While microtubules (MTs) have long been known to be important for delivery of incoming HIV-1 cores to the nucleus, how the virus engages and exploits these filaments remains poorly understood. Our previous work revealed the importance of highly specialized MT regulators that belong to a family called plus-end tracking proteins (+TIPs) in facilitating early stages of infection. These +TIPs perform various functions, such as engaging cargos for transport or engaging peripheral actin to stabilize MTs, suggesting several family members have the potential to contribute to infection in different ways. Here, we reveal that cytoplasmic linker-associated protein 2 (CLASP2), a key regulator of cortical capture and stabilization of MTs, interacts with incoming HIV-1 particles, and we identify a distinct C-terminal domain in CLASP2 that promotes both MT stabilization and early infection. Our findings identify a new +TIP acting as a host cofactor that facilitates early stages of viral infection.
KEYWORDS: +TIPs, CLASP2, HIV-1, capsid, microtubules, trafficking
ABSTRACT
Human immunodeficiency virus type 1 (HIV-1) exploits a number of specialized microtubule (MT) plus-end tracking proteins (commonly known as +TIPs) to induce the formation of stable microtubules soon after virus entry and promote early stages of infection. However, given their functional diversity, the nature of the +TIPs involved and how they facilitate HIV-1 infection remains poorly understood. Here, we identify cytoplasmic linker-associated protein 2 (CLASP2), a +TIP that captures cortical MT plus ends to enable filament stabilization, as a host factor that enables HIV-1 to induce MT stabilization and promote early infection in natural target cell types. Using fixed- and live-cell imaging in human microglia cells, we further show that CLASP2 is required for the trafficking of incoming HIV-1 particles carrying wild-type (WT) envelope. Moreover, both WT CLASP2 and a CLASP2 mutant lacking its C-terminal domain, which mediates its interaction with several host effector proteins, bind to intact HIV-1 cores or in vitro-assembled capsid-nucleocapsid (CA-NC) complexes. However, unlike WT CLASP2, the CLASP2 C-terminal mutant is unable to induce MT stabilization or promote early HIV-1 infection. Our findings identify CLASP2 as a new host cofactor that utilizes distinct regulatory domains to bind incoming HIV-1 particles and facilitate trafficking of incoming viral cores through MT stabilization.
IMPORTANCE While microtubules (MTs) have long been known to be important for delivery of incoming HIV-1 cores to the nucleus, how the virus engages and exploits these filaments remains poorly understood. Our previous work revealed the importance of highly specialized MT regulators that belong to a family called plus-end tracking proteins (+TIPs) in facilitating early stages of infection. These +TIPs perform various functions, such as engaging cargos for transport or engaging peripheral actin to stabilize MTs, suggesting several family members have the potential to contribute to infection in different ways. Here, we reveal that cytoplasmic linker-associated protein 2 (CLASP2), a key regulator of cortical capture and stabilization of MTs, interacts with incoming HIV-1 particles, and we identify a distinct C-terminal domain in CLASP2 that promotes both MT stabilization and early infection. Our findings identify a new +TIP acting as a host cofactor that facilitates early stages of viral infection.
INTRODUCTION
For a successful infection, human immunodeficiency virus type 1 (HIV-1) must undergo a series of early processes, including fusion at the plasma membrane, reverse transcription of the viral RNA into a cDNA copy, disassembly of the viral core (a poorly understood process also known as uncoating), nuclear entry, and integration into the host chromosome. Many aspects of these early processes remain poorly understood.
Like many other viruses, HIV-1 exploits the host cytoskeleton to mediate its intracellular transport. While actin and actin motors mediate entry and early postentry movement of HIV-1 at the cell periphery, subsequent long-range transport to the nucleus involves microtubules (MTs) (1–7). MTs are composed of heteropolymers of α- and β-tubulin that assemble into filaments that first nucleate at perinuclear sites termed MT-organizing centers (MTOCs) and subsequently grow to form a highly dynamic network that radiates throughout the cell (8, 9). These networks mediate the intracellular transport of cargos through minus-end-directed (retrograde) or plus-end-directed (anterograde) movements that are driven by dynein or kinesin motor complexes, respectively (10). Unlike several other viruses that directly associate with MT motors, HIV-1 appears to interact with motors indirectly via the dynein and kinesin motor adaptors, BICD2 and fasciculation and elongation factor 1 (FEZ1), respectively (3, 5, 6, 11).
Although the importance of MTs during infection has long been known, the nature of the filaments and their potential regulation by HIV-1 is only beginning to be understood. We and others have recently shown that HIV-1 causes MT stabilization early after entry in natural target cells (12, 13). Unlike dynamic MT subsets that have a short half-live of several minutes, stable MTs persist for several hours (8). Due to their longevity, stable MTs acquire posttranslational modifications (PTMs), including acetylation and detyrosination (detyrosinated MTs are colloquially referred to as Glu-MTs because detyrosination exposes a glutamic acid at the C terminus of tubulin) (8). While acetylation was recently shown to confer mechanical strength, detyrosinated MTs have a higher affinity for kinesin motors, which, when combined with their longevity, makes stable MT subsets ideal “tracks” for long-range cargo transport in a variety of contexts (14–16). As such, their induction by HIV-1 likely provides stable tracks for virion trafficking and explains the importance not only of the inward motor dynein but also kinesins and FEZ1 during early infection (12). Further advancing our mechanistic understanding, we found that incoming HIV-1 particles induce MT stabilization by binding a number of plus-end tracking proteins (+TIPS) (4, 12), suggesting that +TIPs may be key players in the early stages of infection.
+TIPs play diverse roles in regulating the behavior and function of MTs. Although some +TIPs can localize to MT ends through kinesins, the majority of +TIPs are recruited by end-binding (EB) proteins to the ends of MTs as they grow toward the cell periphery (17). Once at the MT plus end, +TIPs perform various functions, such as promoting polymerization, engaging cargos for transport, or engaging peripheral actin to stabilize MTs. We found that the matrix (MA) protein of incoming HIV-1 particles targets the +TIP Kif4 to induce MT stabilization soon after entry (12). We subsequently discovered that HIV-1 capsid (CA) also induces MT stabilization by binding to another family of +TIPs, the formins Diaphanous 1 and 2 (4). Moreover, microtubule-associated protein 1 (MAP1) has also been reported to interact with incoming HIV-1 CA and facilitate MT stabilization (13). This suggests that targeting multiple MT regulatory proteins likely enables HIV-1 to rapidly amplify the levels of stable MT networks during early infection, but whether other +TIP family members contribute to this process remains unknown.
Given that HIV-1 particles must transition from peripheral actin to stable MTs in order to traffic to the nucleus, here we tested whether cytoplasmic linker protein (CLIP)-associated protein 2 (CLASP2), a key regulator of cortical capture and stabilization of MTs (18, 19), functioned during early HIV-1 infection. In doing so, we found that incoming HIV-1 particles bind CLASP2γ through its N-terminal half while, independently, a critical C-terminal domain in CLASP2γ facilitates the induction of acetylated MTs and trafficking of incoming cores.
RESULTS
CLASP2 is required for HIV-1-induced MT acetylation and early infection.
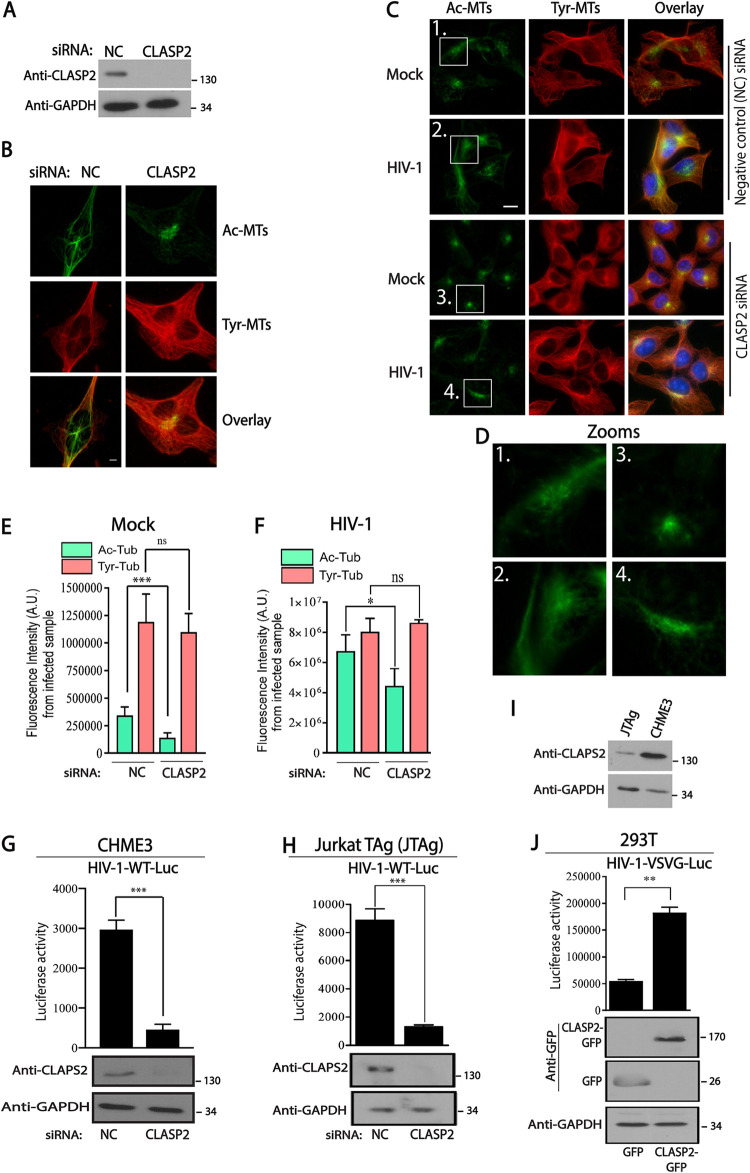
Given its central role in capturing MT plus ends at cortical regions to enable their stabilization (18, 19), we tested the potential role of CLASP2 in early HIV-1 infection in different cell types. To do this, we first used RNA interference (RNAi) to deplete CLASP2 in microglia CHME3 cells (Fig. 1A), a natural target cell type for HIV-1 infection. Immunofluorescence (IF) imaging of fixed cells showed that in control small interfering RNA (siRNA)-treated samples, low levels of acetylated MT (Ac-MT) arrays radiated toward the periphery of CHME3 cells (Fig. 1B). However, in line with its reported functions, depletion of CLASP2 resulted in bundling of Ac-MTs at perinuclear regions.
FIG 1.
CLASP2 is required for HIV-1-induced MT acetylation and early infection. (A and B) CHME3 cells treated with negative-control (NC) or CLASP2 siRNA were either lysed and analyzed by WB to confirm knockdown levels (A) or fixed and stained for tyrosinated (Tyr-MTs) or acetylated (Ac-MTs) tubulin (B). Scale bar = 10 μm. Results are representative of three independent experiments. (C to F) CHME3 cells treated with negative-control or CLASP2 siRNA as described for panel B were infected with mock or HIV-1-WT-Luc virus and stained for Ac-MTs or Tyr-MTs at 6 h postinfection (hpi). Hoechst stain was used to stain nuclei (C). Representative fields are shown. Scale bar = 10 μm. (D) Zoomed images show extended Ac-MT arrays in controls that are confined to perinuclear regions in CLASP2-depleted cells. (E and F) Quantification of the fluorescence intensity of Ac-MTs or Tyr-MTs in siRNA-treated mock-infected (E) or HIV-1-infected (F) samples from the experiment whose results are shown in panel C. Similar results were obtained in three independent experiments. AU, arbitrary units. (G and H) CHME3 (G) or JTAg (H) cells treated with negative-control or CLASP2 siRNA were infected with HIV-1-WT-Luc. Levels of infection were determined by luciferase activity. WB analysis shows levels of CLASP2 or of GAPDH as the loading control. (I) WB analysis showing endogenous levels of CLASP2 in JTAg or CHME3 lines. (J) 293T cells transfected with control GFP alone or CLASP2-GFP were infected with HIV-1-VSVG-Luc followed by measurements of luciferase activity. WB analysis shows protein expression levels. GAPDH was used as the loading control. Results in panels E to J are representative of three experimental replicates. Molecular-weight markers (in kDa) are shown to the right of WBs. **, P ≤ 0.01; ***, P ≤ 0.001.
To determine whether CLASP2 was required for HIV-1-induced MT acetylation or infection, control or CLASP2 siRNA-treated CHME3 cells were mock infected or infected with HIV-1 carrying a luciferase reporter and pseudotyped with wild-type (WT) envelope (HIV-1-WT-Luc). Effects on MTs were assessed by IF imaging following staining for acetylated MTs to label stable filaments, as well as for tyrosinated MTs, to label the overall MT network. Staining and fluorescence intensity measurements showed that HIV-1 infection greatly increased the levels of stable Ac-MT arrays (approximately 20-fold), in line with prior reports (Fig. 1C to F). However, CLASP2 depletion significantly suppressed the formation of Ac-MT arrays in HIV-1-infected cells without affecting the broader tyrosinated MT network (Fig. 1C to F), suggesting that CLASP2 was an important +TIP that mediated HIV-1-induced formation of Ac-MT subsets. In addition, luciferase assays further showed the functional importance of CLASP2 for infection of microglia CHME3 cells with HIV-1-WT-Luc (Fig. 1G). Even though Jurkat TAg (JTAg) T cells express lower levels of CLASP2 than CHME3 microglia, RNA-mediated depletion showed that CLASP2 was also functionally important in JTAg cells for infection with HIV-1-WT-Luc (Fig. 1H and I).
To validate these siRNA-based observations, we also tested the effects of CLASP2 overexpression on infection. Transient expression of green fluorescent protein (GFP)-tagged CLASP2, but not control GFP alone, resulted in a significant increase in the susceptibility of 293T cells to infection with HIV-1 carrying a luciferase reporter and pseudotyped with vesicular stomatitis virus (VSV) G envelope glycoprotein (HIV-1-VSVG-Luc), which mediates entry by endocytosis (Fig. 1J). Similar enhancement of infection by CLASP2 overexpression was also observed in CHME3 cells infected with HIV-1-WT-Luc, as described below. These results indicated that CLASP2 promotes early HIV-1 infection in several different cell types and does so regardless of the route of viral entry. Taken together, these findings established that CLASP2 is required for HIV-1 to induce Ac-MT arrays and for early postentry stages of HIV-1 infection.
CLASP2 regulates trafficking of incoming HIV-1 cores.
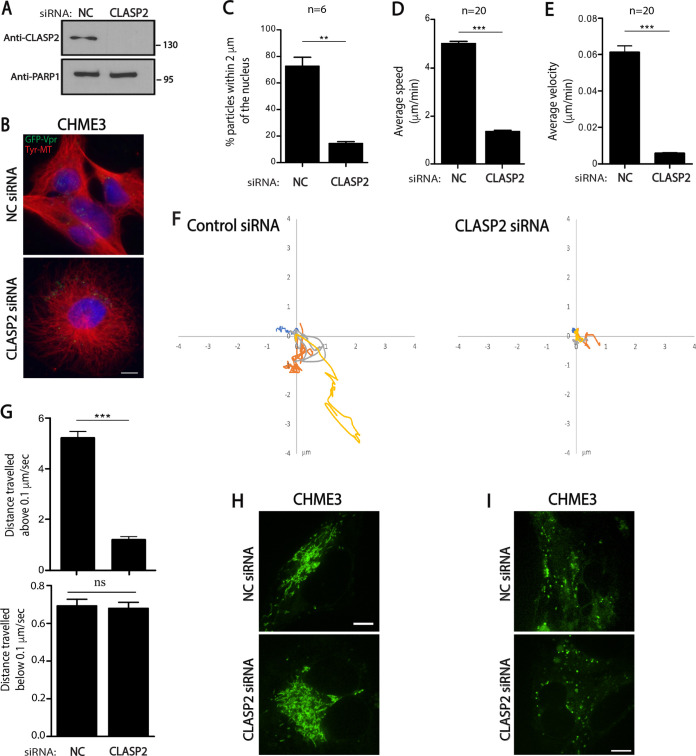
Given that loss of CLASP2 impaired stable MT induction and early HIV-1 infection, we next tested whether this reflected effects on the trafficking of incoming virus particles. To do this, we first used fixed imaging approaches. CHME3 cells treated with control or CLASP2 siRNAs were infected with HIV-1 carrying GFP-tagged viral protein R (Vpr) and pseudotyped with WT envelope (HIV-1-WT-GFP-Vpr). Samples were fixed and stained for GFP (viral particles), tyrosinated MTs, and nuclei at 2 h postinfection (hpi), followed by measurements of the number of viral particles within 2 μm of nuclei, as described previously (3). While >60% of HIV-1 particles translocated to perinuclear regions during early infection in control siRNA-treated cultures, the majority of particles remained at the cell periphery in CLASP2-depleted cells (Fig. 2A to C). To further confirm that virus motility was indeed suppressed in CLASP2-depleted cultures, cells treated with siRNAs and infected as described above were subjected to live-cell microscopy and measurements of particle behaviors (Fig. 2D to G) (3). In line with defects in viral translocation, live-cell imaging showed that, unlike the results for control siRNA-treated cells, wherein HIV-1 particles exhibited rapid, long-range movement, depletion of CLASP2 potently suppressed viral motility. Measurements of individual viral trajectories illustrate these defects and show how, relative to the results for control siRNA-treated cultures, viruses exhibited significant reductions in long-range movement, average speed, and average velocity in CLASP2-depleted cells (Fig. 2D and E, respectively). Particle tracking also showed that, while the majority of viral particles in control siRNA-treated cells moved at speeds greater than 0.1 μm/s, indicative of MT-based transport, in CLASP2-depleted cultures, viruses exhibited slower, short-range movements characteristic of either actin-based motility or free diffusion (Fig. 2F and G). Importantly, depletion of CLASP2 did not noticeably affect the shape or motility of cellular cargos, such as mitochondria or lysosomes (Fig. 2H and I, respectively), demonstrating that CLASP2 depletion did not result in gross defects in host cargo transport and specifically affected early HIV-1 trafficking. Indeed, depletion of CLASP2 did not affect the broader tyrosinated MT network (Fig. 1B to E and 2B). Taken together, fixed- and live-cell imaging approaches established that CLASP2 facilitates MT-based transport of incoming viral cores toward the nucleus.
FIG 2.
CLASP2 facilitates retrograde trafficking of HIV-1 particles. (A to C) CHME3 cells treated with negative-control (NC) or CLASP2 siRNA were either lysed and subjected to WB analysis (A) to confirm knockdown levels or infected with HIV-1-WT-GFP-Vpr followed by IF assay (B and C). (A) Molecular-weight markers (in kDa) are shown to the right of WBs. (B) Representative staining images for tyrosinated tubulin (Tyr-MTs), viral particles (GFP), and nuclei (Hoechst stain) from 3 independent experiments. Scale bar = 10 μm. (C) Quantification of the percentages of viral particles within 2 μm of the nuclei at 2 hpi. A mean of ≥150 virus particles was quantified in ≥20 cells from 3 independent experiments. (D to G) Negative-control or CLASP2 siRNA-treated CHME3 cells were infected with HIV-1-WT-GFP-Vpr followed by live imaging at 1 frame per second (fps) for 5 to 10 min using a Zeiss motorized spinning-disk confocal microscope. Viral particles were tracked to quantify virus movement in trajectories. (D and E) The average cumulative distance traveled over time (speed) (D) and the average displacement tracked over time (velocity) (E) in ≥120 cells. A total of 120 trajectories were analyzed from 10 movies. (F) Four representative viral trajectories are depicted on xy axes. (G) Percentages of movement of virus particles in the experiment whose results are shown in panels D and E that are above or below 0.1 μm/s. Statistical analysis was determined by Mann-Whitney t test. Similar results were obtained in three independent experiments. (H and I) Negative-control or CLASP2 siRNA-treated CHME3 cells were subjected to mitochondrial (H) or lysosomal (I) staining. Representative (n = 3) still images are shown in each case. **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant.
CLASP2 binds HIV-1 cores.
Our data thus far showed that depletion of CLASP2 resulted in perinuclear bundles of Ac-MTs that failed to extend to the cell periphery in uninfected cells and reduced Ac-MTs induced by HIV-1. This phenotype is characteristic of those that occur when specific +TIPs that regulate MT stabilization, such as CLASP2, fail to reach their targets at the cell periphery, such as cortical actin. It is therefore likely that MTs fail to engage the incoming HIV-1 particles at the cell periphery in CLASP2-depleted cells, resulting in impaired MT stabilization.
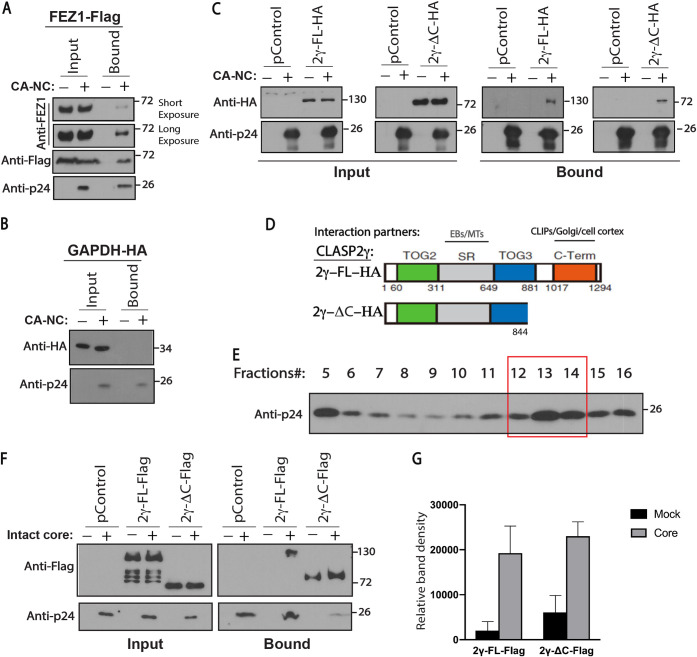
To test this idea, we next determined whether CLASP2 bound to HIV-1 capsid cores, using two approaches. In the first, extracts from cells expressing empty vector control (pControl) or hemagglutinin (HA)-tagged full-length CLASP2γ (2γ-FL-HA) were incubated with in vitro-assembled HIV-1 capsid-nucleocapsid (CA-NC) complexes and were then sedimented over a 70% sucrose cushion. The binding specificity of CA-NC assemblies was confirmed using extracts from cells expressing FEZ1 (a known HIV-1 capsid interactor) or the housekeeping protein glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as positive and negative controls, respectively (Fig. 3A and B, respectively) (3, 20). Similar to FEZ1, CLASP2 pelleted with CA-NC complexes but not in their absence, indicating that these proteins bound HIV-1 cores (Fig. 3C).
FIG 3.
CLASP2 binds to incoming HIV-1 cores. (A to C) 293T cells were transfected with plasmids expressing Flag-tagged FEZ1 (FEZ1-Flag) as the positive control (A), HA-tagged GAPDH (GAPDH-HA) (B) or empty vector (pControl) (C) as negative controls, and HA-tagged full-length CLASP2γ (2γ-FL-HA) or C-terminally truncated CLASP2γ (2γ-ΔC-HA) (C). Thirty-six hours posttransfection, cells were lysed and incubated with mock or in vitro-assembled HIV-1 capsid-nucleocapsid (CA-NC) complexes for 1 h, followed by WB analysis of samples before (Input) or after (Bound) sedimentation through a sucrose cushion. Anti-HA and anti-p24 antibodies were used for detection of CLASP2 and HIV-1 CA, respectively. Similar results were obtained in three independent experiments. (D) Domain organization of HA-tagged full-length CLASP2γ (2γ-FL-HA) and C-terminally truncated CLASP2γ (2γ-ΔC-HA). TOG2 and -3, tumor overexpressed genes 2 and 3; SR, serine-arginine-rich sequence; C-Term, C-terminal domain. Interaction partners with SR-rich and C-terminal domains are indicated. (E and F) CLASP2 binds to intact HIV-1 cores. (E) Representative WB analysis (n = 3) showing the levels of HIV-1 p24 CA in virus fractions (only fractions 5 to 16 are shown) separated by sucrose gradient centrifugation. Fractions 5 to 7 contain free p24 not associated with the intact cores. Fractions 12 to 14 harboring intact HIV-1 cores were pooled and used in the experiment whose results are shown in panel F. (F and G) Lysates from 293T cells expressing empty vector control (pControl), Flag-tagged full-length CLASP2γ (2γ-FL-Flag), or C-terminally truncated CLASP2γ (2γ-ΔC-Flag) were incubated with fractions harboring mock (no intact core, indicated by minus signs above the gels) or intact HIV-1 cores for 2 h followed by WB analysis (F) of input and bound samples using anti-Flag and anti-p24 CA. (G) Quantification of results presented in panel F. Molecular-weight markers (in kDa) are shown to the right of WBs. Similar results were obtained in three independent experiments.
The N-terminal half of CLASP2 contains microtubule interaction domains, while a C-terminal region mediates CLASP2’s interactions with a range of host effector proteins (17) (Fig. 3D). To test whether CLASP2 binding to HIV-1 might be mediated by one or more of these host effector proteins or through the C-terminal domain itself, we also determined whether an HA-tagged CLASP2γ mutant that lacked the C-terminal domain (2γ-ΔC-HA) bound CA-NC complexes. Similar to FEZ1 and full-length CLASP2γ, the C-terminal deletion mutant of CLASP2γ also pelleted with CA-NC complexes in sedimentation assays (Fig. 3C). To validate these findings using CA-NC assemblies, we also tested binding of full-length or C-terminally truncated CLASP2γ to intact HIV-1 cores, which were generated by pooling sedimentation fractions 12 to 14 from infected cells that harbored pelletable viral cores (Fig. 3E). Equivalent fractions from uninfected cells were included as controls in binding assays. Similar to CA-NC complexes, full-length CLASP2γ specifically sedimented in the presence of HIV-1 cores (Fig. 3F and G). In the case of the C-terminal mutant of CLASP2γ, some degree of pelleting was observed in control samples, suggesting that it bound pelletable material present in the fractions from these mock controls. However, we consistently observed higher levels of this C-terminal mutant pelleting with HIV-1 cores, despite lower levels of HIV-1 cores being present than in control or full-length CLASP2γ samples in the experiment whose results are shown in Fig. 3F and G, suggesting that this mutant of CLASP2γ also bound viral cores. Overall, these findings were in line with those using CA-NC assemblies and suggested that CLASP2γ binds HIV-1 particles independently of its C-terminal domain, which mediates many of its effects on microtubules.
The C terminus of CLASP2γ is required for Ac-MT induction and early HIV-1 infection.
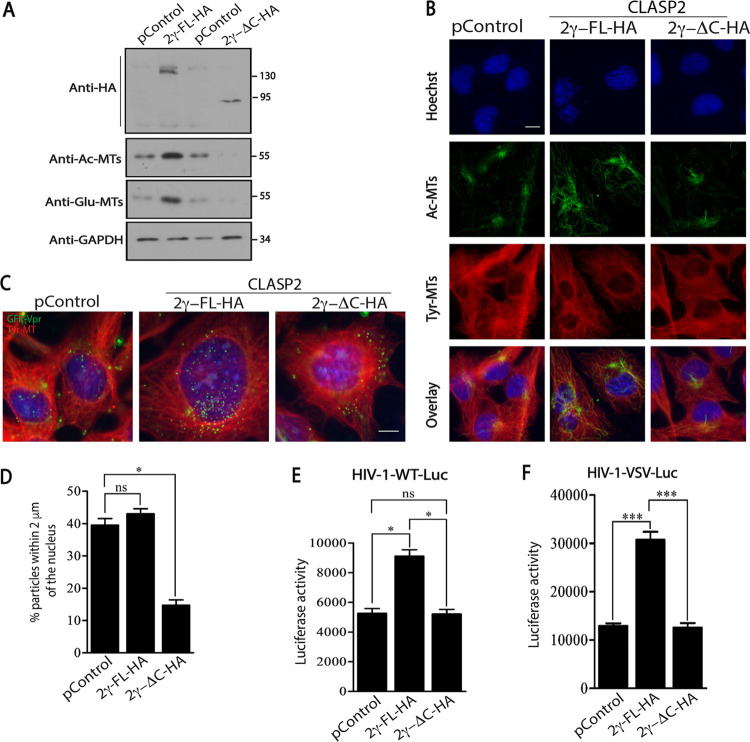
We next tested whether CLASP2 or the CLASP2-ΔC mutant affected MT stabilization and/or infection. To do this, we first measured overall MT levels (Tyr-MTs) and stable MTs (Ac-MTs or Glu-MTs) in CHME3 cells expressing empty vector control, HA-tagged full-length CLASP2γ (2γ-FL-HA), or the CLASP2γ mutant that lacks the C-terminal domain (2γ-ΔC-HA) by Western blot (WB) analysis and IF assay (Fig. 4A and B, respectively). In line with the known function of CLASP2 and reported functions of its C-terminal domain in other cell types, we observed increases in the levels of both Ac-MTs and Glu-MTs in CHME3 cells expressing full-length CLASP2 compared to the levels in control samples (Fig. 4A and B). In contrast, the CLASP2 C-terminal mutant did not induce the formation of posttranslationally modified stable MTs. In line with the WB analysis, IF assay also showed an increase in stable Ac-MTs and further revealed how these arrays extended to the periphery of cells expressing full-length CLASP2 (Fig. 4B). In contrast, not only were the levels of Ac-MTs reduced, but filament arrays were largely confined to perinuclear regions in cells expressing the C-terminal mutant of CLASP2 (Fig. 4B). Similar to our observations described above, exogenous expression of CLASP2 did not have an impact on the broader tyrosinated MT network, suggesting that the C terminus specifically regulated stable MT formation in CHME3 cells.
FIG 4.
C terminus of CLASP2γ induces stable MTs to promote early HIV-1 infection. (A and B) The levels of tyrosinated tubulin (Tyr-MTs), acetylated tubulin (Ac-MTs), or detyrosinated tubulin (also known as Glu-MTs) detected in CHME3 cells stably expressing empty vector control (pControl), 2γ-FL-HA, or 2γ-ΔC-HA by WB analysis (A) or IF assay (B). Molecular-weight markers (in kDa) are shown to the right of WBs. Representative (n = 3) staining images for Tyr-MTs, Ac-MTs, or nuclei (Hoechst stain) are shown. Scale bar = 10 μm. (C and D) CHME3 cells expressing pControl, 2γ-FL-HA, or 2γ-ΔC-HA were infected with HIV-1-WT-GFP-Vpr, followed by IF assay at 6 hpi. (C) Representative (n = 3) staining images for tyrosinated tubulin (Tyr-MTs), viral particles (GFP), and nuclei (Hoechst stain). Scale bar = 10 μm. (D) Quantification of the percentages of viral particles within 2 μm of the nuclei at 6 hpi. A mean of ≥100 virus particles was quantified in ≥20 cells. Statistical analysis was performed by using Mann-Whitney Student’s t test. *, P ≤ 0.05; ns, nonsignificant. (E and F) CHME3 (E) or 293T (F) cells stably expressing pControl, 2γ-FL-HA, or 2γ-ΔC-HA were infected with HIV-1-WT-Luc (E) or HIV-1-VSVG-Luc (F) followed by measurements of luciferase activity. Similar results were obtained in three independent experiments. Statistical analysis was performed by using Mann-Whitney Student’s t test. *, P ≤ 0.05; ***, P ≤ 0.001; ns, nonsignificant.
To determine whether the C terminus of CLASP2 was involved in the translocation of incoming viral cores toward the nucleus, control or CHME3 cells expressing full-length CLASP2-HA or CLASP2-ΔC-HA were infected with HIV-1-WT-GFP-Vpr. Cells were then fixed and stained for tyrosinated MTs, GFP (viral particles), and nuclei, followed by measurements of the numbers of viral particles within 2 μm of the nuclei (3). Imaging showed that the majority of HIV-1 particles translocated to perinuclear regions during early infection in control cultures and that this was moderately enhanced by full-length CLASP2-HA (Fig. 4C and D). In contrast, and similar to the results for CLASP2-depleted cells described above, the majority of particles remained at the periphery of CHME3 cells expressing CLASP2-ΔC-HA (Fig. 4C and D). Moreover, correlating with these effects on stable MTs, expression of full-length CLASP2-HA increased infection of CHME3 or 293T cells by HIV-1-WT-Luc or HIV-1-VSVG-Luc, respectively (Fig. 4E and F). In contrast, expression of CLASP2-ΔC-HA failed to enhance infection in either cell type. Collectively, these findings suggest that CLASP2 binds HIV-1 capsids through N-terminal domains while its C-terminal domain regulates MT stabilization and early HIV-1 trafficking independently of the route of viral entry.
DISCUSSION
Recent work has begun to reveal the importance of highly specialized +TIPs in infection by various viruses (4, 12, 21–23). Here, we identify CLASP2γ as a novel +TIP or host cofactor that utilizes distinct domains to bind HIV-1 particles and regulate their MT-based transport to the nucleus.
Although MTs have long been known to mediate infection by a wide range of viruses, the importance of +TIPs has only recently begun to be appreciated (9, 11). Our earlier work discovered that HIV-1 actively stabilizes MTs by targeting two distinct +TIPs to control retrograde trafficking of incoming viral cores (4, 12). Specifically, we found that after entry, the release of HIV-1 MA targets the EB1-associated +TIP Kif4 to rapidly induce MT stabilization (12). We subsequently discovered that this initial induction of stable MTs is further enhanced by the viral capsid through targeting of a second +TIP complex consisting of the formins Diaphanous 1 and 2, amplifying the levels of stable MTs as the virus proceeds through early infection (4). Others have also shown that HIV-1 CA targets MAP1 to stabilize MTs (13). Given their longevity and higher affinity for kinesin motors, stable MTs have been suggested as ideal tracks for long-range cargo trafficking in a variety of contexts (8, 24). Indeed, we have recently shown the importance of kinesin-1 and its adaptor FEZ1 in retrograde transport of HIV-1 cores to the nucleus (3). Collectively, these findings highlight that HIV-1 exploits +TIPs and MAPs to induce MT stabilization during early infection. CLASPs are key regulators of cortical capture and stabilization of MTs (18, 19), prompting us to test CLASP2γ, one of the most well characterized CLASPs, and its potential contribution to early HIV-1 infection. CLASP2γ was found to be required for HIV-1-induced MT stabilization and MT-based transport of incoming HIV-1 cores. Depletion of CLASP2γ did not affect the broader MT network or the motility of cellular cargoes, such as mitochondria or lysosomes. Moreover, we have shown previously that CLASP2 depletion does not affect early infection by herpes simplex virus 1 (HSV-1) (21), a virus that uses dynamic MTs and does not induce stable MT formation for early infection. As such, CLASP2γ specifically affected the stable MT-based transport mechanisms utilized by HIV-1.
The CLASP family of proteins consists of CLASP1 and CLASP2, which are highly conserved. To date, a single isoform of CLASP1 (CLASP1α) and three isoforms of CLASP2 (CLASP2α, -β, and -γ) have been described (18). The fact that the shortest of these isoforms, CLASP2γ, regulates infection helped us instantly rule out the TOG1 domain found in other CLASP family members as a potential regulator of infection, and using CLASP2γ, we further suggest two distinct domains as functioning in HIV-1 infection. Interestingly, the N-terminal half mediates binding to HIV-1 cores, while the C-terminal portion mediates effects on MT stabilization and HIV-1 trafficking. CLASPs selectively stabilize MTs at the cell periphery, while they promote MT nucleation and growth at the Golgi apparatus. While CLASPs differ at their N terminus, they all harbor a repetitive Ser-Arg (SR)-rich domain that contains two short SXIP motifs required for interaction with EB1-related proteins and MT ends (Fig. 3D). This domain is also responsible for interaction with a number of other cytoskeletal regulatory proteins (25) and likely mediates interactions with HIV-1 cores, given their ability to bind several +TIPs. Although the SR-rich domain mediates interactions with EB1 and MTs, growing evidence suggests that the C-terminal domain is the major effector of CLASP2 localization and function, through interactions with several host effector proteins (25, 26). Our findings further support this notion, as a C-terminal mutant of CLASP2 failed to induce MT stabilization or promote early HIV-1 infection. This suggests that while viral particles engage CLASP2 independently of this domain, it is also a major effector of CLASP2’s function in promoting infection. Indeed, CLASP2 may act as an adaptor for HIV-1 particles to control MT stabilization upon infection, either through C-terminally bound host cofactors or direct functions of the C-terminal domain itself.
Cumulatively, our data suggest that CLASP2γ utilizes distinct domains to engage incoming viral particles and facilitate their induction of the stable MT subsets that they utilize to traffic to the nucleus. These findings add to our understanding of the nature of +TIPs, which are functionally important to early HIV-1 infection, and the underlying mechanisms by which they contribute to this process.
MATERIALS AND METHODS
Cells and viruses.
CHME3, Jurkat TAg (JTAg; derivative of Jurkat E6.1, contains SV40 large T antigen), 293T, and NHDF cells were described previously (3, 27). Viral particles carrying a luciferase reporter and pseudotyped with wild-type (WT) HIV-1 envelope (HIV-1-WT-Luc) or vesicular stomatitis virus (VSV) G envelope (HIV-1-VSVG-Luc) were generated by transfection of HEK293T cells with pNL4-3.Luc.R-.E- (AIDS Reagent Repository catalog number 3418) along with HIV-1 CCR5-tropic (pCl-env) or pVSV-G envelope, respectively (4). HIV-1 particles containing GFP-Vpr were prepared by cotransfecting pNL4-3.Luc.R-.E- along with HIV-1 WT envelope and GFP-Vpr constructs (3). Viral infections were carried out in medium supplemented with 10 μg/ml Polybrene (Santa Cruz Biotechnology), and infected cells were either lysed 48 to 72 h postinfection (hpi) and assayed for luciferase activity or used for immunofluorescence (IF) assay and imaging as described below.
Expression constructs and transfections.
The GFP-tagged CLASP2γ expression construct (CLASP2γ-GFP) (kindly provided by Niels Galjart) (18) was transferred into pQCXIN (Clontech) using the NotI restriction site, and HA tag was added at the N terminus (2γ-FL-HA) using the following primers: Fw-CLASP2-NotI-F (5′-ATAAGAATGCGGCCGCATGTACCCATACGATGTTCCAGATTACGCTGCTATGGGAGATGATAAAAGC-3′) and Re-CLASP2-Not1R (5′-ATAGTTTAGCGGCCGCCTAACTTTGTCCAGAAACATC-3′). Truncated HA-tagged CLASP2γ spanning from position 844 to 1294 (2γ-ΔC-HA) was generated using the following primers: CLASP2-NotI-F (5′-ATAAGAATGCGGCCGCATGTACCCATACGATGTTCCAGATTACGCTGCTATGGGAGATGATAAAAGC-3′) and Pqcxin-CLASP2-1-843-Reverse (5′-ATAGTTTAGCGGCCGCCTATGACTGTGCTGCCTTCCGAA-3′). Both constructs were further used to generate Flag-tagged full-length (2γ-FL-Flag) or truncated (2γ-ΔC-Flag) CLASP2γ. The following primers were used to amplify the 2γ-FL-Flag sequence: Fw-NotFlagClasp2 (5′-ATAAGAATGCGGCCGCATGGATTACAAGGATGACGACGATAAGGCTATGGGAGATGATAAAAGC-3′) and Re-EcoClasp2 (5′-TCACCAGGAAAGTGGTTTC-3′). Intermediary plasmids were generated by subcloning these sequences into NotI-EcoRI-digested 2γ-FL-HA or 2γ-ΔC-HA. Eventually, 2γ-FL-Flag and 2γ-ΔC-Flag were obtained by cloning the EcoRI-digested CLASP2γ sequence into both intermediated plasmids. Each construct was confirmed by sequencing. CHME3 cells stably expressing 2γ-FL-HA, 2γ-ΔC-HA, 2γ-FL-Flag, or 2γ-ΔC-Flag were generated using pQCXIN-based retroviruses followed by treatment with the appropriate antibiotic selection (3). Knockdowns were achieved using the following siRNAs purchased from Ambion: CLASP2 siRNA (number 261146, targeting CLASP2γ) and negative-control nontargeting siRNA (catalog number AM4635). CHME3, 293T, and NHDF cells were transfected with 100 pmol siRNA using Oligofectamine RNAiMax transfection reagent (Invitrogen) as previously described (3). siRNA-mediated knockdown in JTAg cells was achieved using Lonza 4D-Nucleofector. To do this, 1 × 105 JtAg cells/ml were seeded onto 12-well plates for 24 h. The next day, culture medium was replaced with 1.5 ml fresh RPMI medium and the cells were incubated at 37°C prior to transfection. Meanwhile, 150 pmol negative-control or CLASP2 siRNA was mixed with 82 μl of nucleofector solution and 18 μl of supplement solution (Lonza catalog number V4XCP-1024) (mixture A). Cells (1 × 106 cells/transfection) were pelleted and resuspended in mixture A. The whole mixture was transferred to a 100-μl single cuvette and electropulsed using the Lonza 4D-Nucleofector device (program CL-120). After run completion, 500 μl of prewarmed RPMI medium was added to the cuvette, and cells were gently mixed with pipette tips and transferred onto the respective plates. For transient overexpression, 293T cells were plated in 10-cm dishes and transfected with 10 μg DNA and 22.5 μl polyethyleneimine (PEI) for 48 h (3).
Western blotting.
Samples were lysed with Laemmli buffer (0.1 M Tris-HCl [pH 6.8], 20% glycerol, 4% SDS, 0.2 M βME, 0.1% bromophenol blue) and run on 10% SDS-PAGE gels. Primary antibodies were used at a dilution of 1:1,000 in 3% bovine serum albumin (BSA) Tris-buffered saline with Tween 20 (TBS-T) buffer and detected using the appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies. The antibodies to the following proteins were used for Western blot (WB) analysis: CLASP2 (product number 14629), GFP (product number 2555), and PARP-1 (product number 9542) (Cell Signaling); FEZ1 (product number HPA038490), Flag (product number F7425), HA (product number H3663), and acetylated tubulin (product number T7451) (Sigma); GAPDH (catalog number sc-25778) and EB1 (catalog number sc-47704) (Santa Cruz Biotechnology); detyrosinated tubulin (AB3201) (catalog number Millipore); and tyrosinated tubulin (product code ab6160) (Abcam).
Immunofluorescence and imaging.
Antibodies to the following proteins were used for immunofluorescence staining: Ac-MTs (product number T7451) (Sigma), Tyr-MTs (product code ab6160) (Abcam), and GFP (product code ab13970) (Abcam). CHME3 cells were grown on glass coverslips, fixed with ice-cold methanol, blocked, and permeabilized as previously described (4). Samples were incubated with primary antibodies overnight at 4°C. Samples were then washed and incubated with Alexa Fluor-conjugated secondary antibodies for 1 h at room temperature. Nuclei were stained using Hoechst stain for 15 min. FluorSave reagent (Calbiochem) was used to mount the coverslips. Images were acquired by confocal microscopy using a Leica DMI6000 B motorized spinning disk with a Yokogawa CSU-X1 A1 confocal head and run by MetaMorph imaging software (Molecular Devices). Live imaging and viral particle tracking were performed as previously described (3). Briefly, cells were grown on 35-mm glass dishes (MatTek) coated with gelatin. Cells were incubated with HIV-1-WT-GFP-Vpr in CO2-independent medium containing 5% FBS and supplemented with sodium pyruvate and Polybrene, followed by spinoculation at 1,200 × g for 30 min at 16°C. Samples were washed twice with fresh medium prior to incubation with Polybrene, sodium dl-lactate (Sigma), and Oxyrase (Oxyrase Company). Dishes were imaged in an environmental chamber from InVivo at 37°C using a 100× oil objective with a Hamamatsu electron-multiplying charge-coupled-device (EM-CCD) camera and a Yokogawa confocal head on a Leica DMI6000B motorized spinning-disk confocal microscope run by MetaMorph. Images were captured every second for a period of 10 min. Particle tracking was analyzed using the Mosaic plugin in Fiji software. The average speed and velocity of viral particles were calculated as the total distance over time tracked and total displacement over time tracked, respectively. Only viral particles tracked longer than 30 s were included in the calculations. Ten cells were imaged for each condition. Mitochondria were imaged using 100 nM MitoTracker (Thermo Fisher) 15 min prior to imaging at 0.3 frame per second (fps). Lysosomes were imaged using 50 nM Lysotracker (Cell Signaling) immediately before imaging at 0.3 fps.
In vitro-assembled HIV-1 CA-NC binding assay.
Expression and purification of HIV-1 CA-NC were performed as previously described (28). HIV-1 CA-NC proteins were assembled in vitro by incubating CA-NC in 50 mM Tris-HCl (pH 8.0), 0.5 M NaCl, and 2 mg/ml DNA oligo(TG)50 (3). For binding experiments, 293T cells were transfected with expression constructs for 48 h and then lysed in 800 μl hypotonic buffer (10 mM Tris-HCl [pH 7.4], 1.5 mM MgCl2, 10 mM KCl, and 0.5 mM dithiothreitol [DTT] in phosphate-buffered saline [PBS]). Five hundred microliters of lysates was clarified for 10 min at maximum speed prior to incubation with 5 μl of in vitro-assembled CA-NC (labeled “input”) for 2 h at room temperature. The mixture was spun down on a 70% sucrose cushion prepared in 1× PBS and containing 0.5 mM DTT for 1 h at 100,000 × g at 4°C in a SW 55 Ti rotor (Beckman Coulter). The supernatant was discarded, and the pellet was resuspended in Laemmli buffer (labeled “bound”) for further WB analysis.
Intact HIV-1 core binding assay.
The isolation of HIV-1 cores was performed as previously described by Shah and Aiken (29) with a slight modification. Briefly, mock or HIV-1 particles were produced by transient transfection of 293T cells with empty vector or HIV-1 proviral DNA, respectively, using PEI. The virus-containing culture supernatant was collected 48 h posttransfection and filtered using 0.45-μm-pore-size syringe filters. To concentrate the HIV-1 virions, the supernatant was underlaid with 5 ml of a solution of 20% (wt/vol) sucrose in PBS, and ultracentrifugation was performed using an SW 32 Ti Beckman Coulter rotor for 3 h at 32,000 RPM at 4°C. After removing the supernatant, the pelleted viral particles were resuspended in 1× STE buffer (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1 mM EDTA) and spun through a sucrose gradient (30% to 70%) overlaid with 15% sucrose solution containing 1% Triton X-100 and 7.5% sucrose solution in STE using an SW 41 Ti Beckman Coulter rotor overnight at 32,000 RPM at 4°C. Fractions of 750 μl were collected from top to bottom of the gradient and placed on ice upon collection. Meanwhile, 30 μl of each fraction was collected and lysed in Laemmli buffer for WB analysis to determine which fractions contained intact HIV-1 cores. Fractions containing intact cores were then pooled and aliquoted into 0.2-ml aliquots, which were used for binding assay or flash-frozen and stored at −80°C.
The protocol for the intact core binding assay was adopted from a previously described CA-NC assay (4, 28). Briefly, 293T cells were transiently transfected with 10 μg of plasmid expressing Flag or Flag-tagged full-length CLASP2γ or Flag-tagged mutant CLASP2γ using PEI. The cells were lysed in hypotonic lysis buffer (10 mM Tris-HCl [pH 7.4], 1.5 mM MgCl2, 10 mM KCl, and 0.5 mM DTT in PBS) 48 h posttransfection. The lysates were flash-frozen, thawed, and clarified at maximum speed for 10 min. The clarified lysates expressing Flag or full-length CLASP2γ or mutant CLASP2γ were incubated with the mock- or core-containing fraction diluted in cold 1× STE buffer containing BSA (10 μg/ml) for 2 h on ice, with occasional mixing of contents by inverting the tubes. After 2 h of incubation, 10 μl of each sample was collected and lysed with Laemmli buffer (input) for the subsequent WB analysis. The remaining samples were carefully overlaid on 40% sucrose in STE and spun for 1 h at 100,000 × g at 4°C using an SW 55 Ti Beckman Coulter rotor. The pellet was then lysed in 30 μl of Laemmli buffer (bound) for WB analysis.
ACKNOWLEDGMENTS
We thank Niels Galjart at Erasmus University, The Netherlands, and Derek Walsh at Northwestern University for reagents. pNL4-3.Luc.R.-.E- was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from Nathaniel Landau.
This work was supported by NIH grants P01GM105536 and R01AI150559 (to M.H.N.).
REFERENCES
- 1.McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. 2002. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol 159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arhel N, Genovesio A, Kim KA, Miko S, Perret E, Olivo-Marin JC, Shorte S, Charneau P. 2006. Quantitative four-dimensional tracking of cytoplasmic and nuclear HIV-1 complexes. Nat Methods 3:817–824. doi: 10.1038/nmeth928. [DOI] [PubMed] [Google Scholar]
- 3.Malikov V, da Silva ES, Jovasevic V, Bennett G, de Souza Aranha Vieira DA, Schulte B, Diaz-Griffero F, Walsh D, Naghavi MH. 2015. HIV-1 capsids bind and exploit the kinesin-1 adaptor FEZ1 for inward movement to the nucleus. Nat Commun 6:6660. doi: 10.1038/ncomms7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delaney MK, Malikov V, Chai Q, Zhao G, Naghavi MH. 2017. Distinct functions of diaphanous-related formins regulate HIV-1 uncoating and transport. Proc Natl Acad Sci U S A 114:E6932–E6941. doi: 10.1073/pnas.1700247114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dharan A, Opp S, Abdel-Rahim O, Keceli SK, Imam S, Diaz-Griffero F, Campbell EM. 2017. Bicaudal D2 facilitates the cytoplasmic trafficking and nuclear import of HIV-1 genomes during infection. Proc Natl Acad Sci U S A 114:E10707–E10716. doi: 10.1073/pnas.1712033114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carnes SK, Zhou J, Aiken C. 2018. HIV-1 engages a dynein-dynactin-BICD2 complex for infection and transport to the nucleus. J Virol 92:e00358-18. doi: 10.1128/JVI.00358-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh D, Naghavi MH. 2019. Exploitation of cytoskeletal networks during early viral infection. Trends Microbiol 27:39–50. doi: 10.1016/j.tim.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janke C, Bulinski JC. 2011. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol 12:773–786. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- 9.Naghavi MH, Walsh D. 2017. Microtubule regulation and function during virus infection. J Virol 91:e00538-17. doi: 10.1128/JVI.00538-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welte MA. 2004. Bidirectional transport along microtubules. Curr Biol 14:R525–R537. doi: 10.1016/j.cub.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 11.Dodding MP, Way M. 2011. Coupling viruses to dynein and kinesin-1. EMBO J 30:3527–3539. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabo Y, Walsh D, Barry DS, Tinaztepe S, de Los Santos K, Goff SP, Gundersen GG, Naghavi MH. 2013. HIV-1 induces the formation of stable microtubules to enhance early infection. Cell Host Microbe 14:535–546. doi: 10.1016/j.chom.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez J, Portilho DM, Danckaert A, Munier S, Becker A, Roux P, Zambo A, Shorte S, Jacob Y, Vidalain PO, Charneau P, Clavel F, Arhel NJ. 2015. Microtubule-associated proteins 1 (MAP1) promote human immunodeficiency virus type I (HIV-1) intracytoplasmic routing to the nucleus. J Biol Chem 290:4631–4646. doi: 10.1074/jbc.M114.613133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gundersen GG. 2002. Evolutionary conservation of microtubule-capture mechanisms. Nat Rev Mol Cell Biol 3:296–304. doi: 10.1038/nrm777. [DOI] [PubMed] [Google Scholar]
- 15.Portran D, Schaedel L, Xu Z, Thery M, Nachury MV. 2017. Tubulin acetylation protects long-lived microtubules against mechanical ageing. Nat Cell Biol 19:391–398. doi: 10.1038/ncb3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Z, Schaedel L, Portran D, Aguilar A, Gaillard J, Marinkovich MP, Thery M, Nachury MV. 2017. Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science 356:328–332. doi: 10.1126/science.aai8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akhmanova A, Steinmetz MO. 2015. Control of microtubule organization and dynamics: two ends in the limelight. Nat Rev Mol Cell Biol 16:711–726. doi: 10.1038/nrm4084. [DOI] [PubMed] [Google Scholar]
- 18.Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, Galjart N. 2001. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell 104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- 19.Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, Yates JR III, Maiato H, Khodjakov A, Akhmanova A, Kaverina I. 2007. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell 12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang PT, Summers BJ, Xu C, Perilla JR, Malikov V, Naghavi MH, Xiong Y. 2019. FEZ1 is recruited to a conserved cofactor site on capsid to promote HIV-1 trafficking. Cell Rep 28:2373–2385.e7. doi: 10.1016/j.celrep.2019.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naghavi MH, Gundersen GG, Walsh D. 2013. Plus-end tracking proteins, CLASPs, and a viral Akt mimic regulate herpesvirus-induced stable microtubule formation and virus spread. Proc Natl Acad Sci U S A 110:18268–18273. doi: 10.1073/pnas.1310760110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jovasevic V, Naghavi MH, Walsh D. 2015. Microtubule plus end-associated CLIP-170 initiates HSV-1 retrograde transport in primary human cells. J Cell Biol 211:323–337. doi: 10.1083/jcb.201505123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Procter DJ, Banerjee A, Nukui M, Kruse K, Gaponenko V, Murphy EA, Komarova Y, Walsh D. 2018. The HCMV assembly compartment is a dynamic Golgi-derived MTOC that controls nuclear rotation and virus spread. Dev Cell 45:83–100.e7. doi: 10.1016/j.devcel.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li R, Gundersen GG. 2008. Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat Rev Mol Cell Biol 9:860–873. doi: 10.1038/nrm2522. [DOI] [PubMed] [Google Scholar]
- 25.Sayas CL, Basu S, van der Reijden M, Bustos-Morán E, Liz M, Sousa M, van IJcken WFJ, Avila J, Galjart N. 2019. Distinct functions for mammalian CLASP1 and -2 during neurite and axon elongation. Front Cell Neurosci 13:5. doi: 10.3389/fncel.2019.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grimaldi AD, Maki T, Fitton BP, Roth D, Yampolsky D, Davidson MW, Svitkina T, Straube A, Hayashi I, Kaverina I. 2014. CLASPs are required for proper microtubule localization of end-binding proteins. Dev Cell 30:343–352. doi: 10.1016/j.devcel.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Usami Y, Wu Y, Gottlinger HG. 2015. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526:218–223. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 29.Shah VB, Aiken C. 2011. In vitro uncoating of HIV-1 cores. J Vis Exp 2011:3384. doi: 10.3791/3384. [DOI] [PMC free article] [PubMed] [Google Scholar]