Abstract
For centuries, people believed that bats possessed sinister powers. Bats are thought to be ancestral hosts to many deadly viruses affecting humans including Ebola, rabies, and most recently SARS-CoV-2 coronavirus. However, bats themselves tolerate these viruses without ill effects. The second power that bats have is their longevity. Bats live much longer than similar-sized land mammals. Here we review how bats’ ability to control inflammation may be contributing to their longevity. The underlying mechanisms may hold clues to developing new treatments for age-related diseases. Now may be the time to use science to exploit the secret powers of bats for human benefit.
Keywords: Bats, Aging, Longevity, Inflammation, Inflammaging
Bats host and tolerate a wide range of viruses and live considerably longer than similar-sized land mammals. Here, Gorbunova et al. review mechanisms shaping the exceptional longevity and virus resistance of bats. The authors discuss how the bat’s ability to downregulate inflammation may ameliorate age-related diseases, and they propose anti-aging interventions for humans based upon the evolutionary adaptations of bats.
Graphical Abstract
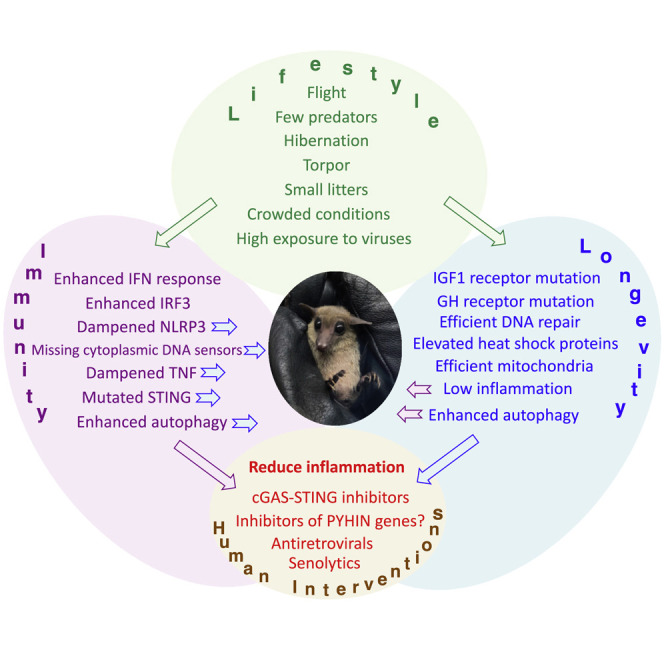
Main Text
Bat Natural History
There are more than 1,400 species of bats (Figure 1 A) that inhabit all continents except Antarctica. A majority of bats are active at night and inhabit “spooky” places such as caves, wells, attics, and hollow trees, which has caused people to (falsely) attribute devilish properties to bats. Many bats live in large colonies, which promotes transmission of viruses and other pathogens. Bats are diverse in their feeding habits. Many bats feed on flying insects, others feed on fruits and nectar, but there are also carnivorous bats that catch fish and small crustaceans such as scorpions and, of course, vampire bats that feed on blood. Bats are the only species of mammal capable of powered flight, and some species can reach speeds of up to 100 miles per hour, making them the fastest mammals on earth (McCracken et al., 2016).
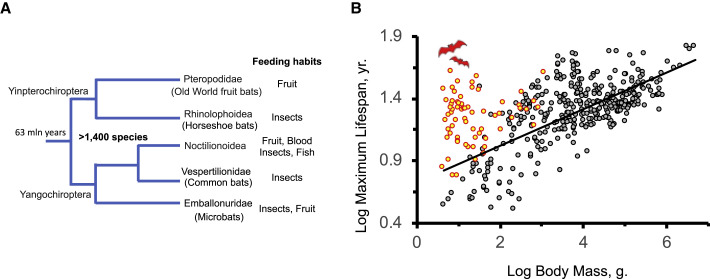
Figure 1.
Bats Live Longer Than Similar-Sized Land Mammals
(A) Major lineages of bats.
(B) Relationship between lifespan and body mass in mammals. Bats are indicated by red circles; all other species of mammals are indicated by black circles. The lifespan and body mass data are from Healy et al. (2014).
Bats play important role in many ecosystems. Insectivorous bats provide pest control by consuming large quantities of insects. Bats suppress crop pests and decrease pesticide use in farming (Maine and Boyles, 2015). Bats also consume mosquitoes that spread human diseases. Furthermore, many plants depend on fruit bats for pollination and seed dispersal.
One of the most amazing properties of bats is their longevity. Many bat species such as little brown bat, Brandt’s bat, mouse-eared bat, and Indian flying fox have maximum lifespans of 30–40 years (Tacutu et al., 2018). Other bat species have maximum lifespans around 20 years, which is still very long for species of this size (Tacutu et al., 2018). In general, species maximum lifespan correlates positively with body mass (Austad and Fischer, 1991). Larger species tend to be longer lived. However, all bats fall above the regression line for mammals as they live longer than other mammals of similar size (Austad and Fischer, 1991; Healy et al., 2014) (Figure 1B).
Resistance to Viruses
As mentioned earlier, bats live in large dense colonies and they stay around for many years, which creates an ideal ground for transmitting pathogens. Indeed, bats are believed to be ancestral hosts for many deadly viruses including rabies, Ebola, Marburg, Nipah, and Hendra viruses (reviewed in Banerjee et al., 2020a). The recent zoonotic transmissions of SARS-CoV (Li et al., 2005), MERS-CoV (Anthony et al., 2017), and SARS-CoV-2 (Lu et al., 2020; Zhou et al., 2020) coronaviruses are believed to have originated in bats, or were transmitted by bats to other species and then passed on to humans. Remarkably, these viruses are tolerated by bats and do not cause clinical symptoms. Even experimental inoculation of bats with some of the deadliest viruses only produced subclinical infections (Halpin et al., 2011; Middleton et al., 2007; Munster et al., 2016; Paweska et al., 2016; Schuh et al., 2017). What biological mechanisms make bats so special that they can tolerate such scourges and innocently transmit them to humans (Figure 2 )?
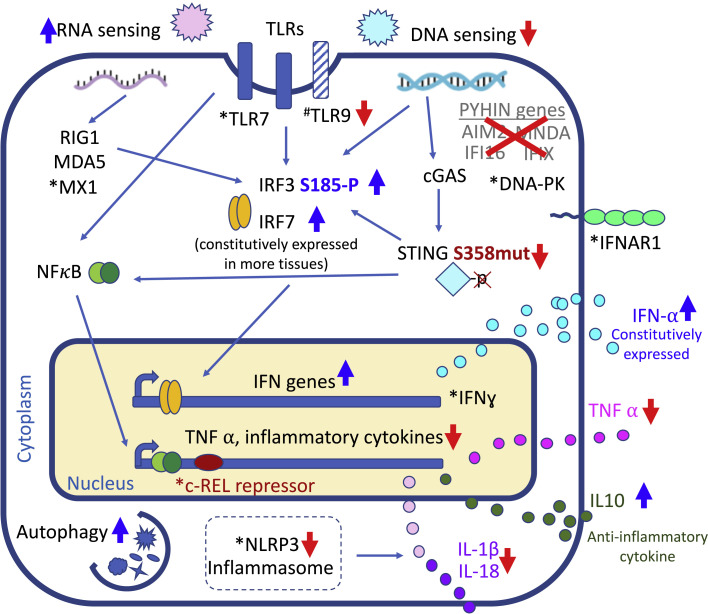
Figure 2.
Unique Aspects of Immunity in Bats
The innate immune response senses cytoplasmic DNA and foreign RNA, responding by upregulation of interferon genes and production of TNFα and other inflammatory cytokines. Bat species have undergone evolutionary adaptations whereby sensing of cytoplasmic DNA is dampened through loss of PYHIN genes and a regulatory site mutation in STING. This leads to reduced expression of TNFα and other inflammatory cytokines, while anti-inflammatory cytokines like IL-10 are upregulated. Additionally, activation of the NLRP3 inflammasome is dampened in bats, leading to reduced production of IL-1β and IL-18. Several aspects of RNA sensing are enhanced in bats. Bats also constitutively express IFN-α, show enhanced expression of interferon genes, and have more active autophagy. Taken together, these adaptations allow bats to tolerate viruses while keeping inflammatory response in check. Processes enhanced in bats are marked by upward blue arrows, while processes dampened in bats are marked by downward red arrows. ∗Genes under positive selection in bats. #Genes under purifying selection in bats.
It is important to point out that bats are a very diverse group, so a mechanism found in one species does not necessarily apply to all bats. Here, we will discuss these adaptations as they apply to individual species and the common trends that emerge. The innate immune response is the first line of defense against viruses. Cells express pattern recognition receptors, such as Toll-like receptors, that recognize pathogen-associated molecular patterns originating from viruses, bacteria, or parasites. These receptors then initiate signaling cascades that lead to expression of antiviral and proinflammatory cytokines. Antiviral cytokines include interferons (IFN), which activate expression of downstream genes that inhibit viral replication or induce death of infected cells.
Bats have a robust interferon response to RNA viruses. In the Australian black flying fox Pteropus alecto, IFN regulatory factor 7 (IRF7) is expressed in a wider range of tissues compared to other mammals (Zhou et al., 2014). In multiple bat species, IFN regulatory factor 3 (IRF3) has evolved a potential novel phosphorylation site, S185, that enhances activation of the downstream IFN response (Banerjee et al., 2020b). Bats also constitutively express IFN-α (Zhou et al., 2016), which in other mammals would lead to widespread inflammation. However, bats have evolved unique adaptations that counteract inflammation. For instance, they show significantly dampened activation of the NLRP3 inflammasome in primary immune cells compared to their human or mouse counterparts (Ahn et al., 2019). NLRP3 is an important sensor that recognizes both endogenous cellular damage and infections of either bacterial or viral origin. NLRP3 over-activation has been linked to the inflammatory state and to age-related diseases (Guo et al., 2015; Youm et al., 2013). Remarkably, dampened NLRP3 activity was observed in distant bat species, such as the fruit bat P. alecto and an insectivorous bat Myotis davidii (Ahn et al., 2019), suggesting that this is a common mechanism across species. Treating kidney cells from the big brown bat Eptesicus fuscus and human with poly(I:C), a viral double-stranded RNA surrogate, showed that although cells from both species induced expression of IFN-β, only human cells expressed tumor necrosis factor α (TNF-α) (Banerjee et al., 2017), a cell signaling protein involved in systemic inflammation. Analysis of the TNF promoter in the big brown bat revealed a potential repressor c-REL binding motif (Banerjee et al., 2017). Thus, downregulated TNF-α expression may be yet another strategy bats use to suppress inflammation.
Whole-genome sequencing revealed that several genes involved in innate immunity, including c-REL and NLRP3, are under positive selection in P. alecto and Myotis davidii (Zhang et al., 2013). Bats may also express unique sets of interferon stimulated genes (ISGs). Some of these genes, such as Myxovirus resistance 1 (Mx1), evolved under positive selection in bats and are reported to reduce viral replication when expressed in human cells (Fuchs et al., 2017).
Although bats mount a strong response to RNA viruses, they exhibit remarkably dampened DNA sensing. The entire PYHIN gene family was found to be missing in 10 bat species, including both fruit- and insect-eating bats (Ahn et al., 2016; Zhang et al., 2013). This gene family includes cytoplasmic DNA sensors AIM2 and IFI16 that activate the inflammasome and interferon pathways (reviewed in Schattgen and Fitzgerald, 2011). AIM2 recognizes bacterial and host DNA in the cytoplasm (Muruve et al., 2008), forming the AIM2 inflammasome that mediates maturation of proinflammatory cytokines (IL-1β and IL-18) (reviewed in Broz and Dixit, 2016). Although loss of the PYHIN locus appears to be universal, genomic analysis of the remaining sequences suggests different evolutionary processes leading to gene loss, rather than a single ancestral event (Ahn et al., 2016). Additionally, bats have dampened STING-dependent IFN activation (Xie et al., 2018). Upon binding to cytosolic DNA, cGAS binds and activates STING, leading to its phosphorylation on S358. Phosphorylated STING ultimately triggers the type I IFN response. Remarkably, an analysis of 30 bat species revealed that while the S358 residue is absolutely conserved among all known non-bat mammalian STING proteins, none of the bat STING proteins retain S358, leading to a weakened IFN response.
The TLR9 receptor, which preferentially recognizes DNA, appears to have evolved under purifying selection in bats and contains multiple mutations in the ligand-binding domain (Escalera-Zamudio et al., 2015). TLR9 in bats shows reduced activation by CpG-containing oligonucleotides compared to human TLR9 (Banerjee et al., 2017). Interestingly, the DNA repair protein DNA-PK, which also serves as cytoplasmic DNA sensor, is positively selected in bats (Zhang et al., 2013), possibly compensating for the lack of other DNA sensors.
Macrophages from the greater mouse-eared bat, Myotis myotis, challenged with various inflammatory stimuli upregulated interferon β (INF-β), TNF, and interleukin-1β (Il-1β). However, in bat macrophages, but not mice, this antiviral, proinflammatory response was associated with high-level transcription of the anti-inflammatory cytokine Il-10, which may help neutralize proinflammatory responses (Kacprzyk et al., 2017).
The picture that emerges from the large number of studies of bat immunity is somewhat counterintuitive. A majority of immune system adaptations found in bats dampen the immune response rather than activating it. We can learn from bats that to co-exist with viruses, controlling inflammation is more important than ramping up the immune system to combat the virus, which could lead to an inflammatory cytokine storm, exacerbating disease phenotypes and contributing to mortality. Perhaps in certain contexts, controlling inflammation is more important than ramping up the immune system to combat the virus, which could lead to an inflammatory cytokine storm, exacerbating disease phenotypes and contributing to mortality.
Outside of the immune system, autophagy is a process that may help bats fight viruses. Autophagy, or “self-eating,” is a cellular process that destroys and recycles cellular proteins and organelles (Saha et al., 2018). Autophagy can remove damaged proteins and can also help rid the cell of viruses. Cells from the black flying fox P. alecto are less susceptible than human cells to death induced by Australian bat lyssavirus, a virus related to rabies. P. alecto cells showed elevated basal autophagic levels, which are further induced in response to high doses of virus (Laing et al., 2019).
Why did bats evolve such tolerance to viruses? It has been proposed that unique evolution of immunity in bats is driven by flight (Banerjee et al., 2020a; Kacprzyk et al., 2017; O’Shea et al., 2014; Zhang et al., 2013). Bats are the only flying mammals and flight requires metabolic adaptation to sudden surges in activity, rapid increases in body temperature (O’Shea et al., 2014), and perhaps, dealing with the molecular damage, such as misfolded proteins and damaged DNA, that arise (Banerjee et al., 2020a). Thus, bats may have downregulated their inflammatory pathways in order not to suffer from bouts of inflammation every time they fly. Flight is indeed a unique feature of bats, and it is plausible that some of the adaptations are related to flight. However, the fastest evolving genes, in general, are genes related to the host-pathogen arms race (Lazzaro and Clark, 2012). This evolution is driven by the presence of pathogens and happens much faster than evolution of such complex functions as flight, which require major changes to body structures. We speculate that the driving force in the evolution of bat immunity has been their lifestyle, which promotes rapid transmission of viruses. Many bat species live in gigantic colonies, where individuals spend resting periods hanging very close together on a cave ceiling or in a tree. Bat colony size may range from a few individuals to hundreds of thousands. This is the highest density among mammals, with the exception perhaps of humans in large metropolises, and considering their high mobility and foraging behavior, bats are exposed to an exceptionally high variety of viruses.
Longevity Adaptations
The long lifespan of bats is quite striking. A recent study by Wilkinson and Adams was able to reconstruct longevity across a wide range of bat species, proposing that the ancestral bat species likely lived 2.6 times longer than a similar-sized placental mammal (Wilkinson and Adams, 2019). Moreover, extreme longevity has arisen at least four separate times during bat speciation, with the longest-lived bat on record, the Brandt’s bat, surviving 41 years (Podlutsky et al., 2005). These findings suggest that long lifespan is no accident; it either arose because long lifespan has fitness benefits for bats or because some other phenotype is selected that also precipitates longevity, one of which being a dampened immune response.
The reasons behind the long lifespan of bats remain debated, with scientists developing hypotheses based either on evolutionary life history or molecular studies testing known longevity pathways. Bats have several features that would favor selection for low mortality rates, including small litters (Barclay and Harder, 2003; Racey and Entwhistle, 1999; Speakman, 2008), the capacity of flight (which permits escape from predators; Austad and Fischer, 1991; Holmes and Austad, 1994), and (in many species) the ability to hibernate, or enter into a low-energy torpor state. Torpor is linked to longevity in bats and other species (Turbill et al., 2011; Turbill and Prior, 2016; Wilkinson and South, 2002) and may protect the animal from bouts of starvation and/or promote homeostatic maintenance during periods of low metabolic rate. Consistent with a beneficial role for hibernation, other species that can enter hibernation, such as gray mouse lemurs and 13-lined squirrels, have longer lifespan than mice of similar size (Al-Attar and Storey, 2020). In the Wilkinson and Adams study, enhanced longevity was associated with body mass, hibernation, and cave use, which presumably provides a safer environment (Wilkinson and Adams, 2019).
Recent reviews have described pathways modulating aging, termed hallmarks or pillars (Figure 3 ) (Kennedy et al., 2014; López-Otín et al., 2013). The overlapping pathways these reviews describe are all intimately linked to aging in a variety of eukaryotic models. Yet it remains unclear to what extent each contributes to different aspects of aging and whether they are connected in a hierarchical structure. Almost certainly they are connected in some kind of homeostatic network that functions to maintain health in the face of damage that accrues with age. One piece of evidence for this is that genetic, dietary, or pharmacological interventions that delay aging typically affect many or all aging pillars. While an intervention likely has a direct target, the net effect is network preservation, and this affects many or all pillars. If this concept extends across species, then long-lived bats may expect to have alterations in many longevity pillars. Early evidence suggests that this is the case, and here we describe evidence connecting longevity to specific hallmarks of aging.
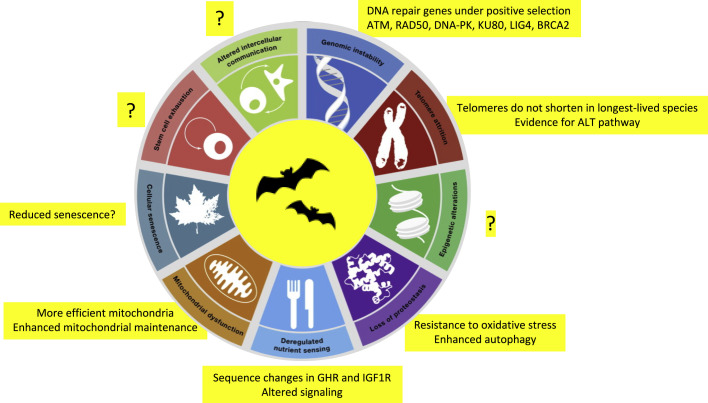
Figure 3.
Longevity Adaptations in Bats
Many hallmarks of aging are altered in bats (yellow rectangles), any of which could underlie their enhanced longevity. Importantly, other hallmarks such as intercellular communication, stem cell exhaustion, and epigenetic alterations are yet to be interrogated. Modified from López-Otín et al. (2013).
Genomic studies have pointed to some longevity clues. For instance, the genome of the Brandt’s bat and several other species has a mutation in the growth hormone receptor gene that may interfere with transmembrane domain function (Seim et al., 2013). Growth hormone receptor hypomorphic mutations are associated with protection from diabetes and cancer in humans and long lifespan in mice (Coschigano et al., 2000; David et al., 2011; Guevara-Aguirre et al., 2011). Indeed, bats have physiologic (e.g., pancreatic structure) and transcriptomic changes that resemble growth hormone receptor knockout mice (Liu et al., 2004; Seim et al., 2013; Swindell, 2007). There are also intriguing changes in the transmembrane region of the IGF1 receptor, which is associated with longevity in a range of model organisms and in centenarians (Kim and Lee, 2019; Seim et al., 2013). Both of these hormonal signaling pathways are intimately linked to nutrient signaling, one of the most robust pillars of aging (Figure 3). A more extensive study compared genomic data from 26 bat species to that of naked mole rats and a variety of other mammals (Davies et al., 2014). While other genomic changes were identified in both the growth hormone receptor and the IGF receptor, no evidence for positive selection was found in either gene, causing the authors to speculate that enhanced longevity was in large part linked to other genetic differences.
Genome maintenance is an important longevity assurance mechanism and another recognized pillar of aging (reviewed in Gorbunova et al., 2007). In an 8-year longitudinal study of blood samples from free-living greater mouse-eared bats (Myotis myotis), Huang et al. reported that DNA repair and DNA damage signaling pathways are maintained throughout lifespan, consistent with the low levels of cancer in bat species (Huang et al., 2016, 2019). Among DNA repair pathways, DNA double-strand break repair shows the strongest correlation with longevity (Tian et al., 2019). Remarkably several DNA double-strand break repair genes such as ATM, Rad50, Lig4, DNA-PK, Ku80, and BRCA2 were shown to be under positive selection in two species of bats (Zhang et al., 2013). Interestingly, some of these DNA repair genes, such as DNA-PK and Rad50, also function as DNA sensors in innate immune response (Burleigh et al., 2020; Ferguson et al., 2012; Kondo et al., 2013; Roth et al., 2014). Hence, the genetic changes that evolved in bats may modulate both processes simultaneously and the innate immune response may be an evolutionary driver of positive selection. However, further studies are needed to test whether positive selection on DNA-PK and Rad50 increases or decreases their activity as immune sensors. These changes may either compensate for dampened STING signaling or, alternatively, further decrease DNA sensing in bats.
Mitochondrial dysfunction is a feature of aging across the evolutionary spectrum and another highly supported pillar of aging (Figure 3) (Pulliam et al., 2013). Energetic demands associated with flight in bats require enhanced mitochondrial respiratory metabolism, which is expected to generate excess oxidative damage. To counteract this damage, bats have evolved more efficient mitochondria, producing less H2O2 per unit oxygen consumed (Brunet-Rossinni, 2004). Bat fibroblasts have also been shown to have lower levels of oxidative damage to proteins and to be resistant to acute oxidative stress (Salmon et al., 2009). Limited data are available on antioxidants in vivo (Dammann, 2017), with superoxide dismutase (SOD) levels reported to be similar in the brain but with a trend toward higher levels in the heart. A different study compared two bats with different lifespans, finding that the longer-lived species (Desmodus rotundus) had higher levels of antioxidant activity across multiple tissues than its shorter-lived counterpart (Myotis velifer) (Conde-Pérezprina et al., 2012). This corresponded to reduced levels of DNA damage.
To help maintain proteostasis upon oxidative stress, bats express major heat shock proteins at higher levels (Chionh et al., 2019). This may simultaneously permit bats to endure high temperatures with flight and maintain protein homeostasis with age. Bats also exhibit enhanced autophagy activity with advancing age (Huang et al., 2019), suggesting that their cells are better able to clear damaged proteins and organelles. This latter finding is consistent with studies in bat fibroblasts, which detect higher levels of macroautophagy (Pride et al., 2015). Interestingly, enhanced autophagy also provides bats with enhanced protection against viruses, as described above (Laing et al., 2019).
Increased mitochondrial oxidative stress would also be expected to generate mitochondrial DNA alterations, or heteroplasmy. However, oxidative lesions in M. myotis are found only at low rates in an age-independent manner, suggesting better repair or removal of damaged mitochondria (Jebb et al., 2018). One recent study suggests that a system involved in limiting mitochondrial membrane potential by tethering of ATP-consuming kinases to mitochondrial membranes may be a mechanism that limits reactive oxygen species production. This system is lost in mice with age but is maintained in Seba’s short-tailed bat (Carollia perspicillata) (Vyssokikh et al., 2020).
As they age, bats avoid upregulation of genes involved in chronic inflammation, which is typically not observed in mammals (Huang et al., 2019). This likely results from the multitude of mechanisms that evolved to suppress inflammation due to viral infections discussed above. Microbiome studies indicate that Myotis myotis may have stable microbiome composition that does not change over time in contrast to mice and humans, where the microbiome undergoes significant changes with age (Hughes et al., 2018). As aging-related gut dysbiosis triggers inflammation (reviewed in Kim and Jazwinski, 2018), the ability of bats to maintain stable microbiome may contribute to the lack of age-related inflammation, or by contrast, low levels of inflammation may promote a more stable microbiome. One major unanswered question is the extent to which cell senescence occurs with age in bats. Since cell senescence may be a major driver of chronic inflammation during mammalian aging, it will be important to determine whether cell senescence, another pillar, is altered with age in bat species. In-depth studies are needed to address this question in vivo and in cell culture.
Among other hallmarks of aging, telomere attrition has been addressed to a limited extent, with mixed results. The shorter-lived bat species, Rhinolophus ferrumequinum and Miniopterus schreibersii, do exhibit telomere shortening, but no evidence was found in the longest-lived species, Myotis myotis (Foley et al., 2018). This bat apparently does not express telomerase but exhibits differential expression of genes involved in telomere maintenance and the alternative lengthening of telomeres (ALT) pathway (Foley et al., 2018). Other pillars of aging, including epigenetic alterations, intercellular communication, and adult stem cell exhaustion, remain unaddressed and are also fertile grounds for further research.
Inflammation and Longevity
A majority of the mechanisms that have evolved to protect bats from viruses likely contribute to their longevity (Figure 4 ). Bats evolved multiple strategies to combat inflammation (discussed above), such as dampened NLRP3 inflammasome activity (Ahn et al., 2019), dampened TNF signaling (Banerjee et al., 2017), and a dampened response to cytoplasmic DNA (Ahn et al., 2016; Xie et al., 2018; Zhang et al., 2013). Inflammation has emerged as a driver of multiple age-related pathologies, including cardiovascular diseases, cancer, Alzheimer’s disease, and diabetes. This led to the concept of inflammaging, defined as the long-term result of the chronic physiological stimulation of the innate immune system, which becomes damaging during aging (Franceschi et al., 2018). Factors that trigger inflammaging include viruses, microbiome bacteria, senescent cells, and self-products of cellular damage such as debris containing cellular DNA and proteins. Reducing inflammation due to any of these factors can be beneficial for longevity; however, bat evolution seems to have attenuated mechanisms of cytoplasmic DNA sensing specifically.
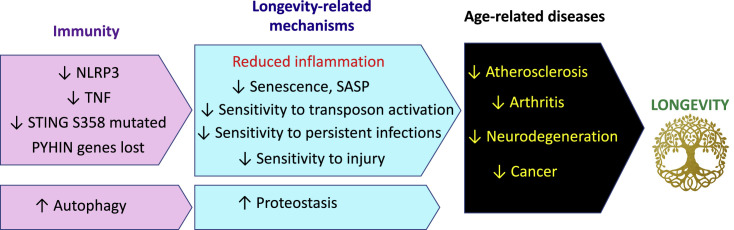
Figure 4.
Changes in Immune Function in Bats that Can Promote Longevity
While many hallmarks of aging are altered in bats, we speculate that altered immunity, which leads to virus tolerance and reduced inflammation, underlies the longevity changes. The downstream consequences of the changes likely include a reduction in age-related diseases, which are fueled by inflammation such as atherosclerosis, arthritis, neurodegeneration, and cancer. This, in turn, promotes longevity in bats.
Free DNA is not supposed to be in the cytoplasm, and when it does go there it signals alarm. Cytoplasmic DNA sensors have evolved to protect the cell from invading viruses. However, viruses are not the only source of cytoplasmic DNA. Mitochondrial DNA can be recognized by pattern recognition receptors such as TLR9, absent in melanoma 2 (AIM2) and cGAS and trigger activation of interferon response (reviewed in Riley and Tait, 2020; West and Shadel, 2017). Mitochondrial DNA-associated inflammasome activation has been linked to such age-related conditions as atherosclerosis (Tumurkhuu et al., 2016) and macular degeneration (Dib et al., 2015). Mitochondrial DNA release into cytoplasm has been linked to neurodegeneration and Parkinson’s disease. Interestingly, in Parkin- or Pink-deficient mice mitochondrial DNA release was precipitated by exercise, while blocking INF-α receptors or deleting STING rescued inflammation (Sliter et al., 2018). Other types of mitochondrial stress, such as oxidative damage and endoplasmic reticulum stress, may also result in release of mitochondrial DNA. Thus, bats displaying improved mitochondrial maintenance and dampened cytoplasmic DNA sensing may be protected from these age-related conditions.
Recent studies demonstrated that nuclear DNA can find its way into the cytoplasm during aging and cellular senescence. In senescent cells, fragments of chromosomal DNA are expelled into the cytoplasm (Dou et al., 2017). Furthermore, nuclear DNA contains endogenous parasites in the form of transposable elements, some of which originated from viruses in the evolutionary past. Long interspersed nuclear elements (LINE1s) are retrotransposable elements that comprise 17% of the human genome. LINE1s encode a reverse-transcriptase enzyme and transpose via a copy-and-paste mechanism involving an RNA intermediate. Recent studies demonstrated that LINE1s become de-repressed in aged tissues of humans and mice, leading to increased transcription, followed by reverse transcription and formation of cytoplasmic DNA copies of LINE1 (De Cecco et al., 2019; Simon et al., 2019). These cytoplasmic DNAs trigger the IFN response via activation of the cGAS/STING pathway, providing a new pathway for age-related sterile inflammation (De Cecco et al., 2019; Simon et al., 2019). The cGAS/STING pathway has also been implicated in cellular senescence, further linking senescence to cytoplasmic DNA (Glück et al., 2017; Yang et al., 2017a). In addition to aging, activation of retrotransposable elements and reactivity to DNA have also been associated with a variety of autoimmune conditions in humans (reviewed in Volkman and Stetson, 2014).
Remarkably, bats are unique in their ability to tolerate DNA transposable elements (Pritham and Feschotte, 2007). DNA transposons move in the genome via a cut-and-paste mechanism involving DNA intermediates. Such transposons are found in invertebrates but are generally inactivated and fossilized in the genomes of mammals. Only the vespertilionid family of bats is known to harbor significant levels of active DNA transposable elements. This bat family includes genus Myotis, which contains the longest-lived bats and is discussed in several contexts above, which suggests that these animals are exceptionally healthy. The ability to tolerate active DNA transposons is likely linked to dampened cytoplasmic DNA sensing.
Increasing evidence links cGAS/STING sensor to aging and age-related disease. In mouse models, STING deficiency alleviated phenotypes of prion-induced neurodegeneration and neuroinflammation (Nazmi et al., 2019). Premature aging syndrome ataxia-telangiectasia (A-T) caused by mutation in ATM kinase involved in DNA damage signaling is characterized by cancer predisposition, radiosensitivity, and neurodegeneration. Both A-T patients and Atm −/− animal models have higher serum levels of proinflammatory cytokines and other inflammatory markers than control subjects (Westbrook and Schiestl, 2010). Neuroinflammation in A-T is associated with activation of STING and AIM2, and STING inhibition blocks the overproduction of neurotoxic cytokines (Song et al., 2019). Interestingly, it has also been reported that progerin expression in Hutchinson-Gilford syndrome fibroblasts, which is known to confer genome instability and replication stress, leads to cytoplasmic DNA and STING activation (Kreienkamp et al., 2018).
Strikingly, in a recent study of an aging Polish cohort, a SNP leading to reduced STING expression was associated with protection from age-related disease, particularly for chronic lung disease (Hamann et al., 2019), where inflammation is a major factor. The same SNP is also reported to be protective from obesity-induced cardiovascular disease (Hamann et al., 2020). In summary, while STING activation is linked to age-related conditions, dampened STING signaling seems to confer protection from age-related pathologies in both bats and humans.
The PYHIN gene family is completely lost in multiple bat lineages (Ahn et al., 2016; Zhang et al., 2013). While PYHINs are reported to be upregulated in astrocytes and glial cells during neuroinflammation (Cox et al., 2015), the role of the PYHIN gene family in aging and age-related pathologies needs to be explored in more detail.
NLRP3 inflammasome activity is dampened in bats (Ahn et al., 2019). NRLP3 activates caspase-1 to mediate processing proinflammatory cytokines. The NLRP3 inflammasome has been linked to metabolic and cognitive diseases, including obesity, type 2 diabetes mellitus, and Alzheimer’s disease (Choi and Ryter, 2014). Mice lacking NLRP3 are resistant to developing Parkinson’s disease (Yan et al., 2015). IL-18, a product of inflammasome activation, may have crucial roles in the initiation and progression of atherosclerosis (Duewell et al., 2010). Multiple studies have found that NLRP3- and/or caspase-1-deficient mice show improved glucose tolerance and insulin sensitivity when exposed to a high-fat diet (Lee et al., 2013; Stienstra et al., 2011; Vandanmagsar et al., 2011; Wen et al., 2011), implicating NLRP3 in type 2 diabetes. Remarkably, ablation of the NLRP3 inflammasome in mice extends the healthspan and attenuates multiple age-related degenerative changes in the brain and in peripheral tissues (Youm et al., 2013). These studies suggest the NLRP3 inflammasome as an upstream target that controls age-related inflammation and that lowering NLRP3 activity may delay multiple age-related diseases driven by inflammation, contributing to the long lifespan of bats and serving as a target for lifespan and healthspan extension in humans.
Bats show dampened TNF-α expression in response to inflammatory stimuli (Banerjee et al., 2017). TNF-α is implicated in a multitude of disease conditions, with enhanced activity linked to a range of age-related diseases. TNF-α expression plays a direct role in obesity-linked insulin resistance (Hotamisligil et al., 1993). Activation of TNF-α is reported to contribute to disease progression in atherosclerosis (Zhang et al., 2009); neurodegenerative conditions such as Alzheimer’s disease, Huntington’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis (Frankola et al., 2011; Greig et al., 2004; Yang et al., 2017b); osteoarthritis (Malemud et al., 2003); sarcopenia (Fan et al., 2016); and a range of other conditions. Thus, reduced TNF-α response may contribute to bat longevity.
In the continuing arms race against pathogens, evolutionary fitness requires a functional immune system. However, a highly active immune system may increase fitness in young age but limit longevity. One example is the ApoE4 allele in humans that improves pathogen resistance in children but is associated with significantly increased risk of dementia and atherosclerosis in the elderly (reviewed in Abondio et al., 2019). Why did bat evolution result in adjusted immune system functions in a way that favors longevity? We speculate that bats’ exceptionally high exposure to viral pathogens forced them to develop ways to co-exist with viruses rather than to fight them. Bats are unique among mammals in the size and density of their colonies, and in their ability to fly long distances, a trait that further increases pathogen exposure (Hayman et al., 2013). Modern humans living in large metropolises and enjoying air travel may be coming close to the bat level of viral exposure. However, humans have only been enjoying this lifestyle for less than 100 years, while bats evolved 60–70 million years ago (Figure 5 ). We speculate that human immune systems have not evolved to deal with such frequent exposure to diverse viruses, which may manifest in a rise in autoimmune diseases and our susceptibility to pandemics. It would take an unacceptable amount of time for humans to naturally evolve adaptations similar to those of bats that would allow us to live long and healthy in these conditions; however, pharmacological interventions might be developed to bring bat-like adaptations to humans in a shorter time frame, reducing disease burden due to viral infections and delaying age-associated diseases.
Figure 5.
Adaptation to High Population Density and Air Travel
Humans evolved to live in small family groups. The first cities were settled 3000 to 4000 B.C., but significant numbers of people moved to live in large cities only over the last several centuries. Air travel is an even more recent development and became available in the last 100 years. Bats, however, have evolved to deal with these challenges over the course of 60–70 million years.
Treatments Based on Bat Strategies
Bats have evolved multiple mechanisms to suppress inflammation, in particular by dampening nucleic acid sensing pathways. A number of pharmacological interventions targeting nucleic acid sensing pathways had already been developed. Historically, the focus has been on developing activators of these pathways to serve as antiviral or anticancer drugs (reviewed in Vanpouille-Box et al., 2019). However, with the realization that inflammation contributes to a wide range of diseases from autoimmunity to age-related conditions, the interest has shifted to developing antagonists of nucleic acid sensors (Sheridan, 2019). This proved to be a challenging task due to high level of redundancy within nucleic acid sensing pathways and the danger of increasing vulnerability to infections. Here, the information obtained from the studies of bats can assist in drug discovery (Table 1 ).
Table 1.
Bat Adaptations that Reduce Inflammation and Human Treatments
| Bat Adaptations |
Human Treatments |
||
|---|---|---|---|
| Dampened Inflammation | Target, Drug | Indications |
|
| Current | Future | ||
| Dampened TNF | TNF, TNFR antagonists | autoimmune diseases | age-related diseases |
| Dampened NLRP3 | NLRP3 inflammasome inhibitors | autoimmune diseases and age-related diseases | |
| Dampened STING signaling | STING inhibitors and cGAS inhibitors | autoimmune diseases and age-related diseases | |
| chloroquine | malaria | ||
| aspirin | inflammation, fever, and age-related diseases | ||
| Deleted PYHIN genes | inhibitors of AIM2, IFI16, IFIX, and MNDA | autoimmune diseases and age-related diseases | |
| Tolerance of transposons | HIV reverse transcriptase, nucleoside analogs, LINE1 reverse transcriptase, nuclease, ORF1 protein | HIV, other viral infections | age-related diseases |
| Reduced senescence? | senolytics | age-related diseases | |
TNF signaling is inhibited in bats (Banerjee et al., 2017) and anti-TNF therapies are successfully used in the treatment of a wide variety of autoimmune diseases such as rheumatoid arthritis, psoriasis, ulcerative colitis, and Crohn disease (reviewed in Croft and Siegel, 2017). Anti-TNF therapeutics are typically monoclonal antibodies to either TNF or TNF receptors. These drugs effectively alleviate inflammation, prevent tissue destruction, and reduce patient mortality (Monaco et al., 2015). Interestingly, arthritis patients treated with TNF antagonists also showed improvements in cardiovascular conditions (Tam et al., 2014), suggesting that TNF inhibition may confer a broader benefit for age-related diseases.
NLRP3 inflammasome is dampened in bats (Ahn et al., 2019), and aberrant activation of NLRP3 is linked to a wide range of age-related diseases. Several direct NLRP3 inhibitors have been reported that alleviate inflammatory conditions in mice (Coll et al., 2015; He et al., 2014; Jiang et al., 2017). Many new inhibitors with improved potency and safety profiles are being developed to treat inflammatory conditions including age-related diseases such as Alzheimer’s, Parkinson’s, and cardiovascular diseases (reviewed in Zahid et al., 2019).
Bats show a dampened cGAS-STING pathway (Xie et al., 2018). Remarkably, recent progress in drug development has been achieved precisely in targeting these two proteins (reviewed in Ding et al., 2020). Small molecule inhibitors of STING have recently been reported (Haag et al., 2018; Li et al., 2018). These molecules reduced STING-mediated inflammatory cytokine production in both human and mouse cells and were effective in ameliorating inflammatory conditions in mouse models (Haag et al., 2018). cGAS inhibitors are also being developed (Lama et al., 2019). The first application of these compounds will be for the treatment of autoimmune conditions, but ultimately regimens may be developed where these compounds target aging and age-related diseases. Remarkably, several old and widely used drugs have been found to attenuate cGAS. This includes aspirin (Dai et al., 2019) and antimalarial drugs quinacrine and chloroquine (An et al., 2015; Piscianz et al., 2018), supporting the idea that targeting pathways inhibited in bats confers health benefits to humans. Interestingly, aspirin is widely used, although controversially, in small doses to reduce cardiovascular disease risk (Ittaman et al., 2014; McNeil et al., 2018), and it has been shown to extend lifespan modestly in male mice (Strong et al., 2008).
Bats may also help identify novel therapeutic targets. As discussed above, bats lost the entire PYHIN family of genes responsible for nucleic acid sensing (Ahn et al., 2016; Zhang et al., 2013). It will be interesting to explore pharmacological targeting of PYHIN genes, such as AIM2, IFI16, and others, for the treatment of age-related and autoimmune conditions. Interestingly, AIM2-deficient mice do not show severe adverse phenotypes, but display enhanced resistance to ionizing radiation (Hu et al., 2016).
Recent studies showed that retrotransposons become activated with age and drive sterile inflammation via cGAS-STING pathway (De Cecco et al., 2019; Simon et al., 2019). Bats are also particularly good at handling transposable elements (Pritham and Feschotte, 2007). Remarkably, inhibiting retrotransposition activity using nucleoside reverse-transcriptase inhibitors developed to target HIV reverse transcriptase ameliorated age-related inflammation in mouse models (De Cecco et al., 2019; Simon et al., 2019). Mice showed reduced expression of inflammatory cytokines; lower expression of the p16INK4A gene, which is a marker of cellular senescence and aging; and reduced methylation age. Treatment of progeroid SIRT6-deficient mice with nucleoside reverse-transcriptase inhibitors doubled the mouse lifespan, reduced inflammation, and alleviated pathology across multiple tissues (Simon et al., 2019). Therefore, targeting retrotransposable elements may be a promising strategy to ameliorate age-related inflammation. In addition to existing nucleoside analogs that serve as chain terminators for generic reverse transcriptases, specific inhibitors targeting LINE1 reverse transcriptase or endonuclease protein can be developed, which would have lower toxicity for other cellular polymerases. Such compounds may even be used to promote heathy aging.
Senescent cells have been shown to contribute to development of a variety of age-related conditions by promoting sterile inflammation (reviewed in Freund et al., 2010). Little is known about cellular senescence in bats. However, since senescence requires cGAS (Glück et al., 2017; Yang et al., 2017a), and bats dampen cGAS-STING signaling (Xie et al., 2018), one can speculate that the senescence response is attenuated in bats. Thus, senolytic compounds that selectively eliminate senescent cells (Wissler Gerdes et al., 2020) may be another way to pharmacologically promote bat-like adaptations in humans. It is already reported that genetic strategies to ablate senescent cells lead to healthspan and lifespan extension in mice (Baker et al., 2016; Jeon et al., 2017). Many different senolytic molecules have been identified (Fuhrmann-Stroissnigg et al., 2018; Kirkland and Tchkonia, 2017), and several are reported to delay aging and/or protect against age-related diseases (Chang et al., 2016; Xu et al., 2018; Yousefzadeh et al., 2018).
Future Perspectives
In summary, besides serving as a source of deadly diseases and harboring viruses similar to SARS-CoV-2, which caused the current pandemics, bats have a lot to offer humanity by illuminating the pathways to develop novel therapeutics to treat age-related conditions and promote longevity. Already studies of the altered innate immune responses in bats point to several classes of small molecules, some of which have links to aging. However, we can only see the tip of the iceberg when it comes to understanding how bats deal with inflammation, and clearly more studies are warranted. This is also true for other hallmarks of aging, which are still minimally explored. In addition to finding small molecules targeting specific pathways that can be tested in humans, it will be of interest to engineer specific bat alterations in mice and determine whether this leads to enhanced lifespan and healthspan.
Bats have evolved skewed, and ultimately successful, strategies to experience longer and healthier lives, even if this is a secondary outcome of selection for responses to viral infections and/or the dramatic range of metabolic states that accompany periods of flight and torpor. Humans in the last century have created a lifestyle that has gone bats; we live in high densities and (many of us) travel extensively, enhancing exposure to and spread of pathogens. By embracing “batty” strategies to deal with the challenges that our new lifestyle presents, we may be able to solve what look to be the two biggest medical challenges of the 21st century: the rise of viral pandemics and the ever-increasing prevalence of chronic diseases that all share aging as their biggest risk factor.
Acknowledgments
This perspective was conceived when the authors were quarantined together for potential exposure to COVID-19. Research in authors’ laboratories is supported by grants from the National Institutes of Health, United States.
References
- Abondio P., Sazzini M., Garagnani P., Boattini A., Monti D., Franceschi C., Luiselli D., Giuliani C. The genetic variability of APOE in different human populations and its implications for longevity. Genes (Basel) 2019;10:222. doi: 10.3390/genes10030222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn M., Cui J., Irving A.T., Wang L.F. Unique loss of the PYHIN gene family in bats amongst mammals: implications for inflammasome sensing. Sci. Rep. 2016;6:21722. doi: 10.1038/srep21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn M., Anderson D.E., Zhang Q., Tan C.W., Lim B.L., Luko K., Wen M., Chia W.N., Mani S., Wang L.C., et al. Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat. Microbiol. 2019;4:789–799. doi: 10.1038/s41564-019-0371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Attar R., Storey K.B. Suspended in time: molecular responses to hibernation also promote longevity. Exp. Gerontol. 2020;134:110889. doi: 10.1016/j.exger.2020.110889. [DOI] [PubMed] [Google Scholar]
- An J., Woodward J.J., Sasaki T., Minie M., Elkon K.B. Cutting edge: antimalarial drugs inhibit IFN-β production through blockade of cyclic GMP-AMP synthase-DNA interaction. J. Immunol. 2015;194:4089–4093. doi: 10.4049/jimmunol.1402793. [DOI] [PubMed] [Google Scholar]
- Anthony S.J., Gilardi K., Menachery V.D., Goldstein T., Ssebide B., Mbabazi R., Navarrete-Macias I., Liang E., Wells H., Hicks A., et al. Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. MBio. 2017;8:e00373-17. doi: 10.1128/mBio.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austad S.N., Fischer K.E. Mammalian aging, metabolism, and ecology: evidence from the bats and marsupials. J. Gerontol. 1991;46:B47–B53. doi: 10.1093/geronj/46.2.b47. [DOI] [PubMed] [Google Scholar]
- Baker D.J., Childs B.G., Durik M., Wijers M.E., Sieben C.J., Zhong J., Saltness R.A., Jeganathan K.B., Verzosa G.C., Pezeshki A., et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530:184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Rapin N., Bollinger T., Misra V. Lack of inflammatory gene expression in bats: a unique role for a transcription repressor. Sci. Rep. 2017;7:2232. doi: 10.1038/s41598-017-01513-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Baker M.L., Kulcsar K., Misra V., Plowright R., Mossman K. Novel insights into immune systems of bats. Front. Immunol. 2020;11:26. doi: 10.3389/fimmu.2020.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Zhang X., Yip A., Schulz K.S., Irving A.T., Bowdish D., Golding B., Wang L.F., Mossman K. Positive selection of a serine residue in bat IRF3 confers enhanced antiviral protection. iScience. 2020;23:100958. doi: 10.1016/j.isci.2020.100958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay R.M.R., Harder L.D. In: Ecology of Bats. Kunz T.H., Fenton M.B., editors. Chicago University Press; 2003. Life histories of bats: life in the slow lane; pp. 209–253. [Google Scholar]
- Broz P., Dixit V.M. Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- Brunet-Rossinni A.K. Reduced free-radical production and extreme longevity in the little brown bat (Myotis lucifugus) versus two non-flying mammals. Mech. Ageing Dev. 2004;125:11–20. doi: 10.1016/j.mad.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Burleigh K., Maltbaek J.H., Cambier S., Green R., Gale M., Jr., James R.C., Stetson D.B. Human DNA-PK activates a STING-independent DNA sensing pathway. Sci. Immunol. 2020;5:eaba4219. doi: 10.1126/sciimmunol.aba4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Wang Y., Shao L., Laberge R.M., Demaria M., Campisi J., Janakiraman K., Sharpless N.E., Ding S., Feng W., et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 2016;22:78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chionh Y.T., Cui J., Koh J., Mendenhall I.H., Ng J.H.J., Low D., Itahana K., Irving A.T., Wang L.F. High basal heat-shock protein expression in bats confers resistance to cellular heat/oxidative stress. Cell Stress Chaperones. 2019;24:835–849. doi: 10.1007/s12192-019-01013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi A.J., Ryter S.W. Inflammasomes: molecular regulation and implications for metabolic and cognitive diseases. Mol. Cells. 2014;37:441–448. doi: 10.14348/molcells.2014.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll R.C., Robertson A.A., Chae J.J., Higgins S.C., Muñoz-Planillo R., Inserra M.C., Vetter I., Dungan L.S., Monks B.G., Stutz A., et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 2015;21:248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde-Pérezprina J.C., Luna-López A., González-Puertos V.Y., Zenteno-Savín T., León-Galván M.A., Königsberg M. DNA MMR systems, microsatellite instability and antioxidant activity variations in two species of wild bats: Myotis velifer and Desmodus rotundus, as possible factors associated with longevity. Age (Dordr.) 2012;34:1473–1492. doi: 10.1007/s11357-012-9399-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschigano K.T., Clemmons D., Bellush L.L., Kopchick J.J. Assessment of growth parameters and life span of GHR/BP gene-disrupted mice. Endocrinology. 2000;141:2608–2613. doi: 10.1210/endo.141.7.7586. [DOI] [PubMed] [Google Scholar]
- Cox D.J., Field R.H., Williams D.G., Baran M., Bowie A.G., Cunningham C., Dunne A. DNA sensors are expressed in astrocytes and microglia in vitro and are upregulated during gliosis in neurodegenerative disease. Glia. 2015;63:812–825. doi: 10.1002/glia.22786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft M., Siegel R.M. Beyond TNF: TNF superfamily cytokines as targets for the treatment of rheumatic diseases. Nat. Rev. Rheumatol. 2017;13:217–233. doi: 10.1038/nrrheum.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Huang Y.J., He X., Zhao M., Wang X., Liu Z.S., Xue W., Cai H., Zhan X.Y., Huang S.Y., et al. Acetylation blocks cGAS activity and inhibits self-DNA-induced autoimmunity. Cell. 2019;176:1447–1460.e14. doi: 10.1016/j.cell.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann P. Slow aging in mammals-lessons from African mole-rats and bats. Semin. Cell Dev. Biol. 2017;70:154–163. doi: 10.1016/j.semcdb.2017.07.006. [DOI] [PubMed] [Google Scholar]
- David A., Hwa V., Metherell L.A., Netchine I., Camacho-Hübner C., Clark A.J., Rosenfeld R.G., Savage M.O. Evidence for a continuum of genetic, phenotypic, and biochemical abnormalities in children with growth hormone insensitivity. Endocr. Rev. 2011;32:472–497. doi: 10.1210/er.2010-0023. [DOI] [PubMed] [Google Scholar]
- Davies K.T., Tsagkogeorga G., Bennett N.C., Dávalos L.M., Faulkes C.G., Rossiter S.J. Molecular evolution of growth hormone and insulin-like growth factor 1 receptors in long-lived, small-bodied mammals. Gene. 2014;549:228–236. doi: 10.1016/j.gene.2014.07.061. [DOI] [PubMed] [Google Scholar]
- De Cecco M., Ito T., Petrashen A.P., Elias A.E., Skvir N.J., Criscione S.W., Caligiana A., Brocculi G., Adney E.M., Boeke J.D., et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature. 2019;566:73–78. doi: 10.1038/s41586-018-0784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib B., Lin H., Maidana D.E., Tian B., Miller J.B., Bouzika P., Miller J.W., Vavvas D.G. Mitochondrial DNA has a pro-inflammatory role in AMD. Biochim. Biophys. Acta. 2015;1853(11 Pt A):2897–2906. doi: 10.1016/j.bbamcr.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C., Song Z., Shen A., Chen T., Zhang A. Small molecules targeting the innate immune cGAS‒STING‒TBK1 signaling pathway. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.03.001. Published online March 13, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z., Ghosh K., Vizioli M.G., Zhu J., Sen P., Wangensteen K.J., Simithy J., Lan Y., Lin Y., Zhou Z., et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature. 2017;550:402–406. doi: 10.1038/nature24050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duewell P., Kono H., Rayner K.J., Sirois C.M., Vladimer G., Bauernfeind F.G., Abela G.S., Franchi L., Nuñez G., Schnurr M., et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalera-Zamudio M., Zepeda-Mendoza M.L., Loza-Rubio E., Rojas-Anaya E., Méndez-Ojeda M.L., Arias C.F., Greenwood A.D. The evolution of bat nucleic acid-sensing Toll-like receptors. Mol. Ecol. 2015;24:5899–5909. doi: 10.1111/mec.13431. [DOI] [PubMed] [Google Scholar]
- Fan J., Kou X., Yang Y., Chen N. MicroRNA-regulated proinflammatory cytokines in sarcopenia. Mediators Inflamm. 2016;2016:1438686. doi: 10.1155/2016/1438686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B.J., Mansur D.S., Peters N.E., Ren H., Smith G.L. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. eLife. 2012;1:e00047. doi: 10.7554/eLife.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley N.M., Hughes G.M., Huang Z., Clarke M., Jebb D., Whelan C.V., Petit E.J., Touzalin F., Farcy O., Jones G., et al. Growing old, yet staying young: the role of telomeres in bats’ exceptional longevity. Sci. Adv. 2018;4:o0926. doi: 10.1126/sciadv.aao0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C., Garagnani P., Parini P., Giuliani C., Santoro A. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 2018;14:576–590. doi: 10.1038/s41574-018-0059-4. [DOI] [PubMed] [Google Scholar]
- Frankola K.A., Greig N.H., Luo W., Tweedie D. Targeting TNF-α to elucidate and ameliorate neuroinflammation in neurodegenerative diseases. CNS Neurol. Disord. Drug Targets. 2011;10:391–403. doi: 10.2174/187152711794653751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund A., Orjalo A.V., Desprez P.Y., Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol. Med. 2010;16:238–246. doi: 10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs J., Hölzer M., Schilling M., Patzina C., Schoen A., Hoenen T., Zimmer G., Marz M., Weber F., Müller M.A., Kochs G. Evolution and antiviral specificities of interferon-induced Mx proteins of bats against Ebola, influenza, and other RNA viruses. J. Virol. 2017;91:e00361-17. doi: 10.1128/JVI.00361-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann-Stroissnigg H., Niedernhofer L.J., Robbins P.D. Hsp90 inhibitors as senolytic drugs to extend healthy aging. Cell Cycle. 2018;17:1048–1055. doi: 10.1080/15384101.2018.1475828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glück S., Guey B., Gulen M.F., Wolter K., Kang T.W., Schmacke N.A., Bridgeman A., Rehwinkel J., Zender L., Ablasser A. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 2017;19:1061–1070. doi: 10.1038/ncb3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V., Seluanov A., Mao Z., Hine C. Changes in DNA repair during aging. Nucleic Acids Res. 2007;35:7466–7474. doi: 10.1093/nar/gkm756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig N.H., Mattson M.P., Perry T., Chan S.L., Giordano T., Sambamurti K., Rogers J.T., Ovadia H., Lahiri D.K. New therapeutic strategies and drug candidates for neurodegenerative diseases: p53 and TNF-alpha inhibitors, and GLP-1 receptor agonists. Ann. N Y Acad. Sci. 2004;1035:290–315. doi: 10.1196/annals.1332.018. [DOI] [PubMed] [Google Scholar]
- Guevara-Aguirre J., Balasubramanian P., Guevara-Aguirre M., Wei M., Madia F., Cheng C.W., Hwang D., Martin-Montalvo A., Saavedra J., Ingles S., et al. Growth hormone receptor deficiency is associated with a major reduction in pro-aging signaling, cancer, and diabetes in humans. Sci. Transl. Med. 2011;3:70ra13. doi: 10.1126/scitranslmed.3001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Callaway J.B., Ting J.P. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat. Med. 2015;21:677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag S.M., Gulen M.F., Reymond L., Gibelin A., Abrami L., Decout A., Heymann M., van der Goot F.G., Turcatti G., Behrendt R., Ablasser A. Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559:269–273. doi: 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
- Halpin K., Hyatt A.D., Fogarty R., Middleton D., Bingham J., Epstein J.H., Rahman S.A., Hughes T., Smith C., Field H.E., Daszak P., Henipavirus Ecology Research Group Pteropid bats are confirmed as the reservoir hosts of henipaviruses: a comprehensive experimental study of virus transmission. Am. J. Trop. Med. Hyg. 2011;85:946–951. doi: 10.4269/ajtmh.2011.10-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann L., Ruiz-Moreno J.S., Szwed M., Mossakowska M., Lundvall L., Schumann R.R., Opitz B., Puzianowska-Kuznicka M. STING SNP R293Q is associated with a decreased risk of aging-related diseases. Gerontology. 2019;65:145–154. doi: 10.1159/000492972. [DOI] [PubMed] [Google Scholar]
- Hamann L., Szwed M., Mossakowska M., Chudek J., Puzianowska-Kuznicka M. First evidence for STING SNP R293Q being protective regarding obesity-associated cardiovascular disease in age-advanced subjects - a cohort study. Immun. Ageing. 2020;17:7. doi: 10.1186/s12979-020-00176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman D.T., Bowen R.A., Cryan P.M., McCracken G.F., O’Shea T.J., Peel A.J., Gilbert A., Webb C.T., Wood J.L. Ecology of zoonotic infectious diseases in bats: current knowledge and future directions. Zoonoses Public Health. 2013;60:2–21. doi: 10.1111/zph.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Varadarajan S., Muñoz-Planillo R., Burberry A., Nakamura Y., Núñez G. 3,4-methylenedioxy-β-nitrostyrene inhibits NLRP3 inflammasome activation by blocking assembly of the inflammasome. J. Biol. Chem. 2014;289:1142–1150. doi: 10.1074/jbc.M113.515080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy K., Guillerme T., Finlay S., Kane A., Kelly S.B., McClean D., Kelly D.J., Donohue I., Jackson A.L., Cooper N. Ecology and mode-of-life explain lifespan variation in birds and mammals. Proc. Biol. Sci. 2014;281:20140298. doi: 10.1098/rspb.2014.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D.J., Austad S.N. Fly now, die later: life-history correlates of gliding and flying in mammals. J. Mammal. 1994;75:224–226. [Google Scholar]
- Hotamisligil G.S., Shargill N.S., Spiegelman B.M. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- Hu B., Jin C., Li H.B., Tong J., Ouyang X., Cetinbas N.M., Zhu S., Strowig T., Lam F.C., Zhao C., et al. The DNA-sensing AIM2 inflammasome controls radiation-induced cell death and tissue injury. Science. 2016;354:765–768. doi: 10.1126/science.aaf7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Jebb D., Teeling E.C. Blood miRNomes and transcriptomes reveal novel longevity mechanisms in the long-lived bat, Myotis myotis. BMC Genomics. 2016;17:906. doi: 10.1186/s12864-016-3227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Whelan C.V., Foley N.M., Jebb D., Touzalin F., Petit E.J., Puechmaille S.J., Teeling E.C. Longitudinal comparative transcriptomics reveals unique mechanisms underlying extended healthspan in bats. Nat. Ecol. Evol. 2019;3:1110–1120. doi: 10.1038/s41559-019-0913-3. [DOI] [PubMed] [Google Scholar]
- Hughes G.M., Leech J., Puechmaille S.J., Lopez J.V., Teeling E.C. Is there a link between aging and microbiome diversity in exceptional mammalian longevity? PeerJ. 2018;6:e4174. doi: 10.7717/peerj.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittaman S.V., VanWormer J.J., Rezkalla S.H. The role of aspirin in the prevention of cardiovascular disease. Clin. Med. Res. 2014;12:147–154. doi: 10.3121/cmr.2013.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jebb D., Foley N.M., Whelan C.V., Touzalin F., Puechmaille S.J., Teeling E.C. Population level mitogenomics of long-lived bats reveals dynamic heteroplasmy and challenges the free radical theory of ageing. Sci. Rep. 2018;8:13634. doi: 10.1038/s41598-018-31093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon O.H., Kim C., Laberge R.M., Demaria M., Rathod S., Vasserot A.P., Chung J.W., Kim D.H., Poon Y., David N., et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat. Med. 2017;23:775–781. doi: 10.1038/nm.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., He H., Chen Y., Huang W., Cheng J., Ye J., Wang A., Tao J., Wang C., Liu Q., et al. Identification of a selective and direct NLRP3 inhibitor to treat inflammatory disorders. J. Exp. Med. 2017;214:3219–3238. doi: 10.1084/jem.20171419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacprzyk J., Hughes G.M., Palsson-McDermott E.M., Quinn S.R., Puechmaille S.J., O’Neill L.A.J., Teeling E.C. A potent anti-inflammatory response in bat macrophages may be linked to extended longevity and viral tolerance. Acta Chiropt. 2017;19:219–228. [Google Scholar]
- Kennedy B.K., Berger S.L., Brunet A., Campisi J., Cuervo A.M., Epel E.S., Franceschi C., Lithgow G.J., Morimoto R.I., Pessin J.E., et al. Geroscience: linking aging to chronic disease. Cell. 2014;159:709–713. doi: 10.1016/j.cell.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Jazwinski S.M. The gut microbiota and healthy aging: a mini-review. Gerontology. 2018;64:513–520. doi: 10.1159/000490615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.S., Lee C.K. Growth signaling and longevity in mouse models. BMB Rep. 2019;52:70–85. doi: 10.5483/BMBRep.2019.52.1.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland J.L., Tchkonia T. Cellular senescence: a translational perspective. EBioMedicine. 2017;21:21–28. doi: 10.1016/j.ebiom.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Kobayashi J., Saitoh T., Maruyama K., Ishii K.J., Barber G.N., Komatsu K., Akira S., Kawai T. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc. Natl. Acad. Sci. USA. 2013;110:2969–2974. doi: 10.1073/pnas.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreienkamp R., Graziano S., Coll-Bonfill N., Bedia-Diaz G., Cybulla E., Vindigni A., Dorsett D., Kubben N., Batista L.F.Z., Gonzalo S. A cell-intrinsic interferon-like response links replication stress to cellular aging caused by progerin. Cell Rep. 2018;22:2006–2015. doi: 10.1016/j.celrep.2018.01.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing E.D., Sterling S.L., Weir D.L., Beauregard C.R., Smith I.L., Larsen S.E., Wang L.F., Snow A.L., Schaefer B.C., Broder C.C. Enhanced autophagy contributes to reduced viral infection in black flying fox cells. Viruses. 2019;11:260. doi: 10.3390/v11030260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lama L., Adura C., Xie W., Tomita D., Kamei T., Kuryavyi V., Gogakos T., Steinberg J.I., Miller M., Ramos-Espiritu L., et al. Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression. Nat. Commun. 2019;10:2261. doi: 10.1038/s41467-019-08620-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro B.P., Clark A. In: Rapidly Evolving Genes and Genetic Systems. Singh R.S., Xu J., Kulathinal R.J., editors. Oxford Scholarship Online; 2012. Rapid evolution of innate immune response genes. [Google Scholar]
- Lee H.M., Kim J.J., Kim H.J., Shong M., Ku B.J., Jo E.K. Upregulated NLRP3 inflammasome activation in patients with type 2 diabetes. Diabetes. 2013;62:194–204. doi: 10.2337/db12-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li S., Hong Z., Wang Z., Li F., Mei J., Huang L., Lou X., Zhao S., Song L., Chen W., et al. The cyclopeptide astin C specifically inhibits the innate immune CDN sensor STING. Cell Rep. 2018;25:3405–3421.e7. doi: 10.1016/j.celrep.2018.11.097. [DOI] [PubMed] [Google Scholar]
- Liu J.L., Coschigano K.T., Robertson K., Lipsett M., Guo Y., Kopchick J.J., Kumar U., Liu Y.L. Disruption of growth hormone receptor gene causes diminished pancreatic islet size and increased insulin sensitivity in mice. Am. J. Physiol. Endocrinol. Metab. 2004;287:E405–E413. doi: 10.1152/ajpendo.00423.2003. [DOI] [PubMed] [Google Scholar]
- López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine J.J., Boyles J.G. Bats initiate vital agroecological interactions in corn. Proc. Natl. Acad. Sci. USA. 2015;112:12438–12443. doi: 10.1073/pnas.1505413112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malemud C.J., Islam N., Haqqi T.M. Pathophysiological mechanisms in osteoarthritis lead to novel therapeutic strategies. Cells Tissues Organs. 2003;174:34–48. doi: 10.1159/000070573. [DOI] [PubMed] [Google Scholar]
- McCracken G.F., Safi K., Kunz T.H., Dechmann D.K., Swartz S.M., Wikelski M. Airplane tracking documents the fastest flight speeds recorded for bats. R. Soc. Open Sci. 2016;3:160398. doi: 10.1098/rsos.160398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil J.J., Wolfe R., Woods R.L., Tonkin A.M., Donnan G.A., Nelson M.R., Reid C.M., Lockery J.E., Kirpach B., Storey E., et al. ASPREE Investigator Group Effect of aspirin on cardiovascular events and bleeding in the healthy elderly. N. Engl. J. Med. 2018;379:1509–1518. doi: 10.1056/NEJMoa1805819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton D.J., Morrissy C.J., van der Heide B.M., Russell G.M., Braun M.A., Westbury H.A., Halpin K., Daniels P.W. Experimental Nipah virus infection in pteropid bats (Pteropus poliocephalus) J. Comp. Pathol. 2007;136:266–272. doi: 10.1016/j.jcpa.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Monaco C., Nanchahal J., Taylor P., Feldmann M. Anti-TNF therapy: past, present and future. Int. Immunol. 2015;27:55–62. doi: 10.1093/intimm/dxu102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Adney D.R., van Doremalen N., Brown V.R., Miazgowicz K.L., Milne-Price S., Bushmaker T., Rosenke R., Scott D., Hawkinson A., et al. Replication and shedding of MERS-CoV in Jamaican fruit bats (Artibeus jamaicensis) Sci. Rep. 2016;6:21878. doi: 10.1038/srep21878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruve D.A., Pétrilli V., Zaiss A.K., White L.R., Clark S.A., Ross P.J., Parks R.J., Tschopp J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- Nazmi A., Field R.H., Griffin E.W., Haugh O., Hennessy E., Cox D., Reis R., Tortorelli L., Murray C.L., Lopez-Rodriguez A.B., et al. Chronic neurodegeneration induces type I interferon synthesis via STING, shaping microglial phenotype and accelerating disease progression. Glia. 2019;67:1254–1276. doi: 10.1002/glia.23592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea T.J., Cryan P.M., Cunningham A.A., Fooks A.R., Hayman D.T., Luis A.D., Peel A.J., Plowright R.K., Wood J.L. Bat flight and zoonotic viruses. Emerg. Infect. Dis. 2014;20:741–745. doi: 10.3201/eid2005.130539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paweska J.T., Storm N., Grobbelaar A.A., Markotter W., Kemp A., Jansen van Vuren P. Experimental inoculation of Egyptian fruit bats (Rousettus aegyptiacus) with Ebola virus. Viruses. 2016;8:29. doi: 10.3390/v8020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piscianz E., Cuzzoni E., Sharma R., Tesser A., Sapra P., Tommasini A. Reappraisal of antimalarials in interferonopathies: new perspectives for old drugs. Curr. Med. Chem. 2018;25:2797–2810. doi: 10.2174/0929867324666170911162331. [DOI] [PubMed] [Google Scholar]
- Podlutsky A.J., Khritankov A.M., Ovodov N.D., Austad S.N. A new field record for bat longevity. J. Gerontol. A Biol. Sci. Med. Sci. 2005;60:1366–1368. doi: 10.1093/gerona/60.11.1366. [DOI] [PubMed] [Google Scholar]
- Pride H., Yu Z., Sunchu B., Mochnick J., Coles A., Zhang Y., Buffenstein R., Hornsby P.J., Austad S.N., Pérez V.I. Long-lived species have improved proteostasis compared to phylogenetically-related shorter-lived species. Biochem. Biophys. Res. Commun. 2015;457:669–675. doi: 10.1016/j.bbrc.2015.01.046. [DOI] [PubMed] [Google Scholar]
- Pritham E.J., Feschotte C. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc. Natl. Acad. Sci. USA. 2007;104:1895–1900. doi: 10.1073/pnas.0609601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam D.A., Bhattacharya A., Van Remmen H. Mitochondrial dysfunction in aging and longevity: a causal or protective role? Antioxid. Redox Signal. 2013;19:1373–1387. doi: 10.1089/ars.2012.4950. [DOI] [PubMed] [Google Scholar]
- Racey P.A., Entwhistle A.C. In: Reproductive Biology of Bats. Krutzsch P.H., Crichton E.G., editors. Academic Press; 1999. Life history and reproductive strategies of bats. [Google Scholar]
- Riley J.S., Tait S.W. Mitochondrial DNA in inflammation and immunity. EMBO Rep. 2020;21:e49799. doi: 10.15252/embr.201949799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S., Rottach A., Lotz-Havla A.S., Laux V., Muschaweckh A., Gersting S.W., Muntau A.C., Hopfner K.P., Jin L., Vanness K., et al. Rad50-CARD9 interactions link cytosolic DNA sensing to IL-1β production. Nat. Immunol. 2014;15:538–545. doi: 10.1038/ni.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Panigrahi D.P., Patil S., Bhutia S.K. Autophagy in health and disease: a comprehensive review. Biomed. Pharmacother. 2018;104:485–495. doi: 10.1016/j.biopha.2018.05.007. [DOI] [PubMed] [Google Scholar]
- Salmon A.B., Leonard S., Masamsetti V., Pierce A., Podlutsky A.J., Podlutskaya N., Richardson A., Austad S.N., Chaudhuri A.R. The long lifespan of two bat species is correlated with resistance to protein oxidation and enhanced protein homeostasis. FASEB J. 2009;23:2317–2326. doi: 10.1096/fj.08-122523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattgen S.A., Fitzgerald K.A. The PYHIN protein family as mediators of host defenses. Immunol. Rev. 2011;243:109–118. doi: 10.1111/j.1600-065X.2011.01053.x. [DOI] [PubMed] [Google Scholar]
- Schuh A.J., Amman B.R., Sealy T.K., Spengler J.R., Nichol S.T., Towner J.S. Egyptian rousette bats maintain long-term protective immunity against Marburg virus infection despite diminished antibody levels. Sci. Rep. 2017;7:8763. doi: 10.1038/s41598-017-07824-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seim I., Fang X., Xiong Z., Lobanov A.V., Huang Z., Ma S., Feng Y., Turanov A.A., Zhu Y., Lenz T.L., et al. Genome analysis reveals insights into physiology and longevity of the Brandt’s bat Myotis brandtii. Nat. Commun. 2013;4:2212. doi: 10.1038/ncomms3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C. Drug developers switch gears to inhibit STING. Nat. Biotechnol. 2019;37:199–201. doi: 10.1038/s41587-019-0060-z. [DOI] [PubMed] [Google Scholar]
- Simon M., Van Meter M., Ablaeva J., Ke Z., Gonzalez R.S., Taguchi T., De Cecco M., Leonova K.I., Kogan V., Helfand S.L., et al. LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab. 2019;29:871–885.e5. doi: 10.1016/j.cmet.2019.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliter D.A., Martinez J., Hao L., Chen X., Sun N., Fischer T.D., Burman J.L., Li Y., Zhang Z., Narendra D.P., et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature. 2018;561:258–262. doi: 10.1038/s41586-018-0448-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Song X., Ma F., Herrup K. Accumulation of cytoplasmic DNA due to ATM deficiency activates the microglial viral response system with neurotoxic consequences. J. Neurosci. 2019;39:6378–6394. doi: 10.1523/JNEUROSCI.0774-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman J.R. The physiological costs of reproduction in small mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363:375–398. doi: 10.1098/rstb.2007.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stienstra R., van Diepen J.A., Tack C.J., Zaki M.H., van de Veerdonk F.L., Perera D., Neale G.A., Hooiveld G.J., Hijmans A., Vroegrijk I., et al. Inflammasome is a central player in the induction of obesity and insulin resistance. Proc. Natl. Acad. Sci. USA. 2011;108:15324–15329. doi: 10.1073/pnas.1100255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong R., Miller R.A., Astle C.M., Floyd R.A., Flurkey K., Hensley K.L., Javors M.A., Leeuwenburgh C., Nelson J.F., Ongini E., et al. Nordihydroguaiaretic acid and aspirin increase lifespan of genetically heterogeneous male mice. Aging Cell. 2008;7:641–650. doi: 10.1111/j.1474-9726.2008.00414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell W.R. Gene expression profiling of long-lived dwarf mice: longevity-associated genes and relationships with diet, gender and aging. BMC Genomics. 2007;8:353. doi: 10.1186/1471-2164-8-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacutu R., Thornton D., Johnson E., Budovsky A., Barardo D., Craig T., Diana E., Lehmann G., Toren D., Wang J., et al. Human ageing genomic resources: new and updated databases. Nucleic Acids Res. 2018;46(D1):D1083–D1090. doi: 10.1093/nar/gkx1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam L.S., Kitas G.D., González-Gay M.A. Can suppression of inflammation by anti-TNF prevent progression of subclinical atherosclerosis in inflammatory arthritis? Rheumatology (Oxford) 2014;53:1108–1119. doi: 10.1093/rheumatology/ket454. [DOI] [PubMed] [Google Scholar]
- Tian X., Firsanov D., Zhang Z., Cheng Y., Luo L., Tombline G., Tan R., Simon M., Henderson S., Steffan J., et al. SIRT6 is responsible for more efficient DNA double-strand break repair in long-lived species. Cell. 2019;177:622–638.e22. doi: 10.1016/j.cell.2019.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumurkhuu G., Shimada K., Dagvadorj J., Crother T.R., Zhang W., Luthringer D., Gottlieb R.A., Chen S., Arditi M. Ogg1-dependent DNA repair regulates NLRP3 inflammasome and prevents atherosclerosis. Circ. Res. 2016;119:e76–e90. doi: 10.1161/CIRCRESAHA.116.308362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turbill C., Bieber C., Ruf T. Hibernation is associated with increased survival and the evolution of slow life histories among mammals. Proc. Biol. Sci. 2011;278:3355–3363. doi: 10.1098/rspb.2011.0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turbill C., Prior S. Thermal climate-linked variation in annual survival rate of hibernating rodents: shorter winter dormancy and lower survival in warmer climates. Funct. Ecol. 2016;30:1366–1372. [Google Scholar]
- Vandanmagsar B., Youm Y.H., Ravussin A., Galgani J.E., Stadler K., Mynatt R.L., Ravussin E., Stephens J.M., Dixit V.D. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat. Med. 2011;17:179–188. doi: 10.1038/nm.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanpouille-Box C., Hoffmann J.A., Galluzzi L. Pharmacological modulation of nucleic acid sensors - therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 2019;18:845–867. doi: 10.1038/s41573-019-0043-2. [DOI] [PubMed] [Google Scholar]
- Volkman H.E., Stetson D.B. The enemy within: endogenous retroelements and autoimmune disease. Nat. Immunol. 2014;15:415–422. doi: 10.1038/ni.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyssokikh M.Y., Holtze S., Averina O.A., Lyamzaev K.G., Panteleeva A.A., Marey M.V., Zinovkin R.A., Severin F.F., Skulachev M.V., Fasel N., et al. Mild depolarization of the inner mitochondrial membrane is a crucial component of an anti-aging program. Proc. Natl. Acad. Sci. USA. 2020;117:6491–6501. doi: 10.1073/pnas.1916414117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H., Gris D., Lei Y., Jha S., Zhang L., Huang M.T., Brickey W.J., Ting J.P. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat. Immunol. 2011;12:408–415. doi: 10.1038/ni.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.P., Shadel G.S. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol. 2017;17:363–375. doi: 10.1038/nri.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook A.M., Schiestl R.H. Atm-deficient mice exhibit increased sensitivity to dextran sulfate sodium-induced colitis characterized by elevated DNA damage and persistent immune activation. Cancer Res. 2010;70:1875–1884. doi: 10.1158/0008-5472.CAN-09-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson G.S., Adams D.M. Recurrent evolution of extreme longevity in bats. Biol. Lett. 2019;15:20180860. doi: 10.1098/rsbl.2018.0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson G.S., South J.M. Life history, ecology and longevity in bats. Aging Cell. 2002;1:124–131. doi: 10.1046/j.1474-9728.2002.00020.x. [DOI] [PubMed] [Google Scholar]
- Wissler Gerdes E.O., Zhu Y., Tchkonia T., Kirkland J.L. Discovery, development, and future application of senolytics: theories and predictions. FEBS J. 2020;287:2418–2427. doi: 10.1111/febs.15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Li Y., Shen X., Goh G., Zhu Y., Cui J., Wang L.F., Shi Z.L., Zhou P. Dampened STING-dependent interferon activation in bats. Cell Host Microbe. 2018;23:297–301.e4. doi: 10.1016/j.chom.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Pirtskhalava T., Farr J.N., Weigand B.M., Palmer A.K., Weivoda M.M., Inman C.L., Ogrodnik M.B., Hachfeld C.M., Fraser D.G., et al. Senolytics improve physical function and increase lifespan in old age. Nat. Med. 2018;24:1246–1256. doi: 10.1038/s41591-018-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Jiang W., Liu L., Wang X., Ding C., Tian Z., Zhou R. Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell. 2015;160:62–73. doi: 10.1016/j.cell.2014.11.047. [DOI] [PubMed] [Google Scholar]
- Yang H., Wang H., Ren J., Chen Q., Chen Z.J. cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. USA. 2017;114:E4612–E4620. doi: 10.1073/pnas.1705499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.M., Yang S., Huang S.S., Tang B.S., Guo J.F. Microglial activation in the pathogenesis of Huntington’s disease. Front. Aging Neurosci. 2017;9:193. doi: 10.3389/fnagi.2017.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youm Y.H., Grant R.W., McCabe L.R., Albarado D.C., Nguyen K.Y., Ravussin A., Pistell P., Newman S., Carter R., Laque A., et al. Canonical Nlrp3 inflammasome links systemic low-grade inflammation to functional decline in aging. Cell Metab. 2013;18:519–532. doi: 10.1016/j.cmet.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefzadeh M.J., Zhu Y., McGowan S.J., Angelini L., Fuhrmann-Stroissnigg H., Xu M., Ling Y.Y., Melos K.I., Pirtskhalava T., Inman C.L., et al. Fisetin is a senotherapeutic that extends health and lifespan. EBioMedicine. 2018;36:18–28. doi: 10.1016/j.ebiom.2018.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahid A., Li B., Kombe A.J.K., Jin T., Tao J. Pharmacological inhibitors of the NLRP3 inflammasome. Front. Immunol. 2019;10:2538. doi: 10.3389/fimmu.2019.02538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Park Y., Wu J., Chen X.p., Lee S., Yang J., Dellsperger K.C., Zhang C. Role of TNF-alpha in vascular dysfunction. Clin. Sci. (Lond.) 2009;116:219–230. doi: 10.1042/CS20080196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Cowled C., Shi Z., Huang Z., Bishop-Lilly K.A., Fang X., Wynne J.W., Xiong Z., Baker M.L., Zhao W., et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Cowled C., Mansell A., Monaghan P., Green D., Wu L., Shi Z., Wang L.F., Baker M.L. IRF7 in the Australian black flying fox, Pteropus alecto: evidence for a unique expression pattern and functional conservation. PLoS One. 2014;9:e103875. doi: 10.1371/journal.pone.0103875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Tachedjian M., Wynne J.W., Boyd V., Cui J., Smith I., Cowled C., Ng J.H., Mok L., Michalski W.P., et al. Contraction of the type I IFN locus and unusual constitutive expression of IFN-α in bats. Proc. Natl. Acad. Sci. USA. 2016;113:2696–2701. doi: 10.1073/pnas.1518240113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]