Abstract
Viruses are linked to a multitude of human illnesses and can disseminate widely in urbanized environments causing global adverse impacts on communities and healthcare infrastructures. Wastewater-based epidemiology was employed using metagenomics and quantitative polymerase chain reaction (qPCR) assays to identify enteric and non-enteric viruses collected from a large urban area for potential public health monitoring and outbreak analysis. Untreated wastewater samples were collected from November 2017 to February 2018 (n = 54) to evaluate the diversity of human viral pathogens in collected samples. Viruses were classified into virus types based on primary transmission routes and reviewed against viral associated diseases reported in the catchment area. Metagenomics detected the presence of viral pathogens that cause clinically significant diseases reported within the study area during the sampling year. Detected viruses belong to the Adenoviridae, Astroviridae, Caliciviridae, Coronaviridae, Flaviviridae, Hepeviridae, Herpesviridae, Matonaviridae, Papillomaviridae, Parvoviridae, Picornaviridae, Poxviridae, Retroviridae, and Togaviridae families. Furthermore, concentrations of adenovirus, norovirus GII, sapovirus, hepatitis A virus, human herpesvirus 6, and human herpesvirus 8 were measured in wastewater samples and compared to metagenomic findings to confirm detected viral genus. Hepatitis A virus obtained the greatest average viral load (1.86 × 107 genome copies/L) in wastewater samples compared to other viruses quantified using qPCR with a 100% detection rate in metagenomic samples. Average concentration of sapovirus (1.36 × 106 genome copies/L) was significantly greater than norovirus GII (2.94 × 104 genome copies/L) indicating a higher burden within the study area. Findings obtained from this study aid in evaluating the utility of wastewater-based epidemiology for identification and routine monitoring of various viruses in large communities. This methodology has the potential to improve public health responses to large scale outbreaks and viral pandemics.
Keywords: Wastewater-based epidemiology, Public health, Viral diseases, Metagenomics, Virus diversity, qPCR
Graphical abstract
1. Introduction
Viruses are linked to a host of illness related to respiratory infections, diarrheal illness, autoimmune diseases, meningitis, hepatitis, cancer, viral hemorrhagic fevers and others. Infections often disseminate quickly in urbanized regions due to densely populated areas and could reach thousands of inhabitants before health care facilities are notified. Since viruses do not replicate outside of a host and can remain stable in the environment for significant periods of time, wastewater-based epidemiology (WBE) can be used to capture a near real-time picture of the viral disease burden within a community. Viruses can enter waste streams through multiple routes including stool, urine, skin, saliva, and blood, thus wastewater has the potential to assess the burden of a variety of viruses.
It is well known that confirmed enteric viruses, such as rotaviruses, adenoviruses, enteroviruses, hepatitis A and E viruses, caliciviruses, and others can be detected in wastewater. While it is logical to investigate the applicability of enteric viruses to WBE, it is also important to demonstrate the potential for other viruses to fit into this methodology. Indeed, it has been shown that multiple, non-enteric viruses such as coronaviruses, herpesviruses, influenza, zika, West Nile, yellow fever, dengue and others have been detected in stool and urine samples or wastewater (Barzon et al., 2013; Gourinat et al., 2015; Gundy et al., 2009; Heijnen and Medema, 2011; Hirayama et al., 2012; Hirose et al., 2016; O’Brien and Xagoraraki, 2019; Poloni et al., 2010; Tonry et al., 2005; Xagoraraki and O’Brien, 2020). These observations confirm that the concept of wastewater-based-epidemiology can be applied to a wide range of viruses beyond the confirmed enteric viruses.
Enteric viruses are commonly discovered in untreated wastewater (Farkas et al., 2018a; Ng et al., 2012; Victoria et al., 2014) and recent studies have confirmed corresponding disease prevalence in the surrounding community (Bisseux et al., 2018). Particularly, picornaviruses constitute an important group of enteric viruses that cause a host of illnesses including diseases of the central nervous systems, respiratory tract, liver, and gastrointestinal tract. Moreover, caliciviruses are common causes of diarrheal illnesses, which were among the top causes of death worldwide in 2017 (GBD 2017 Causes of Death Collaborators, 2018). With that, noroviruses are among the leading causes of gastroenteritis and a significant agent in global viral outbreak of acute gastroenteritis (Glass et al., 2009; Hall et al., 2013). Routine monitoring of picornaviruses and caliciviruses in wastewater can provide insight into the transmission of clinically important diseases, prevent widespread outbreaks, and reduce deaths linked to such viruses.
Molecular and sequencing approaches provide qualitative and quantitative insights into wastewater environments (McCall and Xagoraraki, 2019). Metagenomics allows for the screening of a large panel of viruses in environmental systems that would otherwise prove time consuming using traditional laboratory techniques. Metagenomics has identified co-infecting organisms during outbreak conditions (Li et al., 2019), novel pathogens (Cantalupo et al., 2011), and viral compositions in complex matrices (Fancello et al., 2013; Miranda et al., 2016; O’Brien et al., 2017). Despite its breakthroughs, the sensitivity of metagenomics to identify vial pathogens is confounded by the presence of bacteria, sequencing limitations, and errors imposed by sequence analysis and alignment tools. For these reasons sensitive PCR techniques are commonly used to corroborate results obtained from metagenomic analysis.
Here, metagenomic analysis was utilized to assess the diversity of human viral pathogens in untreated wastewater collected from a large urban center over the course of four months. Detected viral pathogens were further classified according to virus type and compared with health data, with an emphasis on picornaviruses, from the associated community to evaluate the application of wastewater-based epidemiology for identification of endemic disease and potential upcoming viral outbreaks. Quantitative PCR and RT-qPCR assays were performed on select viruses commonly present in sewage to corroborate results obtained through metagenomic approaches. Finally, norovirus and sapovirus were compared in wastewater to assess the burden of caliciviruses on gastrointestinal illness cases reported in the community.
2. Methods
2.1. Study area and wastewater sample collection
Wastewater surveillance was conducted at the Water Resource Recovery Facility (WRRF) located in Detroit, Michigan. The Detroit WRRF is the largest single site wastewater treatment plant in the U.S. and treats wastewater from an estimated 3 million inhabitants with an average daily flow of 650 MGD (GLWA, 2018). It services the three largest counties, by population, in Michigan. These are Wayne, Oakland, and Macomb counties (Jones et al., 2015). The WRRF receives wastewater from its service municipalities via three main interceptors: North Interceptor-East Arm (NI-EA), Detroit River Interceptor (DRI), and Oakwood-Northwest-Wayne County Interceptor (O-NWI). These interceptors are large sewers that collect and transports wastewater from smaller sewers to the WRRF.
Untreated wastewater samples were collected at the WRRF from sampling points located at each of the three interceptors approximately bi-weekly between November 2017 and February 2018 (n = 54). Viruses were isolated from untreated wastewater using electropositive NanoCeram column filters following the EPA’s virus adsorption-elution protocol (U.S. EPA, 2001). Samples were collected in triplicates for each interceptor per sampling date where wastewater was passed through a column filter until fouling occurred. Average filtered sample volumes range between 24 and 44 L per interceptor. Each interceptor was sampled with its own filter house, tubing, and vacuum pump to minimize cross contamination. Virus filters were immediately stored on ice and transported to the Environmental Virology Laboratory at Michigan State University (MSU) and stored in −20 °C until further processing.
2.2. Sample processing and virus isolation
Following wastewater sampling, NanoCeram cartridge filters were eluted within 24 h with 1.5% w/v beef extract (0.05 M glycine, pH 9.5) according to the EPA’s protocol (U.S. EPA, 2001). In short, filters were eluted with 1 L of beef extract for a total of 2 min. The pH of the solution was adjusted to 3.5 ± 0.1 and flocculated for 30 min before centrifugation at 2500g for 15 min at 4 °C. Supernatant was discarded and pellets were resuspended in 30 mL of 0.15 M sodium phosphate (pH 9.0–9.5) followed by a second round of centrifugation carried out at 7000 g for 10 min at 4 °C. The supernatant was neutralized (pH ∼7.25) and subjected to filtration using to 0.45 μm and 0.22 μm syringe filters to eliminate bacterial contamination. Extraction of nucleic acid was performed on 140 μL of purified virus concentrate using the QIAamp Viral RNA Mini Kit (Qiagen) following the manufacturer’s protocol and eluted in 80 μL of elution buffer. Nucleic acid was stored at −80 °C until further processing.
2.3. Metagenomic analysis
2.3.1. Sampling processing and random amplification
To explore human virus diversity between sampling locations and dates, purified nucleic acid from each biological replicate was pooled together for a total of 18 samples. These samples represent genetic material from all three interceptors during each of the six sampling dates. Nucleic acid from each sample was reverse transcribed and subjected to random amplification as previously described (Wang et al., 2003) to evaluate both RNA and DNA viruses.
2.3.2. Next generation sequence processing
Eighteen samples of viral cDNA were sent to the Research Technology Support Facility Genomics Core at Michigan State University for whole-genome shotgun sequencing (WGS). The Illumina TruSeq Nano DNA Library Preparation Kit was used for all cDNA samples. Library preparation was performed on a PerkinElmer Sciclone G3 robot according to the manufacturer’s recommendations. This was followed by sequencing on an Illumina HiSeq4000 platform generating 150 bp paired-end reads.
2.3.3. Sequence analysis and taxonomic annotation
Sequencing reads generated from WGS were processed on a Unix system through the MSU High Performance Computing Center (HPCC). Raw sequences were analyzed for quality using FastQC, a quality control tool for sequencing data (Andrews, 2010). Sequencing adapters and reads with an average quality score below 20 were removed using Trimmomatic (Bolger et al., 2014). Trimmed reads were assembled with IDBA-UD, a short-read de novo sequence aligner for metagenomic data. Reads were assembled into contigs using an iterative k-mer approach with k-mer sizes ranging between 40 and 120 in increments of 10. The remaining parameters were run at default conditions.
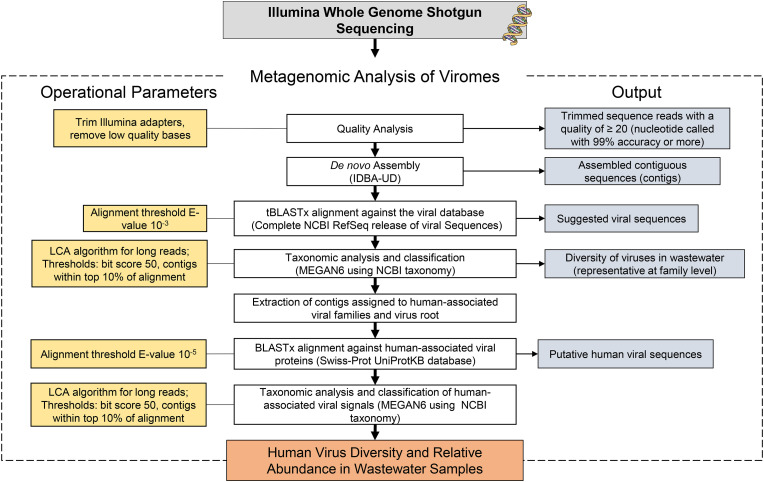
Human virus genomes are relatively small and less abundant in wastewater and therefore may be masked by more dominant bacteria or plant virus genomes. It can also be a challenge for reference databases to maintain updated sequence information for viruses possessing high mutation rates, particularly RNA viruses, leaving room for false negatives and a large percentage of unaffiliated contigs (McCall and Xagoraraki, 2019). To compensate for some of these limitations, an optimized multi-alignment approach was used to improve alignment and annotation of human viral contigs. First, contigs were aligned against the Viral RefSeq database using tBLASTx with an e-value of 10−3. This approach has been known to increase human viral discovery in metagenomic datasets (Bibby et al., 2011). Aligned contigs were assigned to the lowest common ancestor (LCA) according to the NCBI’s taxonomy with MEGAN (v. 6.15.0). The top 10 percent of BLAST alignments with a minimum bit score of 50 and contig coverage of at least 80% were considered in taxonomic analysis. The remaining parameters were run at default conditions. Reads assigned to virus families containing known human pathogens were extracted and aligned with BLASTx with an e-value of 10−5 against a custom human virus database containing 5979 human viral proteins in Swiss-Prot database (Boeckmann et al., 2003). These sequences represented all human viral proteins in the Swiss-Prot database at the time of retrieval (September 2019). Target specific databases can reduce ambiguity and improve pathogens discovery. Furthermore, protein searches are more effective at capturing remote homology as compared to nucleotide searches (Breitwieser et al., 2018). These optimized detection approaches are important for human viral pathogens, which are often difficult to detect in environmental samples due to their low abundance, small genomes, and high mutation rates. To further increase the potential for pathogen discovery and minimize false negatives, contigs assigned to the virus root were also aligned against the Swiss-Prot database. This method was more effective at capturing select viruses confirmed through qPCR and RT-qPCR compared to contigs solely extracted from human viral families (data not shown). Fig. 1 displays the metagenomics workflow for human virus identification in wastewater samples.
Fig. 1.
Metagenomic workflow for human virus identification in wastewater samples.
2.4. Quantification of select viruses
Quantitative PCR or RT-qPCR was performed on six viruses, namely, sapovirus (SaV), norovirus (NoV) GII, human adenovirus (HAdV) 40 and 41, hepatitis A virus (HAV), and human herpesvirus 6 (HHV-6) and 8 (HHV-8).
2.4.1. Preparation of standard curves
HAV and HAdV were obtained from ATCC for preparation of standard controls. Nucleic acid was extracted from each virus isolate as detailed in the previous section and transformed into One Shot TOP10 chemically competent Escherichia coli cells using the TOPO Cloning kit (Invitrogen) following the manufacturer’s protocol. Plasmid DNA containing cloned HAV and HAdV was extracted and quantified according to a previous method (Munir et al., 2011). The protocol detailed in step two of the subsequent section was utilized to prepare a standard curve with 10-fold serial dilutions of positive HAV and HAdV controls.
Quantitative synthetic NoV GII RNA, SaV RNA, HHV-6 DNA, and HHV-8 DNA was obtained from ATCC. RNA or DNA was diluted 10-fold and analyzed as described in the following section. Standard curves for HAV, HAdV, NoV GII, SaV, HHV-6, and HHV-8 obtained R-squared values of >99% and slopes of −3.61, −3.66, −3.82, −3.35, −3.88, and −3.59, respectively. The limit of detection (LOD) for each virus was determined by the lowest point on the standard curve or by the lowest dilution with a 95% positive detection rate in at least 10 replicates (Burns and Valdivia, 2008). HAV, HAdV, NoV, SaV, and HHVs obtained a LOD of 101 genome copies/ul.
2.4.2. qPCR and RT-qPCR
Quantitative PCR or RT-qPCR assays were used to establish the concentration of HAV, HAdV, NoV GII, SaV, HHV-6, and HHV-8 in wastewater samples. All assays were performed in triplicates on a Mastercycler ep realplex2 (Eppendorf) in 96-well optical plates. Amplification of cDNA was mediated using Lightcycler 480 Probes Master (Roche) at a concentration of 1 × in all reactions. Sterile nuclease free water was used to meet volume requirements in all reactions. Primers and probes used are shown in Table A1.
Table 1.
Total number of contigs per virus family with associated host for 18 sequenced samples.
| Family | Host | Total No. of Contigs |
|---|---|---|
| Siphoviridae | Prokaryotes | 627615 |
| Myoviridae | Prokaryotes | 626034 |
| Podoviridae | Prokaryotes | 360840 |
| Microviridae | Prokaryotes | 62058 |
| Herelleviridae | Prokaryotes | 22218 |
| unclassified bacterial viruses | Prokaryotes | 18327 |
| unclassified Caudovirales | Prokaryotes | 13681 |
| Inoviridae | Prokaryotes | 9085 |
| Ackermannviridae | Prokaryotes | 6670 |
| Leviviridae | Prokaryotes | 2362 |
| unclassified archaeal viruses | Prokaryotes | 87 |
| Lipothrixviridae | Prokaryotes | 18 |
| Bicaudaviridae | Prokaryotes | 28 |
| Iridoviridae | Animal Invertebrates | 3874 |
| Baculoviridae | Animal Invertebrates | 849 |
| Ascoviridae | Animal Invertebrate | 299 |
| Polydnaviridae | Animals Invertebrates | 29 |
| Dicistroviridae | Animals Invertebrates | 28 |
| Nudiviridae | Animals Invertebrates | 17 |
| Herpesviridae | Animal Vertebrates (includes Humans) | 6742 |
| Poxviridae | Animal Vertebrates (includes Humans) | 5854 |
| Parvoviridae | Animal Vertebrates (includes Humans) | 875 |
| Retroviridae | Animal Vertebrates (includes Humans) | 50 |
| Circoviridae | Animal Vertebrates | 1297 |
| Alloherpesviridae | Animal Vertebrates | 388 |
| Phycodnaviridae | Plants | 56073 |
| Virgaviridae | Plants | 168 |
| Potyviridae | Plants | 108 |
| Caulimoviridae | Plants | 27 |
| Mimiviridae | Protists | 26077 |
| Marseilleviridae | Protists | 7009 |
| Lavidaviridae | Other | 1623 |
| unclassified DNA viruses | Other | 35010 |
| unclassified Riboviria | Other | 9353 |
| Unclassified viruses | Other | 4672 |
| Genomoviridae | Other | 362 |
HAV and SaV was quantified using a two-step RT-qPCR based on a previously described methods (Jothikumar et al., 2005; Oka et al., 2006). Briefly, viral RNA was reverse transcribed using iScript RT-qPCR Supermix (Bio-Rad) according to the manufacturer’s protocol. For HAV, 5 μL of cDNA, negative control, or positive control was transferred to a 15 μl reaction mix containing HAV primers and TaqMan probe. Reactions were performed with the following conditions: 95 °C for 15 min, followed by 45 cycles of 95 °C for 15 s, 55 °C for 20 s, and 72 °C for 15 s. SaV quantification was carried out in a 25 μL reaction containing each primer and probe. Reactions were performed with the following conditions: 95 °C for 15 min, followed by 45 cycles of 94 °C for 15 s, 62 °C for 1 min, and 72 °C for 15 s.
Norovirus GII was quantified using a one-step RT-qPCR as previously described (Le Guyader et al., 2009). In short, the RT-qPCR was carried out in a 25 μL reaction mixture containing primers and probe, 2 μL of iScript RT-qPCR Supermix, and 5 μL of viral RNA, negative control, or positive control. Reactions were performed with the following conditions: reverse transcription at 25 °C for 5 min, 46 °C for 20 min, and 95 °C for 1 min, followed by 45 cycles of 95 °C for 15 s, 60 °C for 1 min, 65 °C for 1 min.
DNA viruses HAdV, HHV-6, and HHV-8 were quantified according to previously established methods (Gautheret-Dejean et al., 2002; Lallemand et al., 2000; Xagoraraki et al., 2007). HAdV and HHVs were qualified in 20 μL reactions containing 5 μL of DNA or standard control. Denaturation was carried out at 95 °C for 15 min for all DNA viruses followed by 45 cycles at 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 10 s for HAdV; 45 cycles of 95 °C for 15 s and 60 °C for 1 min for HHV-6; and 45 cycles of 95 °C for 15 s and 65 °C for 1 min for HHV-8.
2.5. Health data collection
Disease data for all reportable viral diseases for each service county was obtained from the Michigan Department of Health and Human Services (MDHHS). Probable and confirmed case counts were extracted from the Michigan Disease and Surveillance System (MDSS) weekly surveillance reports (WSR). The MDSS is a communicable disease reporting system used to facilitate coordination and sharing of disease surveillance data among multiple shareholders including healthcare providers and medical laboratories (MDHHS, 2020a). Each weekly surveillance report accounts for disease cases reported from Sunday-Saturday of the corresponding week. It is important to note that the WSR uses gastrointestinal illness (GI) and influenza-like illness (ILI) to represent any disease displaying symptoms of this nature. The etiological agent of the disease is unspecified, but could be of viral, bacterial, or parasitic origin. ILI was defined according to the U.S. influenza surveillance system (CDC, 2019a). GI is defined as symptoms related to diarrhea and/or vomiting (MDHHS, 2018a). Diseases reported in WSRs were considered if is the primary disease of viruses detected in metagenomic samples and if there was at least one case of that disease reported within Macomb, Oakland, or Wayne Counties during the sampling year (2017–2018).
2.6. Statistical and cluster analysis
Cluster analysis was performed using the Bray-Curtis dissimilarity index in MEGAN on metagenomic samples at the family level within the Swiss-Prot taxonomic analysis to determine similarity between samples. A one-way analysis of variance (ANOVA) and Tukey’s HSD post hoc tests were used to investigate significance between mean concentrations of select viruses in wastewater samples. Finally, Spearman’s correlation analysis was used to determine significant associations between select caliciviruses and the number of norovirus and GI reported cases within the service community. All statistical analyses were performed in R (R Core Team, 2019).
3. Results
Illumina sequencing was performed on 18 untreated wastewater samples collected from Detroit’s WRRF from November 2017 to February 2018. A total of 624.4 million reads were subject to quality trimming resulting in 595.2 million reads. Reads were assembled and aligned using tBLASTx against the Viral RefSeq database. The proportion of contigs assigned to viral taxonomic groups range between 72 and 83% (Table A2). As expected, viruses infecting prokaryotes constituted the greatest proportion of viral reads (Table 1 ).
Table 2.
Summary of human viral pathogens detected in wastewater and their associated disease reported in the Michigan Disease Surveillance System (MDSS) Weekly Surveillance Reports (WSR). Associated disease is considered if at least one case was reported during the sampling year (2017–2018).
| Measurements |
MDSS Reportsa |
Other |
||||
|---|---|---|---|---|---|---|
| Virus Family | Virus Genus | Specific Primary Reported Disease | Non-specific Reported Illness | Primary Virus Type (Transmission Route) | Oncogenic? | References |
| Adenoviridae | Mastadenovirus | GI; IFI; Encephalitis, Primary | Enteric; Respiratory | Ghebremedhin (2014) | ||
| Astroviridae | Mamastrovirus | GI | Enteric | Bosch et al. (2014) | ||
| Caliciviridae | Norovirus | Norovirus | GI | Enteric | Glass et al. (2009) | |
| Sapovirus | GI | Enteric | Oka et al. (2015) | |||
| Coronaviridae | Betacoronavirus | Novel Coronavirus | ILI; GI | Respiratory | Chan et al. (2015); Kuiken et al. (2003); Lai et al. (2020) | |
| Flaviviridae | Hepacivirus | Hepatitis C | Bloodborne | Y | Chen and Morgan (2006) | |
| Hepeviridae | Orthohepevirus | Hepatitis E | Guillain-Barre Syndrome | Enteric | Kamar et al. (2014); Van Den Berg et al. (2014) | |
| Herpesviridae | Lymphocryptovirus | Encephalitis, Primary; Guillain-Barre Syndrome; ILI | Bloodborne; Other | Y | Tsao et al. (2015); Van Den Berg et al. (2014) | |
| Roseolovirus | Encephalitis, Primary; GI; ILI | Other; Respiratory | De Bolle et al. (2005); Hall et al. (1994) | |||
| Simplexvirus | Encephalitis, Primary; ILI; Meningitis - Aseptic | Other | Whitley (2011); Widener and Whitley (2014) | |||
| Varicellovirus | Chickenpox; Shingles; VZ infection unspecified; Encephalitis, Post Chickenpox | Respiratory | Arvin (1996) | |||
| Matonaviridae | Rubivirus | Rubella | Guillain-Barre Syndrome; Encephalitis, Primary; ILI | Respiratory | Mawson and Croft (2019) | |
| Papillomaviridae | Alphapapillomavirus | Other | Y | Doorbar et al. (2015) | ||
| Parvoviridae | Bocaparvovirus | ILI; GI | Enteric; Respiratory | Qiu et al. (2017) | ||
| Erythroparvovirus | ILI | Bloodborne; Respiratory | Qiu et al. (2017) | |||
| Picornaviridae | Cardiovirus | ILI; GI; Encephalitis, Primary | Enteric | Tan et al. (2017) | ||
| Enterovirus | Encephalitis, Primary; ILI; GI; Acute Flaccid Myelitis (AFM); Meningitis - Aseptic | Enteric; Respiratory | Wells and Coyne (2019) | |||
| Hepatovirus | Hepatitis A | Guillain-Barré syndrome | Enteric | Lemon et al. (2018) | ||
| Parechovirus | ILI; GI; Meningitis - Aseptic | Enteric; Respiratory | de Crom et al. (2016) | |||
| Poxviridae | Orthopoxvirus | ILI | Other; Respiratory | Buller and Palumbo (1991); Haller et al. (2014) | ||
| Parapoxvirus | Other | Buller and Palumbo (1991); Fox et al. (2002) | ||||
| Retroviridae | Deltaretrovirus | Bloodborne | Y | Gonçalves et al. (2010); Ishitsuka and Tamura (2014) | ||
| Gammaretrovirus | Denner (2010) | |||||
| Lentivirus | HIV | Bloodborne | del Rio (2017) | |||
| Togaviridae | Alphavirus | Eastern equine encephalitis; Chikungunya | Vector-borne | Armstrong and Andreadis (2013); Weaver et al. (2018) | ||
a: Influenza-like illness (ILI) is defined according to the U.S. influenza surveillance systems (CDC, 2019a). Gastrointestinal illness (GI) is defined as symptoms related to diarrhea and/or vomiting (MDHHS, 2018a). ILI and GI were reported if the virus’s primary clinical manifestation is related to that condition. Disease information is not documented for viruses related to non-reportable diseases.
3.1. Classification of human viral pathogens in wastewater
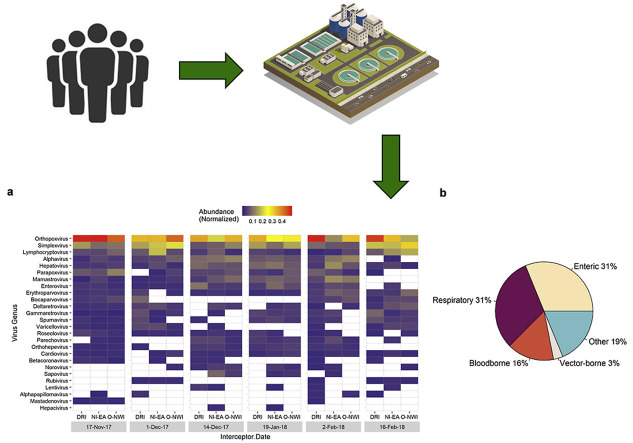
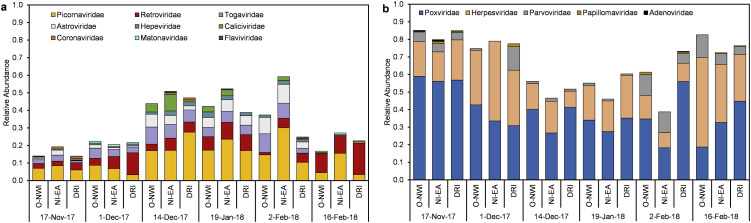
Putative human viral contigs were extracted via MEGAN taxonomic bins and aligned against all human viral proteins in the Swiss-Prot database using BLASTx. An average of approximately 0.18% (0.05–0.78%) of viral affiliated contigs were aligned to human viral proteins. Fourteen different human viral families were identified with the greatest number of contigs largely assigned to Poxviridae, Herpesviridae, and Picornaviridae (Fig. 2 ). Of the fourteen families, nine were classified at ssRNA viruses and the remaining five as DNA viruses. Fig. 3 shows the proportion of ssRNA and DNA viral families in each sample. Comparable relative abundances of ssRNA and DNA viruses were observed during the 14 Dec, 19 Jan, and 2 Feb sampling dates with DNA viruses dominating sequenced samples during the remaining three sampling dates.
Fig. 2.
ssRNA (a) and DNA (b) virus diversity and relative abundance in wastewater samples.
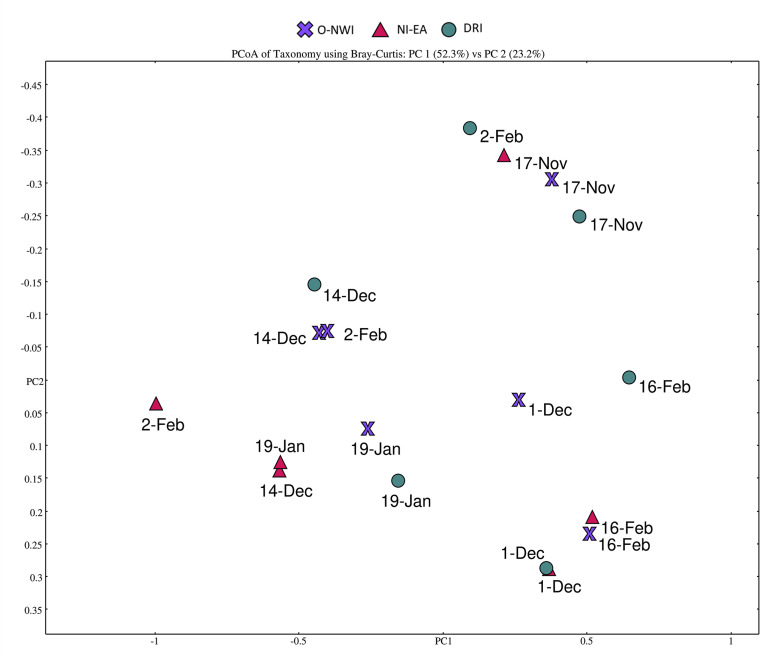
Fig. 3.
Principal component analysis (PCoA) of human viral pathogen presence. PCoA was produced in MEGAN at the family level using Bray-Curtis dissimilarity index.
Bray-Curtis dissimilarity analysis was used to determine the similarity between samples at the family taxonomic level after alignment against human viral protein sequences. According to the Bray-Curtis analysis, there were more similarities within sampling dates rather than sampling locations with samples collected during three consecutive sampling dates (14-Dec.,19-Jan., and 2-Feb.) clustering together (Fig. 3). Poxviridae, Parvoviridae, and Herpesviridae families were the most influential viral families when discriminating between samples (data not shown).
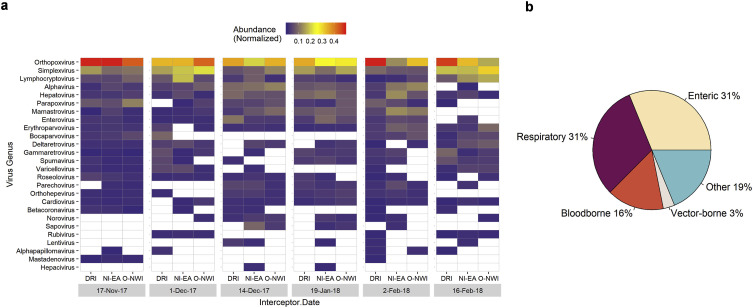
Of the 14 human viral families identified, contigs were assigned to 26 human virus genera with DNA viruses, orthopoxivirus, simplex viruses, and lymphocrytovirus obtaining the greatest number of hits. Alphavirus, a ssRNA virus containing vector-borne viruses was the fourth most abundant genus (Fig. 4 a).
Fig. 4.
(a) Heatmap of human genus virus diversity and normalized abundance in each sample. White cells indicate absence of associated virus in related sample. Virus genera are in descending order according to abundance. Heatmap was produced in R. (b) Proportion of human virus types detected in wastewater samples.
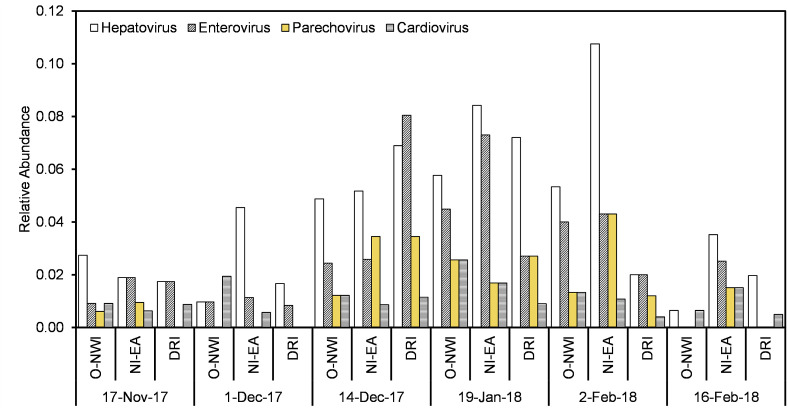
The most frequently detected virus type was enteric and respiratory, followed by other, bloodborne, and vector-borne (Fig. 4b, Table 2). Four of ten enteric viruses detected belong to the Picornaviridae family, namely, hepatovirus, enterovirus, parechovirus, and cardiovirus. Fig. 5 illustrates the temporal and spatial relative abundance of picornaviruses during the study period. Proteins associated with hepatovirus, mamastrovius, and enterovirus obtained the greatest number of assigned contigs within the enteric virus group (Fig. 4a). Within the mamastrovirus genus, classic human astroviruses (HAstV) were detected with >90% homology to the reference sequence in all positive samples (15/18). Enteroviruses, containing respiratory and enteric pathogens, revealed human coxsackievirus and poliovirus species detected in positive samples with >80% identity to the reference gene.
Fig. 5.
Picornaviruses detected in wastewater samples.
Orthopoxivirus constituted the greatest number of hits within the respiratory group with vaccinia virus being the most frequently identified species. Additionally, metagenomic analysis detected the presence of respiratory pathogen betacoronavirus in 8 of the 18 collected samples with human coronavirus HKU1 being the primary species detected with >60% percent identity (Fig. 4a). Other enteric and respiratory viruses detected include mastadenovirus, sapovirus, norovirus, orthohepevirus, varicellovirus, rubivirus, bocaparvovirus, roseolovirus, and erythroparvovirus (Table 2 ).
Viruses solely grouped in the “Other” category consists of viruses transmitted via skin, saliva, or other bodily fluids such as simplex virus, parapoxvirus, and alphapapillomavirus. Furthermore, alphavirus was the only vector-borne virus discovered in metagenomic samples with a detection rate of 89%. Important bloodborne viruses hepacivirus, lentivirus, deltaretrovirus, and lymphocryptovirus were also discovered in metagenomic samples (Fig. 4, Table 2).
3.2. Comparison of human viruses and clinical data
Weekly surveillance reports from the MDHHS MDSS were utilized to evaluate the potential association between the 26 human viruses detected in metagenomic samples and the presence of primary associated diseases within the surrounding community during the study period. Mastadenovirus, mamastrovirus, sapovirus, bocaparvovirus, cardiovirus, enterovirus, and parechovirus were linked to solely non-specific diseases including flu-like, gastrointestinal illnesses, aseptic meningitis, and acute flaccid myelitis. Norovirus, betacoronavirus, and rubivirus were related to at least one non-specific illnesses along with virus-specific diseases norovirus, novel coronavirus, and rubella, respectively. Virus-specific diseases for hepacivirus, orthohepevirus, varicellovirus, hepatovirus, lentivirus, and alphavirus were present in WSRs during the sampling year. During the time of data collection, mandatory reporting was not required for diseases related to several viruses including parapoxvirus, deltaretrovirus, gammaretrovirus, and alphapapillomavirus (Table 2).
3.3. Quantitative screening for select human viral pathogens
To confirm results obtained from metagenomic analysis, select viruses were quantified in wastewater samples. Namely, SaV, NoV GII, HAdV 40/41, HAV, HHV-6, and HHV-8. HAV, NoV, HAdV, and HHV-8 were quantified in all samples and obtained positive detection rate of 100%, 50%, 22%, and 0% in metagenomic samples, respectively. SaV and HHV-6 were quantified in 94% and 39% of the 18 samples considered with a 28% and 83% detection in sequenced samples. All select viruses except HHV-6 were detected during each sampling date using qPCR or RT-qPCR (Fig. 6 ). There were significant differences in average concentrations for some viruses where HAV > HAdV > NoV (p < 0.0001). Mean SaV concentrations were significantly less than HAV (p < 0.0001) and greater than NoV (p < 0.0001). There was no significant difference between SaV and HAdV (p > 0.05). Concentrations of HHV-6 and HHV-8 were significantly lower than HAV, HAdV, SaV, and NoV (p < 0.0001). There was no significant difference between mean concentrations of HHV-6 and HHV-8 (p > 0.05) in collected samples.
Fig. 6.
Boxplot for select viral concentrations per sampling date.
3.4. Calicivirus detection in wastewater
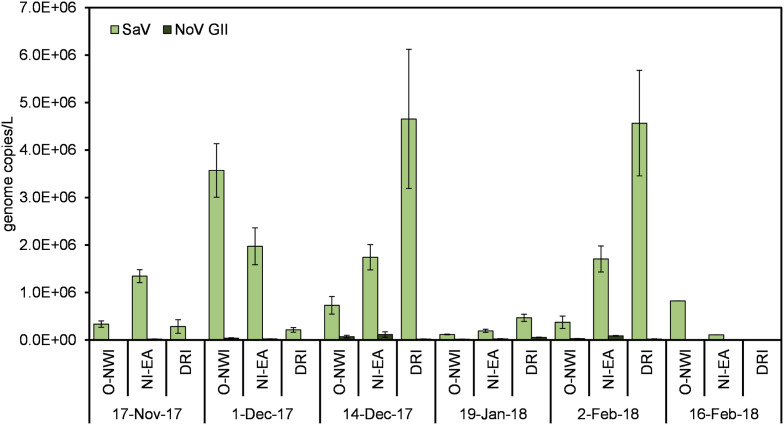
A temporal investigation of NoV GII and SaV concentrations in wastewater samples was carried out to assess the potential burden of these viruses and their contribution to gastrointestinal illnesses within the service community. Average concentrations of SaV and NoV GII were 1.36 × 106 and 2.94 × 104 genome copies/L, respectively. The highest average concentration for both viruses occurred during the 14-Dec sampling date (Fig. 7 ). However, according to spearman’s correlation analysis, there was no significant association between the number of reported GI cases and concentrations of NoV GII and SaV per sampling week (Table 3 ).
Fig. 7.
Temporal comparison of NoV GII and SaV concentrations in collected samples. Error bars represent standard error.
Table 3.
Average concentrations of NoV GII and SaV per sampling date along with number of GI and noroviruses cases reported within the service community. P-value and correlation coefficient were obtained from Spearman’s correlation analysis.
| Sampling Date | Week’s Total No. of Norovirus and GI Cases | Average SaV concentration (genome copies/L) | Spearman (SaV) | Average NoV GII concentration (genome copies/L) | Spearman (NoV GII) |
|---|---|---|---|---|---|
| 17-Nov-17 | 0 | 6.57 E+05 | P = 0.42 | 1.06 E+04 | P = 0.54 |
| 1-Dec-17 | 5 | 2.18 E+06 | rho (ρ) = −0.41 | 2.03 E+04 | rho (ρ) = −0.32 |
| 14-Dec-17 | 0 | 2.38 E+06 | 6.86 E+04 | ||
| 19-Jan-18 | 23 | 2.40 E+05 | 3.14 E+04 | ||
| 2-Feb-18 | 25 | 2.28 E+06 | 4.88 E+04 | ||
| 16-Feb-18 | 48 | 4.66 E+05 | 3.93 E+03 |
4. Discussion
A WBE study for viral diseases was carried out on wastewater samples collected from a large urban municipal wastewater treatment facility. Samples were subjected to metagenomic analysis and qPCR/RT-qPCR assays to identify human viral pathogens circulating within the community. More than half of all aligned contigs were assigned to viral taxonomic groups. The use of virus enrichment techniques has been known to decrease the presence of non-targeted organisms, such as bacteria and improve virus detection within metagenomes (McCall and Xagoraraki, 2019).
Of the virus affiliated contigs, less than 1% were identified as putative human viral pathogens. Due to their relatively small genomes and low abundance in water reservoirs, human viral reads are known to constitute a small portion of metagenomic datasets (McCall and Xagoraraki, 2019). To compensate for this, a second stage of alignment was carried out on contigs identified as potential human viruses against a custom dataset of human viral proteins. Protein-based alignments are effective at detecting remote homology and therefore allows for the discovery of rapidly evolving viral pathogens (Breitwieser et al., 2018). BLASTx alignment facilitated the taxonomic classification of several different virus types including enteric, respiratory, bloodborne and vector-borne viruses.
4.1. Classification of human viral pathogens in wastewater and clinical data comparison
Enteric and respiratory pathogens were the most frequently detected viruses in sewage samples. Enteric viruses are viruses that mainly infect the intestinal tract and can be transmitted via the fecal-to-oral route. Respiratory viruses generally replicate in the respiratory tract and spread via respiratory secretions. Picornaviridae obtained 4 of the 10 enteric viruses detected. Picornaviruses, including hepatovirus, enterovirus, and cardiovirus, are an important group of viruses that display a diverse range of human infections and clinical symptoms. Hepatovirus was detected in all samples with the highest relative abundance among enteric viruses. Hepatitis A virus (HAV) is the type species of the hepatovirus genus and is the causative agent of hepatitis A (Lemon et al., 2018). Over 700 probable and confirmed cases of hepatitis A were reported in the service community during the 2017–2018 sampling years as a result of the 2016 multi-state hepatitis A outbreak (CDC, 2020a; MDHHS, 2020b). The prevalence of HAV in sequenced samples suggest the use of WBE for routine surveillance of hepatitis A outbreaks in communities.
Enteroviruses include enteric (poliovirus, coxsackievirus, echovirus) and respiratory (rhinovirus, enterovirus D68) pathogens (Wells and Coyne, 2019). Enteroviruses cause a host of illnesses including common cold, hand-foot-mouth disease, poliomyelitis, acute flaccid myelitis (AFM), and aseptic meningitis (Wells and Coyne, 2019). In 2018, there was a spike in AFM cases in the U.S. with Michigan obtaining 5 cases in that same year (CDC, 2020b). Although enteroviruses are not the only viruses that cause AFM, there have been well-established links (Dyda et al., 2018). Metagenomic analysis suggest the presence of polioviruses and other enterovirus species in wastewater samples during the 2017–2018 sampling period. This indicates the potential circulation of clinically important enteroviruses in the environment and potential connection to interruptions in community health.
Cardioviruses were believed to mainly infect rodents until 2007 when Saffold virus (SAFV), a novel cardiovirus, was identified in human stool. Since then SAFV has been identified in stool and nasopharyngeal aspirates of patients suffering from gastrointestinal or respiratory illnesses. Since SAFV is commonly present in patients with coinfections, further investigations are needed to determine virus pathology. Moreover, SAFV has also been detected in Cerebrospinal fluid (CSF) of children, but these findings were not consistent across studies, even those focusing on patients with neurological disruptions (Tan et al., 2017). Nonetheless, SAFV associations with neuropathogenesis is of importance given its close relation to Theiler’s Murine encephalomyelitis virus (TMEV), which causes neuropathogenesis in mice (Tan et al., 2017). Cardioviruses have been previously isolated from wastewater (Blinkova et al., 2009; Bonanno Ferraro et al., 2020) and were identified in 17 of 18 samples.
Apart from picornaviruses, mamastrovirus was the most prominent enteric virus genus detected in wastewater samples that primarily causes gastrointestinal illness (Bosch et al., 2014). Astroviruses are commonly detected in the environment during winter months (Bosch et al., 2014) and suggest the prevalence of astrovirus infections within the community during the sampling period. Although there were a significant number of cases linked to gastrointestinal illness during the sampling year (Table A3) it is difficult to assess the burden of astroviruses within the community given the presence of other viruses promoting similar clinical manifestations like norovirus and sapovirus.
Similar to enteric viruses, respiratory viruses were abundant in metagenomic samples with orthopoxivirus obtaining the greatest number of hits among the 26 human viral pathogens identified. Orthopoxiviruses contain respiratory pathogens like variola virus and pathogens transmitted through vaccination, zoonoses, or close contact such as vaccinia virus (Buller and Palumbo, 1991; Haller et al., 2014). Vaccinia virus (VACV) was the most prevalent species detected within the orthopoxivirus genus. VACV has been used widely in human immunization against smallpox (Haller et al., 2014). Although routine vaccination against smallpox is no longer performed in the U.S., recommended vaccination is suggested for individuals who are at risk of exposure, for example, laboratory workers (CDC, 2017). The presence of VACV could be a result of viral shedding from recently vaccinated individuals, silent community spread, or environmental prevalence. Further investigation is needed to determine potential sources of VACV prevalence in the environment.
Moreover, betacoronavirus (BCoV) was detected 8 of 18 wastewater samples. BCoVs are known to cause respiratory illnesses in humans ranging from common cold to more severe diseases like Severe Acute Respiratory Syndrome (SARS), Middle East Respiratory Syndrome (MERS), and Coronavirus Disease 2019 (COVID-19). Frequent mixing of human and animal reservoirs in densely populated areas facilitated outbreaks of viruses from this group in previous years including SARS-CoV in 2003 (Kuiken et al., 2003), MERS-CoV in 2012 (Chan et al., 2015), and SARS-CoV-2 in 2019 (Lai et al., 2020). Along with respiratory ailments, gastrointestinal illnesses have also been reported in patients with BCoV infections (Lai et al., 2020). Albeit viral and clinical data investigations for this study were conducted prior to the COVID-19 pandemic, two possible novel coronavirus cases were reported in the service community during the 2018 sampling year. Additionally, BCoV are known to cause flu-like symptoms and have therefore been linked to ILI reported in the service community. Like GI, the reporting of non-specific diseases poses limitations when determining the burden of such viral pathogens within communities. Routine testing for BCoVs in individuals displaying ILI infections is necessary to prevent large-scale outbreaks of known and novel coronaviruses.
Several clinically important bloodborne pathogens were also detected in sequenced samples. Bloodborne pathogens are often transmitted through contact with infected blood, bodily fluids, or indirect contact with contaminated fomites. Despite the introduction of vaccines and effective medical interventions, bloodborne pathogen hepatitis C virus (HCV) is the leading cause of liver disease worldwide (Chen and Morgan, 2006). HCV is the only species in the Hepacivirus genus known to infect humans. More than 9000 probable cases of hepatitis C were reported in the service community during the 2017–2018 sampling year (Table A3). Despite the high number of disease cases, HCV obtained a low detection rate (2/18). The primary route of transmission, location of viral shedding, duration of infection, and presence of environmental inhibitors can affect the presence of HCV in wastewater. Nonetheless, to our knowledge, this is the first study to detect hepacivirus in untreated wastewater from an urban community.
Along with hepacivirus, human associated lentivirus contigs were detected in 5 of 18 untreated wastewater samples. Lentiviruses are a group of retroviruses that cause chronic and often deadly diseases in vertebrates and are known to have long incubation periods. Human immunodeficiency virus (HIV) 1 and 2 are the only viruses contained in this group that cause infections in humans. HIV is the causative agent of Acquired immunodeficiency syndrome (AIDS). HIV has accounted for more than 30 million deaths since 1981 with the highest burden in southern Africa (del Rio, 2017). The virus is said to have originated from non-human primates through zoonosis and has since evolved to spread through human-to-human contact (Fox et al., 2002). HIV infections are more persistent within the study area as compared other counties (MDHHS, 2018b).
Other bloodborne pathogens include lymphocryptovirus, erythroparvovirus, and deltaretrovirus belonging to the Herpesviridae, Parvoviridae, and Retroviridae families, respectively. Lymphocryptovirus, containing the Epstein–Barrvirus (EBV), was the third most abundant genus in metagenomic samples. EBV is most known for causing mononucleosis (mono) commonly called the kissing disease. The virus is prevalent in >90% of the world’s population and is commonly transmitted through saliva but can also be spread via blood (Tsao et al., 2015). EBV infections are also known to cause epithelial cancers such as nasopharyngeal carcinoma (NPC) and EBV-associated gastric cancers (Tsao et al., 2015). Erythroparvovirus (B19V) is associated with fifth disease, which causes a rash primarily in children. It is transmitted mainly by the respiratory route often causing outbreaks in schools and day care centers. The virus can also be transmitted via blood and blood-contaminated fomites (Qiu et al., 2017). Viruses within the deltaretrovirus genus are recognized for causing Adult T-cell leukemia/lymphoma (ATL) and can be transmitted mother-to-child, sexual intercourse, and blood transfusions (Ishitsuka and Tamura, 2014). No mandatory reporting was required for primary infections associated with the above-mentioned viruses during the study period.
Most samples contained contigs associated with vector-borne genus alphavirus. Alphaviruses consists of mainly viruses transmitted through insect bites (arthropod-borne). Nearly all arthropod-borne viruses are zoonotic (Weaver et al., 2018). Zoonotic viruses, viruses that spread from animals to humans, constitutes an important reservoir of viruses. Many clinically relevant vector-borne viruses are transmitted via mosquitoes including Zika, West Nile, Yellow Fever, Eastern equine encephalitis virus (EEEV), dengue, and chikungunya. Alphavirus consists of several vector-borne viruses including chikungunya virus (CHIKV), EEEV, and Venezuelan equine encephalitis virus (VEEV). Contigs assigned to human viral associated proteins in the alphavirus genus were detected in 16 of 18 samples. EEEV and VEEV were the primary species in 3 and 12 samples, respectively.
EEEV and VEEV cause encephalitis and neurological impairment mainly in equine species and humans and were first identified in the United States in 1933 and in Venezuela in 1938, respectively (Armstrong and Andreadis, 2013; Weaver et al., 2004). Although EEEV infections in humans are rare compared to other clinically relevant vector-borne infections, it is the deadliest with a fatality rate of 35–75% (Armstrong and Andreadis, 2013). This contrasts with VEEV infections where fatalities rates of less than 1% have been reported (Weaver et al., 2004).
One case of EEE was reported in the service area during the 2018 sampling year. Similarly, the CDC reported a spiked in the number of EEE cases in 2019 with 38 confirmed cases and 15 deaths as of December 2019. Among the states affected were Michigan with 10 of the 38 cases (CDC, 2019b). Re-emerging infections of this nature are expected to rise in the future due to urbanization and climate change (McMichael, 2004). This study highlights the potential utility of wastewater to be used as a surveillance tool for vector-borne viruses. To our knowledge, no previous study has reported alphaviruses in raw wastewater samples. Potential exposure routes of alphaviruses into wastewater reservoirs could be facilitated through stormwater collection during rainfall events or viral shedding from infected individuals. Further studies are warranted to examine the fate of vector-borne viruses in wastewater systems.
4.2. Screening of select human viral pathogens
Quantitative analysis of select viruses was performed to strengthen metagenomic findings and assess viral concentrations in collected samples. HAV was detected in all samples during metagenomic analysis with significantly high viral loads in wastewater as compared to other viruses tested. The high occurrence of HAV concentration in wastewater could signify outbreak conditions and critical locations with increased spatial sampling. Along with HAV, HAdV, SaV, NoV GII, and HHV-6 were quantified in wastewater samples with positive detection rates in metagenomic samples. HHVs obtained the lowest concentrations among select viruses. HHV-6 and -8, also referred to as roseolovirus and Kaposi sarcoma (KS) associated herpesvirus, respectively, are non-enteric viruses which could explain their low concentrations in sewage samples as compared to the other viruses screened. HHV-8 was quantified in untreated wastewater using qPCR but went undetected in metagenomic analysis. HHV-8 is commonly transmitted through saliva and is said to be more pervasive among men who have sex with men (MSM) (Engels et al., 2007). Those with HIV are at greater risk for developing KS (Engels et al., 2007). Results here illustrate the utility of WBE to be used for enteric and non-enteric viruses. Furthermore, the integration of NGS with qPCR techniques provides a more sensitive application for low abundant viruses in wastewater systems.
4.3. Calicivirus detection in wastewater
Lastly, a quantitative temporal comparison of NoV GII and SaV was carried out on wastewater samples to determine presence and burden of these viruses within the community. NoV and SaV are genetically similar producing near identical clinical manifestations proving difficult to distinguish without testing. NoV is the leading causes of acute gastroenteritis among adults and children in the U.S. with GII being the responsible genotype in most outbreaks and pandemics (Glass et al., 2009). Nonetheless, results here highlight the burden of sapovirus infections within the surrounding community. Sapovirus cases are not routinely reported in public health databases and therefore are linked to gastrointestinal illnesses in this study. To assess the burden of NoV and SaV within the community one should considered the severity of illness, incubation period, duration of viral shedding, and viral shedding load.
Sapovirus disease symptoms tend to be milder than those produced by norovirus with a similar incubation period of 12–48 h (Lee et al., 2013; Oka et al., 2015). Milder symptoms cause by SaV infection likely impacts the clinical presence of the virus as compared to norovirus infections. Like norovirus, sapovirus infections result in post-symptomatic viral shredding, which can persist for several weeks. Prolonged shedding has been observed in those with compromised immune systems and the elderly in SaV and NoV infections (Glass et al., 2009; Oka et al., 2015). Such information can distort the immediate burden of NoV and SaV infections but can provide insight into other community health indicators. These include prevalence of HIV, other autoimmune infections, and help to identify the outbreak population. Additionally, viral shedding concentrations play a key role in loads captured in wastewater. Previous studies have observed viral shedding loads in feces up to 1011 genome copies/g stool for SaV (Oka et al., 2015) and 1010 genome copies/g stool for NoV (Lee et al., 2007). Likewise, previously reported detection of viral loads in raw wastewater reveal similar concentrations between SaV and NoV (Haramoto et al., 2018).
A recent study observed high concentrations of NoV GII in wastewater following several NoV GII outbreaks within the surrounding community (Iwai et al., 2009) and higher concentration as compared to SaV (Farkas et al., 2018b). Although frequent wastewater sampling is required to confirm the burden of NoV and SaV infections within the community, results here indicate a higher burden of SaV infections during the time of sampling. The lack of correlation between NoV and SaV concentrations and disease cases for norovirus and gastrointestinal illness may indicate interference from wastewater inhibitors, rapid changes in viral concentrations due to the short incubation period, or the presence of other viruses such as astrovirus, adenovirus, or rotavirus that may contribute more significantly to symptomatic illnesses that result in clinical cases.
5. Conclusion
-
•
Metagenomics detected the presence of enteric and non-enteric viruses that cause clinically important diseases that were reported within the study area during the sampling year.
-
•
Findings reveal evidence of re-emerging vector-borne viruses.
-
•
Frequent and rigorous wastewater sampling along with integrative sample processing strategies can be employed to identify the etiological agent of non-specific diseases and viruses that poses a significant burden among inhabitants.
-
•
Results presented in this study suggests that WBE has the potential to advance the area of disease outbreak mitigation and improve public health responses to large scale outbreaks and viral pandemics.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was funded by National Science Foundation Award #1752773. We thank Anil Gosine (Detroit Water and Sewerage Department) and Michael Jurban (Great Lakes Water Authority) for allowing access to the Detroit wastewater treatment utility and assisting with sampling. We thank the Research Technology Support Facility (Michigan State University) for assisting with sequencing and Professor Shinhan Shiu ( Michigan State University) for assisting with bioinformatics analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.watres.2020.116160.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Andrews S. 2010. FastQC. Babraham Bioinforma.https://search.crossref.org/?q=Andrews%2C+S.%2C+2010.+FastQC.+Babraham+Bioinforma [DOI] [Google Scholar]
- Armstrong P.M., Andreadis T.G. Eastern equine encephalitis virus - old enemy, new threat. N. Engl. J. Med. 2013 doi: 10.1056/NEJMp1213696. [DOI] [PubMed] [Google Scholar]
- Arvin A.N.N.M. Varicella-Zoster Virus. 1996;9:361–381. doi: 10.1128/cmr.9.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzon L., Pacenti M., Franchin E., Pagni S., Martello T., Cattai M., Cusinato R., Palù G. Excretion of west nile virus in urine during acute infection. J. Infect. Dis. 2013 doi: 10.1093/infdis/jit290. [DOI] [PubMed] [Google Scholar]
- Bibby K., Viau E., Peccia J. Viral metagenome analysis to guide human pathogen monitoring in environmental samples. Lett. Appl. Microbiol. 2011 doi: 10.1111/j.1472-765X.2011.03014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisseux M., Colombet J., Mirand A., Roque-Afonso A.M., Abravanel F., Izopet J., Archimbaud C., Peigue-Lafeuille H., Debroas D., Bailly J.L., Henquell C. Monitoring human enteric viruses in wastewater and relevance to infections encountered in the clinical setting: a one-year experiment in central France, 2014 to 2015. Euro Surveill. 2018;23:1–11. doi: 10.2807/1560-7917.ES.2018.23.7.17-00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinkova O., Rosario K., Li L., Kapoor A., Slikas B., Bernardin F., Breitbart M., Delwart E. Frequent detection of highly diverse variants of Cardiovirus, Cosavirus, Bocavirus, and Circovirus in sewage samples collected in the United States. J. Clin. Microbiol. 2009;47:3507–3513. doi: 10.1128/JCM.01062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O’Donovan C., Phan I., Pilbout S., Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanno Ferraro G., Mancini P., Veneri C., Iaconelli M., Suffredini E., Brandtner D., La Rosa G. Evidence of Saffold virus circulation in Italy provided through environmental surveillance. Lett. Appl. Microbiol. 2020;70:102–108. doi: 10.1111/lam.13249. [DOI] [PubMed] [Google Scholar]
- Bosch A., Pintó R.M., Guix S. Human astroviruses. Clin. Microbiol. Rev. 2014;27:1048–1074. doi: 10.1128/CMR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitwieser F.P., Lu J., Salzberg S.L. A review of methods and databases for metagenomic classification and assembly. Briefings Bioinf. 2018;20:1125–1139. doi: 10.1093/bib/bbx120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buller R.M.L., Palumbo G.J. Poxvirus pathogenesis. Microbiol. Rev. 1991;55:80–122. doi: 10.1128/mmbr.55.1.80-122.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns M., Valdivia H. Modelling the limit of detection in real-time quantitative PCR. Eur. Food Res. Technol. 2008;226:1513–1524. doi: 10.1007/s00217-007-0683-z. [DOI] [Google Scholar]
- Cantalupo P.G., Calgua B., Zhao G., Hundesa A., Wier A.D., Katz J.P., Grabe M., Hendrix R.W., Girones R., Wang D., Pipas J.M. Raw sewage harbors diverse viral populations. mBio. 2011;2:1–11. doi: 10.1128/mBio.00180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC Widespread person-to-person outbreaks of hepatitis A across the United States. 2020. https://www.cdc.gov/hepatitis/outbreaks/2017March-HepatitisA.htm [WWW Document]. Viral Hepat. (accessed 4.27.20)
- CDC AFM cases and outbreaks. 2020. https://www.cdc.gov/acute-flaccid-myelitis/cases-in-us.html CDC [WWW Document] (accessed 4.27.20)
- CDC U.S. Influenza surveillance system: purpose and methods. 2019. https://www.cdc.gov/flu/weekly/overview.htm [WWW Document]. FluView. (accessed 4.28.20)
- CDC Eastern equine encephalitis. 2019. https://www.cdc.gov/easternequineencephalitis/index.html (accessed 4.27.20)
- CDC Who should get vaccination. 2017. https://www.cdc.gov/smallpox/vaccine-basics/who-gets-vaccination.html#care-for (accessed 4.27.20)
- Chan J.F.W., Lau S.K.P., To K.K.W., Cheng V.C.C., Woo P.C.Y., Yue K.Y. Middle East Respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.L., Morgan T.R. The natural history of hepatitis C virus (HCV) infection. Int. J. Med. Sci. 2006;3:47–52. doi: 10.7150/ijms.3.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bolle L., Naesens L., De Clercq E. Update on human herpesvirus 6 biology, clinical features, and therapy. Clin. Microbiol. Rev. 2005;18:217–245. doi: 10.1128/CMR.18.1.217-245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crom S.C.M., Rossen J.W.A., van Furth A.M., Obihara C.C. Enterovirus and parechovirus infection in children: a brief overview. Eur. J. Pediatr. 2016;175:1023–1029. doi: 10.1007/s00431-016-2725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio C. The global HIV epidemic: what the pathologist needs to know. Semin. Diagn. Pathol. 2017;34:314–317. doi: 10.1053/j.semdp.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denner J. Detection of a gammaretrovirus, XMRV, in the human population: Open questions and implications for xenotransplantation. Retrovirology. 2010 doi: 10.1186/1742-4690-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorbar J., Egawa N., Griffin H., Kranjec C., Murakami I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015 doi: 10.1002/rmv.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyda A., Stelzer-Braid S., Adam D., Chughtai A.A., Macintyre C.R. The association between acute flaccid myelitis (AFM) and enterovirus D68 (EV-D68) – what is the evidence for causation? Euro Surveill. 2018;23:1–9. doi: 10.2807/1560-7917.ES.2018.23.3.17-00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels E.A., Atkinson J.O., Graubard B.I., McQuillan G.M., Gamache C., Mbisa G., Cohn S., Whitby D., Goedert J.J. Risk factors for human herpesvirus 8 infection among adults in the United States and evidence for sexual transmission. J. Infect. Dis. 2007;196:199–207. doi: 10.1086/518791. [DOI] [PubMed] [Google Scholar]
- Fancello L., Trape S., Robert C., Boyer M., Popgeorgiev N., Raoult D., Desnues C. Viruses in the desert: a metagenomic survey of viral communities in four perennial ponds of the Mauritanian Sahara. ISME J. 2013;7:359–369. doi: 10.1038/ismej.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Cooper D.M., McDonald J.E., Malham S.K., de Rougemont A., Jones D.L. Seasonal and spatial dynamics of enteric viruses in wastewater and in riverine and estuarine receiving waters. Sci. Total Environ. 2018;634:1174–1183. doi: 10.1016/j.scitotenv.2018.04.038. [DOI] [PubMed] [Google Scholar]
- Farkas K., Marshall M., Cooper D., McDonald J.E., Malham S.K., Peters D.E., Maloney J.D., Jones D.L. Seasonal and diurnal surveillance of treated and untreated wastewater for human enteric viruses. Environ. Sci. Pollut. Res. 2018;25:33391–33401. doi: 10.1007/s11356-018-3261-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J.G., Newcomer C.E., Rozmiarek H. second ed. Laboratory Animal Medicine. Elsevier Inc; 2002. Selected Zoonoses. [DOI] [Google Scholar]
- Gautheret-Dejean A., Manichanh C., Thien-Ah-Koon F., Fillet A.M., Mangeney N., Vidaud M., Dhedin N., Vernant J.P., Agut H. Development of a real-time polymerase chain reaction assay for the diagnosis of human herpesvirus-6 infection and application to bone marrow transplant patients. J. Virol. Methods. 2002;100:27–35. doi: 10.1016/S0166-0934(01)00390-1. [DOI] [PubMed] [Google Scholar]
- GBD 2017 Causes of Death Collaborators Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1736–1788. doi: 10.1016/S0140-6736(18)32203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghebremedhin B. Human adenovirus: Viral pathogen with increasing importance. Eur. J. Microbiol. Immunol. 2014;4:26–33. doi: 10.1556/eujmi.4.2014.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass R.I., Parashar U.D., Estes M.K. Norovirus gastroenteritis. N. Engl. J. Med. 2009 doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GLWA . 2018. Capital Improvement Plan 2019-2023. [Google Scholar]
- Gonçalves D.U., Proietti F.A., Ribas J.G.R., Araújo M.G., Pinheiro S.R., Guedes A.C., Carneiro-Proietti A.B.F. Epidemiology, treatment, and prevention of human T-cell leukemia virus type 1-associated diseases. Clin. Microbiol. Rev. 2010 doi: 10.1128/CMR.00063-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourinat A.C., O’Connor O., Calvez E., Goarant C., Dupont-Rouzeyrol M. Detection of zika virus in urine. Emerg. Infect. Dis. 2015 doi: 10.3201/eid2101.140894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2009 doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- Hall C.B., Long C.E., Schnabel K.C., Caserta M.T., Mcintyre K.M., Costanzo M.A., Knott A., Dewhurst S., Insel R.A., Epstein L.G. Human herpesvirus-6 infection in children — A prospective study of complications and reactivation. N. Engl. J. Med. 1994 doi: 10.1056/NEJM199408183310703. [DOI] [PubMed] [Google Scholar]
- Hall A.J., Lopman B.A., Payne D.C., Patel M.M., Gastañaduy P.A., Vinjé J., Parashar U.D. Norovirus disease in the United States. Emerg. Infect. Dis. 2013;19:1198–1205. doi: 10.3201/eid1908.130465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller S.L., Peng C., McFadden G., Rothenburg S. Poxviruses and the evolution of host range and virulence. Infect. Genet. Evol. 2014 doi: 10.1016/j.meegid.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Heijnen L., Medema G. Surveillance of influenza A and the pandemic influenza A (H1N1) 2009 in sewage and surface water in The Netherlands. J. Water Health. 2011;9:434–442. doi: 10.2166/wh.2011.019. [DOI] [PubMed] [Google Scholar]
- Hirayama T., Mizuno Y., Takeshita N., Kotaki A., Tajima S., Omatsu T., Sano K., Kurane I., Takasaki T. Detection of dengue virus genome in urine by real-time reverse transcriptase PCR: a laboratory diagnostic method useful after disappearance of the genome in serum. J. Clin. Microbiol. 2012 doi: 10.1128/JCM.06557-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose R., Daidoji T., Naito Y., Watanabe Y., Arai Y., Oda T., Konishi H., Yamawaki M., Itoh Y., Nakaya T. Long-term detection of seasonal influenza RNA in faeces and intestine. Clin. Microbiol. Infect. 2016;22 doi: 10.1016/j.cmi.2016.06.015. 813.e1-813.e7. [DOI] [PubMed] [Google Scholar]
- Ishitsuka K., Tamura K. Human T-cell leukaemia virus type I and adult T-cell leukaemia-lymphoma. Lancet Oncol. 2014;15:e517–e526. doi: 10.1016/S1470-2045(14)70202-5. [DOI] [PubMed] [Google Scholar]
- Iwai M., Hasegawa S., Obara M., Nakamura K., Horimoto E., Takizawa T., Kurata T., Sogen S.I., Shiraki K. Continuous presence of noroviruses and sapoviruses in raw sewage reflects infections among inhabitants of Toyama, Japan (2006 to 2008) Appl. Environ. Microbiol. 2009;75:1264–1270. doi: 10.1128/AEM.01166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B., Cushingberry G., Jr., Ayers J., Benson S., Castaneda-Lopez R., Leland G., Sheffield M.A. 2015. Rehabilitation of the Rectangular Primary Clarifiers, Electrical/mechanical Buildings and Pipe Gallery. (Detroit) [Google Scholar]
- Jothikumar N., Cromeans T.L., Sobsey M.D., Robertson B.H. Development and evaluation of a broadly reactive TaqMan assay for rapid detection of hepatitis A virus. Appl. Environ. Microbiol. 2005;71:3359–3363. doi: 10.1128/AEM.71.6.3359-3363.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamar N., Dalton H.R., Abravanel F., Izopet J. Hepatitis E virus infection. Clin. Microbiol. Rev. 2014;27:116–138. doi: 10.1128/CMR.00057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T., Fouchier R.A.M., Schutten M., Rimmelzwaan G.F., Van Amerongen G., Van Riel D., Laman J.D., De Jong T., Van Doornum G., Lim W., Ling A.E., Chan P.K.S., Tam J.S., Zambon M.C., Gopal R., Drosten C., Van Der Werf S., Escriou N., Manuguerra J.C., Stöhr K., Peiris J.S.M., Osterhaus A.D.M.E. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand F., Desire N., Rozenbaum W., Nicolas J.C., Marechal V. Quantitative analysis of human herpesvirus 8 viral load using a real- time PCR assay. J. Clin. Microbiol. 2000;38:1404–1408. doi: 10.1128/JCM.38.4.1404-1408.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guyader F.S., Parnaudeau S., Schaeffer J., Bosch A., Loisy F., Pommepuy M., Atmar R.L. Detection and quantification of noroviruses in shellfish. Appl. Environ. Microbiol. 2009;75:618–624. doi: 10.1128/AEM.01507-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Chan M.C.W., Wong B., Choi K.W., Sin W., Lui G., Chan P.K.S., Lai R.W.M., Cockram C.S., Sung J.J.Y., Leung W.K. Fecal viral concentration and diarrhea in norovirus gastroenteritis. Emerg. Infect. Dis. 2007;13:1399–1401. doi: 10.3201/eid1309.061535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.M., Lessler J., Lee R.A., Rudolph K.E., Reich N.G., Perl T.M., Cummings D.A.T. Incubation periods of viral gastroenteritis: a systematic review. BMC Infect. Dis. 2013;13:1. doi: 10.1186/1471-2334-13-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon S.M., Ott J.J., Van Damme P., Shouval D. Type A viral hepatitis: a summary and update on the molecular virology, epidemiology, pathogenesis and prevention. J. Hepatol. 2018;68:167–184. doi: 10.1016/j.jhep.2017.08.034. [DOI] [PubMed] [Google Scholar]
- Li T., Mbala-Kingebeni P., Naccache S.N., Thézé J., Bouquet J., Federman S., Somasekar S., Yu G., Martin C.S.S., Achari A., Schneider B.S., Rimoin A.W., Rambaut A., Nsio J., Mulembakani P., Ahuka-Mundeke S., Kapetshi J., Pybus O.G., Muyembe-Tamfum J.J., Chiu C.Y. Metagenomic next-generation sequencing of the 2014 ebola virus disease outbreak in the democratic republic of the Congo. J. Clin. Microbiol. 2019 doi: 10.1128/JCM.00827-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawson A.R., Croft A.M. Rubella virus infection, the congenital rubella syndrome, and the link to autism. Int. J. Environ. Res. Public Health. 2019;16 doi: 10.3390/ijerph16193543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall C., Xagoraraki I. Metagenomic approaches for detecting viral diversity in water environments. J. Environ. Eng. (United States) 2019 doi: 10.1061/(ASCE)EE.1943-7870.0001548. [DOI] [Google Scholar]
- McMichael A.J. Philosophical Transactions of the Royal Society B: Biological Sciences. 2004. Environmental and social influences on emerging infectious diseases: past, present and future. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MDHHS Michigan disease surveillance system background. 2020. https://www.michigan.gov/mdhhs/0,5885,7-339-71550_5104_31274-96814--,00.html (accessed 4.28.20)
- MDHHS Michigan hepatitis A outbreak: Update. 2020. https://www.michigan.gov/mdhhs/0,5885,7-339-71550_2955_2976_82305_82310-447907--,00.html (accessed 4.27.20)
- MDHHS Managing communicable diseases in schools. 2018. https://www.michigan.gov/documents/mdch/Managing_CD_in_Schools_FINAL_469824_7.PDF (accessed 4.28.20)
- MDHHS . 2018. Epidemiologic Profile of HIV in Michigan. [Google Scholar]
- Miranda J.A., Culley A.I., Schvarcz C.R., Steward G.F. RNA viruses as major contributors to Antarctic virioplankton. Environ. Microbiol. 2016;18:3714–3727. doi: 10.1111/1462-2920.13291. [DOI] [PubMed] [Google Scholar]
- Munir M., Wong K., Xagoraraki I. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res. 2011 doi: 10.1016/j.watres.2010.08.033. [DOI] [PubMed] [Google Scholar]
- Ng T.F.F., Marine R., Wang C., Simmonds P., Kapusinszky B., Bodhidatta L., Oderinde B.S., Wommack K.E., Delwart E. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J. Virol. 2012;86:12161–12175. doi: 10.1128/jvi.00869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien E., Nakyazze J., Wu H., Kiwanuka N., Cunningham W., Kaneene J.B., Xagoraraki I. Viral diversity and abundance in polluted waters in Kampala, Uganda. Water Res. 2017 doi: 10.1016/j.watres.2017.09.063. [DOI] [PubMed] [Google Scholar]
- O’Brien E., Xagoraraki I. A water-focused one-health approach for early detection and prevention of viral outbreaks. One Heal. 2019 doi: 10.1016/j.onehlt.2019.100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Katayama K., Hansman G.S., Kageyama T., Ogawa S., Wu F.T., White P.A., Takeda N. Detection of human sapovirus by real-time reverse transcription-polymerase chain reaction. J. Med. Virol. 2006 doi: 10.1002/jmv.20699. [DOI] [PubMed] [Google Scholar]
- Oka T., Wang Q., Katayama K., Saifb L.J. Comprehensive review of human sapoviruses. Clin. Microbiol. Rev. 2015;28:32–53. doi: 10.1128/CMR.00011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poloni T.R., Oliveira A.S., Alfonso H.L., Galvo L.R., Amarilla A.A., Poloni D.F., Figueiredo L.T., Aquino V.H. Detection of dengue virus in saliva and urine by real time RT-PCR. Virol. J. 2010 doi: 10.1186/1743-422X-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J., Söderlund-Venermo M., Young N.S. Human parvoviruses. Clin. Microbiol. Rev. 2017 doi: 10.1128/CMR.00040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2019. R: A Language and Environment for Statistical Computing. (Vienna, Austria) [Google Scholar]
- Tan S.Z.K., Tan M.Z.Y., Prabakaran M. Saffold virus, an emerging human cardiovirus. Rev. Med. Virol. 2017;27 doi: 10.1002/rmv.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonry J.H., Brown C.B., Cropp C.B., Co J.K.G., Bennett S.N., Nerurkar V.R., Kuberski T., Gubler D.J. West Nile virus detection in urine. Emerg. Infect. Dis. 2005 doi: 10.3201/eid1108.050238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao S.W., Tsang C.M., To K.F., Lo K.W. The role of Epstein-Barr virus in epithelial malignancies. J. Pathol. 2015;235:323–333. doi: 10.1002/path.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA . 2001. Manual of Methods for Virology (Chapter 14) [Google Scholar]
- Van Den Berg B., Walgaard C., Drenthen J., Fokke C., Jacobs B.C., Van Doorn P.A. Guillain-Barré syndrome: Pathogenesis, diagnosis, treatment and prognosis. Nat. Rev. Neurol. 2014;10:469–482. doi: 10.1038/nrneurol.2014.121. [DOI] [PubMed] [Google Scholar]
- Victoria M., Tort L.F.L., García M., Lizasoain A., Maya L., Leite J.P.G., Miagostovich M.P., Cristina J., Colina R. Assessment of gastroenteric viruses from wastewater directly discharged into Uruguay River, Uruguay. Food Environ. Virol. 2014;6:116–124. doi: 10.1007/s12560-014-9143-7. [DOI] [PubMed] [Google Scholar]
- Wang D., Urisman A., Liu Y.T., Springer M., Ksiazek T.G., Erdman D.D., Mardis E.R., Hickenbotham M., Magrini V., Eldred J., Latreille J.P., Wilson R.K., Ganem D., DeRisi J.L. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003 doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S.C., Charlier C., Vasilakis N., Lecuit M. Zika, chikungunya, and other emerging vector-borne viral diseases. Annu. Rev. Med. 2018 doi: 10.1146/annurev-med-050715-105122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S.C., Ferro C., Barrera R., Boshell J., Navarro J.-C. V enezuelan E quine E ncephalitis. Annu. Rev. Entomol. 2004;49:141–174. doi: 10.1146/annurev.ento.49.061802.123422. [DOI] [PubMed] [Google Scholar]
- Wells A.I., Coyne C.B. Enteroviruses: a gut-wrenching game of entry, detection, and evasion. Viruses. 2019;11:1–20. doi: 10.3390/v11050460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley R.J. Herpes Simplex Virus Infections. Goldman’s Cecil Med. Twenty Fourth Ed. 2011;2:2125–2128. doi: 10.1016/B978-1-4377-1604-7.00382-1. [DOI] [Google Scholar]
- Widener R.W., Whitley R.J. Handbook of Clinical Neurology. 1st ed. Elsevier B.V; 2014. Herpes simplex virus. [DOI] [PubMed] [Google Scholar]
- Xagoraraki I., Kuo D.H.W., Wong K., Wong M., Rose J.B. Occurrence of human adenoviruses at two recreational beaches of the great lakes. Appl. Environ. Microbiol. 2007 doi: 10.1128/AEM.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xagoraraki I., O’Brien E. 2020. Wastewater-based Epidemiology for Early Detection of Viral Outbreaks. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.