Abstract
We investigated the effects of exposure at ecologically relevant levels of dim light at night (dLAN) on sleep and the 24 h hypothalamic expression pattern of genes involved in the circadian timing (per2, bmal1, reverb-β, cry1, ror-α, clock) and sleep regulatory pathways (cytokines: tlr4, tnf-α, il-1β, nos; Ca2+-dependent pathway: camk2, sik3, nr3a; cholinergic receptor, achm3) in diurnal female zebra finches. Birds were exposed to 12 h light (150 lux) coupled with 12 h of absolute darkness or of 5 lux dim light for three weeks. dLAN fragmented the nocturnal sleep in reduced bouts, and caused sleep loss as evidenced by reduced plasma oxalate levels. Under dLAN, the 24 h rhythm of per2, but not bmal1 or reverb-β, showed a reduced amplitude and altered peak expression time; however, clock, ror-α and cry1 expressions showed an abolition of the 24 h rhythm. Decreased tlr4, il-1β and nos, and the lack of diurnal difference in achm3 messenger RNA levels suggested an attenuated inhibition of the arousal system (hence, awake state promotion) under dLAN. Similarly, changes in camk2, sik3 and nr3a expressions suggested dLAN-effects on Ca2+-dependent sleep-inducing pathways. These results demonstrate dLAN-induced negative effects on sleep and associated hypothalamic molecular pathways, and provide insights into health risks of illuminated night exposures to diurnal animals.
Keywords: bird, dim light at night, gene expression, hypothalamus, sleep, zebra finch
1. Introduction
The major environmental consequence of urbanization is the transformation of the temporal (day-night) environment, with largely unexplained health consequences [1]. The evidence, largely accumulated from nocturnal rodents, suggests that artificial light at night (LAN) impacts a wide range of biological functions including sleep [1–3]. Similar LAN-induced negative effects have been found on daily cycles of the activity-rest and melatonin secretion, and on metabolism, reproduction, depression and cognitive performance in several songbirds including Indian weaver birds, Ploceus phillipinus [4]; blackbirds, Turdus merula [5]; great tits, Parus major [6–8]; Indian house crows, Corvus splendens [9]; zebra finches, Taeniopygia guttata [10,11] and tree sparrows, Passer montanus [12]. In particular, female great tits were awake for a greater portion of the night inside the nest-box when it was dimly illuminated [6]. Similarly, free-living blue tits (Cyanistes caeruleus) adjusted their awakening time to the prevailing light condition [13], and Indian house crows showed increased sleep and decreased awakening latency under a LAN environment [9].
In diurnal vertebrates, a crucial adaptation is the daily sleep-wake cycle, with sleep as the night (hence, awake as day) component of the 24 h temporal environment. The daily sleep-wake pattern is governed by the internal circadian clock, which operates in a closed transcriptional-translational feedback loop (TTFL) formed by a set of core clock genes [14]. TTFL is functionally arranged in positive (brain muscle arnt like (bmal1); circadian locomotor output cycles kaput (clock)) and negative (period (per); cryptochrome (cry)) limbs [14]. Furthermore, the orphan nuclear receptor genes (retinoid-related orphan receptors (rors) and reverse transcript of erythroblastosis (rev-erbs)) stabilize TTFL by acting as activator and repressor, respectively, of bmal1 transcription [14].
The disruption of the daily sleep-wake pattern results in sleep deprivation (=sleep debt), which can be assessed by the measurement of the circulating oxalate (oxalic acid) levels [15]. However, there are conflicting reports on the reliability of plasma oxalate levels as a biomarker of sleep debt in birds. Under a LAN environment, plasma oxalate levels were significantly dropped in sleep-deprived adult great tits [8], like in rats and humans [15]; however, the oxalate levels were found elevated in developing great tits [16]. Under a LAN environment, even in the absence of an immune challenge, changes in the brain cytokines parallel the sleep disruption, suggesting their involvement in the regulation of sleep [1]. A recent study found tissue-specific 24 h rhythms under 12 L : 12 D (LD) and its disruption under dLAN in genes coding for the pro- (IL-1β, IL-6) and anti-inflammatory (IL-10) cytokines in the hypothalamus and liver of zebra finches [17]. In particular, however, there is evidence for interleukin-1β (IL-1β) and tumour necrosis factor-α (TNF-α), which are synthesized and released via activated toll-like receptor 4 (TLR4), playing roles as the promoter of sleep and regulator of sleep duration [18]. The involvement (at least partially) of TLR4 in sleep loss is also suggested by the lack of cerebral reaction in TLR4-deficient mice [18]. Reciprocally induced IL-1β and TNF-α activate the nuclear factor-κB (NF-ΚB), which via the nitric oxide pathway (shown by nitric oxide synthase (NOS) activity) promotes sleep by acting directly on the hypothalamic preoptic neurons [19,20]. This involves the CamK2 hyperpolarization pathway: camk2 knockout (KO) shortened, and sik3 knockin (encoded SIK3 protein kinase gene acting downstream to CamK2) enhanced sleep duration in mice [21,22]. Similarly, KO mice for NR3A, a Ca2+-dependent N-methyl-D-aspartate receptor subunit, showed a significantly shortened sleep and disturbed sleep homeostasis [21,23,24]. Likewise, ACHM3 (a cholinergic muscarinic receptor subunit and expressed in the ventrolateral preoptic area) was shown to contribute significantly to sleep-wake homeostasis in pigeons, mainly by the activation of hypothalamic mechanisms that maintain the awake state [25].
The mechanism(s) underlying LAN-induced effects on sleep-wake homeostasis is (are) poorly understood in diurnal vertebrates. To this end, songbirds can be a good experimental system because they share many sleep traits with mammals [26]. Like in mammals, sleep allows birds to recover from daily stress, and to consolidate memory, conserve energy and maintain the body temperature and homeostasis [26–28]. Here, therefore, we investigated LAN-effects on sleep and associated molecular correlates in diurnal zebra finches, which show profound LAN-induced negative behavioural and physiological effects [10,11]. We first performed behavioural assays to monitor the sleep and activity-rest pattern, and assayed plasma oxalate levels as a biomarker of sleep effects in adult female zebra finches that were exposed daily to 12 h light (150 lux) coupled with 12 h of absolute darkness or of dim light (5 lux). Then, we measured the hypothalamic expression of genes involved in the circadian timing (per2, bmal1, reverb-β, cry1, ror-α and clock), and in the promotion of sleep (cytokines: tlr4, tnf-α, il-1β and nos; Ca2+ dependent hyperpolarization pathway: camk2, sik3 and nr3a) and maintenance of the awake state (muscarinic cholinergic receptor, achm3) in diurnal zebra finches.
2. Material and methods
(a). Animal maintenance and experiment
This study was approved by the Institutional Animal Ethics Committee (IAEC) of the Department of Zoology, University of Delhi, India (DU/ZOOL/IAEC-R/2018/03). The experiment was carried out on adult female zebra finches (T. guttata) that were born and raised in our indoor facility under 12 h daily photoperiod (12 L : 12 D; L = ∼150 lux; D = 0 lux; temperature = 24 ± 2°C). For this study, we chose females to avoid any potential sex-dependent response to an altered light environment. We also expected a larger response in female birds, based on a previous songbird study in which, when compared with males, female great tits spent a greater proportion of the night awake in response to the artificial LAN environment [6]. The experimental protocol has been described in detail in a previous publication [11]. Briefly, 72 females (age: 8–10 months) were individually housed (cage size = 42 × 30 × 54 cm) kept in separate light-proof wooden boxes (size = 58 × 52 × 68 cm); hence, birds were isolated such that they could not see and hear their neighbours. After a week of acclimation to 12 L : 12 D, as before, birds were randomly separated in two equal groups. For the next three weeks, half of the birds remained on 12 L : 12 D (LD group: L = ∼150 lux; D = 0 lux); however, for the other half of birds, the absolute darkness of 12 h night was replaced by 5 lux dim light (dLAN group: L = 150 ± 5 lux; D = 5 lux ± 1 lux). We chose 5 lux for dLAN based on the average night-time illumination intensity of three different locations in a 6 km2 area around Delhi University that we had measured, and this was consistent with previous dLAN studies in songbirds [7,9]. The white light illumination was provided by Philips (India, 220 V-240 V) compact fluorescent bulbs emitting a radiance of 220 lumens. Each cage was fitted with two such bulbs, one of which was used for providing dLAN at 5 lux light intensity to birds under dLAN. Reduction in the light intensity at night was achieved by covering the bulb with a black paper sheet having multiple slits. We checked and verified intermittently the light intensity both during the day and night by using the Macam Q203 radiometer. To all birds, food (Setaria italica seeds) and water were available ad libitum.
(b). Behavioural assay of 24 h activity and sleep patterns
Two perches placed in each cage at unequal heights facilitated the perching activity of the bird within the cage environment. The cage was mounted with a passive infrared motion sensor (DSC, LC100 PI Digital PIR motion detector, Canada) that continuously monitored general activity (movement) of the bird in its cage. The movements collected in 5 min bins for each individual were stored separately in a computerized recording system. The collection, collation, graphics and analysis of activity were done by using the ‘The Chronobiology Kit' software program from Stanford Software Systems, Stanford (USA), as described in several previous publications from our laboratory [9]. Double plotted activity records were graphed and presented as an actogram for each bird, wherein successive days were plotted sideways and underneath to show a better visual illustration of the timing and pattern of 24 h activity-rest behaviour. For statistical analysis of the effect of the light condition of activity behaviour, we extracted activity in 20 min bins for seven consecutive days for each bird. We first averaged activity for 20 min bins over 24 h for each individual, and then calculated and presented the mean (±s.e.) 24 h activity profile.
We monitored the birds' postures in the cage by using a night vision camera and recorded it as sleep or awake state (figure 1), as per a behavioural assay standardized and used in other bird studies [6,9,13,29]. Videographs were analysed for sleep and awake state by using Observer XT 10 software from Noldus Information Technology (Wageningen, The Netherlands), and sleep states were identified by the resting postures, as described in other birds [6,9,13]. We scored sleep mainly in two behavioural postures when bird had its eyes closed (=front sleep) and when it had tucked its beak on the scapula (=back sleep) [29]. We further described and scored nocturnal sleep in four states (parameters): sleep onset (first sleep bout lasting ≥30 s) and offset (last sleep bout lasting ≥10 s), sleep latency (interval from light off to sleep onset), awakening latency (interval from sleep offset to light on), and sleep frequency (sleep bouts per night). The scores obtained from videographs over 2 days were averaged, and from this mean (±s.e.), bout and duration of sleep both for each hour and total 12 h ‘night' was calculated.
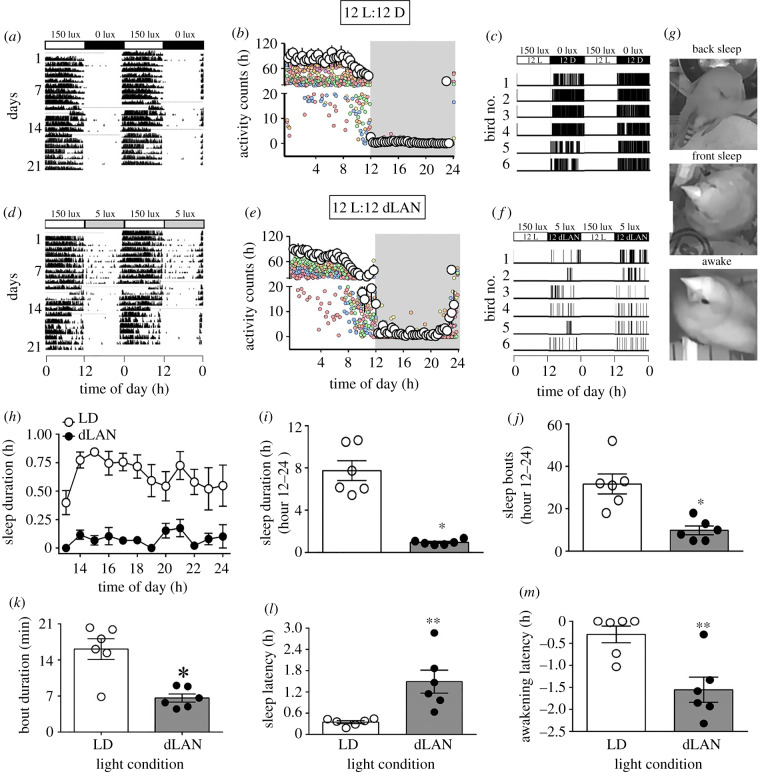
Figure 1.
Effects on activity and sleep behaviours. Adult female zebra finches were exposed to 12 h light (150 ± 5 lux) coupled with 12 h of absolute darkness (0 lux) or of dim light (dLAN, 5 ± 1 lux). Representative double plotted actogram show a 24 h activity-rest pattern under LD (a) and dLAN (d). Black and white portions denote the active and rest states, respectively. Twenty four hour activity-rest profile in 20 min bins (mean ± s.e.) is also shown for both LD (b) and dLAN (e). Representative sleepogram (24 h distribution of sleep-wake pattern over 2 days) under LD (c) and dLAN (f), with sleep (black) and awake (white) states. (g) illustrates representative images of behavioural postures showing back sleep, front sleep and awake states. Mean (± s.e.) hourly profile of the sleep duration (h), total sleep duration (i), total sleep bouts (j), sleep bout length (k), and sleep and awakening latency relative to lights on and lights off, respectively (l, m) during 12 h of the night period. The asterisk (*) indicates a significant difference, as determined by GLMM (h) or by Student's t-test (i–m). p < 0.05 was considered a statistically significant difference. (*p < 0.05; **p < 0.01).
The activity-rest pattern was recorded throughout the experiment, but for the behavioural assay of sleep birds were videographed over 2 days ending a day before the last day of the experiment. Here, we present data on both activity and sleep behaviours for the same six birds from each condition. Although we monitored behavioural responses of only 6 of 36 birds in this study, we believe that by using an identical experimental procedure, the data are representative of the overall behavioural response pattern of birds in each light condition.
(c). Assay of plasma oxalate levels
As a biomarker of the sleep loss, we measured circulating oxalic acid levels in nine blood samples collected by puncturing the wing-vein in a heparinized capillary tube in the middle of the light period. The handling and blood collection were completed in less than 1 min to avoid any stress-induced effect. Blood was centrifuged for 10 min at 3381 g at 4°C, and plasma was separated and stored at −20°C. Plasma oxalate levels were measured by using a commercially available colorimetric assay kit (Cat. no. K663–100; Biovision Inc., Milpitas, CA, USA), as per the manufacturer's protocol and as detailed in the electronic supplementary material, methods section. These nine blood samples included six birds for which we recorded data on activity and sleep behaviours.
(d). Measurement of messenger RNA expression of candidate genes
At the end of the experiment, birds were sacrificed by decapitation at 4 h intervals beginning just before hour 0 (=light on; n = 6 per time point; hour 0 sampling was completed before light on). Because each bird was separately housed in a light-proof box, it could be taken out for sacrifice without disturbing its neighbour that was still inside its own box; this could be easily verified by activity recordings. Brain from the head kept on ice post-decapitation was carefully removed, snap frozen in dry ice and stored at −80°C until processed for messenger RNA (mRNA) assays. The hypothalamus was excised out as described in Sharma et al. [30], and total RNA was extracted using Tri reagent (AM9738; Ambion), as per the manufacturer's protocol. One microgram of total RNA was treated with RQ1 RNase-free DNase (M6101, Promega, Madison, WI, USA) and reverse transcribed using a Revert Aid first strand cDNA synthesis Kit (Thermoscientific, K1622). At all six times of the day, we measured mRNA expression of six genes of the negative and positive limbs of circadian TTFL (per2, bmal1, reverb-β, ror-α, cry1 and clock), and of eight genes involved in the promotion of sleep (cytokines: tlr4, tnf-α, il-1β and nos; Ca2+-dependent hyperpolarization pathway: camk2, sik3 and nr3a) and the maintenance of awake state (muscarinic cholinergic receptor, achm3) Using gene-specific primers (electronic supplementary material, table S1) and SYBR green chemistry, both target and reference (β-actin) genes were run in duplicates on the Applied Biosystems Step One plus system, and relative mRNA expression levels were calculated by the 2−ΔΔCt method [31], as validated and reported in several publications from our laboratory [11,30]. We used β-actin as a reference (control) gene, which was found to be most stable between three genes that were tested as controls earlier by our laboratory [30]. Further details are given in the electronic supplementary material, methods section.
(e). Statistics
All statistical analyses were performed using GraphPad Prism v. 6.0 and IBM SPSS statistics software v. 20, as appropriate. Student's t-test compared data at one time point between two light conditions. We constructed general linear mixed effect models (GLMM) for behavioural responses. Light condition and time of day were included as fixed effects, and the identity of study subjects was included as a random effect. We also fitted general linear models (GLM) to test the effects of the light condition, time of day, and their interactions on gene expressions. If there was a significant interaction effect, we ran Bonferroni post-test for multiple comparisons. Furthermore, the persistence of a 24 h rhythm in gene expressions was tested by unimodal cosinor regression {y = A+ [B*cos (2*pi (X-C)/24)]}; A, B and C are the mesor (mean value for 24 h expression), the amplitude (maximum change in mRNA expression levels relative to the mesor) and the acrophase (the estimated time of peak mRNA expression) of 24 h (daily) rhythm, respectively. The significance of cosinor regression analysis was calculated using the number of samples, r2 values and predictors—the mesor, amplitude and acrophase (https://www.danielsoper.com/statcalc/calculator.aspx?id=15 [32]). If 24 h variation in mRNA expressions showed a significant daily rhythm, then we used an extra sum of squares F-test to determine the significant difference in the rhythm waveform parameters. For statistical significance, α was set at 0.05.
3. Results
(a). Effects on activity, sleep and plasma oxalate levels
All birds showed a diurnal pattern in the activity-rest behaviour, with activity consolidated during light phase of 12 L : 12 D. However, 24 h distribution of activity showed an overall significant effect of the light condition (F1,720 = 20.102, p < 0.0001) and time of day (F71,720 = 28.446, p < 0.0001), but not of the light condition × time of day interaction (F71,720 = 1.143, p = 0.205; GLMM test). Similarly, sleep was restricted to largely at night (black regions figure 1a); sleep bouts were absent during the daytime (white blank regions, figure 1a) in both LD (dark night) and dLAN (dimly illuminated night) conditions (figure 1). However, we found dLAN-induced alteration in the sleep behaviour. To begin with, the duration of nocturnal sleep bouts showed a significant effect of the light condition (F1,120 = 221.36; p < 0.0001) but not of the time of day (F11,120 = 1.1482; p = 0.147) or the light condition × time of day interaction (F11, 120 = 0.888; p = 0.554) (GLMM test; figure 1d). Particularly, the length (p = 0.0014) and frequency (p = 0.0017) of sleep bouts as well as the total duration of nocturnal sleep (p < 0.0001) were decreased in dLAN birds, compared to LD controls (Student's unpaired t-test; figure 1e–g). Under dLAN, the delayed sleep onsets led to a significant increase in the sleep latency (p = 0.0058), and a significant decrease in the awakening latency (p = 0.0043; Student's unpaired t-test; figure 1h–i). Plasma oxalate concentration faithfully reflected differences in the nocturnal sleep; the levels were significantly decreased indicating sleep debt in dLAN birds, compared to LD controls (p = 0.0286; Student's unpaired t-test; figure 2).
Figure 2.
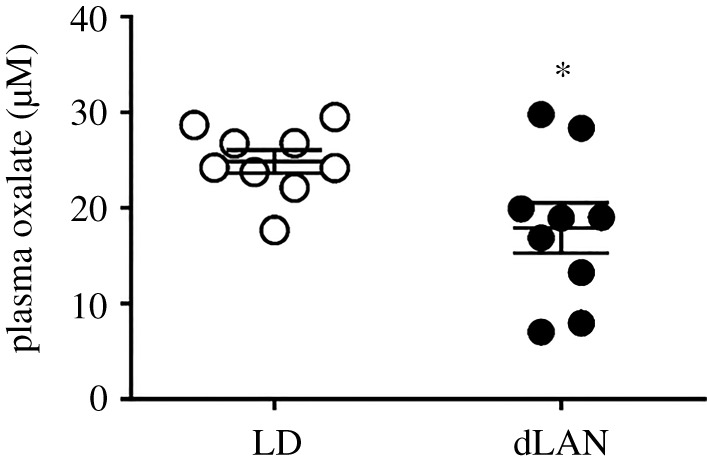
Effect of plasma oxalate levels. Mean (±s.e.) plasma oxalate levels in the middle of the day in zebra finches exposed to 12 h light (150 ± 5 lux) coupled with 12 h of absolute darkness (0 lux) or of dim light (dLAN, 5 ± 1 lux). The asterisk (*) indicates a significant difference between the LD and dLAN. p < 0.05 was considered a statistically significant difference.
(b). Effect on mRNA expressions of candidate genes
(i). Circadian clock genes
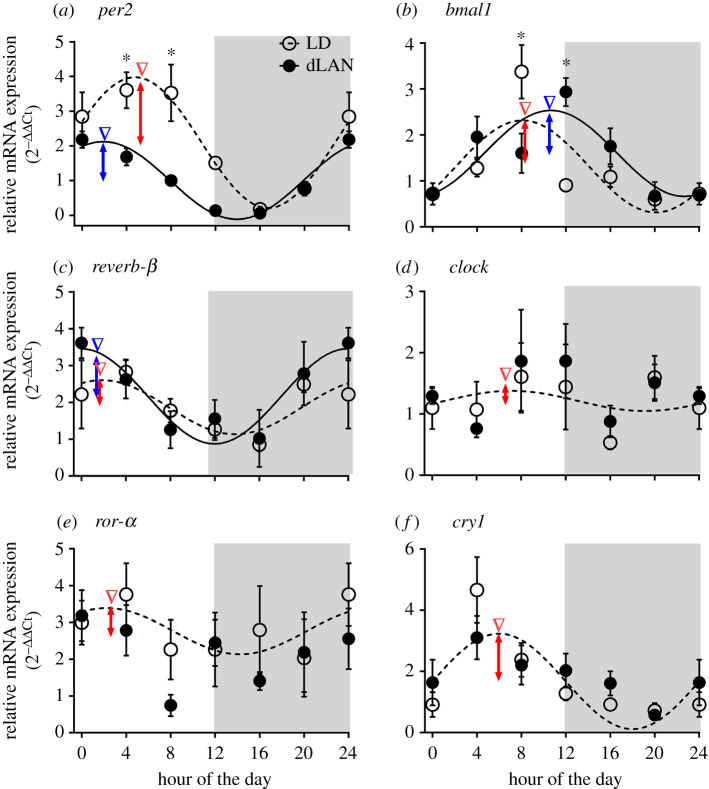
Figure 3 shows the results. There were significant changes in the level and 24 h rhythm of circadian clock gene expressions (electronic supplementary material, tables S2 and S3). per2 mRNA expression showed a significant effect of the light condition (χ2 = 33.438, p < 0.0001), the time of day (χ2 = 106.920, p < 0.0001) and of the light condition × time of day interaction (χ2 = 23.58, p < 0.0001; GLM test, electronic supplementary material, table S3). In particular, per2 mRNA levels were significantly reduced at hour 4 and hour 8 under dLAN, compared to LD (p < 0.05; Bonferroni's post hoc test). However, reverb-β, cry1 and clock expressions showed a significant effect of the time of day (reverb-β: χ2 = 30.474, p < 0.0001; cry1: χ2 = 56.473, p < 0.0001; clock: χ2 = 14.696, p = 0.012), but not of the light condition or the light condition × time of day interaction (GLM test; electronic supplementary material, table S3). Similarly, bmal1 expression showed a significant effect of the time of day (χ2 = 62.438, p < 0.0001) and light condition × time of day interaction (χ2 = 47.568, p < 0.0001), but not of the light condition (GLM test; electronic supplementary material, table S3). Particularly, as compared to LD, bmal1 mRNA levels were significantly decreased and increased at hour 8 and hour 12, respectively, under dLAN (p < 0.05; Bonferroni's post hoc test). Intriguingly, we found neither the effect of the light condition, nor of the time of day or light condition × time of day interaction on ror-α expression. Most interestingly, cosinor analysis revealed a significant 24 h rhythm in per2, bmal1 and reverb-β expressions in both LD and dLAN, and in cry1, clock and ror-α expressions in LD, but not in dLAN (Fischer's test, electronic supplementary material, table S2). However, we found significant changes in the 24 h rhythm waveform of per2, but not of bmal1 and reverb-β genes. There was a significantly decreased mesor and amplitude, and altered acrophase of per2 mRNA rhythm in birds under dLAN, compared to LD controls (Fischer's test, electronic supplementary material, table S2 and figure 3).
Figure 3.
Effect on mRNA expression of clock genes. Mean (±s.e.) 24 h mRNA expression of per2 (a), bmal1 (b), reverb-β (c), clock (d), ror-α (e) and cry1 (f) genes measured at 4 h intervals beginning before light on (hour 0 = light on) in the hypothalamus of zebra finches exposed to 12 h light (150 ± 5 lux) coupled with 12 h of absolute darkness (0 lux) or of dim light (dLAN, 5 ± 1 lux). Broken and solid line curves through six time points indicate a significant daily rhythm under LD and dLAN, respectively, as determined by the cosinor analysis. Grey-shaded areas in graphs mark the 12 h night period. The asterisk indicates a significant difference as determined by Bonferroni's post-test after a significant interaction effect indicated by the GLM test (electronic supplementary material, table S3). p < 0.05 was considered statistically significant. Whereas inverted arrow heads indicate the time of peak mRNA expression during the day, the double arrow-headed vertical lines denote the amplitude of daily mRNA oscillations (LD: red; dLAN: blue), as determined by the rhythmometric test (electronic supplementary material, table S2). (Online version in colour.)
(ii). Cytokine-induced and Ca2+-dependent gene pathways
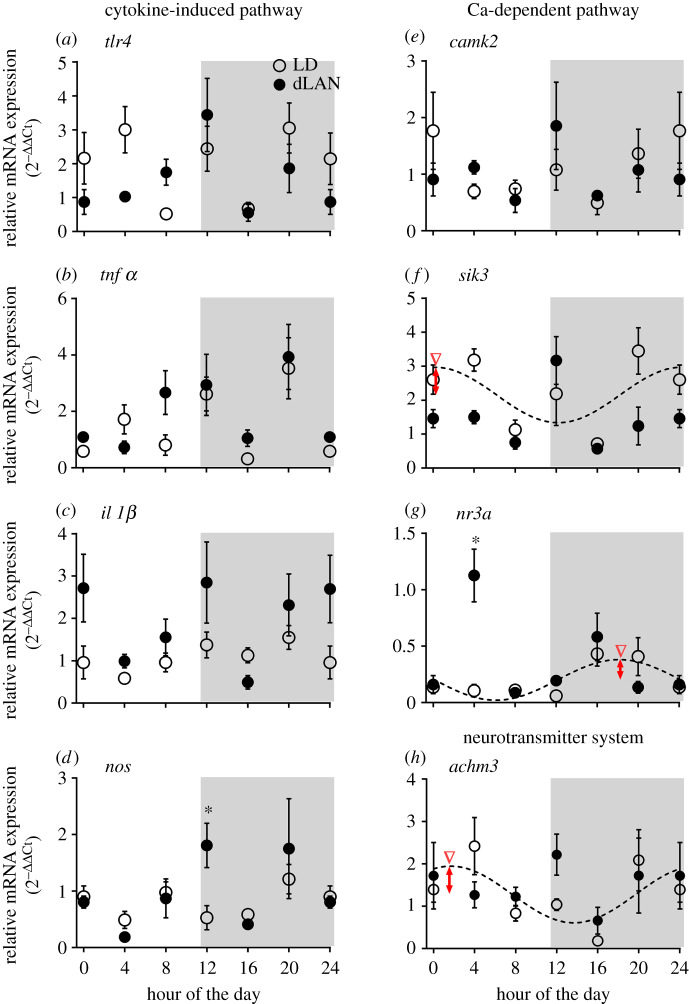
Of eight genes that we measured, the cosinor analysis revealed a significant rhythm under LD alone in the mRNA expression of sik3, nr3a and achm3 genes (figure 4; electronic supplementary material, table S3). However, there were changes over 24 h and/or between light conditions in several genes (figure 4). For example, mRNA expression of il-1β showed a significant effect of the light condition (χ2 = 9.069, p = 0.003) and time of day (χ2 = 19.58, p = 0.001; GLM, electronic supplementary material, table S3), but not of the light condition × time of day interaction (figure 4c). Similarly, there was a significant effect of the time of day (tnf-α: χ2 = 50.04, p < 0.0001; camk2: χ2 = 12.33, p = 0.031; achm3: χ2 = 15.79, p = 0.007), but not of the light condition or the light condition × time of day interaction, on tnf-α, camk2 and achm3 mRNA expression (GLM, electronic supplementary material, table S3; figure 4b,e,h). Likewise, tlr4 and nos showed a significant effect of the time of day (tlr4: χ2 = 28.79, p < 0.0001; nos: χ2 = 26.14, p < 0.0001) and light condition × time of day interaction (tlr4: χ2 = 16.79, p = 0.005; nos: χ2 = 13.39, p = 0.02; GLM test, electronic supplementary material, table S3; figure 4a,d), but not of the light condition. sik3 and nr3a expressions also showed a significant effect of the light condition (sik3: χ2 = 9.89, p = 0.002; nr3a: χ2 = 7.39, p = 0.007), time of day (sik3: χ2 = 39.92, p < 0.0001; nr3a: χ2 = 43.39, p < 0.0001) and light condition × time of day interaction (sik3: χ2 = 18.53, p = 0.002; nr3a: χ2 = 49.13, p < 0.0001; GLM, electronic supplementary material, table S3 and figure 4f,g). Particularly, at hour 20, sik3 mRNA levels were significantly reduced in dLAN, compared to LD (p < 0.05; Bonferroni's post hoc test; figure 4f), whereas at hour 12 and hour 4, respectively, nos and nr3a levels were significantly higher under dLAN, when compared with the levels under LD (p < 0.05; Bonferroni's post hoc test; figure 4d,g).
Figure 4.
Effect on mRNA expression of genes involved in cytokine-induced and Ca2+-dependent gene pathways. Mean (±s.e.) 24 h hypothalamic mRNA expression of tlr4 (a), tnfα (b), il-1β (c), nos (d), camk2 (e), sik3 (f), nr3a (g) and achm3 (h) genes measured at 4 h intervals beginning before light on (hour 0 = light on) in zebra finches exposed to 12 h light (150 ± 5 lux) coupled with 12 h of absolute darkness (0 lux) or of dim light (dLAN, 5 ± 1 lux). The asterisk indicates a significant difference as determined by Bonferroni's post-test after a significant interaction effect indicated by the GLM test (electronic supplementary material, table S3). p < 0.05 was considered a statistically significant difference. Whereas the inverted arrow heads indicate the time of peak mRNA expression during the day, the double arrow-headed vertical lines denote the amplitude of daily mRNA oscillations (LD: red; dLAN: blue), as determined by the rhythmometric test (electronic supplementary material, table S2). (Online version in colour.)
4. Discussion
We demonstrate that dLAN at ecologically relevant light intensity levels affected activity and sleep behaviours in zebra finches, similar to those reported in great tits and Indian house crows [6,9]. There was a disruption in the distribution of activity, and significantly delayed sleep onsets and advanced wake ups (hence, a reduction in the duration of nocturnal sleep) in zebra finches under dLAN. In particular, reduced frequency and decreased length of sleep bouts suggested dLAN effects on the consolidation of sleep in zebra finches. Clearly, considering the enhanced sleep latency, zebra finches took a longer time to fall asleep, and spent a greater proportion of the night awake under dLAN, compared to LD. This is consistent with the prolonged daytime activity, and delayed onset of nocturnal sleep in European blackbirds in response to street lights at night [33]. Reduced plasma oxalate levels further evidenced disturbed nocturnal sleep (=sleep debt) in zebra finches in response to dLAN, as has been reported in great tits, rats and humans [8,15]. Because we did not measure plama oxalate levels before the experiment began, we cannot comment whether the large individual variation in levels at the end of the experiment reflected base line differences in oxalate levels or individual responsiveness to dLAN. Nonetheless, the overall significant difference in circulating oxalate levels between the light conditions does suggest the impact of dLAN on sleep. Notably, plasma oxalate levels were decreased in adults but increased in developing great tits in response to the artificial LAN environment [6,8]. It needs to be investigated if effects of dLAN would vary with development stage of the bird. Furthermore, dLAN-induced effect in sleep disruption could be associated with an attenuated nocturnal melatonin peak, because midnight melatonin levels were reduced to almost daytime levels in female zebra finches [11]. Concomitant dLAN-induced negative effects on the sleep-wake pattern and loss of nocturnal melatonin peak secretion have been found in great tits, pigeons and Indian house crows [6,7,9].
We further suggest that dLAN-induced negative sleep effects in female zebra finches involved the endogenous circadian clock, because hypothalamic clock gene oscillations were negatively affected under dLAN. In particular, 24 h changes in cry1, clock and ror-α mRNA levels were arrhythmic, and per2 24 h mRNA rhythm showed an earlier peak and a reduced amplitude under dLAN. This is consistent with an earlier peak expression time of bmal1, per2 and clock, and a delayed peak expression time of cry1 found in tree sparrows (P. montanus) from the urban (artificial LAN), when compared with the rural (no LAN) environment [34]. The abolition of 24 h mRNA rhythm in per2 expression was found correlated with disrupted locomotor activity rhythms and caused sleep loss in mice [35,36]. dLAN also attenuated rhythmic expression of per2 in the hypothalamus, and of bmal1, per2 and cry1 in the liver of nocturnal mice [37]. However, it cannot be ascertained from the present study if dLAN-induced circadian rhythm impairment was causal to sleep disruption or it was the consequence of dLAN-induced sleep disruption in zebra finches. This might represent a self-reinforcing physiological process in which a strongly self-sustained circadian rhythm improves the sleep quality, and sleep homeostasis influences the amplitude (and perhaps phase) of the endogenous circadian rhythm [38].
In general, changes in hypothalamic mRNA levels of genes are consistent with the involvement of multiple hypothalamic neuronal pathways for the promotion of sleep and the maintenance of awake (arousal) state [39]. For instance, the overall effects on tlr4, il-1b and nos mRNA levels are consistent with reduced inhibition of the arousal systems. This, in turn, justifies the awake state promotion under dLAN. Reduced nocturnal sleep and altered tlr4 mRNA levels in zebra finches under dLAN were, in particular, consistent with reduced sleep and enhanced wakefulness in tlr4-deficient mice [18]. With parallel dLAN-induced decrease in il-1β and nos mRNA expressions, we propose a close linkage of the pro-inflammatory cytokines with dLAN-induced sleep fragmentation in zebra finches. Indeed, tlr4 activation induces the synthesis and release of IL-1β and TNF-α, which promote sleep by acting directly on the hypothalamic preoptic neurons, and via nos, cause nitric oxide production [19,20]. The diurnal difference in camk2 expression further suggests the involvement of Ca2+-dependent pathways in dLAN-induced sleep disruption in zebra finches. Changes in camk2, sik3 and nr3a expressions could also suggest dLAN-induced effects on Ca2+-influx and consequently influences sleep in zebra finches, consistent with increased sleep duration in sik3 mutant mice [22]. Concurrently, the lack of a diurnal difference in achm3 mRNA levels under dLAN (opposed to LD) could suggest that the maintenance of awake state was associated with activation of the hypothalamic preoptic M-cholinoreceptors in zebra finches, as reported in pigeons [25]. We caution though that these genes are involved in multiple functions and pathways including sleep, glutamate signalling, calcium influx, thermogenesis and feeding in vertebrates [18,21–24].
5. Conclusion
dLAN affected the nocturnal sleep and associated hypothalamic molecular pathways in diurnal female zebra finches. Based on changes in mRNA expression of genes comprising the circadian clock circuitry, cytokine and Ca2+-dependent pathways, we suggest an integrated hypothalamic control of sleep-wake state in zebra finches. Thus, by using a diurnal species and ecologically relevant levels of LAN, we closely replicated the prevailing urban night environment, and demonstrated that illuminated nights could negatively impact the molecular underpinnings of hypothalamic sleep-associated pathways in diurnal animals.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Ethics
All procedures were approved and carried out in accordance to guidelines of the Institutional Animal Ethics Committee (IAEC) of the Department of Zoology, University of Delhi, India (Institutional Ethical Approval number: DU/ZOOL/IAEC-R/2018/03).
Data accessibility
The mRNA sequences with their partial CDS can be accessed using GenBank accession numbers as provided in the electronic supplementary material, table S1.
Authors' contributions
V.K. conceived the idea. V.K. and T.B. designed the study. T.B., I.M. and A.P. performed experiments and carried out sampling. T.B. and I.M. performed assays. T.B., I.M. analysed data, and T.B. and S.K.B. prepared final figures. V.K. and T.B. wrote the manuscript. V.K. and S.K.B. provided all chemical and other laboratory resources. All authors gave final approval for publication.
Competing interests
Authors have no competing interests.
Funding
Financial support from a research grant no. (EMR/2015/002158) by the Science and Engineering Research Board, New Delhi, to V.K. is gratefully acknowledged. T.B., I.M. and A.P. received research fellowships from University Grants Commission of India.
References
- 1.Navara KJ, Nelson RJ. 2007. The dark side of light at night: physiological, epidemiological, and ecological consequences. J. Pineal Res. 43, 215–224. ( 10.1111/j.1600-079X.2007.00473.x) [DOI] [PubMed] [Google Scholar]
- 2.Fonken LK, Kitsmiller E, Smale L, Nelson RJ. 2012. Dim night time light impairs cognition and provokes depressive-like responses in a diurnal rodent. J. Biol. Rhythms 27, 319–327. ( 10.1177/0748730412448324) [DOI] [PubMed] [Google Scholar]
- 3.Stenvers DJ, et al. 2016. Dim light at night disturbs the daily sleep-wake cycle in the rat. Sci. Rep. 6, 35662 ( 10.1038/srep35662) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh J, Rani S, Kumar V. 2012. Functional similarity in relation to the external environment between circadian behavioural and melatonin rhythms in the subtropical Indian weaver bird. Horm. Behav. 61, 527–534. ( 10.1016/j.yhbeh.2012.01.015) [DOI] [PubMed] [Google Scholar]
- 5.Dominoni DM, Goymann W, Helm B, Partecke J. 2013. Urban-like night illumination reduces melatonin release in European blackbirds (Turdus merula): implications of city life for biological time-keeping of songbirds. Front. Zool. 10, 60 ( 10.1186/1742-9994-10-60) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raap T, Pinxten R, Eens M. 2015. Light pollution disrupts sleep in free-living animals. Sci. Rep. 5, 13557 ( 10.1038/srep13557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Jong M, Ouyang JQ, van Grunsven RH, Visser ME, Spoelstra K. 2016. Do wild great tits avoid exposure to light at night? PLoS ONE 11, e0157357 ( 10.1371/journal.pone.0157357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ouyang JQ, de Jong M, van Grunsven RH, Matson KD, Haussmann MF, Meerlo P, Visser ME, Spoelstra K. 2017. Restless roosts: light pollution affects behaviour, sleep, and physiology in a free-living songbird. Global Change Biol. 23, 4987–4994. ( 10.1111/gcb.13756) [DOI] [PubMed] [Google Scholar]
- 9.Taufique ST, Prabhat A, Kumar V. 2018. Illuminated night alters hippocampal gene expressions and induces depressive-like responses in diurnal corvids. Eur. J. Neurosci. 48, 3005–3018. ( 10.1111/ejn.14157) [DOI] [PubMed] [Google Scholar]
- 10.Jha NA, Kumar V. 2017. Effect of no-night light environment on behaviour, learning performance and personality in zebra finches. Anim. Behav. 132, 29–47. ( 10.1016/j.anbehav.2017.07.017) [DOI] [Google Scholar]
- 11.Batra T, Malik I, Kumar V. 2019. Illuminated night alters behaviour and negatively affects physiology and metabolism in diurnal zebra finches. Environ. Pollut. 254, 112916 ( 10.1016/j.envpol.2019.07.084) [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Yang W, Liang W, Wang Y, Zhang S. 2019. Intensity dependent disruptive effects of light at night on activation of the HPG axis of tree sparrows (Passer montanus). Environ. Pollut. 249, 904–909. ( 10.1016/j.envpol.2019.03.008) [DOI] [PubMed] [Google Scholar]
- 13.Steinmeyer C, Schielzeth H, Mueller JC, Kempenaers B. 2010. Variation in sleep behaviour in free-living blue tits, Cyanistes caeruleus: effects of sex, age and environment. Anim. Behav. 80, 853–864. ( 10.1016/j.anbehav.2010.08.005) [DOI] [Google Scholar]
- 14.Kumar V, Sharma A. 2018. Common features of circadian timekeeping in diverse organisms. Curr. Opin. Physiol. 5, 58–67. ( 10.1016/j.cophys.2018.07.004) [DOI] [Google Scholar]
- 15.Weljie AM, Meerlo P, Goel N, Sengupta A, Kayser MS, Abel T, Birnbaum MJ, Dinges DF, Sehgal A. 2015. Oxalic acid and diacylglycerol 36: 3 are cross-species markers of sleep debt. Proc. Natl Acad. Sci. USA 112, 2569–2574. ( 10.1073/pnas.1417432112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raap T, Pinxten R, Eens M. 2018. Artificial light at night causes an unexpected increase in oxalate in developing male songbirds. Conserv. Physiol. 6, coy005 ( 10.1093/conphys/coy005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishra I, Knerr RM, Stewart AA, Payette WI, Richter MM, Ashley NT. 2019. Light at night disrupts diel patterns of cytokine gene expression and endocrine profiles in zebra finch (Taeniopygia guttata). Sci. Rep. 9, 1–2. ( 10.1038/s41598-018-37186-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wisor JP, Clegern WC, Schmidt MA. 2011. Toll-like receptor 4 is a regulator of monocyte and electroencephalographic responses to sleep loss. Sleep 34, 1335–1345. ( 10.5665/SLEEP.1274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krueger JM, Rector DM, Roy S, Van Dongen HPA, Belenky G, Panksepp J. 2008. Sleep as a fundamental property of neuronal assemblies. Nat. Rev. Neurosci. 9, 910 ( 10.1038/nrn2521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obal F Jr, Krueger JM. 2003. Biochemical regulation of non-rapid-eye-movement sleep. Front Biosci. 8, d520–d550. ( 10.2741/1033) [DOI] [PubMed] [Google Scholar]
- 21.Tatsuki F, Ode KL, Ueda HR. 2017. Ca2+-dependent hyperpolarization hypothesis for mammalian sleep. Neurosci. Res. 118, 48–55. ( 10.1016/j.neures.2017.03.012) [DOI] [PubMed] [Google Scholar]
- 22.Funato H, et al. 2016. Forward-genetics analysis of sleep in randomly mutagenized mice. Nature 539, 378 ( 10.1038/nature20142) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunagawa GA, et al. 2016. Mammalian reverse genetics without crossing reveals Nr3a as a short-sleeper gene. Cell Rep. 14, 662–677. ( 10.1016/j.celrep.2015.12.052) [DOI] [PubMed] [Google Scholar]
- 24.Shi S, Ueda HR. 2018. Ca2+-dependent hyperpolarization pathways in sleep homeostasis and mental disorders. Bioessays 40, e1700105 ( 10.1002/bies.201700105) [DOI] [PubMed] [Google Scholar]
- 25.Komarova TG, Ekimova IV, Pastukhov YF. 2008. Role of the cholinergic mechanisms of the ventrolateral preoptic area of the hypothalamus in regulating the state of sleep and waking in pigeons. Neurosci. Behav. Physiol. 38, 245–252. ( 10.1007/s11055-008-0036-9) [DOI] [PubMed] [Google Scholar]
- 26.Rattenborg NC, Martinez-Gonzalez D. 2015. Avian versus mammalian sleep: the fruits of comparing apples and oranges. Current Sleep Med. Rep. 1, 55–63. ( 10.1007/s40675-014-0001-9) [DOI] [Google Scholar]
- 27.Cirelli C, Tononi G. 2008. Is sleep essential? PLoS Biol. 6, e216 ( 10.1371/journal.pbio.0060216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roth TC, Rattenborg NC, Pravosudov VV. 2010. The ecological relevance of sleep: the trade-off between sleep, memory and energy conservation. Phil. Trans. R. Soc. B 365, 945–959. ( 10.1098/rstb.2009.0209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yadav A, Kumar R, Tiwari J, Kumar V, Rani S. 2017. Sleep in birds: lying on the continuum of activity and rest. Biol. Rhythm Res. 48, 805–814. ( 10.1080/09291016.2017.1346850) [DOI] [Google Scholar]
- 30.Sharma A, Singh D, Das S, Kumar V. 2018. Hypothalamic and liver transcriptome from two crucial life-history stages in a migratory songbird. Exp. Physiol. 103, 559–569. ( 10.1113/EP086831) [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25, 402–408. ( 10.1006/meth.2001.1262) [DOI] [PubMed] [Google Scholar]
- 32.Soper DS. 2013. F-value and p-value calculator for multiple regression (online software). See http://www.danielsoper.com/statcalc.
- 33.Da Silva A, Samplonius JM, Schlicht E, Valcu M, Kempenaers B. 2014. Artificial night lighting rather than traffic noise affects the daily timing of dawn and dusk singing in common European songbirds. Behav. Ecol. 25, 1037–1047. ( 10.1093/beheco/aru103) [DOI] [Google Scholar]
- 34.Renthlei Z, Trivedi AK. 2019. Effect of urban environment on pineal machinery and clock genes expression of tree sparrow (Passer montanus). Environ. Pollut. 255, 113278 ( 10.1016/j.envpol.2019.113278) [DOI] [PubMed] [Google Scholar]
- 35.Franken P, Thomason R, Heller HC, O'hara BF. 2007. A non-circadian role for clock-genes in sleep homeostasis: a strain comparison. BMC Neurosci. 8, 87 ( 10.1186/1471-2202-8-87) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wisor JP, Pasumarthi RK, Gerashchenko D, Thompson CL, Pathak S, Sancar A, Franken P, Lein ES, Kilduff TS. 2008. Sleep deprivation effects on circadian clock gene expression in the cerebral cortex parallel electroencephalographic differences among mouse strains. J. Neurosci. 28, 7193–7201. ( 10.1523/JNEUROSCI.1150-08.2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fonken LK, Aubrecht TG, Meléndez-Fernández OH, Weil ZM, Nelson RJ. 2013. Dim light at night disrupts molecular circadian rhythms and increases body weight. J. Biol. Rhythms 28, 262–271. ( 10.1177/0748730413493862) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deboer T. 2018. Sleep homeostasis and the circadian clock: do the circadian pacemaker and the sleep homeostat influence each other's functioning? Neurobiol. Sleep Circadian Rhythms 5, 68–77. ( 10.1016/j.nbscr.2018.02.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szymusiak R. 2010. Hypothalamic versus neocortical control of sleep. Curr. Opin Pulm. Med. 16, 530–535. ( 10.1097/MCP.0b013e32833eec92) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mRNA sequences with their partial CDS can be accessed using GenBank accession numbers as provided in the electronic supplementary material, table S1.