Abstract
Positive-strand RNA viruses need to arrogate many cellular resources to support their replication and infection cycles. These viruses co-opt host factors, lipids and subcellular membranes and exploit cellular metabolites to built viral replication organelles in infected cells. However, the host cells have their defensive arsenal of factors to protect themselves from easy exploitation by viruses. In this review, the author discusses an emerging arms race for cellular resources between viruses and hosts, which occur during the early events of virus-host interactions. Recent findings with tomato bushy stunt virus and its hosts revealed that the need of the virus to exploit and co-opt given members of protein families provides an opportunity for the host to deploy additional members of the same or associated protein family to interfere with virus replication. Three examples with well-established heat shock protein 70 and RNA helicase protein families and the ubiquitin network will be described to illustrate this model on the early arms race for cellular resources between tombusviruses and their hosts. We predict that arms race for resources with additional cellular protein families will be discovered with tombusviruses. These advances will fortify research on interactions among other plant and animal viruses and their hosts.
Keywords: Virus–host interaction, Virus replication, Immunity, Tomato bushy stunt virus, Hsp70, Cyclophilin, DEAD-box helicase, Ubiquitin pathway
1. Introduction
Positive-strand (+)RNA viruses of eukaryotes replicate in the cytosol of infected cells by assembling numerous membrane-bound viral replicase complexes (VRCs). The VRCs are usually harbored inside elaborate virus-induced membranous structures, called viral replication organelles (VROs) (Fernandez de Castro et al., 2016; Paul and Bartenschlager, 2015; Shulla and Randall, 2016; Wang, 2015). The VROs consist of single-membrane or double membrane vesicles, convoluted membranes and other structures for a group of (+)RNA viruses that include coronaviruses, enteroviruses and hepatitis C virus (Altan-Bonnet, 2017; Belov and Sztul, 2014; Nagy and Pogany, 2012). Another group of (+)RNA viruses, such as alphaviruses, most flaviviruses, the insect-infecting nodaviruses and the plant-infecting tombusviruses and bromoviruses, deform membranes, creating numerous vesicle-like structures, also called spherules, with small opening toward the cytosol (Brinton, 2013; den Boon and Ahlquist, 2010; Ertel et al., 2017; Ishibashi and Ishikawa, 2016; Neufeldt et al., 2018; Zhang et al., 2019). The spherules could be formed by using membranes derived from the ER, mitochondria, plasma membrane or peroxisomes (Laliberte and Sanfacon, 2010; Shulla and Randall, 2016; Wang, 2015). Overall, the VROs sequester viral and co-opted host components to a confined place to achieve robust viral RNA replication. The VROs organize temporally and spatially the viral replication process and other distinct steps during the infection cycle. The VROs also protect the viral RNA and proteins from degradation and facilitate the avoidance of recognition by the host immune system.
Our knowledge on the biogenesis of VROs and the individual VRCs and overall on the interactions between viruses and their hosts are quickly progressing based on a few model (+)RNA viruses and other important human pathogenic viruses (Altan-Bonnet, 2017; Nagy, 2016; Nagy and Pogany, 2012; Neufeldt et al., 2018; Wang, 2015; Zhang et al., 2019). In this article, we will use the example of the small plant tombusviruses to visualize how the arms race for control of resources between viruses and their hosts include conserved host protein families.
Tomato bushy stunt virus (TBSV) and other tombusviruses have less than 5 kb (+)RNA genomes, which code for five proteins, two of which are the viral replication proteins, termed p33 and p92pol (Nagy, 2008, Nagy, 2011; Nagy and Pogany, 2010). The replication proteins are essential for TBSV replication, the biogenesis of VROs and the formation of spherule structures utilizing peroxisomal membranes in plants and yeast, a model host (Barajas et al., 2009a; McCartney et al., 2005). Several genome-wide and proteome-wide screens in combination with lipidomics and bioinformatics approaches led to the identification of over 500 yeast genes, which affected TBSV replication (Nagy, 2016, Nagy, 2017; Nagy et al., 2014). More detailed, mechanistic studies with cell-free replicase reconstitution assays, yeast knockout and knockdown mutants or temperature-sensitive mutants, and validation experiments in a plant host revealed the intricate interaction of TBSV with the hosts through co-opting numerous mostly-conserved host proteins (Li and Nagy, 2011; Nagy, 2016; Nagy et al., 2012). As discussed below, the hosts also respond by using proteins frequently from the same or associated protein families to greatly interfere with TBSV replication, thus creating a never-ending arms race with the virus.
The article will follow a scheme that presents the idea of how the need of the virus to hijack host factors creates opportunities for the host to use similar host proteins that interfere with virus replication. In other words, the arms race for control of resources between the virus and host could involve different members of the same protein families. Three examples with well-established protein families will be described to illustrate this model on the arms race between tombusviruses and their hosts.
Description of the arms race between viruses and hosts frequently starts with explaining what pathogen associated molecular patterns (PAMPs) are recognized by the host, which is mostly based on specific features present in viral proteins and the viral RNAs. The detection of PAMPs then triggers RNAi and other immune responses in plants and animals (Caplan et al., 2008; Chow et al., 2018; Ding, 2010; Rehwinkel and Gack, 2020; Soosaar et al., 2005). However, viruses well-adapted to their hosts are able to suppress the effective immune responses through expressing suppressor or effector proteins to stave off the antiviral defenses (Chow and Kagan, 2018; Goertz et al., 2018; Kuka and Iannacone, 2018; Li and Ding, 2006). Often the hosts will have another level of antiviral responses in the arms race. For example, plants might select for resistance (R) genes against the most pathogenic viruses or animals produce specialized immune cells guarding against future infections by the same virus (Schwerk et al., 2020; Soosaar et al., 2005; Tassetto et al., 2017). Note that the arms race described in this article is placed prior to the above powerful virus–host interactions, which are rather costly to the hosts and not all individuals are protected by such expensive defensive measures in a population due to genetic diversity. The arms race to exploit cellular resources by the invading virus and the counteractive defensive strategy deployed by the host/host cell to protect those resources from the virus takes place rather early during infections. However, this defensive strategy by the host might be effective against most, poorly or not-well adapted viruses. If the virus succeeds in overtaking this early defensive line of the cells, then the host cell will activate the PAMP-based defensive responses. Taken together, based on the level of adaptation between the virus and hosts, arms races occur at several levels and at various costs for the hosts.
2. The heat shock protein 70 chaperone network
The first example on the arms race between tombusviruses and their hosts will involve the Hsp70 chaperone network. The Hsp70 family of protein chaperones is highly conserved, present in all domains of life forms and required for protein quality control and maintenance of cellular homeostasis (Kim et al., 2013; Moran Luengo et al., 2019; Rosenzweig et al., 2019). Hsp70s are highly abundant, even in the absence of stress. They are ATP-dependent molecular chaperone machines to fold unfolded proteins, refold misfolded protein substrates, protein aggregates or large protein assemblies. Cytosolic Hsp70s are also involved in disassembly of large protein complexes or protein-nucleic acid complexes (Mayer, 2013). These include virions, clathrin coats, or pre-replication complex of bacteriophage λ (Rosenzweig et al., 2019). Hsp70s often cooperate with other accessory co-chaperones or Hsp90 in multiple protein refolding and maturation processes. The co-chaperones include the family of J-domain proteins and tetratricopeptide repeat (TPR) domain-containing proteins, which help determine the fate of client proteins (Genest et al., 2019).
Importantly, Hsp70 and co-chaperones play critical roles in RNA virus replication with ever-increasing numbers of documented cases, such for several flaviviruses (Bozzacco et al., 2016; Taguwa et al., 2015, Taguwa et al., 2019), influenza virus (Cao et al., 2014; Manzoor et al., 2014), and a couple of plant viruses (Chenon et al., 2012; Lamm et al., 2017; Lohmus et al., 2017; Nagy et al., 2011; Verchot, 2012; Wang et al., 2018). Based on these discoveries, the findings described below in case of TBSV and its hosts could be relevant for a wide range of viruses.
3. The pro-viral roles of co-opted protein chaperones in TBSV replication
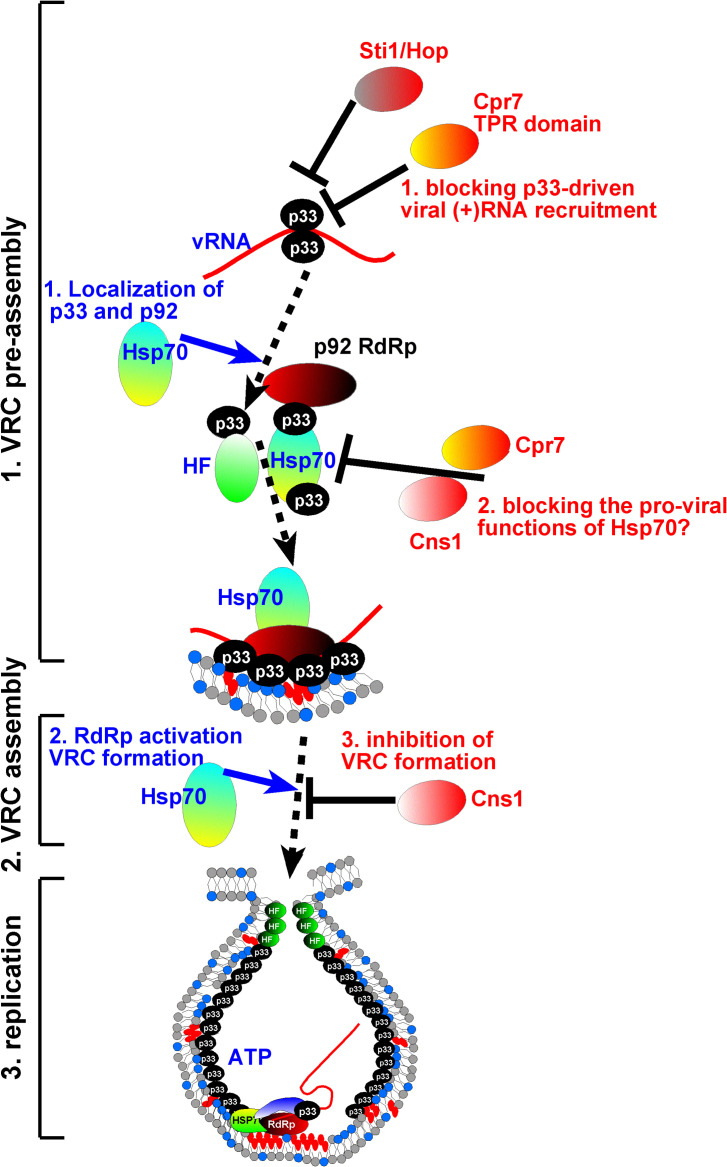
Although viruses are relatively simple parasitic entities, they still require amazing level of coordination and regulation of biological processes during their infection cycles. Therefore, they functionally repurpose co-opted cellular proteins to facilitate viral replication. For example, the p92pol RdRp of TBSV is originally inactive as an RdRp after translation by the host ribosomes (Pogany and Nagy, 2012). The p92pol requires binding to membranes and the viral (+)RNA to become activated as an RdRp. The co-opted Hsp70 chaperone is absolutely essential for this activation process, albeit the p33-domain in p92pol also plays a yet undefined role in the activation process (Pogany and Nagy, 2012, Pogany and Nagy, 2015). Moreover, Hsp70 facilitates the intracellular localization and the proper insertion of p33 and p92pol replication proteins into subcellular membranes, which is needed for the stability of both replication proteins (Wang et al., 2009a, Wang et al., 2009b). In vitro replicase reconstitution experiments unambiguously revealed the absolute essential contribution of Hsp70 members to TBSV replication (Pogany et al., 2008). Hsp70 is also required for the virion assembly process within or close to the VROs (Alam and Rochon, 2015, Alam and Rochon, 2017). Additional members of the Hsp70 chaperone network have also been shown to facilitate TBSV replication (Table 1 ), but the detailed mechanisms are not yet dissected.
Table 1.
Chaperone network genes likely involved in the arms race between TBSV and its hosts.
| Pro-viral effect | |||
|---|---|---|---|
| Gene | Function | Pro-viral function | Homologs |
| SSA1/2 | Hsp70, constitutive | Activation of p92 RdRp | Hsp70-1/2 |
| SSA3/4 | Hsp70, heat-shock induced | Partially redundant with Ssa1/2 | |
| SSB1 | Hsp70, ribosome associated | Binds to p92 | |
| HSC82 | Hsp90, constitutive | Binds to p92 | Hsp90 |
| HSP104 | Hsp104, disaggregase | Binds to p92 | Hsp104 |
| Antiviral effect | |||
|---|---|---|---|
| Gene | Function | Antiviral function | Homologs |
| CPR7 | TPR domain, cyclophilin | Inhibition of VRC assembly | Cyp40 |
| CNS1 | TPR domain, co-chaperone | Inhibition of VRC assembly | TTC4 oncogene |
| STI1 | TPR1 domain, co-chaperone | Inhibition of CIRV VRC assembly | Hop |
| CPR6 | TPR domain, cyclophilin | Weak inhibitor of VRC assembly | Cyp40 |
| Undefined effect/function | |||
|---|---|---|---|
| Gene | Function | Viral target | Homologs |
| SGT2 | TPR domain, co-chaperone | Interacts with p33 | hSGT-like |
| JJJ1 | Co-chaperone | Interacts with p33/p92/(+)RNA over-expression stimulatory | Hsp40 |
| JJJ3 | Co-chaperone | Interacts with p33 | Hsp40 |
| YDJ1 | Co-chaperone | Over-expression inhibitory | Hsp40 |
| CDC37 | Co-chaperone | ts-mutant, reduced viral replication | |
| SEC17 | TPR domain | Inhibition of TBSV replication in vitro | |
Genes shown in bold face are characterized in details in TBSV replication.
These co-opted Hsp70s require lots of ATP molecules for their functions within VROs. Interestingly, the ATP is generated locally by the co-opted glycolytic enzymes within the VROs (Chuang et al., 2017; Nagy and Lin, 2020; Prasanth et al., 2017). TBSV also recruits the fermentation enzymes, possibly to replenish the NAD + pool, which is required for continuous operation of the glycolytic enzymes within VROs (Lin et al., 2019). Through recruiting the aerobic glycolytic pathway into VROs for rapid ATP generation, TBSV might be able to fully exploit the co-opted Hsp70s for efficient VRO formation and robust viral replication to overwhelm the host cells (Nagy and Lin, 2020). Co-opting the Hsp70 chaperone network with the glycolytic enzymes into VROs elegantly illustrates how TBSV could utilize the cell own resources against cellular antiviral responses via the efficient biogenesis of VROs.
4. The antiviral roles of co-opted protein co-chaperones in TBSV replication
Like other powerful molecular machines, Hsp70s and other protein chaperones are also associated with accessory proteins, which help tune their activities (Rosenzweig et al., 2019). Therefore, the host cells could utilize these accessory proteins as antiviral factors. Indeed, genome-wide screens in yeast unearthed protein co-chaperones associated with Hsp70 or Hsp90 chaperones as inhibitors of TBSV replication (Table 1). For example, the Cyp40-like Cpr7 yeast cyclophilin with a TPR domain turned out to be a strong inhibitor of TBSV replication. Cpr7 or its TPR-domain alone bind to the RPR RNA-binding region in p33 and p92pol replication proteins, which likely prevent the replication proteins to recruit the viral (+)RNA into replication and other functions within VRCs (Fig. 1 ) (Lin et al., 2012). Interestingly, a second yeast Cyp40-like cyclophilin Cpr6 with a TPR domain had only weak restriction function on TBSV replication in a cell-free assay or in yeast. However, Cpr6 binds to a different region in the viral replication proteins, not the RPR domain bound by Cpr7 (Lin et al., 2012). Moreover, Cpr6 has a strong PPIase activity, whereas Cpr7 acts as a co-chaperone. These differences between the two yeast Cyp40-like cyclophilins might explain that only Cpr7 is a strong tombusvirus restriction factor. On the contrary, three Cyp40-like cyclophilins from Arabidopsis plants were found to inhibit tombusvirus replication in vitro (Lin et al., 2012). These findings suggest that plants have more Cyp40-like cyclophilins to combat tombusvirus replication than yeast.
Fig. 1.
A model showing the pro-viral function of co-opted Hsp70 and the antiviral functions of the co-chaperones during TBSV replication. TBSV replication is divided into three sequential steps, as shown on the left. Based on published data, we propose that TBSV co-opts the cytosolic Hsp70 chaperones to help perform several pro-viral functions indicated in blue letters on the left. The hosts use several co-chaperones to block the pro-viral function of Hsp70 and the viral replication proteins at several steps (as indicated with red letters), thus reducing viral replication. See further details in the main text.
The human Ttc4 oncogene-like yeast Cns1 co-chaperone also contains a TPR domain in its N-terminal region (Schopf et al., 2019). Cns1 can activate the ATPase function of Hsp70 (Hainzl et al., 2004). Cns1 strongly interacts with the TBSV replication proteins via its extended TPR-domain, which results in blocking VRC assembly (Fig. 1) (Lin and Nagy, 2013). This inhibitory effect of Cns1 on TBSV replication is likely due to binding of Cns1 to the p33-p33/p92 interaction domain in the viral replication proteins. The interaction between the viral replication proteins via the p33-p33/p92 interaction domain is absolutely critical for VRC formation (Panavas et al., 2005a; Rajendran and Nagy, 2006). Unlike Cpr7, which inhibited the replication of both tombusviruses and the unrelated insect-infecting nodaviruses, Cns1 seems to be TBSV-specific restriction factor (Lin et al., 2012; Lin and Nagy, 2013).
Another interesting tombusvirus restriction factor is the Hop-like Sti1 co-chaperone of Hsp70 and Hsp90 (Xu et al., 2014). Sti1 and the plant Hop-1 have three TPR domains and TPR1 is the most important in providing the antiviral effect by binding to the RPR RNA-binding domain in the p36 and p95pol replication proteins of carnation Italian ringspot virus (CIRV), a tombusvirus with mitochondrial membrane-based VROs. Surprisingly, Sti1 and Hop1 only acted as weak restriction factors against TBSV (peroxisomal membrane-based VROs) in yeast or in vitro (Xu et al., 2014). Thus, unlike Cpr7 and Cns1 co-chaperones, which restrict the replication of both TBSV and CIRV, Sti1/Hop seems only to be a strong CIRV restriction factor. Subcellular localization data indicated that the mitochondrial CIRV is targeted more efficiently than the peroxisomal TBSV by Sti1, suggesting that accessibility of Sti1 in various subcellular locations might differ (Lin et al., 2012; Lin and Nagy, 2013; Xu et al., 2014).
Based on our discoveries, we propose that the need of TBSV to hijack Hsp70 functions from the cells during viral replication opens up opportunities for the hosts to fight back by utilizing protein chaperone-associated proteins and co-chaperones. All the three above described TPR-domain containing proteins are co-chaperones of Hsp70 and Hsp90 chaperones. This observation suggests that the co-chaperones might inhibit tombusvirus replication not only by directly binding to the viral replication proteins, but via inhibiting the pro-viral functions of the co-opted Hsp70 chaperone. Indeed, for example, TPR-domain containing proteins are predicted to be present in all eukaryotes in large numbers, including ~ 200 in humans and ~ 250 in Arabidopsis (Haslbeck et al., 2013; Prasad et al., 2010). An emerging picture is that the antiviral co-chaperones, and cyclophilins have to be targeted into action at the very early stage of TBSV infection, before the viral infection overwhelms the host cells defensive potential.
Altogether, this creates an arms race between the virus and its host for the access to the Hsp70 chaperone network. Through utilizing members of the cellular co-chaperones, the host interferes with the “easy” exploitation of Hsp70 chaperone machine for tombusvirus replication.
5. Cellular RNA helicases and RNA chaperones
The second example on the arms race between tombusviruses and their hosts will present the protein families involved in RNA structure modulation. Similar to their protein counterparts, the numerous cellular RNAs also require the guidance of specialized army of proteins that perform their folding, maturation, transport and ultimately degradation. The largest families of proteins involved with cellular RNAs are RNA helicases and RNA chaperones, which often function as ribonucleoprotein complexes (Guenther and Jankowsky, 2009; Jankowsky, 2011; Sloan and Bohnsack, 2018). Whereas RNA helicases use ATP to modulate RNA structures/functions, RNA chaperones perform those functions through binding to RNA targets without using ATP (Gilman et al., 2017; Linder and Jankowsky, 2011; Sharma and Jankowsky, 2014). Similar to cellular processes that depend on RNAs, (+)RNA viruses also require the involvement of RNA helicases and/or RNA chaperones to promote their replication and the infection process (Huang et al., 2010; Li and Nagy, 2011; Ranji and Boris-Lawrie, 2010; Taschuk and Cherry, 2020). The hosts also deploy cellular DEAD-box helicases, such as DICER, RIG-I or MDA5, which are involved in sensing the viral RNAs and initiating antiviral immune signaling or the RNAi pathway to destroy viruses (Ding, 2010; Garcia-Ruiz et al., 2010; Schroder, 2011; Taschuk and Cherry, 2020).
6. The pro-viral roles of co-opted DEAD-box helicases and RNA chaperones in TBSV replication
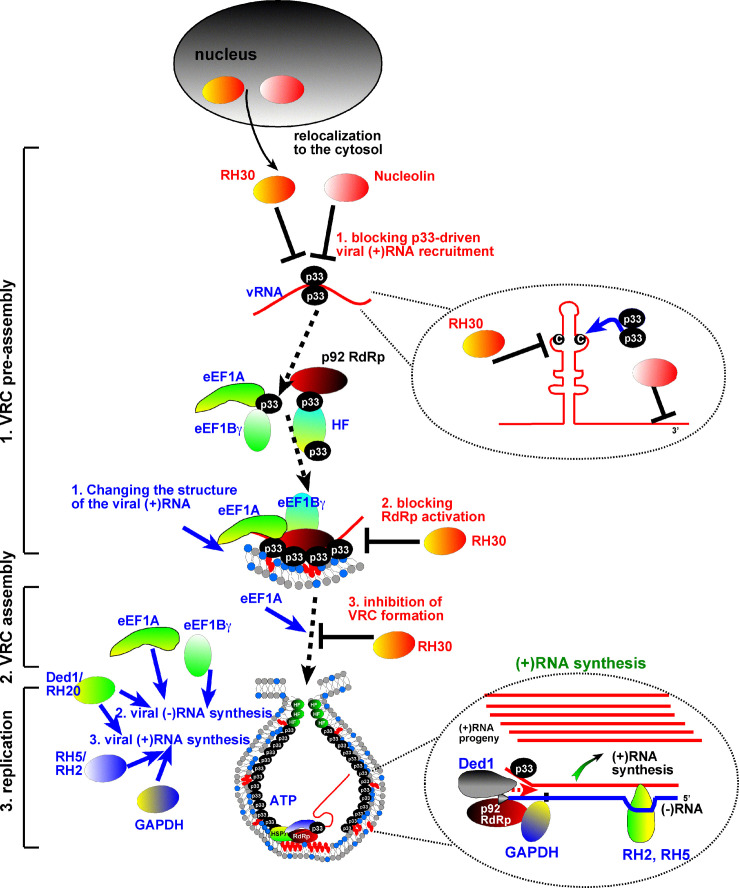
Similar to the viral p92pol replication protein, the TBSV (+)RNA needs to be recruited to the site of replication (Monkewich et al., 2005; Pogany et al., 2005). The (+)RNA template selection for replication is performed by the p33 replication protein via specifically recognizing an internal extended stem-loop structure with a C·C mismatch within an internal loop (Fig. 2 ) (Monkewich et al., 2005; Pogany et al., 2005). The TBSV (+)RNA needs also “be activated” prior to functioning as a template at the sites of replication (Pathak et al., 2011, Pathak et al., 2012). This is because the 3′ end of TBSV (+)RNA is embedded in an RNA secondary structure not readily recognized by the RdRp. An upstream RNA silencer element binds to the 3’terminal promoter, forming a partially dsRNA structure (Pogany et al., 2003). After recruitment into the sites of replication, co-opted RNA chaperones, namely eEF1A and eEF1Bγ, open up the above secondary structures and bring in the p92 RdRp to initiate minus-strand synthesis (Fig. 2) (Li et al., 2010; Sasvari et al., 2011).
Fig. 2.
A model of the arms race between TBSV and its hosts based on RNA helicases and RNA chaperones. The pro-viral functions of the co-opted RNA helicases and RNA chaperones versus the antiviral functions of additional RNA helicases and RNA chaperones during TBSV replication are shown schematically. The co-opted RNA chaperones (translation elongation factors) facilitate refolding of viral (+)RNA, followed by assisting TBSV (−)RNA synthesis as shown on the left. The co-opted RNA helicases facilitate the termination of (−)RNA synthesis, then the following initiation of (+)-strand synthesis and asymmetrical replication with RNA chaperone contribution of Glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Some of the details are enlarged on the right. On the contrary, the cellular DDX17-like RH30 helicase interferes with several steps during TBSV replication as shown in red on the right side of the panel. The RNA chaperone Nucleolin also inhibits the recruitment of the TBSV (+)RNA into replication. The RII(+)-SL cis-acting RNA element with the critical internal C·C mismatch, which specifically binds to the p33 replication protein only when the stem-loop structure is formed, is shown in the circle. The cytosolic pools of RH30 and Nucleolin are essential for the antiviral activity. See further details in the main text.
Dissociation of the p92 RdRp from the template at the end of minus-strand synthesis is greatly affected by a DEAD-box helicase, the DDX3-like Ded1 in yeast, and RH20 in plants (Chuang et al., 2015). The formation of dsRNA during minus-strand synthesis leads to an inactive template (Kovalev et al., 2014), which has to be partially unwound by a DEAD-box helicase, Ded1 and Dbp2 in yeast, and RH20 in plants, prior to initiation of plus-strand synthesis by the p92 RdRp (Kovalev et al., 2012a, Kovalev et al., 2012b). Since, the initiation on dsRNA template has to be repeated by ~ 100 times, a second set of DEAD-box helicases, the eIF4AIII-like RH2 and DDX5-like RH5 in plants, facilitate the “recycling” of the viral RdRp with the help of RNA replication enhancers present on the viral (−)RNA (Kovalev and Nagy, 2014; Panavas and Nagy, 2005).
In contrast with the co-opted eEF1A and eEF1Bγ RNA chaperones, the co-opted DEAD-box helicases require plentiful ATP for their pro-viral functions. As described above for Hsp70, the ATP is generated locally by the co-opted glycolytic enzymes within the membranous VROs (Chuang et al., 2017; Nagy and Lin, 2020; Prasanth et al., 2017).
6.1. The antiviral roles of co-opted DEAD-box helicases and RNA chaperones in TBSV replication
The need for TBSV to recruit conserved cellular RNA chaperones and DEAD-box helicases for viral replication makes the virus “vulnerable” to those cellular DEAD-box helicases, which could inhibit the replication process. This possibility might be really important in plants, which encode more than twice as many helicases as animals and yeast do. Accordingly, the DDX17-like RH30 helicase from Arabidopsis has been shown to be recruited into the VROs and strongly inhibits TBSV replication (Fig. 2) (Wu and Nagy, 2019). The inhibition is due to unwinding of the critical cis-acting secondary element in the viral (+)RNA, which is required for binding to p33 replication protein, followed by selection for replication. Since this is an early step, the effect of RH30 was found to be dominant over the co-opted pro-viral helicases, which act at latter steps in the replication process. By altering the biochemical features of DEAD-box helicases, either by domain analysis or by creating chimeras between pro-viral and antiviral helicases, it might be possible to obtain new antiviral tools by engineering or through evolution, called arms race between viruses and their hosts.
In addition to the well-characterized DEAD-box helicases (see above), plants also deploy RNA chaperones, such as Nucleolin, against TBSV (Jiang et al., 2010). Nucleolin (Nsr1 in yeast) has been shown to be recruited into the VROs and strongly inhibits TBSV replication in yeast and plants (Jiang et al., 2010). The inhibition is due to binding of Nucleolin to a cis-acting secondary element in the viral (+)RNA, which is inhibitory to recruitment of TBSV (+)RNA for replication (Fig. 2). Nucleolin, like other RNA chaperones, modify RNA structures, but without using ATP. Because TBSV has its own RNA chaperone, the p33 replication protein (Stork et al., 2011), and co-opt translation elongation factors, which also have RNA chaperone function, it is possible that the hosts will utilize an arsenal of RNA chaperones against invading viruses. Accordingly, genome-wide screens in yeast did identify numerous RNA binding host proteins that inhibited TBSV replication (Jiang et al., 2006; Li et al., 2009; Panavas et al., 2005b; Prasanth et al., 2016; Shah Nawaz-Ul-Rehman et al., 2012). The antiviral functions of the identified cellular RNA-binding proteins have not been defined (Table 2 ). Further research will highlight whether there is ongoing arms race between viruses and their hosts using RNA chaperones.
Table 2.
DEAD-box helicases and RNA chaperones likely involved in the arms race between TBSV and its hosts.
| Pro-viral effect | |||
|---|---|---|---|
| Gene | Function | Pro-viral function | Homologs |
| DED1 | DEAD-box helicase | (−) and (+)-strand synthesis | DDX3/AtRH20 |
| DBP2 | DEAD-box helicase | (+)-strand synthesis | p68/AtRH20 |
| AtRH2 | DEAD-box helicase | (+)-strand synthesis | eIF4AIII/ScFAL1 |
| AtRH5 | DEAD-box helicase | (+)-strand synthesis | DDX5/ScDBP3 |
| TEF1/2 | Translation elongation factor | (−)-strand synthesis, p92 binding | eEF1A |
| TEF4 | Translation elongation factor | (−)-strand synthesis | eEF1Bγ |
| TDH2/3 | GAPDH, glycolysis | (+)-strand synthesis | GAPDH/AtGAPC |
| Antiviral effect | |||
|---|---|---|---|
| Gene | Function | Antiviral function | Homologs |
| AtRH30 | DEAD-box helicase | Inhibition of (+)RNA recruitment | DDX17 |
| NSP1 | Ribosomal RNA maturation | Inhibition of (+)RNA recruitment | Nucleolin |
| Undefined effect/function | |||
|---|---|---|---|
| Gene | Function | Viral target | Homologs |
| TIF1 | Translation initiation factor eIF4A DEAD-box helicase | Interacts with p33 | eIF4A |
| TIF11 | Translation initiation factor eIF1A DEAD-box helicase | Interacts with p33/p92 | eIF1A |
| PBP2 | heterogeneous nuclear RNP K protein | Over-expression is stimulatory | |
| HAS1 | ATP-dependent RNA helicase | Over-expression is inhibitory | |
Genes shown in bold face are characterized in details in TBSV replication.
An emerging concept is that the antiviral DEAD-box helicases and RNA chaperones have to be targeted into action at the very early stage of TBSV infection to block viral replication (Jiang et al., 2010; Wu and Nagy, 2019). During late stages, the viral infection overwhelms the host cells defensive potential at this level.
Cellular RNA chaperones and DEAD-box helicases affect translation and replication of many RNA viruses (Garbelli et al., 2011; Huang et al., 2010; Ranji and Boris-Lawrie, 2010; Upadya et al., 2014). For example, host DEAD-box helicases have been shown to affect translation of viral proteins (Bolinger et al., 2010; Noueiry et al., 2000; Watanabe et al., 2009); viral RNA replication (Goh et al., 2004; Huang et al., 2010; Lawrence and Rieder, 2009; Morohashi et al., 2011; Upadhyay et al., 2013); subgenomic RNA synthesis (Gimenez-Barcons et al., 2013); and virus assembly (Xu and Hobman, 2012). Rift Valley fever virus is restricted by the DDX17 helicase (Moy et al., 2014). Moreover, DDX3 blocks Dengue virus infections, whereas DDX21 helicase inhibits influenza A virus (Chen et al., 2014; Li et al., 2015; Schroder, 2011; Zhang et al., 2011). Human immunodeficiency virus (HIV-1) infections are affected by several helicases providing either pro-viral or antiviral functions (Lorgeoux et al., 2012; Yasuda-Inoue et al., 2013). Thus, the emerging idea is that cellular DEAD-box helicases are important co-opted host factors for several viruses, whereas other cellular DEAD-box helicases play active roles in restricting RNA virus replication. Taken together, cellular DEAD-box helicases perform critical roles in virus-host interactions with many more discoveries expected in the future.
7. Comparison of protein chaperone and RNA helicase/chaperone functions
It seems that TBSV hijacks two highly-conserved protein family members from the host cells: the protein chaperoning Hsp70 proteins and co-chaperones and the RNA chaperoning DEAD-box helicases. The similarities and parallels between these proteins in function for the virus are interesting: (i) they utilize ATP for their function, which are generated by co-opted glycolytic enzymes within VROs; (ii) they are recruited to the sites of TBSV replication (VROs); (iii) they are directly involved with the viral replication proteins/RNA templates; (iv) and they are permanently part of the VRCs. The major difference between them is the substrate selection: Hsp70s work with the viral replication proteins, whereas the DEAD-box helicases target the viral RNAs as substrates. However, both protein family members contribute pro-viral as well as antiviral functions. This highlights that these proteins families are core members of the arms race between viruses and their hosts. They could be also potential central players in strategies to develop novel antiviral approaches.
8. The ubiquitin network
The last example on the arms race between tombusviruses and their hosts will involve the ubiquitin network. Many cellular proteins have multiple functions at different subcellular locations. Post-translational modifications are frequently being utilized by the host to expand the functionality and stability of host proteins. Ubiquitination, the addition of the conserved 76 amino acids long ubiquitin to the lysine residues in target proteins, are among the most common post-translational modifications (Kwon and Ciechanover, 2017; Nielsen and MacGurn, 2020; Varshavsky, 2017). The protein ubiquitination process consists of multiple enzymes acting sequentially. The process includes an E1 ubiquitin-activating enzyme, then the E2 ubiquitin-conjugating enzyme, in collaboration with an E3 ubiquitin-ligase enzyme. The covalently-linked ubiquitin can be removed by de-ubiquitinases. Interestingly, protein mono-ubiquitination (addition of a single ubiquitin) frequently affects the subcellular localization and other functions of the client proteins, whereas poly-ubiquitination mainly serves as a protein degradation signal for the proteasome machinery (Berner et al., 2018; Isaacson and Ploegh, 2009).
Viruses also utilize their proteins for multiple functions in different subcellular locations, therefore, the ubiquitin network is also central in the arms race between viruses and their hosts. Many examples are documented on the ubiquitin-directed proteasome-based degradation of viral proteins, or viral protein-driven degradation of host proteins in case of animal RNA viruses (Choi et al., 2013; Lee et al., 2019; Liu and Tan, 2020; Llamas-Gonzalez et al., 2019; Luo, 2016; Xu et al., 2017). The role of the ubiquitin network is also drawn major attention in studies of innate immunity against animal RNA viruses (Garcia-Sastre, 2017; Rajsbaum and Garcia-Sastre, 2013). Plant RNA viruses are also capable of taking advantage of the ubiquitin pathways for regulating their replication and interactions with hosts (Camborde et al., 2010; Verchot, 2016). In summary, the ubiquitin network determines many facets of the arms race and it is one of the hot topics in virus-host interactions.
9. The roles of the host ubiquitin network in TBSV replication and in the arms race with hosts
An emerging area in virus–host interaction is the role of the host ubiquitin network. On one hand, TBSV hijacks the yeast Rad6 (Ubc2 in plants) and Cdc34 E2 ubiquitin-conjugating enzymes in order to mono- and bi-ubiquitinate the p33 replication protein (Fig. 3 ) (Imura et al., 2015; Li et al., 2008). In combination with the presence of the so-called late domain sequence in p33 (Fig. 3), the ubiquitination of p33 replication protein promotes the recruitment of cellular Vps23 ESCRT-I protein and Bro1 accessory protein into the VROs (Barajas and Nagy, 2010). This in turn, is needed to further recruit additional ESCRT-I and ESCRT-III proteins and Vps4 AAA ATPase into the VROs (Barajas et al., 2009a, Barajas et al., 2014). The coordinated actions of these ESCRT proteins with p33 and p92pol replication proteins and the viral (+)RNA are needed for membrane deformation and the biogenesis of the spherule-like VRCs (Barajas et al., 2014; Kovalev et al., 2016, Kovalev et al., 2017). This is important to provide a semi-closed membranous structure of VRCs, which is beneficial to protect the viral RNAs from the host ribonucleases (Fig. 3).
Fig. 3.
A model showing the pro-viral functions versus the antiviral functions of the ubiquitin network during TBSV replication. The cellular Rpn11 deubiquitinase is a key co-opted host factor, which helps with the stabilization of the viral replication proteins, and facilitates the subversion of Ded1/RH20 RNA helicases and needed for the correct VRC assembly. In addition, the co-opted Rad6, Ubc2 and Cdc34 E2 ubiquitin conjugating enzymes (shown in blue) bi-ubiquitinate the p33 replication protein as shown in a circle on the right side of the panel. These events together with the late domain in p33 facilitate the hijacking of Vps23 and Bro1 ESCRT proteins, and ultimately the ESCRT machinery to deform intracellular membranes into spherule-like structures, as shown schematically. However, the host also utilize the ubiquitin network for antiviral functions as shown by the action of Rsp5 E3 ubiquitin ligase, resulting in degradation of the viral p92 replication protein, as shown on the right side of the panel. See further details in the main text.
Another characterized member of the ubiquitin network co-opted by TBSV is Rpn11 (Regulatory Particle Non-ATPase) metalloprotease subunit of 19S regulatory particle in the proteasome. Rpn11 is a critical component of the proteasome due to its function in coupling deubiquitination and degradation of proteasome substrates. Multiple pro-viral functions have been documented for the co-opted cellular Rpn11 deubiquitinase, including promotion of the recruitment of the DDX3-like yeast Ded1 and the homologous plant RH20 DEAD-box helicases into the VROs (Prasanth et al., 2016). In addition, Rpn11 contributes to the stability of the viral replication proteins. Overall, Rpn11 is a key co-opted host factor for the correct VRC assembly that greatly affects both viral replication and RNA recombination (Prasanth et al., 2016).
On the other hand, the need to hijack ubiquitination network components, however, renders TBSV vulnerable for the antiviral activities of additional components of this network. Indeed, the NEDD4-like Rsp5 E3 ubiquitin-ligase could enter the VROs based on direct binding to the viral replication proteins, which then leads to the degradation of p92pol RdRp protein (Barajas et al., 2009b). This event greatly inhibits TBSV replication. The previous high throughput screens have identified several additional components of the ubiquitination network, which seem to inhibit TBSV replication in yeast (Table 3 ). Therefore, we propose that the host ubiquitination network could emerge as a major frontier among viruses and their hosts. Further studies are needed to dissect the mechanisms of their anti-TBSV functions in yeast and plants (Table 3). Taken together, the exploitation of the host ubiquitination network seems to be central in the on going arms race between TBSV and its hosts.
Table 3.
Ubiquitin network genes likely involved in the arms race between TBSV and its hosts.
| Pro-viral effect | |||
|---|---|---|---|
| Gene | Function | Pro-viral function | Homologs |
| RAD6 | Ubiquitin-conjugating enzyme | Ubiquitination of p33 | UBC2 |
| CDC34 | Ubiquitin-conjugating enzyme | Ubiquitination of p33 | |
| BRE1 | Ubiquitin-protein ligase | Required for replication | |
| DOA4 | Protein deubiquitination | Required for replication | |
| LGE1 | Protein mono-ubiquitination | Required for replication | |
| RPN11 | Protein deubiquitination | Required for replication | RPN11 |
| Antiviral effect | |||
|---|---|---|---|
| Gene | Function | Antiviral function | Homologs |
| RSP5 | E3 Ubiquitin-protein ligase | Degradation of p92 | NEDD4 |
| Undefined effect/function | |||
|---|---|---|---|
| Gene | Function | Viral target | Homologs |
| ASI1 | E3 Ubiquitin-protein ligase | Over-expression is inhibitory | |
| UBA1 | Ubiquitin activating E1 enzyme | Binds to p33 | |
| UBP3 | Ubiquitin-specific protease | Suppresses recombination | |
| UBP8 | Ubiquitin-specific protease | Over-expression is inhibitory | |
| UBP10 | Ubiquitin-specific protease | Binds to p33/p92/(+)RNA | |
| UBP15 | Ubiquitin-specific protease | Binds to p33 | |
| ATG7 | Ubiquitin activating E1 enzyme Autophagy-related protein |
Over-expression is inhibitory | |
| BUL2 | Rsp5p E3-ubiquitin ligase complex | Over-expression is stimulatory | |
| HUL4 | E3 Ubiquitin-protein ligase-like | Over-expression is stimulatory | |
| PIB1 | E3 Ubiquitin-protein ligase | Over-expression is stimulatory | |
Genes shown in bold face are characterized in details in TBSV replication.
10. Summary
Viruses require resources from the host cells in order to replicate efficiently and complete their infection cycles. Viruses also need to hide from the host antivirus-surveillance network and degradative apparatus. To accomplish all the requirements, viruses co-opt numerous host proteins belonging to several protein families. However, the hosts could use similar members of the same or associated protein families to interfere with the viral replication processes. The rapid changes in the viral genomes in evolutionary terms create a never-ending arms race between the virus and hosts for the exploitation of members of several host protein families. These protein families could be potential central players in strategies to develop novel antiviral approaches that favor the hosts against pathogenic viruses.
Acknowledgments
The authors thank Dr. Z. Feng, Ms. M. Molho and Ms. P. Gonzalez for valuable comments. This work was supported by the National Science Foundation (IOS-1922895), and the USDA hatch grant (KY012042) to P.D.N.
References
- Alam S.B., Rochon D. Cucumber necrosis virus recruits cellular heat shock protein 70 homologs at several stages of infection. J. Virol. 2015;90(7):3302–33017. doi: 10.1128/JVI.02833-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S.B., Rochon D. Evidence that Hsc70 is associated with cucumber necrosis virus particles and plays a role in particle disassembly. J. Virol. 2017;91(2) doi: 10.1128/JVI.01555-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altan-Bonnet N. Lipid tales of viral replication and transmission. Trends Cell Biol. 2017;27(3):201–213. doi: 10.1016/j.tcb.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas D., Nagy P.D. Ubiquitination of tombusvirus p33 replication protein plays a role in virus replication and binding to the host Vps23p ESCRT protein. Virology. 2010;397(2):358–368. doi: 10.1016/j.virol.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas D., Jiang Y., Nagy P.D. A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog. 2009;5(12) doi: 10.1371/journal.ppat.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas D., Li Z., Nagy P.D. The Nedd4-type Rsp5p ubiquitin ligase inhibits tombusvirus replication by regulating degradation of the p92 replication protein and decreasing the activity of the tombusvirus replicase. J. Virol. 2009;83(22):11751–11764. doi: 10.1128/JVI.00789-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas D., Martin I.F., Pogany J., Risco C., Nagy P.D. Noncanonical role for the host Vps4 AAA + ATPase ESCRT protein in the formation of tomato bushy stunt virus replicase. PLoS Pathog. 2014;10(4) doi: 10.1371/journal.ppat.1004087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov G.A., Sztul E. Rewiring of cellular membrane homeostasis by picornaviruses. J. Virol. 2014;88(17):9478–9489. doi: 10.1128/JVI.00922-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berner N., Reutter K.R., Wolf D.H. Protein quality control of the endoplasmic reticulum and ubiquitin-proteasome-triggered degradation of aberrant proteins: yeast pioneers the path. Annu. Rev. Biochem. 2018;87:751–782. doi: 10.1146/annurev-biochem-062917-012749. [DOI] [PubMed] [Google Scholar]
- Bolinger C., Sharma A., Singh D., Yu L., Boris-Lawrie K. RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic Acids Res. 2010;38(5):1686–1696. doi: 10.1093/nar/gkp1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzacco L., Yi Z., Andreo U., Conklin C.R., Li M.M., Rice C.M., MacDonald M.R. Chaperone-assisted protein folding is critical for yellow fever virus NS3/4A cleavage and replication. J. Virol. 2016;90(6):3212–3228. doi: 10.1128/JVI.03077-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton M.A. Replication cycle and molecular biology of the West Nile virus. Viruses. 2013;6(1):13–53. doi: 10.3390/v6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camborde L., Planchais S., Tournier V., Jakubiec A., Drugeon G., Lacassagne E., Pflieger S., Chenon M., Jupin I. The ubiquitin-proteasome system regulates the accumulation of Turnip yellow mosaic virus RNA-dependent RNA polymerase during viral infection. Plant Cell. 2010;22(9):3142–3152. doi: 10.1105/tpc.109.072090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M., Wei C., Zhao L., Wang J., Jia Q., Wang X., Jin Q., Deng T. DnaJA1/Hsp40 is co-opted by influenza A virus to enhance its viral RNA polymerase activity. J. Virol. 2014;88(24):14078–14089. doi: 10.1128/JVI.02475-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan J.L., Mamillapalli P., Burch-Smith T.M., Czymmek K., Dinesh-Kumar S.P. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell. 2008;132(3):449–462. doi: 10.1016/j.cell.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Liu C.H., Zhou L., Krug R.M. Cellular DDX21 RNA helicase inhibits influenza A virus replication but is counteracted by the viral NS1 protein. Cell Host Microbe. 2014;15(4):484–493. doi: 10.1016/j.chom.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenon M., Camborde L., Cheminant S., Jupin I. A viral deubiquitylating enzyme targets viral RNA-dependent RNA polymerase and affects viral infectivity. EMBO J. 2012;31(3):741–753. doi: 10.1038/emboj.2011.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi A.G., Wong J., Marchant D., Luo H. The ubiquitin-proteasome system in positive-strand RNA virus infection. Rev. Med. Virol. 2013;23(2):85–96. doi: 10.1002/rmv.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J., Kagan J.C. The fly way of antiviral resistance and disease tolerance. Adv. Immunol. 2018;140:59–93. doi: 10.1016/bs.ai.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow K.T., Gale M., Jr., Loo Y.M. RIG-I and other RNA sensors in antiviral immunity. Annu. Rev. Immunol. 2018;36:667–694. doi: 10.1146/annurev-immunol-042617-053309. [DOI] [PubMed] [Google Scholar]
- Chuang C., Prasanth K.R., Nagy P.D. Coordinated function of cellular DEAD-box helicases in suppression of viral RNA recombination and maintenance of viral genome integrity. PLoS Pathog. 2015;11(2) doi: 10.1371/journal.ppat.1004680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C., Prasanth K.R., Nagy P.D. The glycolytic pyruvate kinase is recruited directly into the viral replicase complex to generate ATP for RNA synthesis. Cell Host Microbe. 2017;22(5):639–652. doi: 10.1016/j.chom.2017.10.004. e7. [DOI] [PubMed] [Google Scholar]
- den Boon J.A., Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- Ding S.W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010;10(9):632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- Ertel K.J., Benefield D., Castano-Diez D., Pennington J.G., Horswill M., den Boon J.A., Otegui M.S., Ahlquist P. Cryo-electron tomography reveals novel features of a viral RNA replication compartment. Elife. 2017;6:25940. doi: 10.7554/eLife.25940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez de Castro I., Tenorio R., Risco C. Virus assembly factories in a lipid world. Curr. Opin. Virol. 2016;18:20–26. doi: 10.1016/j.coviro.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Garbelli A., Radi M., Falchi F., Beermann S., Zanoli S., Manetti F., Dietrich U., Botta M., Maga G. Targeting the human DEAD-box polypeptide 3 (DDX3) RNA helicase as a novel strategy to inhibit viral replication. Curr. Med. Chem. 2011;18(20):3015–3027. doi: 10.2174/092986711796391688. [DOI] [PubMed] [Google Scholar]
- Garcia-Ruiz H., Takeda A., Chapman E.J., Sullivan C.M., Fahlgren N., Brempelis K.J., Carrington J.C. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell. 2010;22(2):481–496. doi: 10.1105/tpc.109.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A. Ten strategies of interferon evasion by viruses. Cell Host Microbe. 2017;22(2):176–184. doi: 10.1016/j.chom.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest O., Wickner S., Doyle S.M. Hsp90 and Hsp70 chaperones: collaborators in protein remodeling. J. Biol. Chem. 2019;294(6):2109–2120. doi: 10.1074/jbc.REV118.002806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman B., Tijerina P., Russell R. Distinct RNA-unwinding mechanisms of DEAD-box and DEAH-box RNA helicase proteins in remodeling structured RNAs and RNPs. Biochem. Soc. Trans. 2017;45(6):1313–1321. doi: 10.1042/BST20170095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Barcons M., Alves-Rodrigues I., Jungfleisch J., Van Wynsberghe P.M., Ahlquist P., Diez J. The cellular decapping activators LSm1, Pat1, and Dhh1 control the ratio of subgenomic to genomic Flock House virus RNAs. J. Virol. 2013;87(11):6192–6200. doi: 10.1128/JVI.03327-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goertz G.P., McNally K.L., Robertson S.J., Best S.M., Pijlman G.P., Fros J.J. The methyltransferase-like domain of chikungunya virus nsP2 inhibits the interferon response by promoting the nuclear export of STAT1. J. Virol. 2018;92(17) doi: 10.1128/JVI.01008-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh P.Y., Tan Y.J., Lim S.P., Tan Y.H., Lim S.G., Fuller-Pace F., Hong W. Cellular RNA helicase p68 relocalization and interaction with the hepatitis C virus (HCV) NS5B protein and the potential role of p68 in HCV RNA replication. J. Virol. 2004;78(10):5288–5298. doi: 10.1128/JVI.78.10.5288-5298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther U.P., Jankowsky E. Helicase multitasking in ribosome assembly. Mol. Cell. 2009;36(4):537–538. doi: 10.1016/j.molcel.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Hainzl O., Wegele H., Richter K., Buchner J. Cns1 is an activator of the Ssa1 ATPase activity. J. Biol. Chem. 2004;279(22):23267–23273. doi: 10.1074/jbc.M402189200. [DOI] [PubMed] [Google Scholar]
- Haslbeck V., Eckl J.M., Kaiser C.J., Papsdorf K., Hessling M., Richter K. Chaperone-interacting TPR proteins in Caenorhabditis elegans. J. Mol. Biol. 2013;425(16):2922–2939. doi: 10.1016/j.jmb.2013.05.019. [DOI] [PubMed] [Google Scholar]
- Huang T.S., Wei T., Laliberte J.F., Wang A. A host RNA helicase-like protein, AtRH8, interacts with the potyviral genome-linked protein, VPg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol. 2010;152(1):255–266. doi: 10.1104/pp.109.147983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imura Y., Molho M., Chuang C., Nagy P.D. Cellular Ubc2/Rad6 E2 ubiquitin-conjugating enzyme facilitates tombusvirus replication in yeast and plants. Virology. 2015;484:265–275. doi: 10.1016/j.virol.2015.05.022. [DOI] [PubMed] [Google Scholar]
- Isaacson M.K., Ploegh H.L. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe. 2009;5(6):559–570. doi: 10.1016/j.chom.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi K., Ishikawa M. Replication of Tobamovirus RNA. Annu. Rev. Phytopathol. 2016;54:55–78. doi: 10.1146/annurev-phyto-080615-100217. [DOI] [PubMed] [Google Scholar]
- Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem. Sci. 2011;36(1):19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Serviene E., Gal J., Panavas T., Nagy P.D. Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes collection. J. Virol. 2006;80(15):7394–7404. doi: 10.1128/JVI.02686-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Li Z., Nagy P.D. Nucleolin/Nsr1p binds to the 3' noncoding region of the tombusvirus RNA and inhibits replication. Virology. 2010;396(1):10–20. doi: 10.1016/j.virol.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.E., Hipp M.S., Bracher A., Hayer-Hartl M., Hartl F.U. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- Kovalev N., Nagy P.D. The expanding functions of cellular helicases: the tombusvirus RNA replication enhancer co-opts the plant eIF4AIII-like AtRH2 and the DDX5-like AtRH5 DEAD-box RNA helicases to promote viral asymmetric RNA replication. PLoS Pathog. 2014;10(4) doi: 10.1371/journal.ppat.1004051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalev N., Barajas D., Nagy P.D. Similar roles for yeast Dbp2 and Arabidopsis RH20 DEAD-box RNA helicases to Ded1 helicase in tombusvirus plus-strand synthesis. Virology. 2012;432(2):470–484. doi: 10.1016/j.virol.2012.06.030. [DOI] [PubMed] [Google Scholar]
- Kovalev N., Pogany J., Nagy P.D. A co-opted DEAD-box RNA helicase enhances tombusvirus plus-strand synthesis. PLoS Pathog. 2012;8(2) doi: 10.1371/journal.ppat.1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalev N., Pogany J., Nagy P.D. Template role of double-stranded RNA in tombusvirus replication. J. Virol. 2014;88(10):5638–5651. doi: 10.1128/JVI.03842-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalev N., de Castro Martin I.F., Pogany J., Barajas D., Pathak K., Risco C., Nagy P.D. Role of viral RNA and co-opted cellular ESCRT-I and ESCRT-III factors in formation of tombusvirus spherules harboring the tombusvirus replicase. J. Virol. 2016;90(7):3611–3626. doi: 10.1128/JVI.02775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalev N., Inaba J.I., Li Z., Nagy P.D. The role of co-opted ESCRT proteins and lipid factors in protection of tombusviral double-stranded RNA replication intermediate against reconstituted RNAi in yeast. PLoS Pathog. 2017;13(7) doi: 10.1371/journal.ppat.1006520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuka M., Iannacone M. Viral subversion of B cell responses within secondary lymphoid organs. Nat. Rev. Immunol. 2018;18(4):255–265. doi: 10.1038/nri.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y.T., Ciechanover A. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem. Sci. 2017;42(11):873–886. doi: 10.1016/j.tibs.2017.09.002. [DOI] [PubMed] [Google Scholar]
- Laliberte J.F., Sanfacon H. Cellular remodeling during plant virus infection. Annu. Rev. Phytopathol. 2010;48:69–91. doi: 10.1146/annurev-phyto-073009-114239. [DOI] [PubMed] [Google Scholar]
- Lamm C.E., Kraner M.E., Hofmann J., Bornke F., Mock H.P., Sonnewald U. Hop/Sti1—a two-faced cochaperone involved in pattern recognition receptor maturation and viral infection. Front. Plant Sci. 2017;8:1754. doi: 10.3389/fpls.2017.01754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence P., Rieder E. Identification of RNA helicase A as a new host factor in the replication cycle of foot-and-mouth disease virus. J. Virol. 2009;83(21):11356–11366. doi: 10.1128/JVI.02677-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Chan S.T., Kim J.Y., Ou J.J. Hepatitis C virus induces the ubiquitin-editing enzyme A20 via depletion of the transcription factor upstream stimulatory factor 1 to support its replication. MBio. 2019;10(4) doi: 10.1128/mBio.01660-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Ding S.W. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Nagy P.D. Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol. 2011;8(2):305–315. doi: 10.4161/rna.8.2.15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Barajas D., Panavas T., Herbst D.A., Nagy P.D. Cdc34p ubiquitin-conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. J. Virol. 2008;82(14):6911–6926. doi: 10.1128/JVI.00702-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Pogany J., Panavas T., Xu K., Esposito A.M., Kinzy T.G., Nagy P.D. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology. 2009;385(1):245–260. doi: 10.1016/j.virol.2008.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Pogany J., Tupman S., Esposito A.M., Kinzy T.G., Nagy P.D. Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog. 2010;6(11) doi: 10.1371/journal.ppat.1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Feng T., Pan W., Shi X., Dai J. DEAD-box RNA helicase DDX3X inhibits DENV replication via regulating type one interferon pathway. Biochem. Biophys. Res. Commun. 2015;456(1):327–332. doi: 10.1016/j.bbrc.2014.11.080. [DOI] [PubMed] [Google Scholar]
- Lin J.Y., Nagy P.D. Identification of novel host factors via conserved domain search: Cns1 cochaperone is a novel restriction factor of tombusvirus replication in yeast. J. Virol. 2013;87(23):12600–12610. doi: 10.1128/JVI.00196-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.Y., Mendu V., Pogany J., Qin J., Nagy P.D. The TPR domain in the host Cyp40-like cyclophilin binds to the viral replication protein and inhibits the assembly of the tombusviral replicase. PLoS Pathog. 2012;8(2) doi: 10.1371/journal.ppat.1002491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Liu Y., Molho M., Zhang S., Wang L., Xie L., Nagy P.D. Co-opting the fermentation pathway for tombusvirus replication: compartmentalization of cellular metabolic pathways for rapid ATP generation. PLoS Pathog. 2019;15(10) doi: 10.1371/journal.ppat.1008092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P., Jankowsky E. From unwinding to clamping—the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011;12(8):505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- Liu Y., Tan X. Viral manipulations of the cullin-RING ubiquitin ligases. Adv. Exp. Med. Biol. 2020;1217:99–110. doi: 10.1007/978-981-15-1025-0_7. [DOI] [PubMed] [Google Scholar]
- Llamas-Gonzalez Y.Y., Campos D., Pascale J.M., Arbiza J., Gonzalez-Santamaria J. A functional ubiquitin-proteasome system is required for efficient replication of new World Mayaro and Una alphaviruses. Viruses. 2019;11(4):370. doi: 10.3390/v11040370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmus A., Hafren A., Makinen K. Coat protein regulation by CK2, CPIP, HSP70, and CHIP is required for potato virus A replication and coat protein accumulation. J. Virol. 2017;91(3) doi: 10.1128/JVI.01316-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorgeoux R.P., Guo F., Liang C. From promoting to inhibiting: diverse roles of helicases in HIV-1 Replication. Retrovirology. 2012;9:79. doi: 10.1186/1742-4690-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H. Interplay between the virus and the ubiquitin-proteasome system: molecular mechanism of viral pathogenesis. Curr. Opin. Virol. 2016;17:1–10. doi: 10.1016/j.coviro.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoor R., Kuroda K., Yoshida R., Tsuda Y., Fujikura D., Miyamoto H., Kajihara M., Kida H., Takada A. Heat shock protein 70 modulates influenza A virus polymerase activity. J. Biol. Chem. 2014;289(11):7599–7614. doi: 10.1074/jbc.M113.507798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.P. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 2013;38(10):507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- McCartney A.W., Greenwood J.S., Fabian M.R., White K.A., Mullen R.T. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell. 2005;17(12):3513–3531. doi: 10.1105/tpc.105.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monkewich S., Lin H.X., Fabian M.R., Xu W., Na H., Ray D., Chernysheva O.A., Nagy P.D., White K.A. The p92 polymerase coding region contains an internal RNA element required at an early step in tombusvirus genome replication. J. Virol. 2005;79(8):4848–4858. doi: 10.1128/JVI.79.8.4848-4858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran Luengo T., Mayer M.P., Rudiger S.G.D. The Hsp70-Hsp90 chaperone cascade in protein folding. Trends Cell Biol. 2019;29(2):164–177. doi: 10.1016/j.tcb.2018.10.004. [DOI] [PubMed] [Google Scholar]
- Morohashi K., Sahara H., Watashi K., Iwabata K., Sunoki T., Kuramochi K., Takakusagi K., Miyashita H., Sato N., Tanabe A., Shimotohno K., Kobayashi S., Sakaguchi K., Sugawara F. Cyclosporin A associated helicase-like protein facilitates the association of hepatitis C virus RNA polymerase with its cellular cyclophilin B. PLoS One. 2011;6(4) doi: 10.1371/journal.pone.0018285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy R.H., Cole B.S., Yasunaga A., Gold B., Shankarling G., Varble A., Molleston J.M., tenOever B.R., Lynch K.W., Cherry S. Stem-loop recognition by DDX17 facilitates miRNA processing and antiviral defense. Cell. 2014;158(4):764–777. doi: 10.1016/j.cell.2014.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D. Yeast as a model host to explore plant virus-host interactions. Annu. Rev. Phytopathol. 2008;46:217–242. doi: 10.1146/annurev.phyto.121407.093958. [DOI] [PubMed] [Google Scholar]
- Nagy P.D. The roles of host factors in tombusvirus RNA recombination. Adv. Virus Res. 2011;81:63–84. doi: 10.1016/B978-0-12-385885-6.00008-0. [DOI] [PubMed] [Google Scholar]
- Nagy P.D. Tombusvirus-host interactions: co-opted evolutionarily conserved host factors take center court. Annu. Rev. Virol. 2016;3(1):491–515. doi: 10.1146/annurev-virology-110615-042312. [DOI] [PubMed] [Google Scholar]
- Nagy P.D. Exploitation of a surrogate host, Saccharomyces cerevisiae, to identify cellular targets and develop novel antiviral approaches. Curr. Opin. Virol. 2017;26:132–140. doi: 10.1016/j.coviro.2017.07.031. [DOI] [PubMed] [Google Scholar]
- Nagy P.D., Lin W. Taking over cellular energy-metabolism for TBSV replication: the high ATP requirement of an RNA virus within the viral replication organelle. Viruses. 2020;12(1):56. doi: 10.3390/v12010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Pogany J. Global genomics and proteomics approaches to identify host factors as targets to induce resistance against Tomato bushy stunt virus. Adv. Virus Res. 2010;76:123–177. doi: 10.1016/S0065-3527(10)76004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Pogany J. The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 2012;10(2):137–149. doi: 10.1038/nrmicro2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Wang R.Y., Pogany J., Hafren A., Makinen K. Emerging picture of host chaperone and cyclophilin roles in RNA virus replication. Virology. 2011;411(2):374–382. doi: 10.1016/j.virol.2010.12.061. [DOI] [PubMed] [Google Scholar]
- Nagy P.D., Barajas D., Pogany J. Host factors with regulatory roles in tombusvirus replication. Curr. Opin. Virol. 2012;2(6):685–692. doi: 10.1016/j.coviro.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Nagy P.D., Pogany J., Lin J.Y. How yeast can be used as a genetic platform to explore virus-host interactions: from ‘omics’ to functional studies. Trends Microbiol. 2014;22(6):309–316. doi: 10.1016/j.tim.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Neufeldt C.J., Cortese M., Acosta E.G., Bartenschlager R. Rewiring cellular networks by members of the Flaviviridae family. Nat. Rev. Microbiol. 2018;16(3):125–142. doi: 10.1038/nrmicro.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C.P., MacGurn J.A. Coupling conjugation and deconjugation activities to achieve cellular ubiquitin dynamics. Trends Biochem. Sci. 2020;45(5):427–439. doi: 10.1016/j.tibs.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noueiry A.O., Chen J., Ahlquist P. A mutant allele of essential, general translation initiation factor DED1 selectively inhibits translation of a viral mRNA. Proc. Natl. Acad. Sci. U. S. A. 2000;97(24):12985–12990. doi: 10.1073/pnas.240460897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas T., Nagy P.D. Mechanism of stimulation of plus-strand synthesis by an RNA replication enhancer in a tombusvirus. J. Virol. 2005;79(15):9777–9785. doi: 10.1128/JVI.79.15.9777-9785.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas T., Hawkins C.M., Panaviene Z., Nagy P.D. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology. 2005;338(1):81–95. doi: 10.1016/j.virol.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Panavas T., Serviene E., Brasher J., Nagy P.D. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 2005;102(20):7326–7331. doi: 10.1073/pnas.0502604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak K.B., Pogany J., Nagy P.D. Non-template functions of the viral RNA in plant RNA virus replication. Curr. Opin. Virol. 2011;1(5):332–338. doi: 10.1016/j.coviro.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Pathak K.B., Pogany J., Xu K., White K.A., Nagy P.D. Defining the roles of cis-acting RNA elements in tombusvirus replicase assembly in vitro. J. Virol. 2012;86(1):156–171. doi: 10.1128/JVI.00404-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D., Bartenschlager R. Flaviviridae replication organelles: oh, what a tangled web we weave. Annu. Rev. Virol. 2015;2(1):289–310. doi: 10.1146/annurev-virology-100114-055007. [DOI] [PubMed] [Google Scholar]
- Pogany J., Nagy P.D. p33-independent activation of a truncated p92 RNA-dependent RNA polymerase of Tomato bushy stunt virus in yeast cell-free extract. J. Virol. 2012;86(22):12025–12038. doi: 10.1128/JVI.01303-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J., Nagy P.D. Activation of tomato bushy stunt virus RNA-dependent RNA polymerase by cellular heat shock protein 70 is enhanced by phospholipids in vitro. J. Virol. 2015;89(10):5714–5723. doi: 10.1128/JVI.03711-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J., Fabian M.R., White K.A., Nagy P.D. A replication silencer element in a plus-strand RNA virus. EMBO J. 2003;22(20):5602–5611. doi: 10.1093/emboj/cdg523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J., White K.A., Nagy P.D. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 2005;79(8):4859–4869. doi: 10.1128/JVI.79.8.4859-4869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J., Stork J., Li Z., Nagy P.D. In vitro assembly of the Tomato bushy stunt virus replicase requires the host heat shock protein 70. Proc. Natl. Acad. Sci. U. S. A. 2008;105(50):19956–19961. doi: 10.1073/pnas.0810851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B.D., Goel S., Krishna P. In silico identification of carboxylate clamp type tetratricopeptide repeat proteins in Arabidopsis and rice as putative co-chaperones of Hsp90/Hsp70. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0012761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth K.R., Kovalev N., de Castro Martin I.F., Baker J., Nagy P.D. Screening a yeast library of temperature-sensitive mutants reveals a role for actin in tombusvirus RNA recombination. Virology. 2016;489:233–242. doi: 10.1016/j.virol.2015.12.007. [DOI] [PubMed] [Google Scholar]
- Prasanth K.R., Chuang C., Nagy P.D. Co-opting ATP-generating glycolytic enzyme PGK1 phosphoglycerate kinase facilitates the assembly of viral replicase complexes. PLoS Pathog. 2017;13(10) doi: 10.1371/journal.ppat.1006689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran K.S., Nagy P.D. Kinetics and functional studies on interaction between the replicase proteins of Tomato bushy stunt virus: requirement of p33:p92 interaction for replicase assembly. Virology. 2006;345(1):270–279. doi: 10.1016/j.virol.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Rajsbaum R., Garcia-Sastre A. Viral evasion mechanisms of early antiviral responses involving regulation of ubiquitin pathways. Trends Microbiol. 2013;21(8):421–429. doi: 10.1016/j.tim.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranji A., Boris-Lawrie K. RNA helicases: emerging roles in viral replication and the host innate response. RNA Biol. 2010;7(6):775–787. doi: 10.4161/rna.7.6.14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J., Gack M.U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig R., Nillegoda N.B., Mayer M.P., Bukau B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019;20(11):665–680. doi: 10.1038/s41580-019-0133-3. [DOI] [PubMed] [Google Scholar]
- Sasvari Z., Izotova L., Kinzy T.G., Nagy P.D. Synergistic roles of eukaryotic translation elongation factors 1Bgamma and 1A in stimulation of tombusvirus minus-strand synthesis. PLoS Pathog. 2011;7(12) doi: 10.1371/journal.ppat.1002438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopf F.H., Huber E.M., Dodt C., Lopez A., Biebl M.M., Rutz D.A., Muhlhofer M., Richter G., Madl T., Sattler M., Groll M., Buchner J. The co-chaperone Cns1 and the recruiter protein Hgh1 link Hsp90 to translation elongation via chaperoning elongation factor 2. Mol. Cell. 2019;74(1):73–87. doi: 10.1016/j.molcel.2019.02.011. e8. [DOI] [PubMed] [Google Scholar]
- Schroder M. Viruses and the human DEAD-box helicase DDX3: inhibition or exploitation? Biochem. Soc. Trans. 2011;39(2):679–683. doi: 10.1042/BST0390679. [DOI] [PubMed] [Google Scholar]
- Schwerk J., Negash A., Savan R., Gale M., Jr. Innate immunity in hepatitis C virus infection. Cold Spring Harb. Perspect. Med. 2020 doi: 10.1101/cshperspect.a036988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah Nawaz-Ul-Rehman M., Martinez-Ochoa N., Pascal H., Sasvari Z., Herbst C., Xu K., Baker J., Sharma M., Herbst A., Nagy P.D. Proteome-wide overexpression of host proteins for identification of factors affecting tombusvirus RNA replication: an inhibitory role of protein kinase C. J. Virol. 2012;86(17):9384–9395. doi: 10.1128/JVI.00019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D., Jankowsky E. The Ded1/DDX3 subfamily of DEAD-box RNA helicases. Crit. Rev. Biochem. Mol. Biol. 2014;49(4):343–360. doi: 10.3109/10409238.2014.931339. [DOI] [PubMed] [Google Scholar]
- Shulla A., Randall G. (+) RNA virus replication compartments: a safe home for (most) viral replication. Curr. Opin. Microbiol. 2016;32:82–88. doi: 10.1016/j.mib.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan K.E., Bohnsack M.T. Unravelling the mechanisms of RNA helicase regulation. Trends Biochem. Sci. 2018;43(4):237–250. doi: 10.1016/j.tibs.2018.02.001. [DOI] [PubMed] [Google Scholar]
- Soosaar J.L., Burch-Smith T.M., Dinesh-Kumar S.P. Mechanisms of plant resistance to viruses. Nat. Rev. Microbiol. 2005;3(10):789–798. doi: 10.1038/nrmicro1239. [DOI] [PubMed] [Google Scholar]
- Stork J., Kovalev N., Sasvari Z., Nagy P.D. RNA chaperone activity of the tombusviral p33 replication protein facilitates initiation of RNA synthesis by the viral RdRp in vitro. Virology. 2011;409(2):338–347. doi: 10.1016/j.virol.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguwa S., Maringer K., Li X., Bernal-Rubio D., Rauch J.N., Gestwicki J.E., Andino R., Fernandez-Sesma A., Frydman J. Defining Hsp70 subnetworks in dengue virus replication reveals key vulnerability in flavivirus infection. Cell. 2015;163(5):1108–1123. doi: 10.1016/j.cell.2015.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguwa S., Yeh M.T., Rainbolt T.K., Nayak A., Shao H., Gestwicki J.E., Andino R., Frydman J. Zika virus dependence on host Hsp70 provides a protective strategy against infection and disease. Cell Rep. 2019;26(4):906–920. doi: 10.1016/j.celrep.2018.12.095. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschuk F., Cherry S. DEAD-box helicases: sensors, regulators, and effectors for antiviral defense. Viruses. 2020;12(2):181. doi: 10.3390/v12020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassetto M., Kunitomi M., Andino R. Circulating immune cells mediate a systemic RNAi-based adaptive antiviral response in drosophila. Cell. 2017;169(2):314–325. doi: 10.1016/j.cell.2017.03.033. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay A., Dixit U., Manvar D., Chaturvedi N., Pandey V.N. Affinity capture and identification of host cell factors associated with hepatitis C virus (+) strand subgenomic RNA. Mol. Cell. Proteomics. 2013;12(6):1539–1552. doi: 10.1074/mcp.M112.017020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadya M.H., Aweya J.J., Tan Y.J. Understanding the interaction of hepatitis C virus with host DEAD-box RNA helicases. World J. Gastroenterol. 2014;20(11):2913–2926. doi: 10.3748/wjg.v20.i11.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system, autophagy, and regulated protein degradation. Annu. Rev. Biochem. 2017;86:123–128. doi: 10.1146/annurev-biochem-061516-044859. [DOI] [PubMed] [Google Scholar]
- Verchot J. Cellular chaperones and folding enzymes are vital contributors to membrane bound replication and movement complexes during plant RNA virus infection. Front. Plant Sci. 2012;3:275. doi: 10.3389/fpls.2012.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot J. Plant virus infection and the ubiquitin proteasome machinery: arms race along the endoplasmic reticulum. Viruses. 2016;8(11):314. doi: 10.3390/v8110314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu. Rev. Phytopathol. 2015;53:45–66. doi: 10.1146/annurev-phyto-080614-120001. [DOI] [PubMed] [Google Scholar]
- Wang R.Y., Stork J., Nagy P.D. A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J. Virol. 2009;83(7):3276–3287. doi: 10.1128/JVI.02313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.Y., Stork J., Pogany J., Nagy P.D. A temperature sensitive mutant of heat shock protein 70 reveals an essential role during the early steps of tombusvirus replication. Virology. 2009;394(1):28–38. doi: 10.1016/j.virol.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Cao X., Liu M., Zhang R., Zhang X., Gao Z., Zhao X., Xu K., Li D., Zhang Y. Hsc70-2 is required for Beet black scorch virus infection through interaction with replication and capsid proteins. Sci. Rep. 2018;8(1):4526. doi: 10.1038/s41598-018-22778-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Ohtaki N., Hayashi Y., Ikuta K., Tomonaga K. Autogenous translational regulation of the Borna disease virus negative control factor X from polycistronic mRNA using host RNA helicases. PLoS Pathog. 2009;5(11) doi: 10.1371/journal.ppat.1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.Y., Nagy P.D. Blocking tombusvirus replication through the antiviral functions of DDX17-like RH30 DEAD-box helicase. PLoS Pathog. 2019;15(5) doi: 10.1371/journal.ppat.1007771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Hobman T.C. The helicase activity of DDX56 is required for its role in assembly of infectious West Nile virus particles. Virology. 2012;433(1):226–235. doi: 10.1016/j.virol.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Lin J.Y., Nagy P.D. The hop-like stress-induced protein 1 cochaperone is a novel cell-intrinsic restriction factor for mitochondrial tombusvirus replication. J. Virol. 2014;88(16):9361–9378. doi: 10.1128/JVI.00561-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Zhu N., Chen S., Zhao P., Ren H., Zhu S., Tang H., Zhu Y., Qi Z. E3 ubiquitin ligase Nedd4 promotes Japanese encephalitis virus replication by suppressing autophagy in human neuroblastoma cells. Sci. Rep. 2017;7:45375. doi: 10.1038/srep45375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda-Inoue M., Kuroki M., Ariumi Y. Distinct DDX DEAD-box RNA helicases cooperate to modulate the HIV-1 rev function. Biochem. Biophys. Res. Commun. 2013;434(4):803–808. doi: 10.1016/j.bbrc.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Kim T., Bao M., Facchinetti V., Jung S.Y., Ghaffari A.A., Qin J., Cheng G., Liu Y.J. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34(6):866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., He G., Filipowicz N.A., Randall G., Belov G.A., Kopek B.G., Wang X. Host lipids in positive-strand RNA virus genome replication. Front. Microbiol. 2019;10:286. doi: 10.3389/fmicb.2019.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]