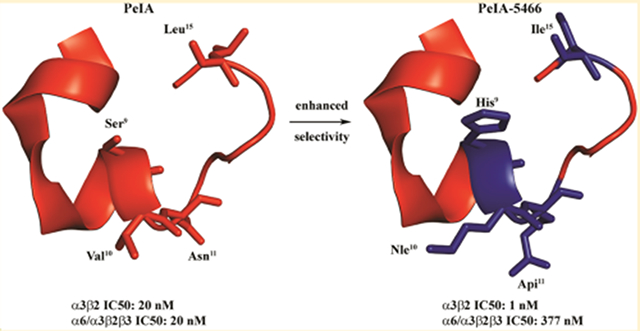
Abstract
Pharmacologically distinguishing α3β2 nicotinic acetylcholine receptors (nAChRs) from closely related subtypes, particularly α6β2, has been challenging due to the lack of subtype-selective ligands. We created analogs of α-conotoxin (α-Ctx) PeIA to identify ligand–receptor interactions that could be exploited to selectively increase potency and selectivity for α3β2 nAChRs. A series of PeIA analogs were synthesized by replacing amino acid residues in the second disulfide loop with standard or nonstandard residues and assessing their activity on α3β2 and α6/α3β2β3 nAChRs heterologously expressed in Xenopus laevis oocytes. Asparagine11 was found to occupy a pivotal position, and when replaced with negatively charged amino acids, selectivity for α3β2 over α6/α3β2β3 nAChRs was substantially increased. Second generation peptides were then designed to further improve both potency and selectivity. One peptide, PeIA-5466, was ~300-fold more potent on α3β2 than α6/α3β2β3 and is the most α3β2-selective antagonist heretofore reported.
Graphical Abstract
INTRODUCTION
Nonmuscle nicotinic acetylcholine receptors (nAChRs) are an extended branch of the Cys-loop superfamily of pentameric ligand-gated ion channels, formed from combinations of α (α2–α10) and β (β2–β4) subunits. These subtypes are important in normal physiology and in a wide range of disease states.1–3 Their pharmacological and functional profiles, as well as expression patterns, frequently overlap, making them difficult to distinguish definitive physiological roles for each nAChR subtype that is expressed in any given location. Because of this, antagonists, which selectively inhibit particular nAChR subtypes, are essential tools for reliably defining causal relationships between receptor function and physiology/behavior. Selective pharmacological tools are also valuable complements to genetically modified animal models.
Within the central nervous system (CNS), nAChRs containing the β2 subunit β2* nAChR (the asterisk denotes the possible or known presence of other subunits in native nAChR complexes4) comprise the majority. Most CNS β2* nAChRs are of the α4β2* subtype, which is expressed across almost all brain regions and frequently at high densities. However, other subtypes are expressed in subsets of brain regions, where they can be very influential.5 Autoradiography studies of mouse brain have indicated that the presence of α3β2* nAChRs is most prominent in the medial habenula–fasciculus retroflexus–interpeduncular tract. Other regions likely to express α3β2* nAChRs are the dopaminergic cell bodies of the substantia nigra (SN) and ventral tegmental area (VTA), dorsal cortex of the inferior colliculus, and the medial vestibular and prepositus hypoglossal nuclei.5,6 Peripherally, α3β2* nAChR subtypes have been shown by immunoprecipitation and gene-knock out studies to be expressed by neurons of autonomic ganglia including those found in superior cervical, nodose, and intracardiac ganglia7,8 as well as in the retina.9 However, pharmacological and functional studies of α3β2 nAChRs have been hampered due to the lack of ligands capable of selectively inhibiting α3β2 over other nAChRs subtypes, particularly α6β2.
α-Conotoxins (α-Ctxs) are small, disulfide-rich peptides produced in the venom glands of carnivorous marine snails of the genus Conus. These snails use the peptides to capture and immobilize their prey and to defend against predators.10 Their small size, usually 13–30 amino acids, ease of de novo synthesis, and amenability to modification make them attractive candidates for the development of novel peptide ligands for drug discovery. One such peptide, PeIA, was isolated from the piscivorous species Conus pergrandis.11 The fully folded peptide is 16 amino acids in length and has two disulfide bonds connecting Cys2 to Cys8 and Cys3 to Cys16, which serve to keep the peptide in its stable and biologically active conformation (see Figure 1).
Figure 1.
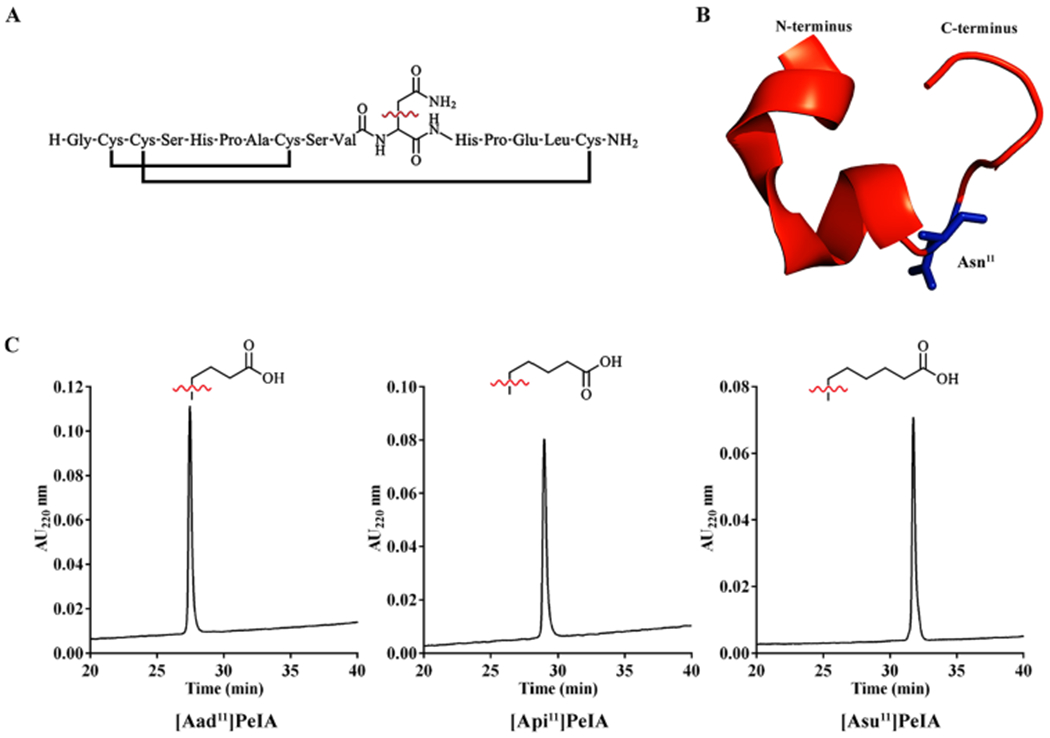
Amino acid sequence and structure of PeIA and RP-HPLC of analogs containing non-natural amino acid residues. (A) The linear sequence of PeIA shown with the disulfide connectivity and Asn11 as the line structure. (B) Cartoon rendition of the crystal structure (PDB: 5JME, chain H)41 of PeIA highlighting the position of Asn11 (blue). (C) Side-chain structures of the non-natural amino acids Aad, Api, and Asu used in analog synthesis and corresponding RP-HPLC traces of the fully folded peptides. [Aad11]PeIA, [Api11]PeIA, and [Asu11]PeIA eluted at 37.5, 39.0, and 42.4% solvent B, respectively, using an ACN gradient of 10–50% in 40 min and a C18 analytical column.
Previous structure–function studies of PeIA identified residues Ser9 and Val10 that when substituted with His and Ala, respectively, increase potency for rat α3β2 nAChRs expressed in Xenopus laevis oocytes.12 However, these changes also substantially increase PeIA activity on rat α6/α3β2β3 nAChRs. A second study identified substitutions that selectively decreased activity on α3β2 but preserved activity on α6/α3β2β3 nAChRs.13 An Asn11 to Arg substitution was found to be critical for exploiting a nonconserved Lys152–Glu152 difference between α3 and α6 subunits, respectively. These two studies provided the basis for using PeIA to interrogate ligand–receptor interactions and to inform the design and synthesis of selective antagonists of α3β2 nAChRs.
Although some α-Ctxs show minor preferences for α3β2 over α6β2 nAChRs (Table 1), there are currently no ligands that can be used to selectively inhibit the α3β2 subtype. Thus, one aim of this study was to develop a potent ligand that can distinguish between these two subtypes. To this end, we synthesized a number of PeIA analogs with single-residue substitutions to identify interactions that increased potency and/or selectivity for α3β2 nAChRs expressed in X. laevis oocytes. Novel peptides were then designed containing these identified amino acids and then assessed for α3β2 over α6/α3β2β3 nAChR selectivity. One peptide, PeIA-5466 (see Table 5), was ~300-fold selective and displayed low nM potency for α3β2 nAChRs. The results of these studies demonstrate that ligands can be developed that are selective for α3β2 over α6β2 nAChRs. The novel peptides described herein may be useful for selectively identifying α3β2* in cells that express multiple nAChR subtypes, and particularly those that express α6β2* nAChRs. This is especially an issue in the MH-IPN tract and VTA dopaminergic projection neurons. In both cases, function within these tracts significantly influences addiction/dependence on nicotine (and other abused drugs), and α3β2* expression is found alongside that of α6β2* nAChRs.
Table 1.
Amino Acid Sequences and Potencies of Select α-Ctxs that Target α3β2 and α6/α3β2β3 nAChRsa
| α3β2 nAChR | α6/α3β2β3 nAChR | |||
|---|---|---|---|---|
| α-Ctx | Sequence | IC50 values (nM) | Ratio | |
| PeIA | GCCSHPACSVNHPELC | 19.4 (15.3–24.6) b | 19.8 (17.9–21.8) b | 1 |
| PnIA | GCCSLPPCAANNPDYC | 7.7 (6.8–8.8) (41) | 10.9 (7.9–14.9) (42) | 1 |
| OmIA | GCCSHPACNVNNPHICG | 11.0 (7.1–17.0) (15) | 201 (101–401) (24) | 18 |
| LvIA | GCCSHPACNVDHPEIC | 8.7 (6.9–11.0) (25) | 108 (54–215) (25) | 12 |
| MII | GCCSNPVCHLEHSNLC | 2.2 (1.2–3.8) (43) | 0.4 (0.3–0.5) (30) | 0.6 |
| LtIA | GCCARAACAGIHQELC | 9.8 (7.3–13.2) (44) | 84.4 (58–123) (44) | 9 |
Residues highlighted in yellow are conserved among these α-Ctxs. IC50 values are from electrophysiology experiments using X. laevis oocytes heterologously expressing cloned rat nAChRs; values in parentheses indicate the 95% confidence interval; selectivity was calculated as the IC50 ratio of (α6/α3β2β3)/(α3β2).
This work.
Table 5.
IC50 Values for α3β2-Selective Peptidesa
|
α3β2 nAChR |
α6/α3β2β3 nAChR |
|||||
|---|---|---|---|---|---|---|
| peptide | residue differences with PeIA | IC50 (nM) | n | IC50 (nM) | n | IC50 ratio |
| PeIA-4769 | His9,Leu10 | 0.29 (0.25–0.34) | 4 | 1.04 (0.921–1.18) | 4 | 3 |
| PeIA-5115 | His9,Leu10,Asp11 | 1.67 (1.44–1.95) | 4 | 19.9 (17.8–22.3) | 4 | 12 |
| PeIA-5080 | His9,Leu10,Glu11 | 1.22 (1.06–1.40) | 4 | 16.2 (13.3–19.8) | 4 | 14 |
| PeIA-5415 | His9,Leu10,Aad11 | 3.01 (2.40–3.78) | 5 | 155 (122–197) | 5 | 52 |
| PeIA-5355 | His9,Leu10,Api11 | 0.38 (0.36–0.44) | 4 | 22.3 (19.9–24.9) | 4 | 59 |
| PeIA-5587 | His9,Leu10,Asu11 | 1.28 (1.06–1.54) | 4 | 50.3 (40.4–62.7) | 4 | 39 |
| PeIA-5106 | His9,Leu10,Ile15 | 0.28 (0.23–0.33) | 5 | 0.86 (0.77–0.96) | 4 | 3 |
| PeIA-5413 | His9,Leu10,Api11,Ile15 | 0.57 (0.44–0.74) | 4 | 58.2 (51.4–65.8) | 5 | 102 |
| PeIA-5416 | His9,Nle10,Api11 | 1.33 (1.17–1.53) | 5 | 286 (220–373) | 4 | 220 |
| PeIA-5466 | His9,Nle10,Api11,Ile15 | 1.34 (1.13–1.60) | 4 | 377 (275–508) | 4 | 290 |
Values in parentheses indicate the 95% CI of the IC50 values. IC50 ratio calculated as (α6/α3β2β3)/(α3β2).
RESULTS
Synthesis of PeIA Analogs Containing Non-natural Amino Acid Residues.
Nonconserved residues of α3 and α6 subunits, including residue 152, have been shown to influence the potencies of certain α-conopeptides for α3β2 and α6/α3β2β3 nAChRs.13,14 In the α3 subunit, residue 152 is a Lys, whereas in α6, a Glu is present. Substituting Asn11 with the positively charged amino acids Lys or Arg substantially reduced PeIA potency on α3β2, probably by creating an unfavorable charge–charge interaction between the ligand and the receptor. Thus, substituting Asn11 with negatively charged amino acid was hypothesized to produce a favorable interaction with Lys152 of the α3 subunit and an unfavorable interaction with Glu152 of the α6 subunit. Therefore, we synthesized a series of PeIA analogs where Asn11 was replaced with amino acids with negatively charged side chains. In addition to Asp and Glu, we also synthesized analogs with non-natural amino acids with longer, negatively charged side chains. These amino acids include α-aminoadipic acid (Aad), α-aminopimelic acid (Api), and α-aminosuberic acid (Asu). The side chains of these amino acids are one, two, and three carbons longer than Glu, respectively (Figure 1A–C). The linear peptides were synthesized using standard solid-phase peptide chemistry and Fmoc amino acids. To ensure that the disulfide bonds were formed correctly, Cys2 and Cys8 side chains were orthogonally protected by trityl and Cys3 and Cys16 by acetamidomethyl groups. The peptides were folded by removing the trityl groups with 20 mM potassium ferricyanide and 0.1 M Tris-HCl, forming the first disulfide bridge (Cys2–Cys8). Closure of the second disulfide bridge (Cys3–Cys16) was accomplished by iodine oxidation. The folded peptides were purified using reverse-phase high-performance liquid chromatography (RP-HPLC) and the masses determined using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) or electrospray ionization (ESI) mass spectrometry (Table 2).
Table 2.
Yield, Purity, and Mass Spectrometry for Synthetic Peptidesa
| peptide | yield (nmol) | % purity | calculated M+ Da | observed M+ Da |
|---|---|---|---|---|
| [Asn9]PeIA | 5569 | >98 | 1678.63 | 1678.65 |
| [Tyr9]PeIA | 3714 | >98 | 1727.65 | 1727.75 |
| [Thr9]PeIA | 3541 | >97 | 1665.64 | 1665.36 |
| [Ile10]PeIA | 2714 | >98 | 1665.54 | 1665.72 |
| [Nle10]PeIA | 3643 | >98 | 1665.54 | 1665.69 |
| [Thr10]PeIA | 2742 | >99 | 1653.60 | 1653.57 |
| [Asp11]PeIA | 5311 | >97 | 1652.61 | 1652.69 |
| [Aadn]PeIA | 2591 | >98 | 1680.63 | 1680.63 |
| [Api11]PeIA | 1637 | >98 | 1694.63 | 1694.82 |
| [Asu11]PeIA | 3957 | >98 | 1708.63 | 1708.68 |
| [Gla14]PeIA | 2014 | >98 | 1695.61 | 1695.62 |
| [Asp14]PeIA | 3440 | >98 | 1637.61 | 1637.65 |
| [Adi14]PeIA | 2244 | >97 | 1665.63 | 1665.66 |
| [Asu14]PeIA | 1809 | >97 | 1693.63 | 1693.68 |
| [Gln14]PeIA | 2571 | >98 | 1650.64 | 1650.63 |
| [Arg14]PeIA | 1650 | >98 | 1678.68 | 1678.80 |
| [Arg15]PeIA | 2027 | >98 | 1694.64 | 1694.79 |
| [Val15]PeIA | 1951 | >98 | 1637.61 | 1637.66 |
| [Ile15]PeIA | 3749 | >99 | 1651.62 | 1651.67 |
| [Nle15]PeIA | 2774 | >98 | 1651.62 | 1651.62 |
| PeIA-4769 | 5289 | >98 | 1715.67 | 1715.69 |
| PeIA-5106 | 5493 | >96 | 1715.67 | 1715.53 |
| PeIA-5115 | 7226 | >97 | 1716.65 | 1716.75 |
| PeIA-5080 | 1145 | >96 | 1730.67 | 1730.81 |
| PeIA-5415 | 2842 | >98 | 1744.67 | 1744.80 |
| PeIA-5355 | 1292 | >98 | 1758.67 | 1758.65 |
| PeIA-5587 | 3644 | >97 | 1772.67 | 1772.78 |
| PeIA-5413 | 3753 | >98 | 1758.67 | 1758.76 |
| PeIA-5416 | 2523 | >98 | 1758.67 | 1758.82 |
| PeIA-5466 | 2603 | >98 | 1758.67 | 1758.84 |
MALDI-TOF mass spectrometry was used to determine the masses of all peptides except for [Gla14]PeIA, which was determined using ESI mass spectrometry. RP-HPLC chromatograms for each of the peptides showing the purity are presented in the SI Figure 2.
Structure–Function Studies Identify Ligand–Receptor Interactions That Are Critical for Activity on α3β2 nAChRs.
To determine if PeIA analogs containing non-natural amino acid residues were active, we tested them on X. laevis oocytes heterologously expressing rat α3β2 or α6/α3β2β3 nAChRs. First, we tested analogs with N11D and N11E substitutions. [Asp11]PeIA showed a ~3-fold loss of activity on α3β2, whereas [Glu11]PeIA retained activity compared to native PeIA (Figure 2A, Table 3). [Asp11]PeIA showed a similar loss of activity on α6/α3β2β3 receptors, but a more substantial loss of ~7-fold was observed with [Glu11]PeIA (Figure 2B, Table 3). Next, we evaluated PeIA analogs containing Aad, Api, or Asu in the 11th position. These substitutions systematically extend the length of the side chain bearing the carboxylic acid group, one carbon at a time. When tested on α3β2 receptors, [Aad11]PeIA, [Api11]PeIA, and [Asu11]PeIA all retained antagonist activity on α3β2 nAChRs but displayed different potencies. [Aad11]PeIA and [Asu11]-PeIA showed minor losses of potency, whereas [Api11]PeIA retained the potency of native PeIA (Figure 2A, Table 3). By contrast, all three analogs showed substantially reduced activity on α6/α3β2β3 receptors and of them, [Aad11]PeIA showed the greatest loss of potency (~500-fold) (Figure 2B, Table 3).
Figure 2.
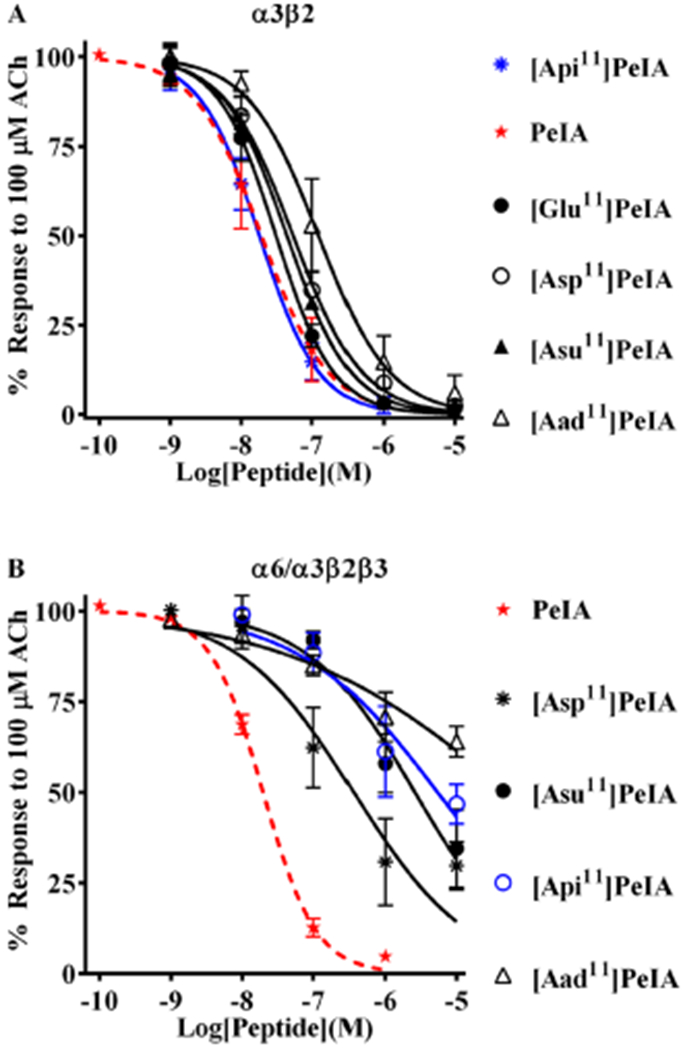
PeIA analogs with negatively charged amino acid residues in position 11 show increased selectivity for α3β2 nAChRs. Analogs with substitutions of Asn11 were synthesized and a concentration–response analysis was performed for each to determine the IC50 values. The value obtained for each analog was compared to that of the native peptide (red dashed curves) to assess the effects of the substitution. (A) Analogs with Asp, Glu, or Api substitutions showed <3-fold change in potency for α3β2 nAChRs. Aad, with a one-carbon longer side chain than Glu and one-carbon shorter than Api, reduced PeIA potency by 6-fold. (B) By contrast, all analogs showed substantially reduced potency (6- to 274-fold) for α6/α3β2β3 nAChRs. The most α3β-selective of these analogs was determined to be [Api11]PeIA (blue curve), which showed a 300-fold preference for the α3β2 subtype. A minimum of four oocytes was used for each IC50 determination, and the error bars indicate the SD. The analogs are listed in rank order of potency from top (most potent) to bottom (least potent); values are presented in Table 3.
Table 3.
IC50 Values for PeIA and Analogs with Negatively Charged Amino Acid Substitutions of Asn11a
|
α3β2 nAChR |
α6/α3β2β3 nAChR |
||||||
|---|---|---|---|---|---|---|---|
| peptide | IC50 (nM) | n | log change in IC50 | IC50 (nM) | n | log change in IC50 | selectivity ratio |
| PeIA | 19.4 (15.3–24.6) | 5 | 19.8 (17.9–21.8) | 4 | 1 | ||
| [Asp11]PeIA | 54.0 (45.7–64.5) | 6 | 0.4 | 362 (215–611) | 6 | 1.3 | 7 |
| [Glu11]PeIA | 31.1 (27.0–36.0) | 4 | 0.2 | 114 (92.7–140)13 | 4 | 0.8 | 4 |
| [Aadn]PeIA | 123 (96.9–157) | 6 | 0.8 | >10 000 | 5 | >2.7 | 81 |
| [Api11]PeIA | 18.1 (14.8–22.2) | 4 | –0.1 | 5432 (2809–10 500) | 4 | 2.4 | 300 |
| [Asu11]PeIA | 44.4 (40.2–49.0) | 4 | 0.4 | 2605 (1628–4168) | 4 | 2.1 | 59 |
Values in parentheses indicate the 95% CI of the IC50 values. Changes in potency were calculated as the log(analog IC50)/(PeIA IC50), negative values indicate increased potency and positive values indicate decreased potency. Selectivity was calculated as the IC50 ratio of (α6/α3β2β3)/(α3β2).
PeIA Analogs with Substitutions of Ser9, Val10, or Leu15 Show Increased Potency for Inhibition of α3β2 nAChRs.
The 300-fold selectivity of [Api11]PeIA greatly exceeds that of α-Ctx OmIA, the most α3β2-selective ligand reported to date.15 The sequences of OmIA and PeIA differ by four residues in positions 9, 12, 14, and 15. In OmIA, these residues are Asn, Asn, His, and Ile. Another α3β2-selective α-Ctx, LvIA, also has Asn in the 9th position and Ile in the 15th position but shares with PeIA an Asn in the 11th position and a Glu in the 14th position. One or more of these amino acid differences probably contribute to the α3β2-selectivity of OmIA and LvIA. A second series of PeIA analogs was synthesized with various substitutions of Ser9, Val10, Glu14, and Leu15 to determine if they might enhance selectivity for α3β2 over α6β2. First, analogs with substitutions of Ser9 including [Thr9]PeIA, [Tyr9]PeIA, and [Asn9]PeIA were synthesized, and their potencies compared to that of the native peptide as well as the previously described analog [His9]PeIA.12 All three of these newly synthesized analogs showed a 2–3-fold change in potency for α3β2 nAChRs, substantially less than the 15-fold change observed for [His9]PeIA (Figure 3A, Table 4). Next, we evaluated a number of Val10-substituted analogs incorporating amino acids with different hydrophobic side-chain lengths or charges. Analogs having amino acid residues with hydrophobic side chains all showed lower IC50 values (3–6-fold) on α3β2 nAChRs, whereas substitution of Val10 with Thr had very little effect (<3-fold change) (Figure 3B, Table 4). Next, Glu14 was substituted with six natural and non-natural amino acids having different chemical properties to evaluate the effects of charge and side-chain length on PeIA potency. All amino acids with negatively charged side chains, except for y-carboxyglutamic acid, produced a substantial loss of potency including Asp, Aad, Api, and Asu (Figure 3C, Table 4). Similar losses of potencies were observed with E14Q and E14R substitutions. These results suggest that Glu14 of PeIA is critical for the activity on α3β2 nAChRs. Finally, we evaluated analogs where Leu15 was substituted with other hydrophobic amino acids or Arg. Both losses and increases in potency were observed with these substitutions. Isoleucine lowered the IC50 value for the inhibition of α3β2 nAChRs by ~3-fold, whereas Val and norleucine (Nle) had little or no effect (<3-fold change) (Figure 3D, Table 4). Alanine or Arg substitution, on the other hand, resulted in ~8- and ~40-fold increases, respectively, in the IC50 value of PeIA. These results suggest that an Ile in position 15 is the most favorable hydrophobic amino acid for activity on α3β2 nAChRs.
Figure 3.
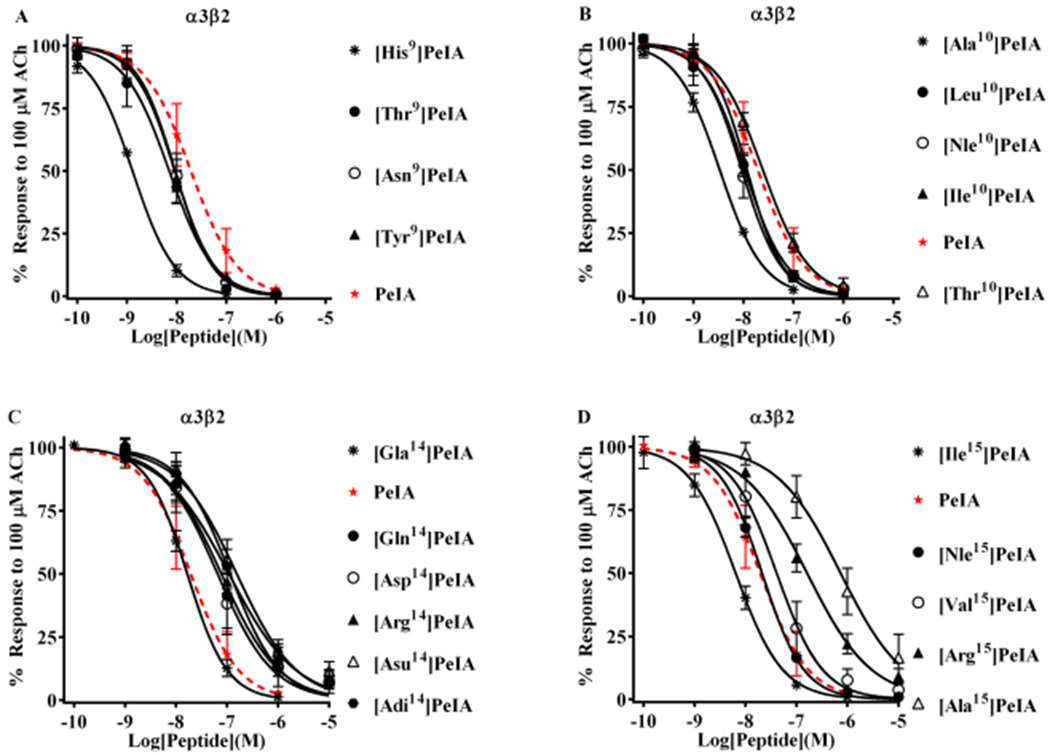
Structure–function analysis of PeIA identifies amino acid substitutions that increase potency for α3β2 nAChRs. PeIA analogs with substitutions of Ser9, Val10, Glu14, or Leu15 were synthesized and a concentration–response analysis was performed for each to determine the IC50 values. The value for each analog was compared to that of the native peptide to evaluate the interaction between the substituted positions and α3β2 nAChRs. (A) Analogs with Asn, Tyr, or Thr substitutions of Ser9 showed a <3-fold change in potency for α3β2 nAChRs. Substitution of Ser9 with His increased the potency of PeIA by 15-fold. (B) Substitution of Val10 with Ala increased (6-fold) PeIA potency, whereas other hydrophobic amino acids showed a <3-fold change but trended toward increased potency. Substitution with Thr also showed a <3-fold change. (C) Substitutions of Glu14 revealed the importance of a Glu in this position for PeIA potency on α3β2 nAChRs. Substitution with any other negatively charged amino acid reduced potency by 3- to 10-fold. A <3-fold change in potency was observed by changing Glu to Gla. Substitution of Glu14 with Gln or Arg reduced PeIA potency by 4- and 6-fold, respectively. (D) Varying effects on PeIA potency were observed when Leu15 was substituted with other hydrophobic amino acids, whereas Ile increased PeIA potency by 3-fold, Ala reduced potency by 38-fold; Nle and Val produced a <3-fold change. Arg reduced PeIA potency by 8-fold. A minimum of four oocytes was used for each IC50 determination and the error bars indicate the SD. The analogs are listed in rank order of potency from top (most potent) to bottom (least potent); values are provided in Table 4. Data for PeIA (red dashed curves) were previously presented in Figure 2 and shown for the ease of comparison to the curves of the analogs.
Table 4.
IC50 Values for PeIA Analogs with Single Substitutions of His9, Val10, Glu14, and Leu15 a
|
α3β2 nAChR |
α6/α3β2β3 nAChR |
||||||
|---|---|---|---|---|---|---|---|
| peptide | IC50 (nM) | n | log change in IC50 | IC50 (nM) | n | log change in IC50 | selectivity ratio |
| PeIA | 19.4 (15.3–24.6) | 5 | 19.8 (17.9–21.8) | 4 | 1 | ||
| [His9]PeIA | 1.30 (1.19–1.41) | 4 | −1.2 | 0.99 (0.93–1.05)12 | 4 | −1.3 | 1 |
| [Asn9]PeIA | 9.32 (8.32–10.4) | 6 | −0.3 | 16.0 (12.8–20.0) | 4 | −0.1 | 2 |
| [Tyr9]PeIA | 9.18 (7.67–11.0) | 5 | −0.3 | 13.0 (9.0–18.9) | 4 | −0.2 | 1 |
| [Thr9]PeIA | 7.10 (5.78–8.72) | 5 | −0.4 | 5.12 (4.51–5.81) | 4 | −0.6 | 1 |
| [Ala10]PeIA | 3.37 (3.05–3.72) | 4 | −0.8 | 2.4 (2.0–2.7)12 | −0.9 | 1 | |
| [Ile10]PeIA | 11.8 (10.2–13.6) | 4 | −0.2 | 13.4 (10.9–16.6) | 6 | −0.2 | 1 |
| [Leu10]PeIA | 10.6 (9.2–12.2) | 4 | −0.3 | 18.8 (16.0–22.1)13 | 0.1 | 2 | |
| [Nle10]PeIA | 9.29 (7.99–10.8) | 6 | −0.3 | 19.9 (17.3–23.0) | 4 | 0.1 | 2 |
| [Thr10]PeIA | 24.5 (21.7–27.8) | 5 | 0.1 | 29.0 (23.7–36.2) | 4 | 0.2 | 1 |
| [Gla14]PeIA | 16.6 (15.1–18.4) | 4 | −0.1 | 28.5 (25.7–31.6) | 4 | 0.2 | 2 |
| [Asp14]PeIA | 65.1 (44.9–94.4) | 4 | 0.5 | 21.0 (16.6–26.5) | 4 | 0.1 | 0.3 |
| [Adi14]PeIA | 115 (88.9–148) | 4 | 0.8 | 29.2 (24.3–35.2) | 5 | 0.2 | 0.3 |
| [Asu14]PeIA | 155 (123–196) | 4 | 0.9 | 135 (107–171) | 4 | 0.8 | 1 |
| [Gln14]PeIA | 77.1 (57.8–1028) | 5 | 0.6 | 14.4 (11.7–17.6) | 5 | −0.1 | 0.2 |
| [Arg14]PeIA | 109 (83.9–141) | 4 | 0.7 | 7.40 (6.06–9.04) | 4 | −0.4 | 0.1 |
| [Arg15]PeIA | 161 (133–196) | 4 | 0.9 | 54.0 (33.1–88.1) | 4 | 0.4 | 0.3 |
| [Ala15]PeIA | 733 (522–1028) | 4 | 1.6 | 82.5 (63.5–107)13 | 0.6 | 0.1 | |
| [Val15]PeIA | 40.9 (32.8–50.9) | 6 | 0.3 | 5.20 (4.40–6.14) | 4 | −0.6 | 0.1 |
| [Ile15]PeIA | 6.41 (5.51–7.46) | 4 | −0.5 | 12.8 (10.5–15.5) | 5 | −0.2 | 2 |
| [Nle15]PeIA | 21.1 (18.5–24.1) | 4 | 0.1 | 13.2 (10.9–15.9) | 5 | −0.2 | 0.6 |
Values in parentheses indicate the 95% CI of the IC50 values. Changes in potency were calculated as the log(analog IC50)/(PeIA IC50), negative values indicate increased potency and positive values indicate decreased potency. Selectivity was calculated as the IC50 ratio of (α6/α3β2β3)/(α3β2).
PeIA Analogs with Substitutions of Ser9, Val10, Glu14, or Leu15 Show Decreased Potency for Inhibition of α6/α3β2β3 nAChRs.
To determine if the PeIA analogs described above might produce a favorable change in the ability of PeIA to discriminate between the α3β2 and α6β2 subtypes, we determined the IC50 values for the inhibition of α6/α3β2β3 and compared the changes to those observed for the α3β2 subtype. Changes in α6/α3β2β3 potency for analogs with substitutions of Ser9 showed patterns similar to those for the α3β2 subtype. The IC50 values of [Asn9]PeIA and [Tyr9]PeIA for α6/α3β2β3 nAChRs were similar to that of the native peptide (Figure 4A, Table 4). Only [Thr9]PeIA showed a substantial change in potency (4-fold increase), relative to PeIA. A lack of change in PeIA potency was observed when analogs with substitutions of Val10 were tested on α6/α3β2β3 nAChRs. The IC50 values for [Leu10]PeIA, [Nle10]PeIA, [Ile10]PeIA, and [Thr10]PeIA were similar to that of native PeIA (Figure 4B, Table 4). Similarly, analogs with substitutions of Glu14 displayed equal potencies with native PeIA with the exception of [Asu14]PeIA, which produced a ~7-fold loss of potency (Figure 4C, Table 4). Analogs where Leu15 was replaced with Val or Ile showed differential changes in potencies for α6/α3β2β3 relative to those for the α3β2 subtype, whereas the L15I substitution increased PeIA potency on α3β2, very little change (<3-fold) was observed for α6/α3β2β3 (Figure 4D, Table 4). The L15V substitution decreased the IC50 value of PeIA for α6/α3β2β3 by 4-fold, whereas an increase was observed for the α3β2 subtype. Finally, like α3β2, α6/α3β2β3 nAChRs were substantially less sensitive to [Arg15]PeIA than the native peptide.
Figure 4.
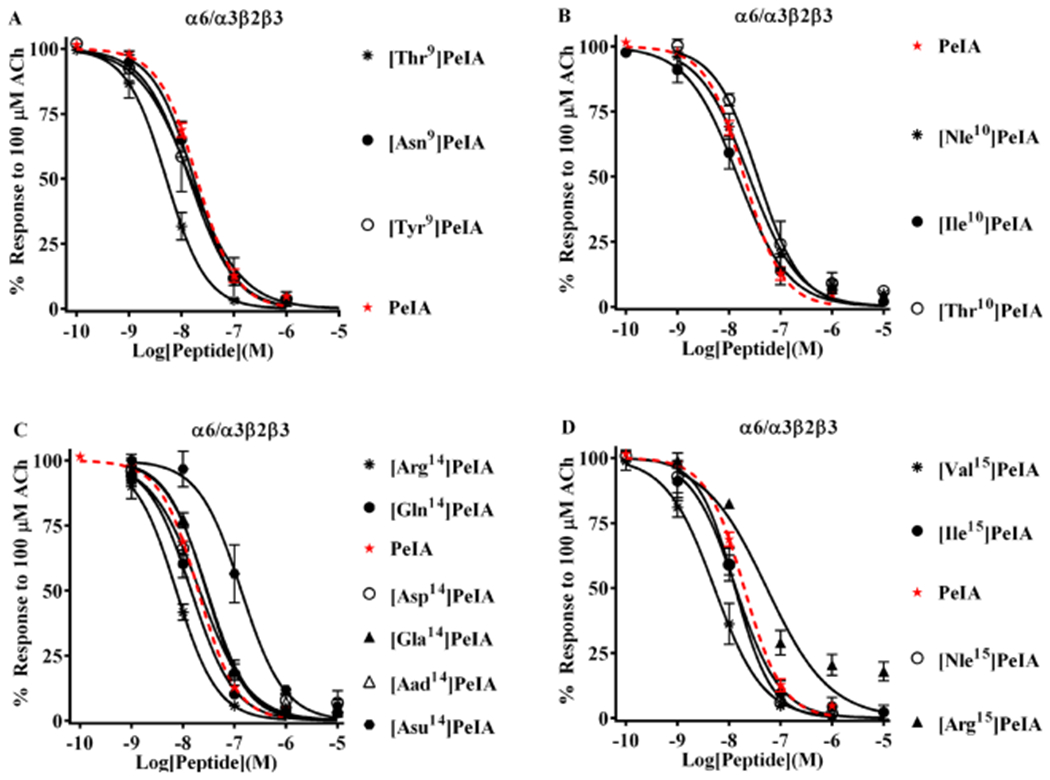
Structure–function analysis of PeIA identifies amino acid substitutions that decrease potency for α6/α3β2β3 nAChRs. A concentration-response analysis was performed for the newly synthesized PeIA analogs, and the IC50 values compared to that of the native peptide and the IC50 values for α3β2 nAChRs. (A) Analogs with substitutions of Ser9 displayed changes in potencies similar to those for α3β2 nAChRs. [Thr9]PeIA displayed increased (4-fold) potency for α6/α3β2β3 nAChRs, whereas all others showed <3-fold change. Note that none of the Ser9 substitutions changed α3β2-α6/α3β2β3 selectivity. (B) Very little change in potency was observed for the newly synthesized analogs with substitutions of Val10. (C) In contrast to the results obtained for α3β2 nAChRs, α6/α3β2β3 nAChRs were less sensitive to perturbations of the interaction between PeIA position 14 and the receptor. Of the analogs with other negatively charged amino acids, only [Asu14]PeIA showed a change in potency (7-fold loss). Substitution of Glu14 with Gln showed a very little effect (<3-fold) on PeIA potency for α6/α3β2β3 nAChRs, whereas Arg reduced the IC50 value by 3-fold. (D) Like α3β2, varying effects on PeIA potency were also observed for α6/α3β2β3 nAChRs when Leu15 was substituted with other hydrophobic amino acids. Whereas the L15I substitutions increased the IC50 value for α3β2, a <3-fold change was observed for α6/α3β2β3 nAChRs. The L15Nle substitution resulted in a <3-fold change in the IC50, whereas L15V decreased the IC50 for α6/α3β2β3 nAChRs by 4-fold. A minimum of four oocytes was used for each IC50 determination and the error bars indicate the SD. The analogs are listed in rank order of potency from top (most potent) to bottom (least potent); values are provided in Table 4. Data for PeIA (red dashed curves) were previously presented in Figure 2 and shown for the ease of comparison to the curves of the analogs.
Novel Peptides Containing Substitutions of Ser9, Val10, Asn14, or Leu15 Show Increased Potency and Selectivity for α3β2 nAChRs.
The results obtained from the single-amino-acid-substituted analogs identified several residue substitutions that were favorable for α3β2 potency and selectivity. When combined, these favorable substitutions might generate peptides with enhanced selectivity for α3β2 over α6β2 nAChRs while maintaining high potency for the α3β2 subtype. Initially, we combined S9H and V10L substitutions to generate the peptide PeIA-4769. These two combined substitutions produced a peptide that was 68-fold more potent than PeIA on α3β2 nAChRs but only 19-fold more potent on α6/α3β2β3 (Figure 5A,B, Table 5). PeIA-4769 was, therefore, used as the initial high potency peptide to incorporate negatively charged amino acids in the 11th position to confer selectivity for α3β2 over α6/α3β2β3 nAChRs. Five derivatives of PeIA-4769 were synthesized containing Asp, Glu, Aad, Api, or Asu in the 11th position and of them, PeIA-5355, containing Api11, showed the most substantial separation (~59-fold) between the IC50 values for α3β2 and α6/α3β2β3 nAChRs (Figure 5A,B, Table 5). PeIA-5415 and PeIA-5587, containing Asu11 and Aad11, respectively, were also substantially more potent on α3β2 nAChRs by 52- and 39-fold, respectively. Next, a 6th PeIA-4769-based peptide was synthesized containing an Ile in the 15th position and evaluated for α3β2-selectivity. The resulting peptide, PeIA-5106, displayed pM potency on both α3β2 and α6/α3β2β3 subtypes (Figure 6A,B, Table 6), but no additional increase in selectivity for α3β2 was found over PeIA-4769. However, when Ile15 was combined with Api11 to produce PeIA-5413, a ~100 fold increase in selectivity for α3β2 over α6/α3β2β3 was obtained, relative to PeIA-5106 (Figure 6A,B, Table 6). These somewhat surprising results with Ile15 in combination with His9, Leu10, and Api11 prompted us to synthesize a final pair of peptides by incorporating a Nle in the 10th position of PeIA-5355. The first Nle10-containing peptide, PeIA-5416, displayed a ~160-fold increase in the selectivity for α3β2 over PeIA-5355. Finally, PeIA-5466 was synthesized containing His9, Nle10, Api11, and Ile15 residues and was found to be ~300-fold more potent on α3β2 than α6/α3β2β3 nAChRs. Furthermore, the peptide displayed high potency for inhibition of α3β2 with an IC50 value of 1.34 (1.13–1.60) nM.
Figure 5.
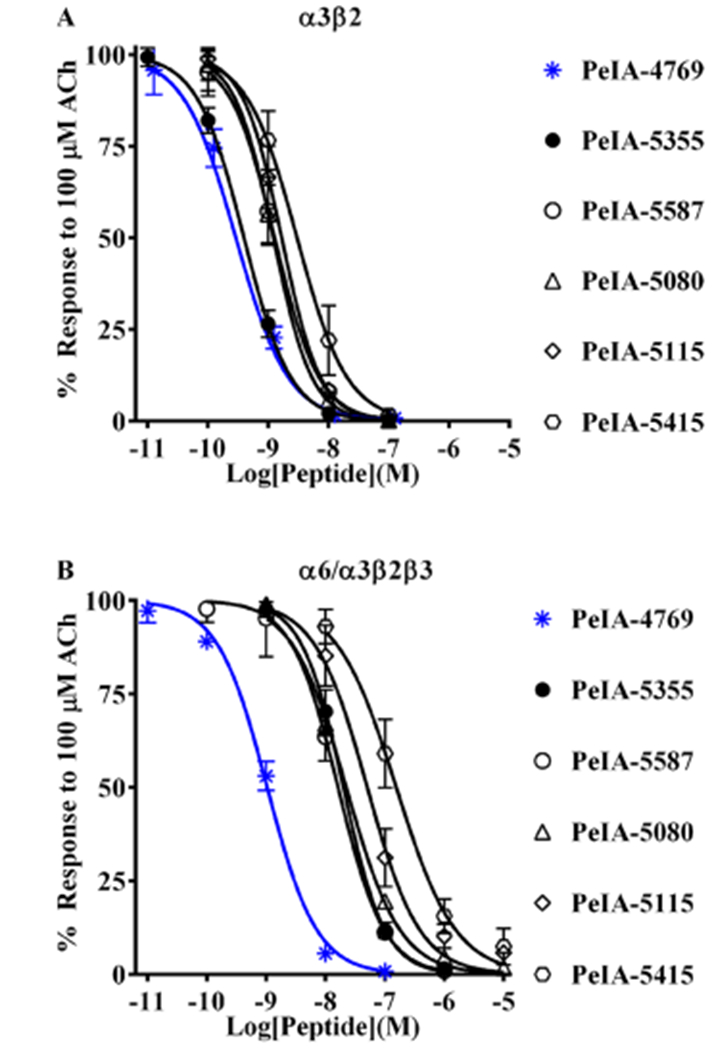
Novel peptides containing the non-natural amino acids Aad, Api, or Asu show enhanced selectivity for α3β2 over α6/α3β2β3 nAChRs. Based on the structure–activity results with PeIA, a series of peptides were synthesized and a concentration–response analysis was performed for each to determine the IC50 values for the inhibition of α3β2 and α6/α3β2β3 nAChRs as well as the selectivity ratio. (A) The first peptide synthesized, PeIA-4769, shares many of the residues found in PeIA (see Table 5) but has His and Leu residues in positions 9 and 10, respectively. PeIA-4769 was 68-fold more potent (blue curve) than PeIA on α3β2 nAChRs. Five additional peptides, based on PeIA-4769, were synthesized containing non-natural amino acid residues in position 11 and of them PeIA-5355, with Api in position 11, was the most potent (pM IC50 value) on α3β2 nAChRs. (B) The IC50 values for the peptides in (A) were determined for the inhibition of α6/α3β2β3 nAChRs and the α3β2-α6/α3β2β3 selectivity ratios calculated. PeIA-4769 was also potent (nM IC50 value, blue curve) α6/α3β2β3 nAChRs. PeIA-5355 displayed the highest selectivity ratio (59-fold) for α3β2 and α6/α3β2β3 nAChRs. Selectivity ratios of 52- and 39-fold were also observed with PeIA-5415 and PeIA-5587, respectively. A minimum of four oocytes was used for each IC50 determination and the error bars indicate the SD; values are provided in Table 5.
Figure 6.
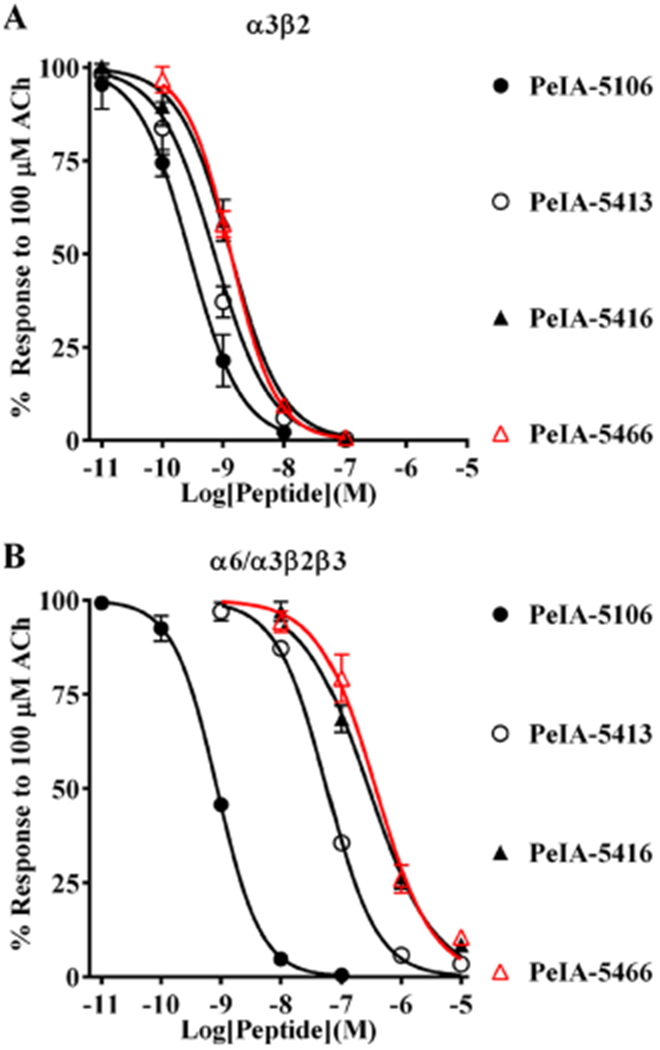
PeIA-5466 is potent and highly selective for α3β2 over α6/α3β2β3 nAChRs. Peptides based on PeIA-4769 and PeIA-5355 were synthesized with different residues in the 10th and 15th positions and a concentration–response analysis was performed for each. The IC50 values for inhibition of α3β2 and α6/α3β2β3 nAChRs for each peptide were compared to those of PeIA and the α3β2-α6/α3β2β3 selectivity ratio calculated. (A, B) PeIA-5106 and PeIA-5416 were synthesized to evaluate the effects of Nle10 and Ile15, respectively, on α3β2-α6/α3β2β3 nAChR selectivity. PeIA-5106 was 69-fold more potent than PeIA on α3β2 nAChRs, but only a minor increase (3fold) in α3β2-α6/α3β2β3 selectivity, over PeIA-4769, was found. PeIA-5416 with Ile15, was 220-fold selective for α3β2 nAChRs. An additional 70-fold enhancement of selectivity was obtained with PeIA-5466 (red curves). A minimum of 4 oocytes was used for each IC50 determination and the error bars indicate the SD; values are provided in Table 6.
Table 6.
IC50 Values for α3β2-Selective Peptides on Other nAChRs Subtypesa
|
α3β4 |
α4β2 |
α6/α3β4 |
α7 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| peptide | IC50 (nM) | n | IC50 ratio | IC50 (nM) | n | IC50 ratio | IC50 (nM) | n | IC50 ratio | IC50 (nM) | n | IC50 ratio |
| PeIA-5355 | 501 (425–592) | 3 | 1329 | 280 (251–313) | 4 | 742 | 43.1 (37.5–49.6) | 4 | 114 | 31.6 (29.2–34.3) | 4 | 84 |
| PeIA-5413 | 341 (317–366) | 3 | 595 | 572 (501–652) | 4 | 998 | 77.3 (57.3–104) | 4 | 134 | 34.5 (28.8–41.4) | 4 | 60 |
| PeIA-5416 | 168 (155–201) | 3 | 129 | 1930 (1520–2451) | 4 | 1461 | 120 (102–141) | 4 | 92 | 18.7 (14.5–24.2) | 4 | 14 |
| PeIA-5466 | 380 (344–419) | 3 | 292 | 2066 (1666–2563) | 8 | 1615 | 205 (164–257) | 4 | 158 | 15.0 (12.1–18.5) | 8 | 12 |
Values in parentheses indicate the 95% CI of the IC50 values. IC50 ratio calculated as (nAChR subtype)/(α3β2).
We also examined the kinetics of PeIA-5466 for the inhibition of the ACh responses and their recovery. Oocytes expressing α3β2 or α6/α3β2β3 nAChRs were continuously perfused with 10 nM PeIA-5466 until a steady-state level of inhibition was observed (Figure 7A,B). The data were analyzed to determine the observed inhibitory rate-constant (kobs) (Figure 7C). The kobs for α3β2 nAChRs was 0.617 ± 0.023 min−1. The kinetics for the inhibition of α6/α3β2β3 nAChRs was more rapid such that kobs could not be determined under the conditions used in this study. The ACh responses in oocytes expressing α3β2 or α6/α3β2β3 nAChRs in the presence of 10 nM PeIA-5466 were inhibited to 8 ± 1% (n = 5) and 90 ± 4% (n = 6) of control values, respectively (Figure 7D). To determine the kinetics for the recovery of the ACh responses, in a separate set of experiments oocytes expressing α3β2 nAChRs were perfused with 100 nM PeIA-5466 until a steady-state level of inhibition was achieved, then the peptide was washed out, and the responses monitored for recovery (Figure 7E). The off-rate constant (koff) was determined to be 0.065 ± 0.001 min−1, and the for recovery was 10.6 (10.2–11.0) min (Figure 7F).
Figure 7.
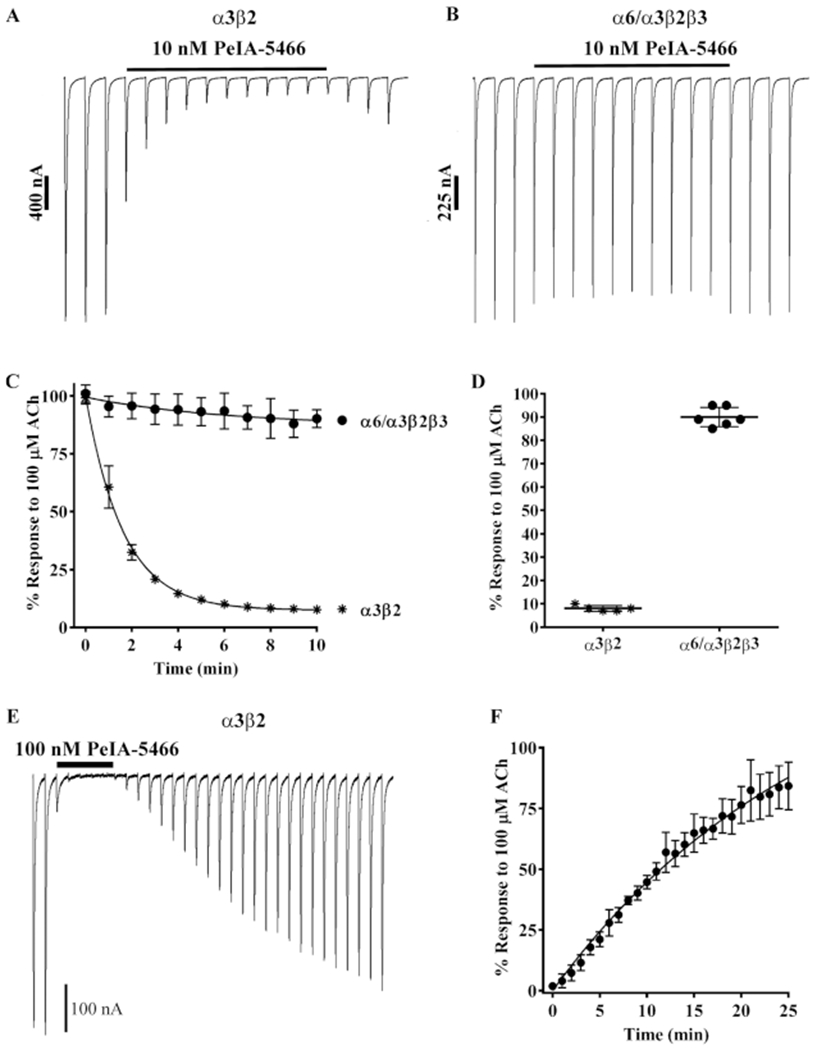
Kinetic analysis of PeIA-5466 for α3β2 and α6/α3β2β3 nAChRs. Xenopus oocytes expressing α3β2 or α6/α3β2β3 nAChRs were subjected to TEVC electrophysiology as described in Methods. (A) Representative current traces from an oocytes expressing α3β2 nAChRs before, during, and after exposure to 10 nM PeIA-5466. (B) Representative current traces from an oocyte expressing α6/α3β2β3 nAChRs before, during, and after exposure to 10 nM PeIA-5466. (C) The exponential fit of the data for the inhibition of α3β2 nAChRs yielded a kobs of 0.617 ± 0.023 min−1 and a t1/2 of 1.1 (1.0–1.2) min. (C, D) The current amplitudes in the presence of 10 nM PeIA-5466 were inhibited to 8 ± 1% (n = 5, SD) of control values for α3β2 nAChRs and to 90 ± 4% (n = 6, SD) of control values for α6/α3β2β3 nAChRs. (E) Representative current traces from an oocyte expressing α3β2 nAChRs showing the inhibition and recovery kinetics during and after exposure to 100 nM PeIA-5466. The current amplitudes in the presence of 100 nM peptide were inhibited to 2 ± 1% (n = 4, SD) of control values. (F) The exponential fit of the data for recovery from inhibition yielded an observed off-rate constant (koff) of 0.065 ± 0.001 min−1 and a t1/2 for recovery of 10.6 (10.2–11.0) min. The Kd was determined to be 1.18 ± 0.47 nM. The 30 s traces in A, B, and E are shown concatenated omitting the 30 s intersweep interval for brevity. The error bars in (C, D, and F) indicate the SD. The Kd was calculated from Kd = koff/kon where kon = (kobs − koff)/[ligand].
Selectivity Profiles of PeIA-5466 and Related Peptides for Other nAChR Subtypes.
To determine if PeIA-5466 and the related Api-containing peptides interacted with other nAChR subtypes, we tested the peptides on a panel of nAChRs subtypes including α3β4, α4β2, α6/α3β4, and α7 nAChRs. The α3β4 and α6β4 subtypes may potentially be expressed with α3β2 in neurons of the PNS and have overlapping sensitivities to ligands, whereas α4β2 and α7 are widely expressed in both the CNS and periphery. All four (PeIA-5355, –5413, –5416, and –5466) of the Api-containing peptides displayed IC50 values for α3β4 that were >100-fold higher than those for α3β2 nAChRs (Figure 8A–D; Table 6). The potency of PeIA-5355, in particular, was 1329-fold lower for α3β4 nAChRs. Similarly, all peptides were substantially less potent on α4β2 nAChRs with PeIA-5466 being the least potent (IC50 1600 higher for α4β2 than that for α3β2 nAChRs). Moderate potency was observed for α6/α3β4 with the least potent being PeIA-5466 (IC50 value ~160-fold higher than for α3β2). Finally, α7 nAChRs displayed varying sensitivities to the four analogs. PeIA-5355 and PeIA-5466 were the least and most potent, respectively.
Figure 8.
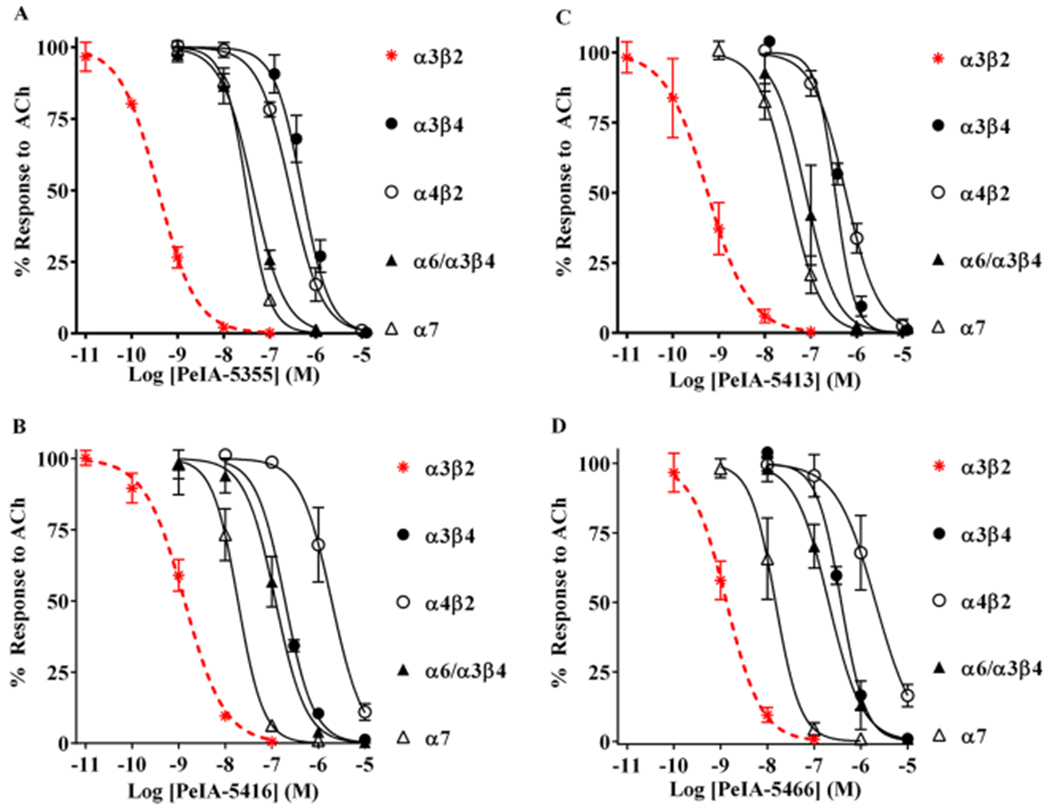
Selectivity profiles of peptides containing non-natural amino acid residues. (A-D) The IC50 values of PeIA-5106, PeIA-5413, PeIA-5416, and PeIA-5466 were determined for a panel of nAChR subtypes including α3β4, α4β2, α6/α3β4, and α7. PeIA-5355 displayed the largest selectivity ratio for α3β2 and α3β4 subtypes (1329-fold). PeIA-5466 displayed the largest selectivity ratio between α3β2 nAChRs and both α4β2 and α6/α3β4 subtypes. A minimum of four oocytes was used for each IC50 curve with the exception of those for α3β4 where three oocytes were used. The exceptionally slow on-rate kinetics of all peptides for α3β4 nAChRs required 45–60 min to reach equilibrium using a concentration of 1 μM (data not shown). Two successive applications of 10 μM were then applied immediately after the equilibrium was achieved with 1 μM. The long perfusion time required for concentrations <300 nM exceeded practical recording times and only a few data points were obtained (<3). The error bars indicate the SD; values are provided in Table 6. Data for α3β2 nAChRs (dashed red curves) were previously presented in Figure 6 and shown for the ease of comparison to the curves for other subtypes.
DISCUSSION AND CONCLUSIONS
The α3β2 nAChR subtype has been challenging to study pharmacologically. To date, no ligands have been reported, either agonists or antagonists, that interact with α3β2 with a sufficient margin of selectivity to allow distinguishing α3β2 from α6β2 nAChRs. In fact, naturally occurring ligands that inhibit α3β2 almost invariably interact with α6β2 nAChRs with similar affinity (Table 1). This lack of α3β2-selective ligands has hampered the investigation of the functional role of this receptor in regions of the CNS where α6β2* subtypes are prominently expressed. To give one example, α-Ctx MII-sensitive nAChRs located on the terminals of SN/VTA dopamine projection neurons are thought to be mostly α6β2* nAChR.16–19 Because of this relative simplicity, the relevance of α6β2*-nAChR in these dopaminergic projections to nicotine (and other drug) use, salience, and movement disorders has been well established using MII in both rodents and non-human primate models.20–22 By contrast, α3β2* nAChR expression overlaps with that of α6β2* nAChR in VTA dopaminergic cell bodies, and the relative roles and/or importance of each subtype in VTA-driven behaviors is not known. To give a second example with relevance to nicotine dependence/drug addiction, the MH-IPN pathway contains the highest density of α3β2* nAChRs within the brain,6 but this compact tract contains a bewildering variety of other nAChR subtypes, including closely related α3β4* and α6* nAChR subtypes. Distinguishing the roles of α3β2* and other nAChR subtypes, and the circuits through which they exert their influence, within the MH, IPN, and the network of regions with which they communicate will require the use of highly selective ligands such as those developed in this study. As a third example, in the PNS superior cervical ganglion neurons have been shown, through gene-knock out and immunoprecipitation studies, to express a number of α3-containing subtypes including α3β4*, α3β4α5, and α3β2β4* subtypes,7,23 but a pharmacological dissection of the role each subtype plays in the physiology of these neurons has been challenging.
α-Conotoxins OmIA and LvIA show some preference for α3β2 over α6/α3β2β3 nAChRs, but the most α3β2-selective of the two, OmIA, shows only an 18-fold preference and, therefore, does not possess a selectivity ratio sufficiently large to permit unequivocal pharmacological identification; an IC50 concentration for α3β2 would be expected to inhibit 10–20% of the response from α6β2 nAChRs.24 OmIA does, however, distinguish well between α3β2 and α3β4 nAChRs. LvIA, on the other hand, does not distinguish α3β2 from α3β4, α6β2, or α6β4 subtypes (selectivity ratios <20).25 Additionally, most pharmacological characterizations of ligands are performed using heterologous receptor expression systems, but the IC50 values for heterologously vs natively expressed nAChRs can differ by 3–5-fold potentially narrowing the selectivity ratio.26 Thus, ligands with selectivity ratios of at least 100-fold are desirable and 300-fold preferred.
Native PeIA is a moderately potent antagonist of α3β2 and α6/α3β2β3 nAChRs (IC50 ~ 20 nM for both) as well as the α9α10 subtype (IC50 ~ 30 nM)11,12 but also inhibits α6β4 and α3β4 subtypes at higher concentrations (IC50 ~ 130 nM and ~ 1.6 μΜ, respectively).12 Like OmIA, PeIA shows some preferences for α3 and α6 subtypes containing β2 subunits over those with β4 subunits, but again the overlapping potencies make it difficult to distinguish α3β2 nAChRs from these other subtypes using PeIA. Previous structure–function analyses of PeIA identified residues that are important for activity, and that can be substituted to enhance potency and selectivity for α3β2 and α6β2 over the closely related α3β4 and α6β4 subtypes.12 A peptide was synthesized containing His9, Ala10, and Asn14 residues that showed a ~65-fold increase in potency for α3β2 and α6/α3β2β3 and a ~230- and ~1600-fold decrease in potency for α6/α3β4 and α3β4 nAChRs, respectively. Subsequent analyses demonstrated that A7V, N11R, and E14A substitutions substantially decreased PeIA activity on α3β2 receptors but had minimal or no impact on α6/α3β2β3 potency.13 Iterative substitution of native α-Ctx sequences has been a successful strategy for generating a number of nAChR subtype-selective peptides12–14,27–36 and could potentially be effective for developing the first α3β2-selective antagonist.
Position 11 of PeIA has been shown to be important for conferring selectivity for α6/α3β2β3 over α3β2 when a positively charged residue is present probably, because this position interacts with a key Lys vs Glu residue difference between α3 and α6 subunits, respectively.13,14 The chemical characteristics (side-chain length and charge) of the non-natural amino acids used in this study have been critically important in allowing us to exploit this α3–α6 subunit difference. Although it is true that the use of non-natural amino acids can allow the introduction of completely different functional groups, this particular study illustrates that the major advantage afforded by this approach is the ability to systematically “fine tune” ligand–receptor interactions at critical positions by incorporating residues with minor differences in chemical properties. In this example, the non-natural amino acids, Adi, Api, and Asu, employed at position 11 differ, by units of one, in the number of carbon atoms in their side chains (Figure 1C). When we replaced Asn11 with negatively charged amino acids, the resulting analogs showed minor reductions in potency for α3β2 with the exception of [Glu11]PeIA and [Api11]PeIA, which retained potency of the native peptide (Figure 2A, Table 3). By contrast, all showed substantially less activity on α6/α3β2β3 nAChRs (Figure 2A,B, Table 3). The most favorable change in selectivity was observed with the analog [Api11]PeIA, which showed a ~300-fold preference for α3β2 over α6/α3β2β3 nAChRs. We note that [Api11]PeIA is substantially more selective than the most α3β2-selective α-Ctxs previously reported (Table 1). These results show definitively that the side-chain length of the residue found in position 11 of PeIA is a critical factor for optimizing charge–charge interaction with Lys152 of the α3 subunit.
Further insights were found from the use of amino acids with minor differences in chemical properties when we substituted residues in the 9th, 10th, 14th, and 15th positions. We found that when Ser9 was substituted with Tyr, Asn, or Thr, favorable effects on PeIA potency for α3β2 were observed (Figure 3A, Table 4). However, only Thr increased potency for α6/α3β2β3 nAChRs (Figure 4A, Table 4). When PeIA Val10 was substituted with other hydrophobic amino acids, small changes, that trended toward increased potency, for α3β2 were observed with [Ala10]PeIA, [Ile10]PeIA, [Leu10]PeIA, and [Nle10]PeIA (Figure 3B, Table 4). None of these analogs substantially changed the IC50 value for α6/α3β2β3 nAChRs (Figure 4B, Table 4) except for Ala, as previously reported.12 In the case of position 14, the substitution of Glu with any other negatively charged amino acid, with the exception of γ-carboxyglutamic acid, resulted in a 3- to 10-fold loss of potency for α3β2 nAChRs. Replacement of Glu14 with Gln and Arg was also detrimental for PeIA potency (Figure 3C, Table 4). These results suggest a critical role for Glu14 in the activity of PeIA on α3β2 nAChRs. By contrast, Glu14 appears to be less important for α6/α3β2β3 nAChRs, which tolerated all amino acid substitutions except Asu (Figure 4C, Table 4). The interaction between PeIA and α3β2 was also more sensitive to perturbation than α6/α3β2β3 nAChRs when Leu15 was replaced with other hydrophobic amino acids. The substitution of Leu15 with Ala resulted in a ~36-fold loss of PeIA potency for α3β2 (Figure 3D, Table 4) but only a ~4-fold loss for α6/α3β2β3 nAChRs (Figure 4D, Table 4).13 The L15V substitution resulted in a 3-fold loss of potency for α3β2 but a ~4-fold increase for α6/α3β2β3 nAChRs. Finally, the L15I substitution selectively decreased the IC50 value for α3β2, whereas L15Nle had no effect on potency for either subtype. The importance of a hydrophobic residue in position 15 was assessed by replacing Leu with Arg, which substantially reduced PeIA potency for both subtypes. These results suggest that the side-chain length as well as hydrophobicity are important determinants of the interaction between position 15 of PeIA and its nAChR targets. In summary, the ability to access expanded chemical spaces using natural and non-natural amino acids allowed us to further identify interactions that could be exploited to improve α3β2* nAChR selectivity of PeIA and related peptides. However, the advantage was (perhaps unsurprisingly) not as substantial in these other positions as at the absolutely critical position 11.
From the substitutions listed in Table 4, we selected those that displayed the most favorable changes in potency or selectivity for α3β2 nAChRs to combine with negatively charged amino acid substitution of Asn11. Two amino acid residues, namely, His9 and Val10, produced the peptide PeIA-4769 that was 68-fold more potent on α3β2 than PeIA (Figure 5A, Table 5). PeIA-4769 was also highly potent on the α6/α3β2β3 subtype (Figure 5B, Table 5). Given the high potency of PeIA-4769, we used the sequence of this peptide as a platform to design novel ligands with non-natural amino acid residues to enhance selectivity for the α3β2 subtype. Substantial gains in selectivity were achieved with each of the PeIA-4769-derived peptides containing non-natural amino acid residues in position 11 (Figure 5A,B, Table 5). Of them, PeIA-5355 displayed the greatest preference (~59-fold) for α3β2 over the α6/α3β2β3 nAChRs. PeIA-5415 and PeIA-5587 also showed ~52- and ~39-fold preferences, respectively, for the α3β2 subtype.
Small differences in the side chains of residues in certain positions can affect the potency of PeIA for α3β2 and α6/α3β2β3 nAChRs. Speculatively, this may suggest that the additional substitutions allow for further fine tuning of the critical interaction between the residue in position 11 and the competitive binding pocket of the α3β2 nAChR. Guided by this principle, we synthesized four additional peptides containing different combinations of His9, Nle10, Api11, and Ile15 residues to gain a more complete picture of the interaction between peptide ligands and α3β2 nAChRs. Interestingly, in contrast to the lower IC50 value of [Ile15]PeIA for α3β2 nAChRs, compared to native PeIA, PeIA-5106, also with Ile15, showed no difference in potency compared to PeIA-4769. However, when we combined His9, Leu10, and Api11 with Ile15, the resulting peptide, PeIA-5413, displayed a ~100-fold increase in the selectivity for α3β2 over α6/α3β2β3 nAChRs, compared to PeIA-4769 (Figure 5A,B, Table 5). Furthermore, PeIA-5413 retained pM potency for α3β2 nAChRs. Therefore, we next reassessed the effects of introducing a Nle in position 10 by synthesizing PeIA-5416. Strikingly, this peptide displayed a ~220-fold preference for α3β2 over α6/α3β2β3 nAChRs, which represents a 100-fold increase in α3β2-selectivity over PeIA-5413. Finally, PeIA-5466 was synthesized containing His9, Nle10, Api11, and Ile15 residues and displayed an IC50 of 1.34 nM for α3β2 nAChRs. This value is ~300-fold lower than that for α6/α3β2β3 nAChRs. These results, obtained by fine tuning the interactions at positions 10 and 15, again demonstrate that minor differences in the chemical properties of residue side chains in key positions can have substantial effects on the ligand activity. Kinetic studies were also performed to examine the binding affinity of PeIA-5466 for α3β2 nAChRs, and a Kd value in the 700–1600 pM range was obtained (Figure 7). Thus, to our knowledge, PeIA-5466 is the most potent and selective α3β2 antagonist heretofore reported.
The pharmacological profiles of the most highly optimized α3β2 nAChR selective peptides are summarized in Table 6. One significant advantage of examining multiple, closely related, peptides is that it is possible to select those with optimal properties for specific applications. For example, PeIA-5355 is exceptionally selective for α3β2 over α3β4 nAChRs, displaying and IC50 value 1329-fold lower for α3β2 (Figure 8A; Table 6). This peptide may be particularly useful for distinguishing α3β2 from α3β4 nAChRs in the PNS where both subtypes are prominently expressed. The same is true in, for example, the MH-IPN tract. In addition to these properties, PeIA-5355 shows the highest selectivity of the final set of peptides against α7 nAChRs (84-fold). This all-round selectivity profile is likely to make it particularly valuable in, for example, in vivo infusion experiments where additional pharmacological manipulations may be difficult or impossible to apply. By contrast, PeIA-5466 has better absolute selectivity for α3β2 over α6/α3β2β3 nAChRs than does PeIA-5355 while retaining nearly 300-fold selectivity against α3β4 nAChR. Although this performance comes at the expense of poor selectivity (only 12-fold) against a7-nAChR, this would be less important in an in vitro assay context where α7 nAChRs can be selectively inhibited by a number of available antagonist including analogs of α-Ctx ArIB31,37 or the three-fingered serpent toxins from Bungurus multicinctus or Naja cobra species.38 As a further example, for distinguishing α3β2 from α4β2 nAChRs, PeIA-5466 is the best ligand, displaying a 1615fold preference for the α3β2 subtype (Figure 8D, Table 6). A high selectivity ratio may be critical for identifying α3β2* nAChRs in the CNS where a significant majority of the nAChR population is of the α4β2* subtype.
In this report, we describe the synthesis and characterization of a series of peptide antagonists with high selectivity for rat α3β2 over α6/α3β2β3 nAChRs. Although some previously identified α-Ctxs show minor preferences for α3β2 nAChRs (Table 1), none of them possesses a selectivity margin sufficiently large enough to facilitate distinguishing α3β2 from α6β2 nAChRs pharmacologically. The development of PeIA-5466 and related peptides represents the first set of ligands, to our knowledge, which display such capabilities and may enable the selective study of α3β2 nAChRs. Importantly, these antagonists and other α3β2-selective ligands such as positive allosteric modulators39 may be useful for defining the functional roles of α3β2* in CNS circuitry involved in nicotine/tobacco dependence where closely related α3β4* and α6β2* subtypes are also present.
EXPERIMENTAL SECTION
Materials.
Acetylcholine chloride (Cat# A6625), potassium chloride (Cat# P3911), and bovine serum albumin (Cat# A2153) were purchased from Sigma Aldrich (St. Louis, MO). Sodium chloride (Cat# S271), calcium chloride dihydrate (Cat# C79), magnesium chloride hexahydrate (Cat# M33), sodium hydroxide (Cat# S313), and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (Cat# BP310) were purchased from Fisher Scientific (Pittsburg, PA).
Peptide Synthesis
Solid-phase Fmoc peptide chemistry and an Apex 396 automated peptide synthesizer (AAPPTec, Louisville, KY) were used to synthesize all peptides. The peptides were initially constructed on a preloaded Fmoc-Rink Amide MBHA resin (substitution: 0.4 mmol/g; Peptides International Inc.; Louisville, KY). All standard amino acids were purchased from AAPPTec except for N-Fmoc-l-γ-carboxyglutamic acid, γ-di-t-butyl ester (γ; Gla; Advanced ChemTech, Louisville, KY), Fmoc-l- and norleucine (Nle) were purchased from EMD Millipore (Burlington, MA). Fmoc-l-2-aminohexanedioic acid δ-tert-butyl ester (Aad) and (S)-Fmoc-2-aminopimelic acid-7-tert-butyl ester (Api) were purchased from Chem-Impex International, Inc. (Wood Dale, IL), (2S)-2-[[(9H-fluoren-9-ylmethoxy)carbonyl]amino]-octanedioic acid-8-(tert-butyl) ester (Asu) was purchased from W&J PharmaChem, Inc. (Silver Spring, MD). Side-chain protection for the following amino acids was as follows: Arg, 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl; Lys, tert-butyloxycarbonyl Hyp; Asn, Gln, and His, trityl; Ser, Gla, Aad, Api, Asu, Asp, and Glu tert-butyl. Cys residues were orthogonally protected by trityl for Cys2 and Cys8 and acetamidomethyl for Cys3 and Cys16. The peptides were synthesized at 50 μmol scale. Coupling activation was achieved with 1 equiv of 0.4 M benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate and 2 equiv of 2 M N,N-diisopropylethylamine in N-methyl-2-pyrrolidone as the solvent. For standard amino acid coupling reactions, a 10-fold molar excess of amino acid was used, and the reaction was carried out for 60 min at room temperature. For nonstandard amino acids, including Asu, Api, and Aad, a 3-fold molar excess was used and a 5-fold excess for Gla and Nle. Coupling reactions for the nonstandard amino acids were extended to 90 min. The Fmoc-deprotection step was performed twice with 20% (v/v) piperidine in dimethylformamide for 5 and then 15 min.
Cleavage and Purification of PeIA-5466.
The following protocol was used for all newly synthesized peptides. PeIA-5466 resin (100 mg) was placed in a 15 mL tube and treated with 2 mL of Reagent K, composed of trifluoroacetic acid (TFA)/phenol/ethanedithiol/thioanisol/H2O (9:0.75:0.25:0.5:0.5 (v/v)). The reaction was gently mixed for 1.5 h at room temperature (for [Arg15]PeIA, the reaction time was extended to 2 h). Next, the cleavage mixture was filtered and precipitated with 100 mL of cold methyl-tert-butyl ether (MTBE). The crude peptide was separated into four 50 mL conical tubes, then precipitated by centrifugation at 7000g for 7 min and washed 2 more times with 30 mL of cold MTBE. The crude peptide pellet was then dissolved in 40 mL of solvent B, and the crude linear peptides were purified by preparative RP-HPLC using a reverse-phase C18-column (Vydac, 5 μM, 250 × 22 mm2) and a gradient of 10–50% solvent B in 40 min (solvent A: 0.1% TFA in water; solvent B: 60% acetonitrile (ACN) 0.092% TFA, 40% water). The purity of linear PeIA-5466 was assessed by RP-HPLC on an analytical C18-column (Vydac, 5 μM, 250 × 4.6 mm2) using the 10–50% solvent B gradient earlier described. Out of 100 mg of resin, 3300 nmol of linear PeIA-5466 were obtained with 76% purity (see the SI Figure 1 for RP-HPLC chromatograms).
Two-Step Oxidative Folding of PeIA-5466.
The following protocol was used to selectively fold the peptides into the correct disulfide configuration for all peptides.
First Disulfide Bond Formation (Cys2–Cys8).
In a solution consisting of 20 mM potassium ferricyanide (0.659 g; 2 mmol) and 0.1 M Tris base (1.21 g; 10 mmol), dissolved in 100 mL of nanopure water, 3300 nmol of linear PeIA-5466, diluted with solvent A to a total volume of 100 mL, was added drop-wise (the peptide final concentration was approximately 20 μM, and the pH was 7.5). The reaction was carried out for 45 min at room temperature, then terminated by dilution with 300 mL of solvent A to lower the pH. The reaction mixture was then passed through a disposable C18 cartridge (Grace Davison Discovery Sciences, Extract Clean C18-HF 1000 mg/8 mL), and the peptide eluted using solvent B. The efficiency of the reaction as well as the purity of the peptide were analyzed by analytical RP-HPLC, as described for the linear peptide. Out of 3300 nmol of the linear peptide, 3150 nmol of monocyclic PeIA-5466 were obtained (95% yield, 84% purity).
Second Disulfide Bond Formation (Cys3–Cys16).
Simultaneous removal of the acetamidomethyl groups and closure of the second disulfide bridge were accomplished by iodine oxidation. Iodine (76 mg, 2 mmols) was added to 15 mL of acetonitrile and stirred until completely dissolved. Then, 45 mL of nanopure water was added followed by 0.18 mL of TFA. The monocyclic PeIA-5466 solution (3150 nmol), diluted with 90 mL with solvent A, was dripped into 60 mL of the 10 mM iodine solution (prepared as described above) and allowed to react for 10 min at room temperature. The peptide final concentration was approximately 20 μM. The reaction was quenched by adding 5–10 drops of 1 M freshly prepared ascorbic acid (0.176 g, 1 mmol) solution in water (1 mL) until the reaction mixture became transparent. The reaction was then diluted 5-fold with solvent A and subsequently purified by RP-HPLC using a preparative C18-column, as described for the linear peptide. The obtained yield was 2603 nmol of the fully folded PeIA-5466. Purity and final yield of PeIA-5466 were determined by RP-HPLC using analytical C18-column using the gradients described earlier for linear peptides and were determined to be >98 and 78% (based on the starting amount of the linear peptide), respectively. The purity of all fully folded peptides was also determined to be >95% (Table 2; SI, Figure 2). The calculated mass of PeIA-5466 was [MH]+1 = 1758.67 and the observed mass was [M + H]+1 = 1758.84, as determined by MALDI-TOF mass spectrometry at the Mass Spectrometry and Proteomics Core Facility at the University of Utah (Table 2).
Two-electrode voltage-clamp electrophysiology of X. laevis oocytes.
Protocols for the isolation of oocytes from female X. laevis frogs were approved by the University of Utah Institutional Animal Care and Use Committee and were strictly adhered to for this study. Methods describing the preparation of cRNA encoding rat nAChR subunits for the expression of nAChRs in oocytes have been previously described.26 Stage IV–V oocytes were injected with equal ratios of cRNA encoding cloned rat α3, α4, α6/α3, α7, β2, β3, or β4 subunits to generate the nAChR subtype of interest and used 1–5 days after injection. The α6/α3 construct was generated by replacing the extracellular ligand-binding domain of the α3 subunit with that of the α6 subunit, as previously described.12,30 This construct was used, because the injection of oocytes with cRNAs encoding α6, β2, and β3 subunits results in few or no functionally expressed receptors.40
To initiate electrophysiology experiments, the oocytes were placed in a 30 μL perfusion chamber constructed from Sylgard 184 (DowDuPont, Midland, MI) and continuously gravity-perfused with saline containing 96 mM NaCl, 2.5 mM KCl, 1.8 mM CaCl2, and 1.0 mM MgCl2. The saline solution was buffered with 5 mM HEPES and had a pH of 7.4. The oocytes were impaled with electrodes pulled from glass capillaries (Cat. # TW100F-4, World Precision Instruments, Sarasota FL), filled with 3 M KCl, and had resistances of 1–10 MΩ. The oocyte membranes were clamped at a holding potential of −70 mV using an Oocyte Clamp OC-725C voltage-clamp amplifier (Warner Instrument Corp., Hamden, CT) and stimulated with 1 s pulses of ACh once every 60 s. The resulting ACh-gated currents were filtered through a 5 Hz low-pass Bessel filter (model F1B1; Frequency Devices, Ottawa, IL) and digitized at 50 Hz using a National Instruments USB-6009 DAQ (National Instruments, Austin, TX). The concentrations of ACh used were 100 μM for receptor subtypes containing β2 subunits and 300 μM for α7 and subtypes containing β4 subunits. The ACh concentrations and brief exposure times were chosen to avoid open-channel block and long-term desensitization of the receptors. The solution changes were controlled through a series of three-way solenoid valves interfaced with a computer via a CoolDrive valve driver (Neptune Research & Development, West Caldwell, NJ) and controlled by the National Instruments LabVIEW 2010 software. The peptides were suspended in saline containing 1 mg/mL of bovine serum albumin, to reduce nonspecific adsorption and either perfusion applied (for concentrations ≤1 μM) or applied in a static bath for 5 min (for concentrations ≥10 μM).
Data Analysis.
Concentration–response data were obtained from a minimum of four oocytes, unless otherwise indicated. The ACh responses in the presence of the peptide were normalized to the average of at least three control responses in the case of perfusion-applied peptides. For peptides applied in a static bath, the current amplitudes in the presence of the peptide were normalized to a single control response in the absence of the peptide. The variance of the data is represented as the ± standard deviation of the mean (SD) and shown with error bars. Peptides were routinely retested on oocytes from different donors to ensure data reproducibility. To estimate the IC50 value for inhibition of the ACh responses by a given peptide, the normalized data were analyzed by nonlinear regression and fit using a four-parameter logistic equation using the Prism statistical analysis software (GraphPad, La Jolla, CA). The IC50 values are presented with corresponding 95% confidence intervals to indicate the precision of the IC50 estimate and displayed in parentheses. For the purposes of this study, changes in IC50 values were considered meaningful if ≥3-fold.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants R01 GM103801 and P01 GM48677 to J.M.M. and R01 DA042749 to P.W.
ABBREVIATIONS
- nAChR
nicotinic acetylcholine receptor
- ACh
acetylcholine
- AChBP
acetylcholine-binding protein
- α-Ctx
α-conotoxin
- Nle
norleucine
- Aad
adipic acid
- Api
pimelic acid
- Asu
suberic acid
- CNS
central nervous system
- PNS
peripheral nervous system
- MH
medial habenula
- IPN
interpeduncular nucleus
- SN
substantia nigra
- MALDI-TOF
matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- ESI
electrospray ionization mass spectrometry
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmedchem.9b00566.
RP-HPLC chromatograms of PeIA-5466 showing the crude linear, purified linear, first- and second-step oxidation products, and the fully folded/purified peptide; RP-HPLC chromatograms of the fully folded peptides (PDF)
The authors declare no competing financial interest
REFERENCES
- (1).Gotti C; Zoli M; Clementi F Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol. Sci 2006, 27, 482–491. [DOI] [PubMed] [Google Scholar]
- (2).Hone AJ; McIntosh JM Nicotinic acetylcholine receptors in neuropathic and inflammatory pain. FEBS Lett. 2018, 592, 1045–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Brunzell DH Preclinical evidence that activation of mesolimbic alpha6 subunit containing nicotinic acetylcholine receptors supports nicotine addiction phenotype. Nicotine Tob. Res 2012, 14, 1258–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Alexander SP; Peters JA; Kelly E; Marrion N; Benson HE; Faccenda E; Pawson AJ; Sharman JL; Southan C; Davies JA CGTP Collaborators. The concise guide to PHARMACOLOGY 2015/16: ligand-gated ion channels. Br. J. Pharmacol 2015, 172, 5870–5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Baddick CG; Marks MJ An autoradiographic survey of mouse brain nicotinic acetylcholine receptors defined by null mutants. Biochem. Pharmacol 2011, 82, 828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Whiteaker P; Peterson CG; Xu W; McIntosh JM; Paylor R; Beaudet AL; Collins AC; Marks MJ Involvement of the alpha3 subunit in central nicotinic binding populations. J. Neurosci 2002, 22, 2522–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).David R; Ciuraszkiewicz A; Simeone X; Orr-Urtreger A; Papke RL; McIntosh JM; Huck S; Scholze P Biochemical and functional properties of distinct nicotinic acetylcholine receptors in the superior cervical ganglion of mice with targeted deletions of nAChR subunit genes. Eur. J. Neurosci 2010, 31, 978–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Mao D; Yasuda RP; Fan H; Wolfe BB; Kellar KJ Heterogeneity of nicotinic cholinergic receptors in rat superior cervical and nodose ganglia. Mol. Pharmacol 2006, 70, 1693–1699. [DOI] [PubMed] [Google Scholar]
- (9).Marritt AM; Cox BC; Yasuda RP; McIntosh JM; Xiao Y; Wolfe BB; Kellar KJ Nicotinic cholinergic receptors in the rat retina: simple and mixed heteromeric subtypes. Mol. Pharmacol 2005, 68, 1656–1668. [DOI] [PubMed] [Google Scholar]
- (10).Prashanth JR; Dutertre S; Lewis RJ Pharmacology of predatory and defensive venom peptides in cone snails. Mol. Biosyst 2017, 13, 2453–2465. [DOI] [PubMed] [Google Scholar]
- (11).McIntosh JM; Plazas PV; Watkins M; Gomez-Casati ME; Olivera BM; Elgoyhen AB A novel alpha-conotoxin, PeIA, cloned from Conus pergrandis, discriminates between rat alpha9alpha10 and alpha7 nicotinic cholinergic receptors. J. Biol. Chem 2005, 280, 30107–30112. [DOI] [PubMed] [Google Scholar]
- (12).Hone AJ; Scadden M; Gajewiak J; Christensen S; Lindstrom J; McIntosh JM alpha-Conotoxin PeIA-[S9H,V10A,E14N] potently and selectively blocks alpha6beta2beta3 versus alpha6beta4 nicotinic acetylcholine receptors. Mol. Pharmacol 2012, 82, 972–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hone AJ; Ruiz M; Scadden M; Christensen S; Gajewiak J; Azam L; McIntosh JM Positional scanning mutagenesis of alpha-conotoxin PeIA identifies critical residues that confer potency and selectivity for alpha6/alpha3beta2beta3 and alpha3beta2 nicotinic acetylcholine receptors. J. Biol. Chem 2013, 288, 25428–25439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Azam L; Yoshikami D; McIntosh JM Amino acid residues that confer high selectivity of the alpha6 nicotinic acetylcholine receptor subunit to alpha-conotoxin MII[S4A,E11A,L15A]. J. Biol. Chem 2008, 283, 11625–11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Talley TT; Olivera BM; Han KH; Christensen SB; Dowell C; Tsigelny I; Ho KY; Taylor P; McIntosh JM Alpha-conotoxin OmIA is a potent ligand for the acetylcholine-binding protein as well as alpha3beta2 and alpha7 nicotinic acetylcholine receptors. J. Biol. Chem 2006, 281, 24678–24686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Zoli M; Moretti M; Zanardi A; McIntosh JM; Clementi F; Gotti C Identification of the nicotinic receptor subtypes expressed on dopaminergic terminals in the rat striatum. J. Neurosci 2002, 22, 8785–8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Champtiaux N; Han ZY; Bessis A; Rossi FM; Zoli M; Marubio L; McIntosh JM; Changeux JP Distribution and pharmacology of alpha 6-containing nicotinic acetylcholine receptors analyzed with mutant mice. J. Neurosci 2002, 22, 1208–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Salminen O; Whiteaker P; Grady SR; Collins AC; McIntosh JM; Marks MJ The subunit composition and pharmacology of alpha-Conotoxin MII-binding nicotinic acetylcholine receptors studied by a novel membrane-binding assay. Neuropharmacology 2005, 48, 696–705. [DOI] [PubMed] [Google Scholar]
- (19).Quik M; Vailati S; Bordia T; Kulak JM; Fan H; McIntosh JM; Clementi F; Gotti C Subunit composition of nicotinic receptors in monkey striatum: effect of treatments with 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine or L-DOPA. Mol. Pharmacol 2005, 67, 32–41. [DOI] [PubMed] [Google Scholar]
- (20).Kulak JM; Nguyen TA; Olivera BM; McIntosh JM Alpha-conotoxin MII blocks nicotine-stimulated dopamine release in rat striatal synaptosomes. J. Neurosci 1997, 17, 5263–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Grady SR; Murphy KL; Cao J; Marks MJ; McIntosh JM; Collins AC Characterization of nicotinic agonist-induced [(3)H]dopamine release from synaptosomes prepared from four mouse brain regions. J. Pharmacol. Exp. Ther 2002, 301, 651–660. [DOI] [PubMed] [Google Scholar]
- (22).Gotti C; Moretti M; Clementi F; Riganti L; McIntosh JM; Collins AC; Marks MJ; Whiteaker P Expression of nigrostriatal alpha6-containing nicotinic acetylcholine receptors is selectively reduced, but not eliminated, by beta3 subunit gene deletion. Mol. Pharmacol 2005, 67, 2007–2015. [DOI] [PubMed] [Google Scholar]
- (23).Simeone X; Karch R; Ciuraszkiewicz A; Orr-Urtreger A; Lemmens-Gruber R; Scholze P; Huck S The role of the nAChR subunits alpha5, beta2, and beta4 on synaptic transmission in the mouse superior cervical ganglion. Physiol. Rep 2019, 7, No. e14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Chi SW; Kim DH; Olivera BM; McIntosh JM; Han KH Solution conformation of a neuronal nicotinic acetylcholine receptor antagonist alpha-conotoxin OmIA that discriminates alpha3 vs. alpha6 nAChR subtypes. Biochem. Biophys. Res. Commun 2006, 345, 248–254. [DOI] [PubMed] [Google Scholar]
- (25).Luo S; Zhangsun D; Schroeder CI; Zhu X; Hu Y; Wu Y; Weltzin MM; Eberhard S; Kaas Q; Craik DJ; McIntosh JM; Whiteaker P A novel alpha4/7-conotoxin LvIA from Conus lividus that selectively blocks alpha3beta2 vs. alpha6/alpha3beta2beta3 nicotinic acetylcholine receptors. FASEB J. 2014, 28, 1842–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Hone AJ; McIntosh JM; Azam L; Lindstrom J; Lucero L; Whiteaker P; Passas J; Blazquez J; Albillos A alpha-Conotoxins identify the alpha3beta4* subtype as the predominant nicotinic acetylcholine receptor expressed in human adrenal chromaffin cells. Mol. Pharmacol 2015, 88, 881–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Pucci L; Grazioso G; Dallanoce C; Rizzi L; De Micheli C; Clementi F; Bertrand S; Bertrand D; Longhi R; De Amici M; Gotti C Engineering of alpha-conotoxin MII-derived peptides with increased selectivity for native alpha6beta2* nicotinic acetylcholine receptors. FASEB J. 2011, 25, 3775–3789. [DOI] [PubMed] [Google Scholar]
- (28).Romero HK; Christensen S; Di Cesare Mannelli L; Gajewiak J; Ramachandra R; Elmslie KS; Vetter DE; Ghelardini C; Ladonato SP; Mercado JL; Olivera BM; McIntosh JM Inhibition of alpha9alpha10 nicotinic acetylcholine receptors prevents chemotherapy-induced neuropathic pain. Proc. Natl. Acad. Sci U.S.A 2017, 114, E1825–E1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Yu J; Zhu X; Harvey PJ; Kaas Q; Zhangsun D; Craik DJ; Luo S Single amino acid substitution in alpha-conotoxin TxID reveals a specific alpha3beta4 nicotinic acetylcholine receptor antagonist. J. Med. Chem 2018, 61, 9256–9265. [DOI] [PubMed] [Google Scholar]
- (30).McIntosh JM; Azam L; Staheli S; Dowell C; Lindstrom JM; Kuryatov A; Garrett JE; Marks MJ; Whiteaker P Analogs of alpha-conotoxin MII are selective for alpha6-containing nicotinic acetylcholine receptors. Mol. Pharmacol 2004, 65, 944–952. [DOI] [PubMed] [Google Scholar]
- (31).Whiteaker P; Christensen S; Yoshikami D; Dowell C; Watkins M; Gulyas J; Rivier J; Olivera BM; McIntosh JM Discovery, synthesis, and structure activity of a highly selective alpha7 nicotinic acetylcholine receptor antagonist. Biochemistry 2007, 46, 6628–6638. [DOI] [PubMed] [Google Scholar]
- (32).Azam L; Maskos U; Changeux JP; Dowell CD; Christensen S; De Biasi M; McIntosh JM alpha-Conotoxin BuIA[T5A;P6O]: a novel ligand that discriminates between alpha6beta4 and alpha6beta2 nicotinic acetylcholine receptors and blocks nicotine-stimulated norepinephrine release. FASEB J. 2010, 24, 5113–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Turner MW; Marquart LA; Phillips PD; McDougal OM Mutagenesis of alpha-conotoxins for enhancing activity and selectivity for nicotinic acetylcholine receptors. Toxins 2019, 11, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Giribaldi J; Dutertre S alpha-Conotoxins to explore the molecular, physiological and pathophysiological functions of neuronal nicotinic acetylcholine receptors. Neurosci. Lett 2018, 679, 24–34. [DOI] [PubMed] [Google Scholar]
- (35).Dutertre S; Nicke A; Tsetlin VI Nicotinic acetylcholine receptor inhibitors derived from snake and snail venoms. Neuropharmacology 2017, 127, 196–223. [DOI] [PubMed] [Google Scholar]
- (36).Wu Y; Zhangsun D; Zhu X; Kaas Q; Zhangsun M; Harvey PJ; Craik DJ; McIntosh JM; Luo S alpha-Conotoxin [S9A]TxID potently discriminates between alpha3beta4 and alpha6/alpha3beta4 nicotinic acetylcholine receptors. J. Med. Chem 2017, 60, 5826–5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Innocent N; Livingstone PD; Hone A; Kimura A; Young T; Whiteaker P; McIntosh JM; Wonnacott S Alpha-conotoxin Arenatus IB[V11L,V16D] [corrected] is a potent and selective antagonist at rat and human native alpha7 nicotinic acetylcholine receptors. J. Pharmacol. Exp. Ther 2008, 327, 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Tsetlin V; Utkin Y; Kasheverov I Polypeptide and peptide toxins, magnifying lenses for binding sites in nicotinic acetylcholine receptors. Biochem. Pharmacol 2009, 78, 720–731. [DOI] [PubMed] [Google Scholar]
- (39).Burgi JJ; Awale M; Boss SD; Schaer T; Marger F; Viveros-Paredes JM; Bertrand S; Gertsch J; Bertrand D; Reymond JL Discovery of potent positive allosteric modulators of the alpha3beta2 nicotinic acetylcholine receptor by a chemical space walk in ChEMBL. ACS Chem. Neurosci 2014, 5, 346–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Kuryatov A; Olale F; Cooper J; Choi C; Lindstrom J Human alpha6 AChR subtypes: subunit composition, assembly, and pharmacological responses. Neuropharmacology 2000, 39, 2570–2590. [DOI] [PubMed] [Google Scholar]
- (41).Hone AJ; Talley TT; Bobango J; Huidobro Melo C; Hararah F; Gajewiak J; Christensen S; Harvey PJ; Craik DJ; McIntosh JM Molecular determinants of alpha-conotoxin potency for inhibition of human and rat alpha6beta4 nicotinic acetylcholine receptors. J. Biol. Chem 2018, 293, 17838–17852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


