(Current Biology 28, 2365–2376.e1–e5; August 6, 2018)
Main Text
In this article, we characterized roles for CLAVATA in the development of a moss, Physcomitrella patens, focusing on the 2D to 3D growth transition. Ongoing work to further characterize mutant phenotypes identified some phenotype discrepancies among the Ppclv1a and Ppclv1b mutant lines published in the original paper. For this reason, we implemented further checks of the published manuscript and fully sequenced both PpCLV1a and PpCLV1b loci in all mutants originally reported in the Methods S1 figure “CRISPR/Cas9 strategy for generating Ppclv1 mutants.” Although the conclusions of the paper remain valid, our investigations revealed errors that we wish to correct.
We found that Ppclv1a line 18 (Figures 4D and 4K; Methods S1, CRISPR figure, panel E) contained an 805 bp deletion at the PpCLV1a locus, but while Ppclv1a line 18 plants had phenotypes resembling WT plants, there was also a 9 bp deletion at PpCLV1b. We found no mutations at PpCLV1a or PpCLV1b in Ppclv1a line 29 or Ppclv1a line 32 (Methods S1, CRISPR figure, panels F and G), and these lines were indistinguishable from WT plants. The genotype of Ppclv1b line 2 (Figures 4E and 4L; Methods S1, CRISPR figure, panel H) was reported as a 2 bp deletion, but genome walking now confirms that there is a 4 bp deletion and >6 kb insertion at PpCLV1b, and the insertion comprises sequence integrated from the pACT::Cas9 expression vector used to engineer the lines [1]. The genotypes of Ppclv1b line 9 and Ppclv1b line 33 (Methods S1, CRISPR figure, panels I and J) were not previously reported, and while Ppclv1b line 9 has a 47 bp deletion at the PpCLV1b locus, Ppclv1b line 33 has no mutation at PpCLV1a or PpCLV1b, and an indistinguishable phenotype from WT plants. The reported genotypes of Ppclv1a1b lines 6, 8, and 12 were accurate (Methods S1, CRISPR figure, panels K–M).
Consequently, we have re-engineered and fully sequenced PpCLV1a and PpCLV1b in three independent Ppclv1a and Ppclv1b mutant lines to verify the mutant phenotypes reported in Figure 4. The Ppclv1a and Ppclv1b mutant phenotypes previously reported hold true, and the genotypes and phenotypes of lines now in use are shown in Figure 1 below.
As we checked our work, we noticed that the genotyping data shown in the Methods S1 figure “Strategy for generating Pprpk2 KO lines” (Methods S1, RPK2 figure, panels B–D) were from different lines to those shown in panel E, and we wish to substitute Figure 2 (below) for this panel.
At a late stage of manuscript preparation, the labels for PpCLV1a and PpCLV1b in Figure S5 and Tables S1 and S3 were transposed from labels elsewhere in the manuscript. The correct identifiers for these genes are as follows:
PpCLV1a Pp1s14_447V6.1 (V1.6 genome [2]) and Pp3c6_21940V1.1 (V3 genome [3])
PpCLV1b Pp1s5_68V6.1 (V1.6 genome [2]) and Pp3c13_13360V1.1 (V3 genome [3]).
The Lead Contact wishes to apologize to the scientific community for the errors above and any consequent confusion, and thanks Zoe Nemec Venza (University of Bristol), Joe Cammarata (Cornell University), and Wei Liu (University of Bristol) for doing the experimental work required for the correction. We also thank two anonymous referees for commenting on a draft correction notice.
Method Details
Genomic DNA Extraction
Genomic DNA was extracted using a CTAB (hexadecyltrimethylammonium bromide) protocol as described in the original paper.
Sequencing
PpCLV1a and PpCLV1b loci were Sanger sequenced using c. 150 ng template DNA with forward and reverse primers listed in Table 1, below. Sequence traces were analyzed by hand using Chromas Lite software, aligned using BioEdit software, and compared to genome sequence data from the V1.6 Physcomitrella genome [2].
Genome Walking for Ppclv1b Line 2
A genome walking protocol was adapted from [4]. Genomic DNA was extracted from WT and Ppclv1b line 2 plants as above, but with an additional phenol-chloroform-isoamyl alcohol (24:24:1) step following extraction. 1 μg DNA was digested to completion using EcoRV, PvuII, or HpaI and cleaned using a Zymoclean Gel DNA Recovery kit. 20 μL each of 50 mM GenomeWalkerAdaptor1 and GenomeWalkerAdaptor2 adaptors were mixed in water, heated to 100°C for 2 min, and annealed by cooling to room temperature. To generate genome walking libraries, digested genomic DNA was ligated to the annealed adaptors using T4 DNA ligase. To isolate the 3′ junction between PpCLV1b and insert sequence, two-step touchdown nested PCR was performed using AP1 + GSP1a primers, and then AP2 + GSP2b primers and Q5 polymerase. To isolate the 5′ junction between PpCLV1b and inserted sequence, an M13 primer against the inserted sequence was used with a PpCLV1b locus-specific primer, CLV1Blocus_reverse_8. Amplicons were gel purified using a QIAquick PCR Purification kit and sent for direct sequencing.
Moss Transformation
New Ppclv1a and Ppclv1b CRISPR lines were generated as described in the original paper except a single guide RNA expression vector, U3::Ppclv1a sgRNA7, was used to generate Ppclv1a crA and Ppclv1a crB lines.
Plant Imaging
Plant imaging was undertaken as described in the original paper.
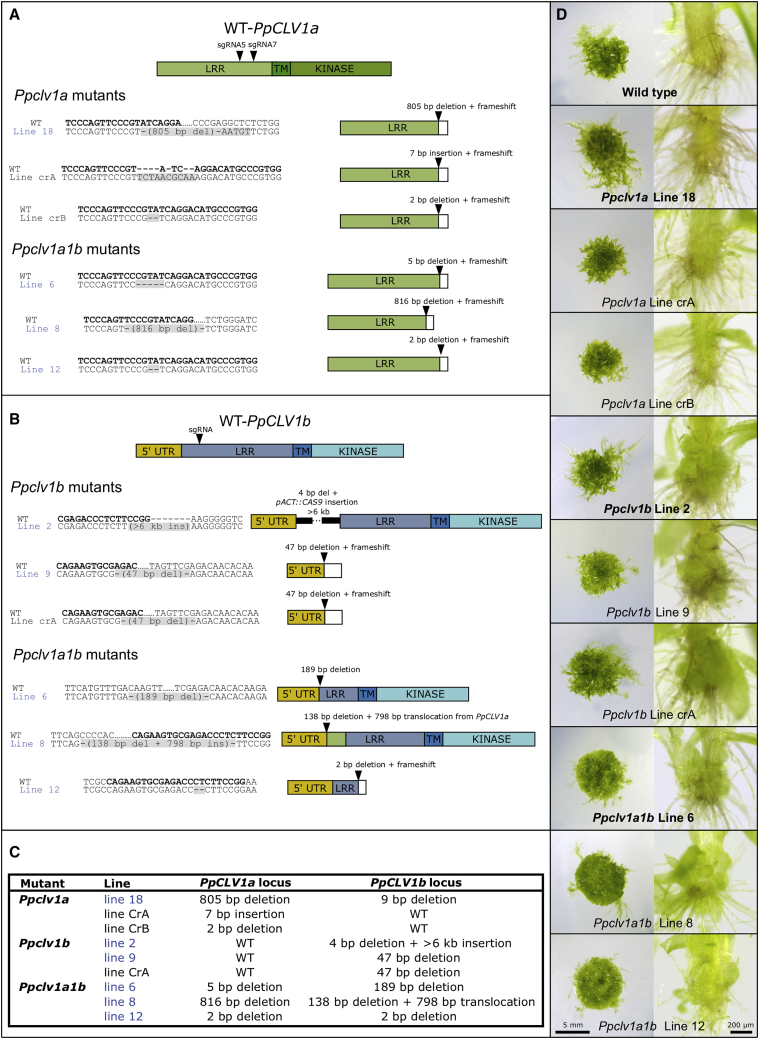
Figure 1.
CRISPR/Cas9 Strategy for Generating Ppclv1 Mutants
(A) Strategy for mutagenizing PpCLV1a to generate Ppclv1a single mutants and Ppclv1a1b double mutants. Two guide RNAs were designed against the LRR domain of PpCLV1a and cloned into pU3::Ppclv1a sgRNA5 and pU3::Ppclv1a sgRNA7 expression vectors (original paper). Plants were co-transformed with pU3::Ppclv1a sgRNA5, pU3::Ppclv1a sgRNA7, pNRF, and pACT::Cas9 (original paper), or with pU3::Ppclv1a sgRNA7, pNRF, and pACT::Cas9 for the Correction. Sanger sequencing of full-length PpCLV1a and PpCLV1b loci in all lines verified the mutations illustrated. Sequences in bold represent sgRNA target sites on WT loci, gray boxes highlight mutations in each line, and schematics to the right illustrate the predicted effect of mutations on the protein. Lines with labels in blue were reported in the original paper.
(B) Strategy for mutagenizing PpCLV1b to generate Ppclv1b single mutants and Ppclv1a1b double mutants. An sgRNA was designed against the LRR domain of PpCLV1b and expressed under the U6 promoter. Plants were co-transformed with pU6::Ppclv1b sgRNA, pNRF, and pACT::Cas9 (original paper). Sanger sequencing of full-length PpCLV1a and PpCLV1b loci in all lines verified mutations shown in three independently generated lines. Sequences in bold represent sgRNA target sites on WT loci, gray boxes highlight mutations arising, and schematics to the right illustrate the predicted effect of the mutation on the protein. Lines with labels in blue were reported in the original paper.
(C) Summary of PpCLV1 mutations at each locus in nine mutant lines currently in use. Lines with labels in blue were reported in the original paper.
(D) Previously described phenotypes of Ppclv1a, Ppclv1b, and Ppclv1a1b mutants are shared by three independently generated lines. Whereas WT plants, Ppclv1a, and Ppclv1b mutants have well-developed gametophores, Ppclv1a1b mutants have a defective 2D-3D growth transition. Ppclv1b and Ppclv1a1b, but not Ppclv1a mutants or WT plants, have tissue outgrowths at the base of gametophores. Lines with labels in bold were included in Figure 4 of the original paper.
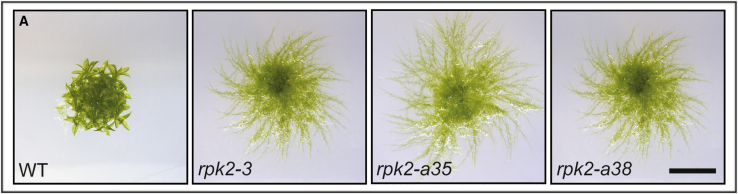
Figure 2.
Strategy for Generating Pprpk2 KO Lines
Multiple knockout lines whose genotyping data are shown in Methods S1 Pprpk2 figure had similar mutant phenotypes.
Table 1.
List of Genome Walking Primers Used to Prepare the Correction Notice
| Primer | Sequence |
|---|---|
| GenomeWalkerAdaptor1 | GTAATACGACTCACTATAGGGCAC GCGTGGTCGACGGCCCGGGCTGGT |
| GenomeWalkerAdaptor2 | 3′-H2N-CCCGACCA-PO4-5′ |
| GenomeWalkerAP1 | GTAATACGACTCACTATAGGGC |
| GenomeWalkerAP2 | ACTATAGGGCACGCGTGGT |
| GSP1a | CAGGAGGAATTGGTCCGGTAAGTGAGTT |
| GSP2b | GCTGGGAAAGATCCATACTGAGAAGGAAT |
| CLV1Blocus_reverse_8 | AAACTCAATCGTCGCAGTGC |
| M13fw | GTAAAACGACGGCCAG |
All primers in 5′-to-3′ orientation unless otherwise stated.
Table 2.
List of Sequencing Primers Used to Prepare the Correction Notice
| Primer | Sequence |
|---|---|
| CLV1Alocus_forward_1 | TGAGCCTGATTGAATCTTAACG |
| CLV1Alocus_forward_2 | AACTCGCTCTCAATGGGCCTCTTC |
| CLV1Alocus_forward_3 | ATCAATCGAATATGTCGTTCCG |
| CLV1Alocus_forward_4 | ATTCCCAGGCTGAGATGAATG |
| CLV1Alocus_forward_5 | ATAGGACCAGAGAGGTTGTTG |
| CLV1Alocus_forward_6 | GATTATCCTGGATCTCTACCATTG |
| CLV1Alocus_forward_7 | CGAGATGATTGTTCATCAAGCTC |
| CLV1Alocus_forward_8 | TCCTCCCAGACTTACGTGTTC |
| CLV1Alocus_reverse_1 | AAAGATGGAGTGCTGGACTTG |
| CLV1Alocus_reverse_2 | AATCCAGGCTGCACATGGTCTTTG |
| CLV1Alocus_reverse_3 | GGTACTTGACTGCTTGGACG |
| CLV1Alocus_reverse_4 | CCGCAATGATGGTGCTCCTTGTAG |
| CLV1Alocus_reverse_5 | ATGAGCGGGAACAATTTATCAG |
| CLV1Alocus_reverse_6 | ATCCTCTGAATCCAATGCCG |
| CLV1Alocus_reverse_7 | CTGGTTGGAGCAATCCCACATGAG |
| CLV1Alocus_reverse_8 | GATAACTTGTCTGAAGCCCATC |
| CLV1Alocus_reverse_9 | GGCCGAAGTGAGGTACATATTTAG |
| CLV1Blocus_forward_1 | CTGAGTGAGAAGAGTGACACATC |
| CLV1Blocus_forward_2 | ACGTCGAGTCTCTACGCAAC |
| CLV1Blocus_forward_3 | CCCTTTATACACAGTTCCAGC |
| CLV1Blocus_forward_4 | GTATGAGAAGTCGAATACGTTGAG |
| CLV1Blocus_forward_5 | GATCGACCCATTCAGAAGATTG |
| CLV1Blocus_forward_6 | TGCAGGAACATGGAGTCGAGATTG |
| CLV1Blocus_forward_7 | CGTGTGCATTACTTCCTGTGTTG |
| CLV1Blocus_forward_8 | TCTTCACCATTCTTGCTTCTG |
| CLV1Blocus_forward_9 | TGTTACTGCGAAGTGTGCTA |
| CLV1Blocus_reverse_1 | TTAGGGTGGTGCATGAACTGCTTG |
| CLV1Blocus_reverse_2 | CGGATTCTCAGCAGAGATTCAAAC |
| CLV1Blocus_reverse_3 | AATAACCTCTCAGGACCAATTCC |
| CLV1Blocus_reverse_4 | CGTTGGGTTTGCTTGGCTTG |
| CLV1Blocus_reverse_5 | ATGAACTCGTAGGTGTCATCCC |
| CLV1Blocus_reverse_6 | CAGGAAGTAATGCACACGCC |
| CLV1Blocus_reverse_7 | ACGCAACTTCCATATAGTCTCTGC |
| CLV1Blocus_reverse_7a | GGAGAGACGCAACTTCCATATAG |
| CLV1Blocus_reverse_8 | AAACTCAATCGTCGCAGTGC |
References
- 1.Lopez-Obando M., Hoffmann B., Géry C., Guyon-Debast A., Téoulé E., Rameau C., Bonhomme S., Nogué F. Simple and efficient targeting of multiple genes through CRISPR-Cas9 in Physcomitrella patens. G3 (Bethesda) 2016;6:3647–3653. doi: 10.1534/g3.116.033266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rensing S.A., Lang D., Zimmer A.D., Terry A., Salamov A., Shapiro H., Nishiyama T., Perroud P.-F., Lindquist E.A., Kamisugi Y. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 3.Lang D., Ullrich K.K., Murat F., Fuchs J., Jenkins J., Haas F.B., Piednoel M., Gundlach H., Van Bel M., Meyberg R. The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J. 2018;93:515–533. doi: 10.1111/tpj.13801. [DOI] [PubMed] [Google Scholar]
- 4.Siebert P.D., Chenchik A., Kellogg D.E., Lukyanov K.A., Lukyanov S.A. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]