Abstract
Psychiatric disorders are the leading cause of disability worldwide while the pathogenesis remains unclear. Genome-wide association studies (GWASs) have made great achievements in detecting disease-related genetic variants. However, functional information on the underlying biological processes is often lacking. Current reports propose the use of metabolic traits as functional intermediate phenotypes (the so-called genetically determined metabotypes or GDMs) to reveal the biological mechanisms of genetics in human diseases. Here we conducted a two-sample Mendelian randomization analysis that uses GDMs to assess the causal effects of 486 human serum metabolites on 5 major psychiatric disorders, which respectively were schizophrenia (SCZ), major depression (MDD), bipolar disorder (BIP), autism spectrum disorder (ASD), and attention-deficit/hyperactivity disorder (ADHD). Using genetic variants as proxies, our study has identified 137 metabolites linked to the risk of psychiatric disorders, including 2-methoxyacetaminophen sulfate, which affects SCZ (P = 1.7 × 10–5) and 1-docosahexaenoylglycerophosphocholine, which affects ADHD (P = 5.6 × 10–5). Fourteen significant metabolic pathways involved in the 5 psychiatric disorders assessed were also detected, such as glycine, serine, and threonine metabolism for SCZ (P = .0238), Aminoacyl-tRNA biosynthesis for both MDD (P = .0144) and ADHD (P = .0029). Our study provided novel insights into integrating metabolomics with genomics in order to understand the mechanisms underlying the pathogenesis of human diseases.
Keywords: serum metabolite, psychiatric disorder, Mendelian randomization, metabolic pathway, 2-methoxyacetaminophen sulfate, 1-docosahexaenoylglycerophosphocholine
Introduction
Psychiatric disorders have become a major driver of the growth of overall morbidity and disability in the past decades, with the rapid transformation of global economic, demographic, and epidemiological conditions.1,2 Major syndromes, such as schizophrenia (SCZ), major depression (MDD), bipolar disorder (BIP), autism spectrum disorder (ASD), and attention-deficit/hyperactivity disorder (ADHD), comprise a large proportion of global health burden worldwide.3 Despite considerable efforts have been undertaken to understand the nature of psychiatric disorders, knowledge of disease mechanisms is still limited and no etiological therapies are available.4 Fortunately, advances in genetics (especially genome-wide association studies [GWASs]) have greatly promoted the progress of etiology research and made remarkable success in identifying genetic risk factors for psychiatric disorders.5–9 However, it is still a major obstacle translating these genetic findings into pathophysiological mechanisms and new therapies.10
Current researches have proposed metabolic traits as functional intermediates to investigate the underlying biological mechanisms of genetics on psychiatric disorders.11–13 Metabolites are the intermediate or end products of metabolism that drive essential biological functions of human bodies.14 The measurement of these markers could reflect the health status of an individual and potentially offer novel insight into the effects of diet, drugs, and disease. Especially the metabolomics, which aims to provide a top-down functional readout of the biochemistry, physiological status, and environmental exposure for human diseases.15 In fact, metabolic strategies have been extensively used to characterize specific metabolic phenotypes associated with psychiatric disorders and a number of metabolites have been identified.16–20 However, a comprehensive analysis is needed to reveal the role of the interactions between genetic variants and metabolites in the pathogenesis of psychiatric disorders.
Mendelian randomization (MR) is a novel genetic epidemiology study design that uses genetic variants as instrumental variables to assess the causal relationships among exposures and clinical outcomes of interest.21 Unlike observational epidemiology, the MR strategy can provide unbiased estimates exploiting the fact that genotypes are determined at conception and not generally susceptible to confounders.22 By taking advantage of these GWAS discoveries, MR has been widely applied to infer causality of exposures on diseases.23–25 Recently, GWAS has been extended to metabolic profiles, from which an atlas of genetically determined metabotypes (GDMs) was established. Based on the generated GDMs, we are able to assess the causal effects of genetically determined nontargeted metabolomics on psychiatric disorders.
Hereby making use of the GDMs and latest GWAS findings on psychiatric disorders, we conducted this two-sample (genetic associations with the exposure and outcome are measured in different samples) MR approach to: (1) assess the causal effects of human blood metabolites on 5 major psychiatric disorders, which respectively were SCZ, MDD, BIP, ASD, and ADHD; (2) identify common metabolites that had causal effects on multi-psychiatric disorders; (3) identify metabolic pathways that might contribute to the development of the 5 major psychiatric disorders; (5) investigate potential genetic variants that lead to both the variation of the metabolites and the progress of psychiatric disorders.
Materials and Methods
Study Design and Data Sources
The design of this MR study is illustrated in supplementary figure S1. We obtained the genome-wide association summary datasets for 486 metabolites from the study by Shin et al,26 which is the most comprehensive investigation of genetic influences on human metabolism. The total sample is comprised of 7824 adult participants from 2 European population studies, including 1768 participants from the German KORA F4 study and 6056 participants from the British Twins UK study. Metabolic profiling was done on fasting serum using ultra-high-performance liquid-phase chromatography and gas-chromatography separations coupled with tandem mass spectrometry.12,27 Standardized processes of identification and relative quantification, data-reduction, and quality-assurance were performed for metabolic analyses by Metabolon, Inc. (https://www.metabolon.com/). After stringent quality control, a total of 486 metabolites were used for genetic analysis, including 309 known and 177 unknown metabolites. The 309 known metabolites could be further assigned to 8 broad metabolic classes (amino acids, carbohydrates, cofactors and vitamins, energy, lipids, nucleotides, peptides, and xenobiotic metabolism) as described in the KEGG (Kyoto Encyclopedia of Genes and Genomes) database.28 Genotyping and imputation steps of the two cohorts are described in detail in previous studies and approximately 2.1 million SNPs were reserved in the GWAS meta-analysis. Full GWAS summary statistics were publicly available through the Metabolomics GWAS Server at http://metabolomics.helmholtz-muenchen.de/gwas/.
Selection of Instrumental Variables for the 486 Metabolites
We implemented uniform criteria for the selection of genetic variants for the 486 serum metabolites. For each metabolite, we first selected SNPs that showed association at P < 1 × 10−5. This relax statistical threshold was usually used in MR research to explain a larger variation when few genome-wide significant SNPs were available for exposures.21,23 After extracting the significant SNPs for each metabolite, we conducted a clumping procedure to retain SNPs with the lowest P-value as independent instruments, setting a linkage disequilibrium threshold of r2 < 0.1 in a 500-kb window in the European 1000G reference panel. We next calculated the explained variation (R2) between the instrumental variable and the corresponding metabolite exposure using the “gtx” package in R (https://www.r-project.org/). An F statistic was also estimated to evaluate the strength of these genetic predictors for the metabolites.29 Generally, an F statistic > 10 was considered as a typical threshold for the selection of strong instrumental variables.30
Primary Outcomes for MR
The latest GWAS summary statistics for SCZ, MDD, BIP, ASD, and ADHD were extracted from the Psychiatric Genomics Consortium (PGC) website (https://www.med.unc.edu/pgc/results-and-downloads/).6–9,31 The PGC has conducted the most influential meta- and mega-analyses of genome-wide genomic data for psychiatric disorders. The samples were collected from multiple cohorts of different ancestry, the majority of whom were of European ancestry. Genotyping, quality control, imputation, and association analysis were performed by the PGC Statistical Analysis Group as described in the primary reports for each study. The GWAS summary datasets for SCZ (36 989 cases and 113 075 controls), MDD (75 607 cases and 231 747 controls), BIP (20 352 cases and 31 358 controls), ASD (18 381 cases and 27 969 controls), and ADHD (20 183 cases and 35 191 controls) were generated from genome-wide meta- or mega-analyses (supplementary table S1).
MR Analysis
Causal effects for two-sample MR analyses were estimated using a standard inverse variance weighted (IVW) method.32 The IVW approach provides a consistent estimate of the causal effect of the exposure under the fundamental assumption that each included variant is a valid instrumental variable. Briefly, the IVW estimate can be equivalently obtained as the slope from a weighted linear regression of the genetic associations with the outcome as a function of the genetic associations with the exposure, weighted by the inverse variance of the genetic associations with the outcome. A fixed-effect meta-analysis model was used for each variant with a fixed intercept of zero. The P-value was calculated as P = 2 × (1 – Φ(Z)), where Φ is the standard normal cumulative distribution function and Z is the ratio of the estimated causal effect and its standard error. We adopted a multiple-testing-adjusted threshold of P < 1.03 × 10−4 (0.05/486) using the Bonferroni correction to declare a statistically significant, causal relationship. We also reported metabolites that had a P < .05, but were above the Bonferroni-corrected threshold, as suggestive risk predictors for psychiatric disorders.
The IVW estimate is an efficient analysis under the basic premise that all genetic variants are valid instrumental variables. This particularly requires the genetic variants to act on the target outcome exclusively through the exposure of interest (“no horizontal pleiotropy” assumption). The IVW estimate can lead to severe bias if the “no horizontal pleiotropy” assumption is violated. To control for the widespread horizontal pleiotropy in MR, we conducted methodologic sensitivity analyses using additional MR methods: the weighted median method, which provided estimates when a subset (< 50%) of the variants came from invalid instrumental variables33; MR-Egger, which provided consistent estimates even when up to 50% of the variants were invalid34; and MR-PRESSO, which provided a correction test by identifying and discarding horizontal pleiotropic outliers.35 Furthermore, we specifically detected the presence of horizontal pleiotropy through the MR-PRESSO Global test. All MR analyses were conducted using R software with the R package “MendelianRandomization” and “MR-PRESSO.”
Metabolic Pathway Analysis
We next conducted a metabolic pathway analysis for the identified metabolites using web-based MetaboAnalyst 4.0 software (https://www.metaboanalyst.ca/MetaboAnalyst/faces/home.xhtml).36 We used both the functional enrichment analysis module and the pathway analysis module to search for potential metabolite sets or pathways that might be involved in the biological processes of psychiatric disorders. A total of 183 human metabolic pathways from two metabolite set libraries, including 99 metabolite sets from The Small Molecule Pathway Database (SMPDB) and 84 metabolite sets from the KEGG database, were tested in the metabolic pathway analysis.28,37 All significant metabolites identified by IVW (PIVW < .05) were extracted for metabolic pathway analysis because we were interested in elucidating plausible metabolic pathways for psychiatric disorders.
Results
Study Overview
We performed a two-sample MR analysis to assess the causal effects of human blood metabolites on 5 major psychiatric disorders using pairs of GWAS summary statistics. For each metabotype, we extracted the genetic variants as instrumental variables to test their causality on the outcome. The number of SNPs in the extracted instrumental variables varied from 3 to 675, with a median number of 17. The instrumental variables explained 8.2%–83.5% of the variance in their respective metabotypes, and the minimum F statistic for validity tests of these genetic predictors was 20.33. All instrumental variables for the 486 metabotypes were sufficiently informative (F statistic >10) for MR analyses.
Causal Effects of the 486 Metabolites on Psychiatric Disorders
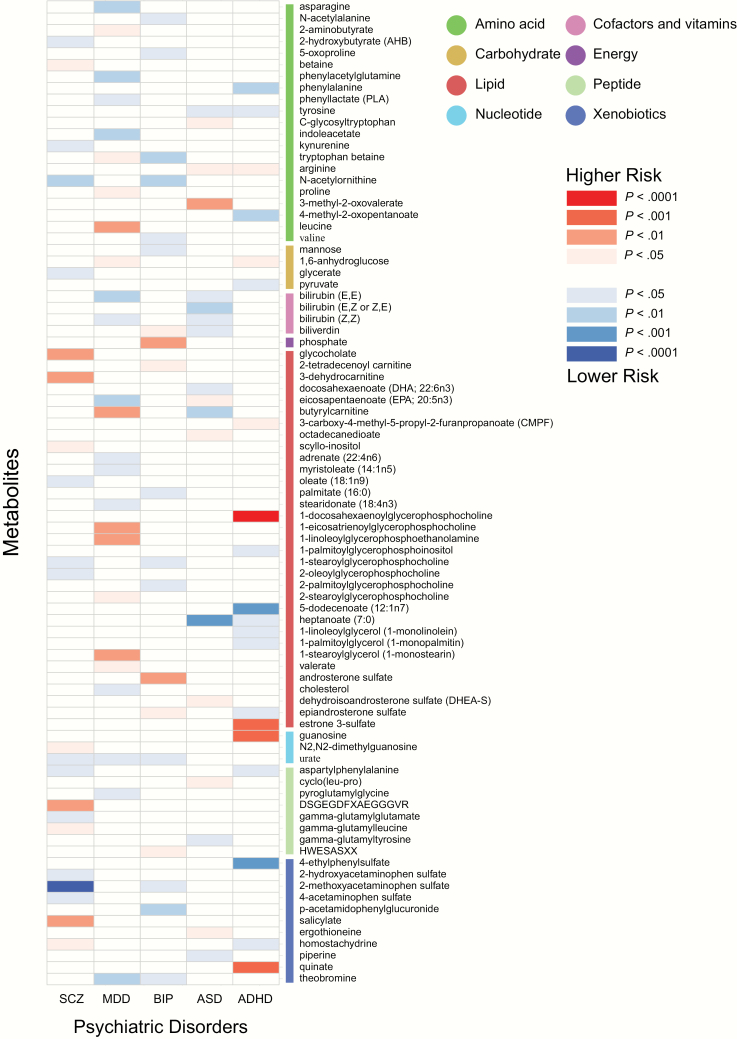
We used the IVW test to identify causal relationships among the 486 metabolites and the 5 major psychiatric disorders using pairs of GWAS summary statistics. A total of 160 significant causative association features (corresponding to 137 unique metabolites) were identified at P < .05, including 104 features for 85 known metabolites and 56 features for 52 unknown metabolites (supplementary tables S2 and S3). Figure 1 presents all significant causative association features between known metabolites with the 5 psychiatric disorders. We further reported the causative association features following a multiple-testing-adjusted threshold of the Bonferroni correction (P < 1.03 × 10−4). We observed two causal influence features of metabolites on psychiatric disorders, which were 2-methoxyacetaminophen sulfate on SCZ (P = 1.73 × 10–5) and 1-docosahexaenoylglycerophosphocholine on ADHD (P = 5.58 × 10–5). It is worth noting that 2-methoxyacetaminophen sulfate also showed suggestive causality on BIP (P = .011), implying that a shared molecular mechanism might exist between SCZ and BIP. Table 1 shows the overlapped metabolites in the 5 psychiatric disorders. These findings provide important information for understanding the molecular mechanism of psychiatric disorders.
Fig. 1.
Mendelian randomization associations of serum metabolites on the risk of 5 major psychiatric disorders. Shown are the results for known metabolites derived from the fixed-effects IVW analysis. IVW, inverse-variance weighted.
Table 1.
Overlapped Metabolites Identified for the 5 Major Psychiatric Disorders
| SCZ | MDD | BIP | ASD | ADHD | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Description | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P |
| 2-Methoxyacetaminophen sulfate | 0.98 (0.98, 0.99) | 1.73E-05 | — | — | 0.99 (0.98, 0.99) | .0114 | — | — | — | — |
| N-Acetylornithine | 0.71 (0.57, 0.89) | .0028 | — | — | 0.79 (0.67, 0.94) | .0061 | — | — | — | — |
| Aspartylphenylalanine | 0.70 (0.52, 0.94) | .0179 | — | — | — | — | — | — | 0.61 (0.38, 0.95) | .0301 |
| 1-Stearoylglycerophosphocholine | 0.67 (0.47, 0.97) | .0316 | — | — | 0.52 (0.28, 0.95) | .0347 | — | — | — | — |
| Homostachydrine | 1.30 (1.02, 1.65) | .0337 | — | — | — | — | — | — | 0.75 (0.56, 0.99) | .0439 |
| Urate | 0.49 (0.24, 0.99) | .0499 | 0.75 (0.57, 0.99) | .0494 | 0.44 (0.21, 0.89) | .0219 | — | — | — | — |
| Butyrylcarnitine | — | — | 1.10 (1.04, 1.17) | .0012 | — | — | 0.80 (0.68, 0.93) | .0031 | — | — |
| Theobromine | — | — | 0.83 (0.73, 0.95) | .0052 | 0.60 (0.37, 0.96) | .0342 | — | — | — | — |
| Eicosapentaenoate (EPA; 20:5n3) | — | — | 0.86 (0.77, 0.96) | .0063 | — | — | 1.46 (1.02, 2.08) | .0377 | — | — |
| Bilirubin (E,E) | — | — | 0.93 (0.88, 0.98) | .0082 | — | — | 0.83 (0.70, 0.99) | .0376 | — | — |
| 1,6-Anhydroglucose | — | — | 1.07 (1.01, 1.13) | .0428 | — | — | — | — | 1.26 (1.05, 1.52) | .0146 |
| Tryptophan betaine | — | — | 1.06 (1.00, 1.13) | .0432 | 0.82 (0.73, 0.94) | .0031 | — | — | — | — |
| Bilirubin (Z,Z) | — | — | 0.95 (0.91, 0.99) | .0477 | — | — | 0.83 (0.71, 0.97) | .0174 | — | — |
| Biliverdin | — | — | — | — | 1.28 (1.06, 1.54) | .0106 | 0.80 (0.67, 0.96) | .0194 | — | — |
| Epiandrosterone sulfate | — | — | — | — | 1.26 (1.03, 1.54) | .0236 | — | — | 0.84 (0.72, 0.98) | .0308 |
| Heptanoate (7:0) | — | — | — | — | — | — | 0.39 (0.24, 0.64) | .0002 | 0.59 (0.35, 0.98) | .0419 |
| Arginine | — | — | — | — | — | — | 1.85 (1.11, 3.11) | .0191 | 1.92 (1.10, 3.33) | .0207 |
| Tyrosine | — | — | — | — | — | — | 0.51 (0.27, 0.96) | .0385 | 0.48 (0.25, 0.93) | .0308 |
Note. SCZ, schizophrenia; MDD, major depression; BIP, bipolar disorder; ASD, autism spectrum disorder; ADHD, attention-deficit/hyperactivity disorder; OR, odds ratio; 95% CI, 95% confidence interval.
Sensitivity and Pleiotropy Analysis
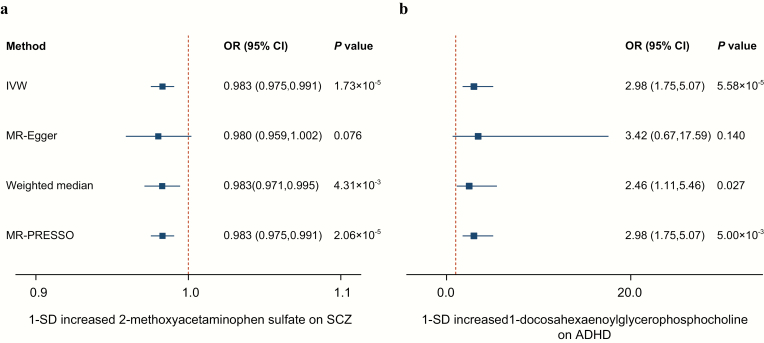
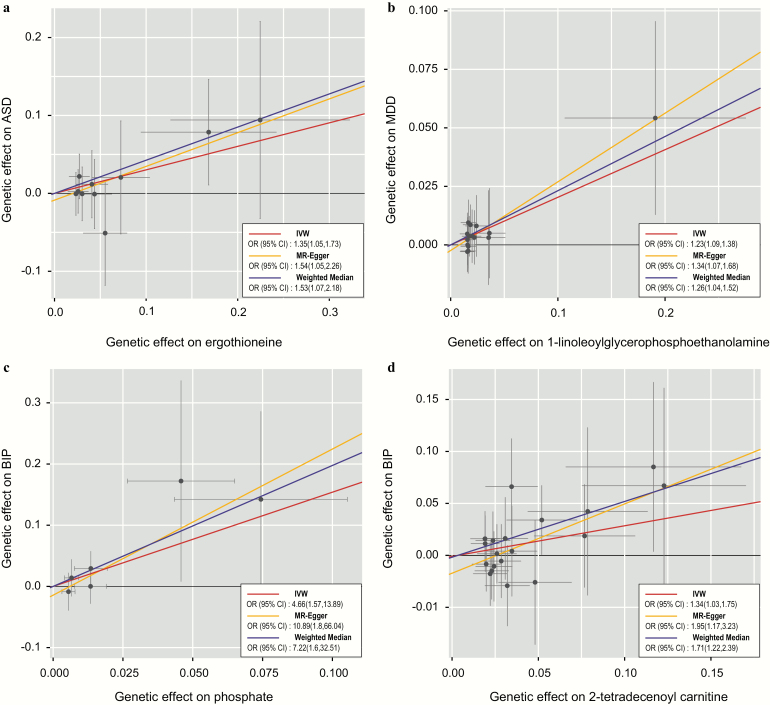
To avoid the horizontal pleiotropy for MR estimate, we further conducted a sensitivity and pleiotropy analysis to evaluate the robustness of the estimates. Figure 2 shows the results of the sensitivity analysis for 2-methoxyacetaminophen sulfate on SCZ and 1-docosahexaenoylglycerophosphocholine on ADHD. The causal relationship was robust when two additional MR tests were performed (PWeighted-median = 4.31 × 10–3 and PMR-PRESSO = 2.06 × 10–5 for 2-methoxyacetaminophen sulfate, PWeighted-median = .027 and PMR-PRESSO = 5.00 × 10–3 for 1-docosahexaenoylglycerophosphocholine), and there was no evidence of horizontal pleiotropy for either metabolite (PMR-PRESSO Global = .134 for 2-methoxyacetaminophen sulfate, PMR-PRESSO Global = .459 for 1-docosahexaenoylglycerophosphocholine). However, both of the two metabolites were nonsignificant in the MR-Egger test. This might be because the MR-Egger method was based on a weaker assumption and was substantially less efficient than the IVW method and weighted median method in inferring causal estimates. We further reported 4 suggestive association features that passed all sensitivity analyses (P < .05) without horizontal pleiotropy, which respectively were ergothioneine on ASD (PIVW = .018, PMR-Egger = .026, PWeighted-median = .018, PMR-PRESSO = .023; PMR-PRESSO Global= .676), 1-linoleoyl glycerophosphoethanolamine on MDD (PIVW = .001, PMR-Egger = .011, PWeighted-median = .018, PMR-PRESSO = .001; PMR-PRESSO Global = .872), phosphate on BIP (PIVW = .006, PMR-Egger = .009, PWeighted-median = .010, PMR-PRESSO = .040; PMR-PRESSO Global =0.426), and 2-tetradecenoyl carnitine on BIP (PIVW = .031, PMR-Egger = .010, PWeighted-median = .002, PMR-PRESSO = .045; PMR-PRESSO Global = .190). The genetically predicted effect sizes of these four additional metabolites on psychiatric disorders are presented in figure 3. The genetic variants for explaining the relationships between the six abovementioned metabolites and psychiatric disorders were listed in supplementary tables S4–S9, respectively.
Fig. 2.
Sensitivity analysis for two significant features passing Bonferroni correction. a. effect of 1-SD genetically determined levels of 2-methoxyacetaminophen sulfate on SCZ; b. effect of 1-SD genetically determined levels of 1-docosahexaenoylglycerophosphocholine on ADHD. SCZ, schizophrenia; ADHD, attention-deficit/hyperactivity disorder.
Fig. 3.
Scatter plot showing the genetic associations of four metabolites on the risk of psychiatric disorders. a. ergothioneine on ASD; b. 1-linoleoylglycerophosphoethanolamine on MDD; c. 4-hydroxyhippurate on BIP; d. 2-tetradecenoyl carnitine on BIP. SNPs showing negative signals with metabolites are plotted after orientation to the exposure-increasing allele. Each of the SNPs associated with metabolites is represented by a black dot with the error bar depicting the standard error of its association with metabolite (horizontal) and the target psychiatric disorder (vertical). The slopes of each line represent the causal association for each method. ASD, autism spectrum disorder; MDD, major depression, BIP, bipolar disorder.
Metabolic Pathway Analysis
The metabolic pathway analysis identified 14 significant metabolic pathways among the 5 psychiatric disorders (table 2). Our results show that the “Glycine, serine, and threonine metabolism” pathway might be associated with the pathogenetic process of SCZ (P = .0238) whereas the “Glycerolipid metabolism” pathway was found to be associated with BIP (P = .0325). We also detected several shared metabolic pathways for different psychiatric disorders, such as the “Alpha-linolenic acid and linoleic acid metabolism” pathway for MDD (P = .0037) and ASD (P = .0225), the “Aminoacyl-tRNA biosynthesis” pathway for MDD (P = .0144) and ADHD (P = .0029), and the “D-arginine and D-ornithine metabolism” pathway for ASD (P = .0393) and ADHD (P = .0328).
Table 2.
Significant Metabolic Pathways Involved in the 5 Major Psychiatric Disorders
| Trait | Metabolic Pathway | Involved Metabolites | P-Value | Database |
|---|---|---|---|---|
| SCZ | Glycine, serine, and threonine metabolism | Glycocholate, Betaine | .0238 | KEGG |
| MDD | Alpha-linolenic acid and linoleic acid metabolism | Eicosapentaenoate, Adrenate, Stearidonate | .0037 | SMPDB |
| MDD | Aminoacyl-tRNA biosynthesis | Asparagine, Leucine, Proline | .0144 | KEGG |
| BIP | Glycerolipid metabolism | 5-oxoproline, Phosphate | .0325 | SMPDB |
| ASD | Alpha-linolenic acid and linoleic acid metabolism | Docosahexaenoate (DHA), Eicosapentaenoate (EPA) | .0225 | SMPDB |
| ASD | D-arginine and D-ornithine metabolism | Arginine | .0393 | KEGG |
| ADHD | Phenylalanine metabolism | Phenylalanine, Tyrosine, Pyruvate | 6.69E-4 | KEGG |
| ADHD | Aminoacyl-tRNA biosynthesis | Phenylalanine, Arginine, Tyrosine | .0029 | KEGG |
| ADHD | Phenylalanine, tyrosine, and tryptophan biosynthesis | Phenylalanine, Tyrosine | .0051 | KEGG |
| ADHD | Valine, leucine, and isoleucine biosynthesis | 4-Methyl-2-oxopentanoate, Pyruvate | .0051 | KEGG |
| ADHD | Nitrogen metabolism | Phenylalanine, Tyrosine | .0106 | KEGG |
| ADHD | D-arginine and D-ornithine metabolism | Arginine | .0328 | KEGG |
| ADHD | Tyrosine metabolism | Tyrosine, Pyruvate | .0376 | KEGG |
| ADHD | Arginine and proline metabolism | Arginine, Pyruvate | .0385 | KEGG |
Note. SCZ, schizophrenia; MDD, major depression; BIP, bipolar disorder; ASD, autism spectrum disorder; ADHD, attention-deficit/hyperactivity disorder; OR, odds ratio; 95% CI, 95% confidence interval; KEGG, kyoto encyclopedia of genes and genomes; SMPDB, small molecule pathway database.
Discussion
This MR study provides unbiased detection of the causal effects of human serum metabolites on 5 major psychiatric disorders. Using genetic variants as proxies, we observed 137 metabolites linked to the risk of psychiatric disorders, including 2-methoxyacetaminophen sulfate, which affects SCZ (P = 1.7 × 10–5) and 1-docosahexaenoylglycerophosphocholine, which affects ADHD (P = 5.6 × 10–5). Fourteen significant metabolic pathways involved in the 5 psychiatric disorders assessed were also detected. To the best of our knowledge, this is the first MR study combining metabolomics with genomics to evaluate the causal effects of serum metabolites on psychiatric disorders. Our study provides novel insight to reveal the role of genetic-environmental interactions in the pathogenesis of human diseases.
Our study detected a group of serum metabolites showing associations with psychiatric disorders, among which 2-methoxyacetaminophen sulfate was observed to have a robust effect on SCZ, as well as BIP. The metabolite 2-methoxyacetaminophen sulfate, also known as 4-(acetylamino)-3-methoxyphenyl hydrogen sulfate, is a member of the acetamide class and has a role as a drug metabolite. The acetamides have long been recognized to be associated with levels of glutathione and N-acetylcysteine, which are known biomarkers for SCZ and BIP.38–41 N-acetylcysteine is widely known for its role as an antidote for acetaminophen overdose. It can modulate multiple pathophysiological processes in psychiatric disorders, including oxidative stress, neurogenesis and apoptosis, mitochondrial dysfunction, and dysregulation of glutamate and dopamine neurotransmitter systems.42 Although 2-methoxyacetaminophen sulfate seems like a promising biomarker for psychiatric disorders, further clarification of the relevant mechanism is required. Another compound, 1-docosahexaenoylglycerophosphocholine, has been recognized to have a causal effect on ADHD. The metabolite 1-docosahexaenoylglycerophosphocholine is a kind of lysophospholipid (LP). The LPs are important signaling molecules that regulate fundamental cellular functions.43 Particularly, the LPs were involved in several aspects of neurodevelopment, such as cortical growth and folding.44 Therefore, 1-docosahexaenoylglycerophosphocholine might play a role in neurodevelopment in ADHD.
The MR analysis also detected several additional metabolites, some of which had already been reported in previous observational studies. McClay et al conducted a neurochemical metabolomics study that found altered levels of N-acetylornithine and leucine in response to the antipsychotic drug haloperidol.45 Scaini et al suggested that phosphate was involved in the mitochondrial dysfunction of BIP.46 Nakamichi et al found that food-derived ergothioneine was distributed to the brain and exerted an antidepressant effect.47 Furthermore, a recent meta-analysis found that patients with ASD had lower eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) levels than those of healthy controls, and a randomized, controlled treatment study of a comprehensive nutritional and dietary intervention suggested that ASD patients could benefit from supplements of EPA and DHA.48,49 Our results further support these findings and highlight the importance of genetics in the progression of psychiatric disorders.
Our metabolic pathway analysis shows that glycine, serine, and threonine metabolism pathways are associated with SCZ. An increasing amount of recent evidence indicates that D-serine and glycine could regulate N-methyl-d-aspartate (NMDA) receptor-dependent synaptic activity, which is one of the core hypotheses explaining the pathophysiology of SCZ.50,51 The alpha-linolenic acid and linoleic acid metabolism pathway is also a known indicator for psychiatric disorders, which participates in multiple pathophysiological processes, including biological stress, inflammation, and brain network structure and function.52 The most significant pathway for ADHD was the phenylalanine metabolism pathway. Symptoms of ADHD, particularly inattention and impairments in executive functioning, have been frequently reported in children, adolescents, and adults with phenylketonuria.53,54 Evidence has also shown that higher blood phenylalanine levels are correlated with ADHD.55 A potential mechanism for phenylalanine involvement in ADHD might lie in its role as a precursor for dopamine; the dopamine and serotonin hypotheses dominate current scientific work on ADHD.56
There are several limitations to our study. First, our study identified a causal relationship between 2-methoxyacetaminophen sulfate and SCZ. However, the very low effect size would limit its potential utility as a biomarker. Second, the metabolic traits of the GWAS analysis were carried out on fasting serum. Because psychiatric disorders mainly manifest as abnormal function of the brain, more data should be collected from cerebrospinal fluid or brain tissues. Third, the accuracy of the MR analysis depended on the explanation of the instrumental variables on exposure. A further expanded sample size might provide a more accurate assessment of the genetic influences on metabolites. Finally, while our study identified multiple metabolites causing a risk of psychiatric disorders, further work should be done to reveal their roles in the pathogenesis of these disorders.
In summary, the MR study identified 137 metabolites that might have causal effects on the development of psychiatric disorders, including 18 metabolites having causal effects on more than one outcome. Our study also identified 14 significant metabolic pathways that might be involved in the development of psychiatric disorders. Our study provides novel insights into combining metabolomics with genomics to reveal the pathogenesis and therapeutic strategies for psychiatric disorders.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81771471) and the Clinical Research Award of the First Affiliated Hospital of Xi’an Jiaotong University (No. XJTU1AF-CRF-2017–025).
Supplementary Material
Acknowledgments
We would like to thank the High-Performance Computing Cluster of the First Affiliated Hospital of Xi’an Jiaotong University for data computing. The authors have nothing to disclose.
References
- 1. Vigo D, Thornicroft G, Atun R. Estimating the true global burden of mental illness. Lancet Psychiatry. 2016;3(2):171–178. [DOI] [PubMed] [Google Scholar]
- 2. Baingana F, al’Absi M, Becker AE, Pringle B. Global research challenges and opportunities for mental health and substance-use disorders. Nature. 2015;527(7578):S172–S177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. DALYs GBD, Collaborators H, Murray CJ, et al. Global, regional, and national disability-adjusted life years (DALYs) for 306 diseases and injuries and healthy life expectancy (HALE) for 188 countries, 1990–2013: quantifying the epidemiological transition. Lancet. 2015;386(10009):2145–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McCarroll SA, Feng G, Hyman SE. Genome-scale neurogenetics: methodology and meaning. Nat Neurosci. 2014;17(6):756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Howard DM, Adams MJ, Clarke TK, et al. ; 23andMe Research Team; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22(3):343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stahl EA, Breen G, Forstner AJ, et al. ; eQTLGen Consortium; BIOS Consortium; Bipolar Disorder Working Group of the Psychiatric Genomics Consortium Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51(5):793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grove J, Ripke S, Als TD, et al. ; Autism Spectrum Disorder Working Group of the Psychiatric Genomics Consortium; BUPGEN; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium; 23andMe Research Team Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51(3):431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Martin J, Walters RK, Demontis D, et al. ; 23andMe Research Team; Psychiatric Genomics Consortium: ADHD Subgroup; iPSYCH–Broad ADHD Workgroup A Genetic investigation of sex bias in the prevalence of attention-deficit/hyperactivity disorder. Biol Psychiatry. 2018;83(12):1044–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Focus on psychiatric disorders. Nat Neurosci. 2016;19(11):1381–1382. [DOI] [PubMed] [Google Scholar]
- 11. Gieger C, Geistlinger L, Altmaier E, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4(11):e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suhre K, Shin SY, Petersen AK, et al. ; CARDIoGRAM Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477(7362):54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kettunen J, Tukiainen T, Sarin AP, et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2012;44(3):269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17(7):451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134(5):714–717. [DOI] [PubMed] [Google Scholar]
- 16. Yang J, Chen T, Sun L, et al. Potential metabolite markers of schizophrenia. Mol Psychiatry. 2013;18(1):67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hashimoto K. Metabolomics of major depressive disorder and bipolar disorder: overview and future perspective. Adv Clin Chem. 2018;84:81–99. [DOI] [PubMed] [Google Scholar]
- 18. Sethi S, Brietzke E. Omics-based biomarkers: application of metabolomics in neuropsychiatric disorders. Int J Neuropsychopharmacol. 2015;19(3):pyv096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guest PC, Guest FL, Martins-de Souza D. Making sense of blood-based proteomics and metabolomics in psychiatric research. Int J Neuropsychopharmacol. 2016;19(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quintero M, Stanisic D, Cruz G, Pontes JGM, Costa TBBC, Tasic L. Metabolomic biomarkers in mental disorders: bipolar disorder and schizophrenia. Adv Exp Med Biol. 2019;1118:271–293. [DOI] [PubMed] [Google Scholar]
- 21. Choi KW, Chen CY, Stein MB, et al. Assessment of bidirectional relationships between physical activity and depression among adults: a 2-sample mendelian randomization study. JAMA Psychiatry. 2019;76(4):399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng J, Baird D, Borges MC, et al. Recent developments in mendelian randomization studies. Curr Epidemiol Rep. 2017;4(4):330–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sanna S, van Zuydam NR, Mahajan A, et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. 2019;51(4):600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hartwig FP, Borges MC, Horta BL, Bowden J, Davey Smith G. Inflammatory biomarkers and risk of schizophrenia: a 2-sample Mendelian Randomization Study. JAMA Psychiatry. 2017;74(12):1226–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Khandaker GM, Zuber V, Rees JMB, et al. Shared mechanisms between coronary heart disease and depression: findings from a large UK general population-based cohort [published online ahead of print March 19, 2019]. Mol Psychiatry. 2019. doi:10.1038/s41380-019-0395-3 [Google Scholar]
- 26. Shin SY, Fauman EB, Petersen AK, et al. ; Multiple Tissue Human Expression Resource (MuTHER) Consortium An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46(6):543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krumsiek J, Suhre K, Evans AM, et al. Mining the unknown: a systems approach to metabolite identification combining genetic and metabolic information. PLoS Genet. 2012;8(10):e1003005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40(3):740–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42(5):1497–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Consortium SGotPG. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chong J, Soufan O, Li C, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46(W1):W486–W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frolkis A, Knox C, Lim E, et al. SMPDB: The Small Molecule Pathway Database. Nucleic Acids Res. 2010;38(Database issue):D480–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nýdlová E, Vrbová M, Cesla P, Jankovičová B, Ventura K, Roušar T. Comparison of inhibitory effects between acetaminophen-glutathione conjugate and reduced glutathione in human glutathione reductase. J Appl Toxicol. 2014;34(9):968–973. [DOI] [PubMed] [Google Scholar]
- 39. Nucifora LG, Tanaka T, Hayes LN, et al. Reduction of plasma glutathione in psychosis associated with schizophrenia and bipolar disorder in translational psychiatry. Transl Psychiatry. 2017;7(8):e1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang AM, Pradhan S, Coughlin JM, et al. Assessing brain metabolism with 7-T proton magnetic resonance spectroscopy in patients with first-episode psychosis [published online ahead of print March 1, 2019]. JAMA Psychiatry. 2019. doi:10.1038/s41380-019-0395-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deepmala, Slattery J, Kumar N, et al. Clinical trials of N-acetylcysteine in psychiatry and neurology: a systematic review. Neurosci Biobehav Rev. 2015;55:294–321. [DOI] [PubMed] [Google Scholar]
- 42. Dean O, Giorlando F, Berk M. N-acetylcysteine in psychiatry: current therapeutic evidence and potential mechanisms of action. J Psychiatry Neurosci. 2011;36(2):78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim EY, Lee JW, Lee MY, et al. Serum lipidomic analysis for the discovery of biomarkers for major depressive disorder in drug-free patients. Psychiatry Res. 2018;265:174–182. [DOI] [PubMed] [Google Scholar]
- 44. Kingsbury MA, Rehen SK, Contos JJ, Higgins CM, Chun J. Non-proliferative effects of lysophosphatidic acid enhance cortical growth and folding. Nat Neurosci. 2003;6(12):1292–1299. [DOI] [PubMed] [Google Scholar]
- 45. McClay JL, Vunck SA, Batman AM, et al. Neurochemical metabolomics reveals disruption to sphingolipid metabolism following chronic haloperidol administration. J Neuroimmune Pharmacol. 2015;10(3):425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Scaini G, Rezin GT, Carvalho AF, Streck EL, Berk M, Quevedo J. Mitochondrial dysfunction in bipolar disorder: Evidence, pathophysiology and translational implications. Neurosci Biobehav Rev. 2016;68:694–713. [DOI] [PubMed] [Google Scholar]
- 47. Nakamichi N, Nakayama K, Ishimoto T, et al. Food-derived hydrophilic antioxidant ergothioneine is distributed to the brain and exerts antidepressant effect in mice. Brain Behav. 2016;6(6):e00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mazahery H, Stonehouse W, Delshad M, et al. Relationship between long chain n-3 polyunsaturated fatty acids and autism spectrum disorder: systematic review and meta-analysis of case-control and randomised controlled trials. Nutrients. 2017;9(2):E115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adams JB, Audhya T, Geis E, et al. Comprehensive nutritional and dietary intervention for autism spectrum disorder-a randomized, controlled 12-month trial. Nutrients. 2018;10(3):E369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rosenberg D, Artoul S, Segal AC, et al. Neuronal D-serine and glycine release via the Asc-1 transporter regulates NMDA receptor-dependent synaptic activity. J Neurosci. 2013;33(8):3533–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cho SE, Na KS, Cho SJ, Kang SG. Low d-serine levels in schizophrenia: a systematic review and meta-analysis. Neurosci Lett. 2016;634:42–51. [DOI] [PubMed] [Google Scholar]
- 52. Mocking RJT, Assies J, Ruhé HG, Schene AH. Focus on fatty acids in the neurometabolic pathophysiology of psychiatric disorders. J Inherit Metab Dis. 2018;41(4):597–611. [DOI] [PubMed] [Google Scholar]
- 53. Arnold GL, Vladutiu CJ, Orlowski CC, Blakely EM, DeLuca J. Prevalence of stimulant use for attentional dysfunction in children with phenylketonuria. J Inherit Metab Dis. 2004;27(2):137–143. [DOI] [PubMed] [Google Scholar]
- 54. Christ SE, Huijbregts SC, de Sonneville LM, White DA. Executive function in early-treated phenylketonuria: profile and underlying mechanisms. Mol Genet Metab. 2010;99(suppl 1):S22–S32. [DOI] [PubMed] [Google Scholar]
- 55. Burton B, Grant M, Feigenbaum A, et al. A randomized, placebo-controlled, double-blind study of sapropterin to treat ADHD symptoms and executive function impairment in children and adults with sapropterin-responsive phenylketonuria. Mol Genet Metab. 2015;114(3):415–424. [DOI] [PubMed] [Google Scholar]
- 56. Bergwerff CE, Luman M, Blom HJ, Oosterlaan J. No tryptophan, tyrosine and phenylalanine abnormalities in children with attention-deficit/hyperactivity disorder. PLoS One. 2016;11(3):e0151100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.