Abstract
COVID-19 pandemic has reshaped our world in a timescale much shorter than what we can understand. Particularities of SARS-CoV-2, such as its persistence in surfaces and the lack of a curative treatment or vaccine against COVID-19, have pushed authorities to apply restrictive policies to control its spreading. As data drove most of the decisions made in this global contingency, their quality is a critical variable for decision-making actors, and therefore should be carefully curated. In this work, we analyze the sources of error in typically reported epidemiological variables and usual tests used for diagnosis, and their impact on our understanding of COVID-19 spreading dynamics. We address the existence of different delays in the report of new cases, induced by the incubation time of the virus and testing-diagnosis time gaps, and other error sources related to the sensitivity/specificity of the tests used to diagnose COVID-19. Using a statistically-based algorithm, we perform a temporal reclassification of cases to avoid delay-induced errors, building up new epidemiologic curves centered in the day where the contagion effectively occurred. We also statistically enhance the robustness behind the discharge/recovery clinical criteria in the absence of a direct test, which is typically the case of non-first world countries, where the limited testing capabilities are fully dedicated to the evaluation of new cases. Finally, we applied our methodology to assess the evolution of the pandemic in Chile through the Effective Reproduction Number Rt, identifying different moments in which data was misleading governmental actions. In doing so, we aim to raise public awareness of the need for proper data reporting and processing protocols for epidemiological modelling and predictions.
Keywords: COVID-19, SARS-CoV-2, Public health, Statistics, ARIMA Models, Data analysis
1. Introduction
Since the outbreak of novel SARS-CoV-2 in late 2019, the spread of COVID-19 has changed nearly every aspect of our daily life, challenging modern society to find a way to function under conditions never seen before. Governmental plans on public health have played a crucial role in its control in the absence of an effective treatment to cure COVID-19 or a vaccine to prevent it. Typically reported variables in the COVID-19 pandemic, available in public repositories [as [13], [48], among others], are the incidence of new cases ΔT, total cases T, discharged/recovered cases R, and deaths D. These variables serve as input for the development and evaluation of governmental plans and to fit the vast variety of SIR-like mathematical models recently proposed [see, e.g., [1], [10], [37], [50], and references therein for a brief review of them]. Among the several factors conditioning the quality/reliability of the variables mentioned above are those related to the sensitivity and specificity of diagnostic tests, delays between sampling and diagnosing, and the delay between contagion, development of symptoms and testing. The latter varies from country to country, depending heavily on the local government’s testing strategy and resources. Even though SIR-like models have been reported to be not-suitable to predict turning points or any other quantitative insight on this pandemic [7], to the best knowledge of the authors, delays have not been pointed out as the cause, as raw data has always been used to fit them.
Different parameters can be used to evaluate the evolution of the SARS-CoV-2 outbreak. Among them we may find the documentation rate [46], secondary infection rate, serological response to infection [44], number of vacancies at ICU [28], and the Basic Reproduction Number R 0 [3], [16], which is one of the most widely used. This parameter (R 0) represents the number of persons a single infected individual might infect before either recovering or dying, when the population is entirely susceptible [38]. Traditional forms to estimate the R 0 are rather complex, heavily depending on the fitting of SIR models to local data [12], [19], [31], [42]. In a previous work [11], we proposed a methodology to obtain real-time estimations of the Effective Reproduction Number Rt directly from raw data, which was satisfactorily applied to evaluate the panorama of the COVID-19 spread in different countries and to forecast its evolution [32]. Nevertheless, its heavy dependence on reported data required the study of common error sources affecting this parameter, and the development of methodologies to control, correct, and quantify their impact [11].
In this work, we analyze the sources of error in the typically reported epidemiological variables and their impact on our understanding of COVID-19 spreading dynamics. We address the existence of different delays in the report of new cases, induced by the incubation time of the virus and testing-diagnosis time gaps, and provide a straightforward methodology to avoid the propagation of delay-induced errors to model-derived parameters. Using our statistically-based algorithm, we perform a temporal reclassification of individuals to the day where they were -statistically- most likely to have acquired the virus, building a new smooth curve with corrected variables. We postulate this new temporally modified curve as the proper one for fitting purposes in SIR-like models. We present an analogous methodology to estimate the number of discharged/recovered individuals, based on the reported evolution of the viral infection, the performance of the different tests for its diagnosis, and the case fatality, which can be easily adapted for a particular country. We used our methodology to assess the evolution of the pandemic in Chile, identifying different moments in which the use of raw data was misleading governmental actions.
2. On the performance of tests for diagnosing COVID-19
Different methods for diagnosing COVID-19 have been developed and reported in the literature, with real-time RT-PCR being the standard applied globally [29]. Nevertheless, techniques such as the IgG and IgM rapid tests, the Chest Computed Tomography (Chest CT), and CRISPR-Cas systems are also being used. In this section we provide a brief analysis of them, highlighting the different characteristics of both the techniques and their basic approach to detect viral infection.
2.1. Real-time RT-PCR
Real-time reverse transcription polymerase chain reaction (RT-PCR) is a mechanism for amplification and detection of RNA in real time [see 17, for an exhaustive description of the technique]. Initially, RNA obtained from samples is retrotranscribed to DNA using a reverse transcriptase enzyme. By applying temperature cycles, the conditions are created for new copies of the DNA to be synthesized from the initial one. The lower the initial DNA concentration, the lower the probability of a synthesis reaction in a given cycle. It is assumed that the minimum time from contagion until testing positive in the RT-PCR test is 2.3days (95% CI, 0.8-3.0days) before the onset of symptoms [18], which typically appear 5.2 days (95% CI, 4.1-7.0days) after contagion [22]. Ganyani et al. [15], He et al. [18] showed that 44% to 62% of the total contagions occur in the pre-symptomatic period. It has been inferred that the viral load reaches a peak value before 0.7 days from the onset of symptoms (95% CI, 0.2-2.0days), when it starts falling monotonically together with the infectivity rate [18]. Finally, the virus has been detected for a median of 20 days after the onset of symptoms [51], but infectivity may decrease significantly eight days after symptom appearance.
A high false-negative rate [26], [49] and a sensitivity of 71 to 83% [14], [30] have been reported for the real-time RT-PCR technique, and several vulnerabilities of it have been identified and quantified [29]. Considering sampling, handling, testing, and reporting, the total time necessary to get RT-PCR results for an individual may range between 2 to 3 days [35]. However, the time it takes to perform the RT-PCR experiment takes about 2 to 3 hours [27].
2.2. IgG And IgM rapid tests
Part of the immune system response to the SARS-CoV-2 infection is the production of specific antibodies against it, including IgG and IgM [25]. Serological tests detect the presence of those antibodies and, unlike the other detection methods, take only 15minutes to produce results [27]. These tests have a sensitivity of 88.6% and a specificity of 90.6% [27]. This technology was developed for the SARS-CoV epidemic, which was caused by a virus belonging to the same family of coronaviruses as SARS-CoV-2, providing satisfactory results after 2–3 days from the onset of symptoms (for IgG), and after eight days (for IgM) [47].
2.3. Chest computed tomography
The principle behind the Chest Computed Tomography (Chest CT) is the analysis of cross-sectional lung images to identify viral pneumonia features, like ground-glass opacity, consolidation, reticulation/thickened interlobular septa or nodules [2]. Chest CT has shown a sensitivity between 97% and 98% [14], [30]. However, due to the similarities between CT images accounting for COVID-19 and CT images for other viral types of pneumonia, false-positives are likely to occur. Compared to RT-PCR, Chest CT tends to be more reliable, practical, and quick to diagnose COVID-19 [2]. Nevertheless, requiring the presence of the potentially infected patient in a health center lacks the flexibility that rapid tests provide, and can backfire on movement restriction measures. COVID-19 pneumonia manifests with abnormalities on computed tomography images of the chest, even in asymptomatic patients [41].
2.4. CRISPR-Cas Systems
In CRISPR-Cas systems, a guide RNA (gRNA) is designed to recognize a specific RNA sequence, like any particular gene or partial ARN sequence of SARS-CoV-2 coronavirus. Endonuclease enzymes of the Cas family and the specific gRNA will search for the sequence match. This match will deliver a signal that confirms the presence of SARS-CoV-2 RNA in the sample [23]. DETECTR and SHERLOCK are two examples of CRISPR-Cas technologies for the detection of SARS-CoV-2, being able to obtain results in less than 1 h at a significantly lower cost compared to the RT-PCR technique. DETECTR showed a 95% positive predictive agreement and 100% negative predictive agreement [6], while SHERLOCK has not been validated using real patient samples and is not suitable for clinical use at this time [20].
3. Methodology
Our work aims to expose and quantify, both theoretically and in a case study, the impact of different sources of error in commonly reported data of the COVID-19 spread, such as newly reported cases ΔT, total cases T, infected I, and recovered R fractions of the population.
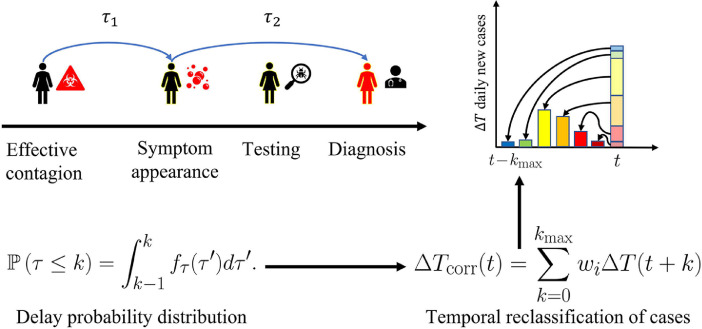
First, we define random variables associated with the delay in both sampling and diagnosing new cases, and by modeling their probability distribution functions, we derive a method to re-classify the newly reported cases accordingly. As reclassification occurs backwardly, for re-evaluating the current scenario through our methodology, we cast predictions on the reported new cases using an ARIMA (Autoregressive Integrated Moving Average) model. Having the corrected variables, we evaluate differences on the effective reproduction number Rt, following the methodology presented by [11]. Assuming the outbreak is well represented with a SIR model in a given timeframe i, Rt can be estimated as which can be directly estimated from the discrete version of the SIR differential equations. Decoupling the “recovered” fraction in the SIR model as clinically recovered individuals R and deaths D, Rt would be given by Eq. (1):
| (1) |
Given the existence of a latent and incubation period for COVID-19, which will be carefully described in the next section, SIR models might not capture all the features of the outbreak and SEIR models would be more suitable. In such models, only infected individuals can propagate the infection and, equation-wise, estimations of Rt remain more or less the same [9]. As data of exposed individuals is not widely available or entirely reliable, its contribution was not included in the estimations of Rt to keep its straightforward use.
Autoregression models for the forecast of ΔT were implemented using the statsmodels Python library [40]. All other calculations and visualizations were made using DMAKit-lib Python library [33] and MATLAB R2018a.
4. Results
4.1. Temporal misclassification of new cases
There exists an incubation period tc for the development of SARS-CoV-2-related symptoms, which average has been reported to range between 5.1 days [18], [21], [24] and 6.4 days [4]. He et al. [18] also reported the existence of a latency period, which is the time required before an infected individual could spread the infection, which was extended until 2.3 days (95% CI, 0.8-3.0days) before the onset of symptoms, which occurred tc days after contagion.
The incubation time tc is especially relevant in the case of a symptoms-based testing strategy or when the spread has reached the non-traceability stage. To model it, we use τ 1 with log-normal distribution, as reported in Lauer et al. [22] and suggested in Nishiura [36]:
| (2) |
According to the values reported in Lauer et al. [22], and retrieve the expected values of 5.1 for the median, and 11.5 for the98th percentile. Even though the required time for performing the test is short [27], delays between testing and diagnosis have been reported [35]. We will sum up secondary delays, such as the symptom-to-testing and testing-to-diagnosis time gaps, into a random variable τ 2, which, for the sake of simplicity, will be assumed to follow a uniform distribution between t minand t max:
| (3) |
Further knowledge on the nature of factors affecting τ 2 could lead to a different distribution. We may postulate a reclassification for obtaining the real number of new contagions occurred in a day t as the sum of contributions of cases reported with a delay of k days:
| (4) |
where wi represents the fraction of patients that were notified at time but had acquired the virus at t. Note that the different delays are referred to the random variable . The probability distribution function for Zis obtained by the convolution method, assuming τ 1 and τ 2 are independent and combining Eqs. (2) and (3):
| (5) |
Assuming that data is reported on a daily basis, we can calculate the probability associated to having a delay of k days:
| (6) |
For practical reasons, we can define a threshold U < 1 for truncating the probability mass distribution (Eq. 6), which otherwise would assign a probability to every . Let n 1be the first positive integer for which Eq. (7) holds,
| (7) |
we may rewrite Eq. (4) as:
| (8) |
The magnitude of the total delay between infection and diagnosis can be estimated through the expected value of Eq. (5) (or equivalently, Eq. (6)). A schematic representation of the proposed methodology is presented in Fig. 1 . Assuming the lowest reported value for the average incubation time, and a conservative timeframe for the delay between the appearance of symptoms, testing and diagnosing, (2–5 days), the expected delay is about 7.5days.
Fig. 1.
Schematic representation of the temporal reclassification methodology proposed herein.
4.2. Case discharge/recovery criteria
As discussed previously, errors in the amount of discharged/recovered patients R are likely to be greater only when no quantitative criteria are applied. In such cases, some countries (like Chile) have adopted the following criteria (officially reported in the [34] report), possibly based on the recommendations published by the WHO [43].
-
•
If there were no previously existing pathologies, a patient should be discharged 14 days after testing positive for COVID-19.
-
•
If there were previously existing pathologies, the patient should be discharged 28 days after testing positive for COVID-19.
This criterion turns out to be quite simplistic, especially considering the existence of uncertainties regarding the diagnosis and contagion days. If we try to model the probability of recovering from COVID-19, some assumptions are necessary. Let τr be the random variable for the time of discharge/recovery such that:
-
•
-
•
Further assumptions are necessary to estimate the probability distribution function as it depends on local diagnosis criteria, testing strategy, and the fraction of the population having preexisting pathologies. In particular, depending on the test applied for diagnosis and its sensitivity/specificity – which were carefully described in Section 2– the probability profile would change. The simplest form that can be assumed, and which, for clarity reasons, is adopted herein, is a triangular distribution:
| (9) |
Further knowledge on the nature of factors affecting τr could lead to a different distribution.
5. Case study: COVID-19 spreading dynamics in chile
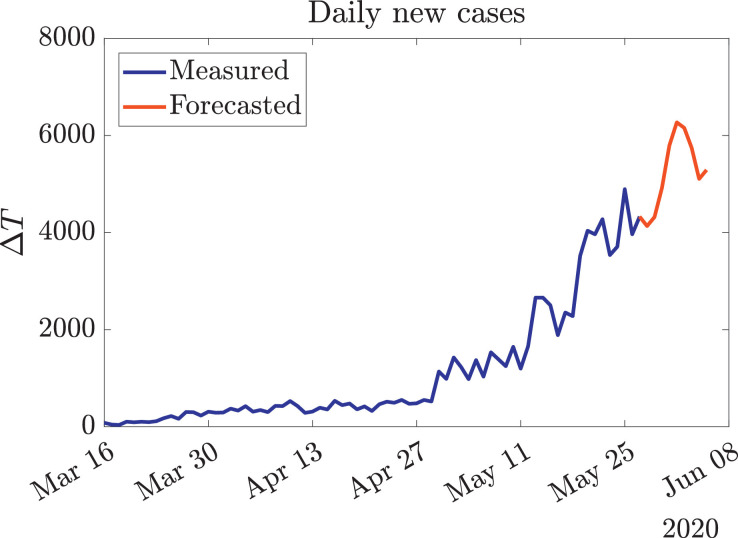
The spread of COVID-19 in Chile is far from being controlled, as shown by the exponential growth that new cases have had in recent weeks [48]. In order to apply our methodology, we need to cast predictions on the trends of ΔT. Fig. 2 presents the current and forecast trends, using an autoregression ARIMA model. These models were chosen over others because of the great success they have had on representing COVID-19 dynamics in other relevant studies [5], [8].
Fig. 2.
Current and forecasted evolution of ΔT in Chile. The official data (blue curve) was obtained from [48], while the red curve was generated using an autoregression ARIMA model. As these reported data are likely to be affected by several exogenous factors [39], the best performance metric for the generated forecast is trend consistency. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
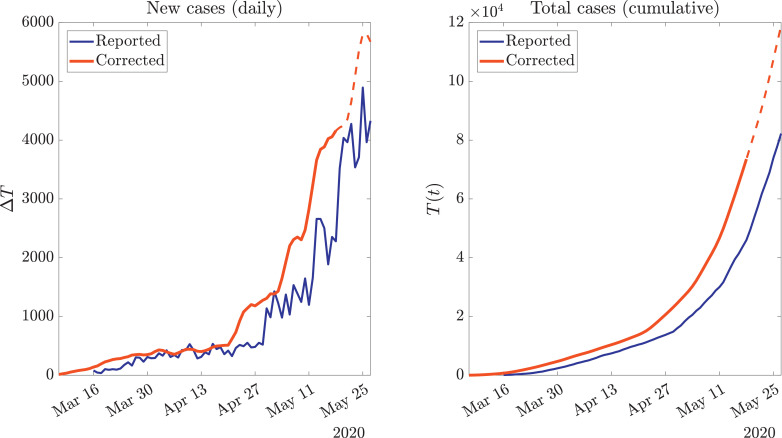
First, we performed a temporal reclassification of new cases to obtain ΔT corr, presented in Fig. 3 . It can be seen that our methodology, besides exhibiting an horizontal semi-displacement, generates a smooth curve, which is due to the inclusion of a probability function that spreads the influence of daily informed cases in previous days (where they were most likely to have occurred). The last part of the red curve is dashed because it partially contains contributions of the forecast of ΔT, and therefore might change in the upcoming days, when the required data for completing the reclassification of cases would be available.
Fig. 3.
Statistically-driven reclassification of new cases (red curve) compared with the raw data (blue curve), both differential a) and cumulative b). The last dashed part of the red curve accounts for the values which depends in the forecast shown in Fig. 2, and therefore might change over time. Official data obtained from [48]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
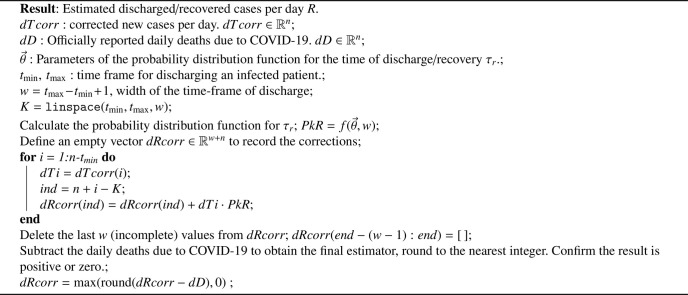
After obtaining statistically-based corrections of both ΔT and ΔR by following Algorithms 1 and 2 , and knowing the daily deaths due to COVID-19 ΔD, we proceed to calculate the variation on the active cases. Based on the closure of the population-balance of the different considered classes, the cumulative new cases T will be the sum of active I, recovered/discharged R, and dead D, therefore their differences would follow:
| (10) |
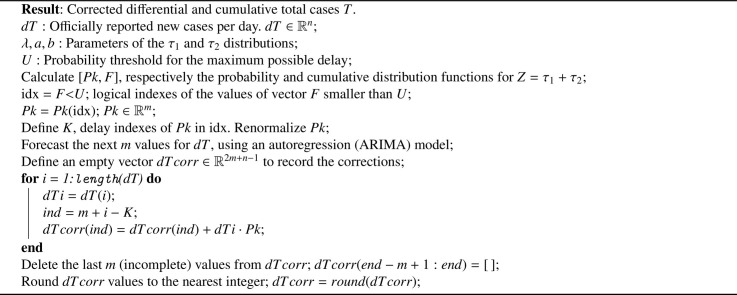
Algorithm 1.
Statistically-based temporal reclassification algorithm for correcting delay-induced errors in the report of new COVID-19 infections.
Algorithm 2.
Statistical estimation of dR, daily patients that have been discharged/recovered from COVID-19.
In the context of SEIR models, it is not possible to explicitly include exposed individuals E, as they are not being recorded separately from COVID-19 positive new cases.
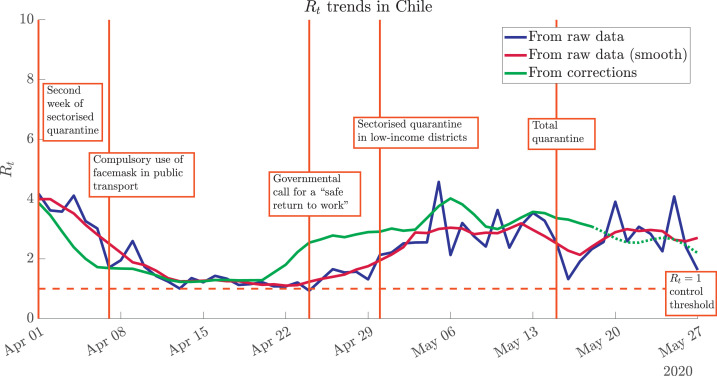
After obtaining ΔI using Eq. (10), we proceed to calculate Rt, using Eq. (1) with raw data, mobile averages of raw data, and the methodology proposed herein. As shown in Fig. 4 , an abrupt growth in Rt was evidenced around April 22nd, consistently with the relaxation of restrictive measures that were applied in Santiago, the capital and most populated city in the country, and the apogee of the governmental plan for a “safe return to work”. Even though different trends seem to stabilise again in the second week of May, a lowering of the trend is not totally clear.
Fig. 4.
Effect of data processing on the evaluation of the spread of COVID-19 through Rt. The noisy raw data (blue curve) can be smoothed through mobile averages (dark red curve), but the trends are the same. A significantly different scenario is shown by the statistically-corrected Rt trend (green curve). Highlighted dates associated with iconic governmental actions in Chile: March 31st (second week of sectorised quarantine for the high-income districts of Santiago, capital of Chile), April 7th (compulsory use of facemask in public transport), April 24th (governmental call for a “safe return to work”), April 30th (sectorised quarantine –low-income districts of Santiago–), May 15th (total quarantine in Santiago). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The different iconic dates highlighted in Fig. 4 were obtained from the chronology presented in [45] and references therein. From this plot, we can assess both successful and unfortunate effects that the different governmental actions have had on the spreading dynamics of COVID-19 in Chile, and specifically in Santiago, by analyzing corrected Rt trends and comparing them with daily reported cases. The various public-health actions were strongly based on locally-observed values from (raw) daily reported cases, yet appear to have been taken too late, or too early, according to the statistically-corrected trend. In particular, the apogee of the governmental plan for a safe return to work occurred in a temporal window where raw-data-driven Rt values were at a minimum, but the corrected contagion rates anticipated a steep growing trend for the following days.
6. Conclusions
We have presented an exhaustive assessment of error sources in reported data of the COVID-19 pandemic and provided a methodology to minimize –and correct– their effect on both reported variables and model-derived parameters by applying a statistically-driven reclassification of newly reported cases, and corrections to discharge/recovery criteria. By using the corrected variables, SIR-like models could be fitted directly, as every value would represent the real contagion dynamics. We present the methodology as a general framework, aiming to provide a useful tool for researchers and decision-making actors looking to adapt it for their particular interests.
In a case study on the spreading dynamics of COVID-19 in Chile, we observed the effects that different iconic public health actions taken by the government had on SARS-CoV-2 spread rate Rt, readily calculated using our previously reported approach, and discussed the reasons behind these effects interpreted under the light of raw data and corrected daily contagion rates, calculated with the methodology presented in this manuscript. The delay-induced error in raw data slowed-down the reaction time, so the actions were taken too late for restrictive actions or too early for the comeback to normality. Our statistically-driven method corrected such reporting errors, exposing the real contagion dynamics at a given time. The proposed methodology also serves as a non-invasive smoothing process, as it only temporally re-sorts daily reported cases according to their most likely report delay from the real contagion day.
We expect our methodology to serve as a valuable input for researchers and public health practitioners trying to add statistical value to their calculations and predictions in order to vanquish the current SARS-CoV-2 pandemic. Finally, we also expect this work to contribute in raising public awareness on the need for a proper (and standardized) strategy for report and curation of data in the COVID-19 pandemic and other highly-contagious diseases.
CRediT authorship contribution statement
Sebastián Contreras: Conceptualization, Methodology, Investigation, Writing - original draft, Visualization, Writing - review & editing, Supervision. Juan Pablo Biron-Lattes: Validation, Investigation, Writing - original draft, Writing - review & editing. H. Andrés Villavicencio: Conceptualization, Investigation, Writing - original draft, Supervision. David Medina-Ortiz: Validation, Writing - review & editing. Nyna Llanovarced-Kawles: Investigation, Writing - original draft. Álvaro Olivera-Nappa: Validation, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
The authors gratefully acknowledge support from the Chilean National Agency for Research and development through ANID PIA Grant AFB180004, and the Centre for Biotechnology and Bioengineering - CeBiB (PIA project FB0001, Conicyt, Chile). DM-O gratefully acknowledges Conicyt, Chile, for PhD fellowship 21181435.
References
- 1.Aguilar J.B., Faust J.S., Westafer L.M., Gutierrez J.B. Investigating the impact of asymptomatic carriers on covid-19 transmission. medRxiv. 2020 [Google Scholar]
- 2.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest ct and rt-pcr testing in coronavirus disease 2019 (covid-19) in china: a report of 1014 cases. Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allieta M., Allieta A., Sebastiano D.R. Covid-19 outbreak in italy: estimation of reproduction numbers over two months toward the phase 2. medRxiv. 2020 doi: 10.1007/s10389-021-01567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backer J., Klinkenberg D., Wallinga J. The incubation period of 2019-ncov infections among travellers from wuhan. China medRxiv. 2020 doi: 10.2807/1560-7917.ES.2020.25.5.2000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benvenuto D., Giovanetti M., Vassallo L., Angeletti S., Ciccozzi M. Application of the arima model on the covid-2019 epidemic dataset. Data Brief. 2020:105340. doi: 10.1016/j.dib.2020.105340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., et al. Crispr–cas12-based detection of sars-cov-2. Nat Biotechnol. 2020:1–5. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castro M., Ares S., Cuesta J.A., Manrubia S.. Predictability: can the turning point and end of an expanding epidemic be precisely forecast?2020. [DOI] [PMC free article] [PubMed]
- 8.Chakraborty T., Ghosh I. Real-time forecasts and risk assessment of novel coronavirus (covid-19) cases: a data-driven analysis. Chaos Soliton Fractals. 2020:109850. doi: 10.1016/j.chaos.2020.109850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowell G., Nishiura H., Bettencourt L.M. Comparative estimation of the reproduction number for pandemic influenza from daily case notification data. J R Soc Interface. 2007;4:155–166. doi: 10.1098/rsif.2006.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Contreras S., Villavicencio H.A., Medina-Ortiz D., Biron-Lattes J.P., Olivera-Nappa A. A multi-group seira model for the spread of covid-19 among heterogeneous populations. Chaos Soliton Fractal. 2020;136:109925. doi: 10.1016/j.chaos.2020.109925. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://www.sciencedirect.com/science/article/pii/S0960077920303246
- 11.Contreras S., Villavicencio H.A., Medina-Ortiz D., Saavedra C.P., Olivera-Nappa A. Real-time estimation of r0 for supporting public-health policies against covid-19. medRxiv. 2020 doi: 10.3389/fpubh.2020.556689. [DOI] [PMC free article] [PubMed] [Google Scholar]; https://www.medrxiv.org/content/early/2020/04/29/2020.04.23.20076984.full.pdf
- 12.Delamater P.L., Street E.J., Leslie T.F., Yang Y.T., Jacobsen K.H. Complexity of the basic reproduction number (r0) Emerging Infect Dis. 2019;25:1. doi: 10.3201/eid2501.171901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong E., Du H., Gardner L. An interactive web-based dashboard to track covid-19 in real time. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest ct for covid-19: comparison to rt-pcr. Radiology. 2020:200432. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganyani T., Kremer C., Chen D., Torneri A., Faes C., Wallinga J., Hens N. Estimating the generation interval for covid-19 based on symptom onset data. medRxiv. 2020 doi: 10.2807/1560-7917.ES.2020.25.29.2001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gevertz J., Greene J., Tapia C.H.S., Sontag E.D. A novel covid-19 epidemiological model with explicit susceptible and asymptomatic isolation compartments reveals unexpected consequences of timing social distancing. medRxiv. 2020 doi: 10.1016/j.jtbi.2020.110539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson U., Heid C.A., Williams P.M. A novel method for real time quantitative rt-pcr. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 18.He X., Lau E.H., Wu P., Deng X., Wang J., Hao X., et al. Temporal dynamics in viral shedding and transmissibility of covid-19. Nat Med. 2020:1–4. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 19.Heesterbeek J.A.P. A brief history of r 0 and a recipe for its calculation. Acta Biotheor. 2002;50:189–204. doi: 10.1023/a:1016599411804. [DOI] [PubMed] [Google Scholar]
- 20.Jin Y., Yang H., Ji W., Wu W., Chen S., Zhang W., Duan G. Virology, epidemiology, pathogenesis, and control of covid-19. Viruses. 2020;12:372. doi: 10.3390/v12040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lauer S.A., Grantz K.H. Qifang bi, forrest k jones, qulu zheng, hannah r meredith, andrew s azman, nicholas g reich, and justin lessler. the incubation period of coronavirus disease 2019 (covid-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;3 doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., et al. The incubation period of coronavirus disease 2019 (covid-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020 doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeMieux J. Covid-19 drives crispr diagnostics: crispr’s role in dna detection, not editing, may fill the gap in covid-19 testing. Genet Eng Biotechnol News. 2020;40:21–22. [Google Scholar]
- 24.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., et al. Early transmission dynamics in wuhan, china, of novel coronavirus–infected pneumonia. N top N Engl J Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of covid-19. J Pharm Anal. 2020 doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., Yao L., Li J., Chen L., Song Y., Cai Z., et al. Stability issues of rt-pcr testing of sars-cov-2 for hospitalized patients clinically diagnosed with covid-19. J Med Virol. 2020 doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., et al. Development and clinical application of a rapid igm-igg combined antibody test for sars-cov-2 infection diagnosis. J Med Virol. 2020 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liew M.F., Siow W.T., MacLaren G., See K.C. Preparing for covid-19: early experience from an intensive care unit in singapore. Critical Care. 2020;24:1–3. doi: 10.1186/s13054-020-2814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lippi G., Simundic A.M., Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (covid-19) Clin Chem Lab Med (CCLM) 2020;1 doi: 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 30.Long C., Xu H., Shen Q., Zhang X., Fan B., Wang C., et al. Diagnosis of the coronavirus disease (covid-19): rrt-pcr or ct? Eur J Radiol. 2020:108961. doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma J. Estimating epidemic exponential growth rate and basic reproduction number. Infect Dis Modell. 2020;5:129–141. doi: 10.1016/j.idm.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medina-Ortiz D., Contreras S., Barrera-Saavedra Y., Cabas-Mora G., Olivera-Nappa A. Country-wise forecast model for the Effective Reproduction Number Rt of COVID-19. Front Phys. 2020 [Google Scholar]; In press. https://www.frontiersin.org/articles/10.3389/fphy.2020.00304. doi:10.3389/fphy.2020.00304.
- 33.Medina-Ortiz D., Contreras S., Quiroz C., Asenjo J.A., Olivera-Nappa A. Dmakit: a user-friendly web platform for bringing state-of-the-art data analysis techniques to non-specific users. Inf Syst. 2020:101557. [Google Scholar]
- 34.MINSAL. Tech report: criteria for discharging a covid-19 infected individual (criterios que se consideran para un paciente covid-19 sin riesgo de contagio). 2020. https://www.minsal.cl/wp-content/uploads/2020/04/2020.04.13_ALTA-DE-CUARENTENA.pdf.
- 35.Nguyen T., Duong Bang D., Wolff A. 2019 Novel coronavirus disease (covid-19): paving the road for rapid detection and point-of-care diagnostics. Micromachines (Basel) 2020;11:306. doi: 10.3390/mi11030306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishiura H. Early efforts in modeling the incubation period of infectious diseases with an acute course of illness. Emerg Themes Epidemiol. 2007;4:2. doi: 10.1186/1742-7622-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pang W. Public health policy: covid-19 epidemic and seir model with asymptomatic viral carriers. arXiv preprint arXiv:200406311. 2020 [Google Scholar]
- 38.Perasso A. An introduction to the basic reproduction number in mathematical epidemiology. ESAIM. 2018;62:123–138. [Google Scholar]
- 39.Ribeiro M.H.D.M., da Silva R.G., Mariani V.C., dos Santos Coelho L. Short-term forecasting covid-19 cumulative confirmed cases: perspectives for brazil. Chaos Soliton Fractal. 2020:109853. doi: 10.1016/j.chaos.2020.109853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seabold S., Perktold J.. Statsmodels: econometric and statistical modeling with python. Scipy; 2010. Proceedings of the 9th Python in Science Conference 61
- 41.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with covid-19 pneumonia in wuhan, china: a descriptive study. Lancet Infectious Dis. 2020 doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., You X., Wang Y., Peng L., Du Z., Gilmour S., et al. [estimating the basic reproduction number of covid-19 in wuhan, china] Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41:476–479. doi: 10.3760/cma.j.cn112338-20200210-00086. [DOI] [PubMed] [Google Scholar]
- 43.WHO . Considerations for quarantine of individuals in the context of containment for coronavirus disease (COVID-19): interim guidance, 19 march 2020. World Health Organization.; 2020. Technical report. [Google Scholar]
- 44.WHO . Protocol for assessment of potential risk factors for coronavirus disease 2019 (COVID-19) among health workers in a health care setting, 23 march 2020. World Health Organization; 2020. Technical report. [Google Scholar]
- 45.Wikipedia.org. Cronología de la pandemia de enfermedad por coronavirus de 2020 en chile2020. https://es.wikipedia.org/wiki/Anexo:Cronolog%C3%ADa_de_la_pandemia_de_enfermedad_por_coronavirus_de_2020_en_Chile.
- 46.Wilder B., Charpignon M., Killian J.A., Ou H.C., Mate A., Jabbari S., et al. The role of age distribution and family structure on covid-19 dynamics: a preliminary modeling assessment for hubei and lombardy. Available at SSRN 3564800. 2020 [Google Scholar]
- 47.Woo P.C., Lau S.K., Wong B.H., Chan K.h., Chu C.m., Tsoi H.w., Huang Y., Peiris J.M., Yuen K.y. Longitudinal profile of immunoglobulin g (igg), igm, and iga antibodies against the severe acute respiratory syndrome (sars) coronavirus nucleocapsid protein in patients with pneumonia due to the sars coronavirus. Clin Diagn Lab Immunol. 2004;11:665–668. doi: 10.1128/CDLI.11.4.665-668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Worldometers.info. Official numbers for the coronavirus outbreak in chile. 2020. https://www.worldometers.info/coronavirus/.
- 49.Xiao A.T., Tong Y.X., Zhang S. False-negative of rt-pcr and prolonged nucleic acid conversion in covid-19: rather than recurrence. J Med Virol. 2020 doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Z., Zeng Z., Wang K., Wong S.S., Liang W., Zanin M., et al. Modified seir and ai prediction of the epidemics trend of covid-19 in china under public health interventions. J Thorac Dis. 2020;12:165. doi: 10.21037/jtd.2020.02.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with covid-19 in wuhan, china: a retrospective cohort study. The Lancet. 2020 doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]