Abstract
Objective
To evaluate the expression and function of long noncoding RNA (lncRNA) Prader-Willi region non-protein coding RNA 1 (PWRN1) in human glioblastoma (GBM).
Materials and Methods
QRT-PCR was applied to assess PWRN1 expression in human GBM tumors and GBM cell lines. PWRN1 was overexpressed by lentiviral infection in LN-229 and U-251 cells to evaluate its effect on GBM cell proliferation and migration in vitro, and xenograft in vivo. The endogenously competing target of PWRN1, human microRNA-21-5p (hsa-miR-21-5p) was evaluated by dual-luciferase activity assay and qRT-PCR. Also, hsa-miR-21-5p was upregulated in PWRN1-overexpressed GBM cells to evaluate the functional involvement of hsa-miR-21-5p in PWRN1-mediated GBM cell proliferation and migration.
Results
PWRN1 was downregulated in both human GBM tumors and GBM cell lines. In LN-229 and U-251, PWRN1 overexpression suppressed cancer cell proliferation and migration in vitro, and xenograft growth in vivo. Hsa-miR-21-5p was demonstrated to be the downstream competing target of PWRN1 in GBM. Conversely, upregulating hsa-miR-21-5p in LN-229 and U-251 cells reversed the tumor-suppressing effects of PWRN1 overexpression.
Conclusion
PWRN1 is markedly downregulated in GBM. Overexpressing PWRN1 has tumor inhibitory effect on GBM cells, likely via reversely suppressing the expression of tumor oncogenic factor of hsa-miR-21-5p.
Keywords: GBM, lncRNAs, PWRN1, microRNA, hsa-miR-21-5p, migration
Introduction
In the United States of America, the very recent cancer statistics showed that brain tumor is the leading cause of cancer death among men aged younger than 40 years and women aged younger than 20 years.1 In China, brain cancer is also the major contributor of cancer deaths in male patients younger than 44 years and female patients younger than 14 years.2 Among all types of brain tumors, glioblastoma multiforme (GBM) is the most malignant subtype, and patients with GBM are often diagnosed at late stages with low survival rates.3–5 In order to find early diagnosis biomarkers and treatment strategies to improve cancer patient prognosis, it is critical to deciphering the genetic code underlying GBM pathology and progression.
Long noncoding RNAs (lncRNAs) are families of long (~>200 n.t.) but noncoding RNAs that do not directly transcribe into proteins. However, recent studies have found that lncRNAs have significant epigenetic roles in regulating both transcriptional and post-transcriptional gene functions in both animal and human cells.6–8 Specifically, emerging evidence had demonstrated that lncRNAs may be aberrantly expressed in human cancer patients, and possibly acting as epigenetic tumor suppressors or oncogenes in regulating cancer cell development, thus making them ideal molecular targets for cancer diagnosis and therapy.9–12 In GBM, lncRNAs were also found to be closely associated with cancer patients’ prognosis and cancer cell progression.13,14 Yet, the intricate genetic network underlying lncRNA-associated regulations in GBM are far from being fully understood.
Like lncRNAs, microRNAs (miRNAs) are another family of non-coding RNAs, but with small-size (~18~22 n.t.). It’s been demonstrated that miRNAs may bind downstream targeting mRNAs to post-transcriptionally suppress gene and protein productions.15 Specifically, studies have shown that cross-link between miRNAs and lncRNAs, namely lncRNAs acting as competing endogenous RNAs to regulate miRNAs and associated signaling pathways, are playing important roles in human cancer progression, maturation, migration and apoptosis.16–18
In the current study, we utilized both GBN tumor samples and immortal GBM cell lines to analyze the expression of a cancer-associated lncRNA, Prader-Willi region non-protein coding RNA 1 (PWRN1) in human GBM. Studies have found that PWRN1was aberrantly expressed in various types of human cancers, including breast, prostate and gastric cancers.19–21 Yet, the expression of PWRN1 in human GBM was never clearly studied. In addition, we applied lentiviral infection technology to genetically upregulate PWRN1in GBM cell lines, to investigate the functional roles of PWRN1in modulating GBM cancer in vitro and in vivo proliferation. Furthermore, the possible epigenetic crosstalk between PWRN1 and its competing candidate, human mature microRNA-21-5p (hsa-miR-21-5p) was investigated in GBM cells. Overall, the goal of this study is to provide an insight on the epigenetic expression and function of PWRN1Cin human GBM.
Materials and Methods
Ethic Statement
The approval to conduct current study was provided by the Clinical Research & Ethic Committee at The Taizhou People’s Hospital in Taizhou, Jiangsu Province, China. Animal study was also reviewed and approved by the Clinical Research & Ethic Committee at The Taizhou People’s Hospital. In addition, all experiments were performed following the guidelines and regulations of the Clinical Research & Ethic Committee at The Taizhou People’s Hospital. Furthermore, written informed consents were signed by all participating patients, and all protocols were performed in full accordance with the World Medical Association Declaration of Helsinki.22
GBM Human Samples and Cell Lines
Between September 2019 and December 2019, human tissue samples were collected from 51 glioblastoma (GBM) patients. None of the participating patients received any radiation therapy or chemotherapy prior to the surgeries. For each patient, samples of both GBM tissue and adjacent normal brain tissues were collected. Then, all human samples were immediately snap-frozen in liquid nitrogen and stored at −75°C prior to RNA extraction.
In addition, nine immortal human GBM cancer cell lines, A172, BT169, BT238, LN-18, LN-229, T98G, U-118MG, U-138MG and U-251 cells, as well as a normal human astrocytes (NHA) cell line were commercially bought from the American Type Culture Collection (ATCC, www.atcc.rog, USA). All cells were maintained in 6-well tissue-culture plates (BD Biosciences, USA) in RPMI-1640 medium (Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific, USA), 1% MEM Non-Essential Amino Acids Solution (100X, Thermo Fisher Scientific, USA), 2 mM L-Glutamine (Thermo Fisher Scientific, USA) and 1% Penicillin-Streptomycin (10,000 U/mL, Thermo Fisher Scientific, USA) in a humidified environment of CO2 /O2 (5%/95%) at 37°C.
RNA Extraction and Quantitative Reverse Transcriptase-Polymerase Chain Reaction (qRT-PCR)
Quantitative assessment on gene expression was conducted using quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR). Briefly, RNA was extracted and purified from GBM cell lines or clinical tissues using a RiboPure™ RNA Purification Kit (Thermo Fisher Scientific, USA) according to the manufacturer’s protocol. Then, purified RNA was reversely transcribed into cDNA using a PrimeScript® RT reagent Kit (Takara, Dalian, China) according to the manufacturer’s protocol. QRT-PCR was performed on an ABI Prism 7900 sequence detection system (Applied Biosystems, USA). To assess gene expression level of PWRN1, a pre-designed TaqMan™ PWRN1 noncoding RNA Assay (Invitrogen, USA) was performed, and an 18S rRNA TaqMan assay (Invitrogen, USA) was used as housekeeping control. In addition, to assess gene expression level of human miR-21-5p (hsa-miR-21-5p), a TaqMan® MicroRNA qPCR assay (Applied Biosystems, USA) was performed and U6 small nuclear RNA was used as housekeeping gene. Relative expression level of each probed gene was then normalized to the expression level of housekeeping gene, using the 2–∆∆Ct method.
PWRN1 Overexpression Assay
A PWRN1-specific lentiviral overexpression plasmid, L/PWRN1, was synthesized, and corresponding lentivirus was generated and tittered by HanBio Biotechnology (HanBio, Shanghai, China). Correspondingly, a non-specific control overexpression lentivirus, L/C, was also purchased from HanBio Biotechnology (HanBio, Shanghai, China). In two GBM cell lines, LN-229 and U-251 cells, lentiviral infection of L/PWRN1or L/C was conducted in the presence of polybrene (8 μg/mL, Thermo Fisher Scientific, USA) for 48 h at multiplicity of infection (MOI) of 3. Then, infected LN-229 and U-251 cells were selected with puromycin (15 μg/mL, Thermo Fisher Scientific, USA) for 10 days. After that, healthy multi-cell colonies were collected and re-seeded in 48-well plates in culture medium supplemented with puromycin (5 μg/mL) to proliferate. After 5~8 passages, qRT-PCR was performed to verify whether PWRN1was efficiently overexpressed/upregulated in LN-229 and U-251 cells.
GBM Cell Proliferation in vitro
After infection, LN-229 and U-251 cells were lifted from 6-well plate and equally re-seeded in 96-well tissue-culture plates (BD Biosciences, USA) (5 X 103 cells/well). Cells were allowed to proliferate for 96 consecutive hours. Every 24 h, a Cell Counting Kit-8 (CCK-8, Dojindo, Japan) was performed according to the manufacturer’s protocol. Then, 96-well plates were mounted onto a Varioskan Flash Microplate reader (Thermo Fisher Scientific, USA). In each well, cancer cell proliferation was characterized by direct measurement of absorbance at optical density (O.D.) of 570 nm.
GBM Cell Migration in vitro
In a 24-well transwell migration assay, infected LN-229 and U-251 cells were equally plated inside the Matrigel (BD Biosciences, USA) pre-coated transwell inserts with a pore size of 8.0 μM (Becton Dickinson, USA) (2.5 X 105 cells/well) in serum-free culture medium. Underneath the transwell inserts, the wells were filled with regular culture medium supplemented with 10% FBS as chemo-attractant. Twenty-four hours later, all medium and transwell inserts were removed. LN-229 and U-251 cells migrated onto the bottoms of 24-well plates were quickly fixed by 70% ethanol (Gibco, USA) and stained with 1% crystal violet (Gibco, USA) for 20 mins at room temperature. Then, 24-well plates were mounted onto an IX71 inverted microscope (Olympus, Japan). For each well, migrated cells were counted in five randomly selected fields using ImageJ software (ImageJ, NIH, USA). Relative migration was then quantified as the percentage of migrated cells in experimental wells against the migrated cells in control wells.
GBM Xenograft in vivo
Nude female BALB/c mice (6-week-old) were purchased from Shanghai SLAC Laboratory Animal Company (Shanghai, China). Infected LN-229 cells were subcutaneously grafted into the dorsal skin of mice (1 million cells/graft, total of 15 mice). The left sides were grafted with cells infected with L/C, and right sides were grafted with cells infected with L/PWRN1. The GBM xenograft assay was carried out for 35 days. Every 7-day, surface dimension of GBM xenograft, including length (L, mm) and width (W, mm) were measured and the total volume (V, mm3) of was estimated using the equation V = L× W × W /2. After 35 days, mice were killed and pairs of left-side and right-side xenografts were retrieved and directly compared under light scope.
Dual-Luciferase Reporter Assay
A pmiR-REPORT luciferase plasmid containing the 3’-untranslated region (3’-UTR) of wild type (WT) human PWRN1 gene, (WT) PWRN1 was commercially synthesized by HanBio Biotechnology (HanBio, Shanghai, China). Alternatively, a pmiR-REPORT luciferase plasmid containing a mutated 3’-UTR of human PWRN1 gene (without hsa-miR-21-5p binding site), (MU) PWRN1, was also synthesized by HanBio Biotechnology (HanBio, Shanghai, China). In addition, A human miRNA hsa-miR-21-5p mimics (mimics_21) and a control human non-specific miRNA mimics (mimics_C) were also purchased from HanBio Biotechnology (HanBio, Shanghai, China).
HEK293T cells were pre-transfected with mimics_C or mimics_21, followed by another infection of (WT) PWRN1 or (MU) PWRN1 for 48 h. After that, a dual-luciferase reporter assay (Promega, USA) was performed according to the manufacturer’s instruction. Relative luciferase activities were then measured, with co-infection of mimics_C and (WT) PWRN1 termed as control condition.
hsa-miR-21-5p Upregulation Assay
In L/PWRN1-infected LN-229 and U-251 cells, they were further transfected with mimics_21 or mimics_C using Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific, China) for 48 h. Then, qRT-PCR was performed in double-infected LN-229 and U-251 cells to verify the upregulation of hsa-miR-21-5p.
Statistical Analysis
In our study, all experiments were independently conducted for biological triplicates or more repeats. Data were presented as means ± standard errors. A windows-based SPSS software (version 13.0, SPSS, USA) was used for all statistical analysis. Comparison among clinical samples, namely between non-cancer and GBM tissues, was conducted using a Mann–Whitney U-test. All other statistical analysis was conducted using a one-way ANOVA test. In all comparisons, P values < 0.05 were determined as significant differences.
Results
LncRNA PWRN1 Is Downregulated in Both GBM Tumors and Cell Lines
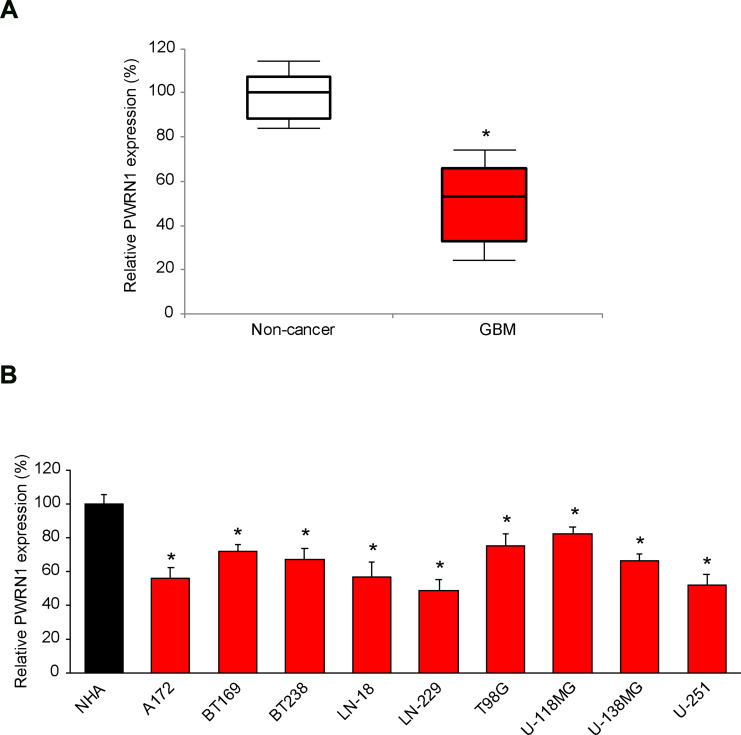
First, we investigated the expression profile of lncRNA PWRN1 in human GBM tumors. Analysis of qRT-PCR demonstrated that PWRN1 was significantly downregulated in samples of GBM tumors, than in samples of adjacent normal (non-cancer) brain tissues (Figure 1A, * P < 0.05, Mann–Whitney U-test).
Figure 1.
Expressing of PWRN1 in human GBM tumors and immortal GBM cell lines. (A) The expression level of human lncRNA PWRN1 was measured in GBM tumor samples obtained from cancer patients, and then compared to the expression level of PWRN1 in adjacent normal brain tissues (* P < 0.05, Mann-Whitney U-test). (B) PWRN1 expression was also assessed by qRT-PCR in nine GBM cell lines, including A172, BT169, BT238, LN-18, LN-229, T98G, U-118MG, U-138MG and U-251 cells, and then compared to the PWRN1 expression in a normal human astrocytes (NHA) cell line (* P < 0.05).
Then, PWRN1 expression levels were investigated in immortal GBM cell lines. As compared to PWRN1 expression level in a normal human astrocytes (NHA) cell line, it was found to be significantly downregulated in all tested GBM cell lines (Figure 1B, * P < 0.05). Specifically, PWRN1 was mostly downregulated in LN-229 and U-251 cells (Figure 1B).
PWRN1 Overexpression Suppressed GBM Cell in vitro Proliferation and Migration
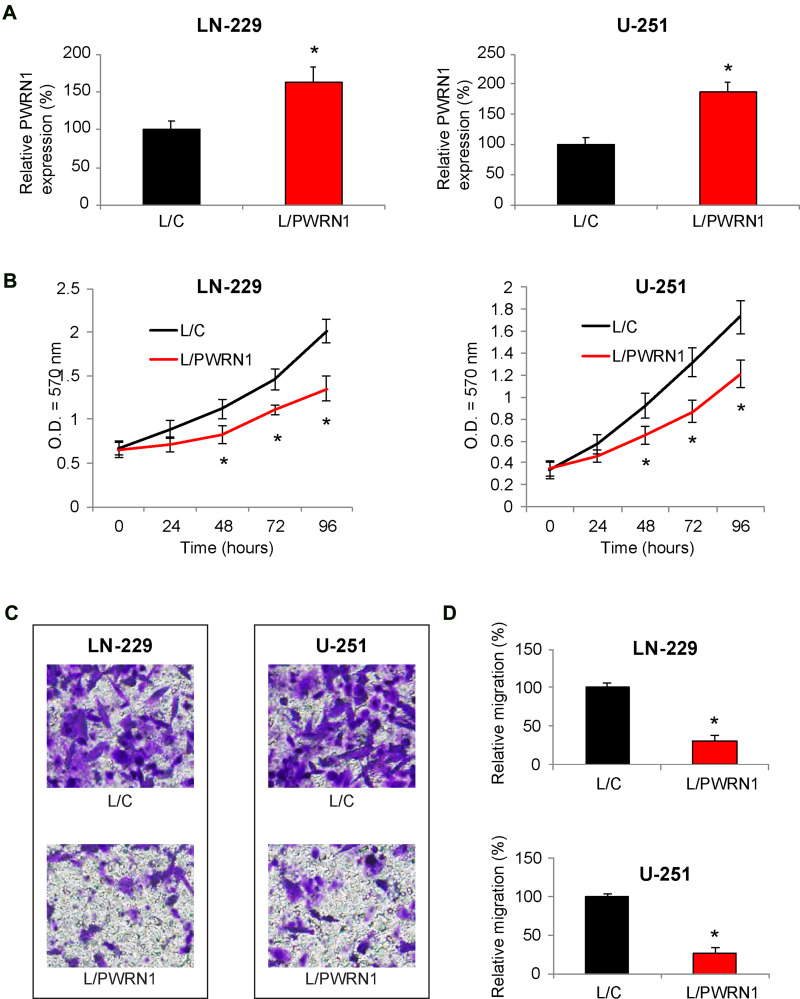
Next, we tried to determine the functional implication of PWRN1 in regulating GBM cells. To do so, LN-229 and U-251 cells were infected by lentiviruses to overexpress endogenous PWRN1. After infection was stabilized, qRT-PCR demonstrated that L/PWRN1-infected LN-229 and U-251 cells had significantly higher PWRN expression levels than L/C-infected cells (Figure 2A, * P < 0.05).
Figure 2.
Inhibitory effects of PWRN1 overexpression on GBM cell proliferation and migration in vitro. (A) LN-229 and U-251 cells were infected with L/C or L/PWRN1. Post infection, cells were passaged 5~8 times, followed by qRT-PCR to assess theirPWRN1expressions (* P < 0.05). (B) InfectedLN-229 and U-251 cells were assessed by a CCK-8 assay for 96 h, to compare in vitro proliferation between L/C- and L/PWRN1- infected GBM cells (* P < 0.05). (C) InfectedLN-229 and U-251 cells were assessed by a 24-well transwell assay. After 24 h, LN-229 and U-251 cells migrated onto the bottoms of wells were stained. Representative images were shown for L/C- and L/PWRN1- infected GBM cells. (D) Formigrating LN-229 and U-251 cells shown in (C), relative migration was quantitatively measured and compared between L/C- and L/PWRN1- infected GBM cells (* P < 0.05).
Infected LN-229 and U-251 cells were then equally plated in 96-well plates, and their in vitro proliferation was assessed using a CCK-8 assay, for 96 hours. The result demonstrated that L/PWRN1-infected LN-229 and U-251 cells had significantly slower proliferating rates then L/C-infected cancer cells (Figure 2B, * P < 0.05).
In addition, cancer cell migrating capability was assessed in infected LN-229 and U-251 cells. Using a 24-h transwell assay, it showed that significantly fewer LN-229 or U-251 cells were discovered at the bottoms of wells, if they were infected with L/PWRN1, rather than L/C (Figure 2C). Correspondingly, quantitative assessment confirmed that migrating capabilities in L/PWRN1 -infected LN-229 and U-251 cells were significantly reduced, as compared to L/C-infected cells (Figure 2D, * P < 0.05).
PWRN1 Overexpression Inhibited GBM Xenograft in vivo
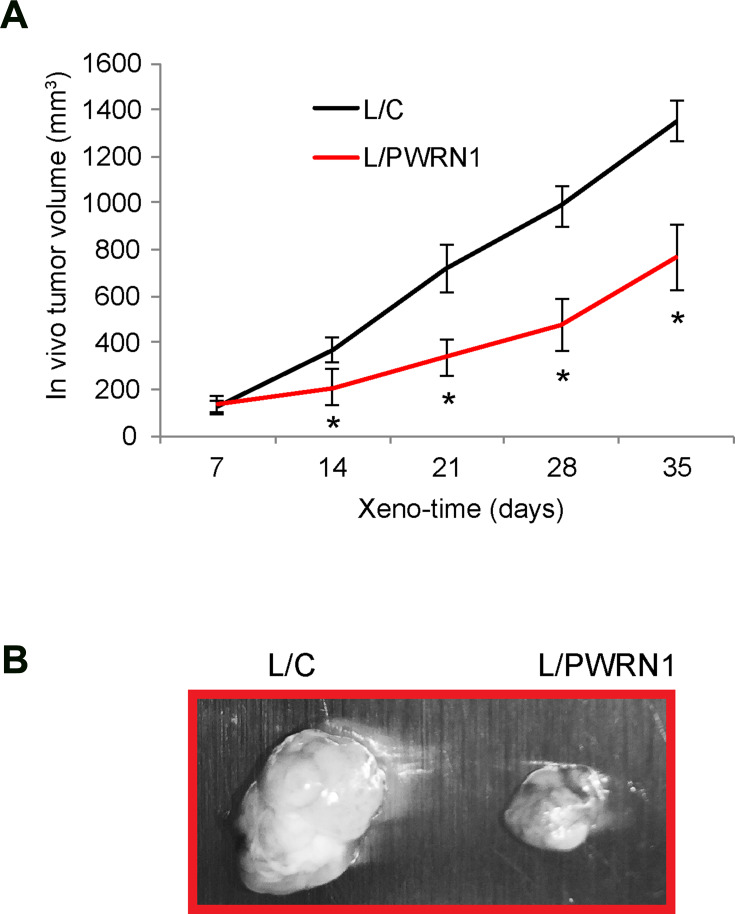
Infected LN-229 cells were subcutaneously grafted into nude BALB/c mice to assess the effect of PWRN1 overexpression on GBM in vivo growth. During the time course of 35 days, the tumor volumes were estimated weekly, and compared between L/C- and L/PWRN1- infected GBM xenografts. It demonstrated that the in vivo growth of GBM xenograft was dramatically inhibited by PWRN1 overexpression (Figure 3A, * P < 0.05). In addition, while mice were sacrificed and GBM xenografts were taken out, imaging results confirmed our in vivo estimation on tumor volumes, demonstrating that L/PWRN1-infected xenografts were significantly smaller than L/C-infected xenografts (Figure 3B).
Figure 3.
Inhibitory effect of PWRN1 overexpression on GBM xenograft growth in vivo. (A) L/C- orL/PWRN1- infectedLN-229 cells were subcutaneously grafted into the dorsal skin compartments of female nude mice (1 million cells / inoculation, total of 15 mice). The left sides were grafted with L/C-infected LN-229 cells, whereas right sides were grafted with L/PWRN1-infected LN-229 cells. The volumes (mm3) of subcutaneous xenograft were measured every 7-day for 35 days (* P < 0.05). (B) After xenograft assay, GBM tumors were taken out and examined under light microscope.
PWRN1 Overexpression Reversely Downregulated hsa-miR-21-5p Expression in GBM Cells
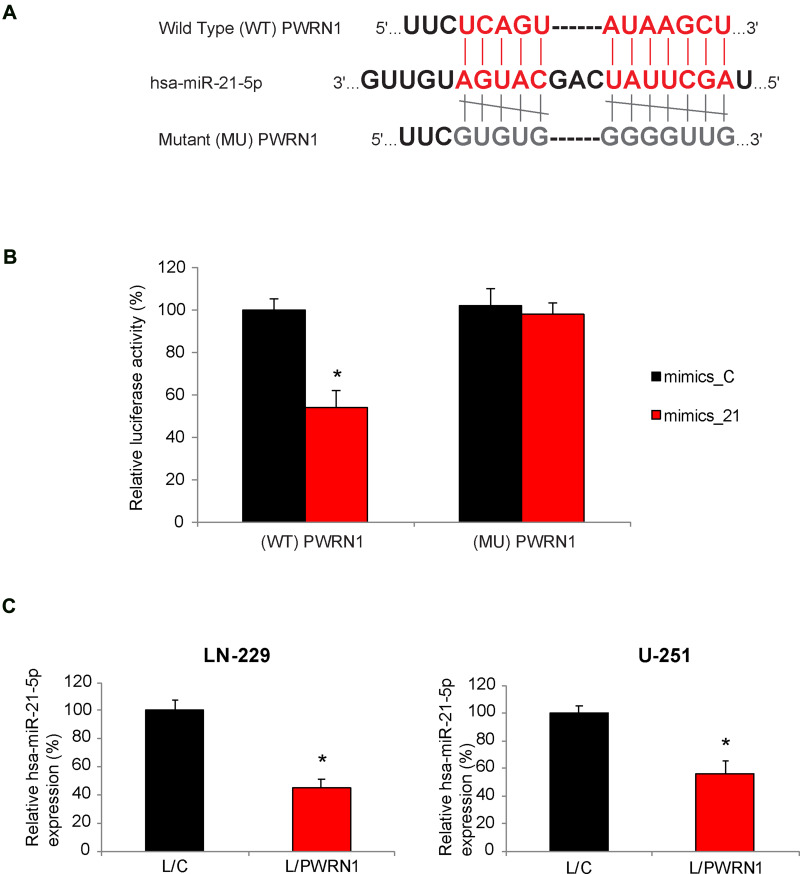
Related signaling pathway of PWRN1 in GBM was further investigated. We firstly used bioinformatic database to search the possible miRNA candidates of PWRN1 (StarBase23,24). Then, we conducted literature search to identify those with known oncogenic factors in human cancers. Finally, we used qRT-PCR and dual-luciferase activity assay to narrow down the candidates in GBM. Using this approach, it was noted that human microRNA-21-5p (hsa-miR-21-5p) was a possible downstream competing candidate of PWRN1 (Figure 4A). Based on this prediction, we created two luciferase reporters. One contained the 3’-untranslated region (3’-UTR) of wild type (WT) PWRN1, (WT) PWRN1. The other contained a mutant PWRN1 3’-UTR without hsa-miR-21-5p binding domain (MU) PWRN1. In HEK293T cells pre-infected with mimics-C or mimics_21, cells were further infected with (WT) PWRN1 or (MU) PWRN1. After that, a dual-luciferase activity assay confirmed that hsa-miR-21-5p was a downstream competing target of PWRN1 (Figure 4B, * P < 0.05). In addition, we discovered, by qRT-PCR, that hsa-miR-21-5p expression levels were significantly downregulated in L/PWRN1-infected LN-229 or U-251 cells, than in L/C-infected cells (Figure 4C, * P < 0.05).
Figure 4.
PWRN1 is endogenously competing against hsa-miR-21-5p in GBM cells. (A) The 3’-UTR of wild type (WT) human PWRN1 was shown to include a hsa-miR-21-5p binding domain. Alternatively, the binding domain was mutated (MU). (B) In a dual-luciferase activity assay, HEK293T cells, pre-infected with mimics_21 or mimics_C, were then infected with L/C or L/PWRN1 for 48 h. Relative luciferase activities were measured for co-infected HEK293T cells (* P < 0.05). (C) Between L/C-infected and L/PWRN1-infected LN-229 or U-251 cells, qRT-PCR was applied to compare their hsa-miR-21-5p expression levels (* P < 0.05).
Upregulating hsa-miR-21-5p Reversed the Tumor-Suppressing Function of PWRN1 Overexpression on GBM Cells
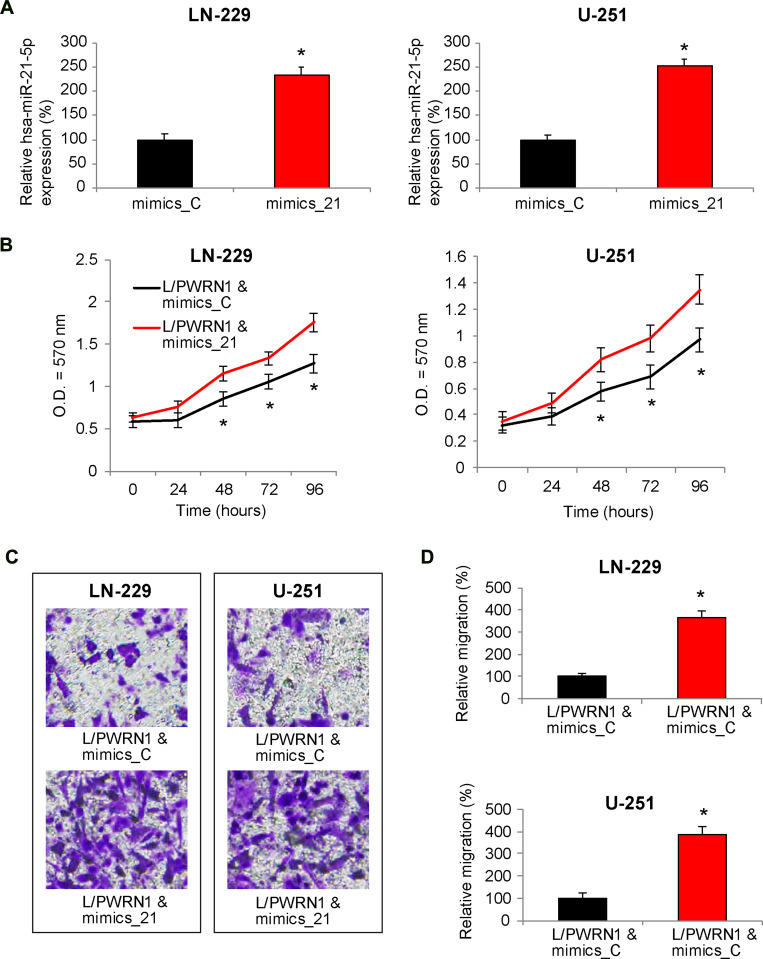
Finally, we upregulated hsa-miR-21-5p in L/PWRN1-infected LN-229 and U-251 cells. QRT-PCR showed that, in GBM cells double-infected with L/PWRN1 & mimics_21, hsa-miR-21-5p expression levels were significantly upregulated, as compared to cells double-infected with L/PWRN1 & mimics_C (Figure 5A, * P < 0.05).
Figure 5.
Reverse effect of hsa-miR-21-5p upregulation on PWRN1-overexpression mediated GBM cell proliferation and migration in vitro. (A) L/PWRN1-infected LN-229 and U-251 cells were further transfected with mimics_21 or mimics_C. Then, qRT-PCR was used to assess theirhsa-miR-21-5p expression levels (* P < 0.05). (B) Double-infectedLN-229 and U-251 cells were assessed by a CCK-8 assay, to compare in vitro proliferation between cells double-infected with L/PWRN1 & mimics_21 and cells double-infected with L/PWRN1 & mimics_C (* P < 0.05). (C) Double-infected LN-229 and U-251 cells were assessed by a 24-well transwell assay. After 24 h, cancer cells migrated onto the bottoms of wells were stained. Representative images were compared between L/PWRN1 & mimics_21- and L/PWRN1 & mimics_C-infected cells. (D) Formigrating GBM cells shown in (C), relative migration was quantitatively measured and compared between L/PWRN1 & mimics_21- and L/PWRN1 & mimics_C-infected cells (* P < 0.05).
Then, double-infected cells were seeded in 96-well plates, and their in vitro proliferation was assessed by a CCK-8 assay for 96 hours. The result of proliferation assay showed that, in LN-229 and U-251 cells infected with L/PWRN1 & mimics_21, cancer cell proliferating rates were significantly augmented, than in cells infected with L/PWRN1 & mimics_C (Figure 5B, * P < 0.05).
In addition, cancer cell migration was assessed in double-infected LN-229 and U-251 cells. Using a 24-h transwell assay, it showed that considerably more GBM cells were observed at the bottoms of wells if they were infected with L/PWRN1 & mimics_21, rather than L/PWRN1 & mimics_C (Figure 5C). Furthermore, quantitative assessment confirmed that, in L/PWRN1 & mimics_21-infected LN-229 and U-251 cells, migrating capability was markedly enhanced, as compared to L/PWRN1 & mimics_C-infected GBM cells (Figure 5D, * P < 0.05).
Discussions
In recent decades, strong evidence has demonstrated that lncRNAs may by aberrantly deregulated in human GBM tumors and closely associated with cancer patients’ prognosis, thus making them potential candidates for diagnostic biomarkers or therapeutic targets for GBM.13,14 In the present study, we suspected lncRNA of PWRN1, which has been shown to be dysregulated in several types of human cancers,19–21,25 is also aberrantly expressed in human GBM, and functionally involved in GBM cell regulation. Based on this hypothesis, we used a systemic approach to assess the expression profile and possible regulatory mechanisms of PWRN1in GBM.
First, qRT-PCR was applied and we discovered that PWRN1 was significantly downregulated in both human GBM tumors and immortal GBM cell lines. This result is consistent with the findings in several previous studies, showing PWRN1 was also lowly expressed in breast cancer,21 prostate adenocarcinoma20 and gastric cancer.19 Based on this information, it seems that PWRN1 is predominantly downregulated in various types of human cancers. Future clinical study may reveal whether PWRN1 may be used as a prognostic biomarker to predict prognosis among patients with GBM.
Then, we sought the functional role of PWRN1 in regulating GBM cancer cell progression and migration. To do so, we used lentivirus to genetically overexpress PWRN1 in two GBM cell lines, LN-299 and U-251. Using several in vitro and in vivo cancer assays, we confirmed that PWRN1 overexpression inhibited BGM cell proliferation and invasion in vitro and xenograft growth in vivo. Recently, it was reported PWRN1 acted as an active factor in suppressing human gastric cancer cell growth.19 However, to the best of our knowledge, discovery in the present study is the first one to report the functional mechanism of PWRN1 in human GBM.
Furthermore, we investigated the downstream signaling pathway associated with PWRN1 in regulating GBM cell functions. Previously, it was demonstrated that hsa-miR-21 acted as an oncogene in GBM.26 Using dual-luciferase and qRT-PCR assessments, we discovered that PWRN1 could bind hsa-miR-21-5p, and PWRN1 overexpression inversely downregulated hsa-miR-21-5p expression in GBM cells. In addition, after we successfully upregulated hsa-miR-21-5p in PWRN1-overexpressed LN-229 and U-251 cells, it was demonstrated that the PWRN1-overexpression-mediated tumor-suppressing effects on GBM cancer cell in vitro proliferation and migration were significantly reversed. Thus, our results strongly suggested that hsa-miR-21-5p was actively associated with PWRN1 in cancer regulation in GBM. It is worth noting that, in human gastric cancer, PWRN1 was acting through miR-425-5p/p53 signaling feedback loop to modulate tumor cell growth and metastasis.19 Thus, it is possible that PWRN1 may activate/inhibit other epigenetic signaling pathways, rather than hsa-miR-21-5p, to regulate cancer cell in GBM.
The limitation of our study was that the downstream mRNA transcripts of hsa-miR-21-5p were not fully elucidated in GBM. In a previous study, it was demonstrated that programmed cell death 4 (Pdcd4), a known tumor-suppressor gene was downstream of miR-21 to modulate proliferation, apoptosis and colony-formation in GBM-derived cell lines.27 In addition, in another study, it was demonstrated that EGFR signaling pathway, including Akt. CyclinD and Bcl-2 were involved in miR-21-mediated GBM cell growth.28 The possible explanation for miR-21 triggering different downstream signaling pathways may be that different upstream activator/inhibitor of miR-21 are initiated under different physical/genetic conditions in GBM. Moreover, it is not clear whether PWRN1 was involved in any of the known miR-21-regulated signaling pathways in GBM. Thus, future experiments may be needed to further define the genetic network associated with PWRN1 regulation in human GBM.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Sun K, Zheng R, et al. Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 2018;30(1):1–12. doi: 10.21147/j.issn.1000-9604.2018.01.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnholtz-Sloan JS, Ostrom QT, Cote D. Epidemiology of brain tumors. Neurol Clin. 2018;36(3):395–419. doi: 10.1016/j.ncl.2018.04.001 [DOI] [PubMed] [Google Scholar]
- 4.Tamimi AF, Juweid M. Epidemiology and outcome of glioblastoma In: De Vleeschouwer S, editor.Glioblastoma.Brisbane (AU);2017. [PubMed] [Google Scholar]
- 5.Tykocki T, Eltayeb M. Ten-year survival in glioblastoma. A systematic review. J Clin Neurosci. 2018;54::7–13. doi: 10.1016/j.jocn.2018.05.002 [DOI] [PubMed] [Google Scholar]
- 6.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu B, Shan G. Functions of long noncoding RNAs in the nucleus. Nucleus. 2016;7(2):155–166. doi: 10.1080/19491034.2016.1179408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001 [DOI] [PubMed] [Google Scholar]
- 9.Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int J Cancer. 2017;140(9):1955–1967. doi: 10.1002/ijc.30546 [DOI] [PubMed] [Google Scholar]
- 10.Lorenzi L, Avila Cobos F, Decock A, et al. Long noncoding RNA expression profiling in cancer: challenges and opportunities. Genes Chromosomes Cancer. 2019;58(4):191–199. doi: 10.1002/gcc.22709 [DOI] [PubMed] [Google Scholar]
- 11.Rafiee A, Riazi-Rad F, Havaskary M, Nuri F. Long noncoding RNAs: regulation, function and cancer. Biotechnol Genet Eng Rev. 2018;34(2):153–180. doi: 10.1080/02648725.2018.1471566 [DOI] [PubMed] [Google Scholar]
- 12.Khan IN, Ullah N, Hussein D, Saini KS. Current and emerging biomarkers in tumors of the central nervous system: possible diagnostic, prognostic and therapeutic applications. Semin Cancer Biol. 2018;52(Pt 1):85–102. doi: 10.1016/j.semcancer.2017.07.004 [DOI] [PubMed] [Google Scholar]
- 13.Han L, Zhang K, Shi Z, et al. LncRNA pro fi le of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol. 2012;40(6):2004–2012. doi: 10.3892/ijo.2012.1413 [DOI] [PubMed] [Google Scholar]
- 14.Zhang XQ, Sun S, Lam KF, et al. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiol Dis. 2013;58::123–131. doi: 10.1016/j.nbd.2013.05.011 [DOI] [PubMed] [Google Scholar]
- 15.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22(3):165–173. doi: 10.1016/j.tig.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 16.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–1109. doi: 10.1016/j.bbagrm.2014.08.012 [DOI] [PubMed] [Google Scholar]
- 17.Cao MX, Jiang YP, Tang YL, Liang XH. The crosstalk between lncRNA and microRNA in cancer metastasis: orchestrating the epithelial-mesenchymal plasticity. Oncotarget. 2017;8(7):12472–12483. doi: 10.18632/oncotarget.13957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Xu Y, Feng L, et al. Comprehensive characterization of lncRNA-mRNA related ceRNA network across 12 major cancers. Oncotarget. 2016;7(39):64148–64167. doi: 10.18632/oncotarget.11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z, Ju H, Yu S, et al. Prader-Willi region non-protein coding RNA 1 suppressed gastric cancer growth as a competing endogenous RNA of miR-425-5p. Clin Sci (Lond). 2018;132(9):1003–1019. doi: 10.1042/CS20171588 [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Yang Z. Analysis of competing endogenous RNA network to identify the key RNAs associated with prostate adenocarcinoma. Pathol Res Pract. 2018;214(11):1811–1817. doi: 10.1016/j.prp.2018.08.029 [DOI] [PubMed] [Google Scholar]
- 21.Tuersong T, Li L, Abulaiti Z, Feng S. Comprehensive analysis of the aberrantly expressed lncRNA-associated ceRNA network in breast cancer. Mol Med Rep. 2019;19(6):4697–4710. doi: 10.3892/mmr.2019.10165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.General Assembly of the World Medical A. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. J Am Coll Dent. 2014;81(3):14–18. [PubMed] [Google Scholar]
- 23.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39(Database issue):D202–209. doi: 10.1093/nar/gkq1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–97. doi: 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu B, Ma T, Li Q, et al. Identification of a lncRNA-associated competing endogenous RNAregulated network in clear cell renal cell carcinoma. Mol Med Rep. 2019;20(1):485–494. doi: 10.3892/mmr.2019.10290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137 [DOI] [PubMed] [Google Scholar]
- 27.Gaur AB, Holbeck SL, Colburn NH, Israel MA. Downregulation of Pdcd4 by mir-21 facilitates glioblastoma proliferation in vivo. Neuro Oncol. 2011;13(6):580–590. doi: 10.1093/neuonc/nor033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou X, Ren Y, Moore L, et al. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90(2):144–155. doi: 10.1038/labinvest.2009.126 [DOI] [PubMed] [Google Scholar]