive traits evolve [4,55,56]; yet studies aimed at understanding IGEs and, particularly, genotype-by-genotype epistasis are still lacking [5,57]. Here, we measured exploratory behaviour in individuals and groups of female D. simulans to uncover how social plasticity in exploratory behaviour depends on genetic similarity among group-mates, in addition to group-mate's behaviour per se. We found substantial differences in exploratory behaviour among genotypes, representing DGEs. Furthermore, we found that female D. simulans adjust their exploratory behaviour in response to the exploratory behaviour of their group-mates, but the direction of this effect depended on the genetic identity of the group-mates considered. The longer a female's same-genotype group-mates remained in the pipette tip, the greater that female's ‘risk’ of leaving the pipette tip, as indicated by a hazard ratio greater than 1. Furthermore, the magnitude of this effect varied among genotypes. However, the same females adjusted their exploratory behaviour to match the behaviour partner-genotype group-mates, as indicated by a hazard ratio less than one in the full model. Thus, the longer a female's partner-genotype group-mates remained in the pipette tip, the lower that female's ‘risk’ of leaving the pipette tip.
Our results represent genotype-by-genotype epistasis at two levels. Within groups, the ways in which individuals altered their exploratory behaviour in response to the behaviour of group members depended both on the focal individual's genotype and on the genotype of the relevant group members, i.e. whether the focal and group member's genotypes were the same, or not. Among groups, we also found that different partner genotypes had different effects on the behaviour of a given focal genotype (figure 1), and that focal genotypes differ in their sensitivity to these social effects.
Our findings of genotype-by-genotype epistasis indicate that flies do not simply follow other flies, but instead, adjust their exploratory behaviour differently depending on the identities of social partners. Flies might ‘know’ who in their social group is genetically similar to them using phenotypic cues, such as heritable variation in cuticular pheromones [58–60]. Previous studies have also suggested that flies can recognize familiar individuals [13,61]. In our experiments, females were isolated prior to testing, so familiarity was unlikely to contribute to our results. Furthermore, familiarity itself may be influenced by the choices flies make early in life [62,63].
Our approach, though powerful, was limited in several respects. First, although our approach is a step forward from summarizing the behaviour of all individuals in the group, we still used summary statistics to describe the social environment experienced by each focal individual (i.e. the median behaviour of same-genotype and partner-genotype individuals). A more nuanced approach, perhaps using social network measurements, would further illuminate the social dynamics that ultimately influence exploratory behaviour and its genetic basis. Second, many flies took longer than 5 h to emerge, suggesting that longer experiments might be needed to understand the decisions made by the least-exploratory individuals in our sample. Finally, although we measured 144 flies in Experiment 2, our experiment only included four genotypes, which may or may not represent the results that would be obtained with different genotypes.
The finding that exploratory behaviour is governed by IGEs and genotype-by-genotype epistasis has implications for how we expect exploratory behaviour to evolve. For example, directional selection on exploratory behaviour is expected to be inefficient under genotype-by-genotype epistasis, because the genotype that is fastest (or slowest) to emerge from a refuge may be different depending on which other genotypes are present [64]. Interestingly, expected evolutionary trajectories under IGEs depend critically on whether social plasticity (described in IGE models as the coefficient of interaction, Ψ) is positive or negative [55,65,66]. In general, positive values of Ψ, in which high trait values in social partners elicit high trait values in the focal individual, are predicted to accelerate evolution, compared to the situation in which IGEs are absent. By contrast, when high trait values in social partners decrease trait values in the focal individual, representing negative Ψ, IGEs can slow evolution [55]. Here, we find that this parameter is negative with respect to same-genotype group members, but positive for partner-genotype group members, suggesting further complexities in predicting evolutionary change under IGEs.
Classical kin selection theory is founded on genotype-by-genotype epistasis, i.e. that individuals should express particular behaviours—such as cooperation, or inbreeding avoidance—when interacting with close relatives, but different behaviours during interactions with non-relatives [67]. Such kin-specific social plasticity is expected to evolve when a behaviour's fitness costs and benefits depend on the relatedness between the interactants. Could this evolutionary mechanism be responsible for the effects of genetic similarity on exploratory behaviour in flies? Females may choose not to explore a new resource with closely related individuals to avoid competition for mates or oviposition substrates. Exploration may also be risky and/or costly [68], so differentiation among same-genotype individuals in exploratory behaviour could represent a bet-hedging strategy [69]. The magnitude of the fitness consequences of exploratory behaviour in groups will also be important for interpreting the small effect sizes of social plasticity that we discovered here. The fitness costs and benefits of exploratory behaviour in flies, and whether these costs and benefits are different when individuals interact with similar and dissimilar partners, require more research.
Overall, these results reinforce the idea that the (genetic or other) identity of social partners matter for social plasticity, complicating our understanding of IGEs in groups. Further integration between kin selection, multilevel evolution and behavioural dynamics in groups is needed to illuminate the proximate and ultimate causes of variation in socially plastic behaviours.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Daniel Matute for providing stocks of D. simulans and two anonymous reviewers for helpful feedback on the manuscript.
Data accessibility
Data are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.m905qftxx [70].
Authors' contributions
A.J., M.P.B. and J.B.S. designed the experiment. A.J. and M.P.B. carried out the experiment. A.J. and J.B.S. analysed the data, and J.B.S. wrote the final manuscript. All authors gave final approval.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the National Science Foundation (grant no. IOS-1856577).
References
- 1.Dingemanse NJ, Wolf M. 2013. Between-individual differences in behavioural plasticity within populations: causes and consequences. Anim. Behav. 85, 1031–1039. ( 10.1016/j.anbehav.2012.12.032) [DOI] [Google Scholar]
- 2.Santostefano F, Wilson AJ, Niemelä PT, Dingemanse NJ. 2017. Indirect genetic effects: a key component of the genetic architecture of behaviour. Sci. Rep. 7, 10235 ( 10.1038/s41598-017-08258-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander RD. 1974. The evolution of social behavior. Annu. Rev. Ecol. Syst. 5, 325–383. ( 10.1146/annurev.es.05.110174.001545) [DOI] [Google Scholar]
- 4.Moore AJ, Brodie ED, Wolf JB. 1997. Interacting phenotypes and the evolutionary process: I. direct and indirect genetic effects of social interactions. Evolution 51, 1352–1362. ( 10.1111/j.1558-5646.1997.tb01458.x) [DOI] [PubMed] [Google Scholar]
- 5.Culumber ZW, Kraft B, Lemakos V, Hoffner E, Travis J, Hughes KA. 2018. GxG epistasis in growth and condition and the maintenance of genetic polymorphism in Gambusia holbrooki. Evolution 72, 1146–1154. ( 10.1111/evo.13474) [DOI] [PubMed] [Google Scholar]
- 6.Clark AG. 1999. Female × male interactions in Drosophila sperm competition. Science 283, 217–220. ( 10.1126/science.283.5399.217) [DOI] [PubMed] [Google Scholar]
- 7.Taborsky B, Oliveira RF. 2012. Social competence: an evolutionary approach. Trends Ecol. Evol. 27, 679–688. ( 10.1016/j.tree.2012.09.003) [DOI] [PubMed] [Google Scholar]
- 8.Rodríguez RL, Rebar D, Fowler-Finn KD. 2013. The evolution and evolutionary consequences of social plasticity in mate preferences. Anim. Behav. 85, 1041–1047. ( 10.1016/j.anbehav.2013.01.006) [DOI] [Google Scholar]
- 9.Sakata JT, Crews D. 2004. Developmental sculpting of social phenotype and plasticity. Neurosci. Biobehav. Rev. 28, 95–112. ( 10.1016/j.neubiorev.2004.01.001) [DOI] [PubMed] [Google Scholar]
- 10.Dingemanse NJ, Kazem AJN, Réale D, Wright J. 2010. Behavioural reaction norms: animal personality meets individual plasticity. Trends Ecol. Evol. 25, 81–89. ( 10.1016/j.tree.2009.07.013) [DOI] [PubMed] [Google Scholar]
- 11.Stamps JA. 2016. Individual differences in behavioural plasticities. Biol. Rev. 91, 534–567. ( 10.1111/brv.12186) [DOI] [PubMed] [Google Scholar]
- 12.Caesar S, Karlsson M, Forsman A. 2010. Diversity and relatedness enhance survival in colour polymorphic grasshoppers. PLoS ONE 5, e10880 ( 10.1371/journal.pone.0010880) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Page S, Sepil I, Flintham E, Pizzari T, Carazo P, Wigby S. 2017. Male relatedness and familiarity are required to modulate male-induced harm to females in Drosophila. Proc. R. Soc. B 284, 20170441 ( 10.1098/rspb.2017.0441) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ward AJW, Krause J. 2001. Body length assortative shoaling in the European minnow, Phoxinus phoxinus. Anim. Behav. 62, 617–621. ( 10.1006/anbe.2001.1785) [DOI] [Google Scholar]
- 15.Croft DP, James R, Ward AJW, Botham MS, Mawdsley D, Krause J. 2005. Assortative interactions and social networks in fish. Oecologia 143, 211–219. ( 10.1007/s00442-004-1796-8) [DOI] [PubMed] [Google Scholar]
- 16.Cote J, Fogarty S, Brodin T, Weinersmith K, Sih A. 2011. Personality-dependent dispersal in the invasive mosquitofish: group composition matters. Proc. R. Soc. B 278, 1670–1678. ( 10.1098/rspb.2010.1892) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y, Bolnick DI, Kirkpatrick M. 2013. Assortative mating in animals. Am. Nat. 181, E125–E138. ( 10.1086/670160) [DOI] [PubMed] [Google Scholar]
- 18.Landeau L, Terborgh J. 1986. Oddity and the ‘confusion effect’ in predation. Anim. Behav. 34, 1372–1380. ( 10.1016/S0003-3472(86)80208-1) [DOI] [Google Scholar]
- 19.Schuett W, Dall SRX, Royle NJ. 2011. Pairs of zebra finches with similar ‘personalities’ make better parents. Anim. Behav. 81, 609–618. ( 10.1016/j.anbehav.2010.12.006) [DOI] [Google Scholar]
- 20.Saltz JB, Alicuben ET, Grubman J, Harkenrider M, Megowan N, Nuzhdin SV. 2012. Nonadditive indirect effects of group genetic diversity on larval viability in Drosophila melanogaster imply key role of maternal decision-making. Mol. Ecol. 21, 2270–2281. ( 10.1111/j.1365-294X.2012.05518.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saltz JB. 2013. Genetic composition of social groups influences male aggressive behaviour and fitness in natural genotypes of Drosophila melanogaster. Proc. R. Soc. B 280, 20131926 ( 10.1098/rspb.2013.1926) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider J, Atallah J, Levine JD. 2017. Social structure and indirect genetic effects: genetics of social behaviour: social structure and indirect genetic effects. Biol. Rev. 92, 1027–1038. ( 10.1111/brv.12267) [DOI] [PubMed] [Google Scholar]
- 23.Bleakley BH, Brodie ED. 2009. Indirect genetic effects influence antipredator behavior in guppies: estimates of the coefficient of interaction psi and the inheritance of reciprocity. Evolution 63, 1796–1806. ( 10.1111/j.1558-5646.2009.00672.x) [DOI] [PubMed] [Google Scholar]
- 24.McGlothlin JW, Wolf JB, Brodie ED, Moore AJ. 2014. Quantitative genetic versions of Hamilton's rule with empirical applications. Phil. Trans. R. Soc. B 369, 20130358 ( 10.1098/rstb.2013.0358) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beckmann C, Biro PA. 2013. On the validity of a single (boldness) assay in personality research. Ethology 119, 937–947. [Google Scholar]
- 26.Bell AM, Sih A. 2007. Exposure to predation generates personality in threespined sticklebacks (Gasterosteus aculeatus). Ecol. Lett. 10, 828–834. ( 10.1111/j.1461-0248.2007.01081.x) [DOI] [PubMed] [Google Scholar]
- 27.Arvidsson LK, Matthysen E. 2016. Individual differences in foraging decisions: information-gathering strategies or flexibility? Behav. Ecol. 27, 1353–1361. ( 10.1093/beheco/arw054) [DOI] [Google Scholar]
- 28.David M, Pinxten R, Martens T, Eens M. 2015. Exploration behavior and parental effort in wild great tits: partners matter. Behav. Ecol. Sociobiol. 69, 1085–1095. ( 10.1007/s00265-015-1921-1) [DOI] [Google Scholar]
- 29.Snijders L, van Rooij EP, Burt JM, Hinde CA, van Oers K, Naguib M. 2014. Social networking in territorial great tits: slow explorers have the least central social network positions. Anim. Behav. 98, 95–102. ( 10.1016/j.anbehav.2014.09.029) [DOI] [Google Scholar]
- 30.Chock RY, Wey TW, Ebensperger LA, Hayes LD. 2017. Evidence for a behavioural syndrome and negative social assortment by exploratory personality in the communally nesting rodent, Octodon degus . Behaviour 154, 541–562. ( 10.1163/1568539X-00003433) [DOI] [Google Scholar]
- 31.Hui A, Pinter-Wollman N. 2014. Individual variation in exploratory behaviour improves speed and accuracy of collective nest selection by Argentine ants. Anim. Behav. 93, 261–266. ( 10.1016/j.anbehav.2014.05.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward AJW. 2012. Social facilitation of exploration in mosquitofish (Gambusia holbrooki). Behav. Ecol. Sociobiol. 66, 223–230. ( 10.1007/s00265-011-1270-7) [DOI] [Google Scholar]
- 33.Soliman MH. 1971. Selection of site of oviposition by Drosophila melanogaster and D. simulans. Am. Midl. Nat. 86, 487 ( 10.2307/2423639) [DOI] [Google Scholar]
- 34.Parsons P, Spence G. 1980. Larval responses to environmental ethanol in three Drosophila species: an indicator of habitat selection. Aust. J. Zool. 28, 543 ( 10.1071/ZO9800543) [DOI] [Google Scholar]
- 35.Via S. 1986. Genetic covariance between oviposition preference and larval performance in an insect herbivore. Evolution 40, 778 ( 10.1111/j.1558-5646.1986.tb00537.x) [DOI] [PubMed] [Google Scholar]
- 36.Gripenberg S, Mayhew PJ, Parnell M, Roslin T. 2010. A meta-analysis of preference–performance relationships in phytophagous insects. Ecol. Lett. 13, 383–393. ( 10.1111/j.1461-0248.2009.01433.x) [DOI] [PubMed] [Google Scholar]
- 37.Miller PM, Saltz JB, Cochrane VA, Marcinkowski CM, Mobin R, Turner TL. 2011. Natural variation in decision-making behavior in Drosophila melanogaster. PLoS ONE 6, e16436 ( 10.1371/journal.pone.0016436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matute DR, Gavin-Smyth J, Liu G. 2014. Variable post-zygotic isolation in Drosophila melanogaster/D. simulans hybrids. J. Evol. Biol. 27, 1691–1705. ( 10.1111/jeb.12422) [DOI] [PubMed] [Google Scholar]
- 39.Schrider DR, Ayroles J, Matute DR, Kern AD. 2018. Supervised machine learning reveals introgressed loci in the genomes of Drosophila simulans and D. sechellia. PLoS Genet. 14, e1007341 ( 10.1371/journal.pgen.1007341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamps J, Buechner M, Alexander K, Davis J, Zuniga N. 2005. Genotypic differences in space use and movement patterns in Drosophila melanogaster. Anim. Behav. 70, 609–618. ( 10.1016/j.anbehav.2004.11.018) [DOI] [Google Scholar]
- 41.Saltz JB. 2017. Genetic variation in social environment construction influences the development of aggressive behavior in Drosophila melanogaster. Heredity (Edinb) 118, 340–347. ( 10.1038/hdy.2016.101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stamps JA, Davis JM. 2006. Adaptive effects of natal experience on habitat selection by dispersers. Anim. Behav. 72, 1279–1289. ( 10.1016/j.anbehav.2006.03.010) [DOI] [Google Scholar]
- 43.Burns MP, Cavallaro FD, Saltz JB. 2020. Does divergence in habitat breadth associate with species differences in decision making in Drosophila sechellia and Drosophila simulans? Genes 11, 528 ( 10.3390/genes11050528) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang W, et al. 2014. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24, 1193–1208. ( 10.1101/gr.171546.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R core team. 2017. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 46.Therneau T. 2018. coxme: mixed effects Cox models See https://CRAN.R-project.org/package=coxme.
- 47.Pankratz VS, de Andrade M, Therneau TM. 2005. Random-effects Cox proportional hazards model: general variance components methods for time-to-event data. Genet. Epidemiol. 28, 97–109. ( 10.1002/gepi.20043) [DOI] [PubMed] [Google Scholar]
- 48.Zwiener I, Blettner M, Hommel G. 2011. Survival analysis. Deutsches Aerzteblatt Online. 108, 163–169. ( 10.3238/arztebl.2011.0163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Austin PC. 2017. A tutorial on multilevel survival analysis: methods, models and applications: multilevel survival analysis. Int. Stat. Rev. 85, 185–203. ( 10.1111/insr.12214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider MdP, Strandberg E, Ducrocq V, Roth A. 2005. Survival analysis applied to genetic evaluation for female fertility in dairy cattle. J. Dairy Sci. 88, 2253–2259. ( 10.3168/jds.S0022-0302(05)72901-5) [DOI] [PubMed] [Google Scholar]
- 51.Anderson CA, Duffy DL, Martin NG, Visscher PM. 2007. Estimation of variance components for age at menarche in twin families. Behav. Genet. 37, 668–677. ( 10.1007/s10519-007-9163-2) [DOI] [PubMed] [Google Scholar]
- 52.Bolker BM, Brooks ME, Clark CJ, Geange SW, Poulsen JR, Stevens MHH, White J-SS. 2009. Generalized linear mixed models: a practical guide for ecology and evolution. Trends Ecol. Evol. 24, 127–135. ( 10.1016/j.tree.2008.10.008) [DOI] [PubMed] [Google Scholar]
- 53.Fox J, Weisberg S. 2011. An {R} companion to applied regression, 2nd edn Thousand Oaks, CA: Sage. [Google Scholar]
- 54.McGlothlin JW, Brodie ED III. 2009. How to measure indirect genetic effects: the congruence of trait-based and variance-partitioning approaches. Evolution 63, 1785–1795. ( 10.1111/j.1558-5646.2009.00676.x) [DOI] [PubMed] [Google Scholar]
- 55.McGlothlin JW, Moore AJ, Wolf JB, Brodie ED III. 2010. Interacting phenotypes and the evolutionary process. III. Social evolution: indirect genetic effects and social selection. Evolution 64, 2558–2574. ( 10.1111/j.1558-5646.2010.01012.x) [DOI] [PubMed] [Google Scholar]
- 56.Bailey NW, Marie-Orleach L, Moore AJ. 2018. Indirect genetic effects in behavioral ecology: does behavior play a special role in evolution? Behav. Ecol. 29, 1–11. ( 10.1093/beheco/arx127) [DOI] [Google Scholar]
- 57.Signor SA, Abbasi M, Marjoram P, Nuzhdin SV. 2017. Social effects for locomotion vary between environments in Drosophila melanogaster females: social effects for locomotion. Evolution 71, 1765–1775. ( 10.1111/evo.13266) [DOI] [PubMed] [Google Scholar]
- 58.Ferveur J-F. 2005. Cuticular hydrocarbons: their evolution and roles in Drosophila pheromonal communication. Behav. Genet. 35, 279–295. ( 10.1007/s10519-005-3220-5) [DOI] [PubMed] [Google Scholar]
- 59.Foley B, Chenoweth SF, Nuzhdin SV, Blows MW. 2006. Natural genetic variation in cuticular hydrocarbon expression in male and female Drosophila melanogaster. Genetics 175, 1465–1477. ( 10.1534/genetics.106.065771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith AA, Hölldober B, Liebig J. 2009. Cuticular hydrocarbons reliably identify cheaters and allow enforcement of altruism in a social insect. Curr. Biol. 19, 78–81. ( 10.1016/j.cub.2008.11.059) [DOI] [PubMed] [Google Scholar]
- 61.Yurkovic A, Wang O, Basu AC, Kravitz EA. 2006. Learning and memory associated with aggression in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 103, 17 519–17 524. ( 10.1073/pnas.0608211103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saltz JB. 2011. Natural genetic variation in social environment choice: context-dependent gene-environment correlation in Drosophila melanogaster. Evolution 65, 2325–2334. ( 10.1111/j.1558-5646.2011.01295.x) [DOI] [PubMed] [Google Scholar]
- 63.Geiger AP, Saltz JB. 2020. Strong and weak cross-sex correlations govern the quantitative-genetic architecture of social group choice in Drosophila melanogaster. Evolution 74, 145–155. ( 10.1111/evo.13887) [DOI] [PubMed] [Google Scholar]
- 64.Turelli M, Barton NH. 2004. Polygenic variation maintained by balancing selection: pleiotropy, sex-dependent allelic effects and G×E interactions. Genetics 166, 1053–1079. ( 10.1534/genetics.166.2.1053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolf JB, Brodie ED III, Cheverud JM, Moore AJ, Wade MJ. 1998. Evolutionary consequences of indirect genetic effects. Trends Ecol. Evol. 13, 64–69. ( 10.1016/S0169-5347(97)01233-0) [DOI] [PubMed] [Google Scholar]
- 66.Bailey NW, Moore AJ. 2012. Runaway sexual selection without genetic correlations: social environments and flexible mate choice initiate and enhance the fisher process: sexual selection, flexible mate choice, and indirect genetic effects. Evolution 66, 2674–2684. ( 10.1111/j.1558-5646.2012.01647.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dawkins R. 1989. The selfish gene. Oxford, UK: Oxford University Press. [Google Scholar]
- 68.Bengston SE, Shin M, Dornhaus A. 2017. Life-history strategy and behavioral type: risk-tolerance reflects growth rate and energy allocation in ant colonies. Oikos 126, 556–564. ( 10.1111/oik.03527) [DOI] [Google Scholar]
- 69.Simons AM. 2011. Modes of response to environmental change and the elusive empirical evidence for bet hedging. Proc. R. Soc. B 278, 1601–1609. ( 10.1098/rspb.2011.0176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jaffe A, Burns MP, Saltz JB. 2020. Data from: Genotype-by-genotype epistasis for exploratory behaviour in D. simulans. Dryad Digital Repository. ( 10.5061/dryad.m905qftxx) [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Jaffe A, Burns MP, Saltz JB. 2020. Data from: Genotype-by-genotype epistasis for exploratory behaviour in D. simulans. Dryad Digital Repository. ( 10.5061/dryad.m905qftxx) [DOI]
Supplementary Materials
Data Availability Statement
Data are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.m905qftxx [70].